Abstract
Mps1 is a dual specificity protein kinase with key roles in regulating the spindle assembly checkpoint and chromosome-microtubule attachments. Consistent with these mitotic functions, Mps1 protein levels fluctuate during the cell cycle, peaking at early mitosis and abruptly declining during mitotic exit and progression into the G1 phase. Although evidence in budding yeast indicates that Mps1 is targeted for degradation at anaphase by the anaphase-promoting complex (APC)-cCdc20 complex, little is known about the regulatory mechanisms that govern Mps1 protein levels in human cells. Here, we provide evidence for the ubiquitin ligase/proteosome pathway in regulating human Mps1 levels during late mitosis through G1 phase. First, we showed that treatment of HEK 293T cells with the proteosome inhibitor MG132 resulted in an increase in both the polyubiquitination and the accumulation of Mps1 protein levels. Next, Mps1 was shown to co-precipitate with APC and its activators Cdc20 and Cdh1 in a cell cycle-dependent manner. Consistent with this, overexpression of Cdc20 or Cdh1 led to a marked reduction of endogenous Mps1 levels during anaphase or G1 phase, respectively. In contrast, depletion of Cdc20 or Cdh1 by RNAi treatment both led to the stabilization of Mps1 protein during mitosis or G1 phase, respectively. Finally, we identified a single D-box motif in human Mps1 that is required for its ubiquitination and degradation. Failure to appropriately degrade Mps1 is sufficient to trigger centrosome amplification and mitotic abnormalities in human cells. Thus, our results suggest that the sequential actions of the APC-cCdc20 and APC-cCdh1 ubiquitin ligases regulate the clearance of Mps1 levels and are critical for Mps1 functions during the cell cycle in human cells.
Keywords: Mitosis, Proteasome, Protein Degradation, siRNA, Ubiquitin, Cdc20, Cdh1, Mps1
Introduction
Ubiquitin- and proteasome-dependent proteolysis is one of the key mechanisms that ensure the uni-directional progression of the cell cycle. There are two main steps involved in the proteolysis of proteins by the ubiquitin/proteosome system as follows: the covalent attachment of multiple ubiquitin molecules to the protein substrate and the degradation of the polyubiquitinated protein by the 26 S proteosome complex (1). Polyubiquitination of proteins is mediated by a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3). Substrate specificity and efficiency are mainly conferred by the E3 ubiquitin ligase. The two main E3 ubiquitin ligases that play a critical role in the cell cycle are the SCF (Skp1, cullin, F-box) complex and the anaphase-promoting complex/cyclosome (APC-c)3 (2). The SCF complex is active throughout the cell cycle. The specificity of its target substrates is regulated by their phosphorylation and by the different phosphate-binding proteins (F box proteins) that associate with SCF (3). The APC-c complex regulates proteolysis of a number of key M phase substrates, including the mitotic cyclins (A and B-type cyclins) and securin, which is critical for cell cycle progression into anaphase and exits from mitosis into the subsequent G1 phase (4, 5). Activation of the APC-c is tightly regulated in a cell cycle-dependent manner through its association with Cdc20/p55CDC/Fizzy or Cdh1/Hct1/Fizzy-related activating proteins that provide substrate specificity and temporal control for polyubiquitination of substrates (1).
The monopolar spindle 1 (Mps1), also called TTK, is a conserved dual specificity protein kinase with various cell cycle functions. In budding yeast, Mps1 controls spindle-pole body duplication (6), spindle-kinetochore attachment (7), and the spindle checkpoint 8). Studies in higher eukaryotes show that Mps1 activity is required for the spindle checkpoint (9–11), correcting errors in spindle-chromosome attachment (12), and is implicated in centrosome duplication (13, 14), although this latter role is still controversial (10). Consistent with these roles, Mps1 protein localizes to centrosomes during G1/S phase (11, 13, 14) and kinetochores from early prophase to metaphase (11, 15). Mps1 mutations in yeast (16), flies (17), and zebrafish (18) cause chromosome segregation errors in meiosis, whereas overexpression of Mps1 has been shown in vertebrate tissue culture cells to induce centrosome amplification and multipolar spindles (13, 14). Therefore, Mps1 has roles in centrosome duplication, the spindle checkpoint, and chromosome segregation whose functions are exquisitely sensitive to dosage.
In human cells, the levels of Mps1 protein and kinase activity fluctuate throughout the cell cycle, peaking at early mitosis and dropping rapidly as cells exit mitosis and re-entered the subsequent G1 phase (10, 11). It was previously shown in budding yeast that Mps1 is degraded at anaphase by the action of APC-cCdc20, which is critical for allowing inactivation of the spindle checkpoint (19). Whether Mps1 is targeted for proteolysis in human proliferating cells and, if so, how this is regulated are currently unknown. In this study, we show that human Mps1 is targeted for degradation by the ubiquitin-proteasome pathway in a cell cycle-dependent manner through the sequential actions of APC-cCdc20 and APC-cCdh1. In addition, we identify a single D-box within the N-terminal region of human Mps1 that is conserved in most mammalian species. Mutation of the D-box (RXXL) residues to alanines (AXXA) suppresses its ubiquitination and degradation and triggers centrosome amplification and mitotic abnormalities in vivo. Thus, we conclude that human Mps1 is a target of the APC-c-ubiquitin-proteasome pathway, which undergoes proteolysis during anaphase through G1 phase to allow for proper centrosome duplication and cell cycle progression.
EXPERIMENTAL PROCEDURES
Plasmids
pHF36-GFP-Mps1 wild type plasmid was generous gift from Dr. Mark Winey (University of Colorado). Site-directed mutation constructs of Mps1 were generated using site-directed mutagenesis kit as described previously. PCR primer sets were custom ordered from Invitrogen to specifically change the consensus sequence RNSL (one putative D-box with RXXL motif within the N-terminal domain of Mps1) to ANSA. HA-hCdh1, HA-hCdc20, and FLAG-Skp2 were generously provided by Dr. Michele Pagano (Howard Hughes Medical Institute at New York University). All constructs were sequenced prior to use.
Cell Cultures
Human embryonic kidney (HEK) 293T cells were grown in Dulbecco modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and penicillin/streptomycin (100 IU/ml and 100 μg/ml, respectively) at 37 °C and in a 5% CO2 atmosphere. To test the in vivo interaction between different proteins with endogenous or exogenous Mps1, the proteasome inhibitor MG132 (25 μm) was added for 6 h prior to harvesting the cells. For experiments investigating the ability of Cdc20 or Cdh1 to induce the degradation of Mps1, Cdc20 or Cdh1 and Mps1 were co-transfected in a 3:1 ratio; cycloheximide (50 μm) was added 6 h prior to harvesting the cells. 293T cells were transfected using the calcium phosphate method as described.
Cell Synchronization
Cells were synchronized at late G1 phase using the thymidine double-blocking method (20). Briefly, 106 cells were plated in 60-mm Petri dishes, and thymidine was added to a final concentration of 2 mm after cell adherence (about 6–8 h). The cells were cultured for 16 h. After removal of the thymidine and incubation for 10 h in the fresh DMEM solution, thymidine was added to a final concentration of 2 mm for an additional 16 h. After removal of thymidine again, synchronized cells were cultured in fresh DMEM and collected at different times for cell cycle analysis and Western blotting. Cells were synchronized in pro-metaphase with 6–12 h of nocodazole treatment as described previously (21) and then released into fresh medium for further incubation (2 h, early G1 phase).
Cell Cycle Analysis Using Flow Cytometry
The thymidine-synchronized cells were collected at different times after release from a G1 block, and the nocodazole-synchronized cells were collected at 2 h after release into fresh medium. After washing twice with PBS solution, cells were fixed with chilled 70% alcohol at −20 °C for 24 h. The cell sediment was collected by centrifugation (1000 rpm, 3 min), washed twice with PBS solution, incubated with 20 μl of RNase A (20 mg/ml) for 30 min at 37 °C, and stained with 25 μg/ml propidium iodide (Sigma) for 30 min at room temperature. The cell cycle distribution was then evaluated using flow cytometry. All experiments were repeated three times.
Protein Stability Experiments
To determine the effects of proteasome inhibitors on Mps1 protein stability, cells were preincubated with 25 μm MG132 or 10 μm clasto-lactacystin (Peptide International, Inc., Louisville, KY) or with the corresponding volume of the vehicle dimethyl sulfoxide (DMSO) and harvested in radioimmunoprecipitation assay (RIPA) buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 10 mg/ml phenylmethylsulfonyl fluoride, aprotinin (2 μg/ml), and 100 mm sodium orthovanadate) at various time intervals indicated in the figures. Western blotting was performed using anti-Mps1 antibody to observe the protein accumulation. Actin was used as loading control. To analyze the stability of the Mps1 protein in anaphase and G1 phase, the cells were treated with nocodazole for 16 h. Nocodazole was then washed out, and the cells were replated for 1 h before cycloheximide (50 μm) was added to the medium. Cells were harvested at different time points after cycloheximide addition.
Gene Silencing by Small Interfering RNA
siRNA duplexes were transfected into cells using Oligofectamine (Invitrogen) according to the manufacturer's instructions and as described previously (34, 36). G1-arrested cells by a double thymidine exposure were transfected with siRNAs targeting hCdh1, whereas cells transfected with siRNAs targeting hCdc20 were synchronized in mitosis by nocodazole treatment and then released into fresh medium for different times. The siRNA oligonucleotide sequence for hCdh1 was 5′-AATGAGAAGTCTCCCAGTCAGTT-3′ (oligo 1, corresponding to nt 199–219 of human Cdh1 cDNA) and 5′-GAAGGGTCTGTTCACGTATT-3′ (oligo 2, corresponding to nt 372–391 of human Cdh1 cDNA). The siRNA oligonucleotide sequences for hCdc20 were 5′-AACGGCAGGACTCCGGGCCGATT-3′ (oligo 1, corresponding to nt 156–170 of human Cdc20 cDNA) and 5′-AATGGCCAGTGGTGGTAATGATT-3′ (oligo 2, corresponding to nt 969–989 of human Cdc20 cDNA). A 21-nt siRNA duplex corresponding to a nonrelevant gene (lacZ) was used as control.
Cell Extraction, Immunoprecipitation, and Immunoblotting
Cells were collected in RIPA buffer (composition given above), allowed to lyse on ice for 5 min, vortexed, and cleared by centrifugation in a microcentrifuge at 14,000 rpm for 8 min at 4 °C. Protein was subjected to Western blot analysis as described previously. Immunoprecipitation experiments were pursued according to standard procedures and as described previously. Briefly, 500 μg of soluble protein was first incubated with primary antibodies for 2 h at room temperature and further incubated overnight at 4 °C after addition of 30 μl of protein A/G-Sepharose beads (Santa Cruz Biotechnology, Santa Cruz, CA). To pull down the immunocomplexes, the beads were washed one time with RIPA buffer and washed three times with 1× PBS at 2,500 rpm for 5 min and finally suspended in 30 μl of 2× SDS- PAGE sample buffer. The immunoprecipitated proteins were separated by SDS-PAGE. Western blot analysis was performed as described previously. The following antibodies were used in this study: mouse monoclonal anti-Mps1 (1:1000-fold dilution) from Abcam (Cambridge, MA); mouse monoclonal anti-cyclin B1 (1:1000-fold dilution) and goat polyclonal anti-actin (1:1500-fold dilution) from Santa Cruz Biotechnology; mouse monoclonal anti-human Cdh1 (anti-hCdh1) and anti-human Cdc20 (hCdc20) (1:1000-fold dilution) from BD Biosciences; mouse monoclonal anti-Skp2 antibody from Invitrogen; and rabbit polyclonal anti-aurora-B antibody from Cell Signaling (Danvers, MA).
In Vivo Ubiquitination Assay
Cells in 100-mm plates were transfected with combinations of 5 μg of His6-ubiquitin expression plasmid with or without 5 μg of Mps1 expression plasmids, adding the proteasome inhibitor MG132 (25 μm) 6 h prior to lysis. Cells were lysed in buffer A (6 m guanidinium HCl, 0.1 m Na2HPO4/NaH2PO4, 0.01 m Tris-HCl, pH 8.0, 5 mm imidazole, 10 mm β-mercaptoethanol) and incubated with Ni2+-nitrilotriacetic acid beads (Qiagen, CA) for 4 h at room temperature. The beads were washed with buffer A (8 m urea, 0.1 m Na2PO4/NaH2PO4, 0.01 m Tris-HCl, pH 8.0, 10 mm β-mercaptoethanol) and buffer B (8 m urea, 0.1 m Na2PO4/NaH2PO4, 0.01 m Tris-HCl, pH 6.3, 10 mm β-mercaptoethanol), and bound proteins were eluted with buffer C (200 mm imidazole, 0.15 m Tris-HCl, pH 6.7, 30% glycerol, 0.72 m β-mercaptoethanol, 5% SDS). The eluted proteins were analyzed by Western blot for the presence of conjugated Mps1 by N1 monoclonal mouse anti-Mps1 antibody. In some experiments, cells were transfected with His6-ubiquitin plasmid and incubated without MG132 treatment.
RESULTS
Degradation of Mps1 Protein Is Targeted by the Ubiquitin-Proteosome Pathway
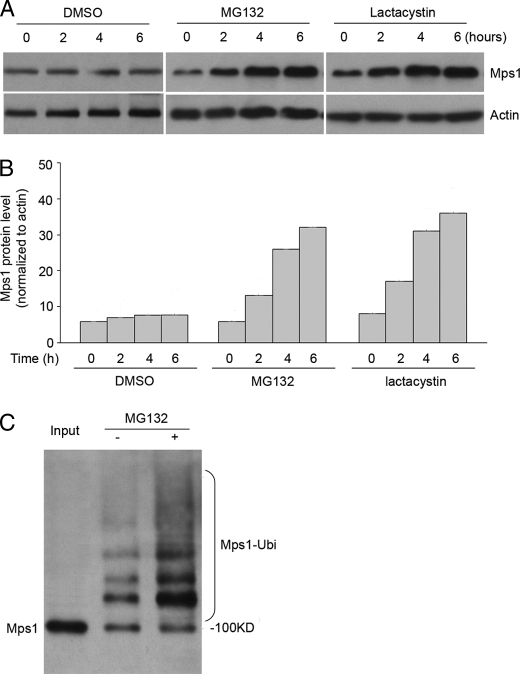
Human Mps1 protein levels fluctuate during the cell cycle peaking during G2/M phase and rapidly declining thereafter (10, 11). Here, we investigated whether the ubiquitin-proteosome system was involved in regulating Mps1 levels in human embryonic kidney (HEK) 293T cells. To do this, HEK 293T cells incubated in the presence or absence of the proteosome inhibitor MG132 were collected at 2-h intervals, and Mps1 levels were assessed by immunoblot analysis. The results showed that Mps1 levels steadily increased in the presence of MG132 compared with DMSO (vehicle control)-treated cells (Fig. 1, A and B). Similar results were obtained with the proteosome inhibitor lactacystin. Next, we determined whether Mps1 is ubiquitinated in vivo. This was accomplished by transfecting 293T cells with a His6-ubiquitin plasmid followed by their incubation in the presence or absence of MG132. His6-ubiquitin complexes were isolated by nickel chelate chromatography and subjected to Western analysis for assessing Mps1 ubiquitination. The results showed a ladder of polyubiquitinated Mps1 forms above 100 kDa that were further enhanced in the presence of MG132 (Fig. 1C). Taken together, these results suggest that Mps1 protein levels are regulated by the ubiquitin-proteasome pathway.
FIGURE 1.
Mps1 protein accumulates and is polyubiquitinated in the presence of a proteasome inhibitor. A, Western blot analysis of Mps1 protein levels at specific times following treatment with a proteosome inhibitor. 293T cells were treated with DMSO (vehicle control), MG132 (20 μm), or lactacystin (10 μm). β-Actin was blotted as a loading control. B, quantitation of Mps1 levels from Western blots normalized to β-actin. C, assessment of Mps1 ubiquitination. 293T cells were transiently transfected with His6-ubiquitin plasmids and incubated with or without the proteasome inhibitor, MG132 (25 μm), for 6 h prior to lysis. 24 h post-transfection, His6-ubiquitin pulldowns were performed, and isolated ubiquitinated complexes were subjected to Western analysis with an Mps1 antibody. Polyubiquitinated Mps1 appears as a ladder of higher molecular weight for Mps1 forms greater than 100 kDa. All results shown are representative of three independent experiments.
APC-c, but Not SCFSkp2, Physically Associates with Mps1
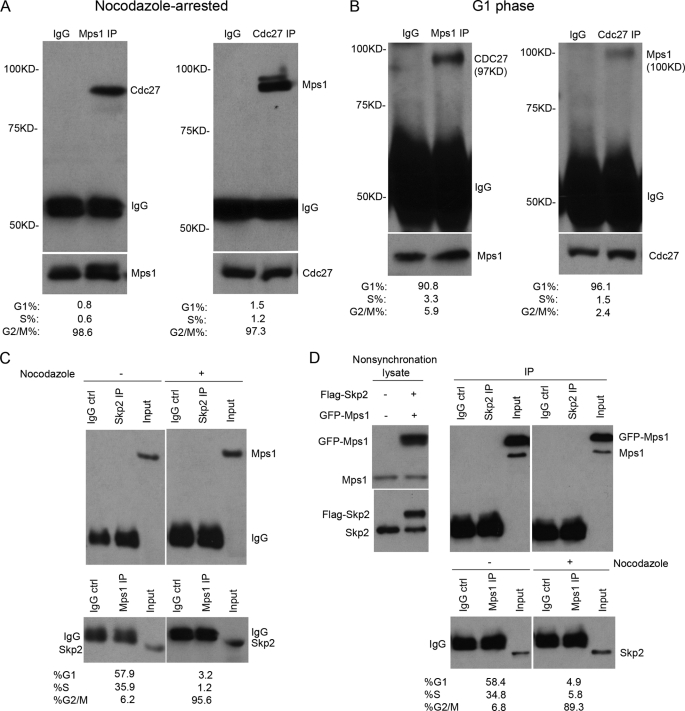
Protein ubiquitination is generally mediated by the SCFSkp2 or APC-c complexes (22). Both function as E3 ligases that facilitate the transfer of ubiquitin from the active site of an E2 enzyme to a substrate that is marked for destruction by the proteasome. To determine whether either of these ubiquitin ligases is involved in the regulation of Mps1 degradation, protein lysates prepared from M phase- or late G1 phase-synchronized cells were subjected to co-immunoprecipitation (IP) assays using antibodies to Mps1, Cdc27 subunit of APC-c, or Skp2. Endogenous Mps1 and Cdc27 were readily detected in co-IP complexes isolated from M phase-arrested cells (Fig. 2A) and to a much lesser extent at late G1 phase (Fig. 2B) due to the lower levels of Mps1 present during this stage of the cell cycle (10). Similar results were observed for exogenous and endogenous interaction in asynchronous cells (supplemental Fig. 1). In contrast, IP complexes of endogenous (Fig. 2C) or exogenously (Fig. 2D) expressed Skp2 and Mps1 failed to detect any physical association between the two proteins in lysates prepared for either asynchronous or synchronous cells (Fig. 2, C and D). Thus, we conclude that the APC-c E3 ligase, but not SCFSkp2 complex, is associated in vivo with Mps1 implying a role in regulating its degradation.
FIGURE 2.
Mps1 associates with the APC-c, but not the SCFSkp2, complex in vivo. Co-IP assays of Mps1 and Cdc27 in nocodazole-arrested (A) and G1-synchronized 293T cells (B). Note that a longer film exposure was required to detect Mps1-Cdc27 complexes in G1-synchronized cells due to the lower levels of Mps1 present. Cells were incubated with nocodazole (40 ng/ml) for 16 h prior to arrest in G2/M. A double thymidine protocol was used for G1 synchronization. The protein expression levels are shown at the bottom. During the last 6 h before harvesting, cells were treated with MG132 (25 μm). C, co-IP assays of endogenous Mps1 and Skp2 (component of SCF) performed with asynchronous and nocodazole-arrested 293T cell lysates. Mps1 did not co-precipitate with endogenous Skp2. D, GFP-Mps1 and FLAG-Skp2 proteins do not associate with each other in vivo following transient expression in 293T cells. Co-IP assays were performed using anti-Mp1 and anti-Skp2 antibodies. IPs using normal mouse IgG were performed as negative controls in all experiments. Cell cycle distribution for asynchronous, G1-synchronized, and nocodazole-arrested cells was assessed by flow cytometry.
Mps1 Is Associated with Cdc20 and Cdh1 in a Cell Cycle-dependent Manner
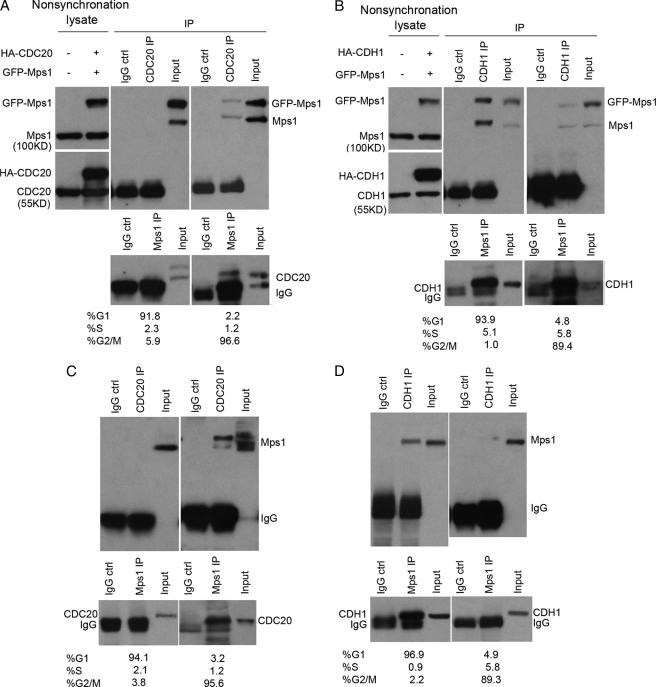
As cells proceed through mitosis, APC is activated through phosphorylation of its subunits and interaction with the regulatory activating protein Cdc20 (5). Upon exiting mitosis, APC remains active but switches from Cdc20 to Cdh1/Hct1 for its activity throughout the G1 phase (4). To determine which of the APC regulators (Cdc20 or Cdh1) are involved in mediating Mps1 degradation, co-IP assays were performed on cell lysates from 293T cells synchronized at the M or G1 phase. 293T cells co-transfected with Mps1 and HA-tagged Cdc20 or Cdh1 plasmids were subjected to immunoblot analysis to verify expression of recombinant proteins (Fig. 3, A and B). As shown in Fig. 3A, anti-Cdc20 IgG co-precipitated Mps1 at the M phase but not at the G1 phase. Equivalent results were obtained from Mps1 IPs (Fig. 3A, lower panel). On the other hand, our analysis for Mps1-Cdh1 associations in vivo showed that Mps1 co-precipitated with Cdh1 IP complexes isolated from G1 phase lysates and, to a much lesser extent, M phase lysates (Fig. 3B). Importantly, Mps1 interactions with Cdc20 and Cdh1 in vivo were also detected for endogenous proteins in nontransfected cells (Fig. 3, C and D). Thus, our data indicate that APC-cCdc20 associates with Mps1 during mitosis, whereas APC-cCdh1 preferentially associates with Mps1 at the G1 phase of the cell cycle.
FIGURE 3.
Human Mps1 associates with Cdc20 and Cdh1 in a cell cycle-dependent manner. 293T cells were transiently co-transfected with GFP-Mps1 and HA-Cdc20 or HA-Cdh1 plasmids, synchronized at mitosis with nocodazole treatment for 16 h or G1 phase by a double thymidine exposure protocol, and incubated in the presence of MG132 (20 μm) for 6 h prior to cell lysis. Co-IPs on cell lysates were performed with anti-Mps1, anti-Cdc20, or anti-Cdh1 antibodies followed by immunoblot analysis. GFP tag was used as a negative control for IPs. A, GFP-Mps1 and endogenous Mps1 co-precipitate with HA-Cdc20 IP complexes isolated from nocodazole-arrested cell lysates. Lysates were collected from 293T cells co-transfected with GFP-Mps1 and HA-Cdc20 plasmids before synchronization. B, GFP-Mps1 and endogenous Mps1 proteins co-precipitate with HA-Cdh1 IP complexes isolated from G1-synchronized cell lysates. Lysates were collected from 293T cells co-transfected with GFP-Mps1 and HA-Cdh1 plasmids before synchronization. C, endogenous Mps1 and Cdc20 proteins are associated with each other at mitosis. D, endogenous Mps1 and Cdh1 proteins are associated with each other at G1 phase. Cell cycle phases were monitored by flow cytometry. Results shown are representative of three experiments.
Overexpression of Cdc20 or Cdh1 Induces Mps1 Degradation
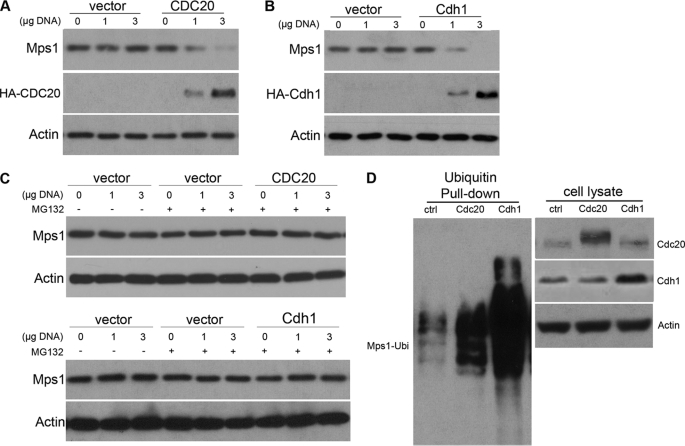
The co-IP results in Fig. 3 implicate a potential role for Cdc20 and Cdh1 in regulating APC-c-mediated degradation of Mps1. To examine this further, HA-tagged Cdc20 or Cdh1 were overexpressed in HEK 293T cells to assess their potential effects on Mps1 degradation. In contrast to vector control cells, overexpression of HA-Cdc20 resulted in a dose-dependent reduction of Mps1 levels in nocodazole-released HEK 293T cells (Fig. 4A) that was blocked in the presence of MG132 (Fig. 4C). Likewise, HA-Cdh1 overexpression in asynchronous growing HEK 293T cells was sufficient to reduce Mps1 levels (Fig. 4B) in a proteosome-dependent manner (Fig. 4C). Furthermore, overexpression of either HA-tagged Cdh1 or Cdc20 greatly enhanced poly-ubiqitination of Mps1 (Fig. 4D). Thus, we conclude that both Cdc20 and Cdh1 stimulate APC-c-mediated ubiquitination and degradation of Mps1.
FIGURE 4.
Overexpression of Cdc20 or Cdh1 induces Mps1 proteolysis. 293T cells were transiently transfected with 0, 1, or 3 μg of HA-hCdc20 or HA-hCdh1 plasmid DNA, arrested at mitosis with nocodazole, and then released into fresh medium for the indicated times. A, Western analysis of Mps1 levels in cells transfected with HA-hCdc20 or empty vector. Cells arrested in mitosis with nocodazole were released into fresh media for 30 min to allow entry into anaphase prior to cell lysis. Progression into anaphase was confirmed by Western analysis of cyclin B levels showing a precipitous drop after 30 min. B, Western analysis of Mps1 levels in 293T cells transfected with HA-hCdh1 or empty vector. Nocodazole-arrested cells were released into fresh medium for 2 h to allow progression into the G1 phase prior to cell lysis. Progression into the G1 phase was monitored by flow cytometry. C, drop in Mps1 by Cdc20 or Cdh1 overexpression is blocked by the proteosome inhibitor MG132 confirming that Mps1 is degraded. Nocodazole-arrested 293T cells were released into fresh media in the absence or presence of MG132 and subjected to cell lysis for either 30 min (Cdc20) or 2 h (Cdh1) later. D, overexpression of Cdc20 or Cdh1 promotes ubiquitination of Mps1 in vivo. 293T cells co-transfected with His6-ubiquitin and HA-Cdc20 or HA-Cdh1 plasmids were incubated in the presence of MG132 (25 μm) for 6 h prior to cell lysis. 24 h post-transfection, His6-ubiquitin pulldowns were performed followed by immunoblotting with a Mps1 antibody (left panel). Exogenous expression of HA-Cdc20 or HA-Cdh1 was assessed by immunoblot analysis of cell lysates (right panel). Actin levels were assessed as a loading for all immunoblots. All data shown are representative of three independent experiments.
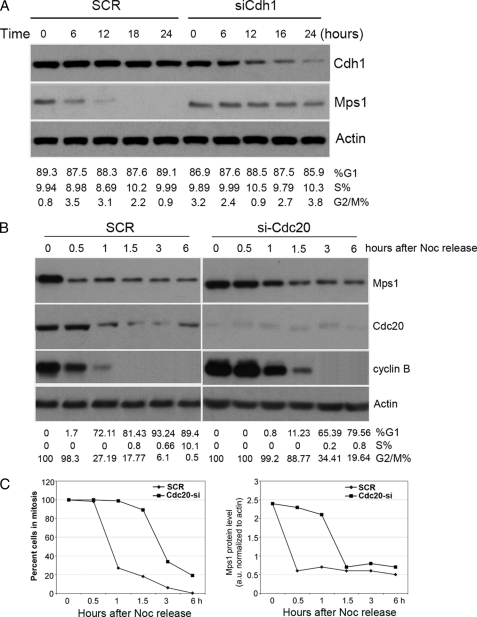
Silencing of Cdh1 or Cdc20 Suppresses Cell Cycle-regulated Mps1 Degradation
To address whether Cdh1 and Cdc20 are required for the degradation of Mps1 in vivo, we used small interfering RNAs (siRNAs) to selectively reduce their expression. Because Cdh1 functions primarily during late mitosis and G1 phase, 293T cells were synchronized in G1 with a double thymidine block and transfected with scrambled (SCR) or Cdh1-specific siRNA. Interestingly, steady-state levels of Mps1 were significantly elevated in Cdh1 siRNA-treated cells compared with SCR controls (Fig. 5A) suggesting that APC-cCdh1 is critical for the degradation of Mps1 protein at the G1 phase. Enrichment of G1 phase cells (>85%) by double thymidine block was verified by FACS analysis of SCR and Cdh1 siRNA-treated cells. Therefore, our data are consistent with the notion that APC-cCdh1 mediates the degradation of Mps1 during the G1 phase.
FIGURE 5.
Silencing of Cdh1 or Cdc20 by RNAi results in a cell cycle-dependent stabilization of Mps1 protein. A, Mps1 levels accumulate in G1/S-synchronized cells transfected with Cdh1 siRNA compared with SCR control. 293T cells synchronized at G1/S with a double thymidine block were treated with SCR or Cdh1 siRNA in the presence of thymidine keeping the cells at G1/S for the duration of the experiment. Whole-cell extracts were Western-blotted with the indicated antibodies at indicated times post siRNA treatment. Cell cycle phases were monitored by flow cytometry. B, Mps1 degradation and progression into anaphase is delayed in Cdc20-depleted 293T cells. 293T cells transfected with SCR or Cdc20 siRNAs were incubated with nocodazole (Noc) for 16 h. Round, prometaphase-like cells were then collected by gentle shake off, replated into fresh medium, and collected at the indicated times for immunoblotting or FACS analysis. C, left panel, delay of mitotic exit in Cdc20-depleted cells. Results from flow cytometry show the percent of mitotic cells at the indicated times following nocodazole release. Right panel, graph of Mps1 protein levels (normalized to β-actin) in SCR and Cdc20 siRNA cells at indicated times following nocodazole release. Results shown are representative of at least three independent experiments.
Next, 293T cells were transfected with siRNA oligonucleotides corresponding to Cdc20 or a scramble control (SCR) and arrested at a prometaphase-like state with nocodazole. Following the removal of nocodazole, SCR or Cdc20 siRNA-treated cells were collected at selected time points to assess for Mps1 protein levels prior to and after anaphase initiation. Cyclin B degradation was assessed in parallel by immunoblot analysis as a marker for anaphase initiation. In SCR control-treated cells, Mps1 levels precipitously dropped by 0.5 h following nocodazole removal, which corresponded to anaphase entry (see Fig. 5, B and C). In contrast, Mps1 levels remained elevated up to 1 h post-nocodazole removal in Cdc20 siRNA-treated cells (Fig. 5, B and C, right panel). This appeared to result in a delay into anaphase as supported by the persistence of high cyclin B levels. Between 1 and 1.5 h post-nocodazole removal, Mps1 levels decreased markedly, which corresponded to the Cdc20-depleted cells progressing into anaphase as evidenced by the precipitous drop in cyclin B (Fig. 5, B and C). Thus, our data suggest that Cdc20 is important for regulating degradation of human Mps1 during the metaphase-anaphase transition.
A Single D Box in Human Mps1 Is Critical for Regulating Its Degradation
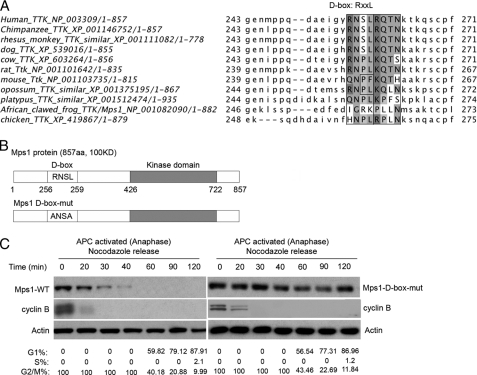
Two important destruction signals have been identified in substrates targeted for proteolysis by the APC-c as follows: the destruction box (D-box, RXXL, where X = any amino acid) and the KEN box (KEN). D-box-containing substrates are recognized by either APC-cCdc20 or APC-cCdh1, whereas KEN box-containing substrates are only ubiquitinated by APC-cCdh1 (23, 24). In addition, a unique sequence in Aurora A, termed the A-box (RXLXPSN), is also required for its efficient ubiquitination by APC-cCdh1 (25). Sequence analysis of the human Mps1 protein reveals a single D box (RNSL) located in its N-terminal region, which is conserved in most mammalian species, including chimpanzee, rhesus monkey, dog, cow, rat, and opossum (see Fig. 6A).
FIGURE 6.
Human Mps1 contains a single destruction D-box that confers instability. A, D-box sequence (RNSLRQTN) located at the N terminus of human Mps1 is conserved in most mammalian species. Conserved amino acids are indicated in black bars. B, schematic representation of full-length human Mps1 wild type denoting its putative D-box (RNSL) versus a Mps1 D-box mutant (ANSA) created by site-directed mutagenesis. C, mutation of the D-box confers resistance to Mps1 degradation at anaphase in human 293T cells. GFP-Mps1 (WT) and (D-box mutant) DNA constructs were transiently transfected into 293T cells and arrested at prometaphase with nocodazole (40 ng/ml) treatment for 16 h. Cells were synchronously released into anaphase by washing twice in fresh media minus nocodazole. At the indicated times, cells were collected for Western analysis of recombinant Mps1 levels using a GFP antibody. Cyclin B degradation was monitored as a marker for anaphase. Actin was used as a loading control. Mitotic synchrony by nocodazole and exit into G1 phase were monitored in parallel by flow cytometry. Results shown are representative of three independent experiments.
To directly test whether the putative D-box sequence in human Mps1 played a role in regulating its degradation, we substituted alanine at the arginine and leucine residues by site-directed mutagenesis (Fig. 6B) and assessed whether the D-box Mps1 mutant (ANSA) was resistant to APC-mediated degradation at anaphase. GFP fusions of wild type or D-box mutant Mps1 proteins were expressed in 293T cells and synchronized in mitosis with nocodazole. Next, the nocodazole-arrested cells were released into anaphase and collected at the indicated time points for up to 2 h for assessing degradation of the wild type and D-box mutant GFP-Mps1 proteins by Western analysis. Cells expressing wild type or D-box mutant Mps1 both initiated anaphase by 20 min, as suggested by cyclin B degradation, and cycled into G1 phase at 60–120 min. Similar to endogenous Mps1, shown in Fig. 5C, wild type GFP-Mps1 was efficiently degraded by 30 min after nocodazole release into anaphase and remained undetectable throughout the remainder of the time course (Fig. 6C, left panel). In contrast, the Mps1 D-box mutant remained stable throughout anaphase and progression into the subsequent G1 phase (Fig. 6C, right panel), indicating that the putative D-box motif is important for regulating degradation of Mps1.
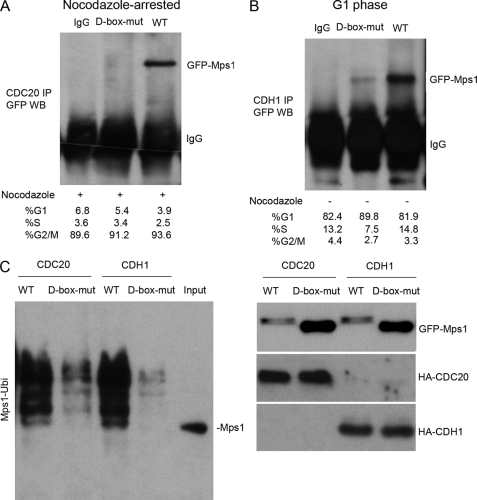
To further evaluate the role of the D-box in regulating Mps1 degradation, we asked whether the D-box mutant was capable of associating with either of the APC activators Cdc20 and Cdh1 in vivo. Compared with recombinant GFP-tagged wild type (WT) Mps1, the D-box mutant did not co-precipitate with Cdc20 IP complexes isolated from 293T cells enriched in mitosis with nocodazole (Fig. 7A). Likewise, the association of Cdh1 with the D-box mutant was greatly reduced in G1 phase-enriched cells compared with co-precipitation of GFP-tagged WT Mps1 (Fig. 7B). In agreement with these results, polyubiquitination of the D-box mutant was greatly diminished compared with WT Mps1 protein (Fig. 7C, left panel). In addition, Western analysis confirmed higher steady-state levels of the Mps1 D-box mutant compared with recombinant WT Mps1 (Fig. 7C, right panel). Collectively, we conclude that the D-box sequence (RNSL) in human Mps1 serves a critical role in regulating its polyubiquitination and degradation by the APC-ubiquitin-proteosome pathway.
FIGURE 7.
D-box mutation suppresses human Mps1 interactions with Cdc20 or Cdh1 and inhibits polyubiquitination of Mps1. GFP-tagged Mps1 proteins (wild type or D-box mutant) were ectopically expressed in 293T cells and subsequently arrested at mitosis by nocodazole treatment or synchronized at late G1 phase as described under “Experimental Procedures.” Cell cycle synchronization was confirmed by flow cytometry. Co-IP assays were performed with antibodies to Cdc20 or Cdh1 proteins to assess whether either protein associates with the Mps1 D-box mutant. A, Mps1 D-box mutant fails to co-precipitate with Cdc20 at mitosis. B, Mps1 D-box mutant fails to co-precipitate with Cdh1 at G1 phase. C, mutation of the D-box inhibits polyubiquitination of Mps1. 293T cells were transiently co-transfected with equal amounts of wild type GFP-Mps1 or D-box-mut-Mps1 plasmids together with HA-tagged Cdc20 or Cdh1 and His6-ubiquitin-containing plasmids. Expression levels of GFP-Mps1 WT or D-box mut and HA-tagged Cdc20 and Cdh1 proteins are shown in the right panel. Polyubiquitination of Mps1 wild type or D-box mutant proteins was assessed as described under “Experimental Procedures.” WB, Western blot.
Preventing Mps1 Degradation Is Sufficient to Trigger Centrosome Amplification and the Mitotic Abnormalities in 293T Cells
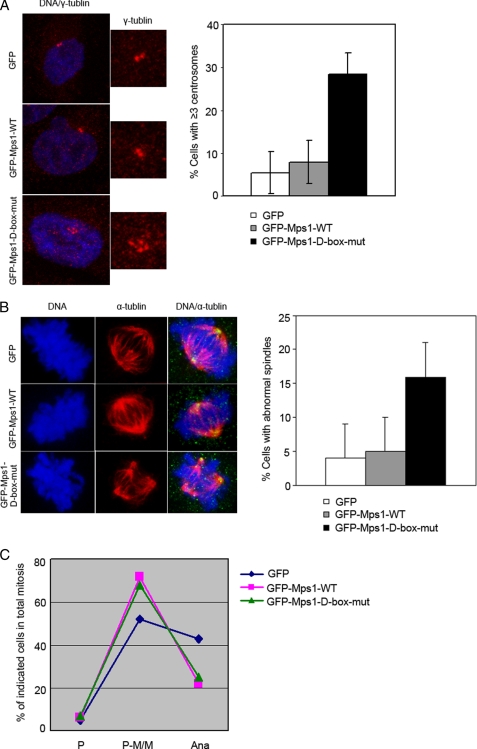
To address whether the D-box mutant has any effect on Mps1 functions on centrosome duplication and mitotic spindle checkpoint, we depleted the endogenous Mps1 from 293T cells by using pSUPER.retro siRNA as described before and compared the ability of ectopically expressed Mps1-WT and D-box mutant in centrosome duplication and mitotic spindle checkpoint. As expected, immunofluorescence staining of γ-tubulin and α-tubulin revealed that overexpression of Mps1-WT is not sufficient to cause centrosome reduplication and the resultant multipolar spindles in 293T cells, which is consist with the Fisk report (34). However, overexpression of Mps1-D-box mutant causes supernumerary centrosomes and multipolar spindles in 293T cells. This suggests that preventing the degradation of Mps1 is sufficient to trigger centrosome amplification (Fig. 8A) and resultant multipolar spindles in 293T cells (Fig. 8B).
FIGURE 8.
D-box mutant preventing Mps1 degradation causes centrosome amplification and mitotic abnormalities. 293T cells were depleted from the endogenous Mps1 by using pSUPER.retro siRNA as described before and then transfected with GFP-tagged Mps1 wild type or D-box mutant constructs. 72–96 h post-transfection, cells were subjected to immunofluorescence by using anti-γ or anti-α antibody. A, representative images and bar graph shows that quantitation of frequency of cells with more than two centrosomes. ∼200 interphase cells were assessed at each treatment. Three independent experiments were performed. B, representative images and bar graph show the frequency of cells with abnormal spindles. ∼200 mitotic cells were assessed at each treatment. Three independent experiments were performed. C, curve graph shows the cell cycle distribution. Three independent experiments were performed. P, prophase; P-M/M, prometaphase/metaphase.
A similar approach was used to determine the effect of D-box mutant on mitotic spindle checkpoint. We found that the majority of the wild type and D-box mutant expression mitotic cells were arrested at prometaphase/metaphase, and only a very small fraction of cells proceeded into anaphase. This is in sharp contrast to GFP control cells, in which cells proceeded into anaphase normally (Fig. 8C). We conclude that overexpressions of Mps1-WT and D-box mutant lead to a prometaphase/metaphase arrest. Together, the D-box mutant could prevent the degradation of Mps1 protein and is sufficient to trigger centrosome amplification and mitotic abnormalities.
DISCUSSION
In this study, the regulatory mechanisms associated with the degradation of human Mps1 were investigated. Our results indicate that human Mps1 is targeted for degradation by the sequential actions of APC-cCdc20 and APC-cCdh1 during anaphase through the G1 phase of the cell cycle. In addition, we identified a single conserved D-box sequence (RXXL) in the N-terminal region of human Mps1 that upon mutation conveys resistance to the polyubquitination and subsequent proteolysis of Mps1. Thus, we propose that Mps1 protein levels in human cells are down-regulated in a cell cycle-dependent manner through the APC-ubiquitin-proteosome system.
Mps1 Is a Substrate of the APC/Ubiquitin/Proteosome Pathway in Human Cells
The Mps1 homologue in S. cerevisiae (budding yeast) was previously shown to be targeted for degradation at anaphase by APC (19). Prior to our study, it was not known how Mps1 levels were regulated in human proliferating cells. Here, we showed that human Mps1 undergoes proteolysis by the APC/ubiquitin/proteosome pathway. This was supported by our data in human HEK 293T cells treated with the proteosome inhibitor MG132 (or lactacystin), which showed the accumulation and increased polyubiquitination of Mps1 compared with nontreated cells (Fig. 1). In addition, we showed that human Mps1 physically associates with the APC-c in vivo during mitosis and, to a lesser extent, at G1 phase confirming similar results previously reported (11). Importantly, our functional data in Figs. 4 and 5 indicate that proteolysis of human Mps1 is regulated by the interactions of the APC-cCdc20 and APC-cCdh1 complexes. Thus, similar to other mitotic substrates of APC-c, including cyclins A and B, securin, Aurora A, and Aurora B (26), the APC/ubiquitin/proteosome system targets human Mps1 for degradation.
The RXXL motif represents the minimal sequence for a “destruction” D box that is recognized by APC-cCdc20 or APC-cCdh1 complexes (23). Unlike Mps1 of budding yeast, which contains three D boxes (19), human Mps1 contains a single RNSL motif (256–259) in its N-terminal region that is conserved in most mammalian species (Fig. 6, A and B). Mutation of the D box rendered the Mps1 protein stable during anaphase and G1 compared with the wild type Mps1 protein (Fig. 6C). Similar results have been reported for mutations made in the D box of mitotic cyclins (27). Hence, we propose that the single RNSL motif in human Mps1 is critical for regulating its proteolysis during anaphase through early G1 phase.
Proteolysis of Human Mps1 Is Cell Cycle-regulated by APC-cCdc20 and APC-cCdh1
APC-c-dependentproteolysis is controlled by its conserved activators Cdc20 and Cdh1. Both of these activating proteins contain WD40 repeats that bind to the APC-c in a cell cycle-dependent manner (1, 5). Our results indicate that APC-cCdc20 and APC-cCdh1 act sequentially during anaphase and G1 phase, respectively, to promote the proteolysis of human Mps1. This is supported in part by our data that show Mps1 co-precipitates with Cdc20 (in addition to APC) during mitosis and with Cdh1 during G1 phase (see Fig. 3). Loss-of-function studies that silenced Cdc20 or Cdh1 expression by RNAi resulted in a cell cycle-dependent stabilization of Mps1 that is consistent with their distinct roles at anaphase (Cdc20) and G1 phase (Cdh1) (see Fig. 5). Indeed, other mitotic substrates such as mitotic cyclins A and B1, securin, and Nek2A are also targeted for degradation by APC-cCdc20 and APC-cCdh1 (4, 28, 29).
Biological Importance of APC-mediated Mps1 Proteolysis
Our data show that Mps1 interacts with Cdc20 in nocodazole-treated cells, and overexpression Cdc20 promotes Mps1 degradation in nocodazole-released cells. How can Mps1 be a target of the APC-c when APC-cCdc20 is normally inhibited by the checkpoint at prometaphase? It is conceivable that a low level of degradation of Mps1 by Cdc20 takes place during early mitosis, leaving an adequate concentration of Mps1 to execute late mitotic functions. There are several examples that APC(Cdc20) is able to interact with in nocodazole-treated cells, such as p21 (30), Aurora-B (24), cyclin A (32), Nek2A (33), and HOXC10 (5). Also, APC(Cdc20) is inactive/active in prometaphase cells in the presence of nocodazole (5, 30–33). However, the reason for a subpopulation of APC(Cdc20) to remain active in the presence of spindle checkpoint activity is not well understood. It is possible that the major degradation of Mps1 by Cdc20 occurs in late mitosis. Once the chromosomes are aligned, the activity of the checkpoint is reduced, allowing APC to associate with its activator Cdc20 and target the degradation of mitotic regulators, including Mps1.
The expression of a nondegradable D-box Mps1 mutant in 293T cells was shown to sustain activation of the spindle checkpoint during anaphase indicating that Mps1 degradation at anaphase is critical for preventing reactivation of the checkpoint (Fig. 8C). Furthermore, degradation of Mps1 during late anaphase and G1 phase by the actions of APC-cCdh1 is important for preventing centrosome overduplication that can be associated with high Mps1 expression in some cell types (13, 14). Our data show that the expression of nondegradable D-box Mps1 mutant in 293T cells leads to centrosome amplification and resultant multipolar spindles suggesting that preventing the degradation of Mps1 at centrosomes is sufficient to cause centrosome re-duplication (Fig. 8, A and B). Consistent with this, the constitutively active B-RafV600E mutant was previously shown to stabilize Mps1 protein levels in human melanoma cells, which resulted in supernumerary centrosomes, multipolar spindles, and hyper-activity of the spindle checkpoint during anaphase (31, 35). Substitution of a phospho-mimetic residue at a Cdk2 phosphorylation site (Thr-468) protects Mps1 from degradation at centrosomes (36). Thus, phosphorylation of Mps1 may be an important mechanism to suppress its proteolysis when its functions are required during the cell cycle.
In summary, our study shows that the decline in human Mps1 levels following entry into anaphase is regulated, at least in part, by the APC-ubiquitin-proteosome pathway during late mitosis and G1 phase. Preventing Mps1 degradation is sufficient to trigger centrosome amplification and mitotic abnormalities in human cells. In the future, it will be interesting to examine whether phosphorylation or other modifications of Mps1 play a role in regulating its stability throughout the cell cycle. Indeed, stabilization of Mps1 may contribute to deregulation of the spindle checkpoint or centrosome duplication in human tumor cells.
Supplementary Material
Acknowledgments
We thank Michele Pagano (Howard Hughes Medical Institute at New York University) and Mark Winey (University of Colorado, Boulder) for providing plasmid DNA constructs. We also thank the Medical Experiment Center of Shanxi Medical University.
This work was supported by Program for New Century Excellent Talents University of China Grant NCET-10-0872, National Nature Science Foundation of China Grant 30872932, Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry Grant 2009-8, Trainee Development Award from Bankhead Coley Program Grant 30-15066-02-07 (to Y. C.), and the Department of Defense, National Functional Genomics Center Project Award DAMD17-02-2-0051 (to T. M. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- APC-c
- anaphase-promoting complex/cyclosome
- SCR
- scrambled
- nt
- nucleotide
- h
- human
- IP
- immunoprecipitation.
REFERENCES
- 1.Acquaviva C., Pines J. (2006) J. Cell Sci. 119, 2401–2404 [DOI] [PubMed] [Google Scholar]
- 2.Murray A. W. (2004) Cell 116, 221–234 [DOI] [PubMed] [Google Scholar]
- 3.Sumara I., Maerki S., Peter M. (2008) Trends Cell Biol. 18, 84–94 [DOI] [PubMed] [Google Scholar]
- 4.Castro A., Bernis C., Vigneron S., Labbé J. C., Lorca T. (2005) Oncogene 24, 314–325 [DOI] [PubMed] [Google Scholar]
- 5.Yu H. (2007) Mol. Cell 27, 3–16 [DOI] [PubMed] [Google Scholar]
- 6.Winey M., Goetsch L., Baum P., Byers B. (1991) J. Cell Biol. 114, 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones M. H., Huneycutt B. J., Pearson C. G., Zhang C., Morgan G., Shokat K., Bloom K., Winey M. (2005) Curr. Biol. 15, 160–165 [DOI] [PubMed] [Google Scholar]
- 8.Weiss E., Winey M. (1996) J. Cell Biol. 132, 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abrieu A., Magnaghi-Jaulin L., Kahana J. A., Peter M., Castro A., Vigneron S., Lorca T., Cleveland D. W., Labbé J. C. (2001) Cell 106, 83–93 [DOI] [PubMed] [Google Scholar]
- 10.Stucke V. M., Silljé H. H., Arnaud L., Nigg E. A. (2002) EMBO J. 21, 1723–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S. T., Chan G. K., Hittle J. C., Fujii G., Lees E., Yen T. J. (2003) Mol. Biol. Cell 14, 1638–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jelluma N., Brenkman A. B., van den Broek N. J., Cruijsen C. W., van Osch M. H., Lens S. M., Medema R. H., Kops G. J. (2008) Cell 132, 233–246 [DOI] [PubMed] [Google Scholar]
- 13.Fisk H. A., Winey M. (2001) Cell 106, 95–104 [DOI] [PubMed] [Google Scholar]
- 14.Fisk H. A., Mattison C. P., Winey M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14875–14880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stucke V. M., Baumann C., Nigg E. A. (2004) Chromosoma 113, 1–15 [DOI] [PubMed] [Google Scholar]
- 16.Straight P. D., Giddings T. H., Jr., Winey M. (2000) Mol. Biol. Cell 11, 3525–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilliland W. D., Wayson S. M., Hawley R. S. (2005) Curr. Biol. 15, 672–677 [DOI] [PubMed] [Google Scholar]
- 18.Poss K. D., Nechiporuk A., Stringer K. F., Lee C., Keating M. T. (2004) Genes Dev. 18, 1527–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palframan W. J., Meehl J. B., Jaspersen S. L., Winey M., Murray A. W. (2006) Science 313, 680–684 [DOI] [PubMed] [Google Scholar]
- 20.An J., Huang Y. C., Xu Q. Z., Zhou L. J., Shang Z. F., Huang B., Wang Y., Liu X. D., Wu D. C., Zhou P. K. (2010) BMC Mol. Biol. 11, 18–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrano A. C., Eytan E., Hershko A., Pagano M. (1999) Nat. Cell Biol. 1, 193–199 [DOI] [PubMed] [Google Scholar]
- 22.Nakayama K. I., Nakayama K. (2006) Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 23.Pfleger C. M., Lee E., Kirschner M. W. (2001) Genes Dev. 15, 2396–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen H. G., Chinnappan D., Urano T., Ravid K. (2005) Mol. Cell. Biol. 25, 4977–4992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Littlepage L. E., Ruderman J. V. (2002) Genes Dev. 16, 2274–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pines J. (2006) Trends Cell Biol. 16, 55–63 [DOI] [PubMed] [Google Scholar]
- 27.King R. W., Deshaies R. J., Peters J. M., Kirschner M. W. (1996) Science 274, 1652–1659 [DOI] [PubMed] [Google Scholar]
- 28.Hames R. S., Wattam S. L., Yamano H., Bacchieri R., Fry A. M. (2001) EMBO J. 20, 7117–7127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zur A., Brandeis M. (2001) EMBO J. 20, 792–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amador V., Ge S., Santamaría P. G., Guardavaccaro D., Pagano M. (2007) Mol. Cell 27, 462–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui Y., Borysova M. K., Johnson J. O., Guadagno T. M. (2010) Cancer Res. 70, 675–684 [DOI] [PubMed] [Google Scholar]
- 32.den Elzen N., Pines J. (2001) J. Cell Biol. 153, 121–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fry A. M., Yamano H. (2006) Cell Cycle 5, 1487–1491 [DOI] [PubMed] [Google Scholar]
- 34.Kasbek C., Yang C. H., Fisk H. A. (2009) Environ. Mol. Mutagen. 50, 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui Y., Guadagno T. M. (2008) Oncogene 27, 3122–3133 [DOI] [PubMed] [Google Scholar]
- 36.Kasbek C., Yang C. H., Yusof A. M., Chapman H. M., Winey M., Fisk H. A. (2007) Mol. Biol. Cell 18, 4457–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.