Abstract
The aggregation of the 37-residue polypeptide IAPP, as either insoluble amyloid or as small oligomers, appears to play a direct role in the death of pancreatic β-islet cells in type II diabetes. While IAPP has been known to be the primary component of type II diabetes amyloid, the molecular interactions responsible for this aggregation have not been identified. To identify the aggregation-prone region(s), we constructed a library of randomly generated point mutants of IAPP. This mutant IAPP library was expressed in E. coli as genetic fusions to the reporter protein enhanced green fluorescent protein (EGFP). Because IAPP aggregates rapidly, both independently and when fused to EGFP, the fusion protein does not yield a functional, fluorescent EGFP. However, mutations of IAPP that result in non-amyloidogenic sequences remain soluble and allow EGFP to fold and fluoresce. Using this screen, we identified 22 single mutations, 4 double mutations and 2 triple mutations of IAPP that appear to be less amyloidogenic than wild type human IAPP. A comparison of these sequences suggests residues 13 and 15–17 comprise an additional aggregation-prone region outside of the main amyloidogenic region of IAPP.
The aggregation of misfolded proteins into toxic oligomers and fibers has been linked to a variety of diseases such as type II diabetes, Alzheimer’s disease and Parkinson’s disease. In type II diabetes the amyloid-forming peptide is islet amyloid polypeptide (IAPP, amylin). This 37 amino acid polypeptide misfolds and forms aggregates within the pancreas. This misfolding into toxic aggregates, such as small soluble oligomers or large fibers, is believed to contribute to the loss of pancreatic β-cells. While the exact role of IAPP in type II diabetes is unclear, it is known that IAPP is found as extracellular deposits of amyloid in approximately 95% of patients afflicted with type II diabetes (1–3). IAPP has also been shown to be a toxic agent in vitro when added to human islet β-cells (4).
Many of the therapeutic strategies for preventing or slowing the progression of amyloid diseases such as type II diabetes, involve slowing or preventing the aggregation of the amyloidogenic proteins. To this end, a great deal of effort has been made in identifying the amino acids responsible for the aggregation-prone nature of amyloidogenic peptides such as IAPP. Preventing IAPP aggregation, and hence the formation of the toxic oligomers and amyloid, could prevent disease progression.
It is well established that rodent IAPP (rIAPP) is nonamyloidogenic and nontoxic. It is also established that rats and mice are not known to develop type II diabetes (5). A comparison of the peptide sequences between human and rodent IAPP showed six amino acid substitutions (Figure 1). Five of the substitutions known to convert amyloidogenic human IAPP into non-amyloidogenic rodent IAPP occur in the 20–29 region of the peptide. Because of these substitutions the 20–29 region has long been described as the amyloidogenic region of IAPP (6–8). While recent studies have indicated that several mutations outside of the 20–29 region can hinder fibrillogenesis, the 20–29 region has been the focus of most studies aimed at preventing IAPP aggregation (9–15). These studies have provided a great deal of information about the aggregation-prone 20–29 region of IAPP. However, no extensive screen has been performed to identify non-amyloidogenic mutations of IAPP. A broad screen would not only identify individual mutants of IAPP that are non-amyloidogenic, but could also pinpoint the interactions responsible for IAPP aggregation.
Figure 1.
Comparison of previously analyzed IAPP variants. Sequences known to aggregate are shown at the top. Sequences known to be non-amyloidogenic are shown in bold at the bottom. Only amino acid differences compared to wild type human IAPP are indicated.
Here, we describe a screen that takes advantage of the slow folding kinetics of enhanced green fluorescent protein (EGFP) to select for IAPP mutations that resist aggregation. Proteins fused to the N-terminus of EGFP can attenuate the extent of EGFP fluorescence within a cell (16). This control over EGFP fluorescence is related to the ability of the fusion protein to remain monomeric and soluble in solution. Fusions proteins that self-assemble into insoluble fibrils prevent the EGFP from folding and fluorescing. In this screen the IAPP peptide was genetically fused to EGFP. When expressed in E. coli, the IAPP-EGFP fusion protein produces virtually no green color or fluorescence due to the amyloidogenic nature of IAPP. Aggregation of IAPP precludes folding, and hence fluorescence, of the EGFP reporter. However, when a soluble and non-amyloidogenic mutant of IAPP is fused to EGFP, the EGFP is capable of folding and fluorescing. Using error-prone PCR, we constructed a library of mutated IAPP sequences genetically fused to EGFP. This mutated IAPP-EGFP library was expressed in E. coli and screened for green fluorescent colonies. Those colonies showing the greatest fluorescence were selected and identified through DNA sequencing. To verify the results, four of the selected IAPP mutant peptides were chemically synthesized and their propensity to aggregate and form fibers was characterized in vitro.
Experimental Procedures
Materials
Synthetic peptides were prepared by GenScript Corporation (Piscataway, NJ), except for A13E IAPP (synthesized by Lifetein, LLC, South Plainfield, NJ). DNA purification kits were from Qiagen Inc (Valencia, CA). Klenow fragment DNA polymerase and restriction endonucleases were from New England Biolabs (Ipswich, MA). Expand High Fidelity DNA Polymerase was from Roche . pET 28a and pCDF-1b plasmids were from Stratagene. DNA sequencing was performed by Davis Sequencing, Inc (Davis, CA).
Construction of the IAPP Gene
The IAPP gene was constructed using overlap gene assembly. Two single-stranded DNA oligonucleotides (Supplemental) were designed to base-pair to each were designed to base-pair to each other with complementary 3’ termini. Oligos were diluted to 10 µM with deionized H2O (dI H2O). 26 µL of oligo 1 and 26 µL of oligo 2 were mixed together in a sterile eppendorf tube along with 25µL 10× Ecopol Buffer (New England Biolabs) and 143 µL dI H2O. This solution was heated at 85°C in a heat block for 15 minutes to fully denature the DNA oligonucleotides. The heat block was turned down to 37°C to allow the mixture to slowly cool and allow proper annealing of the oligos. After the temperature dropped to 37°C, 10 µL of 5mM dNTP’s, 10 µL of 20 mM DTT, and 4 µL Klenow Fragment (5000 U/mL NEB) were added. The solution was incubated at 37°C for 1 hour. Double stranded gene product was purified (Qiagen PCR purification kit). Codons were optimized for expression in E. coli. The full length gene product was PCR amplified (supplemental). The gene product was digested with Nco 1 and Bam H1 restriction endonucleases and ligated into analogously digested pAβ42-EGFP(17) yielding the final plasmid, pIAPP-EGFP.
Construction of the I26P mutant of IAPP
The I26P variant of IAPP is known to be non-amyloidogenic and can inhibit IAPP fibril formation (11). As a positive control, we constructed the I26P mutant of IAPP and genetically fused it to EGFP. We used the Quikchange Site-Directed Mutagenesis System (Stratagene) and the following primers: 5’CAACAACTTTGGCGCGCCGCTGAGCAGCACCAACG3’ and 5’CGTTGGTGCTGCTCAGCGGCGCGCCAAAGTTGTTG3’.
Construction of the mutated IAPP library
The mutated IAPP library was generated using PCR-based random mutagenesis (Stratagene/Agilent Technologies Genemorph II mutagenesis kit). The DNA primers used were 5’-CTTTAATAAGGAGATATACCATGGGC-3’ and 5’-GCGGAGCCAGCGGATCC -3’. These primers maintained the Nco 1 and Bam H1 restrictions sites, but allowed for mutagenesis of each of the 37 codons of the IAPP gene. Four cycles of PCR mutagenesis were performed (following instructions of the manufacturer) on the IAPP gene to yield an amplified gene product containing, on average, one mutation per gene. After the fourth round of amplification, the randomized gene mixture was digested with Nco 1 and Bam H1 restriction endonucleases and ligated into similarly digested pAβ42-EGFP (17). The ligation product was purified with Qiagen PCR purification kit and eluted with dI H2O.
The mutated IAPP-EGFP plasmid library was transformed into electrocompetent XL1-Blue E. coli (18) and plated on LB media plates containing 20 µg/mL of the antibiotic streptomycin. The plates were incubated for 16 hours at 37°C. Approximately 25,000 colonies were produced. Five colonies were randomly chosen for DNA sequencing to verify mutagenesis. Three of the five samples had mutations resulting in changes to the amino acid sequence (K1I, K1N and the triple mutant H18N, A25V, L27N). One sample possessed a silent mutation that yielded the wild type IAPP peptide sequence. The fifth sample was the wild-type IAPP. We estimate that 80% of the 25,000 colonies generated contained at least one mutation. The colonies were scraped from the plates and combined in 100 mL LB media containing 20 µg/mL streptomycin. The mutated IAPP-EGFP plasmid library was purified from this cell suspension using Qiagen’s DNA purification miniprep kit. (Qiagen).
Screening the Mutated IAPP-EGFP Library
The mutated IAPP-EGFP plasmid library DNA was transformed into electrocompetent BL21(DE3) E. coli. The cells were rescued with SOC media and plated on sterile nitrocellulose discs placed on LB media plates containing 20 µg/mL streptomycin. The transformation mixture was plated to yield a density of approximately 500 colonies per plate. The LB plates were incubated for 16 hours at 37°C. The nitrocellulose discs, covered in colonies, were transferred to LB media plates containing 20 µg/mL streptomycin and 2 mM IPTG. The plates were incubated at 37°C for 4 to 6 hours. To select green colonies, the plates were scanned both under white light and under 490 nm light. Green fluorescent colonies were selected based on their visual ability to fluoresce green.
Quantification of Cellular Fluorescence
Selected colonies were grown to an O.D.600 of 0.7 before protein induction with 1 mM IPTG. The induced E. coli were incubated at 37°C with shaking for 4 hours. The cells were transferred to black 96-well fluorometer plates to record the fluorescence emission (Ex490nm and Em516 nm) of each culture using a Hitatchi F-7000 fluorescence spectrophotometer. To ensure expression of each variant, whole cell lysates were analyzed using SDS-PAGE. A 1 mL aliquot of cell culture was pelleted and the supernatant removed. The pellets were washed with 20 mM Phosphate buffer pH 7.2 and pelleted again. The washed pellets were resuspended in 200 µL SDS-PAGE loading buffer and boiled for 10 minutes. Samples were centrifuged for 10 minutes at 13,000 × g. 10 µL of the supernatant for each variant was loaded onto a precast 4–20% gradient polyacrylamide gel (Bio-Rad) and analyzed via SDS-PAGE. Gels were stained with Coomassie Brilliant Blue R250 Dye and destained with a solution of 7% methanol and 7% acetic acid.
Selected colonies were also visually compared by streaking each variant onto a sterile nitrocellulose disc placed on an LB media plate containing streptomycin (20 µg/mL). After incubating the plates for 16 hours at 37°C, the discs were transferred to fresh LB media plates containing streptomycin and 2 mM IPTG. Plates were incubated for 4 to 6 hours at 37°C. The plates were scanned using a Bio-Rad Molecular Imager VersaDoc MP Imaging System.
Preparing Disaggregated IAPP
0.5 mg synthetic IAPP (GenScript Corp) or mutated IAPP (GenScript), was dissolved in 4.0 mL of hexafluoroisopropanol (HFIP) and placed in a sonicating water bath for 10 minutes. The solution was divided into 500 µL aliquots and stored at −80°C.
Thioflavin T Assays
Each aliquot of disaggregated IAPP (from above) was thawed and the HFIP removed over a stream of dry nitrogen gas. The resulting solid was dissolved in 20 mM tris buffer pH 7.40 to 10.6 µM. The samples were incubated at 37°C with shaking (200 rpm). 40 µL of each sample was removed at indicated time points and mixed with 660 µL of 31 µM thioflavin T in 20 mM Tris buffer pH 7.40. The thioflavin T mixture was incubated at room temperature in the dark for 5 minutes before recording the thioflavin T fluorescence (Ex450nm and Em488nm) using a Hitatchi F-7000 fluorescence spectrophotometer 96-well plate reader.
Atomic Force Microscope
Each aliquot (from above) was thawed and the HFIP removed over a stream of dry nitrogen gas. The resulting solid was dissolved in 20 mM Tris buffer pH 7.40 to 106 µM. The samples were incubated at 37°C with shaking (200 rpm). 5 µL was taken from each sample at various time points, diluted 5-fold in filtered dI H2O and added directly to freshly cleaved mica. After 15 minutes of incubation, the mica surface was washed with 200 µL filtered dI H2O and allowed to air dry. Samples were scanned using an MFP-3D atomic force microscope (Asylum Research) set on A/C mode and a 240 µm silicon cantilever (Olympus).
Results
Error-prone PCR was used to construct a library of mutated IAPP sequences genetically fused to the reporter protein EGFP. This library was transformed into electrocompetent BL21(DE3) E. coli. These transformations resulted in approximately 25,000 individual colonies, of which approximately 20,000 possessed at least one mutation. Visual scanning of the plates under both white light and 490 nm wavelength light identified 87 colonies with significant green fluorescence. These bright colonies were selected and their DNA sequenced (Davis Sequencing). 59 of the selected colonies possessed deletions that resulted in truncated IAPP sequences (typically fewer than 15 amino acids fused to EGFP), yet remained in the correct reading frame to yield active EGFP. The remaining samples expressed a full-length IAPP sequence. Twenty-two of the selected full length IAPP sequences possessed a single mutation, four possessed double mutations and two possessed triple mutations (Figure 2).
Figure 2.
Comparison of IAPP variants identified through the use of the mutated IAPP-EGFP screen. Amyloidogenic sequences are listed at the top. Nonamyloidogenic sequences are indicated in bold lettering at the bottom. Only amino acid residues that differ from human IAPP are indicated.
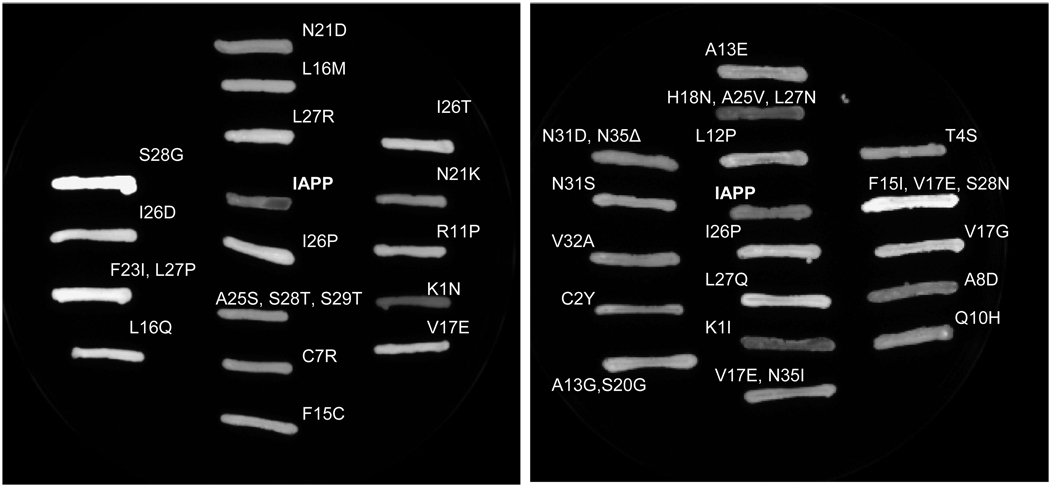
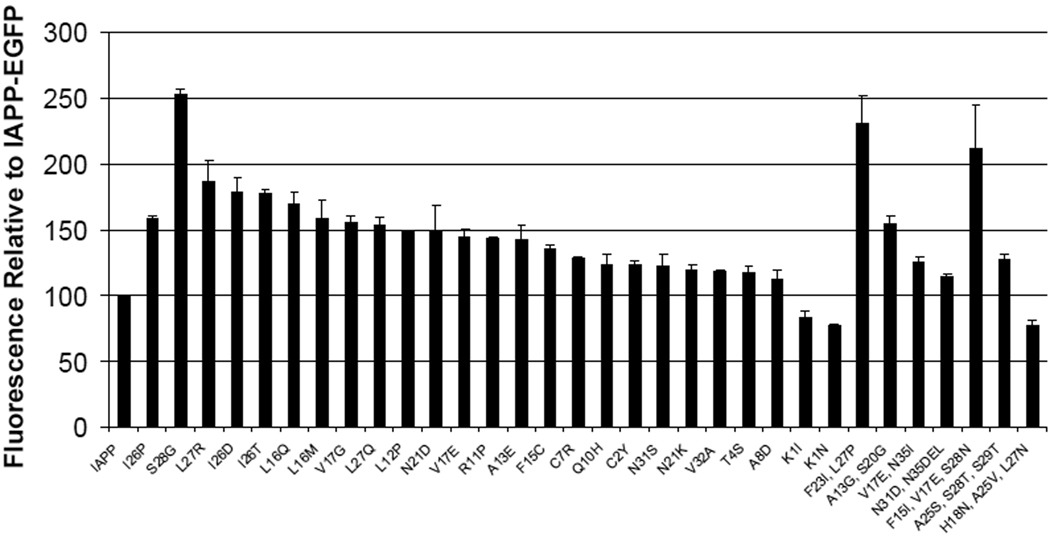
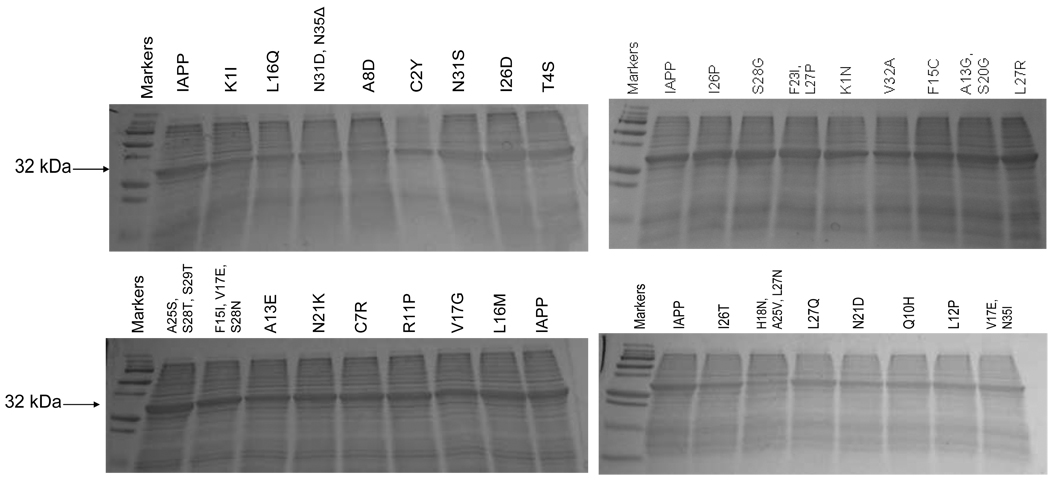
The level of green fluorescence for each of the selected colonies was compared to that of wild type human IAPP fused to EGFP. The selected colonies were also compared to the non-amyloidogenic I26P mutant of IAPP fused to EGFP (11). For a visual comparison, selected variants were grown on nitrocellulose discs and incubated with IPTG to induce protein expression (Figure 3). For a more quantitative comparison, each variant was grown in LB liquid media and induced with IPTG. The resulting EGFP fluorescence was detected using a Hitatchi F-7000 fluorescence spectrophotometer (Figure 4). To ensure comparable expression of each IAPP-EGFP variant, whole cell lysates were analyzed with SDS-PAGE (Figure 5).
Figure 3.
Visual comparison of selected IAPP-EGFP variants to wild-type human IAPP fused to EGFP (IAPP in center of each panel) and to the nonamyloidogenic variant I26P. The CCD camera detector does not show data in color. Data are shown as fluorescence emission intensity.
Figure 4.
Quantitative comparison of selected IAPP-EGFP variants. Fluorescence intensity of wild-type human IAPP fused to EGFP was set at 100% fluorescence. The fluorescence intensity of each variant was plotted as a percentage of the fluorescence of wild-type human IAPP. Variants with a relative fluorescence of 100% or less are considered to be amyloidogenic.
Figure 5.
Comparison of expression of 32 kDa IAPP-EGFP variants in BL21(DE3) E. coli. Each variant showed an expression level comparable to that of wild-type IAPP fused to EGFP (labeled as IAPP in each gel).
The use of fluorescent protein fusions to amyloidogenic proteins has been shown to be an effective method for distinguishing relative levels of amyloidogenicity (17, 19–24). However, it is important to verify the results of any screen to demonstrate the validity of the selected samples. To this end, we chose four of our selected variants, S28G, I26D, A13E and L16Q, to be chemically synthesized and experimentally characterized for their propensity to aggregate. We chose to characterize the S28G and I26D variants due to their elevated EGFP fluorescence. Because several non-amyloidogenic variants were identified having one or more mutations in the A13-V17 region, we chose to also characterize the L16Q and A13E variants.
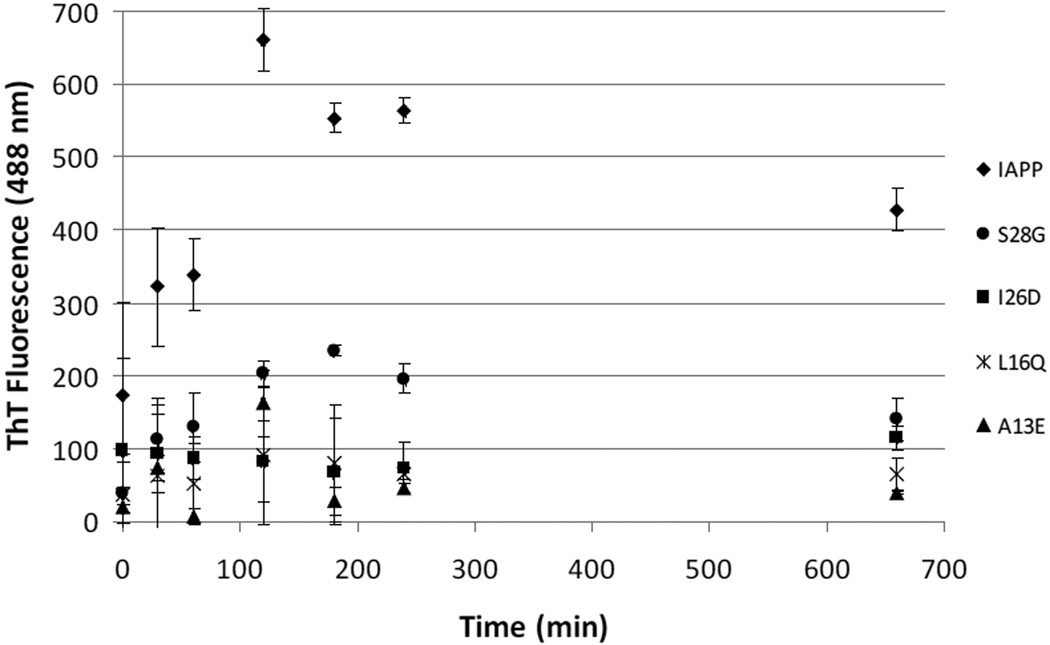
The ability of the four selected variants to bind Thioflavin T (ThT), an indicator of amyloid aggregation, was compared to that of wild type IAPP. Human IAPP is one of the most amyloidogenic peptides known. To characterize the selected variants ability to resist aggregation, we subjected each variant to conditions known to quickly promote aggregation. Samples were incubated at 10 µM concentration (Figure 6) and at 100 µM (Figure 1S), with vigorous shaking at 37°C. Under these conditions, wild type IAPP began to aggregate, and hence bind ThT, within minutes of incubation. The decrease in ThT fluorescence after reaching its maximum is commonly witnessed in these assays and is consistent with previously reported amyloid aggregation reactions (25, 26).
Figure 6.
Comparison of thioflavin T binding time course by selected variants and wild-type human IAPP. One representative time experiment is shown with the error bars representing the standard deviation of all trials (n ≥ 3).
The mutated IAPP samples selected to resist aggregation showed drastically reduced ThT fluorescence compared to IAPP.
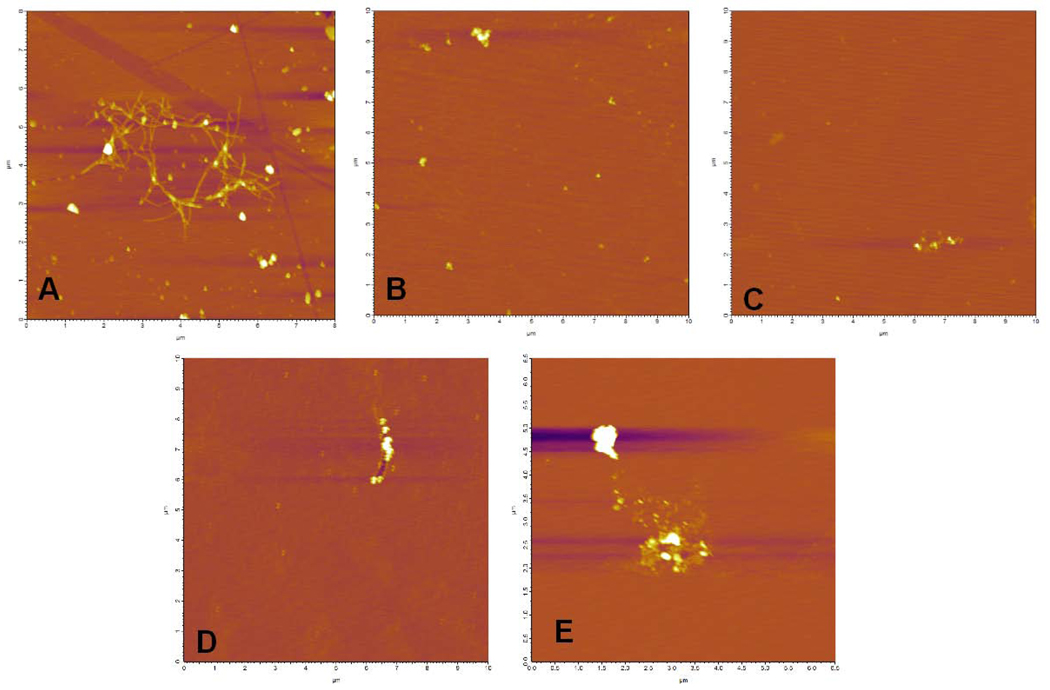
The ability of the synthetic mutant IAPP variants to form fibers was compared to wild-type human IAPP (Figure 7) using atomic force microscopy (AFM). All IAPP samples were incubated under conditions known to promote fiber formation. Only wild-type IAPP was found to form fibers. The selected variants I26D, S28G, A13E and L16Q were not found to form fibers under any of the conditions tested.
Figure 7.
AFM images of (A) IAPP (B) I26D (C) S28G and (D) L16Q after 30 minutes of incubation at 37°C with agitation and (E) A13E after 60 minutes of incubation at 37°C with agitation. Time points shown were selected based on maximum ThT binding as determined in Figure 6. Each variant was tested at multiple time points. Fibers were not detected under any of the conditions tested for the selected mutant variants (while wild type IAPP was found to produce fibers under all conditions tested and at all time points after 15 minutes of incubation).
Discussion
Many of the sequences selected with this screen were found to have mutations within the 20–29 region of IAPP. This is commonly referred to as the amyloidogenic region of IAPP (6, 8, 9, 11). In this screen, 11 of the 28 variants selected had at least one mutation within the 20–29 region of IAPP. Recent work also indicates that the amyloidogenic region could be extended to include the 30–33 region of IAPP (9, 14). Indeed, we find that 14 of the 28 sequences selected had mutations within the 20–33 region of IAPP. These results are consistent with previous studies that indicate the 20–33 region as being important for IAPP aggregation.
Seven of the twenty-eight sequences selected with this screen contained at least one mutation within the 15–17 region of IAPP (the amino acids FLV). There is little evidence within the current literature to indicate these amino acids as being amyloidogenic. Gazit and coworkers make several arguments suggesting a role for the amyloidogenic nature of the 14–19 region (27–29), but the majority of work focuses on the 20–29 region as being responsible for amyloidogenicity. Because 25% of the selected non-amyloidogenic sequences from this screen have at least one FLV mutation, these amino acids may be important for IAPP aggregation. The selected variants with mutations outside the 20–33 region (such as the L16Q mutant) were resistant to aggregation despite the amyloidogenic nature of the 20–33 region of IAPP.
Our results suggest the 15–17 region of IAPP (amino acids FLV) is important for the aggregation of IAPP. Eisenberg and coworkers recently determined the crystal structure of an IAPP fusion to maltose binding protein (MBP) (30). In their model, they show the FLV region of IAPP as the core of a helix thought to be associated with IAPP aggregation (31–33). This model shows the FLV region of IAPP associating with the YLV region of insulin B chain (30). Based on these models, and the results of our screen, the FLV region appears to be an important part of IAPP aggregation. Interestingly, the amyloid peptide associated with Alzheimer’s disease, Aβ, contains an LVF sequence in the putative aggregation region.
Alanine13 was found to be an additional amino acid of interest selected from this screen. Ala13 is in the middle of the putative helical region of IAPP, believed to play a role in aggregation. Our screen identified two Ala13 mutants, A13E and the double mutant A13G, S20G, as being resistant to amyloid aggregation. The lack of aggregation witnessed with the A13E mutation could be attributed to the drastic change of a small alanine for a large negatively charged glutamic acid. However, the double mutant A13G, S20G could be more informative about the role of A13 in IAPP aggregation. Previous studies have shown that S20G (which is found in primates) is a highly amyloidogenic sequence (34). Because the A13G, S20G double mutant was non-amyloidogenic according to our screen, and the S20G single mutation (as found in primates) remains amyloidogenic, we assign the lack of amyloidogenicity of this double mutant as being due to the A13G mutation. It is believed that this region of IAPP folds into an α-helix which may be important for aggregation (33). Because glycine is a strong helix breaker, the observed results with the A13G variant supports the theory that this region of IAPP adopts a helical structure on the pathway to aggregation (33). Decreasing this region’s ability to adopt a helical fold may have resulted in the observed lack of fiber formation and decreased ThT binding of the A13G, S20G variant.
Previous studies have shown that amyloid aggregation and fiber formation have a pH dependence (35–38). Many amyloid proteins become more aggregation prone as the solution pH nears the isoelectric point (pI) of the protein. Wild-type IAPP, L16Q and S28G have an identical pI of 8.90, yet the L16Q and S28G variants were resistant to aggregation. This suggests that the resistance to aggregation is not due to pH effects of the solution, but rather to specific protein-protein interactions. The pI of the A13E and I26D variants are both 8.05. Because the pI of A13E and I26D is closer to the pH of the solution (pH = 7.4) than that of wild type IAPP, we would expect those variants to be more prone to aggregate. Again, the lack of aggregation of these two peptides can not be attributed solely to the pH of the solution, but are likely due to protein-protein interactions.
Several programs have been developed to identify in silico the amyloidogenic regions of proteins. We used one such program, TANGO (http://tango.crg.es) (38–40), to analyze the IAPP and mutant IAPP sequences. Tango identified two aggregation-prone regions in the IAPP peptide sequence, amino acids L12-V17 and amino acids F23-L27. According to TANGO, both the A13E and L16Q mutations eliminate the L12-V17 amyloidogenic region (but have no effect on the F23-L27 region). Likewise, TANGO indicated that the I26D mutation eliminates the F23-L27 amyloidogenic region (but has no effect on the L12-V17 region). The results of the TANGO program support our experimental data in indicating that A13 and F15-V17 are important regions for the overall amyloidogenicity of IAPP.
Several mutant variants are consistent with previously published reports. In IAPP, the disulfide bond between Cys2 and Cys7 has been identified as an important contributor of fiber formation (41). Two variants identified as non-amyloidogenic in our screen were C2Y and C7R. These mutations would both result in the loss of this intramolecular disulfide bond, and hence would be expected to be less amyloidogenic.
This EGFP based screen provided a facile method for sampling large numbers of sequence variants to identify those that resist amyloid aggregation. The screen is predicated on the fact that EGFP is a relatively slow-folding enzyme in prokaryotic expression (42). This slow EGFP folding time allows the amyloidogenic peptides to aggregate prior to EGFP folding and fluorescence. Sequences selected through this screen should prove to be non-amyloidogenic, or at the least, should take longer to aggregate than EGFP takes to fold and fluoresce. The four synthetic variants tested (I26D, S28G, A13E and L16Q) showed decreased levels of ThT fluorescence and did not form fibers under the conditions tested. We believe these results are consistent with previously reported results that the 20–33 region of IAPP is amyloidogenic. However, this work indicates that other regions of the IAPP sequence, FLV (residues 15–17) and Ala13, should be considered important for amyloidogenesis.
Supplementary Material
Abbreviations
- IAPP
Islet Amyloid Polypeptide
- hIAPP
human IAPP
- ThT
thioflavin T
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- EGFP
enhanced green fluorescent protein
- HFIP
hexafluoroisopropanol
- DTT
dithiotreitol
- AFM
atomic force microscope
- NEB
New England Biolabs
- SOC media
super optimal broth with catabolite repression
- LB media
Luria-Bertani media
Footnotes
Funding by NIH grant R15AG032582-01A1
Supplemental Information
Supplemental materials may be accessed free of charge online at http://pubs.acs.org.
References
- 1.Apostolidou M, Jayasinghe SA, Langen R. Structure of alpha-helical membrane-bound human islet amyloid polypeptide and its implications for membrane-mediated misfolding. J Biol Chem. 2008;283:17205–17210. doi: 10.1074/jbc.M801383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hull RL, Westermark GT, Westermark P, Kahn SE. Islet amyloid: a critical entity in the pathogenesis of type 2 diabetes. J Clin Endocrinol Metab. 2004;89:3629–3643. doi: 10.1210/jc.2004-0405. [DOI] [PubMed] [Google Scholar]
- 3.Kahn SE, Andrikopoulos S, Verchere CB. Islet amyloid: A long-recognized but underappreciated pathological feature of type 2 diabetes. Diabetes. 1999;48:241–253. doi: 10.2337/diabetes.48.2.241. [DOI] [PubMed] [Google Scholar]
- 4.Ritzel RA, Meier JJ, Lin CY, Veldhuis JD, Butler PC. Human islet amyloid polypeptide oligomers disrupt cell coupling, induce apoptosis, and impair insulin secretion in isolated human islets. Diabetes. 2007;56:65–71. doi: 10.2337/db06-0734. [DOI] [PubMed] [Google Scholar]
- 5.Matveyenko AV, Butler PC. Islet amyloid polypeptide (IAPP) transgenic rodents as models for type 2 diabetes. Ilar J. 2006;47:225–233. doi: 10.1093/ilar.47.3.225. [DOI] [PubMed] [Google Scholar]
- 6.Westermark P, Engstrom U, Johnson KH, Westermark GT, Betsholtz C. Islet Amyloid Polypeptide - Pinpointing Amino-Acid-Residues Linked to Amyloid Fibril Formation. PNAS. 1990;87:5036–5040. doi: 10.1073/pnas.87.13.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nanga RP, Brender JR, Xu J, Veglia G, Ramamoorthy A. Structures of rat and human islet amyloid polypeptide IAPP(1–19) in micelles by NMR spectroscopy. Biochemistry. 2008;47:12689–12697. doi: 10.1021/bi8014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gazit E. Mechanisms of amyloid fibril self-assembly and inhibition. Febs Journal. 2005;272:5971–5978. doi: 10.1111/j.1742-4658.2005.05022.x. [DOI] [PubMed] [Google Scholar]
- 9.Abedini A, Raleigh DP. Destabilization of human IAPP amyloid fibrils by proline mutations outside of the putative amyloidogenic domain: Is there a critical amyloidogenic domain in human IAPP? J Mol Biol. 2006;355:274–281. doi: 10.1016/j.jmb.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 10.Marek P, Abedini A, Song BB, Kanungo M, Johnson ME, Gupta R, Zaman W, Wong SS, Raleigh DP. Aromatic interactions are not required for amyloid fibril formation by islet amyloid polypeptide but do influence the rate of fibril formation and fibril morphology. Biochemistry. 2007;46:3255–3261. doi: 10.1021/bi0621967. [DOI] [PubMed] [Google Scholar]
- 11.Abedini A, Meng FL, Raleigh DP. A single-point mutation converts the highly amyloidogenic human islet amyloid polypeptide into a potent fibrillization inhibitor. Journal of the American Chemical Society. 2007;129:11300. doi: 10.1021/ja072157y. + [DOI] [PubMed] [Google Scholar]
- 12.Abedini A, Raleigh DP. The role of His-18 in amyloid formation by human islet amyloid polypeptide. Biochemistry. 2005;44:16284–16291. doi: 10.1021/bi051432v. [DOI] [PubMed] [Google Scholar]
- 13.Koo BW, Hebda JA, Miranker AD. Amide inequivalence in the fibrillar assembly of islet amyloid polypeptide. Protein Engineering Design & Selection. 2008;21:147–154. doi: 10.1093/protein/gzm076. [DOI] [PubMed] [Google Scholar]
- 14.Scrocchi LA, Chen Y, Waschuk S, Wang F, Cheung S, Darabie AA, McLaurin J, Fraser PE. Design of peptide-based inhibitors of human islet amyloid polypeptide fibrillogenesis. Journal of Molecular Biology. 2002;318:697–706. doi: 10.1016/S0022-2836(02)00164-X. [DOI] [PubMed] [Google Scholar]
- 15.Tracz SM, Abedini A, Driscoll M, Raleigh DP. Role of aromatic interactions in amyloid formation by peptides derived from human Amylin. Biochemistry. 2004;43:15901–15908. doi: 10.1021/bi048812l. [DOI] [PubMed] [Google Scholar]
- 16.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 17.Baine M, Georgie DS, Shiferraw EZ, Nguyen TPT, Nogaj LA, Moffet DA. Inhibition of A beta 42 aggregation using peptides selected from combinatorial libraries. Journal of Peptide Science. 2009;15:499–503. doi: 10.1002/psc.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma RC, Schimke RT. Preparation of electro-competent E-coli using salt-free growth medium. Biotechniques. 1996;20:42–44. doi: 10.2144/96201bm08. [DOI] [PubMed] [Google Scholar]
- 19.de Groot NS, Aviles FX, Vendrell J, Ventura S. Mutagenesis of the central hydrophobic cluster in A beta 42 Alzheimer's pepticle - Side-chain properties correlate with aggregation propensities. Febs Journal. 2006;273:658–668. doi: 10.1111/j.1742-4658.2005.05102.x. [DOI] [PubMed] [Google Scholar]
- 20.Kim W, Kim Y, Min J, Kim DJ, Chang YT, Hecht MH. A high-throughput screen for compounds that inhibit aggregation of the Alzheimer's peptide. Acs Chemical Biology. 2006;1:461–469. doi: 10.1021/cb600135w. [DOI] [PubMed] [Google Scholar]
- 21.Wurth C, Guimard NK, Hecht MH. Mutations that reduce aggregation of the Alzheimer's Abeta42 peptide: an unbiased search for the sequence determinants of Abeta amyloidogenesis. J Mol Biol. 2002;319:1279–1290. doi: 10.1016/S0022-2836(02)00399-6. [DOI] [PubMed] [Google Scholar]
- 22.Wurth C, Kim W, Hecht MH. Combinatorial approaches to probe the sequence determinants of protein aggregation and amyloidogenicity. Protein Pept Lett. 2006;13:279–286. doi: 10.2174/092986606775338506. [DOI] [PubMed] [Google Scholar]
- 23.Kim W, Hecht MH. Generic hydrophobic residues are sufficient to promote aggregation of the Alzheimer's A beta 42 peptide. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15824–15829. doi: 10.1073/pnas.0605629103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim W, Hecht MH. Mutations enhance the aggregation propensity of the Alzheimer's A beta peptide. Journal of Molecular Biology. 2008;377:565–574. doi: 10.1016/j.jmb.2007.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamin G, Ruchala P, Teplow DB. A Peptide Hairpin Inhibitor of Amyloid beta-Protein Oligomerization and Fibrillogenesis. Biochemistry. 2009;48:11329–11331. doi: 10.1021/bi901325g. [DOI] [PubMed] [Google Scholar]
- 26.Ahn JS, Lee JH, Kim JH, Paik SR. Novel method for quantitative determination of amyloid fibrils of alpha-synuclein and amyloid beta/A4 protein by using resveratrol. Analytical Biochemistry. 2007;367:259–265. doi: 10.1016/j.ab.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 27.Gilead S, Gazit E. The role of the 14–20 domain of the islet amyloid polypeptide in amyloid formation. Exp Diabetes Res. 2008;2008 doi: 10.1155/2008/256954. 256954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilead S, Wolfenson H, Gazit E. Molecular mapping of the recognition interface between the islet amyloid polypeptide and insulin. Angew Chem Int Ed Engl. 2006;45:6476–6480. doi: 10.1002/anie.200602034. [DOI] [PubMed] [Google Scholar]
- 29.Mazor Y, Gilead S, Benhar I, Gazit E. Identification and characterization of a novel molecular-recognition and self-assembly domain within the islet amyloid polypeptide. J Mol Biol. 2002;322:1013–1024. doi: 10.1016/s0022-2836(02)00887-2. [DOI] [PubMed] [Google Scholar]
- 30.Wiltzius JJW, Sievers SA, Sawaya MR, Eisenberg D. Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process. Protein Science. 2009;18:1521–1530. doi: 10.1002/pro.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williamson JA, Loria JP, Miranker AD. Helix stabilization precedes aqueous and bilayer-catalyzed fiber formation in islet amyloid polypeptide. J Mol Biol. 2009;393:383–396. doi: 10.1016/j.jmb.2009.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williamson JA, Miranker AD. Direct detection of transient alpha-helical states in islet amyloid polypeptide. Protein Sci. 2007;16:110–117. doi: 10.1110/ps.062486907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yonemoto IT, Kroon GJ, Dyson HJ, Balch WE, Kelly JW. Amylin proprotein processing generates progressively more amyloidogenic peptides that initially sample the helical state. Biochemistry. 2008;47:9900–9910. doi: 10.1021/bi800828u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakagashira S, Hiddinga HJ, Tateishi K, Sanke T, Hanabusa T, Nanjo K, Eberhardt NL. S20G mutant amylin exhibits increased in vitro amyloidogenicity and increased intracellular cytotoxicity compared to wild-type amylin. American Journal of Pathology. 2000;157:2101–2109. doi: 10.1016/S0002-9440(10)64848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hortschansky P, Schroeckh V, Christopeit T, Zandomeneghi G, Fandrich M. The aggregation kinetics of Alzheimer's beta-amyloid peptide is controlled by stochastic nucleation. Protein Sci. 2005;14:1753–1759. doi: 10.1110/ps.041266605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen L, Khurana R, Coats A, Frokjaer S, Brange J, Vyas S, Uversky VN, Fink AL. Effect of environmental factors on the kinetics of insulin fibril formation: elucidation of the molecular mechanism. Biochemistry. 2001;40:6036–6046. doi: 10.1021/bi002555c. [DOI] [PubMed] [Google Scholar]
- 38.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 39.Linding R, Schymkowitz J, Rousseau F, Diella F, Serrano L. A comparative study of the relationship between protein structure and beta-aggregation in globular and intrinsically disordered proteins. J Mol Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- 40.Rousseau F, Schymkowitz J, Serrano L. Protein aggregation and amyloidosis: confusion of the kinds? Curr Opin Struct Biol. 2006;16:118–126. doi: 10.1016/j.sbi.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 41.Koo BW, Miranker AD. Contribution of the intrinsic disulfide to the assembly mechanism of islet amyloid. Protein Sci. 2005;14:231–239. doi: 10.1110/ps.041051205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang HC, Kaiser CM, Hartl FU, Barral JM. De novo folding of GFP fusion proteins: High efficiency in eukaryotes but not in bacteria. Journal of Molecular Biology. 2005;353:397–409. doi: 10.1016/j.jmb.2005.08.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.