Abstract
Background & Aims
Gastroesophageal reflux causes inflammation, intestinal metaplasia and its downstream sequelum adenocarcinoma in the distal esophagus. The incidence of esophageal adenocarcinoma has increased approximately 6-fold in the U.S. since the 1970s, accompanied with a significant increase in prevalence of gastroesophageal reflux disease (GERD). Despite extensive epidemiological study, the cause for GERD and the unexpected increases remain unexplainable. Microbes are among the environmental factors that may contribute to the etiology of GERD but very little research has been done on the esophageal microbiome, particularly in its relation to GERD. This is the first reported correlation between a change in the esophageal microbiome and esophageal diseases.
Methods
Biopsies of the distal esophagus were collected from 34 patients. Host phenotypes were histologically defined as normal, esophagitis, or Barrett’s esophagus (intestinal metaplasia). Microbiomes from the biopsies were analyzed by bacterial 16S rRNA gene survey and classified into types using unsupervised cluster analysis and phenotype-guided analyses. Independence between host phenotypes and microbiome types were analyzed by Fisher Exact test.
Results
Esophageal microbiomes can be classified into two types. The type I microbiome was dominated by the genus Streptococcus and concentrated in the phenotypically normal esophagus. Conversely, the type II microbiome contained a greater proportion of Gram-negative anaerobes/microaerophiles and primarily correlated with esophagitis (Odds Ratio: 15.4) and BE (Odds Ratio: 16.5).
Conclusions
In the human distal esophagus, inflammation and intestinal metaplasia are associated with global alteration of the microbiome. These findings raise the issue of a possible role for dysbiosis in the pathogenesis of reflux-related disorders.
INTRODUCTION
The distal esophagus is an important anatomic locus where gastric acid reflux causes reflux esophagitis, Barrett’s esophagus (BE), and its downstream sequelum adenocarcinoma (EA).1 Incidence of EA has increased approximately 6-fold in the U.S. since the 1970s following significant increase in prevalence of gastroesophageal reflux disease (GERD).2,3 Although specific host factors might predispose to disease risk, such rapid increase in incidence must be predominantly environmental. Among the environmental agents that have been considered are microbes. The human body can be viewed as a superorganism composed of an amalgam of both microbial and human cells.4,5 Our relationships with bacteria can be considered to span a broad spectrum, from mutualism to pathogenicity.6 Currently, two theories explain bacterial diseases. The classic pathogen theory, attributed to Koch, requires the presence of specific pathogens, i.e. Mycobacterium tuberculosis and Bacillus anthracis.7 Alternatively, the microecological disease or “pathogenic microbial community” theory is a new concept where the entire community contributes to pathogenicity although no individual community members can be categorized as classic pathogens.8 In mouse models mimicking inflammatory bowel diseases, development of mucosal inflammation and adenocarcinoma requires both a trigger (chemical or genetic), and presence of commensal bacteria.9,10
Dysbiosis refers to an abnormal state of the microbial ecosystem in a host.11,12 It further divides commensal bacteria into “protective” and “harmful" species, attributing the causes of certain chronic diseases to alterations of balance between the two species.11,12 Gut microbiome in ob/ob mice (which have a mutation in the leptin gene causing obesity13), for example, has an increased capacity to harvest energy from the diet and might contribute to pathophysiology of obesity.13
Recent studies of a small number of hosts have shown that nearly 100 commensal bacterial species reside in the normal distal esophagus.14,15 Although human exposure to many exogenous pathogens has been monitored, little attention has been paid to change in the indigenous microbiome, partly due to complexity and difficulties in culture and analysis.16 This is exemplified by the number of attempts to date in studying the esophageal microbiome with only limited success (Supplemental Table 1). In the present study, we examined whether pathological findings in the esophageal mucosa are related to change in the overlaying microbiome (Supplemental Figure 1).
MATERIALS AND METHODS
DNA extraction, bacterial 16S rDNA amplification, cloning and sequencing of the PCR products was performed as previously described.14, 15 In brief, the distal esophageal microbiome was sampled by endoscopic biopsy and DNA extracted from the biopsies. From each sample, a 16S rRNA gene cloning library was constructed with 16S rRNA genes amplified using broad range primers 8F and 1510R and 200 clones were sequenced by single-pass Sanger sequencing. The average length of the sequencing reads were 912 nt, ranging between 813 and 970 nt. For assignment of species level taxonomic unit (SLOTU), each sequence was analyzed using Sequence Match at Ribosomal Database Project II (RDP II, release 9.39, http://rdp.cme.msu.edu),17 as previously described.14, 15 Compared with the RDP database of 16S rRNA genes, a sequence that had a similarity score >0.8725 (equivalent to 97% sequence identity14, 15) with a best matched sequence in the database was assigned to the species assigned by RDP II, while a sequence that had a best similarity score <0.8725 was assigned as unclassified species. We used the experimentally defined 97% sequence identity as the species boundary to minimize subjective influence on defining a species.18 Because no single identity cutoff can reliably classify all natural bacterial species, 97% threshold used in this study was for operational purpose to approximate species diversity. Ranks at genus or above were defined by using CLASSIFIER at RDP II,17 with a confidence threshold of 80%, which has been established as one of the most suitable methods for taxonomic assignment of 16S rRNA genes of human gastrointestinal microbiome.19 The classification was verified by phylogenetic analysis (see Supplementary data). The genus level assignment for a sequence was confirmed by inferring from its species assignment if a discrepancy occurred between the assignments by CLASSIFIER and phylogenetic analysis. Each of the species identified was assigned into anaerobic, microaerophilic, or aerobic group, or Gram-positive or Gram negative group by empirically inferring from their taxonomic identity and known culture conditions as described in the American Type Culture Collection (ATCC) instruction and Bergey’s manual as well as original publications describing the species. A sequence was assigned to unclassified if knowledge about it was unavailable or the taxon it belongs to was heterogeneous in these properties. The total number of SLOTUs that could be present in the 16S rRNA gene datasets from the distal esophageal microbiome, as a whole or within phenotypic groups, was predicted by a nonparametric richness estimator, Chao1.20 The Shannon-Wiener diversity and Shannon index of evenness were calculated by using EstimateS at http://purl.oclc.org/estimates. To avoid a type-I error in multiple comparisons on a single dataset, an Omnibus test was first performed to determine whether there is an overall group difference. The Omnibus test was performed for categorical data using the Fisher exact R × C frequency table and for continuous data using the one-way analysis of variance (ANOVA), with the statistical significance level set at P < 0.05, for two-tailed analysis. Follow-up analyses for between-group differences were performed using Fisher exact tests or t-tests with the false discovery rate (FDR) controlled at 5%.21 Data used in the t-test was examined for distribution relative to a normal curve by Kolmogorov-Smirnoff goodness-of-fit test, with critical value set at 0.05. Fisher exact test was performed using StatXact 8 (Cytel Inc., Cambridge, MA), ANOVA and t-test using SPSS 13.0 and SigmaPlot 8.0 (SPSS Inc., Chicago, IL). Regression and correlation were performed using the online statistical tools hosted at http://fonsg3.let.uva.nl/Service/Statistics.html.
RESULTS
Esophageal microbiome differs in health and diseases
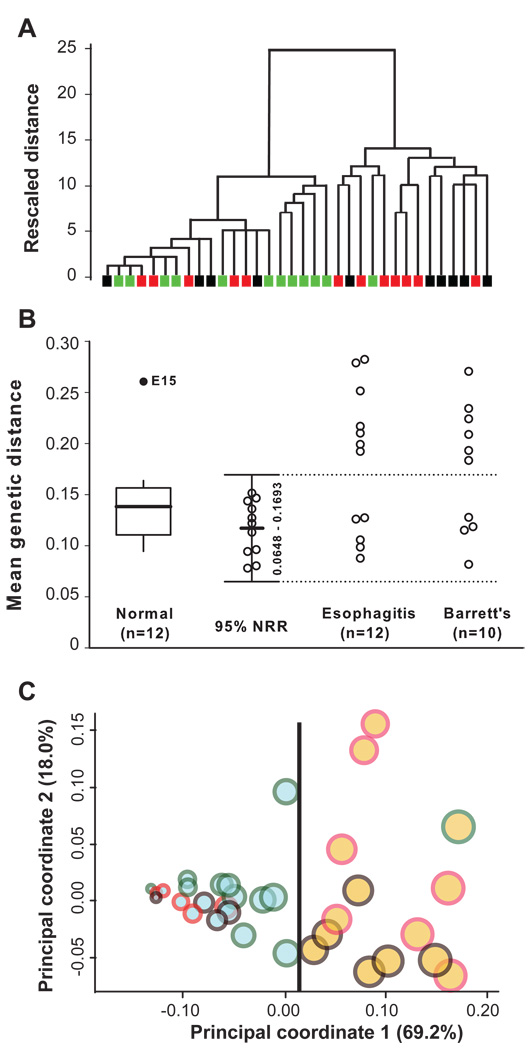
We obtained esophageal samples from 34 subjects and classified them to one of three histological phenotypes based on histopathological changes in the human tissue (Supplemental Figure 1): normal (n = 12), esophagitis (n = 12), or BE (n = 10) (Supplemental Table 2). In total, 6,800 (200/sample) 16S rRNA gene sequences were analyzed. We started with an unsupervised approach by asking whether samples of the microbiome form natural groups, independent of histopathological phenotypes associated with each sample. Hierarchical clustering analysis using combined genetic distance between samples revealed two distinct clusters (Figure 1A), that we designated as two microbiome types. We then asked whether the naturally occurring microbiome types correlate with host phenotypes. Although none of the microbiome types exclusively correlated with the three phenotypes, nearly all normal samples (11/12) were located in one cluster while the majority of abnormal samples (13/22) in another cluster. This type of distribution suggests that the association between the host phenotypes and microbiome types is non-random. To validate this association, we calculated the normal reference range (NRR) (Equations 1–4 in Supplemental Materials and Methods) based on the mean genetic distance among phenotypically normal samples. The 11 normal samples (with normal distribution after exclusion of one outlier) had a mean distance of 0.1170 between themselves and a 95% NRR (Mean ± 1.96 S.D.) between 0.0648 and 0.1693 (Figure 1B). The mean distance between each of the other 22 samples and the 11 normal samples then was calculated (Equation 5 in Supplemental Materials and Methods). Based on the mean distance and the NRR, 20 of the 34 samples were classified as type I, and the remaining 14 samples as type II microbiome, identical to the two clusters identified by the unsupervised clustering analysis. The type I microbiome was more closely associated with normal esophagus (11/12, 91.7%) while the type II microbiome was mainly associated with abnormal esophagus (13/22, 59.1%), including 7 of the 12 esophagitis and 6 of the 10 BE samples. An Omnibus test using the Fisher exact 2 × 3 probability table revealed an overall difference among the three phenotypes in relation to the two types of microbiome (P = 0.0173) (Table 1). Follow-up tests with the false discovery rate21 controlled at < 5% indicated that the esophagitis (7/12) and BE (6/10) samples both were more likely than normal samples (1/12) to exhibit the type II microbiome (OR: 15.4, 95% C.I. 1.5–161.0, and OR: 16.5, 95% C.I. 1.5–183.1, respectively), but were indistinguishable from one another. These data indicated that although the histologically defined phenotypic groups were heterogeneous in microbiome types, strong associations were nevertheless present.
Figure 1.
Typing of esophageal microbiome. (A) Detection of natural microbiome groups by unsupervised cluster analysis. The dendrogram was constructed using the average linkage algorithm and cosine measure of the genetic distance calculated from samples of the microbiome. Samples are represented by colored rectangles (green for normal, red for esophagitis, and black for Barrett’s esophagus). (B) Phenotype-directed classification of the microbiome by genetic distance-based normal reference range. First, the mean genetic distance between each normal sample and other 11 normal samples were calculated. A single outlier was identified and the mean distance for each remaining 11 normal samples was recalculated after excluding the outlier. The 95% NRR was defined as mean distance ± 1.96 S.D. based on the 11 normal samples. The mean distance for each sample in the esophagitis and BE groups is the mean distance between the sample and the 11 normal samples. The dotted line (0.1693) is the upper limit of the 95% NRR, which separates the 34 samples into the normal (inside the NRR) and abnormal microbiome (outside the NRR). (C) Double principal coordinate analysis (DPCoA) of the microbiome. Samples are represented by circles. Microbiome types are indicated by fill colors (blue for type I and brown for type II). Host phenotypes are indicated by edge colors (green for normal, red for esophagitis, and black for Barrett’s esophagus). Within-sample diversity is proportional to circle size, determined by Rao’s analysis. The location of a sample in the plot was determined by the first two orthogonal principal axes. The percentages shown for each axis represents the percent of total dissimilarity captured by the axis. The samples from the two types of microbiome are separable along the first principal coordinate, as indicated by the dividing line at x ≈ 0.015.
Table 1.
Association between host phenotypes and microbiome types in the distal esophagus
| Omnibus testA | |||||||
|---|---|---|---|---|---|---|---|
| Groups compared | Phenotype | ||||||
| Normal | Esophagitis | BEB | |||||
| Microbiome type | I | 11 | 5 | 4 | |||
| II | 1 | 7 | 6 | ||||
| P value | 0.0173 | ||||||
| Follow-up testsC | |||||||
| Groups compared | Phenotype | ||||||
| Normal | Esophagitis | Normal | BE | Esophagitis | BE | ||
| Microbiome type | I | 11 | 5 | 11 | 4 | 5 | 4 |
| II | 1 | 7 | 1 | 6 | 7 | 6 | |
| P value | 0.027* | 0.020* | 1.000 | ||||
| Odds ratio (95% C.I.) | 15.4 (1.5–161.0) | 16.5 (1.5–183.1) | 1.1 (0.2–5.9) | ||||
The Omnibus test was performed using the two-tailed Fisher-Freeman-Halton 3 × 2 probability test.
BE, Barrett’s esophagus.
The follow-up tests were performed with the two-tailed Fisher exact 2 × 2 probability test.
Tests that are statistically different at the false discovery rate < 5% 21 are marked by *.
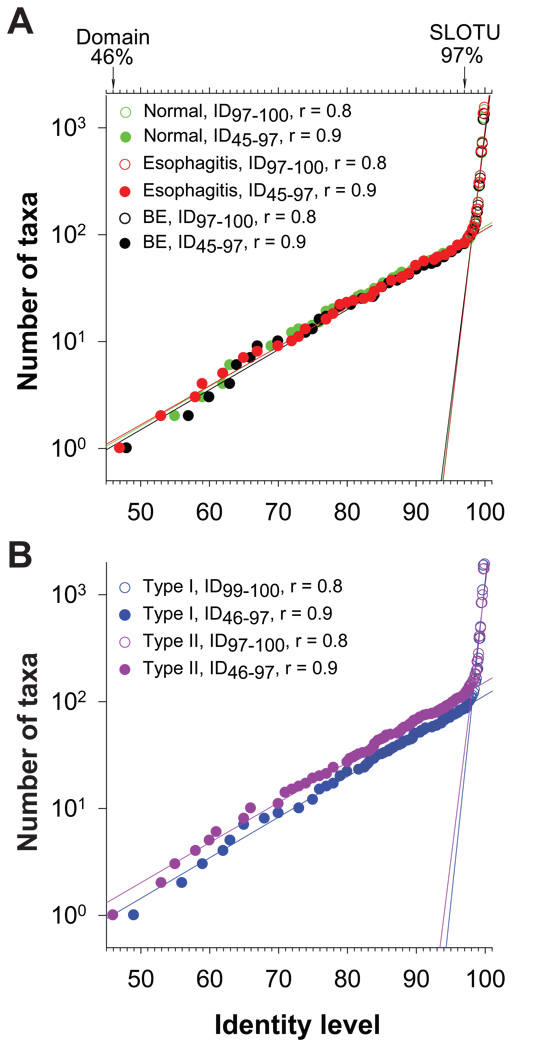
We also examined the relationship between microbiome types and host phenotypes, using the UniFrac Significance Test, with 1000 permutations and Bonferroni correction for multiple comparisons.22,23 UniFrac tests the hypothesis that there has been more phylogenetic diversity unique to a single environment than would be expected if the sequences were randomly distributed among environments. UniFrac revealed across the board differences between the two microbiome types (sequences pooled by type: 4,000 for type I and 2,400 for type II), among and between microbiomes associated with the three phenotypes (sequences pooled by phenotype: 2,400 for normal, 2,400 for esophagitis, and 2,000 for Barrett’s esophagus), as well as among samples (n=34, 200 sequences per sample) (Supplemental Table 3). Further analyses for between-samples difference were not informative because it requires more than 10,000 permutations to reveal any difference, which exceeds the software’s limit of 1,000 permutations. To assess the effect of the intra-community diversity on the difference between the two types of microbiomes, we performed FST test and found that the average within-community diversity is significantly less than the diversity when the two type microbiomes are combined (P < 0.001). Analyses of the lineage through time curves in the dataset revealed constant rates of birth and extinction of operational taxonomic units (OTUs) since the bacterial domain (ID46) was formed through the species level (ID97) when the rates abruptly accelerate, with the result that a limited number of lineage ancestors burst into numerous, closely related OTUs (Figure 2A and 2B), suggesting the majority of differences observed was concentrated below the species level.
Figure 2.
Bacterial phylogenesis in the distal esophagus. (A) DNA sequences were pooled according to host phenotypes: normal, esophagitis, and Barrett’s esophagus. The number of taxa at a specific identity level (ID), every 1% between ID46 and ID80 and every 0.1% between ID80 and ID100, were calculated using DOTUR, showing changing in the number of taxa with increasing mutations over the entire hierarchy of domain Bacteria. The rates of the changes and their turning points were analyzed by linear regressions. Values from the domain level to the species level are represented by filled circles while those at the species level and below are indicated by open circles. (B) DNA sequences were pooled according to microbiome types: type I and type II using the same methods as Figure 2A.
Streptococcus determines microbiome types
Further analysis using double principal coordinate analysis (DPCoA)24 indicated that the maximal separation of the phenotypically normal samples from the abnormal ones could be obtained along the first principal coordinate (PC1) (69.2% of total diversity) by an empirical dividing line (x ≈ 0.015) (Figure 1C), which assigned the 34 samples to two groups, completely in agreement with the two types of microbiome. The reducibility of the complex microbiome typing scheme to PC1 by DPCoA suggests that the microbiome types might be primarily determined by specific bacterial subpopulations within the whole microbiome. This finding led us to examine whether the microbiome types are determined by the relative abundance of one or a few specific taxa (Supplemental Figure 1). We approached this question by classifying the 16S rRNA sequences at various taxonomic ranks, correlating the relative abundances of all main taxonomic groups with PC1, defining normal and abnormal taxonomic types by using abundance-based NRR of the bacterial groups, and validating the taxonomic types by comparing them with microbiome types in assignment of individual samples.
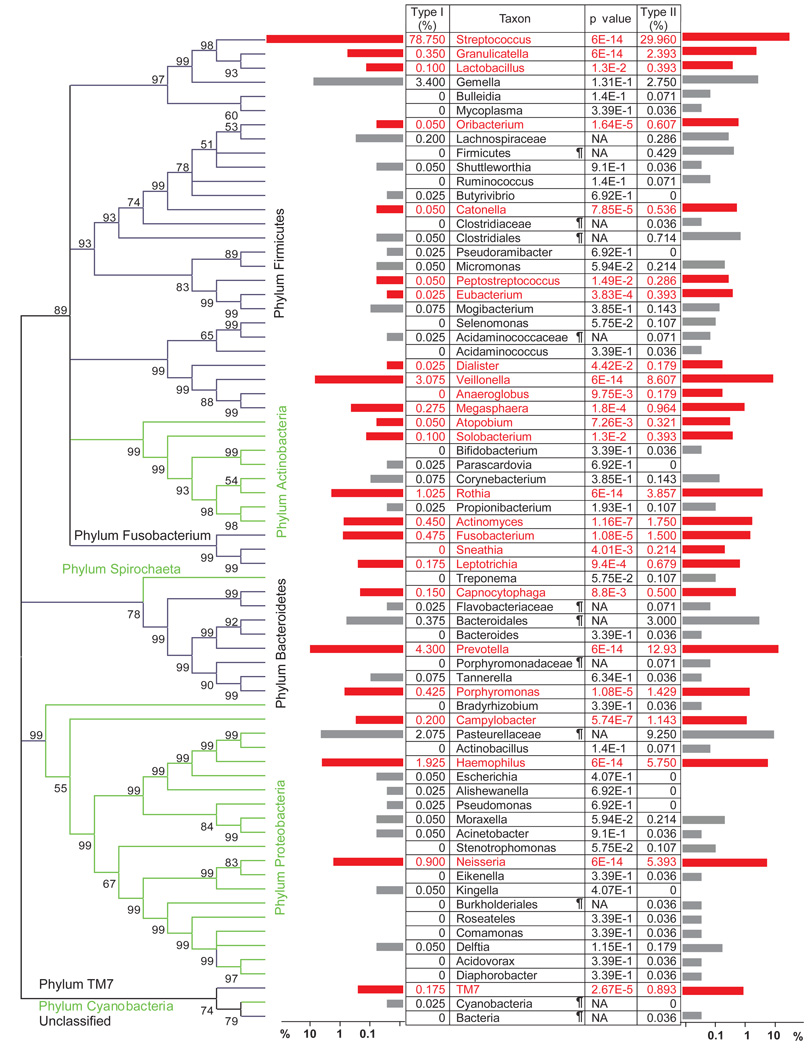
First, we binned the 6,800 PCR clones into taxonomic groups at the phylum, genus, and species levels. In total, nine phyla, 70 genera, and 166 species-level operational taxonomic units (SLOTU) were represented (Figure 3 and Supplemental Figure 2). We used Chao1 estimation to assess the relative depth of coverage.20 The analysis indicates that the human distal esophagus may harbor ~213 (95% C.I. 191–254) SLOTUs and suggests that 77.9% of the SLOTUs (166/213) have been identified in this study (Supplemental Figure 3). Firmicutes (4868 clones) was the only phylum consistently detected in all 34 samples, while the other eight phyla, Bacteroidetes (720, 33/34 samples), Proteobacteria (843, 31/34), Actinobacteria (240, 28/34), Fusobacteria (92, 26/34), TM7 (32, 13/34), Spirochaetes (3, 2/14), Cyanobacteria (1, 1/34), and unclassified bacteria (1, 1/34) were less common. The six most abundant phyla were shared by the two types of microbiome.
Figure 3.
Differential representation of genera between the two types of microbiome. Pooled 16S rRNA gene sequences from type I samples were compared at the genus level (or the lowest classifiable rank above genus) with those from type II samples using LIBRARY COMPARE in RDP II.25 Relative abundances of a genus in the two types are shown in the table and by the horizontal bars, with genera that are significantly different between the two types of microbiome highlighted in red. Unclassified taxa are marked with a ¶. Distribution of the genera in the taxonomic hierarchy of domain bacteria is shown in the phylogenetic tree, with alternating black and green brackets to contrast neighboring phyla. Bootstrap values were based on 500 replicates.
Streptococcus was the predominant (3989 clones) genus, represented by 17 SLOTUs, in particular, S. mitis (2173) and S. pseudopneumoniae (1119), both being members of the mitis group.26 Streptococcus and S. mitis were the only taxa found in all 34 samples at the genus and species levels, respectively.
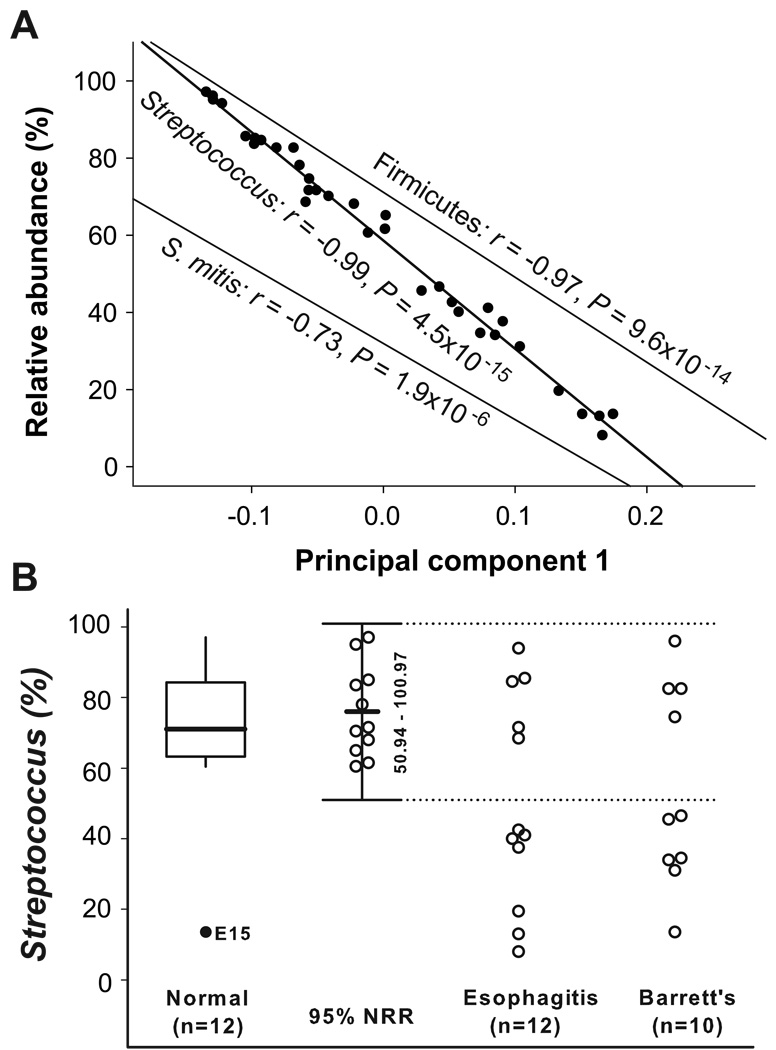
Next, we designed an analysis, which we called microbiome-abundance correlation (MAC) to facilitate identifying taxonomic groups whose relative abundance in the 34 samples significantly correlated with their PC1 in the DPCoA (Figure 1C) (Equation 6 in Supplemental Materials and Methods). In the direction from the type I to type II microbiome, the PC1 significantly correlated with decreasing abundance of Firmicutes (r = −0.97, P = 9.6 × 10−14), Streptococcus (r = −0.99, P = 4.5 × 10−15), and S. mitis (r = −0.73, P = 1.9 × 10−6) (Figure 4A).
Figure 4.
Taxonomic definition of microbiome types. (A) Microbiome-abundance correlation (MAC) analysis. The first principal coordinates (PC1) in DPCoA were correlated with the relative abundance of Firmicutes, Streptococcus, or S. mitis, for every sample, by linear regressions. (B) Classification of microbiome by the relative abundance of Streptococcus. An outlier (solid circle) was excluded using a box plot in which the upper whisker length is 1.5*IQR. The 95% normal reference range (NRR) (mean ± 1.96 S.D.) was calculated by the relative abundance of Streptococcus after excluding the outlier. The dotted line (50.3%) is the upper limit of the 95% NRR, which separates the 34 samples into normal (inside the NRR) and abnormal taxonomic types (outside the NRR).
Due to its stronger correlation with PC1, we considered that the relative abundance of the genus Streptococcus probably determined the two types of microbiome. To test this hypothesis, we calculated a 95% NRR based on the relative abundance of Streptococcus. After excluding the one outlier, the remaining 11 samples from phenotypically normal esophagus had a mean ± S.D. of 75.9 ± 12.8% Streptococcus and a 95% NRR (mean ± 1.96 S.D.) of 50.8–100%. Use of the lower limit of the NRR (50.8%) as a threshold separated the 24 samples from the esophagitis and BE groups into two taxonomic types (Figure 4B). All 13 cases classified as abnormal by the NRR corresponded to the type II microbiome, while all 9 samples classified as normal (n = 9) belonged to the type I microbiome, without ambiguity. These 9 samples had a mean Streptococcus abundance similar to that of the 11 normal samples (82.2% vs. 75.9%, P = 0.238), while the outlier sample from the normal esophagus group that was categorized as type II microbiome had a low Streptococcus abundance (13.5%). Overall, the 20 type I samples had a mean of 78.8% Streptococcus (range 60.5–97.0%), while the 14 type II samples had a mean of 30.0% (range 8.0–46.5%) (P < 1 × 10−10, t-test). The mean of relative abundance of Streptococcus in the normal esophagus group (75.9%, n = 11) was significantly higher than that in the esophagitis (50.5%, n = 12) and BE (54.1%, n = 10) groups (Supplemental Table 4).
Gram-negative anaerobes prevail in the type II microbiome
In addition to Streptococcus, MAC analyses also revealed significant, but weaker, correlations of PC1 with the relative abundances of Bacteroidetes (r = 0.81, P = 4.0 × 10−8), Proteobacteria (r = 0.65, P = 3.7 × 10−5), and Fusobacteria (r = 0.63, P = 8.2 × 10−5). Unlike for Streptococcus, further analyses using abundance-based 95% NRR for each of these phyla or their predominant genera could not clearly assign all samples into the two types of microbiome. Since the majority of PCR clones from these three phyla were Gram-negative and/or anaerobic/microaerophilic bacteria, we hypothesized that these broad properties also could be used to determine the microbiome types.
MAC analysis showed a strong correlation of PC1 with the relative abundance of anaerobic/microaerophilic bacteria (r = −0.98, P = −2.3 × 10−14) (Figure 5A). Anaerobic (type I: 11.0% vs. type II: 38.2%, P = 1.2 × 10−5, t-test) and microaerophilic bacteria (5.4% vs. 23.0%, P = 1.1 × 10−4) were more abundant in the 14 type II samples than in the 20 type I samples (Figure 5E). In combination, anaerobic and microaerophilic bacteria comprised an average of 61.1% of the sampled clones in type II samples but only 16.3% in type I samples (P < 1 × 10−10, t-test). The 95% NRR based on the relative abundance of aerobic/microaerophilic bacteria in the phenotypically normal samples correctly identified the microbiome types for all but one of the 34 samples (Supplemental Figure 4A).
Figure 5.
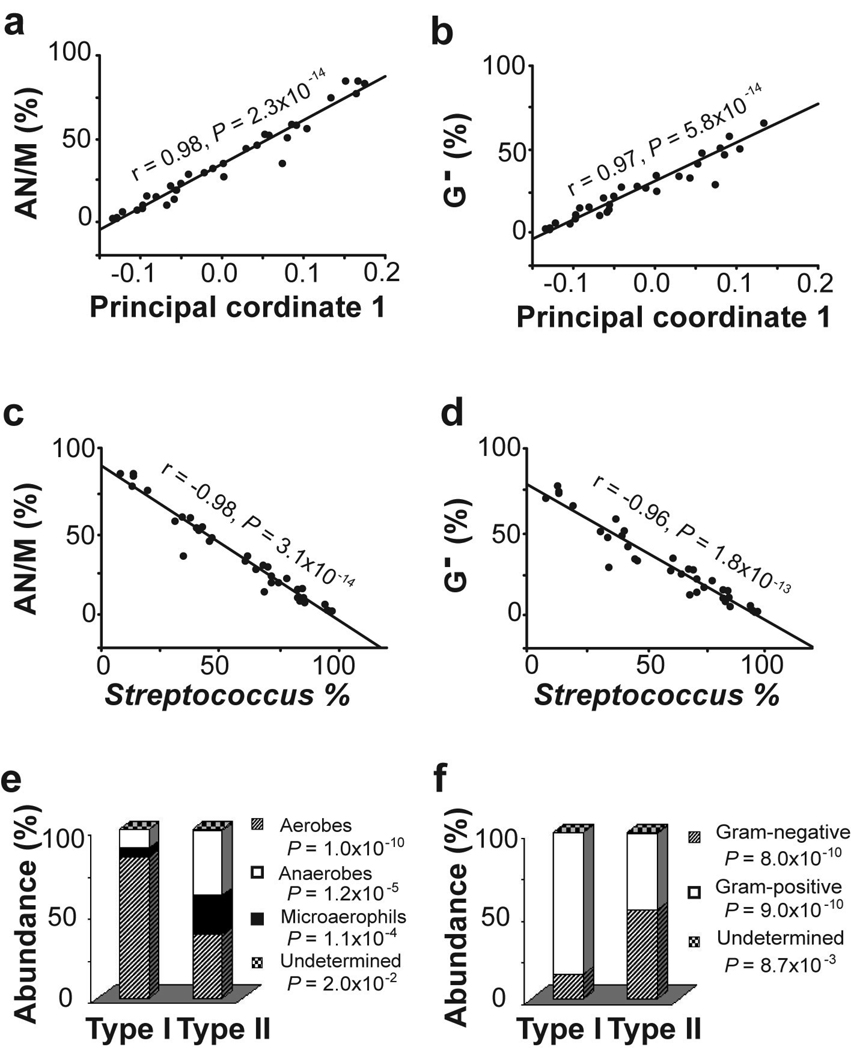
Taxonomic characterization of microbiome by population of main bacterial groups. (A) Microbiome-abundance correlation analyses between the first principal coordinates (PC1) in DPCoA and the relative abundance of anaerobic/microaerophilic bacteria (AN/M). (B) Correlation between the first principal coordinates (PC1) in DPCoA and the relative abundance of Gram-negative (G-) bacteria. (C) Correlations between the relative abundance of Streptococcus and that of anaerobic/microaerophilic bacteria. (D) Correlations between the relative abundance of Streptococcus and that of Gram-negative bacteria. (F) Comparisons of microbiome types according to culture conditions. (E) Comparisons of microbiome types according and staining properties.
MAC analysis also showed a significant correlation of PC1 with the relative abundance of Gram-negative bacteria (r = −0.97, P = 5.8 × 10−14) (Figure 5B). Gram negative bacteria comprised an average of 53.4% of sampled clones in the 20 type II samples but only 14.9% in the 14 type I samples (P = 8.0 × 10−10, t-test) (Figure 5F). The 95% NRR based on the relative abundance of Gram-negative bacteria in the phenotypically normal samples correctly identified the microbiome types of all but three of the 34 samples (Supplemental Figure 4B).
The strong correlations between PC1 and the relative abundance of the predominant bacterial groups in the type I (Streptococcus) and type II microbiome (Gram-negative bacteria and anaerobes/microaerophils) suggest an inverse relationship between the two groups of bacteria. Testing this hypothesis by correlation analysis indicated that the change in the relative abundance of Streptococcus is tightly associated with the inverse changes in anaerobes/microaerophils (r = −0.98, P = 3.1 × 10−14, linear regression, Figure 5C) and Gram-negative bacteria (r = −0.96, P = 1.8 × 10−13, Figure 5D).
To further identify significant taxonomic differences between the two types of microbiome, we compared the pooled 4,000 sequences from the 20 type I samples and 2800 sequences from the 14 type II samples, using LIBCOMPARE (http://rdp.cme.msu.edu/comparison/comp_help.jsp). The two types of microbiome differed in 25 of the 70 OTUs at the genus or higher ranks. The type II microbiome had less abundant Streptococcus than did the type I microbiome, but was more abundant in 24 other genera (Figure 3). Specifically, the more abundant genera that accounted for > 1% of the total bacterial population in the type II microbiome included Veillonella, Prevotella, Haemophilus, Neisseria, Rothia, Granulicatella, Campylobacter, Porphyromonas, Fusobacterium, and Actinomyces, most being Gram-negative anaerobes or microaerophiles. These data suggest a shift from a Gram-positive aerobic microbiome to a Gram-negative anaerobic microbiome in the microenvironment of the histologically abnormal distal esophagus.
Overall, the type II microbiome was significantly more diverse than type I microbiome (Shannon-Wiener diversity index27 mean of 2.69 vs. 1.51, P = 1.3 × 10 −7) (Figure 6A) and had greater SLOTU evenness (Shannon-Wiener evenness index mean 0.78 vs. 0.51, P = 4.2 × 10−8) (Figure 6B).27 It harbored significantly more SLOTUs than the type I microbiome, for both the observed (mean 32.5 vs. 19.3, P = 0.0001) as well as the predicted numbers of SLOTUs per sample (mean 48.9 vs. 27.8, P = 0.0018) by a nonparametric richness estimator, Chao1 (Figure 6C).20
Figure 6.
Difference between the two types of microbiome in biological diversity. (A) Shannon-Wiener diversity index. (B) Shannon-Wiener evenness index. (C) Richness by observed and estimated SLOTUs.20 Mean ± 1.96 S.D. is indicated by horizontal lines.
DISCUSSION
The present study has provided two new contributions to the field of human microbial ecology. First, we have performed a comprehensive study of the human distal esophagus microbiome and demonstrated the presence of a complex microbiome about which little prior knowledge was available. The esophageal microbiome is comparable in complexity to those found in the mouth, stomach, colon, vagina, and skin.14–15, 28–32 Collectively, nine phyla were observed, represented by 166 species. The distal esophagus could harbor >200 species, as predicted by the Chao1 richness estimator.20 Second, we have demonstrated by both unsupervised and phenotype-directed analyses that the esophageal microbiome can be classified into two types and that the type II microbiome is the strongest (OR >15) amongst all known environmental factors that are associated with the pathological changes related to GERD (Supplemental Table 5). Overall, the findings have opened a new approach to understanding the recent surge in the incidence/prevalence of GERD and EA, and suggest the possible role of dysbiosis in their pathogenesis.
The need to compare with risk factors identified by conventional studies promoted us to design a dual assignment scheme for the esophageal samples. This approach enabled testing the independency of two categorical variables, host phenotype and microbiome type. The microbiome types determined from the present study are relevant to pathology in the distal esophagus. There are two possible explanations for the significant association between the type II microbiome and the abnormal histological phenotype. First, the type II microbiome might play a causative role in GERD, which has a complex and not yet completely understood pathophysiology. Abnormal lower esophageal sphincter (LES) pressure and esophageal acidification during transient LES relaxation are believed to be critical, but the etiology of the abnormal LES function is unknown.33 One possibility is that the esophageal microbiome could be intrinsic – each individual might harbor either a stable type I or type II microbiome. Distinct microbiome can be inherited via kinship from mother or caregivers, as suggested in studies of mouse colonic microbiome,34 which can be modified by exposure to antibiotics during or after the postnatal development of microbiome is complete.6 The Gram-negative predominant type II microbiome could serve as a primary or synergistic mechanism in promoting gastric reflux, since lipopolysaccharides (LPS), mainly produced by Gram-negative bacteria induce abnormal relaxation of the lower esophageal sphincter via activation of the inducible nitric oxide synthase (iNOS) pathway.35 Second, the type II microbiome might be secondary to changes caused by gastric reflux. The esophageal microbiome could be transitory: the type I microbiome could represent a direct extension of the normal oral flora via saliva while the type II microbiome could represent regurgitated bacteria in gastric juice. Alternatively, gastroesophageal reflux might modify the esophageal microbiome by selecting against acid-sensitive bacteria in the esophagus. Testing these hypotheses might shed light on the pathogenesis of GERD and lead to new biomarkers for GERD.
Although the present study yielded one of the largest data sets from a single study of microbiome in human diseases,16,29 our understanding of the esophageal microbiome is far from complete. The power of this study might be limited by unrecognized factors unrelated to GERD, but that potentially affect the bacterial microbiome, including diet, medications, and oral and gastric diseases. Similarly, any interpretation of our data in relation to GERD should be cautious, since GERD has a complex definition.36 The GERD phenotype is composed of three heterogenous factors: symptoms, abnormal acid exposure, and mucosal damage.36 While not optimal, this definition is necessary because of the substantial overlap between normal subjects and those with GERD for any of the single factors. Significant acid reflux can occur in 19% of (normal) subjects without reflux-related symptoms.37 Similarly, erosive esophagitis may be found in patients with normal acid exposure.38 Not all patients with abnormal acid exposure have esophagitis, as defined by histology, which can present in subjects without reflux;39 a substantial proportion of patients with BE lack reflux symptoms.40 The significant but nonexclusive association of the type II microbiome with histologically defined changes related to GERD is consistent with the complex GERD phenotype. Notably, our study subjects were generally elderly male veterans. This relatively homogenous cohort helped limit confounding, but also might limit the application of our findings to a more general population. Despite the complex host and environmental factors, the findings in the present analyses make possible design of further studies to directly examine possible causal roles of the type II microbiome in GERD development. If GERD represents a microecological disease, a new type of treatment for reflux might become possible, for example, by converting the type II to type I microbiome through use of antibiotics, probiotics, or prebiotics.
Supplementary Material
Acknowledgments
Grant Support: This study was supported by grants from the National Cancer Institute and the National Institute for Allergy and Infectious Diseases, (UH2CA140233, R01CA97946, R01AI063477) awarded to Z.P. and by the Medical Research Service of the Department of Veterans Affairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Shaheen N, Ransohoff DF. Gastroesophageal reflux, Barrett’s esophagus, and esophageal cancer: scientific review. JAMA. 2002;287:1972–1981. doi: 10.1001/jama.287.15.1972. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Time trends of gastroesophageal reflux disease: a systematic review. Clin Gastroenterol Hepatol. 2007;5:17–26. doi: 10.1016/j.cgh.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 3.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142–146. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 4.Lederberg J. Infectious history. Science. 2000;288:287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 5.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 6.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 7.Kaufmann SHE, Schaible UE. 100th anniversary of Robert Koch's Nobel Prize for the discovery of the tubercle bacillus. Trends in Microbiology. 2005;13:469–475. doi: 10.1016/j.tim.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 9.Yang L, Pei Z. Bacteria, inflammation, and colon cancer. World Journal of Gastroenterology. 2006;12:6741–6746. doi: 10.3748/wjg.v12.i42.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kado S, Uchida K, Funabashi H, et al. Intestinal microflora are necessary for development of spontaneous adenocarcinoma of the large intestine in T-cell receptor beta chain and p53 double-knockout mice. Cancer Res. 2001;61:2395–2398. [PubMed] [Google Scholar]
- 11.Dunne C, O'Mahony L, Murphy L, et al. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr. 2001;73:86S–92S. doi: 10.1093/ajcn/73.2.386s. [DOI] [PubMed] [Google Scholar]
- 12.O'Mahony L, Feeney M, O'Halloran S, et al. Probiotic impact on microbial flora, inflammation and tumour development in IL-10 knockout mice. Aliment Pharmacol Ther. 2001;15:1219–1225. doi: 10.1046/j.1365-2036.2001.01027.x. [DOI] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 14.Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. Bacterial microbiome in the human distal esophagus. Proc Natl Acad Sci. U S A. 2004;101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pei Z, Yang L, Peek RM, Jr, Levine SM, Pride DT, Blaser MJ. Bacterial microbiome in reflux esophagitis and Barrett's esophagus. World J Gastroenterol. 2005;11:7277–7283. doi: 10.3748/wjg.v11.i46.7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:635–638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental microbiology. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stackebrandt E, Goebel BM. Taxonomic Note: A Place for DNA-DNA Reassociation and 16S rRNA Sequence Analysis in the Present Species Definition in Bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 19.Liu Z, DeSantis TZ, Andersen GL, Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 2008;36:e120. doi: 10.1093/nar/gkn491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao A. Nonparametric estimation of the number of classes in a population. Scand J Statist. 1984;11:265–270. [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:89–300. [Google Scholar]
- 22.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pavoine S, Dufour AB, Chessel D. From dissimilarities among species to dissimilarities among communities: a double principal coordinate analysis. J Theor Biol. 2004;228:523–537. doi: 10.1016/j.jtbi.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 25.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucl Acids Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawamura Y, Hou XG, Todome Y, et al. Streptococcus peroris sp. nov. and Streptococcus infantis sp. nov., new members of the Streptococcus mitis group, isolated from human clinical specimens. Int J Syst Bacteriol. 1998;48:921–927. doi: 10.1099/00207713-48-3-921. [DOI] [PubMed] [Google Scholar]
- 27.Cronholm JN. A General Method of Obtaining Exact Sampling Probabilities of the Shannon-Wiener Measure of Information, H. Rep 575. Rep US Army Med Res Lab. 1963;26:1–9. doi: 10.21236/ad0421157. [DOI] [PubMed] [Google Scholar]
- 28.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bik EM, Eckburg PB, Gill SR, et al. Molecular analysis of the bacterial microbiome in the human stomach. Proc Natl Acad Sci U S A. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial microbiome. Proc Natl Acad Sci U S A. 2007;104:927–932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:899–911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 32.Paster BJ, Boches SK, Galvin JL, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falk GW. Gastroesophageal reflux disease. Curr Opin Gastroenterol. 1999;15(33) doi: 10.1097/00001574-199907000-00011. [DOI] [PubMed] [Google Scholar]
- 34.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alter gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan YP, Chakder S, Gao F, Rattan S. Inducible and neuronal nitric oxide synthase involvement in lipopolysaccharide-induced sphincteric dysfunction. Am J Physiol Gastrointest Liver Physiol. 2001;280:G32–G42. doi: 10.1152/ajpgi.2001.280.1.G32. [DOI] [PubMed] [Google Scholar]
- 36.DeVault KR, Castell DO. Updated guidelines for the diagnosis and treatment of gastroesophageal reflux disease. The Practice Parameters Committee of the American College of Gastroenterology. Am J Gastroenterol. 1999;94:1434–1442. doi: 10.1111/j.1572-0241.1999.1123_a.x. [DOI] [PubMed] [Google Scholar]
- 37.Kiljander TO, Laitinen JO. The prevalence of gastroesophageal reflux disease in adult asthmatics. Chest. 2004;126:1490–1494. doi: 10.1378/chest.126.5.1490. [DOI] [PubMed] [Google Scholar]
- 38.Schlesinger PK, Donahue PE, Schmid B, Layden TJ. Limitations of 24-hour intraesophageal pH monitoring in the hospital setting. Gastroenterology. 1985;89:797–804. doi: 10.1016/0016-5085(85)90575-x. [DOI] [PubMed] [Google Scholar]
- 39.Zentilin P, Savarino V, Mastracci L, et al. Reassessment of the diagnostic value of histology in patients with GERD, using multiple biopsy sites and an appropriate control group. Am J Gastroenterol. 2005;100:2299–2306. doi: 10.1111/j.1572-0241.2005.50209.x. [DOI] [PubMed] [Google Scholar]
- 40.Gerson LB, Shetler K, Triadafilopoulos G. Prevalence of Barrett's esophagus in asymptomatic individuals. Gastroenterology. 2002;123:461–467. doi: 10.1053/gast.2002.34748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.