Abstract
The GacS/GacA two-component system regulates the expression of bacterial traits during host association. Although the importance of GacS/GacA as a regulator of virulence is well established, its role in benign associations is not clear, as mutations in either the gacS or gacA gene have little impact on the success of colonization in nonpathogenic associations studied thus far. Using as a model the symbiotic association of the bioluminescent marine bacterium Vibrio fischeri with its animal host, the Hawaiian bobtail squid, Euprymna scolopes, we investigated the role of GacA in this beneficial animal-microbe interaction. When grown in culture, gacA mutants were defective in several traits important for symbiosis, including luminescence, growth in defined media, growth yield, siderophore activity, and motility. However, gacA mutants were not deficient in production of acylated homoserine lactone signals or catalase activity. The ability of the gacA mutants to initiate squid colonization was impaired but not abolished, and they reached lower-than-wild-type population densities within the host light organ. In contrast to their dark phenotype in culture, gacA mutants that reached population densities above the luminescence detection limit had normal levels of luminescence per bacterial cell in squid light organs, indicating that GacA is not required for light production within the host. The gacA mutants were impaired at competitive colonization and could only successfully cocolonize squid light organs when present in the seawater at higher inoculum densities than wild-type bacteria. Although severely impaired during colonization initiation, gacA mutants were not displaced by the wild-type strain in light organs that were colonized with both strains. This study establishes the role of GacA as a regulator of a beneficial animal-microbe association and indicates that GacA regulates utilization of growth substrates as well as other colonization traits.
During colonization of animal or plant tissue, bacteria must adapt to the requirements of these environments and prevail over host defenses. There is great interest in understanding how signaling between beneficial bacteria and their hosts is initiated and a stable association is permitted, while at the same time detrimental pathogens are detected and infection is prevented. Although long-term, benign bacterial associations with animals are ubiquitous, studies of these associations are often confounded by the complexity of consortial populations and our inability to culture obligate symbionts. Elucidating the mechanisms underlying recognition and persistence by bacteria in beneficial associations with animal hosts will both add to our understanding of health and aid in the successful treatment of disease.
Vibrio fischeri is a bioluminescent marine bacterium that forms long-term, beneficial associations with certain fishes and sepiolid squid, such as the Hawaiian bobtail squid, Euprymna scolopes. In this association, newly hatched squid acquire V. fischeri from the surrounding seawater in which they are present at a few hundred CFU per ml (30) in a total background of about 106 other marine bacteria per ml. Only V. fischeri colonizes the nascent light-emitting organs of the juvenile squid, forming an essentially monospecific culture (44). The specificity of the association suggests that specialized colonization mechanisms in the bacterial symbiont have coevolved with cognate recognition mechanisms in the squid host (53).
Squid colonization is both spatially and temporally dynamic. Only motile V. fischeri cells can migrate specifically through ducts before they reach the crypt spaces of the light organ (18). During colonization, bacterial symbionts that reach the crypt grow to a population density of 105 to 106 CFU, using as growth substrates host-derived nutrients, including small peptides (20). The increase in population size allows the density-dependent induction of luminescence gene (lux) expression via the accumulation of acylated homoserine lactone (acyl-HSL) quorum-sensing molecules (7). Two different acyl-HSL signals, N-(3-oxohexanoyl) homoserine lactone (C6-HSL) and N-octanoyl homoserine lactone (C8-HSL), work in concert to activate the lux operon (32), which contains both the structural genes for luciferase and the aldehyde synthetase genes (34). Although the squid host expels an estimated 95% of the bacterial contents of its light organ daily (29), creating a new level of selective pressure, regrowth of the remaining bacterial cells results in their persistent association with the host. Mutants defective or reduced in luminescence effectively initiate colonization and grow to a normal cell density but are impaired at longer-term host association during these subsequent regrowth periods (32, 52).
Bacteria often coordinately express multiple traits that are generally important for host association, including motility, attachment, and stress defense, together with other traits that are important to their interaction with specific hosts or host tissues, such as the production and secretion of effectors, secondary metabolites, or virulence determinants. For instance, in the genus Pseudomonas, both host association traits and virulence are globally controlled by a two-component regulatory system composed of the sensor kinase GacS and the response regulator GacA (22). Early studies with plant-pathogenic Pseudomonas spp. revealed that GacS/GacA controls the production of exoenzymes and is required for virulence in a number of host-microbe systems (22). In the opportunistic pathogen Pseudomonas aeruginosa, the production of acyl-HSL signal molecules and factors necessary for virulence on both plant and animal hosts are GacA controlled (22). More recent studies have also linked the GacA homologs ExpA, SirA, UvrY, VarA, and LetA, respectively, to (i) the regulation of extracellular enzymes and acyl-HSL signals in Erwinia carotovora, (ii) motility and invasion gene expression in Salmonella spp., (iii) colonization and stress response of Escherichia coli, (iv) colonization and virulence of Vibrio cholerae, and (v) motility, transmission, and stress response in Legionella pneumophila (21, 22, 40, 49).
The GacS/GacA system also regulates the expression of traits during benign host association. For example, in biocontrol Pseudomonas spp., GacA controls the production of antifungal secondary metabolites that contribute to the health of their host plants, although the production of these compounds has little impact on host-microbe association (22). Additionally, GacS/GacA mutants of these biocontrol bacteria have an enhanced fluorescence typical of overproduction of fluorescent siderophore compounds that function in iron sequestration and bacterial competition (9-11, 55). Thus, GacS/GacA regulation of host association in these superficial and nonspecific benign associations appears to function indirectly and predominately influences microbe-microbe interactions.
We cloned the gacA gene from V. fischeri and investigated its role in global regulation of symbiotic colonization of the squid E. scolopes to establish what role GacA plays in colonization in this intimate and specific bacterium-host association. Our studies showed that GacA controlled multiple traits important for successful squid colonization, including motility, growth-substrate utilization, and luminescence. However, the effect of gacA on luminescence was not accomplished through a deficiency in acyl-HSL accumulation but instead involved depression of luminescence via an undefined mechanism. Animal studies with the gacA mutant alone, or in competition with the wild-type strain, revealed that GacA facilitates but is not required for host colonization. Although GacA was required for luminescence in culture, gacA strain-colonized squid became luminous, demonstrating that GacA effects differed in the host and in culture. The results of this study indicate that GacA is a global regulator of colonization traits and that mutations result in defects in symbiotic colonization, most notably during colonization initiation and growth within the light organ.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacteria and plasmids are listed in Table 1. Chemicals used for culture media preparation were from Sigma (St. Louis, Mo.) unless otherwise noted. Wild-type V. fischeri strain ES114, isolated from an adult specimen of E. scolopes (6), and its derivatives were routinely grown at 28°C in either liquid seawater-tryptone broth (SWT) (6) with shaking at 200 rpm or on nutrient agar medium with added salt (LBS) (18). V. fischeri was also grown on minimal agar plates supplemented with either 24 mM ribose (20), 50 mM fumarate, or 0.5% Casamino Acids (CA; Difco) as a carbon source. E. coli strains were routinely grown in Luria-Bertani (LB) broth (45) or in brain heart infusion medium (Difco). When required, media were supplemented with antibiotics at the following concentrations: for V. fischeri, kanamycin at 50 μg/ml, chloramphenicol (Ch) at 5 μg/ml for multiple copies of the resistance gene in plasmids and at 2.5 μg/ml for a single copy on the chromosome, and erythromycin at 5 μg/ml; for E. coli, kanamycin at 50 μg/ml, Ch at 25 μg/ml, ampicillin at 100 μg/ml, and erythromycin at 150 μg/ml. Plates were supplemented with 40 mg of 5-bromo-4-chloro-3-indolyl-β-galactopyranosidase/ml for visualization of β-galactosidase activity. Where appropriate, C6-HSL and C8-HSL (Aurora Biosciences, San Diego, Calif.) were added to media at 120 nM before inoculation with bacteria as previously described (32). Conditioned broth was prepared by combining fresh broth with spent broth at a ratio of 1:1. Spent broth was prepared by growing bacterial cultures to a final optical density at 600 nm (OD600) of 1.8, pelleting cells by centrifugation at 12,000 × g, and filter sterilizing the cleared broth by passage through a 0.2-μm-pore-sized filter.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F−recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 Δ(argF-lacZYA) U169 φ80lacZΔM15 λ− | Gibco-BRL, Inc. |
| CC118λpir | Δ(arg-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1, lysogenized with λpir dam dcm | 24 |
| V. fischeri strains | ||
| ES114 | Wild-type E. scolopes light organ isolate | 6 |
| KV433 | ES114 Rfr derivative ESR1; katA::erm | 54 |
| SP301 | ES114 Rfr derivative ESR1; glnD::mini-Tn5Cm | 19 |
| DM66 | Spontaneously arising, hyperflagellated, hyperswimmer variant of ES114 | 36 |
| KV495 | ES114 Rfr derivative ESR1; iucA::mini-Tn10 | 50 |
| JRM200 | Chr derivative of ES114 | 33 |
| VCW2A1 | ES114 derivative with a random EZ::TN<KAN-2> insertion in the gacA gene disrupting the codon for P58; Kmr; gacA::EZ::TN<KAN> | This study |
| VCW2E1 | ES114 derivative with a random EZ::TN<KAN-2> insertion in uvrC; Kmr; uvrC | This study |
| VCW2F5 | ES114 derivative with an internal in-frame AatII deletion resulting in loss of coding region between I27 and G194; ΔgacA | This study |
| VCW2H7 | ES114 derivative with a frameshift mutation in luxI; C6-HSL−; luxI | 32 |
| Plasmids | ||
| pVO8 | pACYC, Chr, Emr; multicopy vector stably maintained by V. fischeri | 51 |
| pEVS79 | pBCSK derivative, Mob+, Chr | 47 |
| pEVS104 | R6Kγ derivative of pRK2013; ΔColE1, oriT tra trb ΔTn903 Kmr | 47 |
| pHV200I− | Entire lux operon from V. fischeri ES114 with a frameshift mutation in the luxI gene cloned into pBR322; Apr | 39 |
| p395B | aidA::lacZ fusion; Spr Tcr | 16 |
| pVCW1A7 | pEVS79, containing a 4.0-kb HindIII genomic insert from V. fischeri strain ES114 including the intact gacA gene and partial uvrC gene; Chr | This study |
| pVCW1C7 | Random EZ::TN<KAN> insertion in the gacA gene in pVCW1A7, Chr Kmr | This study |
| pVCW1D7 | Random EZ::TN<KAN> insertion in the uvrC gene in pVCW1A7; Chr Kmr | This study |
| pVCW2A5 | Plasmid pVCW1A7 with a single nucleotide substitution from T to C, resulting in the generation of an AatII restriction site; Chr | This study |
| pVCW2D5 | Plasmid pVCW2A5 with a 519-bp AatII deletion within the gacA gene; Chr | This study |
| pVCW2A6 | Plasmid pEVS79 with an 8-kb SalI fragment from KV29 containing frameshifted luxI; Chr | 32 |
| pVCW3C3 | Plasmid pVO8 with a 4.0-kb fragment containing wild-type gacA from pVCW1A7; Emr | This study |
Ap, ampicillin; Ch, chloramphenicol; Em, erythromycin; Km, kanamycin; Rf, rifampin; Sp, spectinomycin; Tc, tetracycline.
Recombinant DNA techniques.
Standard molecular methods were used for transformations, restriction enzyme digestions, gel electrophoresis, Southern analysis, and PCR (45). Restriction enzymes were from New England BioLabs (Beverly, Mass.). Gel purification of restriction enzyme-digested DNA was performed using the QiaQuick gel extraction kit (Qiagen, Valencia, Calif.). Plasmids for laboratory procedures were purified using the Qiaprep Spin Miniprep kit (Qiagen). Plasmid DNA for sequence analysis was prepared using the Perfect Prep plasmid mini kit (Eppendorf Scientific Inc., Westbury, N.Y.). Ligations were performed by the thermal cycling method (31). Genomic DNA was isolated by a cetyltrimethylammonium bromide method (3). Digoxigenin-11-dUTP-labeled probes for Southern blotting were generated by PCR using materials and protocols supplied by the manufacturer (Boehringer Mannheim Corporation, Indianapolis, Ind.). Oligonucleotide primers were synthesized by Integrated DNA Technologies (Coralville, Iowa).
Cloning and sequence analysis of gacA.
Alignments of the predicted amino acid sequence from various GacA homologs were used to identify conserved regions to which degenerate primers were designed. The forward (5′-GARGCNGAYWSNGGNGARGA-3′) and reverse (5′-NARYTTYTCNGTRTCNARDATNCC-3′) primers were generated and used to amplify a 550-bp fragment from the genome of V. fischeri strain ES114 by PCR. After confirmation that the sequence identity was similar to that of the GacA homolog VarA from V. cholerae (56) by directly sequencing the PCR product, gene-specific primers (forward primer Vgac2, ATGAGTTAATTCAACGTCTCAC; reverse primer Vgac3, TTATGGATATGAACATGCCTGG) were designed. These primers were used to amplify an internal fragment of GacA (525 bp in length) which served as a gene-specific probe to identify and isolate a genomic clone containing the intact gene. Southern analysis of genomic fragments from V. fischeri generated by digestion with various restriction enzymes revealed that the gene was contained on a 4.0-kb HindIII fragment. A genomic sublibrary of 4.0-kb HindIII-digested fragments of ES114 was generated by gel purification of the restriction fragments and subsequent ligation of ES114 DNA to HindIII-digested pEVS79 plasmid DNA. The library DNA was transformed into E. coli cells which were plated on LB with Ch and 5-bromo-4-chloro-3-indoyl-β-galactopyranosidase. White colonies containing plasmids with inserted DNA were directly screened by PCR using the Vgac2 and Vgac3 primers. From a single clone, pVCW1A7, we amplified the predicted 525-bp fragment. Sequencing of the entire gacA gene was performed at the Biotechnology Molecular Biology Instrumentation Facility, University of Hawaii, using primers designed to the sequence obtained from the PCR-amplified fragment. Comparisons of the cloned DNA with sequences in GenBank were performed using the BLAST software package (2).
Generation of mutants.
Plasmid pVCW1A7 that contained the wild-type gacA region was mutagenized using the EZ::TN<KAN-2> insertion kit following protocols supplied by the manufacturer (Epicentre, Madison, Wis.). Insertions in gacA were identified by PCR amplification and confirmed to be in the gacA open reading frame (ORF) by sequence analysis of the clones. One random EZ::TN<KAN-2> insertion was identified within the sequence encoding amino acid residue P58 of GacA. The mutagenized gene was recombined with the chromosomal copy of gacA by marker exchange mutagenesis as described previously (47). A single colony, designated strain VCW2A1, was confirmed to have the gacA::EZ::TN<KAN-2> mutation by Southern analysis. A random EZ::TN<KAN-2> insertion in the uvrC gene was generated by a similar approach, creating strain VCW2E1.
To make an in-frame deletion of gacA, we created an AatII restriction site within the gacA ORF. A single nucleotide conversion from T to C at position 600 in the nucleotide sequence was generated using the QuikChange site-directed mutagenesis kit and protocols supplied by the manufacturer (Stratagene, La Jolla, Calif.). Two overlapping primers, VGA-AATF (3′-AAGTGGAGACGTCGAATTAACTCATCTAGCTATTCGTC-5′) and VGA-AATR (5′-ATCACGAATAGCTAGATGAGTTAATTCGACGTCTCCAC-3′), were used for mutagenesis. An in-frame deletion of 89% of the gacA ORF was generated within the predicted protein sequence from amino acid residue I27 to G194 by digesting the resulting plasmid, pVCW2A5, with AatII and self-ligating it, creating plasmid pVCW2D5. The mutation was exchanged with the wild-type gacA gene by marker-exchange mutagenesis, creating strain VCW2F5, and gene replacement was confirmed by Southern analysis.
A LuxI mutation in strain ES114 constructed for these studies was generated similarly to that mutation in V. fischeri strain ESR1 (52) and was described in a previous publication (32).
Luminescence detection.
A 1-ml aliquot of V. fischeri cells grown in broth culture was removed at regular intervals to determine the luminescence and optical density (OD600). Luminescence levels were measured with a Turner 20/20 luminometer (Turner Designs, Sunnyvale, Calif.) calibrated with a light standard. Where appropriate, decanal suspended in 95% ethyl alcohol was added at a final concentration of 0.01% to an aliquot of culture immediately prior to measurement of luminescence (32).
Luminescence of V. fischeri within colonized squid was routinely measured at 24, 48, and 72 h postinoculation. The luminescence detection limit was determined on squid monitored continuously between 7 and 9 h postinoculation with the wild-type strain ES114. Squid with a luminescence level between 1 and 5 luminescence units (LU) were immediately frozen, and the number of bacteria contributing to luminescence was quantified subsequently by homogenization of the squid in seawater (SW) that was formerly sterilized by autoclaving, plating the contents onto LBS agar plates, and enumerating the colonies of V. fischeri that grew following overnight incubation. The experiment was done twice with similar results, and the data from both experiments were combined and reported as the mean CFU.
Quantification of acyl-HSLs.
Published methods for the purification and quantification of C6-HSL and C8-HSL were used (46). Briefly, acyl-HSLs were extracted twice with an equal volume of acidified ethyl acetate from cell-free supernatants of ES114 and derivative cultures grown in SWT broth to a final OD600 of 1.6. The samples were concentrated by evaporation before analysis. Because C8-HSL is produced at a higher level (micromolar) than C6-HSL (nanomolar) in strain ES114 (A. Schaefer, personal communication), C6-HSL cross-reaction with the biological reporter strain for C8-HSL does not interfere with quantification of C8-HSL; therefore, C8-HSL was directly quantified from extracts of 15-ml cultures. However, C8-HSL cross-reaction with the biological reporter strains for C6-HSL could interfere with its quantification. Therefore, we first purified C6-HSL from 500-ml cultures using a C18 reverse-phase high-performance liquid chromatography column and a linear, 10-to-100% (vol/vol) methanol-water gradient at 0.5 ml/min. The elution profile of synthetic C6-HSL was determined to identify which fractions contained activity, and these and flanking 1-ml fractions from extracts were assayed for activity. C6-HSL was quantified using the reporter strain E. coli VJS533 harboring plasmid pHV200I−, which does not produce its own acyl-HSL but which responds to C6-HSL by producing luminescence (39). C8-HSL was quantified using the reporter strain Ralstonia solanacearum AW1-AI8 harboring plasmid p395B, which expresses lacZ in response to exogenous C8-HSL (16). LacZ activity was measured by a standard assay (35). The amounts of C6-HSL and C8-HSL were determined by comparing the activity measured from a dilution series of the extracted and purified samples to the linear range of each standard curve.
Siderophore and catalase activities.
The ability of mutant strains to produce siderophores was assessed qualitatively with chrom-azurol S indicator (CAS) agar plates and compared to that of ES114, which produces an orange halo around bacterial colonies indicative of sequestration of the iron from CAS, and two strains defective in siderophore production, KV495 and SP301, which produce no halo indicative of the absence of siderophore secretion or activity, as negative controls. The CAS was added to artificial seawater medium (6) supplemented with 0.3% CA and buffered with piperazine-N,N′-bis(ethanesulfonic acid) (pH 6.8) as previously described (19, 28) to make CAS agar plates.
Published methods for quantification of catalase activity were used without modifications, using a strain that is defective in catalase production, KV433, as a control (4, 54). Protein concentrations were determined using the Bio-Rad DC protein assay kit with protocols supplied by the manufacturer (Bio-Rad, Hercules, Calif.). The experiment was repeated with similar results.
Motility, flagellation, and chemotaxis.
Motility of exponentially growing (OD600 of 0.2 to 0.4) cells of V. fischeri in liquid cultures was assessed by light microscopy. Flagella were examined and the number of flagella per cell was determined from a total of 75 cells per treatment from three separate experiments by transmission electron microscopy (36). The data were combined and reported as the mean number of flagella ± the standard error (SE).
Swimming motility in soft agar was determined using SWT containing concentrations between 0.3 and 0.7% of Bacto Agar (Difco, Detroit, Mich.). At these agar concentrations, the polarly flagellated V. fischeri ES114 is presumed to swim, as the pattern of movement is not typical of the swarming motility seen for peritrichously flagellated Vibrio spp. (48). Three microliters of an exponentially growing culture (OD600 of 0.4) was spotted on the surface of duplicate agar plates, and the movement of the cells in the agar as a concentric circle away from the spotted culture was periodically measured at the leading edge. The ability of strains to move toward attractants was assessed by spotting 10 μl of an exponentially growing culture (OD600 of 0.4) on soft agar plates made with 0.25% Bacto Agar in 70% artificial seawater, 1% tryptone (Difco) with or without the addition of 0.5% CA or 1.6 mM serine and observing the bands of cells migrating up a concentration gradient created by the degradation of each band's attractant (13). The experiments were repeated with similar results, and the data from one representative experiment are reported.
Animal colonization.
The ability of V. fischeri strains to colonize juvenile E. scolopes squid was determined as previously described (36, 44) with the following modifications. Exponentially growing bacteria (OD600 between 0.2 and 0.4) from cultures grown with shaking at 200 rpm were suspended in a volume of between 50 and 250 ml of filtered-sterilized seawater (FSW) at a final concentration of between 110 and 20,000 CFU/ml. Squid were placed collectively into bowls in a volume that allowed a minimum of 2 ml of SW/squid for either 3 h or overnight and then transferred to fresh FSW before being placed in individual vials containing 4 ml of FSW. Each morning, squid were aseptically transferred to fresh vials containing 4 ml of FSW. Colonization of squid light organs based on bacterial cell counts recovered from squid was routinely assessed at 24, 48, and 72 h postinoculation by rinsing squid in FSW and then freezing animals at −70°C before homogenizing, serially diluting, and plating the homogenate on LBS agar plates to determine the number of CFU of V. fischeri/light organ. Aposymbiotic animals placed in SW without bacteria and otherwise treated identically were also plated to confirm the absence of contaminating V. fischeri bacteria. These experiments were repeated a minimum of two times with both strains VCW2A1 and VCW2F5, which were comparable to each other, and one representative experiment with VCW2F5 was reported.
The ability of bacteria expelled from luminous squid to colonize previously uncolonized squid was also determined. Previously uncolonized juveniles were placed in vials containing serial dilutions of FSW that contained bacteria expelled from luminous VCW2F5- or ES114-colonized animals. The number of V. fischeri cells in the SW could not be determined by direct plating due to a high background of other bacteria. After 3 h, squid were placed in fresh vials and luminescence and colonization were determined at 30 h postcolonization.
Cocolonization experiments were performed by placing squid overnight in FSW containing both VCW2F5 and a wild-type ES114 derivative, JRM200 (33), containing a Ch resistance gene inserted in the genome in single copy, at various concentrations. In cocolonization experiments, the identity of the light organ symbionts plated on LBS agar was assessed by replica plating colonies onto LBS agar containing antibiotic (Ch) selection and by visual assessment of colony morphology.
The ability to complement the colonization defects of VCW2F5 was tested with the gacA-containing plasmid, pVCW3C3, or with a vector control, pVO8. Squid inoculated with bacteria at 3,000 CFU/ml of SW were subsequently maintained in FSW containing Ch (2 μg/ml) to select for the plasmids. At 24 h postinoculation, luminescence and colonization levels were determined.
Nucleotide sequence accession number.
The nucleotide sequence of the gacA gene from V. fischeri along with flanking DNA has been submitted to the GenBank databases under accession number AY377390.
RESULTS
Cloning, characterization, and mutagenesis of the gacA gene in V. fischeri.
Amplification of ES114 (wild-type) genomic DNA using fully degenerate primers to various gacA homologs (see Materials and Methods) generated a PCR product with high sequence similarity to the genes encoding GacA homologs. Subsequently, a genomic clone containing the intact gacA gene was isolated and sequenced. Sequence analysis of the clone revealed an ORF that was 642 bp in length, encoding a predicted protein of 214 amino acids with the alternative start codon GTG. The predicted sequence of the protein was 85% identical to VarA from V. cholerae (56). Within the amino-terminal receiver domain, between amino acids 1 and 123, we identified the putative phosphate-accepting aspartate residue (D54) involved in phosphorelay and, located in the carboxy-terminal region of the protein, between amino acids 146 and 203, was a conserved helix-turn-helix domain. A partial ORF 276 bp downstream of the gacA ORF, also beginning with GTG, was homologous to UvrC. The uvrC gene is also located downstream of the gacA genes in several other bacterial species.
Three mutants were generated to study the role of GacA in V. fischeri (Table 1). Two gacA mutants included a marked-insertion mutant strain, VCW2A1 (gacA::EZ::TN<KAN-2>), and an unmarked, in-frame deletion mutant strain, VCW2F5 (ΔgacA). In other bacterial species, gacA and the downstream gene uvrC are cotranscribed; thus, insertions can cause polar loss of UvrC. To control for potential polar effects of the insertion on UvrC, a third mutant harboring an insertion in the uvrC gene was generated in strain VCW2E1 (uvrC). When grown on LBS agar plates, the two gacA mutants had colonies that were smaller, less yellow, and had a translucent morphology when compared to the wild-type strain; however, the uvrC mutant strain colony morphology was indistinguishable from that of the wild type, suggesting colony morphology was affected by GacA and not polar loss of UvrC.
The gacA mutants have a growth yield defect that is relieved by the addition of Casamino Acids.
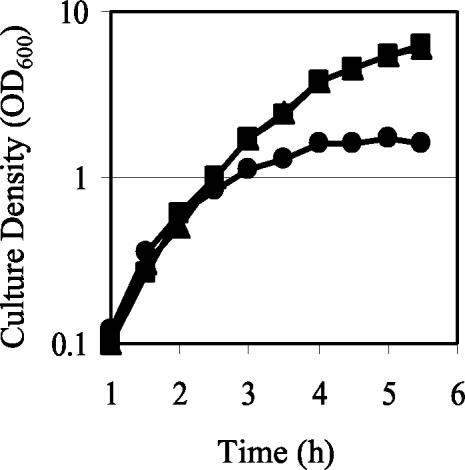
The small colony size of gacA mutants led us to evaluate whether growth was influenced by GacA. The exponential growth rates of V. fischeri mutant and wild-type strains in SWT broth did not differ (Fig. 1). However, the gacA mutants had a growth yield defect (Fig. 1) and reached a lower final cell density (OD600 of 1.8) compared to the wild type (OD600 of >5.0). No growth yield defect was observed in the uvrC mutant, which reached a final cell density similar to the wild type. When cultured on minimal agar plates with either ribose or fumarate as the sole carbon source, the ΔgacA mutant did not grow, although the wild type grew on these media, indicating that the ΔgacA mutant was unable to adapt to the metabolic requirements of prototrophic growth on these sole carbon compounds. The addition of 0.5% CA, which can serve as a source of nitrogen, carbon, and vitamins, improved but did not restore the growth of the mutant to the level seen for the wild type, indicating that an amino acid auxotrophy alone could not account for the growth defect.
FIG. 1.
Growth of wild-type V. fischeri and derivatives in culture. The optical densities (OD600) of wild-type (▪), luxI (▴), and ΔgacA (•) cultures grown in SWT were determined throughout the growth cycle. One representative experiment is presented.
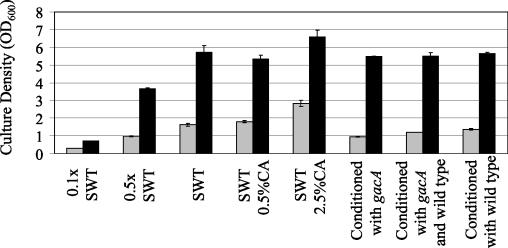
To assess whether the growth yield defect of the ΔgacA mutant was caused by either insufficient nutrients or accumulation of repressive compounds in the supernatant, we measured the growth rate and final growth yield of the wild type and the ΔgacA mutant in complex media of different composition. The media included (i) SWT diluted to different extents with 70% SW, (ii) SWT supplemented with CA, or (iii) SWT conditioned with an equal volume of cell-free supernatants from either the wild-type or ΔgacA strain or both strains grown to a final OD600 of 1.8 (Fig. 2). In all media tested, the ΔgacA mutant attained the same exponential growth rate as the wild type (data not shown), but it reached a lower cell density than the wild type (Fig. 2). Although the addition of 0.5% CA (the same amount that improved but did not restore growth of the mutant in minimal medium) to SWT did not substantially improve the growth of either strain, the addition of 2.5% CA to SWT increased the yield of both strains. Additionally, the ΔgacA mutant reached the same final cell density in 0.5× SWT as it did in SWT conditioned with ΔgacA broth. This cell density was lower than the final cell density that the mutant reached in broth conditioned with either a mixture of both wild type and ΔgacA supernatants at a 1:1 ratio or wild type alone. In contrast, the wild type reached the same cell density in all conditioned broths. These data support the hypothesis that the growth yield defect of the mutant was caused by a limitation of growth substrates rather than the generation of growth-restrictive compounds by the ΔgacA mutant. The growth yield of the mutant was fully restored by carrying the gacA gene in trans on pVCW3C3 (data not shown).
FIG. 2.
Growth yield of wild-type V. fischeri (black bars) and the ΔgacA mutant (gray bars) in various diluted and amended complex media (SWT) after 18 h of incubation. Conditioned broth was prepared by mixing SWT with cell-free supernatants of either the wild type or the ΔgacA mutant at a ratio of 1:1, or by combining SWT with cell-free supernatants of both the wild type and the ΔgacA mutant at a ratio of 2:1:1. Bars indicate the SE.
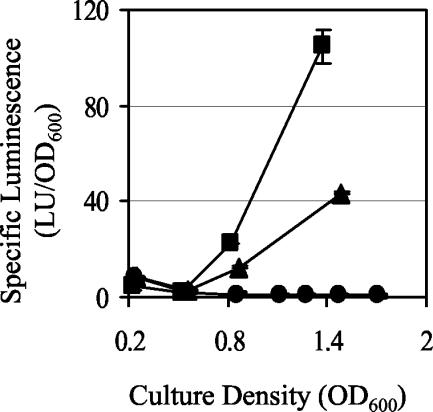
The gacA mutants have a luminescence defect that is not complemented by the addition of acyl-HSLs or aldehyde.
Luminescence was not detected from broth cultures either of the gacA::EZ::TN<KAN-2> mutant (data not shown) or of the ΔgacA mutant (Fig. 3) unless the gacA gene was restored in trans on pVCW3C3 (data not shown). In contrast, luminescence was detected from both the uvrC mutant (data not shown) and the luxI mutant (Fig. 3), which harbors a mutation in the C6-HSL synthetase and therefore is defective in the production of one of the two activating signals of the lux biosynthetic operon. Although the luxI mutant produces less luminescence than the wild type, the luxI mutant was significantly more luminous than the ΔgacA mutant. This finding implies that GacA did not simply affect LuxI activity, but instead influenced luminescence by another mechanism. One hypothesis is that the gacA mutation affected C8-HSL production, as mutants in its synthetase, AinS, produce no luminescence in culture (32).
FIG. 3.
Luminescence of wild-type V. fischeri and derivatives in culture. The luminescence of wild-type (▪), luxI (▴), and ΔgacA (•) cultures grown in SWT was determined throughout the growth cycle. One representative experiment is presented. Bars indicate the SE and are sometimes obscured by the symbols.
To determine whether the ΔgacA mutant was defective in the synthesis of C8-HSL, which is produced at a relatively high level in culture by the wild type (32), we tested the ability of excess amounts of either acyl-HSL or decanal, a substrate of the luciferase reaction that is limiting in culture, to complement the mutant's luminescence defect. Exponentially growing culture of the wild type or the luxI or ΔgacA mutant that produced no detectable light became luminous after the addition of decanal or either of the acyl HSLs (Table 2). The wild type and the luxI mutant were similar in their luminescence response to all three substances (Table 2). However, with the addition of C6-HSL or decanal, the ΔgacA mutant produced only about 20% of the luminescence of either the wild type or the luxI mutant. With C8-HSL, the ΔgacA mutant produced only 2% of the luminescence of the other two strains (Table 2). The inability of exogenous acyl-HSL to complement its luminescence defect suggests that repression of luminescence in the gacA mutants is not caused solely by an acyl-HSL or decanal deficiency but is effected by another mechanism.
TABLE 2.
Luminescence response to acyl-HSL and decanal
| Strain | Specific luminescence (LU/OD)a
|
|||
|---|---|---|---|---|
| No additions | C6-HSLb | C8-HSLb | Decanalc | |
| Wild type | <0.02 | 252 | 3.5 | 20 |
| luxI mutant | <0.02 | 203 | 2.7 | 16 |
| ΔgacA mutant | <0.01 | 43 | 0.069 | 3.5 |
One LU = 1.3 × 107 quanta/s. Luminescence was determined on aliquots of exponentially growing cultures at an OD600 of 0.25 to 0.35 from which no luminescence was detected prior to additions. One representative time point is shown.
Cells were grown with shaking in the presence of 120 nM C6-HSL or C8-HSL.
Decanal (0.01%) was added to an aliquot of culture prior to measuring luminescence.
Since the ΔgacA mutant was minimally responsive to addition of excess acyl-HSLs, it was still unclear whether the strain produced these compounds. Quantification of acyl-HSLs revealed the ΔgacA mutant produced both C6-HSL (0.07 nM) and C8-HSL (1.2 μM) at the same molarity as the wild type (0.15 nM and 1.3 μM, respectively). The luxI mutant also produced C8-HSL at a similar concentration (1.7 μM); however, as expected, no C6-HSL was detectable (<0.005 nM).
Additional colonization traits are affected by the GacA mutations in culture.
Previous studies have identified additional traits of V. fischeri important during host colonization. These include the production of catalase (32, 54) and siderophore (19), as well as motility (18, 36), all of which have been shown to be regulated by GacA in other bacterial species (22). Therefore, we determined whether GacA from V. fischeri globally controls these colonization phenotypes in culture.
The gacA::EZ::TN<KAN-2> mutant was not defective in catalase activity, as culture extracts were comparable to the wild type in the degradation of hydrogen peroxide. However, both the gacA mutants, but not the uvrC mutant, were defective at siderophore-mediated iron sequestration on CAS agar plates. CAS agar, which is a defined, low-iron medium containing 0.3% CA, did sustain growth of the gacA mutants, although they grew more slowly than other mutants that are also defective in siderophore activity (see Materials and Methods), indicating that iron limitation alone did not cause the gacA growth defect observed on minimal agar plates. An intact gacA gene supplied in trans on pVCW3C3 restored siderophore production to the gacA mutants.
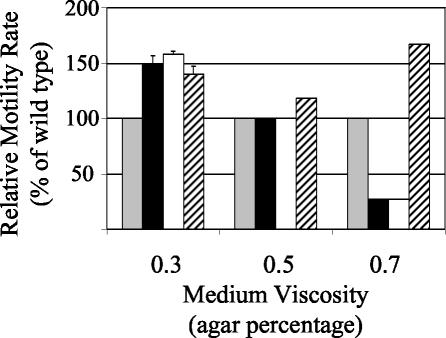
GacA also regulates motility behavior in V. fischeri, but its impact was complex. Exponentially growing gacA mutants of V. fischeri from liquid cultures were motile; however, their ability to swim through various concentrations of soft agar was altered. Although other Vibrio spp. exhibit swarming behavior on higher concentrations of agar due to a lateral flagellar gene system distinct from the polar flagella used for swimming (48), V. fischeri ES114 swims on agar concentrations between 0.25 and 0.7% and has not been reported or observed to be peritrichously flagellated or to exhibit swarming motility. At a relatively low viscosity (0.3% agar), both gacA mutants swam faster than the wild type and were similar to a hyperswimmer strain of V. fischeri, DM66 (Fig. 4). However, at a higher viscosity (0.7% agar), the gacA mutants swam more slowly than the wild type, which swam more slowly than DM66 (Fig. 4). The motility of the gacA mutants when grown at an intermediate viscosity (0.5% agar) (Fig. 4) and at all agar concentrations tested when gacA was supplied in trans on pVCW3C3 (data not shown) was indistinguishable from that of the wild type. The uvrC mutation had no detectable effect on motility (data not shown). Examination by transmission electron microscopy of the ΔgacA mutant grown in broth cultures revealed no apparent differences in flagellum length, width, or appearance; however, it was slightly hyperflagellated (5.2 ± 0.3 flagella per cell) compared to the wild type (3.1 ± 0.2 flagella per cell).
FIG. 4.
Effect of medium viscosity on the motility of wild-type V. fischeri and derivatives. The extent of movement of duplicate samples of wild-type (gray bars), ΔgacA (black bars), gacA::EZ::TN<KAN> (white bars), and hyperswimmer strain DM66 (hatched bars) cells was measured over time. Average values (± 1 standard deviation) were normalized to the wild-type rates at each viscosity. The absence of error bars indicates no variability within treatment.
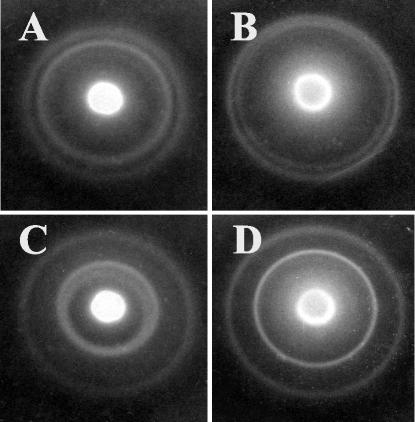
Differences in chemotaxis between gacA mutants and the wild type were also observed. At the leading edge of the migrating front, both the wild type (data not shown) and the hyperswimmer strain DM66 (Fig. 5A) created two distinct concentric bands, representing chemotaxis up a gradient created by degradation of each of two attractants, nucleosides and serine (13). In contrast, the concentric bands of the ΔgacA mutant migrated more closely to each other (Fig. 5B) and often appeared as one diffuse band. With the addition of either 0.5% CA (data not shown) or 1.6 mM serine to the medium, which slows the migration of the inner band of both wild-type (data not shown) and DM66 (Fig. 5C) cells, the two bands generated by migration of the gacA mutant became distinctly separated (Fig. 5D). This observation is consistent with the hypothesis that the gacA mutant depleted serine from the medium more rapidly than the wild type did.
FIG. 5.
Patterns of chemotaxis in soft agar by hyperswimmer derivatives of V. fischeri. The relative migration rates towards serine (inner ring) or nucleosides (outer ring) are indicated by the ring diameters. Shown are the patterns of strain DM66 (A and C), which is the same as the wild-type pattern, and the ΔgacA mutant (B and D) in medium without (A and B) or with (C and D) the addition of 1.6 mM serine.
Symbiotic gacA mutants are impaired in host colonization and growth, but not in luminescence.
Because gacA influenced traits in culture that could affect (i) colonization initiation, e.g., motility (18), (ii) growth within the light organ, e.g., substrate utilization (20), and (iii) persistent association, e.g., luminescence (52) and siderophore production (19), we tested the ability of the ΔgacA mutant to colonize, grow within, and maintain an association with juvenile squid. When newly hatched juvenile squid were placed for 3 h in SW containing 2 × 103 wild-type cells/ml, 100% of the animals became colonized; however, only 51% of animals exposed to the same concentration of ΔgacA cells were successfully colonized. A colonization efficiency of 100% was achieved by gacA mutants only after a 14- to 18-h inoculation with ≥104 CFU/ml, a level that is at least 50-fold higher than that required by the wild type.
Although most ΔgacA mutant-colonized squid produced no detectable luminescence, a subset was luminous (Table 3). These data contrast with what we observed with the gacA mutant grown in culture, which never produced detectable luminescence without the addition of acyl-HSL or decanal (Fig. 3 and Table 2). We confirmed that bacteria isolated from these luminous squid were ΔgacA based on both their colony morphology on LBS and CAS agar and their luminescence and growth yield phenotypes in culture (data not shown); however, the possibility remained that a mutation had occurred that suppressed squid phenotypes or that the strains had adapted in some other way to the light organ environment. We confirmed that ΔgacA mutants from luminous animals had not acquired a mutation that suppressed gacA colonization phenotypes, because such squid isolates retained a comparably low efficiency of colonization and a low proportion of luminescence (10%) characteristic of the original ΔgacA mutant inoculum. Similarly, bacteria directly expelled from luminous gacA mutant-colonized squid and not cultured in medium prior to a subsequent exposure to squid were characteristically impaired at colonizing juveniles, whereas expelled, wild-type bacteria were not impaired, even when diluted 100-fold (data not shown). Thus, there was no evidence that ΔgacA symbionts in luminous animals had adapted to the host, improving their ability to reinfect squid.
TABLE 3.
Luminescence characteristics of colonized squid
| Strain | % Colonized squid that were detectably luminous at:
|
LUa | CFUb | Specific luminescencec | ||
|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | ||||
| Wild type | 100 | 100 | 100 | 247 ± 42 | (1.2 ± 0.1) × 105 | 5.8 ± 3 |
| ΔgacA mutant | 10 | 11 | 14 | 16.4 ± 8.7 | (1.0 ± 0.4) × 104 | 5.0 ± 2 |
One LU = 1.3 × 104 quanta/s and is reported as the mean ± SE. Luminescence is reported from 8 of 72 ΔgacA mutant-colonized squid and 32 wild-type-colonized squid that were detectably luminous on the day cell counts were determined.
Mean CFU of only luminous squid reported ± SE.
The specific luminescence was calculated as the mean of the sum of individual specific luminescence values (LU/103 CFU from each luminous individual) reported ± SE and does not represent the average LU/average CFU.
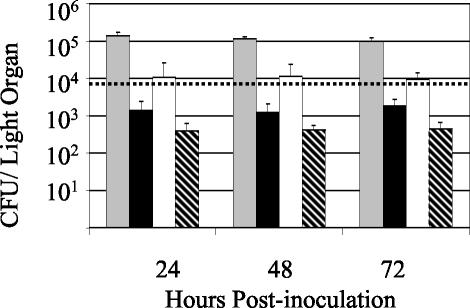
Enumeration of bacteria from the light organs of colonized squid provided insight into why there were differences in luminescence levels between various ΔgacA mutant-colonized squid. Wild-type bacterial populations averaged 1.2 × 105 CFU/squid, whereas ΔgacA populations averaged only 1.5 × 103 CFU/squid, and this density was maintained over several days (Fig. 6), whereas previously characterized derivatives of V. fischeri in which luminescence has been either abolished or reduced do not maintain their initial population levels and their population diminishes by 48 h postinoculation (32). Squid colonized by the wild type became detectably luminous between 7 and 9 h postinoculation, when their populations reached an average of 8 × 103 CFU/squid (Fig. 6). On average, ΔgacA mutant-colonized animals that were dark contained populations of only 4 × 102 cells and, thus, were below this minimum level of luminescence detection. The ΔgacA mutant-colonized animals that were detectably luminous had larger symbiont populations (Table 3) and were above the detection limit (Fig. 6). Therefore, the inability of the ΔgacA mutant to reach a normal colonization level in the light organ most likely prevented the induction of detectable levels of luminescence in these animals. Because a percentage of ΔgacA mutant-colonized animals was detectably luminous and their symbionts did not differ from the wild type in their luminescence per bacterial cell (Table 3), we inferred that GacA was not required to achieve light emission in the squid. Normal (100%) colonization efficiency (data not shown), colonization level, and luminescence of the mutant were fully restored by carrying the gacA gene in trans on pVCW3C3 (Table 4).
FIG. 6.
Colonization levels of wild-type V. fischeri and its ΔgacA derivative. The mean number (± SE) of symbiotic bacteria per colonized squid for each treatment was determined by plating light organ contents at different times following colonization with the wild type (gray bars) or the ΔgacA mutant (black bars). The colonization levels of the subsets of ΔgacA mutant-colonized animals that were either detectably luminous (white bars) or not (hatched bars) are also plotted separately. The dashed line represents the average CFU at the luminescence detection limit for wild-type-colonized squid. The mean CFU level in nonluminous wild-type-colonized squid was below this detection limit.
TABLE 4.
Complementation of ΔgacA symbiotic defects by gacA
| Strain | Plasmid | LUa | CFUb |
|---|---|---|---|
| Wild type | pVO8 | 175 ± 54 | (4.9 ± 1.2) × 105 |
| ΔgacA mutant | pVO8 | 10 ± 8 | (1.3 ± 0.4) × 103 |
| ΔgacA mutant | pVCW3C3 | 117 ± 22 | (3.4 ± 0.6) × 105 |
One LU = 1.3 × 104 quanta/s and is reported as the mean ± SE.
Mean CFU per light organ is reported ± SE.
gacA mutants are severely impaired at competitively initiating colonization but are not displaced by the wild type in cocolonized light organs.
Determining the relative effectiveness of mutant and wild-type bacteria during coinoculation experiments can help elucidate interactions between the strains as they initiate association with the host, because a direct competition can accentuate defects and reveal otherwise subtle differences between strains. Thus, we used such competition experiments to determine (i) whether the wild type either complemented or exacerbated the association defects of the mutant and, conversely, (ii) whether the mutant interfered with colonization by the wild type.
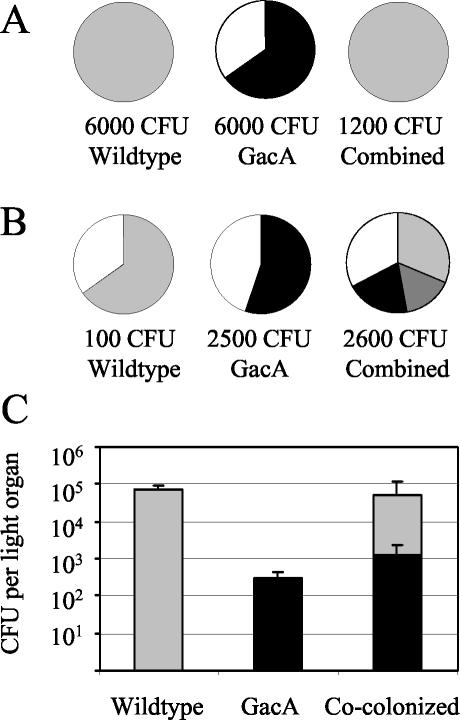
When squid were coinoculated for 3 h with both the ΔgacA mutant and the wild type, each at 6 × 103 CFU/ml, less than 0.05% of the cells present in the symbiotic population at either 24 or 48 h were ΔgacA (Fig. 7A). This result indicated that the ΔgacA mutant was at a competitive disadvantage in colonization in the presence of wild-type cells. Since at an inoculum of 6 × 103 CFU/ml the wild type by itself colonized 100% of the squid, whereas the ΔgacA mutant by itself colonized only 65% of the squid (Fig. 7A), we hypothesized that the mutant's defect was expressed during the initiation of the colonization rather than during competitive growth within the light organ. To test this hypothesis, we adjusted the inoculum so that it would give the two strains an equal chance at initiating symbiosis. To equalize the strains' colonization efficiencies, we combined ΔgacA cells at a concentration of 2.5 × 103 CFU/ml, which by itself resulted in colonization of 55% of the squid, with wild-type cells at a concentration of only 1.1 × 102 CFU/ml, which by itself resulted in colonization of 65% of the squid (Fig. 7B). With this 23:1 advantage, the ΔgacA mutant initiated the cocolonization process with the efficiency expected and successfully cocolonized squid with the wild type (Fig. 7B). In the mixed symbiotic populations that resulted, the mutant and the wild type attained essentially the same levels as they did when they colonized in the absence of the other strain (Fig. 7C). Thus, it appears that the ΔgacA competition defect is important primarily during initiation. Even after initiation, wild-type cells neither complemented the growth defect of the ΔgacA mutant nor, surprisingly, displaced the ΔgacA population after 48 h postinoculation. Similarly, there was no indication that the presence of the ΔgacA mutant affected the ability of the wild type to attain and maintain its normal level of colonization.
FIG. 7.
Colonization of squid by wild-type V. fischeri and its ΔgacA derivative in mixed bacterial inoculations. (A and B) The proportion of wild-type-colonized (light gray), ΔgacA mutant-colonized (black), cocolonized (dark gray), or uncolonized (white) squid after inoculation with either 6,000 CFU of each strain by itself or 12,000 CFU of both strains combined at a 1:1 ratio (wild type/ΔgacA mutant) (n = 20 for each treatment) (A), or with a total of 110 CFU of the wild type by itself (n = 20), 2,500 CFU of the ΔgacA mutant by itself (n = 20), or 2,600 CFU of both strains combined at a 1:23 ratio (wild type/ΔgacA) (n = 70) (B). (C) The mean number (± SE) of wild-type (light gray) and ΔgacA (black) CFU per light organ at 48 h postinoculation is shown from the three subgroups that resulted from the combined treatment presented in panel B (n = 46). For wild-type-colonized or cocolonized squid a minimum of 200 colonies were identified, but for ΔgacA mutant-colonized squid frequently fewer than 100 bacterial colonies were available for assessment due to this mutant's lower colonization level.
DISCUSSION
We show here that in V. fischeri GacA globally controls multiple phenotypes, several of which are generally important to bacteria during host colonization, and at least one of which, luminescence, plays a specific role in this symbiotic association (52). This study also establishes that growth substrate utilization is coregulated by GacA along with other colonization traits. The contribution of each phenotype to the observed colonization defects remains speculative. However, this study confirms the conservation of GacA as a bacterial colonization regulator and indicates that GacA regulates not only pathogenic associations but also a specific, beneficial animal association.
Substrate utilization and growth.
Although GacS/GacA has long been associated with the regulation of secondary metabolism (22), until recently the implication that GacA is an important regulator of growth had been largely overlooked. In several bacterial species, including Pseudomonas fluorescens (55), Azotobacter vinelandii (8), and E. coli (37), GacA controls the production of RpoS, an alternative sigma factor associated with the transition from rapid growth rates to slow or nongrowing states. Consistent with reductions in RpoS, spontaneous gacS and gacA mutants in P. fluorescens are frequently isolated from and can overtake stationary-phase cultures (14), as altered RpoS function can confer a growth advantage in stationary-phase (GASP) phenotype (57, 58). Recent work has confirmed the role of GacS/GacA in growth and substrate utilization, as mutants in the GacS/GacA homologs of E. coli are similar to CsrA mutants and show preference for growth on gluconeogenic substrates, such as amino acids, but not glycolytic growth substrates (41). Enhanced ability to catabolize amino acids can also confer a GASP phenotype (59). In several bacterial species, including P. fluorescens (1, 5, 23), E. carotovora (12, 25), and E. coli (49), GacA antagonizes the repressive activity of CsrA homologs via positive regulation of small regulatory RNA paralogs of csrB. These examples demonstrate that there is a strong link between GacA and growth.
Several phenotypes of the gacA mutants of V. fischeri substantiate the role of gacA in primary metabolism, including (i) their inability to grow on a minimal medium with simple sugars as a carbon source, (ii) their low growth yield in rich medium (Fig. 1), and (iii) a growth yield defect in squid light organs (Table 3; Fig. 6). Further results suggest that as with the homologous mutant of E. coli (41), the gacA mutant of V. fischeri preferentially utilized amino acids as growth substrates. These include (i) growth on minimal medium with CA as a sole carbon source, (ii) improved growth yield in rich medium when supplemented with CA (Fig. 2), and (iii) enhanced chemotaxis toward serine consistent with a more rapid utilization and depletion of this amino acid (Fig. 5). Interestingly, the wild type did not preferentially deplete the substrates that are growth limiting for the gacA mutant; in fact, the gacA mutant reached a higher cell density in wild-type-conditioned medium than in gacA-conditioned medium, whereas the wild type reached the same cell density in both conditioned media (Fig. 2). Such differences between the wild type and the gacA mutant in growth substrate utilization could allow the strains to occupy different nutritional niches during early stages of growth in cocolonized light organs, allowing the gacA mutant to maintain its minority population despite the abundance of competitors (Fig. 7C). Although the extent of the metabolic defects of the gacA mutant of V. fischeri remains unknown, the inability of the mutant to grow on the gluconeogenic substrate fumarate implies that regulation by GacA in V. fischeri may be more complex than a defect in switching between gluconeogenesis and glycolysis, as has been observed with E. coli (41).
The growth defects described both in culture and during symbiotic association imply that the gacA mutation interfered with the ability of V. fischeri to sense and adapt to the nutrient conditions of the light organ. For instance, the limited availability of amino acids in the light organ could underlie the restricted growth of the gacA mutant much as it does for amino acid auxotrophs (20). Recently, it has been reported that pathogenic Salmonella enterica serovar Typhimurium recognizes its location within the enteric tract by sensing the presence of intestinal short-chain fatty acids and, in response, induces invasion genes through a process mediated by the GacS/GacA homologs SirA/BarA (27). A similar inability to respond appropriately to a light organ signal could impair the gacA mutant not only during growth in the light organ (Table 3; Fig. 6) but also during initiation (Fig. 7A and B). Further characterization of the gacA mutant may elucidate which nutrient resources serve as host-specific signals during symbiotic association.
Luminescence regulation.
One of the most striking phenotypes of the GacA mutants in culture was their inability to produce luminescence, a trait that is specifically important for the squid-V. fischeri association (52). Due to the dependence of squid luminescence on acyl-HSL signal accumulation (32, 52) and the linkage of GacS/GacA to acyl-HSL expression in other host-associated bacteria (9, 15, 43), we suspected that the dark phenotype of the GacA mutant of V. fischeri resulted from a deficiency in acyl-HSL synthesis or accumulation. However, the GacA mutant produced typical levels of both C6-HSL and C8-HSL in culture and responded only partially to the addition of excess acyl-HSLs (Table 2), suggesting that luminescence expression could be blocked in the absence of GacA. Surprisingly, whereas GacA was required for luminescence in culture, it was not required for characteristic induced levels of luminescence per bacterial cell in the host light organ (Table 3). In contrast luxI mutants, which are luminous in culture (Fig. 3), are not luminous in the light organ even though they initially reach populations similar to the wild-type strain (32, 52). Although other factors may play a role, these results demonstrate that C6-HSL-mediated induction of luminescence and not GacA is the dominant activating pathway in the squid host.
Other symbiosis-related phenotypes.
The appropriate expression of motility behavior, which is regulated by GacA in other bacteria (17, 26, 56), is critical during early stages of squid-host association (18, 36). Although the hyperflagellation of gacA mutants may explain their hyperswimmer phenotype in low-viscosity medium, it is unclear why they appear less motile than the wild type in high-viscosity medium, since other hyperflagellated strains swim faster than the wild type at all medium viscosities (Fig. 4) (36). Furthermore, because V. fischeri ES114 does not exhibit the swarming motility that other Vibrio spp. exhibit (48), such differences cannot be explained as a defect in lateral flagella. Since nonmotile V. fischeri strains cannot initiate colonization (18), this study implies that the gacA mutants were motile during squid association; however, their hyperflagellation phenotype could lead to a delay in colonization (36).
We investigated two additional colonization traits that are often present in GacA regulons. Catalase production was identified as an important bacterial factor during growth in the squid light organ (54) and indicated that the oxidative environment of the light organ may restrict the growth of certain bacteria. Although bacterial defenses to oxidative damage are controlled by GacA in other bacteria (37, 55), V. fischeri did not require GacA for normal catalase activity in culture. In contrast, GacA was required for the production of another colonization factor, siderophore. The production of siderophores by pathogenic bacteria can contribute to virulence by mediating iron acquisition from host sources, but they can also contribute to protection from oxidative damage by preventing the Fe2+-catalyzed generation of free radicals (42). A recent study determined that the siderophore biosynthetic gene, iucA, is induced by V. fischeri cells within squid light organs (50), supporting the importance of iron sequestration during persistent host association (19).
GacA and symbiont specificity.
This study demonstrated that gacA mutants were not only defective in reaching normal colonization levels but also were severely impaired during initiation and early colonization phases of symbiosis, suggesting that GacA may coregulate defense and communication activities along with nutrient acquisition. During host association, it is postulated that a selective winnowing occurs that eventually allows colonization only by V. fischeri (53). Indeed, while other bacterium species can participate in the initial stages of association, even at these early stages V. fischeri exhibits dominance (38). Such a selection process is likely to involve not only symbiont defense traits but also reciprocal bacterium-host signaling and recognition. Analysis of the GacA regulon in V. fischeri is ultimately aimed at discovering such traits that may elucidate how bacteria colonize and maintain beneficial associations with animals.
Acknowledgments
We thank C. DeLoney-Marino, K. Visick, and J. Graf for protocols and helpful suggestions and A. Schaefer for assistance with acyl-HSL purification and quantification. We further thank A. Schaefer, D. Millikan, C. Lupp, J. McCann, J. Graber and L. Sycuro for technical assistance, helpful conversations, and comments on earlier versions of the manuscript and D. K. Willis for guidance in the initiation of these studies.
This work was supported in part by a postdoctoral fellowship in microbial biology from the National Science Foundation to C.A.W., by National Institutes of Health grant RR12294 to E.G.R. and M. McFall-Ngai, by National Science Foundation grant IBN0211673 to M. McFall-Ngai and E.G.R., and by a W. M. Keck Foundation grant to E.G.R. and others.
REFERENCES
- 1.Aarons, S., A. Abbas, C. Adams, A. Fenton, and F. O'Gara. 2000. A regulatory RNA (prrB RNA) modulates expression of secondary metabolite genes in Pseudomonas fluorescens F113. J. Bacteriol. 182:3913-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S., T. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. Wiley and Sons, Inc., New York, N.Y.
- 4.Beers, R. F., Jr., and I. W. Sizer. 1952. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195:133-140. [PubMed] [Google Scholar]
- 5.Blumer, C., S. Heeb, B. Pessi, and D. Haas. 1999. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc. Natl. Acad. Sci. USA 96:14073-14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boettcher, K. J., and E. G. Ruby. 1995. Detection and quantification of Vibrio fischeri autoinducer from symbiotic squid light organs. J. Bacteriol. 177:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castaneda, M., J. Sanchez, S. Moreno, C. Nunez, and G. Espin. 2001. The global regulators GacA and sigma S form part of a cascade that controls alginate production in Azotobacter vinelandii. J. Bacteriol. 183:6787-6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chancey, S. T., D. W. Wood, and L. S. Pierson III. 1999. Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 65:2294-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chancey, S. T., D. W. Wood, E. A. Pierson, and L. S. Pierson III. 2002. Survival of GacS/GacA mutants of the biological control bacterium Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. Appl. Environ. Microbiol. 68:3308-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbell, N. A., and J. E. Loper. 1995. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J. Bacteriol. 177:6230-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui, Y., A. Chatterjee, and A. K. Chatterjee. 2001. Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and HarpinEcc. Mol. Plant Microbe Interact. 14:516-526. [DOI] [PubMed] [Google Scholar]
- 13.DeLoney-Marino, C. R., C. R. Wolfe, and K. L. Visick. Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 14.Duffy, V. K., and G. Defago. 2000. Controlling instability in gacS-gacA regulatory genes during inoculate production of Pseudomonas fluorescens biocontrol strains. Mol. Plant Microbe Interact. 66:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eriksson, A. R. B., R. A. Andersson, M. Pirhonen, and E. T. Palva. 1998. Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol. Plant Microbe Interact. 8:743-752. [DOI] [PubMed] [Google Scholar]
- 16.Flavier, A. B., L. M. Ganova-Raeva, M. A. Schell, and T. P. Denny. 1997. Hierarchical autoinduction in Ralstonia solanacearum: control of acyl-homoserine lactone production by a novel autoregulatory system responsive to 3-hydroxypalmitic acid methyl ester. J. Bacteriol. 179:7089-7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodier, R. I., and B. M. M. Ahmer. 2001. SirA orthologs affect both motility and virulence. J. Bacteriol. 183:2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graf, J., P. V. Dunlap, and E. G. Ruby. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 176:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graf, J., and E. G. Ruby. 2000. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol. Microbiol. 37:168-179. [DOI] [PubMed] [Google Scholar]
- 20.Graf, J., and E. G. Ruby. 1998. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl. Acad. Sci. USA 95:1818-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44:107-118. [DOI] [PubMed] [Google Scholar]
- 22.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant Microbe Interact. 14:1351-1363. [DOI] [PubMed] [Google Scholar]
- 23.Heeb, S., C. Blumer, and D. Haas. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184:1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyytiainen, H., M. Montesano, and E. T. Palva. 2001. Global regulators ExpA (GacA) and KdgR modulate extracellular enzyme gene expression through the RsmA-RsmB system in Erwinia carotovora subsp. carotovora. Mol. Plant Microbe Interact. 14:931-938. [DOI] [PubMed] [Google Scholar]
- 26.Kinscherf, T. G., and D. K. Willis. 1999. Swarming by Pseudomonas syringea B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J. Bacteriol. 181:4133-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawhon, S. D., R. Maurer, M. Suyemoto, and C. Altier. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46:1451-1464. [DOI] [PubMed] [Google Scholar]
- 28.Lee, K. H., and E. G. Ruby. 1994. Competition between Vibrio fischeri strains during initiation and maintenance of a light organ symbiosis. J. Bacteriol. 176:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, K. H., and E. G. Ruby. 1994. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl. Environ. Microbiol. 60:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, K. H., and E. G. Ruby. 1995. Symbiotic role of the viable but nonculturable state of Vibrio fischeri in Hawaiian coastal seawater. Appl. Environ. Microbiol. 61:278-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lund, A. H., M. Dutch, and F. S. Pedersen. 1996. Increased cloning efficiency by temperature-cycle ligation. Nucleic Acids Res. 24:800-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupp, C., M. Urbanowski, E. P. Greenberg, and E. G. Ruby. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50:319-331. [DOI] [PubMed] [Google Scholar]
- 33.McCann, J., E. V. Stabb, D. S. Millikan, and E. G. Ruby. 2003. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl. Environ. Microbiol. 69:5928-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meighen, E. A., and P. V. Dunlap. 1993. Physiological, biochemical and genetic control of bacterial bioluminescence. Adv. Microb. Physiol. 34:1-67. [DOI] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Millikan, D. S., and E. G. Ruby. 2002. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl. Environ. Microbiol. 68:2519-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukhopadhyay, S., J. P. Audia, R. N. Roy, and H. E. Schellhorn. 2000. Transcriptional induction of the conserved alternative sigma factor RpoS in Escherichia coli is dependent on BarA, a probable two-component regulator. Mol. Microbiol. 37:371-381. [DOI] [PubMed] [Google Scholar]
- 38.Nyholm, S. V., B. Deplancke, H. R. Gaskins, M. A. Apicella, and M. J. McFall-Ngai. 2002. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl. Environ. Microbiol. 68:5113-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pernestig, A. K., S. J. Normark, D. M. Georgellis, and O. Melefors. 2000. The role of the AirS two-component system in uropathogenic Escherichia coli. Adv. Exp. Med. Biol. 485:137-142. [DOI] [PubMed] [Google Scholar]
- 41.Pernestig, A.-K., D. Georgellis, T. Romeo, K. Suzuki, H. Tomenius, S. Normark, and O. Melefors. 2003. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J. Bacteriol. 185:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 43.Reimmann, C., M. Beyeler, M. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Hass. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 44.Ruby, E. G., and L. M. Asato. 1993. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbioses. Arch. Microbiol. 159:160-167. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Schaefer, A. L., B. L. Hanselka, M. R. Parsek, and E. P. Greenberg. 2000. Detection, purification, and structural elucidation of the acylhomoserine lactone inducer of Vibrio fischeri luminescence and other related molecules. Methods Enzymol. 305:288-301. [DOI] [PubMed] [Google Scholar]
- 47.Stabb, E. V., and E. G. Ruby. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 48.Stewart, B. J., and L. L. McCarter. 2003. Lateral flagellar gene systems of Vibrio parahaemolyticus. J. Bacteriol. 185:4508-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki, K., X. Wang, T. Weilbacher, A.-K. Pernestig, O. Melefors, D. Georgellis, P. Babitzke, and T. Romeo. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 184:5130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Visick, K. L., and E. G. Ruby. 1997. New genetic tools for use in the marine bioluminescent bacterium Vibrio fischeri, p. 119-122. In J. W. Hastings, L. J. Kricka, and P. E. Stanley (ed.), Bioluminescence and chemiluminescence. John Wiley & Sons, New York, N.Y.
- 51.Visick, K. L., and E. G. Ruby. TnluxAB insertion mutants of Vibrio fischeri with symbiosis-regulated phenotypes. Nova Acta Leopold., in press.
- 52.Visick, K. L., J. Foster, J. Doino, M. McFall-Ngai, and E. G. Ruby. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Visick, K. L., and M. J. McFall-Ngai. 2000. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J. Bacteriol. 182:1779-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visick, K. L., and E. G. Ruby. 1998. The periplasmic group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J. Bacteriol. 180:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whistler, C. A., N. A. Corbell, A. Sarniguet, W. Ream, and J. E. Loper. 1998. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor σs and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriol. 180:6635-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong, S. M., P. A. Carroll, L. G. Rahme, F. M. Ausubel, and S. B. Calderwood. 1998. Modulation of expression of the ToxR regulon in Vibrio cholerae by a member of the two-component family of response regulators. Infect. Immun. 66:5854-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zambrano, M. M., and R. Kolter. 1996. GASPing for life in stationary phase. Cell 86:181-184. [DOI] [PubMed] [Google Scholar]
- 58.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]
- 59.Zinser, E. R., and R. Kolter. 1999. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 181:5800-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]