Abstract
Exercise induces a pleiotropic adaptive response in skeletal muscle, largely through peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α). PGC-1α enhances lipid oxidation and thereby provides energy for sustained muscle contraction. Its potential implication in promoting muscle refueling remains unresolved, however. Here, we investigated a possible role of elevated PGC-1α levels in skeletal muscle lipogenesis in vivo and the molecular mechanisms that underlie PGC-1α-mediated de novo lipogenesis. To this end, we studied transgenic mice with physiological overexpression of PGC-1α and human muscle biopsies pre- and post-exercise. We demonstrate that PGC-1α enhances lipogenesis in skeletal muscle through liver X receptor α-dependent activation of the fatty acid synthase (FAS) promoter and by increasing FAS activity. Using chromatin immunoprecipitation, we establish a direct interaction between PGC-1α and the liver X receptor-responsive element in the FAS promoter. Moreover, we show for the first time that increased glucose uptake and activation of the pentose phosphate pathway provide substrates for RNA synthesis and cofactors for de novo lipogenesis. Similarly, we observed increased lipogenesis and lipid levels in human muscle biopsies that were obtained post-exercise. Our findings suggest that PGC-1α coordinates lipogenesis, intramyocellular lipid accumulation, and substrate oxidation in exercised skeletal muscle in vivo.
Keywords: Fatty Acid Synthase, Lipogenesis, Nuclear Receptors, Pentose Pathway, Skeletal Muscle Metabolism, Transcription Coactivators, Transcription Regulation, LXRα, PGC-1α
Introduction
Over the last century, people have become increasingly sedentary. This development has resulted in a marked rise in the prevalence of metabolic disorders (1). Inversely, exercise is an excellent intervention in metabolic diseases. However, the molecular mechanisms that underlie the beneficial health effects of exercise are unclear.
Chronic endurance exercise elicits substantial adaptations in skeletal muscle, including changes in metabolic and myofibrillar properties (2). Peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α)2 is a crucial regulator of this adaptive response (3–5). Accordingly, exercise leads to a rapid and marked increase in PGC-1α levels in skeletal muscle (6, 7). Subsequently, PGC-1α interacts with a broad range of different transcription factors (8) and thereby promotes adaptive processes (9, 10). Chronic PGC-1α induction drives fiber-type switching from fast glycolytic toward slow oxidative fibers (11), increases aerobic work performance (12), and confers higher resistance to fatigue in skeletal muscle preparations ex vivo (11). Interestingly, muscle-specific overexpression of PGC-1α per se, even in the absence of physical activity or a functional motor neuron, is sufficient to drive these changes, which are typical of regular endurance training (11–13).
Higher intramyocellular lipid (IMCL) content is observed in endurance athletes and constitutes a typical adaptation to chronic endurance exercise (14). These lipids are recruited as an energy source during prolonged physical activity (15, 16). Although a central role for PGC-1α in enhancing lipid catabolism is well established (9, 10), it is currently not known whether elevated PGC-1α affects lipid anabolism in trained skeletal muscle in vivo. A recent in vitro study in myoblasts suggested that adenoviral overexpression of PGC-1α is paralleled by increased gene expression of fatty acid synthase (FAS) (17). Thus, PGC-1α might play a role in muscle lipid refueling, at least in a cell culture model under hyperglycemic conditions (17). Currently, it is unresolved whether PGC-1α can induce FAS transcription in vivo, whether there is a direct relationship between activation of PGC-1α and lipogenesis, how this could be brought about mechanistically, and how this process is fueled.
We therefore studied the effect of stably elevated muscle PGC-1α levels on de novo lipogenesis in vivo in a transgenic mouse model that allows the dissociation between the consequences of elevated muscle PGC-1α and other confounding factors induced by exercise (11). In particular, we tested the effect of PGC-1α on FAS, the multifunctional enzyme that catalyzes all seven reactions of de novo lipogenesis, as well as on glucose-6-phosphate dehydrogenase, the rate-limiting enzyme of the pentose phosphate pathway. The latter produces NADPH, which serves as reducing agent for lipogenesis. Moreover, we investigated the molecular mechanisms that underlie these adaptations. We focused our efforts on the function of PGC-1α in vivo in the extensor digitorum longus (EDL) muscle, which represents a typical glycolytic muscle that naturally expresses low levels of PGC-1α (7, 11). Gain of function of PGC-1α in EDL muscle results in PGC-1α protein levels that are comparable with those normally observed in oxidative muscles (11). The main findings of this study in the muscle-specific PGC-1α transgenic mouse model were subsequently validated in human muscle biopsies that were obtained pre- and post-exercise.
MATERIALS AND METHODS
Animals
MPGC-1α transgenic (TG) mice (11) and control littermates were maintained in a conventional facility with a fixed 12-h light/dark cycle on a commercial pelleted chow diet with free access to tap water.
Human Muscle Biopsies
Human biopsies were obtained from vastus lateralis muscle in the course of a previously published study (18). In brief, all subjects trained on an electrically braked cycle ergometer at a constant load for 30 min/day, 5 days/week, for a total of 6 weeks with 85% of maximal heart rate after 10 min of exercise and lactate levels between 4 and 6 mmol/liter. Biopsies were taken from vastus lateralis muscle before and after the 6-week training period using the Bergstrom technique. Muscle tissue was immediately frozen in liquid nitrogen and stored at −80 °C for later RNA extraction.
Lipid Quantification by Electron Microscopy
Quantification of muscle lipids was performed as described previously (19). Glutaraldehyde-fixed samples were dehydrated in increasing ethanol concentrations and embedded in Epon. After cutting ultrathin sections (50–70 nm) on an LKB Ultrotome III, uranyl acetate and lead citrate staining was performed. The sections were then photographed and examined. We analyzed 15 micrographs of one section per animal.
RNA Extraction and RT-PCR
Frozen tissues were homogenized under liquid nitrogen, and total RNA was isolated using TRIzol reagent (Invitrogen). RNA concentrations were adjusted, and reverse transcription was carried out using random hexamer primers (Promega). Real-time PCR analysis (Power SYBR Green Master Mix, Applied Biosystems) was performed using an ABI Prism 7000 sequence detector. Relative expression levels for each gene of interest were calculated with the ΔΔCt method and normalized to the expression of the Tata box-binding protein (TBP). Primer sequences are listed in supplemental Table T1.
De Novo Lipogenesis
De novo lipogenesis was assayed according to the method of Stansbie et al. (20). In brief, 3H2O (25 μCi/g of body weight) was injected intraperitoneally, and 1 h later, the animals were killed. Muscles were dissected, saponified in ethanolic KOH, and acidified with H2SO4. Fatty acids were extracted with light petroleum, and radioactivity was quantified using a Beckman liquid scintillation counter. The contralateral muscle was used to assay the incorporation of tritium into different lipid classes as described by Fungwe et al. (21).
Glucose Uptake
Glucose uptake was determined using tritium-labeled 2-deoxyglucose (Amersham Biosciences) according to the protocol of Hansen et al. (22). Radioactivity was quantified using a Beckman liquid scintillation counter. All values were corrected for initial tissue weight and are expressed as -fold change compared with control animals.
Muscle Metabolites (NADP, NADPH, Glutathione, and Glucose 6-Phosphate)
NADP and NADPH levels were assessed colorimetrically using an EnzyChrom NADP+/NADPH assay kit (BioAssay Systems). Reduced, oxidized, and total glutathione levels were determined using a glutathione assay kit (BioVision Research Products). Glucose 6-phosphate was determined as described by Wende et al. (23).
Enzymatic Activities
Muscles were dissected, immediately frozen in liquid nitrogen, and homogenized under liquid nitrogen. FAS activity was measured photometrically according to the method described by Pénicaud et al. (24). Glucose-6-phosphate dehydrogenase (G6PDH) activity was determined according to the method of Ninfali et al. (25).
Incorporation of Glucose into Macromolecules
Incorporation of [6-3H]glucose (Amersham Biosciences) into RNA and DNA was measured according to the method described by Glazer and Weber (26).
Cell Culture Experiments
Mouse C2C12 myoblasts were maintained in DMEM supplemented with 10% fetal bovine serum in an atmosphere of 5% CO2 in a subconfluent culture. For reporter gene assays, C2C12 myoblasts were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Renilla luciferase was used for normalization. Cells were harvested 24 h after transfection, and luciferase activity was measured using the Dual-Glo luciferase assay (Promega). Liver X receptor α (LXRα) siRNA was purchased from Santa Cruz Biotechnology. C2C12 myoblasts were differentiated into myotubes and then infected with adenoviral vectors for murine GFP or GFP-PGC-1α.
ChIP Assay
ChIP assays were performed using ChIP-IT Express (Active Motif) according to the manufacturer's instructions. Chromatin was subjected to immunoprecipitation using anti-PGC-1α antibodies (PGC-1 (H-300), Santa Cruz Biotechnology) or unrelated IgG bound to protein G-coupled magnetic beads. Precipitated DNA was recovered and amplified by RT-PCR with primers flanking the LXR-responsive element (LXRE)- or sterol-responsive element-binding protein (SREBP)-binding site, respectively.
Data Analysis and Statistics
All data are presented as means ± S.E. The data were analyzed by Student's two-tailed unpaired t test or the Mann-Whitney test when the difference between the two S.D. values was significantly different. For the analyses of human samples, Student's two-tailed paired t test was used. For the analysis of reporter gene assays, two-factor analysis of variance was performed. Post hoc pairwise comparison was done by Student's unpaired t test.
RESULTS
Elevated de Novo Lipogenesis and Lipid Accumulation in Glycolytic Muscle of MPGC-1α TG Mice
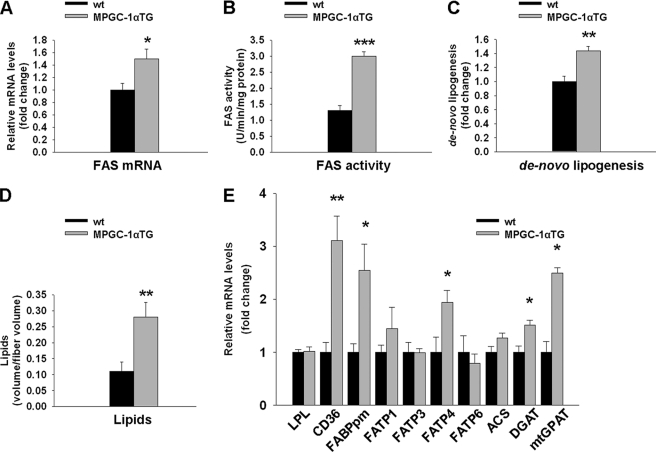
We first tested whether PGC-1α affects gene expression and/or activity of skeletal muscle FAS in vivo. MPGC-1α TG mice showed higher levels of FAS expression (+50%, p < 0.05) (Fig. 1A) and enzymatic activity (+131%, p < 0.001) (Fig. 1B) in skeletal muscle than their control littermates, along with increased de novo lipogenesis (+44%, p < 0.01) (Fig. 1C) and elevated IMCL content (+157%, p < 0.01) (Fig. 1D). Detailed analysis of the different lipid components revealed higher levels of de novo synthesized free fatty acids and triglycerides in skeletal muscle of MPGC-1α TG mice (supplemental Fig. S1, A and B). Because muscle often contains interspersed adipose cells that express high levels of FAS and show high rates of lipogenesis, we validated FAS expression, activity, and de novo lipogenesis in cultured skeletal muscle cells overexpressing PGC-1α. As observed in muscle in vivo, PGC-1α also increased FAS mRNA expression, activity, and de novo lipogenesis directly in muscle cells (supplemental Fig. S2, A–C).
FIGURE 1.
Enhanced lipid anabolism in EDL muscle of MPGC-1α TG animals. A–D, FAS gene expression, activity, de novo lipogenesis, and accumulation of intramyofibrillar lipids, respectively, in glycolytic muscle of MPGC-1α TG versus wild-type animals. E, relative expression of genes involved in fatty acid uptake (lipoprotein lipase (LPL), CD36, plasma membrane fatty acid-binding protein (FABPpm), and fatty acid transporter proteins (FATPs)), fatty acid activation (ACS), and esterification (acetyl-CoA:diacylglycerol acyltransferase (DGAT) and mitochondrial glycerol-3-phosphate acyltransferase (mtGPAT)). All values are expressed as means ± S.E. (n = 6–8/group). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To test whether mRNA levels of genes involved in lipid uptake are concomitantly altered in MPGC-1α TG mice, we quantified the mRNA levels of lipoprotein lipase, CD36 (cluster of differentiation 36), plasma membrane fatty acid-binding protein, and different fatty acid transporter proteins. Of those, only CD36, plasma membrane fatty acid-binding protein, and FATP4 were significantly elevated in TG animals (Fig. 1E). We then determined the level of acetyl-CoA synthase (ACS), which activates imported lipids by linking them to coenzyme A. ACS mRNA expression was not different between control and TG animals (Fig. 1E). Finally, expression of genes implicated in fatty acid esterification, namely DGAT1 (acetyl-CoA:diacylglycerol acyltransferase 1) and mitochondrial glycerol-3-phosphate acyltransferase, was increased in MPGC-1α TG mice (+51 and +150%, respectively; p < 0.05) (Fig. 1E), whereas other mediators of lipid synthesis, such as lipin, remained unaltered (supplemental Fig. S3).
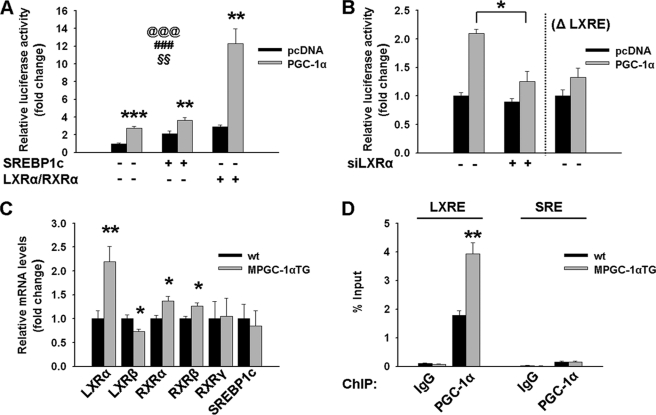
PGC-1α Directly Regulates the FAS Promoter through Coactivation of LXRα/Retinoid X Receptor α (RXRα)
As a consequence of the elevated FAS transcription in MPGC-1α TG animals (Fig. 1A), a possible regulatory effect of PGC-1α on the FAS proximal promoter was investigated. Our data indicate that PGC-1α increased FAS promoter activity and that this effect of PGC-1α was potentiated by the concomitant expression of LXRα/RXRα, but not of SREBP1c (Fig. 2A). Silencing of LXRα or deletion of the LXRE in the FAS promoter (ΔLXRE) abrogated the effect of PGC-1α on FAS transcription (Fig. 2B). In contrast, mutation of the SREBP1c-binding site did not reduce the effect of PGC-1α on LXRα/RXRα-induced FAS transcription (supplemental Fig. S4). Moreover, LXRα, RXRα, and RXRβ expression levels were significantly elevated, whereas SREBP1c was unaltered in skeletal muscle of MPGC-1α TG animals (Fig. 2C). Using ChIP followed by RT-PCR, we demonstrated that in MPGC-1α TG animals, PGC-1α was enriched in the LXRE of the FAS proximal promoter (Fig. 2D). In contrast, PGC-1α was not recruited to the SREBP-binding site in the FAS promoter of MPGC-1α TG and control animals (Fig. 2D).
FIGURE 2.
LXRα-dependent activation of the FAS promoter. A, FAS promoter activity following transfection of myoblasts with pcDNA (empty vector; black bars) or PGC-lα (gray bars) and in response to cotransfection with LXRα/RXRα or SREBP1c expression plasmids. Values are expressed as means ± S.E. (n = 6/group). @@@, effect of PGC-lα (p < 0.001); ###, effect of LXRα/RXRα or SREBP1c (p < 0.001); §§, interaction (p < 0.01) as assessed by two-factor analysis of variance. B, FAS promoter activity following transfection of myoblasts with pcDNA (empty vector; black bars) or PGC-lα (gray bars) and in response to silencing of LXRα or deletion of the LXR-responsive element (ΔLXRE). C, relative expression of genes involved in FAS promoter activation in muscle of MPGC-1α TG mice versus control mice. All values are expressed as means ± S.E. (n = 6–8/group). *, p < 0.05; **, p < 0.01. D, ChIP assays of mouse skeletal muscle: recruiting of PGC-lα to LXRE- and SREBP-responsive element (SRE)-binding sites in the FAS promoter of MPGC-1α TG mice and control animals, respectively. All values are expressed as means ± S.E. (n = 4/group). **, p < 0.01.
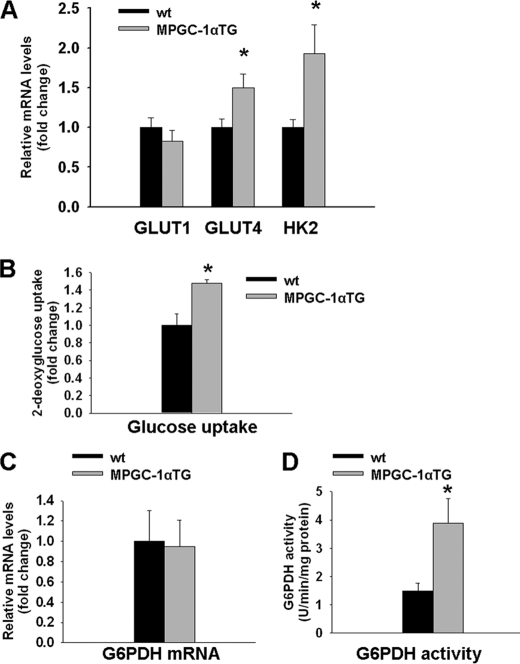
Increased Glucose Utilization in Glycolytic Muscle of MPGC-1α TG Mice
In addition to FAS activation, important cofactors for lipid biosynthesis need to be delivered through glucose metabolism, and sustained glucose uptake is required for lipogenesis to proceed. We analyzed different key steps of glucose uptake at the transcriptional level as well as glucose uptake directly. Gene expression of GLUT4, the main insulin-sensitive glucose transporter in muscle, and hexokinase 2, the enzyme used to trap glucose in muscle, was higher in MPGC-1α TG mice compared with control mice (+50 and +93%, respectively; p < 0.05), whereas expression of the second major glucose transporter in muscle (GLUT1) was unchanged (Fig. 3A). Moreover, glucose uptake as measured by the intracellular accumulation of tritiated deoxyglucose was elevated in MPGC-1α TG animals (+48%, p < 0.05) (Fig. 3B).
FIGURE 3.
Enhanced glucose uptake and pentose phosphate metabolism in EDL muscle of MPGC-1α TG mice. A, relative gene expression of GLUT1, GLUT4, and hexokinase 2 (HK2) in EDL muscle as measured by RT-PCR and expressed as -fold change over controls. B, glucose uptake in EDL muscle. C and D, determination of G6PDH mRNA levels and activity, respectively, in glycolytic muscle of MPGC-1α TG and control mice. All values are means ± S.E. (n = 6–8/group). *, p < 0.05.
Enhanced G6PDH Activity in MPGC-1α TG Mice
We speculated that increased glucose uptake in MPGC-1α TG mice leads to intracellular accumulation of Glc-6-P. Indeed, MPGC-1α TG mice showed higher levels of muscle Glc-6-P than their control littermates (supplemental Fig. S5A). Glc-6-P concomitantly serves as the initial substrate and thereby as an activator of the pentose phosphate pathway. Thus, we measured the expression and enzymatic activity of G6PDH. PGC-1α gain of function did not affect G6PDH gene expression (Fig. 3C) but led to increased enzymatic activity (+158%, p < 0.05) (Fig. 3D). To determine whether PGC-1α increases G6PDH activity in muscle cells, we assessed G6PDH in isolated muscle cells overexpressing PGC-1α. Similar to our findings in muscle in vivo, PGC-1α augmented G6PDH activity directly in muscle cells (supplemental Fig. S5B).
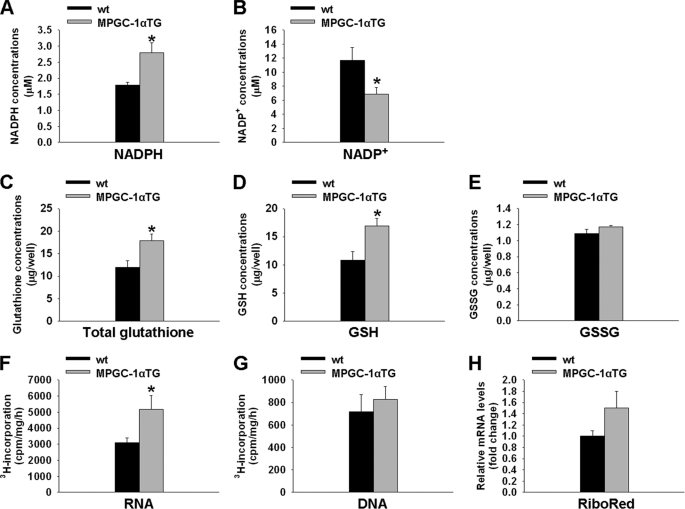
Increased Synthesis of NADPH, GSH, and Ribose 5-Phosphate in MPGC-1α TG Mice
The pentose phosphate pathway is the major anaplerotic pathway of NADPH. NADPH serves as reducing agent for glutathione and is furthermore required for de novo lipogenesis. Thus, consistent with the higher G6PDH activity in muscle of MPGC-1α TG mice, we found significantly elevated NADPH with a parallel reduction in NADP+ concentration compared with controls (p < 0.05 for both) (Fig. 4, A and B). Consequently, we also observed higher levels of total and reduced glutathione (Fig. 4, C and D) in MPGC-1α TG mice (p < 0.05). In contrast, the amount of oxidized glutathione was similar in MPGC-1α TG and control animals (Fig. 4E).
FIGURE 4.
Increased pentose phosphate pathway products in MPGC-1α TG mice. A and B, concentrations of NADPH and NADP+, respectively, in glycolytic muscle of MPGC-1α TG mice versus control mice. C–E, amounts of total, reduced (GSH), and oxidized (GSSG) glutathione, respectively, extracted from glycolytic muscle. F and G, incorporation of tritium-labeled glucose into RNA and DNA, respectively, in EDL muscle of MPGC-1α TG and control mice. H, determination of ribonucleotide reductase (RiboRed) mRNA levels. All values are means ± S.E. (n = 6–8/group). *, p < 0.05.
In addition to its role in NADPH and glutathione production, the pentose phosphate pathway plays a crucial role in nucleotide biosynthesis by producing ribose 5-phosphate. Therefore, tritium-labeled glucose was used to test whether glucose diverted toward the pentose phosphate pathway is partially incorporated into nucleotides as ribose 5-phosphate. Although tritium-labeled glucose was used as a substrate for RNA synthesis in MPGC-1α TG mice (+65%, p < 0.05) (Fig. 4F), no changes were observed for incorporation of glucose metabolites into DNA (Fig. 4G). Furthermore, gene expression of ribonucleotide reductase, a transcriptionally regulated gene involved in converting ribonucleotides to deoxyribonucleotides, was not different in MPGC-1α TG and control littermates (Fig. 4H).
Improved Capacity for de Novo Lipogenesis in Endurance Athletes
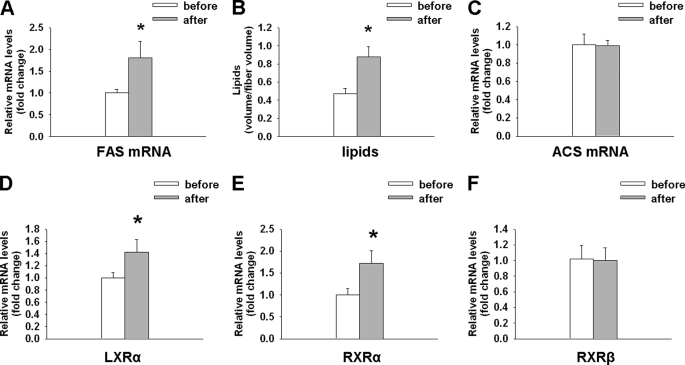
Endurance athletes display high levels of IMCLs; however, the origin of these lipids is unknown. Similar to the MPGC-1α TG animals, FAS gene expression was elevated in human muscle post-exercise (Fig. 5A). Importantly, the induction of FAS gene expression was paralleled by an increased level of IMCLs (Fig. 5B) after 6 weeks of endurance training. In contrast, ACS levels were not altered in human muscle post-exercise (Fig. 5C). Consistent with our mouse date (Fig. 2C), we observed elevated LXRα and RXRα levels in human muscle biopsies post-exercise (Fig. 5, D and E). RXRβ levels were not altered (Fig. 5F). Moreover, in line with our mouse data, GLUT4 and hexokinase 2 levels were elevated post-exercise in human muscle, whereas G6PDH levels were unchanged (supplemental Fig. S6, A–C).
FIGURE 5.
Higher FAS expression and IMCL in endurance athletes. A–C, FAS mRNA expression, IMCLs, and ACS mRNA expression, respectively, in human endurance athletes before and after 6 weeks of high intensity endurance exercise. D–F, relative expression of LXRα, RXRα, and RXRβ, respectively, in human muscle biopsies pre- and post-exercise. Values are expressed as means ± S.E. (n = 6). *, p < 0.05 as assessed by a paired t test.
DISCUSSION
Elevated IMCLs are a hallmark of type 2 diabetic muscle and contribute to harmful effects on glucose homeostasis and insulin sensitivity (27, 28). However, endurance athletes, who are highly insulin-sensitive, also display elevated IMCL levels, a phenomenon referred to as the “athlete's metabolic paradox” (29, 30). The difference in the molecular mechanisms between the IMCL accumulation in exercise and diabetes is poorly defined. We have shown that de novo lipogenesis in skeletal muscle contributes to the elevated IMCL content in muscle-specific PGC-1α TG mice, a genetic model for elevated endurance capacity, and, importantly, also in human endurance athletes. We have demonstrated in vivo that PGC-1α mediates accumulation of IMCL by recruiting the lipogenic machinery in muscle, specifically by inducing the expression and activity of FAS, a multifunctional enzyme that catalyzes all seven reactions required for de novo lipid biosynthesis. In combination, PGC-1α increases the expression of genes encoding lipid esterification proteins (namely acetyl-CoA:diacylglycerol acyltransferase and mitochondrial glycerol-3-phosphate acyltransferase) and thereby allows the coordinate synthesis and subsequent accumulation of intramyofibrillar lipids. Moreover, we have provided novel insights into the molecular mechanisms by which PGC-1α directly promotes FAS transcription by inducing and coactivating the nuclear receptor LXRα on the proximal FAS promoter. Taken together, these findings demonstrate for the first time a direct role of PGC-1α in muscle de novo lipogenesis and IMCL accumulation in vivo.
The absence of a transcriptional response of key genes for lipid uptake and activation implies that the uptake of fatty acids plays a minor role in accumulation of IMCL compared with de novo lipogenesis. Of all the genes that we studied, only CD36 and plasma membrane fatty acid-binding protein were significantly induced in MPGC-1α TG mice. However, in addition to their function at the cell membrane, these two genes have been associated with mitochondrial lipid oxidation and mitochondrial aspartate metabolism, respectively (31, 32), and thus might be more relevant as mitochondrial enzymes in this context. An important role for muscle de novo lipogenesis in the accumulation of IMCLs as opposed to lipolysis of triglycerides from adipose tissue, transport of fatty acids to the muscle, and subsequent lipid import is further substantiated by previous studies showing unaltered fat mass, dietary lipid intake, and circulating lipid levels in MPGC-1α TG mice fed regular chow or high fat-containing diet (12, 33).
Moreover, the mRNA levels of ACS, an enzyme that is indispensable to activate imported lipids but that is not required for de novo lipogenesis, are unaltered in MPGC-1α TG compared with wild-type mice and in human muscle biopsies pre- versus post-exercise, respectively. Finally, a direct functional analysis performed in muscle cells revealed that overexpression of PGC-1α reduces lipid import (17). Together, these data suggest that de novo lipogenesis takes precedence over lipid import for the generation of IMCL in muscle, at least in our experimental setting. Thus, the cause of IMCL accumulation in trained muscle might differ from that in type 2 diabetes, which could be more dominantly driven by lipid uptake (34).
According to the classic Randle hypothesis, lipid oxidation and glucose oxidation mutually regulate each other in a negative manner (35, 36). Consistently, it has been found that the elevation of β-oxidation by PGC-1α in muscle-specific TG animals is accompanied by a reduction in glycolysis and glucose oxidation. Moreover, the increase in glucose uptake mediated by PGC-1α can only partially be explained by glycogen synthesis because even in muscles fully loaded with glycogen, muscle-specific PGC-1α TG mice retain higher glucose uptake (23). Here, we have demonstrated that PGC-1α induces glucose uptake to enhance the glucose flux through the pentose phosphate pathway as a prerequisite to produce the NADPH that is required as the reducing agent for de novo lipogenesis. Furthermore, ribose 5-phosphate is made for nucleotide synthesis and the subsequent increase in mRNA transcription that is strongly promoted by PGC-1α. Although previous studies demonstrated elevated G6PDH activity in skeletal muscle following exercise (37), we now provide evidence that G6PDH activation and the parallel increase in FAS and IMCL are driven by PGC-1α.
Interestingly, muscle-specific PGC-1α TG animals exhibit a higher susceptibility to developing insulin resistance when fed a high-fat diet (33). This seems paradoxical because these animals exhibit all the features of trained animals (11, 12), and exercise is known to improve insulin sensitivity in muscle (38). Our findings could now explain this ostensible discrepancy by the direct effect of elevated PGC-1α on de novo lipogenesis and lipid accumulation. In MPGC-1α TG animals, lipids accumulate due to the high-fat diet and also the de novo lipogenesis that is promoted by ectopic PGC-1α. Moreover, despite their elevated oxidative capacity, these mice are sedentary when kept in standard animal cages, and lipid turnover in skeletal muscle is accordingly low compared with an active animal. Similarly, athletes that undergo training cessation often experience perturbations in insulin sensitivity and glucose homeostasis (39–41). These periods of relative insulin resistance are preceded by a reduction in PGC-1α, carnitine palmitoyltransferase 1b, and citrate synthase gene expression along with reduced mitochondrial lipid oxidation (42). Reduced muscle lipid oxidation in combination with elevated IMCL, i.e. a low turnover of lipids, mirrors the metabolic profile of type 2 diabetes (43). Our findings have highly relevant implications for the development of so-called exercise mimetics, pharmaceutical interventions aimed at mirroring the plastic adaptations of muscle to exercise (44, 45). If these compounds are true exercise mimetics, they will also promote de novo lipogenesis and thus increase the susceptibility to unwanted side effects, in particular with a high-fat Western diet and lack or cessation of actual exercise. Therefore, as a monotherapy, such pharmaceuticals are not likely to constitute a valid alternative to real exercise (46, 47).
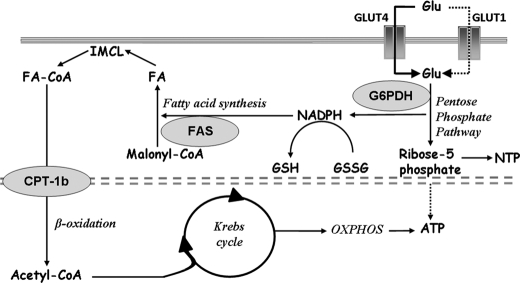
In conclusion, we have provided new evidence that PGC-1α promotes lipid anabolism in vivo by stimulating pentose phosphate pathway activity, de novo lipid biosynthesis, and IMCL accumulation, which can serve as substrates for subsequent physical activity (Fig. 6). In addition, we have provided the first insights into the molecular mechanisms that underlie PGC-1α-driven de novo lipogenesis in skeletal muscle. These metabolic adaptations are highly advantageous for trained muscle due to the tight coordination of energy demand and supply. Our data highlight novel approaches as well as pitfalls for the therapy of metabolic diseases that are associated with a sedentary lifestyle.
FIGURE 6.
PGC-1α coordinates anabolic and catabolic pathways in skeletal muscle. The model integrates the findings of this study and shows the PGC-1α-mediated coordination of de novo lipogenesis, lipid accumulation, and lipid oxidation. Enzymes being activated by PGC-1α and regulating metabolic key steps are indicated in gray ovals (G6PDH, FAS, and carnitine palmitoyltransferase 1b (CPT-1b)). PGC-1α coordinates anabolic processes (lipogenesis and IMCL accumulation) and catabolic processes (β-oxidation, the Krebs cycle, and oxidative phosphorylation (OXPHOS)) in skeletal muscle. PGC-1α enhances glucose (Glu) uptake and flux through the pentose phosphate pathway. Concomitantly, NADP+ is reduced to NADPH, which serves as reducing agent for lipogenesis. De novo synthesized fatty acids (FA) are then stored as IMCL and serve as energy substrate during endurance exercise. By metabolizing lipids through β-oxidation, the Krebs cycle, and oxidative phosphorylation, ATP for muscle contraction is produced. The flux of glucose toward the pentose phosphate pathway also generates ribose 5-phosphate, which constitutes a structural element of ATP and other nucleotides.
Supplementary Material
Acknowledgments
We thank Urs A. Meyer (Biozentrum, Basel, Switzerland) and our colleagues in the laboratory for discussions and comments on the manuscript, and we acknowledge Franziska Graber and Adolfo Odriozola (Institute of Anatomy, University of Bern) for excellent technical assistance. FAS-luciferase constructs were a generous gift from Tim Osborne (Department of Molecular Biology and Biochemistry, University of California).
This work was supported by Swiss National Science Foundation Grant SNF PP00A-110746, the Muscular Dystrophy Association, the SwissLife “Jubiläumsstiftung für Volksgesundheit und Medizinische Forschung,” the Swiss Society for Research on Muscle Diseases, the Swiss Diabetes Association, the Roche Research Foundation, the United Mitochondrial Disease Foundation, the Association Française contre les Myopathies, and the University of Basel.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table T1.
- PGC-1α
- peroxisome proliferator-activated receptor γ coactivator 1α
- IMCL
- intramyocellular lipid
- FAS
- fatty acid synthase
- EDL
- extensor digitorum longus
- MPGC-1α
- muscle PGC-1α
- TG
- transgenic
- G6PDH
- glucose-6-phosphate dehydrogenase
- LXRα
- liver X receptor α
- LXRE
- LXR-responsive element
- SREBP
- sterol-responsive element-binding protein
- ACS
- acetyl-CoA synthase
- RXRα
- retinoid X receptor α.
REFERENCES
- 1.Booth F. W., Laye M. J., Lees S. J., Rector R. S., Thyfault J. P. (2008) Eur. J. Appl. Physiol. 102, 381–390 [DOI] [PubMed] [Google Scholar]
- 2.Flück M., Hoppeler H. (2003) Rev. Physiol. Biochem. Pharmacol. 146, 159–216 [DOI] [PubMed] [Google Scholar]
- 3.Handschin C., Spiegelman B. M. (2006) Endocr. Rev. 27, 728–735 [DOI] [PubMed] [Google Scholar]
- 4.Lin J., Handschin C., Spiegelman B. M. (2005) Cell Metab. 1, 361–370 [DOI] [PubMed] [Google Scholar]
- 5.Handschin C., Spiegelman B. M. (2008) Nature 454, 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baar K., Wende A. R., Jones T. E., Marison M., Nolte L. A., Chen M., Kelly D. P., Holloszy J. O. (2002) FASEB J. 16, 1879–1886 [DOI] [PubMed] [Google Scholar]
- 7.Russell A. P., Feilchenfeldt J., Schreiber S., Praz M., Crettenand A., Gobelet C., Meier C. A., Bell D. R., Kralli A., Giacobino J. P., Dériaz O. (2003) Diabetes 52, 2874–2881 [DOI] [PubMed] [Google Scholar]
- 8.Finck B. N., Kelly D. P. (2006) J. Clin. Invest. 116, 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 10.St-Pierre J., Lin J., Krauss S., Tarr P. T., Yang R., Newgard C. B., Spiegelman B. M. (2003) J. Biol. Chem. 278, 26597–26603 [DOI] [PubMed] [Google Scholar]
- 11.Lin J., Wu H., Tarr P. T., Zhang C. Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., Lowell B. B., Bassel-Duby R., Spiegelman B. M. (2002) Nature 418, 797–801 [DOI] [PubMed] [Google Scholar]
- 12.Calvo J. A., Daniels T. G., Wang X., Paul A., Lin J., Spiegelman B. M., Stevenson S. C., Rangwala S. M. (2008) J. Appl. Physiol. 104, 1304–1312 [DOI] [PubMed] [Google Scholar]
- 13.Sandri M., Lin J., Handschin C., Yang W., Arany Z. P., Lecker S. H., Goldberg A. L., Spiegelman B. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16260–16265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Loon L. J., Koopman R., Manders R., van der Weegen W., van Kranenburg G. P., Keizer H. A. (2004) Am. J. Physiol. Endocrinol. Metab. 287, E558–E565 [DOI] [PubMed] [Google Scholar]
- 15.Stellingwerff T., Boon H., Jonkers R. A., Senden J. M., Spriet L. L., Koopman R., van Loon L. J. (2007) Am. J. Physiol. Endocrinol. Metab. 292, E1715–E1723 [DOI] [PubMed] [Google Scholar]
- 16.Rico-Sanz J., Moosavi M., Thomas E. L., McCarthy J., Coutts G. A., Saeed N., Bell J. D. (2000) Lipids 35, 1313–1318 [DOI] [PubMed] [Google Scholar]
- 17.Espinoza D. O., Boros L. G., Crunkhorn S., Gami H., Patti M. E. (2009) FASEB J. 24, 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geiser J., Vogt M., Billeter R., Zuleger C., Belforti F., Hoppeler H. (2001) Int. J. Sports Med. 22, 579–585 [DOI] [PubMed] [Google Scholar]
- 19.Hoppeler H., Howald H., Conley K., Lindstedt S. L., Claassen H., Vock P., Weibel E. R. (1985) J. Appl. Physiol. 59, 320–327 [DOI] [PubMed] [Google Scholar]
- 20.Stansbie D., Brownsey R. W., Crettaz M., Denton R. M. (1976) Biochem. J. 160, 413–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fungwe T. V., Fox J. E., Cagen L. M., Wilcox H. G., Heimberg M. (1994) J. Lipid Res. 35, 311–318 [PubMed] [Google Scholar]
- 22.Hansen P. A., Gulve E. A., Holloszy J. O. (1994) J. Appl. Physiol. 76, 979–985 [DOI] [PubMed] [Google Scholar]
- 23.Wende A. R., Schaeffer P. J., Parker G. J., Zechner C., Han D. H., Chen M. M., Hancock C. R., Lehman J. J., Huss J. M., McClain D. A., Holloszy J. O., Kelly D. P. (2007) J. Biol. Chem. 282, 36642–36651 [DOI] [PubMed] [Google Scholar]
- 24.Pénicaud L., Ferré P., Assimacopoulos-Jeannet F., Perdereau D., Leturque A., Jeanrenaud B., Picon L., Girard J. (1991) Biochem. J. 279, 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ninfali P., Aluigi G., Pompella A. (1997) Brain Res. 1, 357–363 [DOI] [PubMed] [Google Scholar]
- 26.Glazer R. I., Weber G. (1971) J. Neurochem. 18, 1569–1576 [DOI] [PubMed] [Google Scholar]
- 27.Jacob S., Machann J., Rett K., Brechtel K., Volk A., Renn W., Maerker E., Matthaei S., Schick F., Claussen C. D., Häring H. U. (1999) Diabetes 48, 1113–1119 [DOI] [PubMed] [Google Scholar]
- 28.Perseghin G., Scifo P., De Cobelli F., Pagliato E., Battezzati A., Arcelloni C., Vanzulli A., Testolin G., Pozza G., Del Maschio A., Luzi L. (1999) Diabetes 48, 1600–1606 [DOI] [PubMed] [Google Scholar]
- 29.Goodpaster B. H., He J., Watkins S., Kelley D. E. (2001) J. Clin. Endocrinol. Metab. 86, 5755–5761 [DOI] [PubMed] [Google Scholar]
- 30.Russell A. P. (2004) Int. J. Obes. Relat. Metab. Disord. 28, Suppl. 4, S66–S71 [DOI] [PubMed] [Google Scholar]
- 31.Bezaire V., Bruce C. R., Heigenhauser G. J., Tandon N. N., Glatz J. F., Luiken J. J., Bonen A., Spriet L. L. (2006) Am. J. Physiol. Endocrinol. Metab. 290, E509–E515 [DOI] [PubMed] [Google Scholar]
- 32.Berk P. D., Wada H., Horio Y., Potter B. J., Sorrentino D., Zhou S. L., Isola L. M., Stump D., Kiang C. L., Thung S. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 3484–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi C. S., Befroy D. E., Codella R., Kim S., Reznick R. M., Hwang Y. J., Liu Z. X., Lee H. Y., Distefano A., Samuel V. T., Zhang D., Cline G. W., Handschin C., Lin J., Petersen K. F., Spiegelman B. M., Shulman G. I. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19926–19931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergman B. C., Perreault L., Hunerdosse D. M., Koehler M. C., Samek A. M., Eckel R. H. (2010) J. Appl. Physiol. 108, 1134–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Randle P. J., Garland P. B., Hales C. N., Newsholme E. A. (1963) Lancet 1, 785–789 [DOI] [PubMed] [Google Scholar]
- 36.Sugden M. C. (2007) Br. J. Nutr. 97, 809–813 [DOI] [PubMed] [Google Scholar]
- 37.Morifuji M., Sakai K., Sanbongi C., Sugiura K. (2005) Nutrition 21, 1052–1058 [DOI] [PubMed] [Google Scholar]
- 38.Holloszy J. O. (2005) J. Appl. Physiol. 99, 338–343 [DOI] [PubMed] [Google Scholar]
- 39.Rogers M. A., King D. S., Hagberg J. M., Ehsani A. A., Holloszy J. O. (1990) J. Appl. Physiol. 68, 1833–1837 [DOI] [PubMed] [Google Scholar]
- 40.Chen S. Y., Chen S. M., Chang W. H., Lai C. H., Chen M. C., Chou C. H., Kuo C. H. (2006) Int. J. Obes. 30, 40–44 [DOI] [PubMed] [Google Scholar]
- 41.Liu T. C., Liu Y. Y., Lee S. D., Huang C. Y., Chien K. Y., Cheng I. S., Lin C. Y., Kuo C. H. (2008) J. Sports Sci. 26, 919–925 [DOI] [PubMed] [Google Scholar]
- 42.Rimbert V., Vidal H., Duche P., Debard C., Giraudet C., Boirie Y., Chardigny J. M., Morio B. (2009) FEBS Lett. 17, 2927–2933 [DOI] [PubMed] [Google Scholar]
- 43.Schrauwen P. (2007) Proc. Nutr. Soc. 66, 33–41 [DOI] [PubMed] [Google Scholar]
- 44.Narkar V. A., Downes M., Yu R. T., Embler E., Wang Y. X., Banayo E., Mihaylova M. M., Nelson M. C., Zou Y., Juguilon H., Kang H., Shaw R. J., Evans R. M. (2008) Cell 134, 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. (2006) Cell 127, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 46.Goodyear L. J. (2008) N. Engl. J. Med. 359, 1842–1844 [DOI] [PubMed] [Google Scholar]
- 47.Richter E. A., Kiens B., Wojtaszewski J. F. (2008) Cell Metab. 8, 96–98 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.