Abstract
Mx (myxovirus-resistant) proteins are induced by interferon and inhibit viral replication as part of the innate immune response to viral infection in many vertebrates. Influenza A virus appears to be especially susceptible to Mx antiviral effects. We characterized exon 13 and the 3’ UTR of the Mx gene in wild ducks, the natural reservoir of influenza virus and explored its potential relevance to influenza infection. We observed a wide range of intra- and interspecies variation. Total nucleotide diversity per site was 0.0014, 0.0027, 0.0044, 0.0051, and 0.0061 in mallards, northern shovelers, northern pintails, American wigeon, and American green-winged teals, respectively. There were 61 haplotypes present across all five species and four were shared among species. Additionally, we observed a significant association between Mx haplotype and influenza infection status in northern shovelers. However, we found no evidence of balancing or diversifying selection in this region of the Mx gene. Characterization of the duck Mx gene is an important step in understanding how the gene may affect disease resistance or susceptibility in wild populations. Furthermore, given that waterfowl act as a natural reservoir for influenza virus, the Mx gene could be an important determinant in the ecology of the virus.
Keywords: Mx, Duck, Anas, Population genetics, Influenza, Nucleotide diversity
1. Introduction
Mx (myxovirus-resistant) proteins play a key role in the innate immune response to viral infection. Discovered in 1962 in an inbred mouse strain resistant to influenza infection (Lindenmann, 1962), Mx proteins have since been shown to provide resistance to a wide array of both DNA and RNA viruses (reviewed by Haller et al., 2007). The Mx gene is readily induced by type I and type III interferon (IFN) and where examined, Mx proteins have been found in mammals, birds and fish (reviewed by Haller et al., 2007). Once induced, Mx proteins associate with the smooth endoplasmic reticulum (Stertz et al., 2006) or localize to distinct subnuclear compartments (Engelhardt, 2001; Engelhardt, 2004) depending on the organism and site of Mx expression.
Prevention of viral replication is dependent on the co-localization of Mx proteins and the viral replication stage. In the case of orthomyxoviruses and bunyaviruses, a physical interaction occurs between Mx and the viral nucleoprotein (NP) component of the nucleocapsid (Haller et al., 2008). If both occur in the same location, Mx traps viral nucleoproteins and shuttles them elsewhere in the cell (Haller and Kochs, 2002). It is hypothesized that Mx can also bind and cover nuclear localization signals on the viral nucleoprotein, blocking entrance to the nucleus (Kochs and Haller, 1999). The nucleoprotein of most viruses is a generally well-conserved portion of the virus because of its critical role in the virus’ structure. However, in a recent study of over 2500 influenza A NP amino acid sequences, it was found that sequence conservation varies considerably by protein domain; 80.7% of residues in the head domain are conserved, whereas only 66.1% and 67.5% of residues in the body and tail loop domains, respectively, are conserved (Ng et al., 2009). It is unknown at this time with which domain Mx interacts, but by targeting the nucleoprotein rather than other viral proteins, it is thought that the Mx protein is better able to maintain viral recognition and prevent viral immune evasion (Sadler and Williams, 2008).
Interaction with viral components is mediated by the effector domain of the Mx protein (Johannes et al., 1997). The effector domain, also called the leucine zipper region, is coded for in exon 13 of the Mx gene. This region is characterized by a series of leucines that appear to be involved in self-assembly and oligomerization (Schumacher and Staeheli, 1998). It is thought the formation of oligomers creates a stable pool from which monomeric Mx proteins can be recruited (Janzen et al., 2000; Kochs et al., 2002a). The effector domain also contains amino acids important for viral target recognition. Glutamic acid at position 645 in the human Mx protein (MxA) was mutated to arginine and the protein became ineffective against vesicular stomatitis virus and LaCrosse virus, but still inhibited influenza and Thogoto virus (Zürcher et al., 1992; Kochs et al., 2002b).
Antiviral activity of Mx protein varies among organisms. Only the cytoplasmic form (MxA) in humans is antivirally active (Pavlovic et al., 1990), while both forms (nuclear, Mx1; cytoplasmic, Mx2) are active in mice (Arnheiter et al., 1980). Although chicken Mx proteins were not thought to be antivirally active (Bernasconi et al., 1995), work has shown a single amino acid change (S631N) determines antiviral activity against influenza virus and vesicular stomatitis virus in vitro (Ko et al., 2004). However, this finding remains controversial as additional researchers have not successfully replicated the experiment (Benfield et al., 2008). The demonstration that single amino acid changes may determine antiviral activity of some forms of the Mx protein against many different types of virus suggests that genetic variation in the domains of the protein that interact with viral proteins for innate defense may be critical to co-evolving host-pathogen relationships such as what appears to exist with dabbling ducks and influenza.
Duck Mx was also reported as inactive against influenza infection (Bazzigher et al., 1993). However, that study was limited by the extremely small sample size and effectively neglected the possible effects of inter-individual variation in the innate immune response. In addition, to test for resistance to influenza, chicken and turkey-adapted influenza virus strains were used in mouse and chicken-derived cell lines, but it is now well established that the immune response to influenza is strain dependent in both ducks and chickens (reviewed by Cardona et al., 2009). This fact makes it equally likely that the effects observed in a limited number of tissues and a single species were highly dependent on the lab adapted strain of virus used. Taken together, prior studies leave the function of the Mx protein in wild duck populations in question.
Wild duck populations are an important part of the ecology of the influenza virus. Prevalence of low pathogenic avian influenza (LPAI) is highest in ducks compared to other birds and in juveniles compared to other age classes (Webster et al., 1992; Olsen et al., 2006; Runstadler et al., 2007). Dabbling ducks (genus Anas) are infected with influenza more often than diving ducks (Olsen et al., 2006). This difference in prevalence may be due to dabbling ducks’ feeding and migratory behavior (Olsen et al., 2006). Dabbling ducks serve as the natural reservoir for low pathogenic strains and it appears they likely carry and shed the virus without overt symptoms (Kida et al., 1980; Webster et al., 1992; Jourdain et al., 2010). However, when experimentally infected with LPAI virus, pekin ducks and mallards produce a relatively weak antibody response, but are still protected against reinfection with the same virus (Kida et al., 1980; Jourdain et al., 2010). The role of the primary innate immune system in response to LPAI infection in ducks is worth further exploration.
Multiple studies suggest the Mx protein is fundamental to the innate immune response to influenza infection in mice, rats, humans, and pigs (Lindenmann and Klein, 1966; Meier et al., 1988; Pavlovic et al., 1990; Nakajima et al., 2007). Given the differing level of conservation in the influenza NP and the ability of Mx to recognize the NP of many different viruses, it could be advantageous for a population to harbor multiple Mx alleles. In this study, we sequenced and analyzed a portion of the Mx gene which codes for the leucine zipper region of the translated protein which is thought to direct the antiviral effects of the Mx protein in other species. Using those sequences, we were also able to test for an association between Mx allele and influenza infection status.
We used samples from multiple individuals with a known infection status representing five dabbling duck species: American green-winged teal (Anas crecca carolinensis, hereafter teal), American wigeon (Anas americana, hereafter wigeon), mallard (Anas platyrhynchos), northern pintail (Anas acuta, hereafter pintail) and northern shoveler (Anas clypeata, hereafter shoveler). We sampled at summer breeding grounds in Interior Alaska where large numbers of these birds converge and where, historically, dabbling ducks in these sites have a high prevalence of LPAI infection (Ito et al., 1995; Runstadler et al., 2007). Nucleotide diversity at the Mx locus would support further investigation of the functional differences between alleles and the impact on influenza infection in wild duck populations, the major reservoir of influenza in nature.
2. Materials and methods
2.1. Sample Collection
We used samples from three existing collections: genomic DNA extracted from blood and feathers collected from ducks at sites in the Yukon Flats National Wildlife Refuge (66°20’N, 147°58’W) in May and June 2006, genomic DNA extracted from blood and feathers collected from live-trapped ducks at sites in the Minto Flats State Game Refuge (64°53′N, 148°46′W) in August 2006, and blood collected from ducks post-mortem at Minto Flats in September 2006 and 2007. Additional feathers taken at Minto Flats in 2006 were processed and used to supplement the existing collection. Minto Flats and Yukon Flats are both located in interior Alaska and are important breeding areas for more than 20 waterfowl species (Clausen et al., 1992; Petrula, 1994). Table 1 lists samples used in laboratory analysis.
Table 1.
Summary of samples collected from five species of wild ducksa
| Species | n | Type of sample | Location | Year |
|---|---|---|---|---|
| American green-winged teal (Anas crecca carolinensis) | 13 | Genomic DNA | Minto Flats | 2006 |
| 1 | Feathers | Minto Flats | 2006 | |
| 12 | Blood | Minto Flats | 2007 | |
| Total | 26 | |||
| American wigeon (Anas americana) | 5 | Genomic DNA | Yukon Flats | 2006 |
| 10 | Blood | Minto Flats | 2007 | |
| Total | 15 | |||
| Mallard (Anas platyrynchos) | 140 | Genomic DNA | Minto Flats | 2006 |
| 11 | Feathers | Minto Flats | 2006 | |
| 53 | Blood | Minto Flats | 2007 | |
| Total | 204 | |||
| Northern pintail (Anas acuta) | 34 | Genomic DNA | Minto Flats | 2006 |
| 5 | Feathers | Minto Flats | 2006 | |
| 26 | Blood | Minto Flats | 2007 | |
| Total | 65 | |||
| Northern shoveler (Anas clypeata) | 7 | Genomic DNA | Yukon Flats | 2006 |
| 36 | Blood | Minto Flats | 2007 | |
| Total | 43 | |||
Genomic DNA samples were from an archived collection originally extracted from blood and feathers. Additional feathers and blood were processed as described in the text.
2.1.1. Yukon Flats
Briefly, wigeon and shovelers were live-captured in traps during May and June 2006. Blood and feathers were collected as described by Welsh (2008).
2.1.2. Minto Flats
Wigeon, teals, mallards, pintails, and shovelers were live-captured in traps at Minto Flats in cooperation with Alaska Department of Fish and Game banding operations during August 2006. Blood and feathers were collected as described by Welsh (2008). Briefly, 0.5 to 1.5 mL of blood were collected from the brachial or jugular vein, transferred to 2 mL EDTA tubes (BD Vacutainer), and stored at -80 °C. During September 2006 and 2007, blood was collected post-mortem via heart puncture from hunter-killed ducks. The blood samples were stored in 2 mL EDTA tubes (BD Vacutainer) at -80 °C.
2.2. Determination of influenza infection status
In addition to blood and feather samples, cloacal swabs were taken from all ducks using methods described by Runstadler et al. (2007). RNA was extracted and used for the creation of cDNA for real-time RT-PCR (RRT-PCR) as previously described (Runstadler et al., 2007). The cDNA product was then screened for the avian influenza virus matrix gene using primers developed by Spackman et al. (2003) and modified by Runstadler et al. (2007). A threshold cutoff (Ct) value of <40 was used to assign infection status. The overall prevalence in 2006 was 16.3% (173/1062) and in 2007 was 7.0% (120/1714).
2.3. Chicken and duck Mx sequences and primer design
The NCBI Genbank database was searched in June 2007 for chicken and duck Mx mRNA sequences. The two available duck sequences (Genbank accession numbers Z21549 and Z21550) and a chicken sequence (Genbank accession number NM_204609) were aligned using the ClustalW algorithm in DNADynamo (Blue Tractor Software Ltd, UK). The area of duck sequence that best aligned with exon 13 of the chicken Mx mRNA was used to design primers in Primer3 (Rozen and Skaletsky, 2000). The forward (F) and reverse (R) primers are: Mx-F 5’ –AGCAAGTAAACGCCTCTCCA – 3’ and Mx-Rs 5’ – GAAACTGGCAGTAAAGGTCAGC – 3’.
2.4. Genomic DNA extraction from whole blood and plucked body feathers
DNA was extracted from 5 - 10 μl of duck whole blood using the Qiagen Inc DNeasy Blood and Tissue Kit (Qiagen Inc., California, USA) according to the manufacturer’s instructions. The DNeasy Blood and Tissue kit was also used to extract DNA from plucked body feathers as described by Bush et al. (2005). Briefly, one to six feather tips were placed in a 1.5 ml microfuge tube with 25 μl of 100 mg/ml DTT solution, 20 μl Proteinase K solution (600 mAU/ml) and 180 μl Buffer ATL. The mixture was incubated at 55 °C for 48 hours in a shaking incubator. After incubation, DNA was extracted according to the manufacturer’s instructions regarding tissue. DNA concentration was quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Delaware, USA).
2.5. Mx gene amplification and sequencing
The Mx gene fragment was amplified through polymerase chain reaction (PCR) with the following reaction mixture: 80 ng of genomic DNA, each primer at 0.4 μM, 1 unit Platinum Taq DNA polymerase (Invitrogen, California, USA), 0.4 mM each dNTP, 2 mM MgCl2, and 1X Platinum Taq polymerase buffer in a final volume of 25 μl. The PCR conditions were: 94 °C for 5 min followed by 35 cycles of 94 °C for 45 s, 56 °C for 45 s, 72 °C for 1 min, and a single incubation at 72 °C for 10 min followed by a hold at 4 °C. PCR products were run on a 1.5% agarose gel and then visualized using ethidium bromide staining. The expected PCR product size was approximately 340 bp. PCR products were cleaned with the QIAquick PCR purification kit (Qiagen Inc.) and sequenced with the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems; Foster City, CA) according to the manufacturer’s recommendations. The sequencing reactions were analyzed on an ABI 3100 sequencer at the University of Alaska Fairbanks DNA Core Facility and/or an ABI 3730XL sequencer at Macrogen Inc., Seoul, Korea.
2.6. DNA sequence alignment and analysis
Chromatograms were edited using Sequencher 4.7 (Gene Codes Corporation, USA), sequences aligned with the MUSCLE alignment algorithm (Edgar, 2004), and allelic phase was determined using PHASE v.2 (Stephens et al., 2001). Results from PHASE were verified by cloning and sequencing 50 heterozygous individuals across all five species. Additionally, samples with rare mutations were amplified and sequenced at least twice to ensure the mutations were not PCR artifacts. Summary statistics including π (Tajima, 1983) and Tajima’s D (Tajima, 1989) were estimated using DnaSp v 4.50 (Rozas and Rozas, 1999) and SITES (Hey and Wakeley, 1997). OmegaMap was used to search for evidence of diversifying selection within species (Wilson, 2005). Codon-based models contained within MEGA4.1 (Tamura et al., 2007) were also used to test for evidence of selection within species. Unrooted median-joining haplotype networks were constructed with the program Network v 4.5.1.0 in order to visualize how alleles were related within and between species (Bandelt et al., 1999). In order to evaluate how genetic variation was partitioned within and among species, an analysis of molecular variance (AMOVA) was performed using Arlequin v 3.11 (Excoffier et al., 2005).
2.7. Amino acid translation and analysis
The coding region was translated using web-based EMBOSS Transeq (http://bioweb2.pasteur.fr/docs/EMBOSS/transeq.html) and the variable sites were located using FaBox (http://www.birc.au.dk/~biopv/php/fabox/). SIFT was used to predict the effects of amino acid substitutions on the functionality of the protein (Ng, 2003).
2.8. Disease association analysis
The number of influenza-positive and negative individuals in each haplotype was counted using a Perl script (written by Dillon). In order to assess whether infection status was significantly associated with Mx haplotype, the program CLUMP was used (Sham and Curtis, 1995). A set of 100,000 simulations was run for each species and all species combined. The data were also stratified by year to investigate possible effects of time on disease association. The p-value for the normal chi-squared (T1) table was used to determine statistical significance.
3. Results
3.1. Nucleotide diversity
We sequenced a 336 bp region of the duck Mx gene that encompasses exon 13 and a portion of the 3’ UTR. Our sample consisted of 353 individuals from five species (mean = 71, range = 15 - 204). The total number of sequences (n) and haplotypes (H) from each species can be seen in Table 2.
Table 2.
Summary statistics for Mx exon 13 and 3’ UTR from five duck species
| Species | na | H | S | DTaj | dN/dS |
|---|---|---|---|---|---|
| American green-winged teal (Anas crecca carolinensis) | 52 | 9 | 8 | 0.41 | 0.86 |
| American wigeon (Anas americana) | 30 | 9 | 12 | -1.41 | 0.06 |
| Mallard (Anas platyrynchos) | 408 | 24 | 22 | -2.22b | 0.19 |
| Northern pintail (Anas acuta) | 130 | 17 | 16 | -1.35 | 0.84 |
| Northern shoveler (Anas clypeata) | 86 | 9 | 9 | -1.24 | 0.47 |
n, number of alleles sampled; H, number of haplotypes; S, number of segregating sites; DTaj, Tajima’s D; dN/dS, the ratio of Πnonsyn to Πsyn
P < 0.01
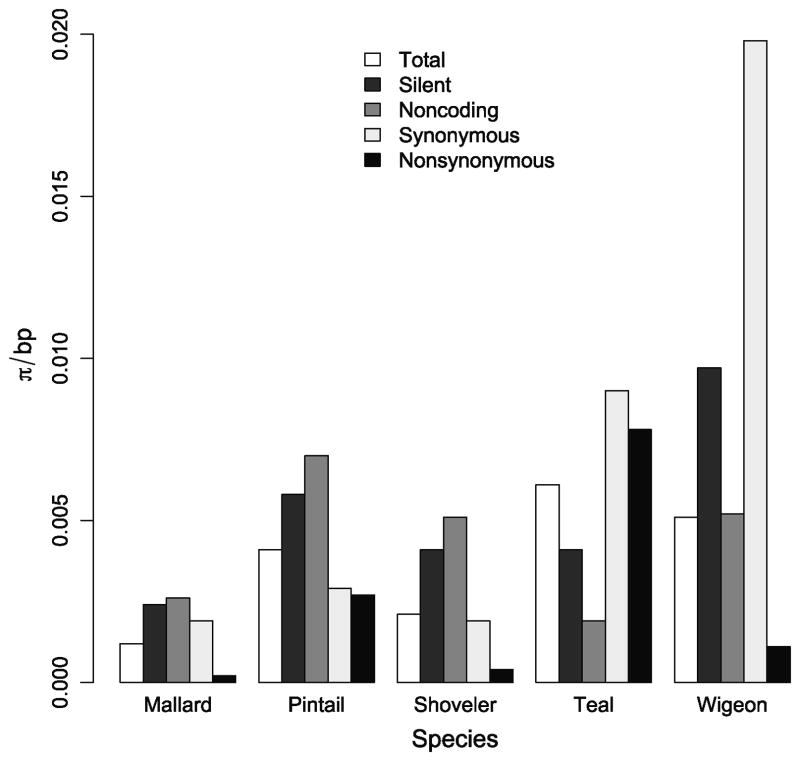
Mean pair-wise differences per site (π/bp) varied widely among species (Fig. 1). The greatest overall diversity (πtotal) was found in teals (0.0061). Teals also had the highest π for nonsynonymous sites (πs; 0.0078). Wigeons had the highest π for both synonymous sites (πn; 0.0198) and silent sites (synonymous and noncoding sites; πsilent: 0.0097). Mallards were the least diverse in all categories with the exception of noncoding sites (πnoncoding: 0.0026).
Figure 1. Mean pairwise differences (π) per base pair (bp) for exon 13 and 3’ UTR of the Mx gene in five duck species.
Total diversity: all sites (336 bp), silent: noncoding sites and synonymous sites of coding region (156 bp), noncoding: 3’ UTR (108 bp), synonymous: synonymous sites in coding region (48 bp), nonsynonymous: nonsynonymous sites in coding region (180 bp)
3.2. Selection
We did not find evidence of long-term positive selection on our region of interest in any of the species studied. All species had dN/dS ratio less than one (more synonymous mutations than nonsynonymous mutations; Table 2). Mallards showed a significantly negative Tajima’s D value while all other species had non-significant values (Table 2). No significant posterior probabilities of positive selection were found using omegaMap (data not shown).
3.3. Nucleotide haplotype network and disease association
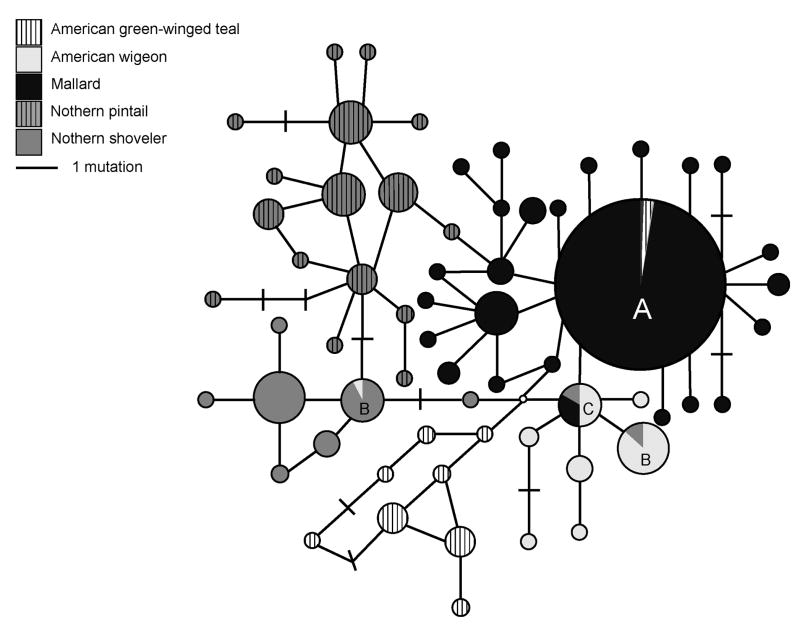
Constructing a haplotype network allowed us to visualize the relationship between the nucleotide sequences from the five duck species (Fig. 2). We found 61 haplotypes, 32 of which were represented by two or more sequences from sampled individuals. One haplotype accounted for 49% of all sequences and 83% of all mallard sequences (‘A,’ Fig. 2). It is possible that more haplotypes could exist, as our primers were designed based on three available avian Mx sequences and may not have captured all duck Mx alleles. Overall, we observed a large degree of interdigitation and some sharing of sequences; two haplotypes were shared by two species (‘B’, Fig. 2) and two haplotypes were shared by three or more species (‘A,’ ‘C’, Fig. 2). With the exception of the four shared haplotypes, sequences grouped by species. Eighty-two percent of observed variation was explained by differences among species (AMOVA; p < 0.00001).
Figure 2. Unrooted median joining network showing the relationship among Mx nucleotide sequences (336 bp) from five duck species.
Species are denoted in the legend. The area of circles is proportional to the number of sequences in that haplotype. A - Largest haplotype, haplotype contains 49% of all sequences and 83% of all mallard sequences. B - Haplotype is shared by two or more species. C - Haplotype is shared by three or more species.
We used CLUMP to evaluate whether a statistical association exists between Mx allele and influenza infection status (Table 3). Northern shovelers showed a suggestive association (χ2 = 12.1, p = 0.09), with five unique alleles contributing to this association. When data was stratified by year, there was a significant association in shovelers during 2006 (p = 0.029) and in all species combined during 2007 (p = 0.001).
Table 3.
| Species | All years p-value | 2006 p-value | 2007 p-value |
|---|---|---|---|
| American green-winged teal (Anas crecca carolinensis) | 0.20 | 0.25 | 0.59 |
| American wigeon (Anas americana) | 0.23 | 1.00 | 0.29 |
| Mallard (Anas platyrynchos) | 0.27 | 0.53 | 0.089 |
| Northern pintail (Anas acuta) | 0.29 | 0.11 | 0.47 |
| Northern shoveler (Anas clypeata) | 0.09 | 0.029 | 0.18 |
| All species | 0.16 | 0.089 | 0.001 |
Nucleotide sequences from five species of wild ducks were tested for an association between influenza infection state and Mx haplotype using CLUMP. The data were also evaluated after stratification by sample year. A p-value < 0.05 indicates a significant association.
Overall prevalence of infection by species and year (prevalence, positive/total sampled): 2006, teal (32.5%, 13/40), wigeon (1.2%, 1/81), mallard (10.5%, 62/592), pintail (9.1%, 28/307), shoveler (18.2%, 4/22); 2007, teal (7.5%, 10/133), wigeon (3.5%, 6/241), mallard (9.6%, 48/499), pintail (2.6%, 25/978), shoveler (25.4%, 30/118).
3.4. Amino acid diversity
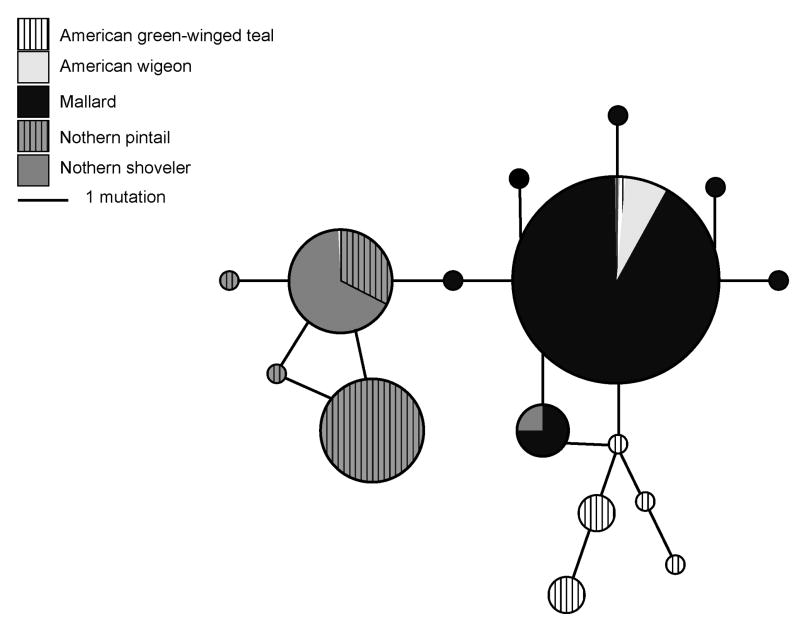
The coding region translated to 76 amino acids. We found 11 nonsynonymous substitutions and one stop codon; four substitutions, including the stop codon, were predicted to affect protein function based on sequence homology and the physical properties of amino acids (Fig. 4). Relative to the entire protein, the four substitutions were E682STOP, L701F, S702G, and D717A. The L701F and S702G substitutions only occurred in teals; L701F appeared in four teal sequences, but S702G was present in 79% (41/52) of all teal sequences. The majority (92%; 24/26) of teal individuals were homozygous at position 702 and both individuals exhibiting the L701F substitution were homozygotes. The stop codon and D718A substitution each occurred in one mallard sequence. Overall, there were 16 unique protein sequences among the five duck species, eight of those were singletons, four were shared by multiple species, and five were only present in the teal population (Fig. 3).
Figure 4. Unique Mx protein amino acid sequences (pos 646 – 721).
Dashed lines indicate residues are identical to the top sequence. Boxed residues are predicted to affect protein function. Dots denote a leucine zipper motif. Species names indicate if that sequence is present in the sample population.
Figure 3. Unrooted median joining network showing the relationship among Mx protein sequences (76 amino acids) from five duck species.
Species are denoted in the legend. The area of circles is proportional to the number of sequences in that haplotype.
4. Discussion
We examined diversity and tested for evidence of selective pressure in a region of the Mx gene in five duck species: American green-winged teal, American wigeon, mallard, northern pintail, and northern shoveler. Additionally, we investigated whether an association exists between influenza infection status and Mx haplotype in these species. Innate immune mechanisms, like that of Mx, may play a major role in limiting infection in ducks as it does in other species (Lindenmann, 1962; Pavlovic et al., 1990; Ko et al., 2002; Nakajima et al., 2007).
A high degree of variability exists among duck species at the Mx locus; total nucleotide diversity (πtotal) per site ranges from 0.0014 in mallards to 0.0061 in teals (Fig. 1). Both Mx proteins and toll-like receptor (TLR) proteins show conservation across vertebrate species but diversity is needed in order to interact with a wide array of pathogens. In this study, there were 61 Mx haplotypes across 353 individuals. Palermo et al. (2009) explored variation at the TLR4 locus in 259 pigs and found 74 haplotypes and a total nucleotide diversity of 0.00077. The haplotype diversity is similar to that observed at the duck Mx locus, but there is less nucleotide diversity at the TLR4 locus in pigs. This difference in diversity may be because mammalian TLR4 recognizes bacterial lipopolysaccharide and extensive variation at this locus may significantly alter recognition and signal transduction (Palermo et al., 2009). Allelic diversity at TLR loci in humans is heterogeneous; in 100 individuals the number of alleles ranged from four to 15 across nine TLR genes (Georgel et al., 2009).
Constructing a haplotype network is a useful tool to visualize diversity and sequence relatedness. Figure 2 represents Mx nucleotide sequences from our five duck species and it is clear that, with a few exceptions, the sequences are grouped by species. There are four shared alleles, which could be an artifact of hybridization events or ancestral polymorphisms that are still segregating in these populations.
The low dN/dS ratios, non-positive values of Tajima’s D (Table 2), and non-significant results from omegaMap did not show support of balancing or diversifying selective pressure on our region of interest. In addition, the haplotype network did not show patterns of balancing or diversifying selection. If strong selective pressure from disease or some other force was acting on this region, we might expect to see an advantageous allele at high frequency as a result of a selective sweep or perhaps a few alleles being maintained at stable frequencies driven by balancing selection. Although we did not measure diversity at the Mx locus relative to other genes, our summary statistics did not provide evidence for a selective sweep, with the possible exception of mallards (discussed in detail below).
Berlin et al. (2008) studied Mx sequences from a small group of 14 mallards and did not find indicators of selection in exon 13 or the 3’ UTR. However, when Berlin et al. (2008) considered additional exons, avian Mx did show signatures of positive selection. The majority of positively selected sites were in what the authors designated the N-terminal “avian-specific region.” The significance of this region is unknown, but it could be important to avian Mx expression or protein function, and will be interesting to examine in wild birds.
Given the lack of evidence for selective pressure on our region of interest, it is curious that we discovered a suggestive association between Mx haplotype and infection status in shovelers (p = 0.09; Table 3). This region may contribute to infection susceptibility in ducks. Shovelers are unique among the species in our study due to their specialized bill morphology (dense lamellae) that allows them to feed on small invertebrates filtered out of the water column (Dubowy, 1985). Shovelers devote around 80% of their foraging time to straining, while other dabbling ducks feed in the mud or around vegetation (Dubowy, 1985). Garamszegi and Møller (2007) found that species which employ surface-feeding foraging methods have a higher prevalence of influenza infection and Hill et al. (2009) discovered that lamellar density was significantly correlated with influenza prevalence. Northern shovelers’ feeding strategy may lead them to interact with recently shed influenza virus more frequently than other dabbling ducks. Thus, birds carrying Mx alleles conferring susceptibility to viral infection may be at a higher risk of acquiring infection and revealing an association with disease.
Our data also illustrate temporal effects on the association between Mx haplotype and influenza infection status (Table 3). Northern shovelers, mallards, and pintails showed a significant or suggestive disease association in one year, but not the other (Table 3). In addition, when all species were combined, a significant association was found in 2007 (p = 0.001). Influenza subtypes circulating in wild duck populations change from year to year, so it is possible that certain Mx haplotypes are better adapted to the viral repertoire of a particular year.
Disease-related associations with the Mx gene have been found in other species as well. Nakajima et al. (2007) reported a porcine Mx1 allele that does not confer resistance to influenza infection as the wild type does. In chickens, three SNPs in exon 13 were found to be significantly associated with commercial traits; the SNP previously described to be important in resistance or susceptibility to influenza infection was positively associated with increased antibody titers to vaccination for infectious bursal disease virus (Livant et al., 2007). Additionally, Mx genotype was also associated with differential antibody titers to influenza virus vaccination in chickens (Qu et al., 2009), suggesting that innate immune responses mediated by Mx in chickens, and possibly ducks, are tied to an integrated immune response to the virus. The association seen in our study is not without precedent and warrants further investigation. It is most likely a part of the Mx gene outside of, but still in linkage disequilibrium with, our sequenced region that is driving the association in shovelers. A fine-mapping study of this chromosomal area in ducks with the development of genetic markers may help focus the true association.
Mallards also showed unique features among the species in our study. Total, silent, and nonsynonymous site diversity were lowest in mallards. Parker et al. (1981) also reported low genetic diversity across 20 loci in a wintering population of mallards, in spite of their large population size and ability to hybridize with other duck species. Berlin et al. (2008) studied eight exons (960 bp) of the Mx gene in 14 mallards and reported πsyn (0.0080) and πnonsyn (0.0021). Our values of πsyn and πnonsyn in mallards were lower than those reported by Berlin et al. (2008) which is most likely a reflection of higher conservation in our region of interest relative to the rest of the gene.
The relative lack of diversity in mallards in our study is illustrated by the haplotype denoted ‘A’ in Fig. 2 which represents 84% (342/408) of mallard Mx sequences. The other four species, in contrast, have several large clusters of alleles (Fig. 2) and greater total, silent, and nonsynonymous diversity (Fig. 1). Low genetic diversity, a significantly negative Tajima’s D, and a star-like pattern in the haplotype network could result from a selective sweep, wherein strong positive selection has reduced genetic diversity at this locus, or a recent population expansion. If a selective sweep occurred, this could suggest that the major Mx allele present in mallards confers fitness and may actually be co-adapted to tolerate or control influenza infection better than alleles present in other species. In this case, we would expect a greater fitness effect of infection on populations of species besides mallards. Those fitness effects have only recently begun to be addressed (van Gils et al., 2007; Latorre-Margalef et al., 2009) and more work will shed light on the relative effects of infection in different dabbling duck species. However, we did not examine patterns of diversity at another locus relative to Mx and thus cannot conclusively state if a selective sweep occurred in mallards.
Alternatively, low diversity in mallards may be due to a recent population expansion, which is supported by mtDNA evidence (McCracken et al., 2001; Kulikova et al., 2005). In the context of influenza virus infection, our mallard data support a hypothesis that genetic diversification is important for maintaining immune response that prevents detectable infection from specific pathogens. Mallards are found to be positive for influenza virus more often than other duck species (Olsen et al., 2006) and this could be partly due to low genetic diversity at immune-related loci and an inability to resist infection on a population level.
Without fully understanding the function of the amplified region in duck Mx protein, it is difficult to predict the effect of amino acid substitutions. SIFT, the predictive program used for analysis, is reliable when compared with protein function data (92% agreement; Luoma et al., 2010) and when used in human disease studies (Budny et al., 2010). However, we propose that the S702G mutation is not a deleterious substitution as it appears in 79% of teal sequences and involves substituting small, uncharged glycine for the larger, uncharged serine. Steric interactions would appear to be more of an issue in the L701F substitution wherein a large aromatic ring is inserted in place of a branched hydrocarbon side chain. The structure of the stalk region (middle domain and GTPase effector domain) of human MxA was recently reported (Gao et al., 2010; Haller et al., 2010). This region is involved in oligomerization (Schumacher and Staeheli, 1998) and antiviral activity (Zurcher et al., 1992; Johannes et al., 1997). Our putative protein sequence aligns with the fourth and fifth alpha helices of the crystallized region. Substitutions within those regions, particularly at interfaces between helices, negatively affect oligomerization and subsequently, antiviral activity. These findings support the predictions in our study that certain substitutions in the effector domain could affect protein function, but more investigation is clearly warranted.
This is the first study to characterize the Mx locus in a large number of individuals from multiple wild duck species. Our results demonstrate that nucleotide diversity at this locus is being maintained at different levels in five duck species. We examined the 3’ region of this locus because of its purported functional role, but other regions of the Mx gene may play important species-specific roles as well. In particular, studying diversity and selection in the avian-specific region documented by Berlin et al. (2008) could provide valuable insight into its functional importance. Mx gene expression in ducks in response to viral infection may provide clues to its relative importance in the host-virus interaction, as would investigation of a possible link between the Mx gene and antibody production. An assay of Mx protein antiviral activity in wild ducks in response to influenza and other viruses is also a necessary step.
In this study, we found 16 unique protein sequences with 11 variable sites translated from 336 bp of the Mx locus. Four of those mutations were predicted to affect protein function, but it is difficult to extend that prediction without a more complete understanding of the duck Mx protein. Our results demonstrate a variety of Mx alleles exist in wild ducks and we found evidence in northern shoveler populations of a possible influenza disease association with the Mx locus. It is important to note that resistance or susceptibility to disease is multifactorial and depends on the characteristics of both the host and the virus. We investigated one innate immune response gene and its involvement in resistance to influenza infection, but many more factors likely impact an organism’s ability to resist infection. More work clarifying the role of Mx and other genes in these species may lead to insight concerning influenza viral pathogenicity in other species.
Supplementary Material
Acknowledgments
The authors would like to thank the field crews that collected the samples used in this study, T. Welsh for genomic DNA, N. Takebayashi and D. Wolf for invaluable assistance with data analysis, J. Peters and K. Hueffer for useful discussion and C. Mulder and reviewers in BIOL 693 at the University of Alaska Fairbanks for greatly improving this manuscript. DD was supported by an Institute of Arctic Biology Research Fellowship and an Alaska INBRE Graduate Fellowship. The project described was supported in part by INBRE Grant Number 5P20RR016466 from the National Center for Research Resources (NCRR); and in part by Sub-Award Contract from NIAID Number HHSN266200700009C, a component of the National Institutes of Health (NIH) and its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR, NIAID or NIH.
Footnotes
Note: Nucleotide sequence data reported in this paper are available in the GenBank database under the accession numbers: GU202016 – GU202296.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnheiter H, Haller O, Lindenmann J. Host gene influence on interferon action in adult mouse hepatocytes: specificity for influenza virus. Virology. 1980;1:11–20. doi: 10.1016/0042-6822(80)90122-1. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;1:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bazzigher L, Schwarz A, Staeheli P. No enhanced influenza virus resistance of murine and avian cells expressing cloned duck Mx protein. Virology. 1993;1:100–112. doi: 10.1006/viro.1993.1350. [DOI] [PubMed] [Google Scholar]
- Benfield C, Lyall J, Kochs G, Tiley L. Asparagine 631 variants of the chicken Mx protein do not inhibit influenza virus replication in primary chicken embryo fibroblasts or in vitro surrogate assays. J Virol. 2008;15:7533–7539. doi: 10.1128/JVI.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin S, Qu L, Li X, Yang N, Ellegren H. Positive diversifying selection in avian Mx genes. Immunogenetics. 2008;11:689–697. doi: 10.1007/s00251-008-0324-0. [DOI] [PubMed] [Google Scholar]
- Bernasconi D, Schultz U, Staeheli P. The interferon-induced Mx protein of chickens lacks antiviral activity. J Interferon Cytokine Res. 1995;1:47–53. doi: 10.1089/jir.1995.15.47. [DOI] [PubMed] [Google Scholar]
- Budny B, Badura-Stronka M, Materna-Kiryluk A, Tzschach A, Raynaud M, Latos-Bielenska A, Ropers HH. Novel missense mutations in the ubiquitination-related gene UBE2A cause a recognizable X-linked mental retardation syndrome. Clin Genet. 2010;6:541–551. doi: 10.1111/j.1399-0004.2010.01429.x. [DOI] [PubMed] [Google Scholar]
- Bush K, Vinsky M, Aldridge C, Paszkowski C. A comparison of sample types varying in invasiveness for use in DNA sex determination in an endangered population of greater Sage-Grouse (Centrocercus uropihasianus) Conserv Genet. 2005;5:867–870. [Google Scholar]
- Cardona C, Xing Z, Sandrock CE, Davis CE. Avian influenza in birds and mammals. Comp Immunol Microbiol Infect Dis. 2009;4:255–273. doi: 10.1016/j.cimid.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Clausen D, Robus M, Matthews M. Minto Flats State Game Refuge Management Plan. Alaska Department of Fish and Game 1992 [Google Scholar]
- Dubowy P. Feeding ecology and behavior of postbreeding male blue-winged teal and northern shovelers. Can J Zool. 1985;6:1292–1297. [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt O. Interferon-induced antiviral Mx1 GTPase is associated with components of the SUMO-1 system and promyelocytic leukemia protein nuclear bodies. Exp Cell Res. 2001;2:286–295. doi: 10.1006/excr.2001.5380. [DOI] [PubMed] [Google Scholar]
- Engelhardt O. Mx1 GTPase accumulates in distinct nuclear domains and inhibits influenza A virus in cells that lack promyelocytic leukaemia protein nuclear bodies. J Gen Virol. 2004;8:2315–2326. doi: 10.1099/vir.0.79795-0. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005:47–50. [PMC free article] [PubMed] [Google Scholar]
- Garamszegi LZ, Møller AP. Prevalence of avian influenza and host ecology. Proc Biol Sci. 2007;1621:2003–2012. doi: 10.1098/rspb.2007.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P, Macquin C, Bahram S. The heterogeneous allelic repertoire of human toll-like receptor (TLR) genes. PLoS ONE. 2009;11:e7803. doi: 10.1371/journal.pone.0007803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O, Kochs G. Interferon-induced Mx proteins: dynamin-like GTPases with antiviral activity. Traffic. 2002;10:710–717. doi: 10.1034/j.1600-0854.2002.31003.x. [DOI] [PubMed] [Google Scholar]
- Haller O, Kochs G, Weber F. Interferon, Mx, and viral countermeasures. Cytokine Growth Factor Rev. 2007;(5-6):425–433. doi: 10.1016/j.cytogfr.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O, Staeheli P, Kochs G. Protective role of interferon-induced Mx GTPases against influenza viruses. Rev Sci Tech. 2008;1:219–231. doi: 10.20506/rst.28.1.1867. [DOI] [PubMed] [Google Scholar]
- Hey J, Wakeley J. A coalescent estimator of the population recombination rate. Genetics. 1997;3:833–846. doi: 10.1093/genetics/145.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill NJ, Takekawa JY, Cardona CJ, Ackerman JT, Schultz AK, Spragens KA, Boyce WM. Waterfowl ecology and avian influenza in California: do host traits inform us about viral occurrence? Avian Dis. 2010;54(s1):426–432. doi: 10.1637/8912-043009-Reg.1. [DOI] [PubMed] [Google Scholar]
- Ito T, Okazaki K, Kawaoka Y, Takada A, Webster RG, Kida H. Perpetuation of influenza A viruses in Alaskan waterfowl reservoirs. Arch Virol. 1995;7:1163–1172. doi: 10.1007/BF01322743. [DOI] [PubMed] [Google Scholar]
- Janzen C, Kochs G, Haller O. A monomeric GTPase-negative MxA mutant with antiviral activity. J Virol. 2000;17:8202–8206. doi: 10.1128/jvi.74.17.8202-8206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L, Kambadur R, Lee-Hellmich H, Hodgkinson CA, Arnheiter H, Meier E. Antiviral determinants of rat Mx GTPases map to the carboxy-terminal half. J Virol. 1997;12:9792–9795. doi: 10.1128/jvi.71.12.9792-9795.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain E, Gunnarsson G, Wahlgren J, Latorre-Margalef N, Bröjer C, Sahlin S, Svensson L, Waldenström J, Lundkvist A, Olsen B. Influenza virus in a natural host, the mallard: experimental infection data. PLoS One. 2010;1:e8935. doi: 10.1371/journal.pone.0008935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida H, Yanagawa R, Matsuoka Y. Duck influenza lacking evidence of disease signs and immune response. Infect Immun. 1980;2:547–553. doi: 10.1128/iai.30.2.547-553.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Takada A, Mitsuhashi T, Agui T, Watanabe T. Native antiviral specificity of chicken Mx protein depends on amino acid variation at position 631. Anim Genet. 2004;2:119–122. doi: 10.1111/j.1365-2052.2004.01096.x. [DOI] [PubMed] [Google Scholar]
- Ko JH, Jin HK, Asano A, Takada A, Ninomiya A, Kida H, Hokiyama H, Ohara M, Tsuzuki M, Nishibori M, Mizutani M, Watanabe T. Polymorphisms and the differential antiviral activity of the chicken Mx gene. Genome Res. 2002;4:595–601. doi: 10.1101/gr.210702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochs G, Haener M, Aebi U, Haller O. Self-assembly of human MxA GTPase into highly ordered dynamin-like oligomers. J Biol Chem. 2002a;16:14172–14176. doi: 10.1074/jbc.M200244200. [DOI] [PubMed] [Google Scholar]
- Kochs G, Haller O. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc Natl Acad Sci USA. 1999;5:2082–2086. doi: 10.1073/pnas.96.5.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochs G, Janzen C, Hohenberg H, Haller O. Antivirally active MxA protein sequesters La Crosse virus nucleocapsid protein into perinuclear complexes. Proc Natl Acad Sci USA. 2002b;5:3153–3158. doi: 10.1073/pnas.052430399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikova IV, Drovetski SV, Gibson DD, Harrigan RJ, Rohwer S, Sorenson MD, Winker K, Zhuravlev YN, McCracken KG. Phylogeography of the mallard (Anas platyrhynchos): hybridization, dispersal, and lineage sorting contribute to complex geographic structure. Auk. 2005;3:949–965. [Google Scholar]
- Lindenmann J. Resistance of mice to mouse-adapted influenza A virus. Virology. 1962:203–204. doi: 10.1016/0042-6822(62)90297-0. [DOI] [PubMed] [Google Scholar]
- Lindenmann J, Klein P. Further studies on the resistance of mice to myxoviruses. Arch Virol. 1966;1:1–12. [Google Scholar]
- Livant E, Avendano S, Mcleod S, Ye X, Lamont S, Dekkers J, Ewald S. Mx1 exon 13 polymorphisms in broiler breeder chickens and associations with commercial traits. Anim Genet. 2007;2:177–179. doi: 10.1111/j.1365-2052.2007.01577.x. [DOI] [PubMed] [Google Scholar]
- Luoma LM, Deeb TMM, Macintyre G, Cox DW. Functional analysis of mutations in the ATP loop of the Wilson disease copper transporter, ATP7B. Hum Mutat. 2010;5:569–577. doi: 10.1002/humu.21228. [DOI] [PubMed] [Google Scholar]
- McCracken K, Johnson W, Sheldon F. Molecular population genetics, phylogeography, and conservation biology of the mottled duck (Anas fulvigula) Conserv Genet. 2001;2:87–102. [Google Scholar]
- Meier E, Fäh J, Grob MS, End R, Staeheli P, Haller O. A family of interferon-induced Mx-related mRNAs encodes cytoplasmic and nuclear proteins in rat cells. J Virol. 1988;7:2386–93. doi: 10.1128/jvi.62.7.2386-2393.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima E, Morozumi T, Tsukamoto K, Watanabe T, Plastow G, Mitsuhashi T. A naturally occurring variant of porcine Mx1 associated with increased susceptibility to influenza virus in vitro. Biochem Genet. 2007;(1-2):11–24. doi: 10.1007/s10528-006-9045-y. [DOI] [PubMed] [Google Scholar]
- Ng A, Wang J, Shaw P. Structure and sequence analysis of influenza A virus nucleoprotein. Sci China C Life Sci. 2009;5:439–449. doi: 10.1007/s11427-009-0064-x. [DOI] [PubMed] [Google Scholar]
- Ng P. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;13:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B, Munster V, Wallensten A, Waldenström J, Osterhaus AD, Fouchier RA. Global patterns of influenza A virus in wild birds. Science. 2006;5772:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- Palermo S, Capra E, Torremorell M, Dolzan M, Davoli R, Haley CS, Giuffra E. Toll-like receptor 4 genetic diversity among pig populations. Anim Genet. 2009;3:289–299. doi: 10.1111/j.1365-2052.2008.01833.x. [DOI] [PubMed] [Google Scholar]
- Parker LE, Bolen EG, Baker RJ. Genetic variation in a winter population of mallard ducks. Southwest Nat. 1981;4:425–428. [Google Scholar]
- Pavlovic J, Zurcher T, Haller O, Staeheli P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J Virol. 1990;7:3370–3375. doi: 10.1128/jvi.64.7.3370-3375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrula MJ. M.S. Thesis University of Alaska Fairbanks. 1994. [Google Scholar]
- Qu LJ, Li XY, Xu GY, Ning ZH, Yang N. Lower antibody response in chickens homozygous for the Mx resistant allele to avian influenza. Asian Austral J Anim. 2009;4:465–470. [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;2:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Runstadler J, Happ G, Slemons R, Sheng Z, Gundlach N, Petrula M, Senne D, Nolting J, Evers D, Modrell A, Huson H, Hills S, Rothe T, Marr T, Taubenberger J. Using RRT-PCR analysis and virus isolation to determine the prevalence of avian influenza virus infections in ducks at Minto Flats State Game Refuge, Alaska, during August 2005. Arch Virol. 2007;10:1901–1910. doi: 10.1007/s00705-007-0994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler A, Williams B. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;7:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Curtis D. Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet. 1995;(Pt 1):97–105. doi: 10.1111/j.1469-1809.1995.tb01608.x. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Staeheli P. Domains mediating intramolecular folding and oligomerization of MxA GTPase. J Biol Chem. 1998;43:28365–28370. doi: 10.1074/jbc.273.43.28365. [DOI] [PubMed] [Google Scholar]
- Spackman E, Senne DA, Bulaga LL, Myers TJ, Perdue ML, Garber LP, Lohman K, Daum LT, Suarez DL. Development of real-time RT-PCR for the detection of avian influenza virus. Avian Dis. 2003;3(Suppl):1079–1082. doi: 10.1637/0005-2086-47.s3.1079. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;4:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S, Reichelt M, Krijnse-Locker J, Mackenzie J, Simpson JC, Haller O, Kochs G. Interferon-induced, antiviral human MxA protein localizes to a distinct subcompartment of the smooth endoplasmic reticulum. J Interferon Cytokine Res. 2006;9:650–660. doi: 10.1089/jir.2006.26.650. [DOI] [PubMed] [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;2:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;3:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol. 2007;8:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;1:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh T. M.S. Thesis, University of Alaska Fairbanks. 2008. [Google Scholar]
- Wilson D. Estimating diversifying selection and functional constraint in the presence of recombination. Genetics. 2005;3:1411–1425. doi: 10.1534/genetics.105.044917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zürcher T, Pavlovic J, Staeheli P. Mechanism of human MxA protein action: variants with changed antiviral properties. EMBO J. 1992;4:1657–1661. doi: 10.1002/j.1460-2075.1992.tb05212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.