Abstract
A dynamic cell-matrix interaction is crucial for a rapid cellular response to changes in the environment. Appropriate cell behavior in response to the changing wound environment is required for efficient wound closure. However, the way in which wound keratinocytes modify the wound environment to coordinate with such cellular responses remains less studied. We demonstrated that angiopoietin-like 4 (ANGPTL4) produced by wound keratinocytes coordinates cell-matrix communication. ANGPTL4 interacts with vitronectin and fibronectin in the wound bed, delaying their proteolytic degradation by metalloproteinases. This interaction does not interfere with integrin-matrix protein recognition and directly affects cell-matrix communication by altering the availability of intact matrix proteins. These interactions stimulate integrin- focal adhesion kinase, 14-3-3, and PKC-mediated signaling pathways essential for effective wound healing. The deficiency of ANGPTL4 in mice delays wound re-epithelialization. Further analysis revealed that cell migration was impaired in the ANGPTL4-deficient keratinocytes. Altogether, the findings provide molecular insight into a novel control of wound healing via ANGPTL4-dependent regulation of cell-matrix communication. Given the known role of ANGPTL4 in glucose and lipid homeostasis, it is a prime therapeutic candidate for the treatment of diabetic wounds. It also underscores the importance of cell-matrix communication during angiogenesis and cancer metastasis.
Keywords: Cell Migration, Extracellular Matrix Proteins, Signal Transduction, Skin, Steroid Hormone Receptor, Angiopoietin-like 4
Introduction
Skin repair after an injury proceeds via a finely tuned pattern of integrated biological events aimed at restoration of the epithelial barrier. The inflammatory stage of repair is followed by the proliferation and migration of keratinocytes, a process called re-epithelialization (1). These events are regulated spatiotemporally by several classical growth factors and cytokines, the effects of which have been well documented (2). Less studied are extracellular factors such as matricellular proteins and adipocytokines, both shown to have a profound local impact during wound repair (3, 4). Effective directed cell migration requires constant cellular interaction with the extracellular matrix (ECM)3 in response to the changing wound environment. Although the importance of such cell-matrix communication in wound healing is well recognized, the mechanism that modifies the external wound microenvironment for coordinated keratinocyte behavior remains unclear.
Integrins on the cell surface often function as biosensors to constantly interrogate the wound environment and modulate cell responses accordingly. Binding of integrins to their cognate matrix proteins activates intracellular signaling pathways that modulate a broad range of cellular processes, including cell migration (5). Integrin-mediated signaling requires that integrins bind substrate-anchored matrix proteins. This interaction provides mechanical resistance that permits tensional forces to be generated via the actomyosin system (6). In contrast, small soluble matrix protein fragments generated by the action of proteases during re-epithelialization can compete with substrate-anchored matrix proteins for integrin binding and impair cell signaling (7). Thus, productive integrin signaling depends on the context in which the intact matrix protein is presented to cells. However, the way in which migrating wound keratinocytes coordinate the balance between substrate-anchored and small soluble matrix protein fragments by the specific induction of wound integrins requires further investigation.
Transcriptional regulation plays an important role in the control of keratinocyte behavior at the different phases of wound healing, but little is known about the mechanism that modifies the wound microenvironment to coordinate with changes in cellular behavior for cell-matrix communication. Effective cell-matrix communication is crucial for efficient wound healing. Several nuclear hormone receptors, one of the largest known classes of transcription factors, have been implicated wound repair (8, 9). Of interest, studies have shown that nuclear hormone receptor peroxisome proliferator-activated receptor β/δ (PPARβ/δ) is an early transcription factor that modulates keratinocyte response to inflammation during wound healing (10, 11). Most studies have focused on intracellular signaling or events mediated by PPARβ/δ that were important for keratinocyte survival and migration (10, 12, 13). However, the mechanism by which PPARβ/δ alters the wound microenvironment for effective cell-matrix communication remains unknown. Conceivably, as an intracellular transcription factor, PPARβ/δ is likely to exert such an effect via extracellular factors.
Angiopoietin-like 4 (ANGPTL4) belongs to a group of secreted factors that play important roles in lipid and glucose metabolism (14). Its expression is up-regulated by PPAR (15) and by hypoxia (16). ANGPTL4 is also implicated in breast cancer metastasis via the regulation of vascular integrity (17, 18). The native ANGPTL4 is proteolytically cleaved, giving rise to an N-terminal coiled-coil fragment (nANGPTL4) and a C-terminal fibrinogen-like domain (cANGPTL4). Despite multiple physiological and pathological functions, the significance of the different cleaved fragments of ANGPTL4 is only beginning to be understood. Importantly, the identity of the binding partners for ANGPTL4 and the mechanism by which ANGPTL4 modulates cell migration is unknown, hampering our understanding of its contribution to wound healing and cancer metastasis.
Here, we show that ANGPTL4 is a PPARβ/δ target gene in keratinocytes and that its expression is elevated after injury. We show that ANGPTL4 produced by wound keratinocytes coordinates cell-matrix communication. Specifically, ANGPTL4 interacts with vitronectin and fibronectin in the wound bed, delaying their proteolytic degradation by metalloproteinases and thereby regulating the availability of local extracellular matrix. This interaction does not interfere with the binding of matrix protein to its cognate integrin receptor or with integrin-mediated signaling. Our findings reveal a novel control of the wound environment by keratinocytes that coordinates the dynamic interactions between integrins and components of extracellular matrices.
EXPERIMENTAL PROCEDURES
Reagents
Sensor CM5 chips, amine coupling kits, and Immobiline pK buffers were from GE Healthcare. Purified vitronectin, fibronectin, and laminin were from Calbiochem. Transfection reagent ExGen 500 was from Fermentas. Real-time PCR KAPA SYBR Fast Master mix was from KAPABiosystem. DUOlink proximity ligation assay was from Olink Bioscience. Drosophila Schneider 2 (S2) expression vector harboring a proprietary secretory signal pSSAc5.1/V5-His A was as previously described (19). Double promoter pFIV-U1/H6-Puro lentivirus-based siRNA vector (catalog #SI110A-1) and pPACKF1 packaging plasmid kit were from System Biosciences. Purified matrix proteins were purchased from Sigma. All chemicals were from Sigma unless otherwise stated.
Antibodies
p21-activated kinase (PAK), LIMK1 (LIM kinase 2), PKBα, and their cognate phosphorylated forms were from Cell Signaling. Rac1 and cdc42 were from Cytoskeleton. β-Tubulin, His tag, laminin, fibronectin, matrix metalloproteinases (MMPs), and HRP-conjugated secondary antibodies were from Santa Cruz Biotechnology; vitronectin and integrin αvβ5 were from Chemicon; keratin 6 for wound keratinocytes and hair follicle, α-smooth muscle actin for myofibroblasts, F4/80 for macrophages, and CD31 for endothelial cells were from BioLegend. Anti-human PPARβ/δ monoclonal antibodies were from Perseus Proteomics Inc., Japan. Rabbit polyclonal antibodies against the C-terminal region of human (186–406 amino acid) and mouse (190–410 amino acids) ANGPTL4 were produced in-house. Briefly, female rabbits (New Zealand White, 2–2.5 kg) were injected intramuscularly with 300 μg of recombinant proteins homogenized with 500 μl of complete Freund's adjuvant solution. First and second booster immunization with the same immunization dose were performed 3 and 6 weeks after priming immunization using incomplete Freund's adjuvant, respectively. Final harvest was done by bleeding a whole blood volume, and the rabbits were then culled with injection of euthanasia into the marginal ear vein. The carcasses were disposed after confirming no heart beat and corneal reflex, pedal reflex reactions. Preimmune blood sampling was collected as the negative control. Antibodies were purified by Protein A affinity chromatography as recommended by manufacturer (GE Healthcare).
Keratinocyte Culture
Primary human keratinocytes (Cascade Biologics) were cultured in Quantum 153 medium supplemented with insulin, transferrin, EGF, cholera toxin, and 5% FBS (PAA Laboratories) in a 5% CO2, 37 °C humidified incubator. This medium is a modification of the keratinocyte medium previously described (20, 21). Medium was changed every 3 days. Cells were subcultured upon reaching 70% confluence. Briefly, medium was removed, and the cells washed with PBS. Trypsin (0.25%), EDTA (1 mm) in PBS was added to the culture (0.08 ml/cm2) and incubated at room temperature for 15 min. The flask was rapped gently to dislodge cells from the surface of the flask. PBS containing 1% dialyzed FBS was added, and the cells were collected by centrifugation. The cell pellet was resuspended with fresh medium and subcultured in new flask at 2.5 × 103 cells/cm2.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed according to the manufacturer's (Upstate Biotechnology) instructions with some modifications. Briefly, ChIP assay was performed using the monoclonal PPARβ/δ antibody. Cells were treated with 1% formaldehyde at 37 °C for 15 min. Cross-linked DNA was sonicated to form fragments ranging from 200 to 500 bp in length. DNA fragments were reverse cross-linked at 65 °C for 6 h. The DNA was subsequently purified using Qiaquick column (Qiagen). DNA was amplified by PCR for 20–23 cycles. The ChIP primers for the amplification of the PPAR-response element of the human ANGPTL4 gene were as previously described (22).
Skin Wounding Experiment
Wounding was performed as previously described (23, 24). Briefly, the hair follicle cycle of each mouse was synchronized by shaving the back of the animal 2 weeks before the start of the experiment. After anesthetizing, the mice were shaven. A full thickness mid-dorsal wound (0.5-cm2, square-shaped) was created by excising the skin and the underlying panniculus carnosus. Wound closure was measured daily in a double-blinded fashion until it was complete. At indicated days post-wounding, the entire wound, including a 5-mm margin, was excised. Wounds were dissected for immunohistochemistry, RNA, and protein analyses (23, 25). Six-week-old PPARβ/δ+/+ and −/− (24) and pure bred wild type (ANGPTL4+/+) and ANGPTL4-knock-out (ANGPTL4−/−) male mice were used (26). All mice used in this study had a C57BL/6 background and were individually caged, housed in a temperature-controlled room (23 °C) on a 10-h dark/14-h light cycle, and fed with the standard mouse chow diet. Animal experiments were approved by the University Institutional Animal Care and Use Committee (ARF-SBS/NIE-A-0093, ARF SBS/NIE-A-0078 and ARF SBS/NIE-A-004).
Western Blot and Immunofluorescence Assays
Cells or tissues were lysed in ice-cold lysis buffer (20 mm NaH2PO4, 250 mm NaCl, 1% Triton X-100, 0.1% SDS) supplemented with complete protease inhibitors (Roche Applied Science). Equal amounts of protein extracts were resolved by SDS-PAGE and electroblotted onto polyvinylidene difluoride membranes for Western analysis. Membranes were processed according to standard protocol and developed using chemiluminescence (Millipore). Equal loading/transfer was verified by Coomassie staining of gels or by immunodetection of β-tubulin. Wound biopsies were fixed with 4% paraformaldehyde in PBS for 2 h at 25 °C. The fixed tissues were centrally bisected, washed twice with PBS, and embedded in Tissue-Tek OCT compound medium (Sakura) overnight at 4 °C. The tissues were subsequently frozen at −70 °C for cryosectioning. Cryostat sections (8 μm) mounted on SuperFrost Plus slides were analyzed by immunofluorescence as previously described, except that anti-ANGPTL4 antibodies were used (23). The slides presenting the largest wound diameter was defined as the wound center. As a control for immunofluorescence staining, 10-fold more peptide antigen was preincubated with anti-ANGPTL4 at 4 °C for 1 h before use. Images were taken using a LSM710 confocal laser scanning microscope with a Plan-Apochromat 40×/1.40 oil objective and ZEN software (Carl Zeiss).
Flow Cytometry (FACS)
Wound tissues were subjected to FACS analysis as previously described (27). Entire excised skin wounds were dispersed enzymatically into single cell suspensions. The tissue was incubated with dispase I (1 mg/ml) overnight at 4 °C, minced, and incubated in digestion buffer containing hyaluronidase (1 mg/ml), collagenase D (1 mg/ml), and DNase (100 unit/ml) (Sigma) in a 37 °C shaking incubator for 2 h. The dispase and hyaluronidase digests were pooled and filtered through a 70-μm Nylon cell strainer. Cells were washed, pelleted, and resuspended in equal volume of PBS containing 3% FBS. For staining of surface marker, cells were first blocked with mouse BD Fc Block and then incubated with either phyoerythrin- or FITC-conjugated monoclonal antibodies specific for F4/80 (macrophages) and CD31 (endothelial cells) or control isotype IgG on ice for 30 min. After washing with PBS, the samples were subjected to flow cytometry on a FACSCalibur system (BD Biosciences). Data were analyzed using the CellQuest software (BD Biosciences). The analyzer threshold was adjusted on the flow cytometer channel to exclude most of the subcellular debris to reduce the background noise.
Laser-capture Microdissection (LCM)
Paraffin-embedded sections of PPARβ/δ+/+ and -−/− wounds (10 μm) were mounted onto MembraneSlides (Carl Zeiss). Hematoxylin- and eosin-stained sections were then subjected to LCM using PALM MicroBeam according to the manufacturer's instructions (Carl Zeiss). LCM tissues were collected into microcentrifuge tubes with opaque AdhesiveCaps (Carl Zeiss). RNA was extracted using OptimumTM FFPE RNA Isolation kit (Ambion) pooled from eight LCM tissues. RNA was reverse-transcribed using random primers, and the resulting cDNA was used for real-time PCR.
Expression and Purification of Recombinant ANGPTL4 Proteins
The cDNA sequences encoding human full-length ANGPTL4, nANGPTL4, and cANGPTL4 were amplified by Pfu polymerase and subcloned into pSSAc5.1/V5-His-A (19). A histidine tag was introduced between the secretory signal and the ANGPTL4 cDNA. All ligated products were transformed into competent Escherichia coli Top 10 bacteria and selected on Luria broth agar plates containing 80 μg/ml ampicillin. Positive clones were confirmed by DNA sequencing. Positive constructs were co-transfected with hygromycin expression vector pCoHygro (Invitrogen) into S2 cells. Recombinant ANGPTL4 proteins were purified from the conditioned medium of stable ANGPTL4-expressing S2 cells by preparative isoelectric membrane electrophoresis as described (28).
Surface Plasmon Resonance (SPR) Coupled to Liquid Chromatography-Tandem Mass Spectrometry
Purified cANGPTL4 was immobilized onto a CM5-carboxylated dextran sensor chip by amine coupling using the Surface Prep Module of BIACORE 3000 as recommended by the manufacturer (BIAcore). Acute wound fluid/exudate (WF) was collected from two patients undergoing split-thickness skin grafting. The acute wound fluid was collected daily under sterile conditions from beneath a vapor-permeable membrane applied to the donor site and changed every 24 h for 3 days postoperatively. WF was centrifuged, aliquoted, and frozen at −70 °C. WF buffered with 50 mm Tris, pH 8.0, was introduced into the cANGPTL4-conjugated CM5 chip at a flow rate of 5 μl/min for 10 min with running buffer (50 mm Tris, pH 8.0, 100 mm NaCl). After incubation for 45 s, the chamber was washed with the same buffer, and the bound molecules were subsequently eluted using 10 mm glycine, pH 6.0, and collected in a recovery vial. The CM5 chip was reused to pool more samples after washing with running buffer for 10 min at 20 μl/min. The recovered cANGPTL4-binding proteins were digested with trypsin, reduced, alkylated, and then analyzed with a Finnigan Surveyor HPLC system coupled online to a LTQ-Orbitrap mass spectrometer (Thermo Electron) equipped with a nanospray source. Proteins were identified using a Mascot search. SPR was used to determine the dissociation constant of the interaction between fibronectin and vitronectin with recombinant cANGPTL4 immobilized onto a CM5 chip. Six concentrations (0.16, 0.32, 0.63, 1.25, 2.50, and 5.0 μm) of various matrix proteins were used. Each sensorgram was corrected by subtracting a sensorgram obtained from a reference flow cell with no immobilized protein. Anti-cANGPTL4 antibodies run against the immobilized cANGPTL4 determined the Rmax value to be 251.8 resonance units. Global fitting of the SPR data to a Langmuir 1:1 model was used to determine the dissociation constant (KD) with Scrubber 2 software. Values are given as the mean ± S.D. of five independent preparations of recombinant proteins.
Affinity Co-precipitation Assay
Purified recombinant His-tagged ANGPTL4, nANGPTL4, or cANGPTL4 was immobilized onto nickel-nitrilotriacetic acid resin (GE Healthcare). The resin was washed with wash buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 0.1% Triton X-100) to remove excess ANGPTL4. An equal amount of ANGPTL4-bound resin was dispensed and incubated with 500 ng of purified matrix protein in PBS at 25 °C for 30 min. The resin was then thoroughly washed with wash buffer. The unbound fractions were pooled, and the bound fractions were released by SDS-PAGE loading dye. Both fractions were analyzed by immunoblotting with their indicated antibodies. Resin treated with Tris-buffered saline was used as a control. In vivo co-immunoprecipitation was performed using corresponding antibodies as previously described (29).
Sucrose Gradient Sedimentation Assay
Proteins (1 μg) were allowed to interact at 4 °C for 2 h in 150 μl of 50 mm Tris, pH 8.0, and 100 mm NaCl. The protein mixture was size-fractionated by ultracentrifugation for 16 h at 132,000 × g at 18 °C through a 5-ml sucrose density gradient (25–40%). Fractions of 300 μl were collected, chloroform/ethanol-precipitated, and analyzed by Western blot using their respective antibodies.
In Situ Proximity Ligation Assay (PLA)
Keratinocytes subcultured overnight on glass chamber slides (Lab-Tek) or cryosections of wound biopsies were fixed with 4% paraformaldehyde for 15 min. The slides were washed twice with PBS and blocked for 1 h at room temperature with 2% BSA in PBS containing 0.1% Triton X-100 followed by incubation with the indicated antibody pairs overnight at 4 °C. The slides were washed as described above. DUOlinkTM in situ PLA was performed as recommended by the manufacturer (OLink Biosciences) using a slide incubated without primary antibody as a negative control. Triple PLA was performed as previously described with minor modifications (30). Rabbit anti-cANGPTL4 (in-house), mouse anti-integrin αvβ5, and goat anti-vitronectin antibodies (Chemicon) were used as proximity probes. DNA was ligated at 37 °C for 1 h. All probe sequences were as previously described (30) and were synthesized by Proligo (Sigma). As a negative control, rabbit anti-cANGPTL4 and mouse anti-integrin β5 proximity probes were omitted. Images were taken using a LSM710 confocal laser scanning microscope with a Plan-Apochromat 63×/1.40 oil objective and ZEN software (Carl Zeiss).
Knockdown of ANGPTL4 and Real-time PCR
siRNA against human ANGPTL4 and a scrambled sequence control were subcloned into the pFIV-H1/U6-puro siRNA lentivirus system (System Biosciences). An equimolar ratio of sense and antisense oligonucleotide mixture was heated to 95 °C for 5 min and allowed to anneal in 20 mm Tris, pH 7.8, 100 mm NaCl, and 0.2 mm EDTA by slow cooling to room temperature. The annealed oligonucleotide was phosphorylated using polynucleotide kinase before ligation with BbsI-linearized pFIV-H1/U6-puro siRNA vector. Ligated products were transformed into competent E. coli Top 10 bacteria and selected on Luria broth agar plates containing 80 μg/ml ampicillin. Positive clones were confirmed by DNA sequencing. Positive constructs were co-transfected with pPACK packing plasmids into 293TN cells using ExGen 500. Supernatant was collected 48 h post-transfection, and pseudovirus-containing precipitate was obtained by centrifugation at 50,000 × g for 90 min at 4 °C. Cells were transduced using Polybrene according to the manufacturer's recommendation. Transduced cells were enriched by 350 μg/ml puromycin selection for 2 weeks. Knockdown efficiency of ANGPTL4 and relative expression level of indicated genes were determined by quantitative PCR. All oligonucleotides and Taqman probes sequences were provided in supplemental Table S1. Control and ANGPTL4-knockdown keratinocytes were denoted as KCTRL and KANGPTL4, respectively. The interferon response detection kit was from System Biosciences.
Matrix Protein Degradation Assay
Purified ECM proteins (200 ng) were first allowed to interact with various recombinant ANGPTL4 proteins (200 ng) before incubation at 37 °C with either WF or serum-free KANGPTL4-conditioned medium (CM). At the indicated times, aliquots of the reaction were stopped by the addition of SDS-PAGE loading dye. CM was prepared as follows; 3 × 106 KANGPTL4 cells were subcultured in a 10-cm dish the day before treatment. The next day cells were treated with 50 μg/ml TNF-α in 3 ml of serum-free basal Quantum 153 medium for 12 h. CM was collected, sterile-filtered, and stored at −80 °C for use in assays. Three independent experiments from two WF samples were performed. Protease inhibition assays were performed using the protease inhibitors pepstatin A (8 μm), EDTA (8 mm), and PMSF (1 mm) either alone or in indicated combinations in the CM. The matrix proteins were analyzed by Western blot using the corresponding antibodies.
Statistical Analysis
Data were analyzed statistically by two-tailed Mann-Whitney tests using SPSS software. Values were expressed as the mean ± S.E., and p < 0.05 was considered statistically significant.
RESULTS
ANGPTL4 Expression Is Regulated by PPARβ/δ in Keratinocytes
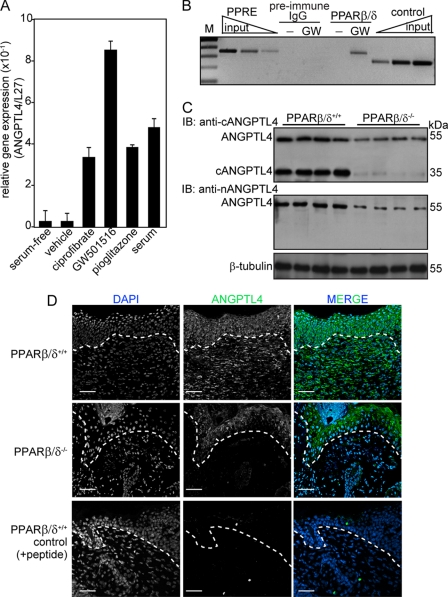
ANGPTL4 is a direct target gene of PPARβ/δ in HaCaT cells, a non-tumorigenic human keratinocyte cell line (22). However, the role, expression, and regulation of ANGPTL4 in skin wound healing is unclear. To this end, we first examined the expression level of ANGPTL4 in human keratinocytes after ligand activation of specific PPAR isotypes. Quantitative PCR revealed that ANGPTL4 mRNA was up-regulated by all three PPAR isotypes (Fig. 1A), with an ∼8.5-fold induction with the specific PPARβ/δ ligand, GW501516. We found that serum, which contains undefined and complex mixture of lipid metabolites that can act as ligands for PPARs, also increased ANGPTL4 expression by ∼5-fold (Fig. 1A). Next, the PPAR response element of the ANGPTL4 (22) from human keratinocytes was analyzed by ChIP using monoclonal anti-PPARβ/δ. Results showed that PPARβ/δ was bound to this site of the ANGPTL4 promoter region (Fig. 1B), indicating that ANGPTL4 is a direct target of PPARβ/δ in keratinocytes. Immunoblot analysis of day 3 wound biopsies using different polyclonal anti-ANGPTL4 antibodies detected the native ANGPTL4 and cANGPTL4 in the PPARβ/δ+/+ mice, whereas the expression of ANGPTL4 was reduced in PPARβ/δ−/− littermates (Fig. 1C). The specificity of anti-cANGTPL4 is shown in supplemental Fig. S1A and specificity of anti-nANGPTL4 was as previously reported (15). Day 3 wound biopsies were used because PPARβ/δ expression peaked at day 3 post-wounding (31). Immunofluorescence staining further confirmed that ANGPTL4 was highly expressed in both the wound epithelia and the wound bed in PPARβ/δ+/+ mice, whereas reduced expression was detected in their PPARβ/δ+/+ littermates (Fig. 1D). Quantitative PCR analysis of the LCM wound epithelium, dermis, and wound bed of day 3 wound biopsies from PPARβ/δ+/+ and PPARβ/δ+/+ mice showed that the wound epithelium was the major producer of ANGPTL4 (supplemental Fig. S1B). These results suggested that ANGPTL4 secreted by wound keratinocytes may play an important role during wound healing.
FIGURE 1.
Reduced expression of ANGPLT4 in PPARβ/δ−/− mice wounds. A, relative mRNA levels of ANGPTL4 in human keratinocytes treated with different agonists selective for each PPAR isotype, ciprofibrate (30 μm, PPARα), GW501516 (2 nm, GW, PPARβ/δ), and pioglitazone (500 nm, PPARγ) are shown. Values are mean ± S.E. of four independent studies. Ribosomal protein L27 used as a normalizing housekeeping gene. B, ChIP was done in keratinocytes using pre-immune IgG or monoclonal anti-PPARβ/δ. Regulatory region with the PPAR response element was immunoprecipitated with anti-PPARβ/δ and specifically amplified. No amplified signal was obtained with preimmune IgG. A control region upstream of PPAR response element served as the negative control. Aliquots of chromatin were analyzed before immunoprecipitation (input). M, 100-bp DNA marker. C, expression is shown of ANGPTL4 protein in day 3 post-wounding mice skin biopsies. Polyclonal antibodies that recognized the N-terminal (anti-nANGPTL4) and C-terminal (anti-cANGPTL4) of ANGPTL4 were used. β-Tubulin served as loading and transfer control. n = 5. IB, immunoblot. D, shown is immunofluorescence staining of ANGPTL4 in PPARβ/δ+/+ and PPARβ/δ−/− day 3 wound biopsies using anti-cANGPTL4. Sections were counterstained with DAPI. Negative control is performed with anti-cANGPTL4 preincubated with antigen peptide. Representative images from wound epithelia and wound beds were shown (n = 5). The dotted white line denotes epidermal-dermal junction. Scale bar, 40 μm.
cANGPTL4 Interacts with Specific Matrix Proteins
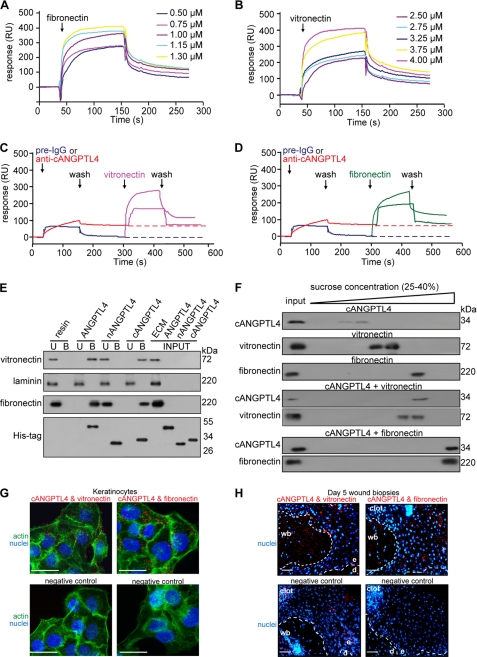
To begin to understand the role of ANGPTL4 during wound healing, we sought to identify ANGPTL4-binding proteins using SPR-MS. Prompted by our initial observation (Fig. 1D), we hypothesized that WF may harbor ANGPTL4-interacting proteins. Using recombinant cANGPTL4 and WF as the bait and lysate, respectively, we identified the ECM proteins, vitronectin and fibronectin, as ANGPTL4 binding partners (supplemental Fig. S1C). Recombinant cANGPTL4 was expressed and purified from Drosophila S2 culture medium (supplemental Fig. S1D). Further analyses using SPR with purified vitronectin and fibronectin and ANGPTL4 revealed binding constants (KD) of ∼10−7 m (ANGPTL4 with fibronectin and vitronectin, 3.80 ± 1.74 × 10−7 and 3.04 ± 1.33 × 10−7 m, respectively; cANGPTL4 with fibronectin and vitronectin, 3.52 ± 1.41 × 10−7 and 5.94 ± 1.79 m, respectively. Fig. 2, A and B). Specific anti-cANGPTL4 antibodies against immobilized cANGPTL4 determined the theoretical Rmax value to be 251.8 resonance units. The experimental Rmax values of fibronectin and vitronectin for cANGPTL4 were 238.6 and 218.5 resonance units, respectively, suggesting a 1:1 stoichiometry of binding. This interaction was specific, as the binding of anti-cANGTPL4 antibody, but not pre-immune IgG, to immobilized-ANGPTL4 blocked its interaction with vitronectin and fibronectin (Fig. 2, C and D). Specific interactions between cANGPTL4 with vitronectin and fibronectin were confirmed by in vitro affinity co-immunoprecipitation (Fig. 2E). In addition, we also examined the formation of the cANGPTL4-matrix protein complex by sedimentation using sucrose gradient ultracentrifugation, which separates proteins and protein complexes according to their native molecular weight, with larger proteins/complexes sedimenting at a higher sucrose density. Consistent with our previous results, immunodetection after sucrose gradient ultracentrifugation showed that cANGPTL4 associated with either vitronectin or fibronectin, which were detected in higher density fractions than the individual protein (Fig. 2F). No interaction between nANGPTL4 with identified matrix proteins was observed by either SPR analysis or co-immunoprecipitation (Fig. 2E).
FIGURE 2.
cANGPTL4 interacts with vitronectin and fibronectin. Sensograms show binding profiles between immobilized-cANGPTL4 and the indicated concentrations of either fibronectin (A) or vitronectin (B). A representative sensorgram (n = 5) shows binding profiles of vitronectin (C, pink) and fibronectin (D, green) with immobilized-cANGPTL4 CM5 chip after preblocking with either pre-immune IgG (blue) or anti-cANGPTL4 (red). E, co-immunoprecipitation assays (n = 5) were carried out by using different forms of His-tagged ANGPTL4 proteins immobilized on nickel-Sepharose and incubated with the indicated purified matrix molecules. Matrix proteins and ANGPTL4 were detected by immunodetected using corresponding antibodies and revealed by chemiluminescence. U and B denote the unbound/washed and bound fractions from the resin, respectively. RU, resonance units. F, shown is immunodetection of the indicated proteins from sucrose density gradient fractions. The proteins were allowed to interact in the indicated combinations before separation by sucrose gradient ultracentrifugation. Blots showed increasing sucrose density from left to right. An aliquot of the indicated protein, prior incubation, and centrifugation was also loaded (input) (n = 5). Detection of various complexes between ANGPTL4 and the indicated binding partners in KCTRL (G) and in Day-5 (H) wound biopsies using PLA is shown. PLA signals (red), nuclei stained with Hoechst dye (blue), and actin stress fiber (green) by Alexa488-phalloidin is shown. The nuclei-image has been acquired in one z-plane using LSM710 confocal microscope. The dotted white line represents epidermal-dermal junction. Negative control is performed without primary antibodies. Representative pictures from wound section with epidermis (e), dermis (d), the adjacent wound bed (wb), and KCTRL from six independent experiments or sections from three mice are shown. Scale bar, 40 μm.
In situ PLAs using indicated antibody pairs on primary human keratinocytes and day 5 wound sections were subsequently used to examine whether these interactions occur in vivo. These experimental PLA signals, visualized as individual red dots, were observed in the wound bed as well as in the wound epithelium (Fig. 2, G and H), confirming that cANGPTL4 interacts with vitronectin and fibronectin in vivo. As a positive control, vitronectin was shown to interact with integrin β5 (supplemental Fig. S2A), whereas the negative control recommended by the manufacturer revealed negligible nonspecific binding of PLA probes (Fig. 2, G and H, and supplemental Fig. S2A). Taken together, these data indicate that ANGPTL4 directly interacts with specific matrix proteins in the wound bed during wound healing.
ANGPTL4-bound Matrix Protein Can Interact with Integrins
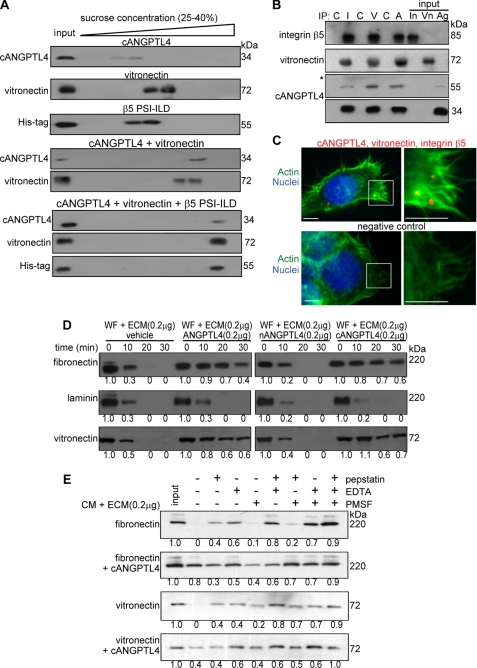
Having shown that ANGPTL4 interacts with vitronectin and fibronectin, it was necessary to determine whether this interaction would interfere with integrin recognition of the matrix protein. To this end, we examined the ability of ANGPTL4-bound vitronectin to interact with the extracellular domain PSI-ILD of integrin β5. Purified recombinant cANGPTL4 with no histidine tag, histidine-tagged integrin β5 PSI-ILD, and vitronectin were allowed to interact in solution and then sedimented using sucrose gradient ultracentrifugation. When the three proteins were present together, all three proteins were detected at a higher sucrose density, suggesting that ANGPTL4-bound matrix protein could still interact with PSI-ILD of integrin β5 (Fig. 3A). In vivo co-immunoprecipitation and triple PLA were used to further confirm this observation. Co-immunoprecipitation from human keratinocyte lysate using antibodies against integrin αvβ5, vitronectin, or cANGPTL4 followed by immunodetection showed that the corresponding two proteins were also found in the immunoprecipitates (Fig. 3B). Finally, triple PLA further revealed the close proximity of ANGPTL4, integrin β5, and vitronectin at focal adhesions (Fig. 3C). In retrospect, the PLA signals from ANGPTL4 and matrix proteins detected in wound epithelium (Fig. 2H) represented ANGPTL4-bound matrix proteins that had interacted with their cognate integrins in keratinocytes. Taken together, our results suggest that the binding of ANGPTL4 to matrix proteins, such as vitronectin, does not prevent the matrix protein from associating with its cognate integrin.
FIGURE 3.
ANGPTL4 modulates matrix protein degradation. A, shown are immunoblots of cANGPTL4, His-tagged integrin β5 PSI-ILD, and vitronectin of sucrose density gradient fractions. The proteins were allowed to interact in the indicated combinations before separation by sucrose density gradient ultracentrifugation. Blots showed increasing sucrose density from left to right. B, in vivo co-immunoprecipitation (IP; n = 3) was performed by using antibodies against integrin β5 (I), vitronectin (V), and cANGPTL4 (A). As control, pre-immune IgG (C) was used. Antibodies were covalently cross-linked to Protein G on agarose beads were incubated with total keratinocyte cell lysate. Immunoprecipitates were detected by immunoblot using corresponding antibodies and revealed by chemiluminescence. Native ANGPTL4 of ∼55 kDa was after longer exposure time as denoted by asterisk. Total cell lysate served as the input. (In, integrin β5; Vn, vitronectin; Ag, ANGPTL4). C, a triple PLA showed the ternary complex in keratinocytes. Triple PLA signals (red) nuclei were stained with Hoechst dye (blue) and Alexa488-phallodin for actin fiber (green). Representative PLA images from three independent experiments are shown. Negative control is without anti-cANGPLT4 and anti-integrin proximity probes. Scale bar, 40 μm. Immunodetection of the matrix proteins, vitronectin, and fibronectin after incubation for indicated time with WF (D) or TNF-α-treated KANGPTL4 (E) CM in the presence of indicated protease inhibitors is shown. Laminin, which does not interact with cANGPTL4, serves as control. Three independent experiments from two wound fluids were performed. Values below denote change in mean -fold expression compared with input.
ANGPTL4 Interacts with Matrix Proteins and Delays Their Degradation
Directed migration of wound keratinocytes over the provisional wound bed requires the controlled turnover of matrix proteins by proteases (32). We next examined the effect of the interaction between ANGPTL4 and matrix proteins on the turnover rate of matrix proteins. We preincubated purified ECM proteins with various recombinant ANGPTL4 proteins and subjected the mixture to WF. Our results revealed that the degradation of vitronectin and fibronectin was slower in the presence of ANGPTL4 and cANGPTL4 compared with either the vehicle control or nANGPTL4 (Fig. 3D). As a control, laminin-5, which does not bind to cANGPTL4, was degraded at a similar rate regardless of the presence of ANGPTL4 (Fig. 3D).
To eliminate the possible contribution of endogenous ANGPTL4 from WF, we performed a similar matrix degradation experiment with serum-free CM. We initially suppressed endogenous ANGPTL4 expression by RNA interference in human keratinocytes. Keratinocytes were transduced with a lentivirus-mediated ANGPTL4 or control scrambled siRNA. The ANGPTL4 expression level in ANGPTL4-knockdown keratinocytes (KANGPTL4) was reduced by 90% compared with control siRNA keratinocytes (KCTRL) (supplemental Fig. S2B). The expression of β-tubulin remained unchanged, as did the transfer and loading control. The expression of ANGPTL3, a closely related member of the family, remained unchanged, indicating the specificity of the knockdown. The induction of interferon responses has been reported as a challenge to the specificity of some RNAi approaches (33). Real-time PCR analysis of key interferon response genes OAS1, OAS2, MX1, and ISGF3γ revealed no significant difference between KANGPTL4 and either wild type non-transduced cells or KCTRL (supplemental Fig. S2C). These data suggest that gene silencing is not associated with nonspecific interferon-response induction, namely, an off-target effect. Next, we stimulated the expression of proteases in KANGPTL4 by TNF-α treatment and used the resulting serum-free CM for a matrix protein degradation assay. Consistent with our previous results, the degradation of vitronectin and fibronectin was slower in the presence of ANGPTL4 and cANGPTL4 (supplemental Fig. S2D). Using different protease inhibitors, we further showed that ANGPTL4 mainly protected the degradation of vitronectin and fibronectin from MMPs (Fig. 3E). SPR analysis failed to detect any interaction between recombinant MMP2 or MMP9 and cANGPTL4, arguing against a direct role of ANGPTL4 in the inhibition of MMPs (supplemental Fig. S2E). Taken together, our results showed a physical interaction between ANGPTL4 with specific matrix proteins that resulted in the selective delay of the degradation of matrix proteins by MMPs during wound healing.
ANGPTL4 Deficiency Delays Wound Re-epithelialization
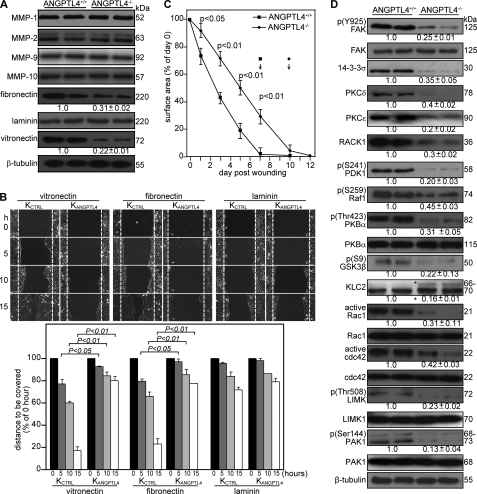
We showed that ANGPTL4 produced by keratinocytes interacts with vitronectin and fibronectin in the wound bed and delays their proteolytic degradation by MMPs. To underscore the in vivo relevance of ANGPTL4 in the degradation of specific matrix proteins by MMPs, we examined the expression of MMPs and matrix proteins in ANGPTL4+/+ and ANGPTL4−/− wound biopsies. Keratinocytes synthesize and secrete mainly MMP-1, -2, -9 and -10, and their expression is required to regenerate the injured tissue (32). Immunoblot analysis of day 5 ANGPTL4+/+ and ANGPTL4−/− wound biopsies showed that the protein levels of vitronectin and fibronectin, but not laminin, was reduced (Fig. 4A). Our analysis did not reveal significant differences in the protein level of major MMPs (Fig. 4A), indicating that the differential matrix protein level was a consequence of increased susceptibility of matrix proteins to proteolytic degradation.
FIGURE 4.
ANGPTL4 knock-out mice displayed impaired wound re-epithelialization. A, shown is an immunoblot analysis of MMP-1, -2, -9, -10, fibronectin, laminins, and vitronectin from day 5 wound biopsies of ANGPTL4+/+ and ANGPTL4−/− mice. Values below the band represent the mean -fold differences in protein expression levels relative to ANGPTL4+/+ from eight wound biopsies for each genotype. β-Tubulin served as loading and transfer controls. B, shown are wound closure kinetics of KCTRL and KANGPTL4 treated with mitomycin C (2 μg/ml) on the indicated matrix protein-coated surfaces. Representative time-lapsed images of wounded cultures are shown. Yellow dotted lines represent the scratch gap at the time of wounding. The graph shows the distance to be covered by the migrating keratinocytes as the percentage of 0 h (=100%) in vitro wound gap distance (±S.E., n = 5, using the Mann-Whitney test). C, shown are the wound closure kinetics of ANGPTL4+/+ and ANGPTL4−/− mice. Wound surface areas are plotted as percentage of day 0 (=100%) wound surface area (±S.E., n = 10, using the Mann-Whitney test). Arrows indicate the mean time for complete wound closure. D, shown are immunoblot analyses of the indicated proteins from ANGPTL4+/+ and ANGPTL4−/− mice day 5 wound biopsies (n = 5). Values below the bands represent the mean -fold differences in protein expression levels when compared with ANGPTL4+/+, which was assigned the value 1. GSK-3, glycogen synthase kinase-3; KLC, kinesin-light chain; PAK, p21-activated kinase; PDK1, 3-phosphoinositide-dependent kinase-1; LIMK, LIM kinase. β-Tubulin served as the loading and transfer control.
We hypothesized that such actions would have a direct impact on wound healing. We first examined keratinocyte migration using in vitro scratch wound assays on surfaces coated with matrix proteins using KCTRL and KANGPTL4 treated with mitomycin C to exclude any effects of proliferation. Our results showed that KANGPTL4 re-populated the in vitro wound significantly more slowly on fibronectin- and vitronectin-coated surfaces compared with KCTRL (Fig. 4B). No significant difference was observed on laminin-coated surfaces. Next, we examined the healing of full-thickness skin wounds in ANGPTL4+/+ and ANGPTL4−/− mice. Our analysis of the day 3–10 wound biopsies showed a delayed re-epithelialization of ANGPTL4−/− wounds compared with ANGPTL4+/+ (Fig. 4C). The length of the wound epidermis measured from the first hair follicle to the tip of the wound epithelial tongue is used as an indicator of keratinocyte migration, and this was also reduced in ANGPTL4−/− wounds (supplemental Fig. S3A). No difference in wound contraction, defined by the distance between the first hair follicle on either side of the wound edge, was observed. Immunohistochemical staining of wound biopsies for keratin 6 identifies the wound epithelia and hair follicles, whereas α-smooth muscle actin reveals the myofibroblasts (supplemental Fig. S3, B and C). We harvested the entire wound along with a 5-mm perimeter of the surrounding tissue. Cell counts indicated cell number per wound at day 3 (ANGPTL4+/+ 3.20 ± 0.39 × 106; ANGPTL4−/− 3.01 ± 0.44 × 106, n = 4) and day 7 (ANGPTL4+/+ 3.54 ± 0.26 × 106; ANGPTL4−/− 3.33 ± 0.54 × 106, n = 4). The cell suspension was analyzed on FACS after staining with antibodies against F4/80 for macrophages showed no significant difference between ANGPTL4−/− and ANGPTL4+/+ biopsies (day 3, ANGPTL4+/+ 15.1 ± 1.2%; ANGPTL4−/− 14.1 ± 2.1%; day 7, ANGPTL4+/+ 18.3 ± 2.6%; ANGPTL4−/− 18.7 ± 3.8% of cells in wound, n = 3) (supplemental Fig. S3D). We observed a consistently lower number of CD31+ endothelial cells in ANGPTL4−/− compared with ANGPTL4+/+ wounds (day 3, ANGPTL4+/+ 15.7 ± 2.4%; ANGPTL4−/− 10.1 ± 1.8%; day 7, ANGPTL4+/+ 19.7 ± 3.6%; ANGPTL4−/− 14.7 ± 2.8% of cells in wound, n = 3) (supplemental Fig. S3E). Our results showed that ANGPTL4 deficiency delays wound re-epithelialization associated, at least in part, with an increase in matrix protein degradation.
ANGPTL4 Deficiency Affects Focal Adhesion Kinase- and 14-3-3σ-dependent Signaling Pathways
Given that integrins are receptors for matrix proteins and having shown that ANGPTL4 deficiency affects matrix protein integrity and wound healing, it is conceivable that the underlying mechanism involves integrin-mediated signaling. Indeed, the expression or phosphorylation of downstream effectors like focal adhesion kinase- and 14-3-3-dependent signaling cascades was reduced in ANGPTL4−/− compared with ANGPTL4+/+ wounds (Fig. 4D). 14-3-3 associates with integrins to modulate cell migration via a focal adhesion kinase-independent mechanism involving protein kinase C (PKC) (34). ANGPTL4−/− wounds also showed decreased expression of RACK1, indicating an attenuated PKC-mediated signal transduction (Fig. 4D) (35). A reduced activation of focal adhesion kinase is also known to converge with a decreased activation of the Raf-MEK-ERK signaling pathway (36).
The downstream mediators of the PI3K cascade such as 3-phosphoinositide-dependent kinase-1 (PDK1), PKBα, and glycogen synthase kinase 3β (GSK-3β) were also altered (Fig. 4D). Glycogen synthase kinase 3β is a target of PKBα known to phosphorylate kinesin light chain and, thus, to negatively regulate kinesin-based motility and integrin recycling (37, 38). We observed hyperphosphorylated kinesin light chain 2 in ANGPTL4−/− wounds, which suggested that integrin recycling may be impaired in ANGPTL4-deficient keratinocytes (Fig. 4D). Small Rho GTPases are effectors of PI3K pathway. Among them, cdc42 and Rac1 are pivotal intracellular mediators for the formation of lamellipodia and cell migration. They activate downstream effectors such as p21-activated kinase, which in turn activate LIM kinases (39). ANGPTL4 deficiency led to a reduction in the phosphorylation of p21-activated kinase 1 (PAK1) and LIM kinase1 (LIMK1) (Fig. 4D). These would have a direct impact on lamellipodia formation and migration, consistent with our earlier observation that wound healing was delayed in KANGPTL4 (Fig. 4B). Taken together, our results show that ANGPTL4 deficiency impairs the activation of numerous integrin-initiated downstream signaling cascades, including focal adhesion kinase and 14-3-3, to mediate gene expression involved in cell migration.
DISCUSSION
Wound healing is a complex process that involves a cascade of overlapping events, including inflammation, re-epithelialization, and remodeling, all directed at the restoration of the epidermal barrier. Re-epithelialization is accomplished by increased keratinocyte proliferation and the guided migration of the keratinocytes over the wound. Cellular interactions with ECM proteins, i.e. cell-matrix communication, among others, coordinates the individual events, enabling temporal and spatial control as well as ordered changes in keratinocyte behavior and phenotype. We revealed a newly discovered cell-matrix communication role for ANGPTL4 in influencing wound re-epithelialization. ANGPTL4 binds and delays the degradation of specific matrix proteins; they are, thus, available as intact components of ECM to regulate cell-matrix communication.
During wound healing, migrating cells must display appropriate cellular behavior in response to the changing wound environment to enable effective wound closure. Integrins on the cell surface function as biosensors to constantly monitor changes in the microenvironment. Simultaneously, the context in which the cognate matrix protein is presented to the cells dictates productive integrin activation. At a low protein ratio of soluble to substrate-anchored matrix that is well below that required for blocking adhesion, one may observe an accelerated turnover of the integrin-matrix protein interaction (40). The deficiency in ANGPTL4 has a dramatic effect on wound closure. ANGPTL4 binds to specific matrix proteins via its C-terminal fibrinogen-like domain and delays their degradation by proteases. This association, however, does not interfere with integrin-matrix protein recognition. Instead, it directly affects integrin-mediated signaling by altering the balance between substrate-anchored matrix proteins and soluble matrix protein fragments, thereby modifying the availability of the local substrate-anchored ECM and consequently modulating cellular behavior.
Numerous studies have shown that inflammation-induced PPARβ/δ is crucial for wound repair. PPARβ/δ confers anti-apoptotic properties to keratinocytes, in part via transcriptional control of the PKBα signaling pathway and by maintaining a sufficient number of viable wound keratinocytes at the wound edge (10, 12). Recently, PPARβ/δ was shown to potentiate cell polarization and directed migration during re-epithelialization (13). Most of these studies focused on intracellular signaling or events mediated by PPARβ/δ that were important for cell survival and migration. Clearly, cell-matrix communication is needed for effective directed cell migration during wound healing. However, the mechanism by which PPARβ/δ modifies the wound microenvironment to coordinate cell-matrix communication remains unknown. Conceivably, as an intracellular transcription factor, PPARβ/δ is likely to exert such an effect via an extracellular factor. The expression of ANGPTL4, only weakly detectable in normal intact skin, was markedly elevated during the re-epithelialization phase of wound healing, as was similarly observed in PPARβ/δ-knock-out mice (31). We provide evidence that PPARβ/δ stimulates the expression of the adipocytokine, ANGPTL4, in keratinocytes, which allows the migrating keratinocytes to modulate the wound microenvironment and to coordinate with cellular responses. Similar to its effect on integrin-matrix interactions, integrin-focal adhesion kinase-mediated signaling and key intracellular signaling cascades involved in actin polymerization and for the establishment of a leading lamellipodium in migrating cells are dependent on ANGPTL4. Besides regulating cell proliferation, PI3K/PKBα, PDK1, and glycogen synthase kinase-3β are involved in the coordinated assembly and disassembly of actin filaments and integrin recycling and contribute to the motility of rapidly migrating cells, such as wound keratinocytes (42). The ANGPTL4-knock-out mice did not have any obvious skin abnormalities but displayed altered epidermal differentiation (data not shown). This suggests that a low level of ANGPTL4 may be required for normal skin homeostasis and may play an important role during wound repair. The role of PPARβ/δ in epidermal differentiation and maintenance of a lipid barrier is well recognized, although the underlying mechanism remains unclear (43). Although not the focus of this study, we observed that the expression of 14-3-3, as assessed by immunoblot, was diminished in ANGPTL4−/− compared with ANGPTL4+/+ mice, and this reduction may play a role in regulating epidermal differentiation. Studies have shown that 14-3-3 associates with PKC, which has a well established role in epidermal differentiation (44).
Emerging works have shown adipocytokines such as leptin, to have a profound local impact on wound healing (45). However, the mechanism for the observed beneficial effect on wound repair is unclear, and research efforts are currently directed toward understanding the molecular regulation. We identified ANGPTL4, an adipocytokine, as having a beneficial effect on wound healing in part due to its effect on the integrity of matrix proteins and cell-matrix communication. Given that ANGPTL4 is involved in lipid and glucose homeostasis and that ANGPTL4 decreases blood glucose and improves glucose tolerance in mice (41), it is a prime therapeutic candidate for diseases such as diabetes and for wound healing. A better understanding of its role in wound healing, especially in diabetic and chronic wounds, would provide better wound management. Altogether, ANGPTL4 modulates cell-matrix communications through its interactions with and effects on matrix proteins. Importantly, it provides a novel means by which migrating wound keratinocytes can scrutinize the changes in the wound ECM and modulate their cell behavior.
Supplementary Material
Acknowledgments
We thank Dr. Samuel Ko and Anna Teo (Carl Zeiss, Singapore Pte Ltd.) for expertise in image acquisition using the LSM710 confocal microscope, Mirax MIDI, and PALM Laser-capture microdissection.
This work was supported by A*STAR BMRC Grant 05/1/22/19/377, Ministry of Education Grant ARC 18/08, and Nanyang Technological University Grants RGD 127/05 and 158/06.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
- ECM
- extracellular matrix
- ANGPTL4
- angiopoietin-like 4
- nANGPTL4
- N-terminal coiled-coil fragment
- KANGPTL4
- human keratinocytes knockdown of ANGPTL4
- cANGPTL4
- C-terminal fibrinogen-like domain
- KCTRL
- human keratinocytes with control scrambled siRNA
- MMP
- matrix metalloproteinase
- PLA
- proximity ligation assay
- PPAR
- peroxisome proliferator-activated receptor
- SPR
- surface plasmon resonance
- WF
- wound fluid/exudate
- LCM
- laser-capture microdissection
- CM
- conditioned medium
- FACS
- fluorescence-activated cell sorter.
REFERENCES
- 1.Hynes R. O. (2002) Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 2.Werner S., Grose R. (2003) Physiol. Rev. 83, 835–870 [DOI] [PubMed] [Google Scholar]
- 3.Bornstein P., Sage E. H. (2002) Curr. Opin Cell Biol. 14, 608–616 [DOI] [PubMed] [Google Scholar]
- 4.Bulcão C., Ferreira S. R., Giuffrida F. M., Ribeiro-Filho F. F. (2006) Curr. Diabetes Rev. 2, 19–28 [DOI] [PubMed] [Google Scholar]
- 5.Caswell P. T., Norman J. C. (2006) Traffic 7, 14–21 [DOI] [PubMed] [Google Scholar]
- 6.Ginsberg M. H., Partridge A., Shattil S. J. (2005) Curr. Opin Cell Biol. 17, 509–516 [DOI] [PubMed] [Google Scholar]
- 7.Giannone G., Sheetz M. P. (2006) Trends Cell Biol. 16, 213–223 [DOI] [PubMed] [Google Scholar]
- 8.Tan N. S., Michalik L., Desvergne B., Wahli W. (2003) Am. J. Clin. Dermatol. 4, 523–530 [DOI] [PubMed] [Google Scholar]
- 9.Grose R., Werner S., Kessler D., Tuckermann J., Huggel K., Durka S., Reichardt H. M., Werner S. (2002) EMBO Rep. 3, 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan N. S., Michalik L., Noy N., Yasmin R., Pacot C., Heim M., Flühmann B., Desvergne B., Wahli W. (2001) Genes Dev. 15, 3263–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan N. S., Michalik L., Desvergne B., Wahli W. (2004) Expert Opin. Ther. Targets. 8, 39–48 [DOI] [PubMed] [Google Scholar]
- 12.Di-Poï N., Tan N. S., Michalik L., Wahli W., Desvergne B. (2002) Mol. Cell 10, 721–733 [DOI] [PubMed] [Google Scholar]
- 13.Tan N. S., Icre G., Montagner A., Bordier-ten-Heggeler B., Wahli W., Michalik L. (2007) Mol. Cell. Biol. 27, 7161–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oike Y., Akao M., Kubota Y., Suda T. (2005) Trends Mol. Med. 11, 473–479 [DOI] [PubMed] [Google Scholar]
- 15.Mandard S., Zandbergen F., Tan N. S., Escher P., Patsouris D., Koenig W., Kleemann R., Bakker A., Veenman F., Wahli W., Müller M., Kersten S. (2004) J. Biol. Chem. 279, 34411–34420 [DOI] [PubMed] [Google Scholar]
- 16.Belanger A. J., Lu H., Date T., Liu L. X., Vincent K. A., Akita G. Y., Cheng S. H., Gregory R. J., Jiang C. (2002) J. Mol. Cell Cardiol. 34, 765–774 [DOI] [PubMed] [Google Scholar]
- 17.Minn A. J., Gupta G. P., Siegel P. M., Bos P. D., Shu W., Giri D. D., Viale A., Olshen A. B., Gerald W. L., Massagué J. (2005) Nature 436, 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padua D., Zhang X. H., Wang Q., Nadal C., Gerald W. L., Gomis R. R., Massagué J. (2008) Cell 133, 66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan N. S., Ho B., Ding J. L. (2002) Protein Eng. 15, 337–345 [DOI] [PubMed] [Google Scholar]
- 20.Rheinwald J. G., Green H. (1977) Nature 265, 421–424 [DOI] [PubMed] [Google Scholar]
- 21.Rheinwald J. G., Green H. (1975) Cell 6, 331–343 [DOI] [PubMed] [Google Scholar]
- 22.Schug T. T., Berry D. C., Shaw N. S., Travis S. N., Noy N. (2007) Cell 129, 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chong H. C., Tan M. J., Philippe V., Tan S. H., Tan C. K., Ku C. W., Goh Y. Y., Wahli W., Michalik L., Tan N. S. (2009) J. Cell Biol. 184, 817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan N. S., Michalik L., Desvergne B., Wahli W. (2005) J. Biol. Chem. 280, 18163–18170 [DOI] [PubMed] [Google Scholar]
- 25.Tan S. H., Pal M., Tan M. J., Wong M. H., Tam F. U., Teo J. W., Chong H. C., Tan C. K., Goh Y. Y., Tang M. B., Cheung P. C., Tan N. S. (2009) J. Biol. Chem. 284, 18047–18058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Köster A., Chao Y. B., Mosior M., Ford A., Gonzalez-DeWhitt P. A., Hale J. E., Li D., Qiu Y., Fraser C. C., Yang D. D., Heuer J. G., Jaskunas S. R., Eacho P. (2005) Endocrinology 146, 4943–4950 [DOI] [PubMed] [Google Scholar]
- 27.Chen L., Tredget E. E., Wu P. Y., Wu Y. (2008) PloS one 3, e1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan N. S., Ho B., Ding J. L. (2000) FASEB J. 14, 859–870 [DOI] [PubMed] [Google Scholar]
- 29.Tan N. S., Michalik L., Di-Poï N., Ng C. Y., Mermod N., Roberts A. B., Desvergne B., Wahli W. (2004) EMBO J. 23, 4211–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. (2006) Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 31.Michalik L., Desvergne B., Tan N. S., Basu-Modak S., Escher P., Rieusset J., Peters J. M., Kaya G., Gonzalez F. J., Zakany J., Metzger D., Chambon P., Duboule D., Wahli W. (2001) J. Cell Biol. 154, 799–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Page-McCaw A., Ewald A. J., Werb Z. (2007) Nat. Rev. Mol. Cell Biol. 8, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bridge A. J., Pebernard S., Ducraux A., Nicoulaz A. L., Iggo R. (2003) Nat. Genet. 34, 263–264 [DOI] [PubMed] [Google Scholar]
- 34.Dellambra E., Patrone M., Sparatore B., Negri A., Ceciliani F., Bondanza S., Molina F., Cancedda F. D., De Luca M. (1995) J. Cell Sci. 108, 3569–3579 [DOI] [PubMed] [Google Scholar]
- 35.Schechtman D., Mochly-Rosen D. (2001) Oncogene 20, 6339–6347 [DOI] [PubMed] [Google Scholar]
- 36.Porter G. W., Khuri F. R., Fu H. (2006) Semin Cancer Biol. 16, 193–202 [DOI] [PubMed] [Google Scholar]
- 37.Morfini G., Szebenyi G., Elluru R., Ratner N., Brady S. T. (2002) EMBO J. 21, 281–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts M. S., Woods A. J., Dale T. C., Van Der Sluijs P., Norman J. C. (2004) Mol. Cell. Biol. 24, 1505–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knaus U. G., Bokoch G. M. (1998) Int. J. Biochem. Cell Biol. 30, 857–862 [DOI] [PubMed] [Google Scholar]
- 40.Legler D. F., Wiedle G., Ross F. P., Imhof B. A. (2001) J. Cell Sci. 114, 1545–1553 [DOI] [PubMed] [Google Scholar]
- 41.Xu A., Lam M. C., Chan K. W., Wang Y., Zhang J., Hoo R. L., Xu J. Y., Chen B., Chow W. S., Tso A. W., Lam K. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6086–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enomoto A., Murakami H., Asai N., Morone N., Watanabe T., Kawai K., Murakumo Y., Usukura J., Kaibuchi K., Takahashi M. (2005) Dev. Cell 9, 389–402 [DOI] [PubMed] [Google Scholar]
- 43.Burdick A. D., Kim D. J., Peraza M. A., Gonzalez F. J., Peters J. M. (2006) Cell. Signal. 18, 9–20 [DOI] [PubMed] [Google Scholar]
- 44.Denning M. F. (2004) Int. J. Biochem. Cell Biol. 36, 1141–1146 [DOI] [PubMed] [Google Scholar]
- 45.Frank S., Stallmeyer B., Kämpfer H., Kolb N., Pfeilschifter J. (2000) J. Clin. Invest. 106, 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.