Abstract
SopE is a bacteriophage-encoded effector protein of Salmonella enterica serovar Typhimurium that is translocated into the cytosol of eukaryotic cells by a type III secretion system (TTSS) (W.-D. Hardt, H. Urlaub, and J. E. Galán, Proc. Natl. Acad. Sci. USA 95:2574-2579, 1998; M. W. Wood, R. Rosqvist, P. B. Mullan, M. H. Edwards, and E. E. Galyov, Mol. Microbiol. 22:327-338, 1996). In this study, we provide evidence that an unlinked gene carried within the Salmonella pathogenicity island 1 (SPI-1), invB (K. Eichelberg, C. Ginocchio, and J. E. Galán, J. Bacteriol. 176:4501-4510, 1994), is required for the secretion of SopE through the SPI-1 TTSS. Furthermore, far-Western blotting analysis shows that SopE directly interacts with InvB through a domain located at its amino terminus. We conclude that InvB is the TTSS-associated chaperone for SopE.
Many gram-negative bacteria that are pathogenic for humans, animals, and plants have evolved a specialized protein secretion system, designated type III, which mediates the delivery of a myriad of virulence effectors into eukaryotic cells (6, 13). Once translocated, these effectors are able to subvert host cellular processes for the benefit of the infecting pathogen. Salmonella enterica is equipped with two type III secretion systems (TTSSs), which contribute to pathogenesis at different stages during infection (12). One of the Salmonella TTSSs, encoded within Salmonella pathogenicity island 1 (SPI-1), mediates the initial interaction of Salmonella with the intestinal epithelium, eventually leading to bacterial internalization and the production of proinflammatory cytokines (15). Central to the stimulation of these responses is SopE, a Cdc42 and Rac1 guanine nucleotide exchange factor encoded within a lysogenic (or for some strains, defective) bacteriophage that is integrated at a chromosomal location away from SPI-1 (17, 18, 22, 33). Many effector proteins destined to be secreted by the type III secretion machinery are often associated with specific chaperones that form a tight complex by binding a discrete domain within the amino terminus of their cognate substrates (24, 26, 32). Although the function of these chaperones is not completely understood, it is clear that they maintain the substrate proteins as unfolded polypeptides within the bacterial cytoplasm, presumably in a secretion-competent state (1, 27). Although poorly conserved at the primary amino acid sequence level, the crystal structures of several TTSS-associated chaperones have revealed a remarkable structural conservation among the members of this protein family (1, 27). A chaperone for SopE has not yet been identified. However, several biochemical properties of this protein suggest that it must have a chaperone. (i) Full-length SopE, but not a deletion mutant version lacking the first 78 amino acids, is insoluble when expressed in Escherichia coli (3, 17). (ii) The catalytic effector domain of SopE has been mapped to amino acid residues 78 to 240 (3). (iii) The first ∼100 amino acids of SopE are sufficient to mediate the translocation of heterologous proteins into host cells (10). TTSS-associated chaperones are often, though not always, encoded in the vicinity of their cognate substrate proteins (32). Inspection of the chromosomal region in the vicinity of SopE did not reveal the presence of any open reading frame capable of encoding a protein that could constitute a candidate for its putative chaperone (i.e., a protein of small molecular weight, acidic pI, and propensity to form amphipathic α-helices). We hypothesized that since SopE is specifically secreted by the SPI-1 TTSS, a protein encoded within this pathogenicity island may serve as its cognate chaperone.
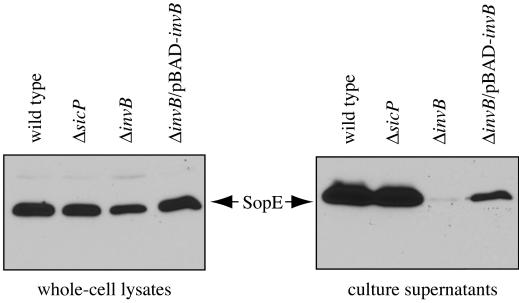
Two TTSS-associated chaperones are encoded within SPI-1: SicP, the chaperone for SptP (11), and InvB, the chaperone for SipA (2, 9). It has been previously shown that some TTSS-associated chaperones can exert their function on more than one substrate (21, 29). Absence of the cognate chaperones most often leads to deficiency of secretion and/or expression of the cognate effector proteins (24). We therefore examined the effect of loss-of-function mutations in either sicP or invB on the expression and secretion of SopE. In-frame deletions of sicP or invB were introduced into an S. enterica serovar Typhimurium strain carrying an M45 epitope-tagged SopE in the chromosome. Strains were grown under SPI-1-TTSS-inducing conditions (0.3 M NaCl) (5); whole cells and culture supernatants were harvested when cultures reached an optical density measured at 600 Å of 0.8 and were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The samples were then transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore) and immunoblotted with a monoclonal antibody directed to the M45 epitope tag (23). Neither secretion nor expression of SopE was altered in the strain carrying a sicP deletion (Fig. 1). In contrast, the level of SopE was drastically reduced in culture supernatants of a strain harboring an invB deletion (Fig. 1, right panel), suggesting that InvB is required for efficient SopE secretion. The secretion defect associated with the invB mutation could be complemented by expression of invB on an arabinose-inducible plasmid (16) (Fig. 1, right panel). Secretion of other TTSS-secreted proteins such as SptP and SipB was unaffected in the ΔinvB strain (data not shown), indicating that the secretion defect observed in this strain was not the result of an overall effect on TTSS-mediated secretion.
FIG. 1.
InvB is required for SopE secretion. A Salmonella serovar Typhimurium strain carrying a chromosomal copy of M45 epitope-tagged sopE (wild type) and isogenic derivatives carrying deletion mutations in sicP or invB were grown under SPI-1-TTSS-inducing conditions (5). The presence of SopE-M45 in whole-cell lysates and culture supernatants was evaluated by Western immunoblot analysis using a monoclonal antibody directed to the M45 epitope as previously described (11). A complementing arabinose-inducible plasmid, pBAD-invB, was introduced into the ΔinvB mutant strain, and whole-cell lysates and culture supernatants of the strain grown under inducing conditions (in the presence of 0.02% arabinose) were prepared under identical conditions.
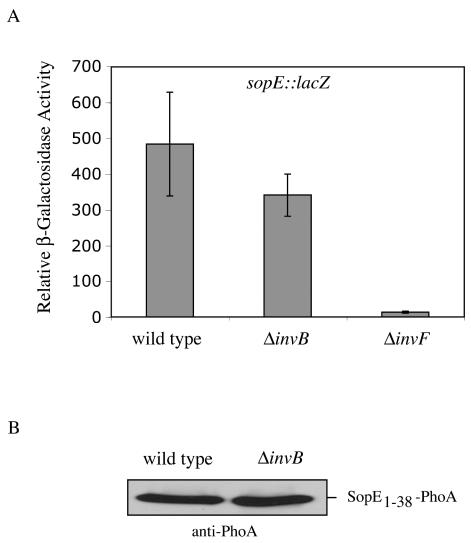
It is often observed that the stability of secreted proteins within the bacterial cytoplasm is compromised in the absence of their cognate chaperones (24). In addition, it has been reported that some chaperones control the transcription or the translation of genes encoding their cognate secreted proteins (7, 29). Despite the drastic defect in secretion, the levels of SopE in whole-cell lysates of the ΔinvB strain were only slightly reduced (Fig. 1, left panel). Furthermore, transcription and translation of SopE in the ΔinvB strain were also equivalent to those of the wild type (Fig. 2). This behavior of SopE is reminiscent of the Yersinia species effector proteins YopH, YscM, and YopN, which in the absence of their chaperones are produced but not secreted (4, 19, 25, 30).
FIG. 2.
InvB does not affect the transcription or translation of sopE. (A) Salmonella serovar Typhimurium carrying a sopE::lacZ transcriptional fusion (wild type) (8) and isogenic derivatives carrying a ΔinvB or ΔinvF (negative control) mutation were grown under SPI-1-TTSS-inducing conditions (5). The ΔinvF strain was included as a negative control since this transcriptional regulator controls the expression of SopE, and in its absence, expression of sopE is abolished(20). β-Galactosidase activities of whole-cell lysates of these strains were measured by using chemiluminescence as indicated by the manufacturer (Roche). Results represent the means ± standard deviations of three independent determinations. (B) A plasmid encoding a translational fusion of amino acids 1 to 38 of SopE to PhoA under the regulation of the native sopE promoter was introduced into wild-type Salmonella serovar Typhimurium or an ΔinvB isogenic derivative. Strains were grown under SPI-1-TTSS-inducing conditions (5), and the levels of the SopE1-38-PhoA chimeric protein in whole bacterial cell lysates were examined by Western immunoblot analysis using a rabbit antibody directed against PhoA.
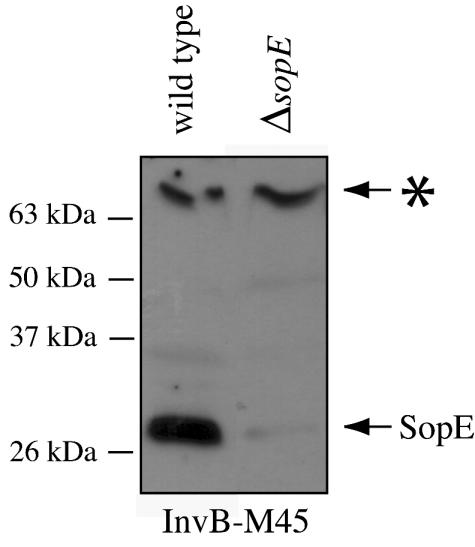
A key characteristic of chaperones is their ability to bind to their cognate substrates (31). To investigate whether InvB is able to bind to SopE, we utilized far-Western blotting analysis as previously described (11). Wild-type Salmonella serovar Typhimurium (SL1344) and its isogenic derivative carrying a nonpolar in-frame deletion of sopE were grown under SPI-1-TTSS-inducing conditions, and proteins in whole-cell lysates were separated by SDS-PAGE and transferred to a PVDF membrane. The membranes were then treated with a soluble extract of an Escherichia coli strain expressing InvB-M45 epitope tag (equivalent to 109 CFU) for 2 h, followed by Western immunoblot analysis using a monoclonal antibody directed to the M45 epitope tag. Far-Western blot analysis revealed an InvB-interacting band corresponding to the molecular mass of SopE (∼28 kDa) (Fig. 3). This band was not observed in the sopE mutant, which strongly suggests that InvB specifically binds to SopE. A high-molecular-mass band (>70 kDa) presumably corresponding to SipA was also detected, in keeping with the reported activity of InvB as a chaperone for SipA (2) (Fig. 3).
FIG. 3.
InvB specifically binds SopE. Whole-cell lysates of wild-type Salmonella serovar Typhimurium or its isogenic ΔsopE mutant were separated by SDS-PAGE and transferred to a PVDF membrane. The blot was treated with a soluble lysate of E. coli expressing M45 epitope-tagged InvB, and the bound InvB-M45 was detected with a monoclonal antibody directed against M45 as previously described (11). Notice that in addition to SopE, InvB binds to a high-molecular-mass band, which has been tentatively identified as SipA, consistent with a previous report (2). More experiments would be required for confirmation of the identity of this protein.
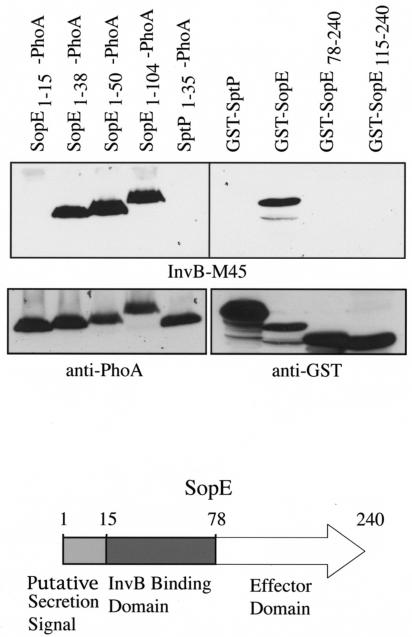
Far-Western blot analysis was also used to dissect the InvB-interacting domain of SopE. Various amino-terminal segments of SopE (amino acid residues 1 to 15, 1 to 38, 1 to 50, and 1 to 104) were fused to PhoA and introduced into serovar Typhimurium carrying an in-frame deletion of sopE. In addition, various carboxy termini of SopE (amino acid residues 78 to 240 and 115 to 240) were fused to glutathione S-transferase (GST) and expressed in E. coli. Whole-cell extracts of these strains were separated by SDS-PAGE and transferred to PVDF membranes, which were then overlaid with a lysate of an E. coli strain expressing InvB-M45 and then immunoblotted with monoclonal antibody directed against the M45 epitope. InvB was unable to bind to the first 15 residues of SopE or to its carboxy terminus (residues 78 to 240 or 115 to 240), which comprises its catalytic guanine nucleotide exchange factor domain (Fig. 4). These results indicate that InvB binds to residues 15 to 78 of SopE, a finding which is consistent with the observation that TTSS-associated chaperones bind to the amino terminus of their cognate substrates (24). The observed binding profile was not due to nonspecific binding either to PhoA or GST, since SptP1-35-PhoA or GST-SptP did not interact with InvB (Fig. 4). Furthermore, the absence of binding was not due to lack of expression of the relevant constructs, since all constructs were shown to be expressed to equivalent levels when subsequently probed with antibodies directed against PhoA or GST (Fig. 4, lower panels). Even though SopE and SipA bind the same chaperone, there is no obvious primary amino acid similarity between these two proteins. However, this is not surprising, since despite the structural similarity of many TTSS-associated chaperones, there is little similarity in the primary amino acid sequence of the binding domains of their cognate binding proteins. Presumably, binding to the chaperones is dictated by a few key amino acids and secondary structural features which are compatible with variations in the primary amino acid sequence (26).
FIG. 4.
Delineation of the InvB-binding domain of SopE. Whole-cell lysates of Salmonella serovar Typhimurium ΔsopE strains carrying different plasmids expressing various segments of the amino terminus of SopE (residues 1 to 15, 1 to 38, 1 to 50, and 1 to 104) fused to PhoA, or whole-cell lysates of E. coli expressing carboxy-terminal domains of SopE (residues 78 to 240 and 115 to 240) fused to GST, were separated by SDS-PAGE and transferred to PVDF membranes. The blots were treated with a soluble lysate of E. coli expressing M45 epitope-tagged InvB, and the bound InvB-M45 was detected with a monoclonal antibody directed against M45 as previously described (upper panels) (2). To confirm the expression of the different constructs, the membranes were reprobed with antibodies directed against PhoA or GST (lower panels).
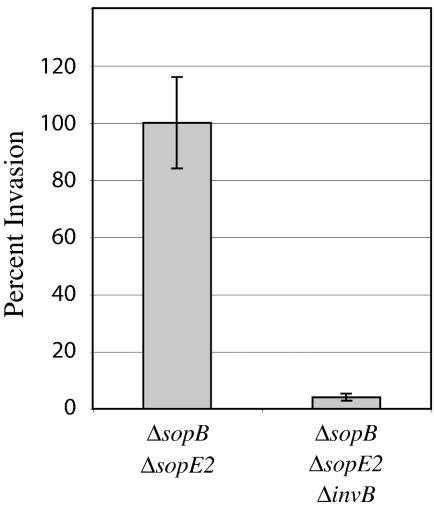
The observation that SopE is not secreted into the culture supernatant in the absence of InvB did not rule out the possibility that InvB may not be required for the translocation of SopE into eukaryotic cells. To address this issue, we examined whether the SopE-mediated invasion phenotype of a Salmonella strain carrying loss-of-function mutations in sopB and sopE2 was affected by the introduction of the invB mutation. In the absence of SopB and SopE2, Salmonella invasion into tissue culture cells is mediated solely by the activity of SopE (34). Therefore, bacterial internalization is a sensitive surrogate measure of SopE translocation. The ability of a Salmonella strain carrying deletion mutations of the sopB and sopE2 genes or that of its isogenic derivative carrying an invB null mutation to enter into cultured intestinal Henle-407 cells was examined by using the gentamicin protection assay as previously described (14). In the absence of InvB, the ΔsopB ΔsopE2 strain was severely defective in its ability to invade cultured intestinal cells (Fig. 5), indicating that InvB is required for the translocation of SopE into host cells.
FIG. 5.
InvB is required for SopE translocation into host cells. Intestinal Henle-407 cells were infected for 30 min with a serovar Typhimurium strain lacking sopB and sopE2 or with its isogenic derivative lacking invB, and the numbers of bacteria that resisted the treatment with gentamicin due to bacterial internalization were enumerated as previously described (14). Notice that in the absence of sopB and sopE2, Salmonella internalization is exclusively the result of the activity of translocated SopE (34). Values represent the means and standard deviations of three determinations of the percentage of the initial inoculum that survived the gentamicin treatment; values have been normalized to that of the ΔsopB ΔsopE2 mutant, which was considered to be 100% (actual value, 12% ± 2%).
In this study, we have identified InvB as the chaperone for the Salmonella type III secreted effector protein SopE. This conclusion is supported by the following pieces of evidence. (i) In the absence of InvB, SopE is not secreted or translocated into cultured host cells. (ii) InvB specifically binds a discrete domain within the amino terminus of SopE. InvB exhibits a number of unique features. Unlike most chaperones identified thus far, InvB is not encoded in the vicinity of its cognate SopE effector protein. Interestingly, the chaperone and its cognate substrate are maintained in two separate genetic elements, a pathogenicity island (SPI-1) and an integrated bacteriophage, which were presumably horizontally acquired independently through evolution. It has been previously shown that InvB is also a chaperone for an SPI-1-encoded secreted protein, SipA (2). Although not specifically examined in this study, it is possible that InvB serves as a chaperone for the highly related protein SopE2 (28). Therefore, InvB serves as a chaperone for two or perhaps even three secreted proteins that are genetically unlinked. SopE, SopE2, and SipA exert their function very early during the infection process (15). It is therefore possible that the utilization of a common chaperone is related to yet-undefined control mechanisms of the secretion process to ensure the rapid and early delivery of these effector proteins.
Acknowledgments
We thank members of the Galán laboratory for critical reading of the manuscript.
S.H.L. was supported by NRSA fellowship number AI52710-01 from the National Institutes of Health. This work was supported by Public Health Service grant number AI30492 from the National Institutes of Health to J.E.G.
REFERENCES
- 1.Birtalan, S. C., R. M. Phillips, and P. Ghosh. 2002. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol. Cell 9:971-980. [DOI] [PubMed] [Google Scholar]
- 2.Bronstein, P. A., E. A. Miao, and S. I. Miller. 2000. InvB is a type III secretion chaperone specific for SspA. J. Bacteriol. 182:6638-6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchwald, G., A. Friebel, J. E. Galán, W. D. Hardt, A. Wittinghofer, and K. Scheffzek. 2002. Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. EMBO J. 21:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cambronne, E. D., L. W. Cheng, and O. Schneewind. 2000. LcrQ/YscM1, regulators of the Yersinia yop virulon, are injected into host cells by a chaperone-dependent mechanism. Mol. Microbiol. 37:263-273. [DOI] [PubMed] [Google Scholar]
- 5.Chen, L. M., K. Kaniga, and J. E. Galán. 1996. Salmonella spp. are cytotoxic for cultured macrophages. Mol. Microbiol. 21:1101-1115. [DOI] [PubMed] [Google Scholar]
- 6.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 7.Darwin, K., and V. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichelberg, K., C. Ginocchio, and J. E. Galán. 1994. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J. Bacteriol. 176:4501-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, D. T., L.-M. Chen, J. Gillis, K.-C. Lin, B. Harty, G. P. Mazzara, R. O. Donis, K. G. Mansfield, J. D. Lifson, R. C. Desrosiers, J. E. Galán, and R. P. Johnson. 2003. Mucosal priming of simian immunodeficiency virus-specific cytotoxic T lymphocyte responses in rhesus macaques by the Salmonella type III secretion antigen delivery system. J. Virol. 77:2400-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu, Y., and J. E. Galán. 1998. Identification of a specific chaperone for SptP, a substrate of the centisome 63 type III secretion system of Salmonella typhimurium. J. Bacteriol. 180:3393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galán, J. E. 2001. Salmonella interaction with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 13.Galán, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 14.Galán, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galán, J. E., and D. Zhou. 2000. Striking a balance: modulation of the actin cytoskeleton by Salmonella. Proc. Natl. Acad. Sci. USA 97:8754-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardt, W.-D., L.-M. Chen, K. E. Schuebel, X. R. Bustelo, and J. E. Galán. 1998. Salmonella typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93:815-826. [DOI] [PubMed] [Google Scholar]
- 18.Hardt, W.-D., H. Urlaub, and J. E. Galán. 1998. A target of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc. Natl. Acad. Sci. USA 95:2574-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson, M., J. Day, and G. Plano. 1998. YscB of Yersinia pestis functions as a specific chaperone for YopN. J. Bacteriol. 180:4912-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaniga, K., J. C. Bossio, and J. E. Galán. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues to the PulD and AraC family of proteins. Mol. Microbiol. 13:555-568. [DOI] [PubMed] [Google Scholar]
- 21.Ménard, R., P. J. Sansonetti, C. Parsot, and T. Vasselon. 1994. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell 79:515-529. [DOI] [PubMed] [Google Scholar]
- 22.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschape, H. Russmann, E. Igwe, and W. D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein. Proc. Natl. Acad. Sci. USA 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obert, S., R. J. O'Connor, S. Schmid, and P. Hearing. 1994. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol. Cell. Biol. 14:1333-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page, A. L., and C. Parsot. 2002. Chaperones of the type III secretion pathway: jacks of all trades. Mol. Microbiol. 46:1-11. [DOI] [PubMed] [Google Scholar]
- 25.Persson, C., R. Nordfelth, A. Holmström, S. Hakansson, R. Rosqvist, and H. Wolf-Watz. 1995. Cell surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol. Microbiol. 18:135-150. [DOI] [PubMed] [Google Scholar]
- 26.Stebbins, C. E., and J. E. Galan. Priming virulence factors for delivery into the host. Nat. Rev. Mol. Biol., in press. [DOI] [PubMed]
- 27.Stebbins, C. E., and J. E. Galán. 2001. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414:77-81. [DOI] [PubMed] [Google Scholar]
- 28.Stender, S., A. Friebel, S. Linder, M. Rohde, S. Mirold, and W. D. Hardt. 2000. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 36:1206-1211. [DOI] [PubMed] [Google Scholar]
- 29.Tucker, S. C., and J. E. Galán. 2000. Complex function for SicA, a Salmonella enterica serovar Typhimurium type III secretion-associated chaperone. J. Bacteriol. 182:2262-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wattiau, P., B. Bernier, P. Deslée, T. Michiels, and G. R. Cornelis. 1994. Individual chaperones required for Yop secretion by Yersinia. Proc. Natl. Acad. Sci. USA 91:10493-10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wattiau, P., and G. R. Cornelis. 1993. SycE, a chaperone-like protein of Yersinia enterocolitica involved in the secretion of YopE. Mol. Microbiol. 8:123-131. [DOI] [PubMed] [Google Scholar]
- 32.Wattiau, P., S. Woestyn, and G. R. Cornelis. 1996. Customized secretion chaperones in pathogenic bacteria. Mol. Microbiol. 20:255-262. [DOI] [PubMed] [Google Scholar]
- 33.Wood, M. W., R. Rosqvist, P. B. Mullan, M. H. Edwards, and E. E. Galyov. 1996. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol. Microbiol. 22:327-338. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, D., L. M. Chen, L. Hernandez, S. B. Shears, and J. E. Galán. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39:248-259. [DOI] [PubMed] [Google Scholar]