Abstract
A quantitative description of the relationship between protein expression levels and open reading frame nucleotide sequences (ORFs) is important for understanding natural systems, designing synthetic systems, and optimizing heterologous expression. Codon identity, mRNA secondary structure, and nucleotide composition within ORFs markedly influence expression levels. Bioinformatic analysis of ORF sequences in 816 bacterial genomes revealed that these features show distinct regional trends. To investigate their effects on protein expression, we designed 285 synthetic genes and determined corresponding expression levels in vitro using E. coli extracts. We developed a mathematical function, parameterized using this synthetic gene dataset, which enables computation of protein expression levels from ORF nucleotide sequences. In addition to its practical application in the design of heterologous expression systems, this equation provides mechanistic insight into the factors that control translation efficiency. We found that expression is strongly dependent on the presence of high AU content and low secondary structure in the ORF 5′ region. Choice of high-frequency codons contributes to a lesser extent. The 3′ terminal AU content makes modest, but detectable contributions. We present a model for the effect of these factors on the three phases of ribosomal function: initiation, elongation, and termination.
Keywords: protein expression, nucleotide composition, mRNA secondary structure, codon usage, synthetic genes, bioinformatic analysis
Introduction
Quantitative description of the factors that determine protein expression levels is central to understanding natural systems 1, designing synthetic systems 2–3, and optimizing heterologous expression 4. Protein expression is a complex, multi-step process involving transcription, mRNA turnover, translation, post-translational processing, and protein stability. Although much of the information controlling expression levels is encoded in untranslated regions (UTRs) of bacterial genes 5–6, sequence variation in open reading frames (ORFs) also can have profound effects 7–9. The latter is mediated through the presence or absence of recognition sequences for stimulatory or inhibitory factors such as RNA-binding proteins 10–11 or non-coding RNAs 12 and, more generally, through variation of three features: codon identity, levels of mRNA secondary structure, and nucleotide composition.
A quantitative description of the relationship between protein expression levels and ORF sequence features has remained elusive. An average ORF in Escherichia coli potentially can adopt ~10159 iso-coding sequences (this study). Experimental exploration of such enormous sequence spaces to define the relationship between sequence and protein expression levels is very challenging 9. Powerful computational algorithms have been developed to solve the class of huge discrete combinatorial searches that arise in optimizing codon choice, and can be applied to design synthetic sequences for testing critical sequence features that contribute to protein expression 13–17. However, before the advent of affordable large-scale DNA synthesis together with gene assembly automation methodologies 18–19, it has been impractical to experimentally test large number of synthetic genes. In an effort to address these issues, we integrated four approaches: we undertook a bioinformatic analysis of available prokaryotic genomic sequences to identify ORF sequence features that potentially influence protein expression; developed a gene sequence design program (OrfOpt) that tunes regional nucleotide composition, codon choice, and mRNA secondary structure to design synthetic sequences that test critical values and combinations of such features; used gene assembly automation to construct synthetic genes; and used coupled in vitro transcription and translation (TnT) in E. coli extracts to measure their protein expression levels.
The genomic frequency distribution of codons that encode the same amino acid is often very uneven (codon bias), and can differ dramatically between organisms 20. The presence of infrequently used codons in an ORF can significantly depress protein expression levels 9 and may even affect the fidelity of translation 21. There has been considerable speculation on the origin and biochemical consequences of codon bias, including correlation with tRNA populations 22 and metabolic load 20. Codon biases can affect translational elongation step rates and ribosomal movement 23, and may provide pausing points during protein folding 24. Optimization of codon bias is a common strategy to improve heterologous protein expression using synthetic genes 4. Nevertheless, the importance of codon bias relative to other factors in optimizing protein expression remains unresolved: one recent study suggests that codon bias does not correlate well with high levels of heterologous protein expression in E. coli 7, whereas another identifies it as a critical determinant by controlling the choice of metabolically available aminoacylated tRNAs 8.
RNA secondary structure at the 5′ end of an ORF also has been recognized to be important for protein expression, acting through a variety of mechanisms. One effect involves ‘masking’ of ribosome binding sites (RBS) by inverted repeats in the mRNA that cover part of the ORF itself 25. A second effect involves mRNA secondary structures encoded entirely within the 5′ end of the ORF, which are likely to hamper loading of the mRNA onto the ribosome at initiation of translation 26. Bioinformatic analysis indicates that mRNA secondary structure content decreases toward the 5′ end of genes in several Eubacteria 27. Furthermore, it has been suggested that absence of secondary structure in the 5′ end of ORFs is more important than codon choice in determining expression levels 7,28.
There is considerable anecdotal evidence that the AU composition within the 5′ end of an ORF may also play a role in expression levels 16,23,29–35. Expression patterns of computationally designed genes with elevated 5′ AU composition suggest that the impact of this parameter can be profound 16. Nevertheless, the influence of regional nucleotide composition in ORFs on protein expression levels remains poorly characterized.
Our bioinformatic analysis of 816 fully sequenced bacterial genomes revealed that there are two readily discernible canonical ORF sequence features within the 5′ and 3′ ends relative to the middle regions of ORFs: AU content is elevated, and secondary structure is depressed. To test the contributions of these features together with high-frequency codon usage, we developed a computational algorithm that enables their systematic variation in synthetic gene sequences. We constructed 285 synthetic genes distributed over three proteins differing in species origin, gene length, and protein fold. We find that the resulting experimental expression levels can be calculated from the ORF mRNA sequence by a non-linear scoring function. This function comprises a set of sigmoidal thresholds in which each feature transitions through a critical region below which it fully or partially inhibits expression, above which its contribution plateaus, and within which expression levels are sensitive to its quantitative value. This mathematical model provides important insights into the relative contributions of the three ORF sequence features: protein expression levels are strongly dependent on the presence of high AU content and low secondary structure in the N-terminal segment, and that codon choice contributes when favorable.
The bioinformatic analysis together with the experimental characterization of synthetic genes and resulting mathematical model provides a quantitative mapping between ORF mRNA sequence and protein expression levels. This quantitative model has also enabled us to develop a predictive gene design method that has yielded synthetic ORF sequences with high levels of protein expression for a variety of proteins (Allert et al., in preparation). Most importantly, it shows that the properties of the 5′ ORF region play a critical role in determining bacterial protein expression levels.
Results
Statistical analysis of bacterial ORFs
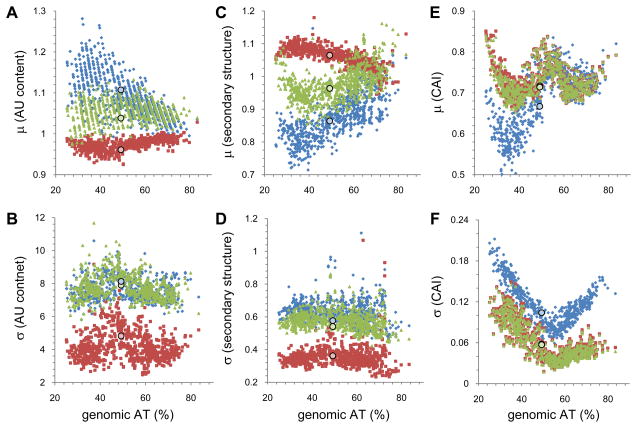
We analyzed regional nucleotide composition, mRNA secondary structure, and codon choice in the 2.5×106 ORFs of 816 fully sequenced bacterial genomes that span genomic AT contents ranging from 25% to 83% (Supplementary Table 1). Computational algorithms and definitions of the statistical measures are described in Experimental Procedures. The mean AU content of the first and last 35 base regions within ORFs is significantly higher than the middle section (Figure 1A). Of the two terminal regions, the 5′ end tends to have a higher AU bias than the 3′ end. mRNA secondary structure content also shows significant regional differences, with the two ends having lower mean structural content and higher variance than the middle (Figures 1C & 1D). Again, the trend is stronger in the 5′ than the 3′ terminus. The trends in these two sequence features are present regardless of genomic nucleotide composition, but the signal is more pronounced in GC- than AT-rich genomes. The variances of the nucleotide composition and secondary structure content also is much higher at the two termini than the middle region (Figures 1B,D). Such increased variance suggests that some aspect of control might be encoded in these regions: an increased level of variance in a parameter indicates that genes differ from one another in this respect, as would be expected for features with regulatory functions.
Figure 1. Genomic averages and variances of regional ORF nucleotide composition, RNA secondary structure, and codon adaptation index.
All parameters are shown as mean values and variances calculated over all ORFs within a genome. Blue, 5′ ORF region (first 35 bases); red, middle region; green, 3′ ORF region (last 35 bases). Circles indicate the values of these parameters calculated for E. coli strain K-12 DH10B. (A) Mean ORF regional nucleotide composition is reported as the ratio of the composition of that region to that of the genome average. (B) Variances of the mean genomic regional nucleotide compositions. (C) Mean ORF regional secondary structure content is reported as the ratio of a region relative to the genome average. (D) Variances of the mean ORF regional secondary structure content. (E) Mean regional codon adaptation indices. (F) Variances of the regional genomic CAI values.
Codon choices can be quantified as the codon adaptation index (CAI), which varies from 0 to 1, reflecting choice of low- or high-frequency codons respectively 36. Codon frequencies were calculated over all the ORFs for each genome individually. The means and variances of CAI values averaged over a genome exhibit a complex, but well-defined pattern (Figure 1E), precluding identification of clear canonical rules governing codon bias by this approach. The CAI pattern shows some regional variation. At genomic AT contents below ~50%, the 5′ regional CAI tends to be significantly lower than that observed in either the middle or 3′ regions, which are indistinguishable from one another. The variance of the 5′ regional CAI values are always higher than that of the other two regions (Figure 1F), again suggesting the presence of regulatory function in this region.
Construction and expression patterns of synthetic genes in E. coli
Our bioinformatic analysis indicates that regional AU content and secondary structure at the beginning and end of ORFs are important features that are conserved across bacterial species. However, this analysis provided no further information on their functional importance, nor does it provide guidance on codon usage. To test experimentally which features effect protein expression levels and to assess their relative contributions, we developed a computer algorithm (OrfOpt; see Experimental Procedures) that enables us to specify quantitatively all six parameters (AU content in the 5′ and 3′ ORF termini; secondary structure in 5′, 3′, and middle segments; and the CAI calculated over the entire ORF) in synthetic genes. We designed and constructed 285 synthetic genes distributed over three test proteins that vary in size, structure, species origin, and heterologous expression levels of their wild-type ORF sequences: asparate aminotransferase (~43 kDa, (αβα)-sandwich, Thermus thermophilus, no expression), fatty acid binding protein (~15 kDa, β-clam, Gallus gallus, good expression), and triose phosphate isomerase (~28 kDa, (αβ)8-barrel, Leishmania mexicana, poor-to-no expression). Genes were assembled from synthetic oligonucleotides by an automated, robust, PCR-mediated gene construction scheme 19. Full-length linear PCR fragments containing synthetic ORFs flanked by invariant, untranslated control regions encoding a T7 promoter, ribosome binding site, and T7 terminator were tested for protein expression using a TnT extract prepared from the E. coli BL21 Star strain. The in vitro expression approach provides a standard, well-defined set of conditions for protein expression, which can correlate with in vivo expression levels 37. Furthermore, the BL21 Star strain lacks the C-terminal portion of RNase E 38, thereby disentangling complexities associated with endonucleolytic mRNA cleavage 39 within the variable portion of the designed ORF sequences from effects on translation.
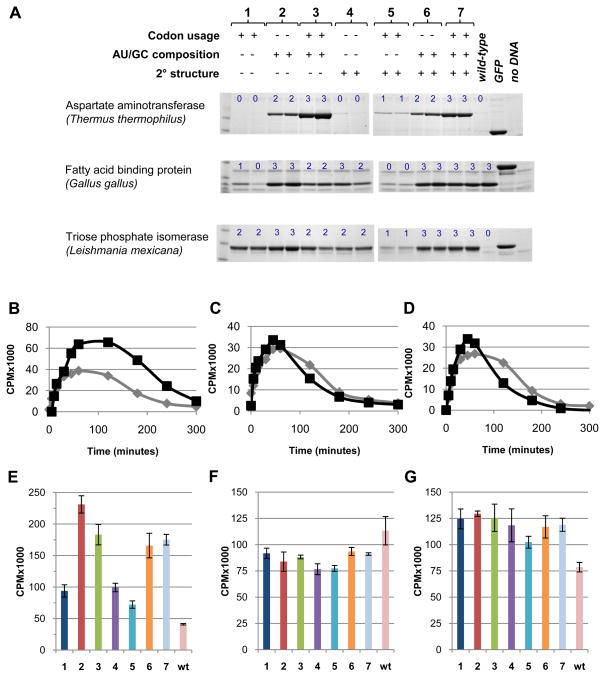
The contributions and interplay of the six parameters are illustrated qualitatively by 42 synthetic alleles (Figure 2 and Figure S1). These alleles were designed using seven different calculation conditions in which values for the six parameters were targeted individually and in combination; the values of parameters that were not explicitly targeted were left unconstrained. The resulting seven conditions were tested in all three proteins. For each condition in vitro expression levels of two alleles differing by at least 10 iso-codon changes were evaluated using Coomassie-stained gels. To distinguish between effects on transcription or translation, we measured radiolabeled RNA levels in the reactions after one hour (at which point the RNA levels are highest; Fig 2B–D), and found that these varied by less than 20–30% in two cases (Figure 2F,G) and less than three-fold in the third case (Figure 2E), whereas protein expression levels in the Coomassie-stained gels showed much greater variation in all three cases (Figure 2A) suggesting that the differences are due to effects on translation. The pattern of observations indicates that expression levels are most strongly influenced by high AU content in the 5′ region, followed in importance by low secondary structure content. Optimization of the CAI by itself is less effectual. Typically, the highest expression levels are obtained if all three factors are optimized simultaneously.
Figure 2. Experimental expression levels of synthetic genes determined using E. coli coupled in vitro transcription and translation reactions.
(A) Synthetic genes were designed by optimizing CAI, mRNA secondary structure, and 5′ ORF regional nucleotide composition singly or in combination giving a total of seven conditions. For each condition, the expression pattern of two alleles differing by at least 10 mutations are shown. Three proteins differing in size, structure, origin and expression of wild-type ORF sequences were used: asparate aminotransferase (ttAST), fatty acid binding protein (ggFABP), and triose phosphate isomerase (lmTIM). Proteins were purified from coupled in vitro transcription and translation (TnT) reactions using immobilized metal affinity chromatography and run on 4–12% SDS-PAGE gradient gels. Green florescent protein template was included as a positive control for protein expression levels and an extract without added DNA as a negative control. Observed expression levels were classified into one of four categories (blue numbers: 0, no band; 1, weak band; 2, medium band; 3, strong band). Full gel images are shown in Figure S1. The identity of the observed protein band was verified by mass spectrometry for each of the three proteins in the first allele of the optimization condition 7 (Figure S2). (B–D) Time course of radiolabeled RNA in TnT reactions containing a high- (black) and low- (grey) expression level allele (background of a reaction without added DNA was substracted): ttAST (B), ggFABP (C), lmTIM (D). (E–G) Total radiolabeled RNA at one hour using one allele for each condition presented in panel A and the wild-type sequences: ttAST (E), ggFABP (F), lmTIM (G).
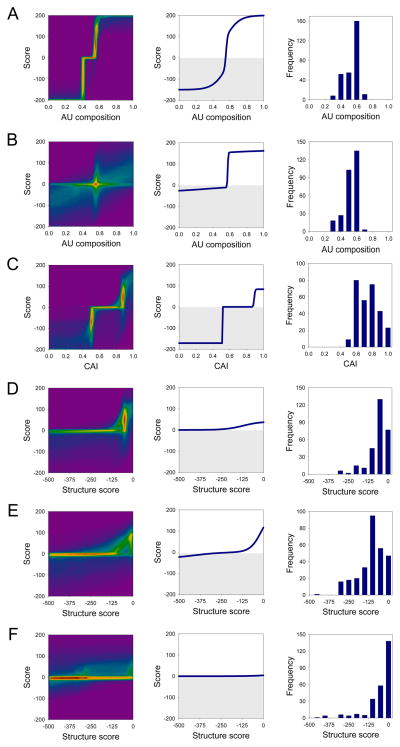
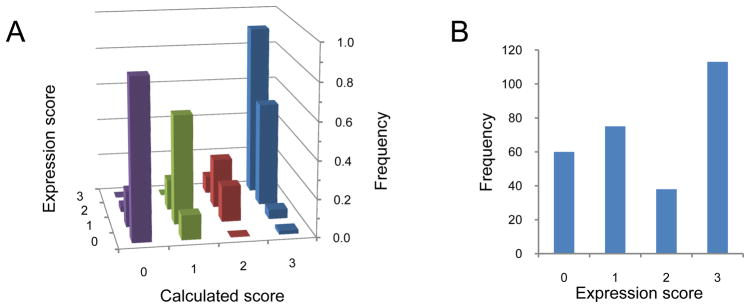
To build a dataset that might enable a quantitative mapping to be established between ORF mRNA sequence and protein expressions, we constructed a total of 285 synthetic genes (Figure S1, Supplementary Table 2). With exception of the CAI, which was not sampled below 0.45, ORF features were sampled over a wide range (Figure 3, right column). The experimental expression levels of all synthetic genes were classified into four categories determined by inspection of band intensities in 4–12% gradient SDS-PAGE gels: zero (no band), one (weak band), two (medium band), three (strong band). The intensities of the strongest bands are comparable across all three proteins, indicating that the categories can be compared directly between different proteins. The dataset samples all four experimental categories (Figure 4B). We found that the experimental observations could not be mapped well to their corresponding ORF mRNA sequences using linear combinations of the ORF parameters. We chose to represent each of the six parameters as a sum of two sigmoidal curves corresponding to penalty (inhibitory) and reward (stimulatory) contributions, respectively. This function was parameterized against the entire 285 synthetic gene dataset, using a simulated annealing algorithm with 10,000 independent optimization calculations (see Experimental Procedures). In addition to obtaining an optimal parameter set, this ensemble provides a Monte Carlo sampling of near-optimal solutions (Figure 3, left column). The resulting function (Figure 3, middle column) accurately calculates experimentally observed expression categories for 69% of the dataset, with the remainder being calculated to their closest neighboring category (Figure 4A). The highest and lowest expression levels fit the most precisely (91% and 85% respectively).
Figure 3. Parameterization of a mathematical function that calculates protein expression levels from ORF sequence.
The function is the sum of six pairs of sigmoids representing reward and penalty contributions of 5′ (A) and 3′ (B) ORF regional AU composition, the ORF codon adaptation index (C), 5′ (D), middle (E) and 3′ (F) ORF regional secondary structure content. The score of each component ranges [−200,200]; their sum is mapped onto the protein expression category as <−100→0 (no expression), [−100,0]→1 (low), [0,100]→2 (medium), >100→3 (high). Left column: density plot of the distribution of sigmoids in the ensemble of near-optimal solutions. False coloring indicates how many sigmoidal curve segments pass through a region (magenta, none < blue < green < yellow < red, high). These distributions give an indication of the uncertainty in the parameter set. For instance, although there are many solutions for the 3′ ORF regional composition (B), it is clear that all have a penalty (lower-left quadrant) and reward (upper-right quadrant) with a critical transition centered at ~56% (red peak). Middle column: sigmoids of the parameters set that best fits the data (grey area: penalty score values). Right column: distribution of parameters in the experimental dataset (note that for the C-terminal segment there are 29 alleles with secondary structure scores <−500, which are not shown).
Figure 4. Correlation between observed and calculated protein expression levels.
(A) correlation between calculated and observed expression levels. The frequencies are normalized to 1 within each predicted category. For 69% of the data calculated and observed expression categories are accurately calculated (diagonal). The remainder is usually off by only one expression level category; (B) distribution of observed protein expression levels (0, no expression; 1, low expression; 2, medium expression; 3, high expression).
The distribution of near-optimal solution values gives an indication of how well the parameters are determined (Figure 3, left column). The 5′ regional AU content (Figure 3A) and secondary structure dependencies (Figure 3D) are well determined. The effect of 5′ regional nucleotide composition is very pronounced and centered around a critical point at 53–55% AU content, above which it is strongly stimulatory and below which it is equally strongly inhibitory. Low secondary structure content in this region is stimulatory, and does not become strongly inhibitory until high levels are reached. Features in the 3′ region are less well defined by this dataset. Nevertheless, AU composition has both a reward and penalty contributions above and below ~57% respectively, but their numerical weights remain ill determined (Figure 3B). By contrast, 3′ regional secondary structure does not appear to play a significant role (Figure 3F). Contributions of secondary structure in the middle region are fairly well determined (Figure 3E): a high structural content has a modest negative impact, whereas near-absence of secondary structure is moderately favorable. The contributions of the CAI are qualitatively well determined, but uncertainty regarding the precise weight of penalty and reward remain (Figure 3C). The CAI can significantly enhance expression levels if above ~0.8; conversely, if below ~0.5, it is inhibitory although the precise numerical value of this latter effect is not well determined by our sampling.
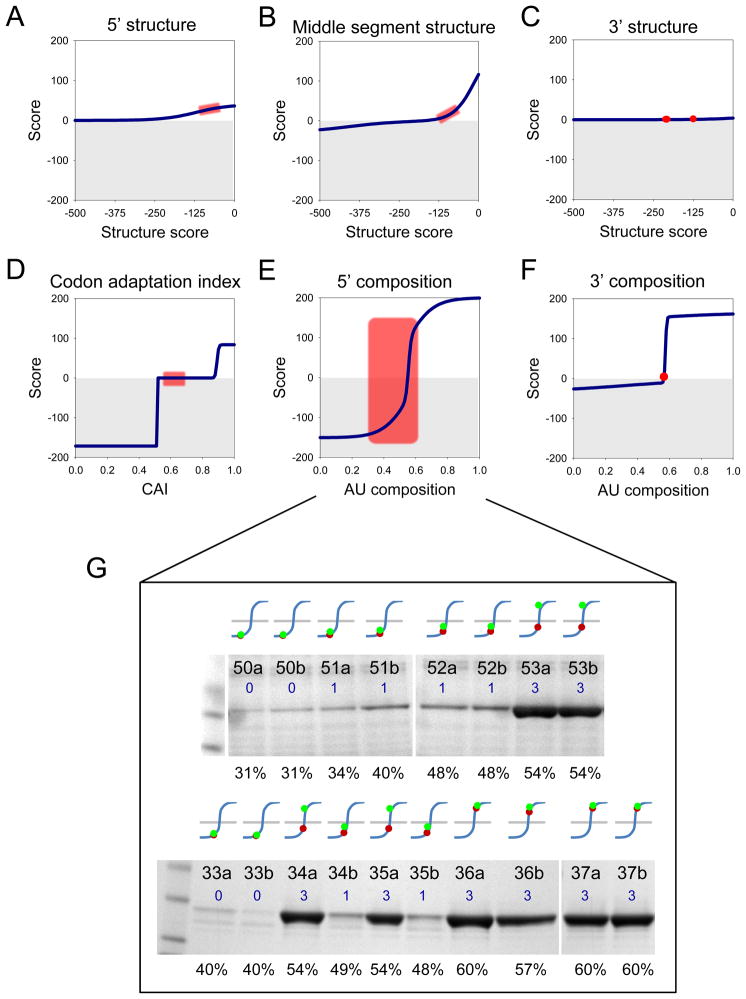
The fits for the dependence on regional nucleotide composition and the CAI behave as thresholds (Figure 3, middle column), in which these parameters below a critical value are inhibitory (e.g. 5′ nucleotide composition), or do not contribute much (e.g. 3′ nucleotide composition), and above which their contributions plateau. The effect of transitioning through a critical value can be very abrupt (e.g. 5′ nucleotide composition), or gradual (e.g. 5′ secondary structure). The overall expression level is the sum of all contributions. For strongly contributory parameters, such as the 5′ regional nucleotide composition, inhibitory threshold effects can dominate, even when other parameters adopt favorable values. Such threshold effects and interplay between parameters are demonstrated by a series of alleles in which the 5′ regional AU content was systematically varied (Figure 5) while maintaining the other five parameters close to constant, slightly favorable values (Figure 5A–F). For the ttAST alleles 52a–53b (Figure 5G top) and the lmTIM alleles 34a–35b (Figure 5G bottom), there is a sudden increase in expression levels on transitioning from 48.6% to 54.3% (compare ttAST alleles 52a,b and 53a,b, lmTIM alleles 34a and 34b, or lmTIM 35a and 35b) as the 5′ regional composition transitions through its critical threshold. The four alleles lmTIM 36a–37b with 57.1% to 60.0% AU content illustrate that if this threshold is exceeded, there is not much apparent further gain in expression.
Figure 5. The effect of varying N-terminal AU content in the presence of (near-) constant other parameters.
Eight alleles of ttAST (50a–53b; G, top) and ten alleles of lmTIM (33a–37b; G, bottom) were constructed in which the 5′ regional composition was varied from 31% to 60%AU content (E); while keeping the other five parameters near-constant in a range where they have little effect on the predicted expression score (A–D, F). Panels A–F show the range of values (red rectangles or circles) of the six parameters for the eighteen alleles, mapped on the scoring function parameterized by the optimal global fit (see Figure 3). Panel G shows the expression levels (blue numbers) of the eighteen alleles (identity indicated at the top of each lane; see Supplementary Table 2 and Supplementary Figure S1) determined in Coomassie-stained gels. The curves above each lane indicate the mapping of the allelic 5′ regional AU content (shown as percentage at the bottom of each lane) onto the scoring function for this parameter (blue line). Mapping of the allelic values is shown for two critical points: red dots, 55% AU content, obtained from the optimal global fit of all the data (see Figure 3A, middle); green dots, 53% AU content, corresponding to the lower limit observed in the range of near-optimal fits (see Figure 3A, left). The latter value exhibits a clear threshold transition for these alleles in these two proteins. In addition to illustrating the effect of transitioning through a threshold, these results show that the value of the nucleotide composition critical point is not yet determined precisely (2% uncertainty).
It is difficult to distinguish between changes in nucleotide composition and RNA secondary structure, because these are inter-related. However, analysis of the ttAST 52a–53b and the lmTIM 33a–34b alleles, which constitute examples of carefully paired low- and high-expression alleles in which N-terminal composition was changed while minimizing changes in secondary structure as much as possible, suggests that nucleotide composition, not secondary structure is primarily responsible for the control of protein expression levels by the 5′ ORF region. The ttAST alleles 52a,b have lower secondary structure content (−82.7, −94.8; see Supplementary Material), lower AT content (48.6%), and lower expression level than ttAST 53a,b (−103.3,−117.3; 54.3%). Thus the expression level changes dramatically (1→3) despite a gain, rather than loss, in secondary structure. The situation for the paired low- (34b,35b) and high-expression (34a,35a) alleles constructed in lmTIM is less clear, because a large increase in protein expression (1→3) is accompanied both by a transitioning through the nucleotide composition critical region (48.6→54.3%), and a slight loss of secondary structure content (−63.8,−59.2→−45.8,−49.2). However the magnitude of the loss of secondary structure score in the lmTIM pair (~15), is less than the gain in the ttAST pair (~20), suggesting that the effects are due to changes in nucleotide composition, not secondary structure. This dominance of compositional effects over secondary structure content is also reflected by the contributions of these terms in the expression function, established using the entire set of synthetic alleles (Figure 3).
Discussion
Our bioinformatic analysis revealed that bacterial genes have distinct regional trends in nucleotide composition and RNA secondary structure content within the first and last 35 bases of ORFs, which are present regardless of genomic nucleotide composition. Our experimental analysis of synthetic genes shows that protein expression levels are strongly dependent on the presence of high AU content and low secondary structure in the 5′ region, and that choice of high-frequency codons contributes to a lesser extent. Furthermore, we find that the 3′ regional AU content makes a modest, but detectable contribution to protein expression, an effect not previously observed.
The size of the synthetic gene data set and the range of values of the parameters enabled us to develop a mathematical function that calculates protein expression levels from ORF sequences. The nonlinear character of this equation emphasizes the complexity of the multifactorial effects, illustrates why it is so difficult to unravel these factors, and further emphasizes that correlation between protein and mRNA levels should be approached with caution 40. Even with the use of computer algorithms, we have not been able to obtain many examples in which variation in one parameter is cleanly separated from changes in others. We note a few caveats to our approach. First, it is likely that as more data becomes available, the parameterization of the equation will change. Second, the OrfOpt computer algorithm addresses neither message transcription levels, nor mRNA lifetimes, nor specific sequence elements that bind factors which affect protein expression. Finally, the method is silent on aspects of post-translational processing and physico-chemical properties of proteins that affect solubility and turnover, as these require optimization of the amino acid sequence.
The universe of mRNA iso-coding sequences is vast, as illustrated by the 10159 variations that can be encoded by an average ORF in E. coli. It is therefore remarkable that quantitatively predictive rules which map mRNA ORF nucleotide sequence to protein expression levels can be obtained in relatively limited experimental explorations of this sequence space. This achievement indicates that the encoding of the factors determining expression levels is based on highly degenerate mRNA sequence features, which can be captured mathematically to a first approximation. Quantitative mapping between ORF mRNA sequences and protein expression enables the development of computer programs for the design of synthetic genes optimized for heterologous protein expression, which is an important goal for biotechnology and synthetic biology 13–17. The success of our approach is illustrated here by the design of well-expressed genes for Leishmana mexicana triosephosphate isomerase and Thermus thermophilus asparate aminotransferase, of which the wild-type sequences express barely, if at all. The OrfOpt program also has been used successfully to predict synthetic gene sequences that express a variety of proteins at high levels (Allert et al. in preparation).
The contributions of the mRNA sequence features in the open reading frame region affect the three phases of translation: initiation, elongation, and termination 41–43. Initiation involves the highly regulated loading of mRNA onto the ribosome 26. Similarly, termination is a carefully orchestrated process, involving recognition of the stop codon by release factors 44. In addition to its practical application in the design of heterologous expression systems, the equation that predicts expression levels from sequence features may provide mechanistic insight into factors that control translation efficiency. Its functional form and the numerical weights of its terms can be qualitatively interpreted in terms of control logic and the importance of individual factors. Depending on its paramerization (σ, μ, W; see Material and Methods) a sigmoidal representation encodes different logic, ranging from a switch (very high sigmoidicity), linear response (no sigmoidicity), to absence of contribution (zero weight). The penalty and reward components of the function could be interpreted as representing inhibitory or stimulatory effects.
With these notions in mind, the empirical function linking sequence and protein expression level (Figure 3, middle column) could be interpreted in terms of a descriptive model for control of prokaryotic protein expression levels through modulation of basal ribosomal activity by sequence features within an ORF mRNA. We distinguish between two effects: passive, inhibiting basal ribosomal activity; and active, stimulating ribosomal activity through recruitment of extrinsic factors or activation of intrinsic ribosomal function. The parameterization of the function suggests that compliance of codon choice with available tRNA populations (set by availability of tRNA transcript 22–23, metabolic control of aminoacylation 8, and which in this study is approximated by the CAI) provides passive control: choice of non-compliant codons is inhibitory, and mildly stimulatory only if very compliant (Figure 3C). Control presumably emerges from the effect of the concentration of the available tRNAs on the rate of their complex formation with the ribosome. Secondary structure also exercises largely passive control, with mild stimulatory effects observed only at low levels of structure (Figure 3D–F). These inhibitory effects presumably arise from the need to unwind secondary structure as the single-stranded mRNA is threaded through the ribosome 41–43. By contrast, the effect of nucleotide composition at the 5′ end (Figure 3A), and, to a lesser extent, at the 3′ end (Figure 3B), appears to have a strong active component, suggestive of recruitment of an extrinsic or intrinsic stimulatory function.
The critical contribution of the 5′ ORF end has been noted also by others 16,23,29–35. The interrelationships between nucleotide composition, secondary structure, and codon compliance remain open to debate and are challenging to dissect. We and others 28 find that elevated secondary structure in this region tends to diminish protein expression, but that this effect does not dominate. It has been proposed that choice of low-compliance codons in this region is a dominant effect universally conserved in all domains of life 23. The hypothesis is that regional low-compliance codons locally slow down ribosome progress, thereby regulating downstream traffic and preventing downstream abortive translation events arising from multiple stalled ribosomes or inter-ribosomal collision. However, regional codon non-compliance and regional nucleotide composition are inter-related, because one affects the other. If regional non-compliance is the dominant cause, and nucleotide composition a side-effect, we should observe regional non-compliance independent of genomic composition, which in turn should result in AT-rich 5′ regions in GC-rich organisms, and GC-rich 5′ regions in AT-rich organisms. We observe a different pattern, however (Figure 1E): regional non-compliance is observed only in GC-rich organisms (up to ~50% genomic AT content), whereas elevated regional AT can be detected even within AT-rich organisms. This suggests strongly that it is nucleotide composition which is the dominant factor and codon non-compliance a side-effect. The latter is observed only in GC-rich organisms where maintenance of elevated AT-content skews codon choice to non-compliance. We note, however, that this conclusion is based on the use of the CAI, which is only a crude estimate of codon compliance; a definitive analysis requires the use of the tRNA adaptation index used in other studies 22–23 (work in progress).
If nucleotide composition is the primary determinant of the stimulatory properties of the 5′ORF, and, to a lesser extent, the 3′ORF regions, what molecular mechanism(s) could account for this effect? As we noted above, the nucleotide composition contributions appear to have the hallmarks of an active rather than passive effect, and therefore are likely to arise from recruitment of an extrinsic factor, or stimulation of intrinsic ribosomal function. One possible mechanism could involve binding of RNA helicases which catalyze the unfolding of secondary structure. Nucleotide composition could encode semi-specific recognition by such a helicase activity, giving rise to threshold effects through binding events. For instance, the DEAD-box protein superfamily includes prokaryotic RNA helicases that recognize AU-rich sequences 45, and in E. coli are involved in the RNA degradosome 39, or ribosomal RNA maturation 46. DEAD-box helicases play a role in eukaryotic initiation of translation, but no such function has been reported in prokaryotes 45. The ribosome itself contains an mRNA helicase activity that acts on a position within eleven bases from the codon that is being read 47. We hypothesize that local nucleotide composition could influence this activity. If this is the case, the patterns of nucleotide composition variance observed in all prokaryotes reflects increased ribosomal helicase activity at the beginning and end of ORFs, respectively to enhance post-initiation threading of the mRNA into the ribosome, and access of release factors at termination. The involvement of a composition-sensitive helicase activity (extrinsic or intrinsic), also could account for the difficulty in separating out contributions from nucleotide composition and RNA secondary structure. Both factors contribute to substrate recognition, and the latter additionally determines catalytic efficiency.
Translation and mRNA unwinding are tightly coupled in the elongation phase 48, accounting for the observed boost in expression levels observed at low secondary structure content for the middle segment. The lack of penalty at intermediate structural content again may reflect RNA helicase activity. The CAI also affects the elongation phase as it tends to reflect the relative concentrations of available tRNA pool 20. In our TnT reactions, the tRNA concentrations relative to each other are probably similar to in vivo ratios, because a total E. coli tRNA extract is added to the reactions, but their absolute concentration is higher than in vivo levels. Even so, we find that favorable CAI values influence protein expression levels, but that this effect drops off at values below ~0.8. The lowering of the regional CAI in the N-terminal region of GC-rich bacteria presumably reflects the dominance of AU content over CAI at initiation, because in those genomes, AU-rich codons are less frequent and therefore have a lower CAI.
Regardless of its detailed mechanistic origin, the sequence of the 5′ORF region plays a dominant role in determining prokaryotic protein expression levels 16,23,29–35. This and other studies 23,28 have proposed different hypotheses for the underlying molecular mechanism(s) of this effect, many of which are open to direct experimental testing. We look forward to learning the answer to this important riddle.
Materials and Methods
Bioinformatics
Annotated sequence files for 816 complete bacterial genomes were downloaded from http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi. Custom software was developed to calculate nucleotide composition, RNA secondary structure, and codon adaptation indices. Limits for the open reading frames were taken from the annotations in the genome sequence files. Regional nucleotide composition is calculated as the ratio of A and T content in the region relative to the segment length. RNA secondary structure is represented by a scoring function based on inverted repeats in a region, weighted by their base-pairing character, and loop or bulge sizes. This score was calculated by centering a recursive search of maximally-sized inverted repeats within 100 bp from a given nucleotide; duplexes are allowed to contain 10 bp bulges, 10 bp gaps, and a maximum 30–base loop. The score for such a maximally-sized inverted repeat is determined using stem-loop base-pairing energies 49–50 and assigned to each nucleotide encompassed within the calculated stem-loop. The secondary structure score of a region is the summed score for all stem-loops assigned to each base, normalized by the region length. Codon adaptation indices 36 are calculated according to codon tables constructed independently for each genome as the frequency distribution of codons in that genome. The CAI value for an ORF or region is the geometric mean of all the codons in such segments. Arithmetic means and standard deviations for genomic CAI values are calculated over the CAI values determined for each ORF in a genome. The number of possible iso-coding sequences for an ORF is calculated as the sum of the logarithm of the number of possible codons at each position. The mean genomic iso-coding sequence diversity is the arithmetic mean of the diversities of all the ORFs.
Computational design of synthetic ORF sequences
A simulated annealing algorithm was used to minimize an objective function capturing sequence features of interest within the available degrees of freedom in the ith trial Ei = Σν wrνr + Σs wrsr + cwc where νwr, swr are the relative weights assigned regional nucleotide composition (νr) and RNA secondary structure (sr) respectively (r: 5′, middle, 3′); cw, c are the ORF codon adaptation index weight and value, respectively. For minimizations in which only subsets of parameters were optimized, the weights for the unconstrained parameters were set to zero. Two types of calculations were run: absolute minimization of the objective function, or achievement of a target value. For target value optimization, the objective function was modified to where it represents weights, pi the parameters, and ti the target values. Sequence trial configurations were generated by randomly choosing two iso-codons per trial. At the beginning of the simulation, a cut-off of 0.45 was applied to remove low-frequency codons (this is the reason why synthetic ORFs with low CAI values were not sampled). Alleles were generated by maintaining a dynamic list of ~100 sequences differing by at least 10 mutations from the current best sequence and each other. A dynamic cooling schedule was used to drive the simulated annealing progress: to determine whether the ith trial is acceptable, ΔEi = Ei − Ei−1 was calculated and i accepted either if ΔEi ≤ 0 or in a Boltzmann decision if pi < e−ΔEi/T where pi is a random number [0,1] and T a control parameter. After 1,000 trials, the acceptance rate r was assessed, and T changed if r > 0.250,Tn+1 = 0.8Tn or r < 0.225, Tn+1 = 1.3Tn.
Final outcomes of such minimizations are critically dependent on the choice of weights. We used two approaches to address this problem. In one method we assigned weights empirically in a successive number of trial and error calculations. In a second method we developed a new Boltzmann decision scheme that circumvents the issue of weights and enables parameters with different numerical magnitudes to be combined. In this method, we used three independent Boltzmann decisions for each of the parameter classes, respectively, resulting in a ‘vote’ vi = νβi+sβi+cβi where βi = {1,0} and captures the outcome of a Boltzmann decision for a given parameter class (ν, regional nucleotide composition; c, codon preference; s, regional secondary structure). Unanimous votes (vi = 3) are always accepted; majority votes (vi = 2) are accepted half the time, and minority rule (vi=1) is accepted only if the overall acceptance rate has dropped below a 5% threshold value. Between two and 20 runs were executed in parallel on a Beowulf cluster and merged to construct the final set of alleles. All synthetic ORFs were optimized within the context of the invariant 5′ and 3′ UTR regions. The resulting sequences were fed into an automated experimental gene assembly pipeline (see below).
Parameterization of a function that predicts protein expression levels from ORF sequence
We developed a function in which an expression score is given as a sum of a series of thresholds applied to the composition, structure, and codon usage values of a sequence E = τν(5′)+τν(3′)+τs(5′)+τs(middle)+τs(3′)+τc where τν, τs are regional thresholds of nucleotide composition and structure respectively, and τc, the CAI threshold. A given threshold score for a feature value x is the sum of two sigmoids τ = Wp(1 + e−3(x−μp)/σp)−1 + Wr(1 + e−3(x−μr)/σr)−1 where p and r denote parameters for penalty and reward phases, respectively; W, weight, μ midpoint, σ sigmoidicity of each curve. The final value of the scoring function is the sum of all six components, and is mapped onto the expression level categories as ≤−100→0 (no expression), [−100, 0]→1 (weak expression), [0,100]→2 (medium expression), >100→3 (high expression). Parameters were fit as a minimization of the sum of the absolute differences between observed and calculated expression categories using a simulated annealing algorithm.
Oligonucleotide synthesis and synthetic gene assembly
Full-length genes (0.65 – 1.40 kb) encoding a synthetic ORF flanked by 5′ (122 or 131 bp) and 3′(103 or 112 bp) regulatory regions were assembled from oligonucleotides (80 – 100 bases) synthesized in-house (Mermade 192 DNA Synthesizer, BioAutomation) using an automated PCR-mediated gene assembly procedure 19,51. Full-length products were verified by agarose gel electrophoresis and reamplified with biotinylated flanking primers that provide some protection against endogenous exonuclease activity in the subsequent TnT reaction. Oligonucleotide synthesis and ORF assembly are detailed in the Supplementary Information.
In vitro coupled transcription and translation reactions
We used a TnT system based on the PANOxSP E. coli S30 lysate system 52–53. Lysate was prepared from BL21 Star (DE3) E. coli cells (Invitrogen) grown to mid-log phase in shaking culture flasks, rinsed of medium, flash-frozen, thawed, lysed in a French press, centrifuged to remove cellular debris, and incubated to facilitate a ‘run-off’ of any mRNA still bound to ribosomal complexes. The lysate was dialyzed, centrifuged to remove precipitate, and stored in flash-frozen aliquots. Reactions were initiated by adding biotinylated linear PCR template (1 μg per 100 μl reaction) to the lysate with magnesium glutamate, ammonium glutamate, potassium glutamate, ribonucleotide triphosphates, folinic acid, total E. coli tRNAs, amino acids, phosphoenolpyruvate, nicotinamide adenine dinucleotide, coenzyme A, oxalic acid, putrescine, spermidine, and rifampicin. The components were mixed gently by repeated pipetting and incubated for 5 hours at 30°C, 500 rpm, in a RTS ProteoMaster (Roche), in reaction tubes sealed with Air Pore membrane (Qiagen). After incubation, expressed protein was purified by affinity chromatography (see below). Additional details can be found in the Supplementary Information.
Purification of proteins encoded by synthetic genes
All proteins were constructed with a C-terminal hexahistidine fusion and purified using EZview Red HIS-Select HC Nickel Affinity Gel (Sigma Aldrich). A suspended gel slurry of 50 μl was washed with 1 ml loading buffer (20 mM MOPS, 7.5 mM imidazole, 500 mM NaCl, pH 7.5). Completed TnT reactions (25, 50, or 100 μl) were combined with the affinity gel and 1 ml loading buffer, captured at 4°C for 1 hour rotating end-over-end in a Mini LabRoller (Labnet International), washed with loading buffer (1 ml twice), and eluted with 100 μl elution buffer (20 mM MOPS, 400 mM imidazole, 500 mM NaCl, pH 7.5) incubating for 30 minutes at 4°C (rotating end-over-end). Each sample was concentrated using Vivaspin 500 centrifugal concentrators (Sartorius Stedim, 5 kDa molecular weight cut-off; pre-incubated with 2 mg/ml bovine serum albumin in PBS buffer for 12–16 hrs at 4°C, and washed with water before use) by centrifugation at 13,500 g for 10–15 minutes. The entire final volume (~25 μl) was loaded onto one lane of a SDS-PAGE gradient gel (NuPAGE 4–12% Bis-Tris, Invitrogen); the gel was stained with GelCode Blue Stain Reagent (Thermo Fisher Scientific). Poly-histidine tagged GFP template was included in each experiment as a positive expression and purification control; a reaction without added DNA template was used as a negative control. The seven condition experiments (Figure 2, Figure S1) for all three scaffolds were tested in at least three independent experiments and the majority of the other synthetic genes were tested in at least two independent experiments.
Protein identification by mass spectrometry
Liquid chromatography (LC) – tandem mass spectrometry (MS/MS) was used to confirm protein identity by analysis of peptides generated from in-gel tryptic digests. Samples were prepared according to the in-gel digestion protocol available at http://www.genome.duke.edu/cores/proteomics/sample-preparation/. Approximately half of the sample from each gel band was analyzed on a nanoAcquity LC and Synapt HDMS system (Waters Corporation) using a 30 minute LC gradient, with the top three precursor ions from each MS scan selected for MS/MS sequencing. Raw data was processed using Mascot Distiller v2.0 and searched against the Swiss-Prot database (v57.11) using Mascot v2.2 (Matrix Sciences), allowing for fixed modification of Cys (carbamidomethylation) and variable modification of Met (oxidation). Scaffold (v2.6) software was used to analyze the data. Sequence coverage obtained from this analysis for each of the proteins is shown in Figure S2.
Determination of mRNA levels
mRNA levels were determined by addition of 10 μCi of α-labeled rATP (Perkin Elmer) to a TnT reaction. Aliquots (10 μl) were removed and mixed with 100 μl Trizol (Invitrogen), incubated (5 minutes, room temperature), followed by addition of 20 μl chloroform (3 minutes at ambient temperature), vortexing (15 seconds), centrifugation at 12,000 g to separate phases (15 minutes), and aspiration of the RNA-containing aqueous phase which was subsequently passed through a NucAway Spin Column (Applied Biosystems) to remove unincorporated label. The resulting ~50 μl eluate was mixed with 200 μl of OptiPhase SuperMix scintillation cocktail (Perkin Elmer) and label incorporation was measured in a MicroBeta Trilux scintillation counter (Perkin Elmer). To determine an optimal assay time point, a time course was constructed for representative poorly and highly expressing DNA templates for each of the three proteins. Near-maximal label incorporation was observed at one hour; this time point was used subsequently to characterize the RNA levels of the alleles.
Supplementary Material
Acknowledgments
We thank J. Will Thompson and the Duke University Proteomics Core Facility for protein identification by mass spectrometry, and Philippe Marguet, Curtis Layton, & Chris Nicchitta for critical reading of the manuscript.
Footnotes
Accession Numbers
Genbank accession numbers for the genes constructed in this experiment are provided in Supplementary Table II.
Conflict of interest
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carothers JM, Goler JA, Keasling JD. Chemical synthesis using synthetic biology. Curr Opin Biotechnol. 2009;20:498–503. doi: 10.1016/j.copbio.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol. 2006;2:2006 0028. doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jana S, Deb JK. Strategies for efficient production of heterologous proteins in Escherichia coli. Appl Microbiol Biotechnol. 2005;67:289–298. doi: 10.1007/s00253-004-1814-0. [DOI] [PubMed] [Google Scholar]
- 5.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 6.Osada Y, Saito R, Tomita M. Analysis of base-pairing potentials between 16S rRNA and 5′ UTR for translation initiation in various prokaryotes. Bioinformatics. 1999;15:578–581. doi: 10.1093/bioinformatics/15.7.578. [DOI] [PubMed] [Google Scholar]
- 7.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C. Design parameters to control synthetic gene expression in Escherichia coli. PLoS One. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welch M, Villalobos A, Gustafsson C, Minshull J. You’re one in a googol: optimizing genes for protein expression. J R Soc Interface. 2009;6(Suppl 4):S467–476. doi: 10.1098/rsif.2008.0520.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattheakis L, Vu L, Sor F, Nomura M. Retroregulation of the synthesis of ribosomal proteins L14 and L24 by feedback repressor S8 in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:448–452. doi: 10.1073/pnas.86.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenner L, Romby P, Rees B, Schulze-Briese C, Springer M, Ehresmann C, Ehresmann B, Moras D, Yusupova G, Yusupov M. Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding. Science. 2005;308:120–123. doi: 10.1126/science.1105639. [DOI] [PubMed] [Google Scholar]
- 12.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 13.Puigbo P, Guzman E, Romeu A, Garcia-Vallve S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007;35:W126–131. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu G, Dress L, Freeland SJ. Optimal encoding rules for synthetic genes: the need for a community effort. Mol Syst Biol. 2007;3:134. doi: 10.1038/msb4100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorimer D, Raymond A, Walchli J, Mixon M, Barrow A, Wallace E, Grice R, Burgin A, Stewart L. Gene composer: database software for protein construct design, codon engineering, and gene synthesis. BMC Biotechnol. 2009;9:36. doi: 10.1186/1472-6750-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voges D, Watzele M, Nemetz C, Wizemann S, Buchberger B. Analyzing and enhancing mRNA translational efficiency in an Escherichia coli in vitro expression system. Biochem Biophys Res Commun. 2004;318:601–614. doi: 10.1016/j.bbrc.2004.04.064. [DOI] [PubMed] [Google Scholar]
- 17.Puigbo P, Romeu A, Garcia-Vallve S. HEG-DB: a database of predicted highly expressed genes in prokaryotic complete genomes under translational selection. Nucleic Acids Res. 2008;36:D524–527. doi: 10.1093/nar/gkm831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Czar MJ, Anderson JC, Bader JS, Peccoud J. Gene synthesis demystified. Trends Biotechnol. 2009;27:63–72. doi: 10.1016/j.tibtech.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Cox JC, Lape J, Sayed MA, Hellinga HW. Protein fabrication automation. Protein Sci. 2007;16:379–390. doi: 10.1110/ps.062591607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hershberg R, Petrov DA. Selection on codon bias. Annu Rev Genet. 2008;42:287–299. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- 21.Calderone TL, Stevens RD, Oas TG. High-level misincorporation of lysine for arginine at AGA codons in a fusion protein expressed in Escherichia coli. J Mol Biol. 1996;262:407–412. doi: 10.1006/jmbi.1996.0524. [DOI] [PubMed] [Google Scholar]
- 22.dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 24.Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem Sci. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 26.Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 27.Katz L, Burge CB. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 2003;13:2042–2051. doi: 10.1101/gr.1257503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci USA. 2010;107:3645–3650. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Etchegaray JP, Inouye M. Translational enhancement by an element downstream of the initiation codon in Escherichia coli. J Biol Chem. 1999;274:10079–10085. doi: 10.1074/jbc.274.15.10079. [DOI] [PubMed] [Google Scholar]
- 30.Qing G, Xia B, Inouye M. Enhancement of translation initiation by A/T-rich sequences downstream of the initiation codon in Escherichia coli. J Mol Microbiol Biotechnol. 2003;6:133–144. doi: 10.1159/000077244. [DOI] [PubMed] [Google Scholar]
- 31.Moll I, Huber M, Grill S, Sairafi P, Mueller F, Brimacombe R, Londei P, Blasi U. Evidence against an Interaction between the mRNA downstream box and 16S rRNA in translation initiation. J Bacteriol. 2001;183:3499–3505. doi: 10.1128/JB.183.11.3499-3505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sprengart ML, Fuchs E, Porter AG. The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J. 1996;15:665–674. [PMC free article] [PubMed] [Google Scholar]
- 33.Rush GJ, Steyn LM. Translation enhancement by optimized downstream box sequences in Escherichia coli and Mycobacterium smegmatis. Biotechnol Lett. 2005;27:173–179. doi: 10.1007/s10529-004-7664-0. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Guo P, Jing G. A vector with the downstream box of the initiation codon can highly enhance protein expression in Escherichia coli. Biotechnol Lett. 2003;25:755–760. doi: 10.1023/a:1023563600459. [DOI] [PubMed] [Google Scholar]
- 35.Keum JW, Ahn JH, Choi CY, Lee KH, Kwon YC, Kim DM. The presence of a common downstream box enables the simultaneous expression of multiple proteins in an E. coli extract. Biochem Biophys Res Commun. 2006;350:562–567. doi: 10.1016/j.bbrc.2006.09.072. [DOI] [PubMed] [Google Scholar]
- 36.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coleman MA, Lao VH, Segelke BW, Beernink PT. High-throughput, fluorescence-based screening for soluble protein expression. J Proteome Res. 2004;3:1024–1032. doi: 10.1021/pr049912g. [DOI] [PubMed] [Google Scholar]
- 38.Lopez PJ, Marchand I, Joyce SA, Dreyfus M. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol Microbiol. 1999;33:188–199. doi: 10.1046/j.1365-2958.1999.01465.x. [DOI] [PubMed] [Google Scholar]
- 39.Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 40.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 42.Bashan A, Yonath A. Correlating ribosome function with high-resolution structures. Trends Microbiol. 2008;16:326–335. doi: 10.1016/j.tim.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Steitz TA. A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol. 2008;9:242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 44.Petry S, Weixlbaumer A, Ramakrishnan V. The termination of translation. Curr Opin Struct Biol. 2008;18:70–77. doi: 10.1016/j.sbi.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 45.Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 46.Iost I, Dreyfus M. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 2006;34:4189–4197. doi: 10.1093/nar/gkl500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 48.Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jaeger JA, Turner DH, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao X, Yo P, Keith A, Ragan TJ, Harris TK. Thermodynamically balanced inside-out (TBIO) PCR-based gene synthesis: a novel method of primer design for high-fidelity assembly of longer gene sequences. Nucleic Acids Res. 2003;31:e143. doi: 10.1093/nar/gng143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jewett MC, Swartz JR. Substrate replenishment extends protein synthesis with an in vitro translation system designed to mimic the cytoplasm. Biotechnol Bioeng. 2004;87:465–472. doi: 10.1002/bit.20139. [DOI] [PubMed] [Google Scholar]
- 53.Jewett MC, Swartz JR. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol Bioeng. 2004;86:19–26. doi: 10.1002/bit.20026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.