Abstract
Histone modification is well established as a fundamental mechanism driving the regulation of transcription, replication, and DNA repair through the control of chromatin structure. Likewise, it is apparent that incorrect targeting of histone modifications contributes to misregulated gene expression and hence to developmental disorders and diseases of genomic instability such as cancer. The KMT2 family of SET domain methyltransferases, typified by mixed lineage leukemia protein-1 (MLL1), is responsible for histone H3 lysine 4 methylation, a marker of active genes. To ensure that this modification is correctly targeted, a multiprotein complex associates with the methyltransferase and directs activity. We have identified a novel interaction site on the core complex protein WD repeat protein-5 (WDR5), and we mapped the complementary site on its partner retinoblastoma-binding protein-5 (RbBP5). We have characterized this interaction by x-ray crystallography and show how it is fundamental to the assembly of the complex and to the regulation of methyltransferase activity. We show which region of RbBP5 contributes directly to mixed lineage leukemia activation, and we combine our structural and biochemical data to produce a model to show how WDR5 and RbBP5 act cooperatively to stimulate activity.
Keywords: Chromatin, Chromatin Regulation, Crystal Structure, Enzyme Mechanisms, Epigenetics, Histone Methylation, Histone Modification, Transcription Regulation, X-ray Crystallography
Introduction
Aberrant regulation of epigenetic networks has been identified as a key driver of many diseases, especially where genomic instability is a factor, such as in developmentally related disorders and many cancers (1, 2). The principal signaling components of these networks are covalent post-translational modifications to the side chains of residues on the histone tails that extend beyond the nucleosome core (3). These modified residues generate specific binding sites for the chromatin-associated proteins and multiprotein complexes responsible for processes such as transcription, replication, and DNA repair and are thought to act in a highly coordinated fashion (4–6). To create and maintain the correct gene transcription profiles and to facilitate the appropriate response to environmental stimulation, it is essential that the enzymes that deposit or remove these marks be accurately targeted and their activity tightly regulated. The best conserved modification is methylation of the histone H3 lysine 4 (H3K4)3 residue that is predominantly, although not exclusively, associated with transcriptionally active genes (7, 8). Lysine side chains can be mono, di-, or trimethylated, and it is essential that the correct level of H3K4 methylation be deposited on the appropriate nucleosomes, as it has been shown that the different levels of methylation are related with differing outcomes for the associated DNA (9). The Set1/MLL (KMT2) family of methyltransferases is the principal enzyme family responsible for H3K4 methylations, and an associated multiprotein complex has evolved to ensure that this activity is tightly regulated (10–13).
The KMT2 family of H3K4 methyltransferases include six members as follows: Set1A, Set1B, and the four mixed lineage leukemia (MLL) proteins. All six share the carboxyl-terminal conserved SET domain, but as the other domains of the Set1A and Set1B proteins resemble the yeast Set1 protein, the MLL subgroup more resembles the Drosophila trithorax protein. The MLL proteins are so named because of the association of rearrangements at the locus of the MLL1 gene with a subset of infant leukemias that have a particularly poor prognosis (14). The MLL1 gene is essential in embryonic development where it is predominantly associated with the expression of HOX genes (15, 16). Disruption of normal MLL1 expression in mice results in changes to normal H3K4 methylation patterns and leads to skeletal defects and issues with the establishment of hematopoiesis (17, 18). All six of the KMT2 family members associate with a four-member protein complex, often termed the MLL core complex. A wide range of cellular and biochemical studies have highlighted the important role that the core multiprotein complex, WDR5, RbBP5, Ash2L, and Dpy30, makes to the regulation of activity of the KMT2 methyltransferases (12, 19–22). Significantly, this core complex is conserved in yeast through to humans indicating that this is an essential factor for preventing misdirected H3K4 methylation. The key common finding of these studies is that deletion or down-regulation of a core complex protein leads to an overall reduction in the observed level of methyltransferase activity, especially trimethylation of H3K4. The impact of disrupting the integrity of this complex can be detected directly by measurement of methyltransferase levels or indirectly by monitoring downstream expression (12, 13, 19, 21).
WDR5, which forms a WD40 repeat β-propeller, is a key protein in the assembly of the MLL core complex. Although the WDR5 protein has been the subject of a number of studies, the molecular basis of its role in complex assembly and MLL regulation is still enigmatic. Initially, its binding to the histone H3 amino-terminal region, which contains the lysine 4 target for KMT2 methylation, leads to the proposal that it might discriminate between different methylation states (21, 23). Subsequently it was shown that the WDR5 axial cavity specifically recognized the histone H3 arginine 2 side chain leaving the target lysine residue exposed on the surface and potentially available for presentation to the methyltransferase (24–26). More recently, a WDR5-binding segment, similar to the H3 tail, has been identified in MLL1 (27, 28). This motif is located in an apparently unstructured region just outside the N-flanking regions of the catalytic SET domain, and it binds in the same way to the axial cavity of WDR5 as described for the H3 peptide. Disruption of this interaction is detrimental to the assembly and overall methylation activity of the complex (27, 29). In the crystal structure of the MLL1 SET domain in complex with a histone tail peptide, the substrate peptide is buried deep in a cleft in the protein making it difficult to envisage a mechanism where the H3 lysine is presented to the catalytic site by WDR5. This supports the hypothesis that binding of WDR5 to the MLL “Win” motif (WDR5 interaction motif) may be the most physiologically relevant model (30).
In addition to the interaction of WDR5 with the histone tail or the Win motif of the methyltransferase, it has been shown that the yeast orthologue Swd3 (Cps30) forms a stable heterodimer with the complex partner and RbBP5 orthologue Swd1 (Cps50) (12). Indeed, we have been able to purify co-expressed recombinant WDR5 and RbBP5 as a heterodimer from insect cells and show that this subcomplex stimulates the activity of a minimal SET domain MLL1 construct (30). However, in this analysis, we observed that the stimulation of methyltransferase activity by the addition of the WDR5-RbBP5 subcomplex was equivalent to the addition of RbBP5 alone, and it is therefore important to delineate the contribution of the two proteins. Here, we present a new analysis of the WDR5/RbBP5 interaction that reveals a novel binding site on the opposite face of the WDR5 β-propeller from that shown to interact with histone H3 or MLL1. We map the interaction to a conserved motif in RbBP5 and describe the role of the different regions of RbBP5 in the stimulation of methyltransferase activity and combine structural and biochemical data to develop a model for the tripartite complex between the two β-propeller proteins and MLL1.
EXPERIMENTAL PROCEDURES
Expression and Purification
The mouse Δ22WDR5 construct (residues 22–334) was cloned into the pOPINJ vector (31) and expressed as a His6-GST fusion protein in Escherichia coli BL21 (DE3) RIL cells. Following induction with 0.2 mm IPTG, cells were incubated for 72 h at 17 °C. Following lysis by sonication, the fusion protein was purified from cell lysate using Ni-NTA-agarose (Qiagen). The tag was removed by cleavage with rhinovirus 3C protease, and Δ22WDR5 was further purified on heparin resin (GE Healthcare) and gel filtration (Superdex 75, GE Healthcare). The gel filtration buffer was 40 mm HEPES, pH 7.5, 300 mm NaCl, 2 mm 2-mercaptoethanol. Mouse RbBP5 constructs containing the WD40-repeat domain (R1 and R2) were prepared by expression in insect cells as described in Ref. 30 for use in co-precipitation experiments (Fig. 2B). Full-length RbBP5 and RbBP5 (residues 1–380) were cloned into the pOPINS vector (32) and expressed as His6-SUMO fusion proteins in BL21 (DE3) RIL cells for use in methyltransferase assays (Fig. 1B) and further co-precipitation experiments (supplemental Fig. 3). RbBP5 mutants were generated by standard PCR methods. Following induction with 0.2 mm IPTG, cells were incubated at 21 °C for 20 h and purified from cell lysate by incubation with Ni-NTA-agarose (Qiagen), anion exchange (Mono Q, GE Healthcare), and gel filtration (Superdex 75, GE Healthcare) in a final buffer consisting of 40 mm HEPES, pH 7.5, 300 mm NaCl, 2 mm 2-mercaptoethanol. For untagged RbBP5 used in activity assays, protein was cleaved with SENP1C protease prior to anion exchange. The mouse RbBP5 constructs (R3 to R8) excluding the WD40-repeat domain were cloned into the vector pTHREE-E (GST fusion vector modified to contain the multiple cloning site of pET-17b, following an encoded rhinovirus 3C-protease site; A. W. Oliver Institute of Cancer Research) and expressed as GST fusion proteins in BL21 (DE3) RIL cells. Following induction, cells were incubated at 21 °C for 20 h and purified from cell lysate by incubation with glutathione-Sepharose resin (Generon) and were subsequently eluted with 20 mm reduced glutathione. Further purification was achieved by gel filtration (Superdex 200, GE Healthcare) in a buffer containing 40 mm HEPES, pH 7.5, 1 m NaCl, 2 mm 2-mercaptoethanol. Human MLL1 constructs (residues 3752–3969 and 3784–3969) were subcloned into vector pTHREE-E and expressed and purified using the protocol described in Southall et al. (30).
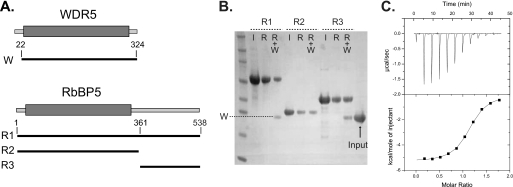
FIGURE 2.
Interaction of the RbBP5 tail with WDR5. A, schematic diagram showing the WDR5 and RbBP5 constructs used in co-precipitation experiments; the β-propeller domains are indicated by a gray box. B, co-precipitation of WDR5 by RbBP5 constructs immobilized on affinity resin (R1 and R2 His-tagged on nickel resin and R3 GST-tagged on glutathione resin) analyzed by polyacrylamide gel and stained by Coomassie Blue. (I = RbBP5 input; R = RbBP5 bound to resin; R+W, RbBP5 resin plus WDR5). C, binding of RbBP5 peptide to WDR5 protein using isothermal calorimetry. Data were fitted to a one-site model, and calculated binding parameters were N = 1.1 ± 0.0, K = 1.8 ± 0.1, ΔH = −5330 ± 34, ΔS = 5.8).
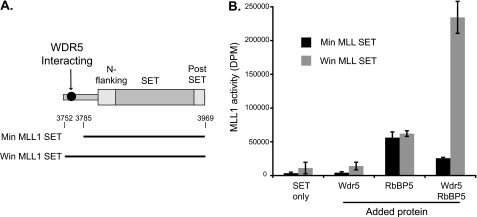
FIGURE 1.
WDR5 contributes to enhancement of MLL activity. A, schematic diagram showing the MLL1 carboxyl-terminal SET domain, the position of the WDR5-interacting motif, and the extent of the constructs used for activity assays. B, effect of addition of WDR5 and RbBP5 separately, or in combination, on the methyltransferase activity of the MLL1 carboxyl-terminal constructs with an unmodified H3 peptide substrate. The slight reduction in activity for WDR5 plus RbBP5 with the shorter MLL1 construct compared with RbBP5 was only robustly observed but was not observed in previous experiments with insect cell purified WDR5-RbBP5 heterodimer (30), and it may reflect the dynamics of the interaction of the three proteins in the absence of the WDR5/MLL1 interaction.
Co-precipitation (Pulldown) Binding Experiments
RbBP5 fusion proteins (50 μg) were incubated with Ni-NTA-agarose (Qiagen), Talon (Clontech), or glutathione-Sepharose (Generon) in a buffer containing 40 mm HEPES, pH 7.5, 150 mm NaCl. After 1 h, the resin was pelleted and washed with buffer by centrifugation. The resin was then resuspended in 1 ml of buffer, and Δ22WDR5 (50 μg) was added and incubated for 1 h at 4 °C. The resin samples were pelleted and washed twice by centrifugation, resuspended in sample loading buffer (Invitrogen), analyzed by SDS-PAGE on 4–12% BisTris gel (Invitrogen), and visualized by staining with Coomassie Blue.
Isothermal Titration Calorimetry
Isothermal titration calorimetry measurements were performed at 20 °C, using an ITC200 microcalorimeter (MicroCal Inc.). Experiments were performed by injecting 3.4 μl of RbBP5(369–381) peptide solution (1.82 mm, sequence YAAEDEEVDVTSVD) into a sample cell containing 187 μm Δ22WDR5 in 40 mm HEPES, pH 7.5, 300 mm NaCl, and 2 mm 2-mercaptoethanol. A total of 11 injections were performed with a reference power of 5 μcal/s. Binding isotherms were plotted and analyzed using Origin software (MicroCal Inc.).
Crystallization
Crystals of the Δ22WDR5-RbBP5 peptide ternary complex were obtained using the microseed matrix screening method (33). Crystals of apo-Δ22WDR5 (stock concentration 300 μm) were obtained by the hanging-drop method in a crystallization condition consisting of 100 mm HEPES, pH 7.0, 70 mm AmSO4, and 30% PEG 3350. The resulting crystals were harvested, diluted in reservoir solution, and mixed with glass microbeads. This mixture was vortexed vigorously, for 2 min, and serial 10-fold dilutions were made in reservoir solution to produce a series of seed stock solutions. A complex of Δ22WDR5 (300 μm), RbBP5 peptide (3.83 mm) was crystallized in a reservoir solution consisting of 0.1 m Tris-HCl, pH 8.0, 25% PEG 8000 seeded with a 1:10,000 dilution of the apoprotein seed stock. For crystals of the ternary complex, histone H3K4me2 peptide, residues 1–8 (3.86 mm), was included in the protein mixture, and crystals were obtained with a reservoir solution consisting of 0.2 m K2SO4 and 20% PEG 3350. The crystals were harvested, transferred to a cryoprotectant solution consisting of reservoir solution with 20% glycerol plus 10% PEG 8000 or PEG 3350 (respectively), and flash-frozen in liquid nitrogen.
Data Collection, Structure Determination
The dataset for the binary complex was collected at the Diamond Light Source (Diamond Light Source Ltd., Harwell Science and Innovation Campus, Oxfordshire, UK) on station I03, and the dataset for the ternary complex was collected at European Synchrotron Research Facility (Grenoble, France) on station ID14-2. The reflections were indexed using the program iMOSFLM (34) and reduced/scaled with programs from the CCP4i suite (35). The structures were solved by molecular replacement using the program PHASER using the coordinates of WDR5 from the H3K4me2 binary complex (Protein Data Bank code 2H13 (26)) as the search model. Difference maps were used to rebuild and extend the initial model using the Coot molecular graphics package (36). Iterative cycles of refinement were carried out using the phenix-refine module within the Phenix program (37). Structural figures in this study were prepared using the CCP4MG program (38).
Methyltransferase Assays
Methyltransferase assays were performed using synthetic peptide substrates based on the histone H3 amino-terminal sequence (ARTKQTARKSTGGKAPR-Y) (University of Bristol) and carried out essentially as described previously (30). In brief, incorporation of tritiated S-adenosylmethionine was monitored by separating peptide from the unincorporated S-adenosylmethionine using “Sep-Pak” C18 cartridges (Waters Associates) followed by scintillation counting. The final peptide concentration was 1 mm, 0.5 mm S-adenosylmethionine (including 0.625 mm S-[lsqb]3H]adenosylmethionine (GE Healthcare or PerkinElmer Life Sciences)), and the assay buffer was 50 mm Tris, pH 8.8. Assays were carried out at 22 °C for 1 h with a final equimolar concentration of both MLL1 and complex members of 25 μm. The final NaCl concentration for assays ranged from 130 to 150 mm. Assays were carried out in triplicate and expressed as means ± S.D.
RESULTS
Enhancement of Methyltransferase Activity by WDR5 and RbBP5
It is well established that WDR5, a conserved protein of the KMT2 core complex, is essential for the MLL1 enzyme to achieve its full methyltransferase activity (21, 25). However, the WDR5-binding site on MLL1 has been mapped to a region outside the catalytic SET domain (27, 28), and this is consistent with our previous findings that addition of WDR5 to a methyltransferase assay containing the minimal catalytic domain did not enhance activity (30). To investigate the role of WDR5 in regulating the catalytic activity of MLL1, we prepared two constructs, our original minimal construct lacking the WDR5 interacting domain and a second slightly longer construct based on that used by the Cosgrove laboratory that contains the Win region for MLL1 binding (Fig. 1A). We investigated the effect of addition of pure recombinant WDR5 and/or RbBP5 protein on the methyltransferase activity of these two SET domains containing constructs using an unmodified histone H3 peptide as substrate (Fig. 1B). The addition of WDR5 alone did not significantly affect the activity of either of the MLL1 constructs, whereas addition of RbBP5 alone did enhance the activity of both constructs. Significantly, addition of WDR5 and RbBP5 together enhances the methyltransferase activity to a level much higher than that seen for RbBP5 alone but only with the longer MLL1 construct. Our interpretation of this result is that the interaction between RbBP5 and the MLL1 SET domain for the longer construct is stabilized by the presence of WDR5 by its binding to both proteins and that both RbBP5 and WDR5 are able to simultaneously bind to both each other and MLL1.
Mapping of the RbBP5/WDR5 Interaction
To investigate the independent binding of MLL1 and RbBP5 to WDR5, we characterized the interaction between the two β-propeller proteins. RbBP5 consists of a predicted β-propeller domain followed by a long, apparently unstructured “tail” of about 200 residues. Initially, a co-precipitation (pulldown) methodology was used to investigate WDR5 binding to either the predicted RbBP5 β-propeller or tail region. Recombinant WDR5 protein was prepared and a series of RbBP5 constructs consisting of the full-length protein (R1) or just the β-propeller domain (R2) or tail region (R3) (Fig. 2A). The RbBP5 constructs were immobilized on affinity resin, and WDR5 binding was monitored by PAGE analysis (Fig. 2B). Robust binding was detected only for full-length RbBP5 or the tail region construct indicating that the RbBP5 β-propeller does not directly interact with WDR5. This WDR5 binding region within the RbBP5 tail was more precisely mapped and eventually limited to a short segment located between residues 361 and 380 (supplemental Fig. 1). Using synthetic peptides and a combination of crystallography and isothermal calorimetry eventually leads to identification of an optimal peptide that corresponded to residues 369–381 (data not shown). In isothermal calorimetry experiments, this peptide binds to WDR5 with a stoichiometry of ∼1:1 and a KD of 5.6 μm (Fig. 2C).
Structural Analysis of RbBP5 Peptide Binding to WDR5
To determine the molecular basis of the interaction, crystals were obtained of a complex of WDR5 and the RbBP5(369–381)-peptide. The complex crystallized in space group P21, and the structure was solved by molecular replacement at 2.4 Å as described under “Experimental Procedures.” The asymmetric unit contains two complexes, and the relevant crystallographic statistics are presented in Table 1. There are no significant conformational changes between the WDR5 molecule in the model and the new structure (a root mean square deviation of 0.42 Å on superposition of all Cα atoms). Examination of the complex shows that the RbBP5 peptide binds in an extended fashion to an uncharacterized site located on one of the faces of the WDR5 β-propeller disc (Fig. 3A). The backbone of the peptide is well ordered between residues equivalent to RbBP5(372–381), as are the side chains of the peptide that mediate the interaction with WDR5. In contrast, many of the acidic side chains in the peptide are solvent-exposed and poorly ordered (Fig. 3B).
TABLE 1.
Crystallographic data collection and refinement
| Protein Data Bank code, binary (2XL2) | Protein Data Bank code, ternary (2XL3) | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c | 47.4, 81.3, 87.5 Å | 47.1, 81.3, 86.3 Å |
| α, β, γ | 90.0, 93.1, 90.0° | 90.0, 91.1, 90.0° |
| Resolution | 59.5-2.4 Å (2.5-2.4 Å) | 86.3-2.7 Å (2.9-2.7 Å) |
| Rmerge | 0.090 (0.534)a | 0.113 (0.458)a |
| Mn I/σI | 9.4 (2.3)a | 7.2 (2.5)a |
| Completeness | 98.4% (98.0%)a | 98.7% (99.0%)a |
| Redundancy | 3.6 (3.6)a | 3.0 (3.0)a |
| Wilson B-factor | 31.2 | 32.0 |
| Refinement | ||
| Resolution | 2.40 Å | 2.70 Å |
| No. of reflections | 23,706 | 16,657 |
| Rworkb/Rfreec | 0.188/0.243 | 0.176/0.235 |
| No. of atoms | ||
| Protein | 4884 | 4902 |
| Ligand/ion | 24 (glycerol) | 18 (glycerol) |
| Water | 189 | 105 |
| B-factors | ||
| Protein | 37.8 | 37.1 |
| Ligand | 21.9 (glycerol) | 56.2 (glycerol) |
| Water | 37.2 | 31.8 |
| Root mean square deviation | ||
| Bond lengths | 0.007 Å | 0.007 Å |
| Bond angles | 1.1° | 1.1° |
a The average value across the resolution range and that in parentheses is the value for the highest resolution bin (2.5-2.4 and 2.9-2.7 Å, respectively).
b Rwork = Σ‖Fo| − |Fc‖/Σ |Fo|.
c Rfree = ΣT ‖Fo| − |Fc‖/ΣT |Fo|, where T is a test data set of 5% of the total reflections randomly chosen and set aside before refinement.
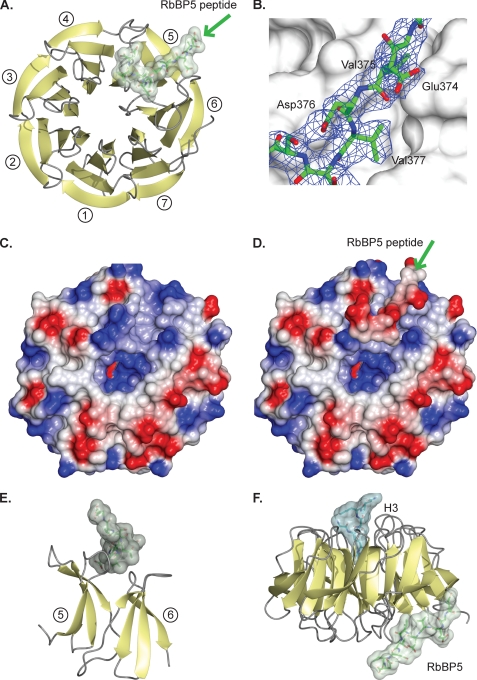
FIGURE 3.
Structural analysis of WDR5 protein complexes. A, schematic of WDR5 (yellow strands, gray loops) and RbBP5 peptide (green stick with transparent surface). The β-propeller blades are numbered from 1 to 7 starting from the second strand. The greens arrow indicates the direction of the peptide. B, RbBP5 peptide (green stick) on the WDR5 surface (gray) with the refined 2Fo – Fc map at 1σ for the peptide in blue. C, surface potential of the WDR5 face, a positively charged feature (blue), stands out against the otherwise negatively charged (red) features. D, surface potential of the WDR5 face with the RbBP5 peptide bound. The green arrow indicate the direction of the peptide. E, side view of blades 5 and 6 of the WDR5 β-propeller (yellow) showing RbBP5 peptide interacting with the connecting loops. F, ternary complex of WDR5 (yellow/gray) shown in the conventional orientation with histone H3 peptide (cyan) bound in the canonical axial binding site at the top and RbBP5 (green) bound to the opposite face.
Analysis of the electrostatic potential surface of WDR5 highlights how the RbBP5-binding site forms a distinct, positively charged feature that contrasts to the dominant negatively charged character of that side of the protein (Fig. 3C). This may have a role in initially capturing the predominantly negatively charged motif in RbBP5. Interestingly, binding of the negatively charged RbBP5 peptide leads to a uniformly negatively charged surface on the complex (Fig. 3D). It is currently unclear if this feature has any further role in the assembly of the core complex or the association of the core complex with any of the other chromatin modification machineries localized at sites of H3K4 methylation. The groove into which the peptide binds is generated by the loops that link the strands that form blades 5 and 6 of the WDR5 β-propeller (Fig. 3E). As the peptide only binds to one edge of the blades, the interacting residues are dispersed throughout the primary structure of WDR5 rather than forming a continuous stretch of residues. Nevertheless, when the sequences of orthologues are threaded onto the WDR5 structure and the predicted electrostatic surfaces compared, this positively charged feature, between blades 5 and 6, is clearly conserved in higher species (supplemental Fig. 2).
The RbBP5-binding site is on the opposite face of WDR5 from the canonical binding site that has been shown to accommodate either the WDR5 amino terminus, the histone H3 tail, or the MLL Win peptide (23, 25–27, 29). To confirm that binding of the RbBP5 peptide was independent of occupation of the H3/MLL-binding site, a ternary complex was crystallized consisting of WDR5 and both the RbBP5 peptide and a histone H3 tail peptide (Fig. 3F). This ternary complex diffracted to 2.7 Å and was solved by molecular replacement, and the relevant crystallographic statistics are presented in Table 1. To our knowledge, this is the first example of a crystal structure in which two independent binding sites on a β-propeller are simultaneously occupied. As expected, the two peptides bind independently to their respective binding sites on opposite faces of WDR5. There are no evident conformational changes in the WDR5 structure between the binary and ternary complex structures (a root mean square deviation of 0.2 Å on all Cα atoms and no significant side chain conformation changes in the binding site). The activity and structural data suggest that WDR5 acts as a binding platform, and its role in the mechanism of enhancing MLL1 methyltransferase activity is to promote the interaction between RbBP5 and the MLL1 SET domain.
RbBP5 Binds to WDR5 through a Conserved Interaction Motif
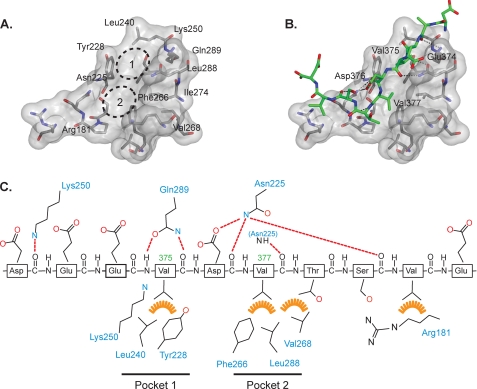
Detailed analysis shows that as well as having the positively charged features described above, the RbBP5-binding site also has a strongly hydrophobic character (Fig. 4A). Two hydrophobic pockets are formed that accommodate two residues, Val-375 and Val-377, on the RbBP5 motif (Fig. 4B). These pockets contribute to the relatively high affinity binding of the peptide for WDR5. Pocket 1 is created by the WDR5 residues Tyr-228 and Leu-240 and the aliphatic part of the Lys-250 side chain, although pocket 2 includes Phe-266, Val-268, and Leu-288 (Fig. 4C). A further interaction occurs between the RbBP5 Val-380 residue and the aliphatic part of WDR5 Arg-181. There are relatively few polar interactions, but the notable ones include hydrogen bonds involving the main chain adjacent to the two principal valine residues and the side chains of the WDR5 residues Asn-225 and Gln-289. These presumably help to lock the valine side chains into their respective hydrophobic pockets and stabilize the interaction.
FIGURE 4.
Details of the WDR5/RbBP5 interaction. A, surface representation of the RbBP5-binding site on WDR5 (transparent gray surface). The residues that interact with the peptide are shown as sticks, and the two valine binding pockets are indicated by a dotted line. B, same surface view as A, but with the RbBP5 peptide (green) bound and hydrogen bonds indicated by dotted lines. C, schematic representation of RbBP5 peptide indicating polar (red dotted lines) and hydrophobic (orange) interactions with WDR5 residues (blue labels).
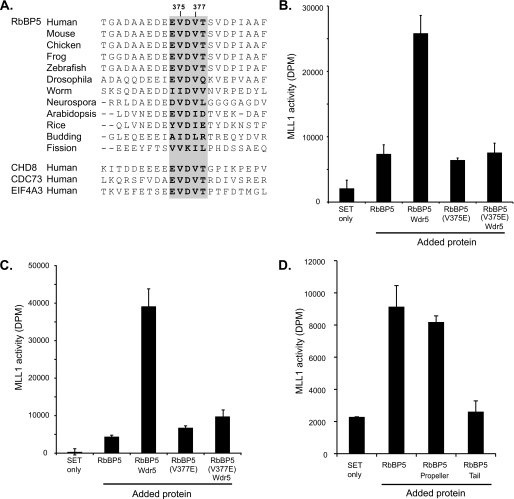
Like WDR5 proteins, the orthologues of RbBP5 have relatively high overall sequence similarity, and the Glu-Val-Asp-Val-Thr pentapeptide motif is evolutionarily well conserved (Fig. 5A). In fact, the pentapeptide motif is identical from humans to frog and zebrafish. Having identified a conserved WDR5-binding motif within RbBP5, we were interested to discover if the same motif occurred in other eukaryotic regulatory proteins. Using the ProSite server, eukaryotic protein sequences on the UniProtKB/Swiss Protein Database were searched with the pattern [GAED]-E-V-D-V-T. In addition to RbBP5, three other potentially significant proteins were identified in which this pattern occurs and is evolutionary conserved: CHD8, CDC73, and EIF4A3 (Fig. 5A). Of these, the chromatin-remodeling enzyme CHD8 is the most immediately noteworthy, as it has recently been shown to associate with the MLL core complex (39). CHD8 is a member of a family of SNF2-like ATP-dependent DNA helicases characterized by tandem chromodomains (40), and the sequence Glu-Glu-Val-Asp-Val-Thr occurs in a region just before the tandem chromodomains.
FIGURE 5.
WDR5 interaction motif. A, multiple sequence alignment of RbBP5 orthologues indicating the WDR5 interaction motif (gray background with boldface type) and flanking sequences. The same motif found in other nuclear proteins identified by motif scanning is shown. B and C, methyltransferase assay of MLL1 SET domain (Win construct) using unmodified H3 peptide substrate with RbBP5 wild-type and mutant proteins alone and in combination with WDR5. D, methyltransferase assay of MLL1 SET domain construct with unmodified H3 substrate alone and with addition of RbBP5 constructs representing the full-length protein, β-propeller through to interaction motif (residues 1–380) and tail (residues 361–538).
To probe the functional role of the WDR5/RbBP5 interaction, we assessed the effect of mutations that replaced the key valine residues with an acidic residue (Val-375 or Val-377 to glutamate). Full-length RbBP5 protein constructs carrying either V375E or V377E mutations were prepared and compared with wild-type protein in terms of binding or the ability to stimulate methyltransferase activity. The mutant proteins expressed at a similar level to the wild-type protein and exhibited equivalent behavior through all steps of purification. In co-precipitation experiments, the ability of these mutant proteins to bind to WDR5 was so severely reduced that recovery of WDR5 on the beads was only equivalent to that of the negative control of WDR5 binding to resin alone (supplemental Fig. 3) or histidine-tagged SUMO control. A series of methyltransferase assays were carried out with the MLL1 Win construct using an unmodified peptide substrate and analyzing the addition of wild-type or mutant RbBP5s (Fig. 5, B and C). In the absence of WDR5, the RbBP5 glutamate mutants stimulate MLL1 SET domain activity to similar levels as the wild-type protein, but the ability of the mutant proteins to further stimulate MLL1 activity in the presence of WDR5 is completely lost. Taken together, our data argue that disruption of the WDR5/RbBP5 interaction destabilizes the tripartite complex, and the ability of WDR5 to stimulate activity by promoting the RbBP5/MLL1 interaction is lost.
RbBP5 Topology and MLL1 Activation
Having established that only a small motif in the RbBP5 carboxyl-terminal tail region binds to WDR5, we wanted to determine which region of the protein is responsible for stimulating MLL1 methyltransferase activity. As noted above, in addition to the β-propeller domain, RbBP5 has a lengthy and apparently unstructured carboxyl-terminal tail. To delineate the role of these two regions in the regulation of MLL1 methyltransferase activity, we prepared three RbBP5 constructs representing the full-length protein, a construct consisting of the β-propeller region, and a short section of tail, including the WDR5 interaction motif (residues 1–380) and the tail region only (residues 361–538). The ability of these constructs to stimulate methyltransferase activity was measured in an assay with the minimal SET domain construct and an H3 tail peptide substrate (Fig. 5D). Consistent with earlier findings, addition of the full-length RbBP5 construct significantly stimulates the activity of the isolated SET domain. Addition of a truncated construct (residues 1–380) containing the β-propeller and a short section of tail is also sufficient to enhance activity to the same degree, whereas the construct representing only the tail region did not enhance activity to levels beyond those observed using the MLL1 SET domain alone. This indicates that the β-propeller domain of RbBP5 interacts with the SET domain and promotes the active complex, whereas the adjacent tail region binds to WDR5 and stabilizes formation of the subcomplex. Here, we infer that one role of the RbBP5 β-propeller is to interact with the MLL1 SET domain, but as we have observed with WDR5, it is possible that it may also have other binding partners with roles in the formation of the core complex. Our mapping and structural analysis suggests that only a small motif in the RbBP5 tail is sufficient for the interaction with WDR5, and the function of the rest of the RbBP5 tail region remains unaccounted for.
DISCUSSION
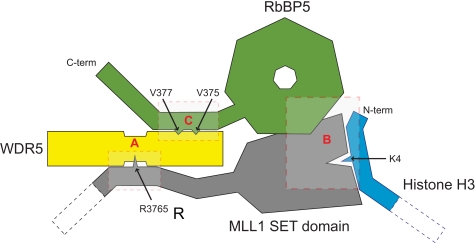
Histone H3K4 methylation is a potent signal that positively regulates transcription, and consequently, an intricate multiprotein system that controls the deposition of this mark has evolved. The core MLL complex proteins, WDR5, RbBP5, Ash2L, and DPY-30, associate with all six of the human H3K4 methyltransferase enzymes (Set1A, Set1B, MLL1, MLL2, MLL3, and MLL4) and control both the specificity and the activity of the partner enzyme. Here, we have focused on the role of the two β-propeller proteins, WDR5 and RbBP5, in the assembly of the complex and the activation of MLL1. Previous analyses of WDR5 interactions have focused on the binding site at the axial cavity (reviewed in Ref. 41). In the WD40 repeat β-propeller proteins, this location has commonly been identified as a protein/protein interaction site (42–44). For WDR5, reported binding partners at this site include histone H3 tail, the MLL1 Win peptide, or even the WDR5 amino terminus (23, 25–27, 29). Although this suggests weak affinity, in fact, in all cases the complementary recognition sequence is a related short arginine-containing motif. Here, we have identified a novel binding site on the opposite face of WDR5 and shown that it binds independently to a conserved motif in RbBP5. This WDR5-binding motif is located in the tail region of RbBP5 and is presumably positioned to facilitate the productive interaction between the RbBP5 β-propeller and the SET domain. We propose that in the active complex WDR5 acts as a hub that secures the methyltransferase enzyme on one side and its effector protein RbBP5 on the other (Fig. 6). This model incorporates the available biochemical data, which indicates that WDR5 is necessary for full methyltransferase activity but is not sufficient to directly enhance activity in the absence of RbBP5.
FIGURE 6.
A schematic model of the active complex between WDR5, RbBP5, and the MLL1 SET domain summarizing the interactions between the methyltransferase and the two β-propeller proteins. Residues that are critical for the interactions and the target histone lysine have been indicated with arrows. The interactions in the shaded boxes are supported by evidence from A, Refs. 26–28; B, this work; C, this work and Ref. 29.
The WDR5-binding pentapeptide motif is absolutely conserved in higher mammal orthologues of RbBP5 and even in zebrafish and frog (Fig. 5A). With further evolutionary distance, there are some conservative substitutions, but an obviously related sequence motif can be identified in worms and plants. Likewise, when the electrostatic surface of WDR5 is analyzed, by threading their sequences on to the WDR5 structure, the characteristic positively charged binding site is also conserved (supplemental Fig. 2). Given that the binding channel forms this conspicuous positively charged feature on WDR5, and the region of RbBP5 in which the pentapeptide motif occurs is predominantly characterized by acidic side chains, the dependence of binding on hydrophobic interactions was particularly surprising. However, in the bound conformation the acidic side chains are oriented so that they point away from the WDR5 surface toward solvent rather than interacting with residues in the binding groove. It is not known if this effective masking of the binding site by presentation of the negative surface has any further significance for the eventual formation of the higher order complexes. In yeast species, it is not clear if the interaction between the WDR5 and RbBP5 orthologues (Swd3 and Swd1) has a similar molecular basis. Neither the sequence conservation of the Schizosaccharomyces pombe nor Saccharomyces cerevisiae Swd1 nor the surface feature on the modeled Swd3 structure is as immediately apparent. To determine whether binding of these orthologues involves an analogous hydrophobically charged hydrophobic motif will require further experimental data.
We think it is significant that an RbBP5-like motif can be identified in other chromatin-associated human proteins (Fig. 5A). We have not yet verified the interaction between these proteins and WDR5, but the chromodomain helicase CHD8, a regulator of HOX gene expression, has been shown to associate directly with the MLL core complex members, including WDR5 (39, 45). In RbBP5 the motif is located in a putative unstructured region just after the β-propeller, and in CHD8 the motif is located in a putative unstructured region prior to the histone-binding tandem chromodomains. It is plausible that there could be an analogous interaction between WDR5 and CHD8 that involves the newly identified binding site and the pentapeptide motif. A second protein in which this motif is conserved is the tumor suppressor CDC73 (Parafibromin or HRPT2), which is a member of the chromatin-associated regulatory Paf1 complex, and this has also been shown to be located at sites of H3K4 trimethylation and therefore may interact with the MLL complex (46, 47). Another helicase EIF4A3, which is a member of the exon splicing junction complex, also contains the pentapeptide motif. In this protein it is located just before the ATP binding domain and so is also potentially located in an exposed unstructured region (48, 49). It is not currently known if either CDC73 or EIF4A3 directly interacts with WDR5, but the presence of the RbBP5-like motif and their association with chromatin-mediated mechanisms indicate that this could be an active area of future research.
It is noteworthy that in addition to being integral to the assembly of the methyltransferase complex, there is evidence that WDR5 may have roles independent of the KMT2 family. For example, it has been reported that WDR5 associates with the histone acetyltransferase males absent on first containing the nonspecific lethal complex (50, 51). Additionally, isolation of complexes containing the nuclear acceptor co-activator interacting factor (NIF-1) identified the core MLL complex members WDR5, Ash2L, and RbBP5 but not a methyltransferase component (52). A recent report indicates that not only is WDR5 an important component of an NF-κB-IRF3 complex assembled in response to viral infection, but it was also predominantly localized to the cytoplasm in a 293 cell line (53). It is plausible that both WDR5 interaction sites may accommodate different binding partners, and the protein has multiple roles involving different complexes depending on the context of these specific binding partners.
The picture that is emerging is that WDR5 functions as a binding platform that helps support the formation of protein complexes and promotes their activity by facilitating molecular interactions between effector molecules. In the KMT2 methyltransferase core complex, WDR5 provides an essential scaffold without which the effective activity of the methyltransferase complex is compromised. In the absence of this platform, the interaction of RbBP5 with the methyltransferase is weakened, and it is not able to achieve appropriate regulatory control. We have demonstrated that simple amino acid substitutions targeting the WDR5/RbBP5 interaction can have a profound effect on the activity of the enzyme complex. It is feasible that the small well defined pockets into which the valine residues of the WDR5-interacting motif on RbBP5 bind could be targeted by small chemical moieties. This may provide an alternative pharmaceutical route to target the activity of the KMT2 family methyltransferases other than by directly targeting the SET domain.
Supplementary Material
Acknowledgments
We thank S. M. Roe for assistance with crystallography in the preliminary stages of this project and La'Verne Rennalls for assistance with protein expression in insect cells. National Institute of Health Research Biomedical Research Centre is recipient of National Health Service funding.
This work was supported by the Career Development Faculty Programme of the Institute of Cancer Research (to J. R. W.) and benefits from infrastructural support for structural biology at ICR by Cancer Research UK.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
The atomic coordinates and structure factors (codes 2XL2 and 2XL3) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- H3K4
- histone H3 lysine 4
- MLL
- mixed-lineage leukemia
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1.Bhaumik S. R., Smith E., Shilatifard A. (2007) Nat. Struct. Mol. Biol. 14, 1008–1016 [DOI] [PubMed] [Google Scholar]
- 2.Albert M., Helin K. (2009) Semin. Cell Dev. Biol. 21, 209–220 [DOI] [PubMed] [Google Scholar]
- 3.Fischle W., Wang Y., Allis C. D. (2003) Curr. Opin. Cell Biol. 15, 172–183 [DOI] [PubMed] [Google Scholar]
- 4.Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007) Nat. Rev. Mol. Cell Biol. 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger S. L., Kouzarides T., Shiekhattar R., Shilatifard A. (2009) Genes Dev. 23, 781–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 7.Sims R. J., 3rd., Reinberg D. (2006) Genes Dev. 20, 2779–2786 [DOI] [PubMed] [Google Scholar]
- 8.Shilatifard A. (2008) Curr. Opin. Cell Biol. 20, 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 10.Roguev A., Schaft D., Shevchenko A., Pijnappel W. W., Wilm M., Aasland R., Stewart A. F. (2001) EMBO J. 20, 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krogan N. J., Dover J., Khorrami S., Greenblatt J. F., Schneider J., Johnston M., Shilatifard A. (2002) J. Biol. Chem. 277, 10753–10755 [DOI] [PubMed] [Google Scholar]
- 12.Dehé P. M., Dichtl B., Schaft D., Roguev A., Pamblanco M., Lebrun R., Rodríguez-Gil A., Mkandawire M., Landsberg K., Shevchenko A., Shevchenko A., Rosaleny L. E., Tordera V., Chávez S., Stewart A. F., Géli V. (2006) J. Biol. Chem. 281, 35404–35412 [DOI] [PubMed] [Google Scholar]
- 13.Dou Y., Milne T. A., Ruthenburg A. J., Lee S., Lee J. W., Verdine G. L., Allis C. D., Roeder R. G. (2006) Nat. Struct. Mol. Biol. 13, 713–719 [DOI] [PubMed] [Google Scholar]
- 14.Bach C., Slany R. K. (2009) Future Oncol. 5, 1271–1281 [DOI] [PubMed] [Google Scholar]
- 15.Hanson R. D., Hess J. L., Yu B. D., Ernst P., van Lohuizen M., Berns A., van der Lugt N. M., Shashikant C. S., Ruddle F. H., Seto M., Korsmeyer S. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14372–14377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu B. D., Hess J. L., Horning S. E., Brown G. A., Korsmeyer S. J. (1995) Nature 378, 505–508 [DOI] [PubMed] [Google Scholar]
- 17.McMahon K. A., Hiew S. Y., Hadjur S., Veiga-Fernandes H., Menzel U., Price A. J., Kioussis D., Williams O., Brady H. J. (2007) Cell. Stem Cell. 1, 338–345 [DOI] [PubMed] [Google Scholar]
- 18.Terranova R., Agherbi H., Boned A., Meresse S., Djabali M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6629–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider J., Wood A., Lee J. S., Schuster R., Dueker J., Maguire C., Swanson S. K., Florens L., Washburn M. P., Shilatifard A. (2005) Mol. Cell 19, 849–856 [DOI] [PubMed] [Google Scholar]
- 20.Dou Y., Milne T. A., Tackett A. J., Smith E. R., Fukuda A., Wysocka J., Allis C. D., Chait B. T., Hess J. L., Roeder R. G. (2005) Cell 121, 873–885 [DOI] [PubMed] [Google Scholar]
- 21.Wysocka J., Swigut T., Milne T. A., Dou Y., Zhang X., Burlingame A. L., Roeder R. G., Brivanlou A. H., Allis C. D. (2005) Cell 121, 859–872 [DOI] [PubMed] [Google Scholar]
- 22.Miller T., Krogan N. J., Dover J., Erdjument-Bromage H., Tempst P., Johnston M., Greenblatt J. F., Shilatifard A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han Z., Guo L., Wang H., Shen Y., Deng X. W., Chai J. (2006) Mol. Cell 22, 137–144 [DOI] [PubMed] [Google Scholar]
- 24.Schuetz A., Allali-Hassani A., Martín F., Loppnau P., Vedadi M., Bochkarev A., Plotnikov A. N., Arrowsmith C. H., Min J. (2006) EMBO J. 25, 4245–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruthenburg A. J., Wang W., Graybosch D. M., Li H., Allis C. D., Patel D. J., Verdine G. L. (2006) Nat. Struct. Mol. Biol. 13, 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couture J. F., Collazo E., Trievel R. C. (2006) Nat. Struct. Mol. Biol. 13, 698–703 [DOI] [PubMed] [Google Scholar]
- 27.Song J. J., Kingston R. E. (2008) J. Biol. Chem. 283, 35258–35264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel A., Vought V. E., Dharmarajan V., Cosgrove M. S. (2008) J. Biol. Chem. 283, 32162–32175 [DOI] [PubMed] [Google Scholar]
- 29.Patel A., Dharmarajan V., Cosgrove M. S. (2008) J. Biol. Chem. 283, 32158–32161 [DOI] [PubMed] [Google Scholar]
- 30.Southall S. M., Wong P. S., Odho Z., Roe S. M., Wilson J. R. (2009) Mol. Cell 33, 181–191 [DOI] [PubMed] [Google Scholar]
- 31.Berrow N. S., Alderton D., Sainsbury S., Nettleship J., Assenberg R., Rahman N., Stuart D. I., Owens R. J. (2007) Nucleic Acids Res. 35, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Assenberg R., Delmas O., Graham S. C., Verma A., Berrow N., Stuart D. I., Owens R. J., Bourhy H., Grimes J. M. (2008) Acta Crystallogr. F Struct. Biol. Crystalliz. Comm. 64, 258–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ireton G. C., Stoddard B. L. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 601–605 [DOI] [PubMed] [Google Scholar]
- 34.Leslie A. G. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26 [Google Scholar]
- 35.Bailey S. (1994) Acta Crystallogr. D Biol. Crystallogr 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 36.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 39.Yates J. A., Menon T., Thompson B. A., Bochar D. A. (2010) FEBS Lett. 584, 689–693 [DOI] [PubMed] [Google Scholar]
- 40.Hall J. A., Georgel P. T. (2007) Biochem. Cell. Biol. 85, 463–476 [DOI] [PubMed] [Google Scholar]
- 41.Trievel R. C., Shilatifard A. (2009) Nat. Struct. Mol. Biol. 16, 678–680 [DOI] [PubMed] [Google Scholar]
- 42.Lodowski D. T., Pitcher J. A., Capel W. D., Lefkowitz R. J., Tesmer J. J. (2003) Science 300, 1256–1262 [DOI] [PubMed] [Google Scholar]
- 43.Jennings B. H., Pickles L. M., Wainwright S. M., Roe S. M., Pearl L. H., Ish-Horowicz D. (2006) Mol. Cell 22, 645–655 [DOI] [PubMed] [Google Scholar]
- 44.Margueron R., Justin N., Ohno K., Sharpe M. L., Son J., Drury W. J., 3rd., Voigt P., Martin S. R., Taylor W. R., De Marco V., Pirrotta V., Reinberg D., Gamblin S. J. (2009) Nature 461, 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson B. A., Tremblay V., Lin G., Bochar D. A. (2008) Mol. Cell. Biol. 28, 3894–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adelman K., Wei W., Ardehali M. B., Werner J., Zhu B., Reinberg D., Lis J. T. (2006) Mol. Cell. Biol. 26, 250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y. J., Han J. W., Youn H. D., Cho E. J. (2010) Nucleic Acids Res. 38, 382–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan C. C., Dostie J., Diem M. D., Feng W., Mann M., Rappsilber J., Dreyfuss G. (2004) RNA 10, 200–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z., Krainer A. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11574–11579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai Y., Jin J., Swanson S. K., Cole M. D., Choi S. H., Florens L., Washburn M. P., Conaway J. W., Conaway R. C. (2010) J. Biol. Chem. 285, 4268–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendjan S., Taipale M., Kind J., Holz H., Gebhardt P., Schelder M., Vermeulen M., Buscaino A., Duncan K., Mueller J., Wilm M., Stunnenberg H. G., Saumweber H., Akhtar A. (2006) Mol. Cell 21, 811–823 [DOI] [PubMed] [Google Scholar]
- 52.Garapaty S., Xu C. F., Trojer P., Mahajan M. A., Neubert T. A., Samuels H. H. (2009) J. Biol. Chem. 284, 7542–7552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y. Y., Liu L. J., Zhong B., Liu T. T., Li Y., Yang Y., Ran Y., Li S., Tien P., Shu H. B. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.