Abstract
Kinetochore attachment to the ends of dynamic microtubules is a conserved feature of mitotic spindle organization that is thought to be critical for proper chromosome segregation. Although kinetochores have been described to transition from lateral to end-on attachments, the phase of lateral attachment has been difficult to study in yeast due to its transient nature. We have previously described a kinetochore mutant, DAM1-765, which exhibits lateral attachments and misregulation of microtubule length. Here we show that the misregulation of microtubule length in DAM1-765 cells occurs despite localization of microtubule associated proteins Bik1, Stu2, Cin8 and Kip3 to microtubules. DAM1-765 kinetochores recruit the spindle checkpoint protein Bub1, however Bub1 localization to DAM1-765 kinetochores is not sufficient to cause a cell cycle arrest. Interestingly, the DAM1-765 mutation rescues the temperature sensitivity of a biorientationdeficient ipl1-321 mutant, and DAM1-765 chromosome loss rates are similar to wild-type cells. the spindle checkpoint in DAM1-765 cells responds properly to unattached kinetochores created by nocodazole treatment and loss of tension caused by a cohesin mutant. progression of DAM1-765 cells through mitosis therefore suggests that satisfaction of the checkpoint depends more highly on biorientation of sister kinetochores than on achievement of a specific interaction between kinetochores and microtubule plus ends.
Key words: spindle assembly checkpoint, kinetochore-microtubule attachments, biorientation, DAM1-765
Introduction
Proper chromosome segregation depends on the assembly of a bipolar spindle, which physically separates each pair of replicated sister chromatids towards opposite poles. Kinetochores assemble onto centromeric DNA and bind to microtubules of the mitotic spindle. Because the interactions between kinetochores and microtubules are stochastic, errors in kinetochore attachment occur and must be corrected prior to cell division in order to prevent chromosome missegregation. Biorientation, i.e. attachment of sister kinetochores to microtubules from opposite spindle poles, generates tension between sister kinetochores and is required for correct segregation. The spindle checkpoint is responsible for detecting errors in kinetochore attachment and preventing the metaphase to anaphase transition until all chromosomes are correctly bioriented and under tension.
The checkpoint proteins Mad1 and Mad2 are sequestered at nuclear pore complexes during interphase and recruited to unattached kinetochores during mitosis, where they function as key components of the spindle checkpoint response.1–3 In mammalian cells, Mad2 is transported away from the kinetochore by dynein once a connection to microtubules is established.4 If microtubule attachments are subsequently lost, Mad2 rebinds kinetochores.5 In budding yeast, Mad1 and Mad2 do not localize to kinetochores during an unperturbed cell cycle, presumably because kinetochores remain in close proximity to the spindle poles and attach rapidly to the forming spindle.6 However, both Mad1 and Mad2 are bound to kinetochores that have been detached by treatment with the microtubule depolymerizing drug nocodazole.6
Mutations in budding yeast that allow attachment to the spindle but prevent establishment of tension between sister kinetochores cause a checkpoint-mediated cell cycle delay that is dependent on Ipl1 (Aurora kinase B) and the three other chromosomal passengers Sli15 (INCENP), Bir1 (survivin) and Nbl1 (borealin).7–10 The latter three proteins colocalize with Ipl1/Aurora kinase B throughout the cell cycle and are required for proper Ipl1/Aurora kinase B localization and function.9–14 Ipl1/Aurora kinase B has been shown to promote correction of kinetochore-microtubule connections that correlate with a lack of tension between sister kinetochores, but is not required for a checkpoint-dependent cell cycle arrest in response to unattached kinetochores.7,15–17
The checkpoint proteins Bub1 and Bub3 localize to unattached kinetochores, but additionally label unaligned kinetochores during the early stages of mitosis and leave kinetochores as metaphase progresses.6,18–23 The correlation between Bub1 kinetochore localization and chromosome alignment could indicate a role for Bub1 in sensing tension or end-on attachment, and there is evidence for both roles. Mammalian Bub1 has been shown to localize to kinetochores in response to reduced tension caused by low levels of microtubule poisons, supporting a role for Bub1 in monitoring tension.18,19 In other experiments, depletion of Bub1 or Bub3 results in an increased number of laterally attached kinetochores, indicating a role for the two checkpoint proteins in the formation of end-on attachments.24,25
In budding yeast, Bub1 is recruited to kinetochores in some, but not all, situations in which tension at kinetochores is absent and is proposed to localize to laterally attached kinetochores.6 However, because yeast chromosomes remain in close proximity to spindle poles and quickly become attached to the spindle, studies of lateral attachment have been restricted to rare natural occurrences or chromosomes that have intentionally been positioned away from the spindle.26 In previous work, we isolated a kinetochore mutant, DAM1-765, which biorients and segregates its chromosomes despite a lack of stable kinetochore attachment to the ends of microtubules.27 Although these attachments may not be identical to the lateral attachments characterized by Tanaka et al.26 for simplicity we will refer to the non-end-on attachments in DAM1-765 as lateral attachments. Here we show that localization of the microtubule-associated proteins Bik1, Stu2, Cin8 and Kip3 to DAM1-765 microtubules is not sufficient for normal microtubule length regulation in the absence of end-on kinetochore attachments. The lateral attachments in DAM1-765 cells are recognized by the spindle checkpoint protein Bub1, but not by Mad1. Consistent with the ability of DAM1-765 cells to biorient their kinetochores under tension, the chromosomal passenger Bir1 is not present at metaphase kinetochores. Furthermore, the DAM1-765 mutation rescues the temperature sensitivity of an ipl1-321 mutant and DAM1-765 cells display normal rates of chromosome loss. Therefore, the novel spindle architecture in the DAM1-765 mutant, where sister kinetochores are bioriented despite being laterally attached, satisfies all checkpoints and allows cell cycle progression with little delay.
Results
DAM1-765 kinetochores contain all examined kinetochore components.
In previous work, we showed that a mutation (DAM1-765; S221F) in the outer kinetochore protein Dam1 allows kinetochore attachment to microtubules but prevents stable interaction with the microtubule end.27 This mutation disrupts a site of phosphorylation by Mps1 kinase and was isolated as a dominant mutation in a synthetic lethal screen with a fragile spindle pole body mutant (spc110-226). In the presence of wild-type SPC110, DAM1-765 cells grow normally and do not require the spindle checkpoint for survival at 30°C, despite the absence of stable end-on kinetochore attachments to microtubules. Both alanine and aspartic acid substitutions at this site reproduce the DAM1-765 kinetochore localization and synthetic lethality with spc110-226. Colocalization of the kinetochore proteins Dam1, Nuf2 and Ndc10 is not affected by the DAM1-765 mutation.27 Here, we examined whether the DAM1-765 mutant kinetochores also contain other known kinetochore complexes. We fluorescently tagged the kinetochore components Mtw1 and Spc105 and showed that they colocalize with Nuf2 at metaphase in both wild-type and DAM1-765 cells (Fig. 1A). Combined with our previous work, we have examined the localization of at least one component from each of the major kinetochore complexes, from Ndc10 in the inner kinetochore to Dam1 in the outer kinetochore.27 Our results suggest that the DAM1-765 kinetochores contain all the major complexes that comprise wild-type kinetochores.
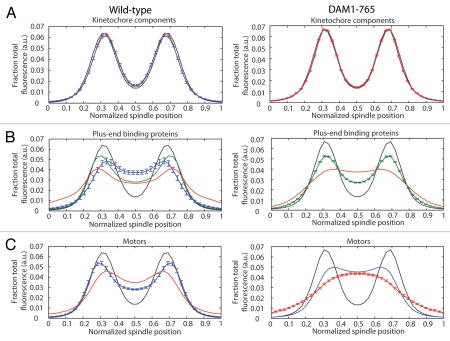
Figure 1.
Localization of kinetochore and microtubule-binding components in wild-type (left) and DAM1-765 mutant spindles (right). Each spindle component was GFP-tagged in a strain expressing Nuf2-mCherry. Fluorescence distributions were analyzed as described in Materials and Methods. The black line in each part represents the average Nuf2 (kinetochore) distribution. The center of the x-axis represents the spindle equator, with the spindle poles approximately at positions 0 and 1. The error bars in each part represent the standard error of the mean for the component with the fewest spindles analyzed. (A) Core kinetochore components: Mtw1 (red, MSY255-1C, MSY255-15C), Spc105 (blue, MSY267-2A, MSY267-7D). (B) Plus-end binding proteins: Bik1 (red, pWY266-8B, MSY58), Bim1 (blue, MSY304-18C), Stu2 (green, MSY307-16C, MSY307-18D). (C) Plus-end directed motors: Cin8 (blue, MSY306-5C, MSY305-14C), Kip3 (red, MSY263-23B, MSY263-46D).
Microtubule associated proteins are not sufficient for microtubule length control in the absence of end-on kinetochore attachments.
In wild-type cells, kinetochore microtubule lengths are maintained such that kinetochores cluster within their respective half-spindles.27–29 In contrast, the kinetochores in DAM1-765 are tightly clustered near the spindle poles, while microtubules are nearly random in length and often extend across the spindle equator.27 We examined whether loss of microtubule length control in the DAM1-765 mutant could be explained by mislocalization of plus-end binding proteins or motors such as Bik1, Cin8, Kip3 and Stu2, which have been described to regulate microtubule dynamics and/or kinetochore position.30–36 In wild-type cells, the plus-end binding proteins (Bik1, Bim1 and Stu2) and plus-end directed motors (Cin8 and Kip3) each localize into two peaks of fluorescence that overlap with the two peaks of kinetochore fluorescence (Fig. 1B and C). These localizations are consistent with an enrichment of these proteins at microtubule plus ends, where the kinetochores attach.30,37–40
In contrast, DAM1-765 kinetochores are uncoupled from microtubule ends and accordingly, some plus-end binding proteins and plus-end directed motors display reduced colocalization with kinetochores (Fig. 1B and C). As shown previously, the plus-end binding protein Bik1 localizes randomly along DAM1-765 spindles instead of colocalizing with kinetochores.27 The plus-end directed motor Kip3, which can bind centromeric DNA independently of microtubules,39 also fails to colocalize with DAM1-765 kinetochores. Instead, Kip3 is randomly distributed along the spindle, consistent with the distribution of microtubule plus ends in DAM1-765 (Fig. 1C; reviewed in ref. 27). Stu2 can be found both colocalized with kinetochores and along the spindle in DAM1-765 (Fig. 1B). This result is consistent with the observed localization in wild-type cells and with previous reports that Stu2 binds the kinetochore independently of microtubules.26,40 Cin8 localizes along the DAM1-765 spindle, and in some cells also colocalizes with kinetochores, as it does in wild-type cells (Fig. 1C). Fluorescently tagging the C-terminus of the plus-end binding protein Bim1 impaired growth of DAM1-765 cells and reduced formation of bipolar spindles, preventing determination of Bim1 localization in the mutant (data not shown). Our data suggest that Stu2, Bik1, Cin8 and Kip3 remain properly associated with microtubules in DAM1-765, but are not sufficient for proper microtubule length control when the kinetochores are not at the microtubule plus-ends.
The checkpoint protein Mad1 does not localize to DAM1-765 kinetochores.
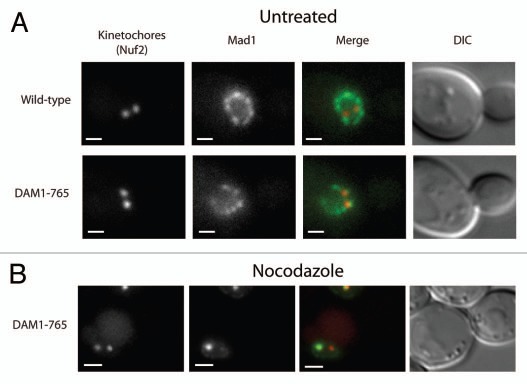
Yeast Mad1 localizes to unattached kinetochores created by the microtubule depolymerizing drug nocodazole, but not to transiently unattached kinetochores that form during a normal cell cycle.6 To test whether the prolonged lateral attachments in DAM1-765 recruit Mad1, we compared Mad1 kinetochore localization in wild-type and DAM1-765 spindles. Mad1-GFP was seen to colocalize with at least one Nuf2-mCherry labeled kinetochore cluster in 5% of both wild-type (N = 39) and DAM1-765 (N = 52) spindles (Fig. 2A). In both strains, Mad1 localization was consistent with previously described nuclear pore localization.2,3 Upon treatment with nocodazole, 46% of DAM1-765 (N = 50) cells showed Mad1 associated with a Nuf2-mCherry focus (Fig. 2B). This is consistent with previous results showing that Mad1 and Mad2 only localize to unattached kinetochores, which often exhibit fainter kinetochore fluorescence than the main kinetochore cluster that remains attached to the depolymerized spindle.6 Thus, Mad1 is recruited to unattached kinetochores caused by nocodazole, even in the DAM1-765 mutant, but is not recruited to laterally attached kinetochores in DAM1-765 cells.
Figure 2.
Mad1 does not localize to kinetochores in unperturbed DAM1-765 cells. Kinetochores are labeled with Nuf2-mCherry (red) and Mad1 is labeled with GFP (green). Bars are 1 µm. (A) Mad1-GFP localizes to the nuclear envelope in both wild-type (MSY198-11D) and DAM1-765 (MSY197-3B) cells. (B) Mad1-GFP localizes to unattached DAM1-765 kinetochore clusters generated by treatment with nocodazole, but not to the main kinetochore cluster, which is presumably still attached to the spindle.
Bub1 is retained on DAM1-765 kinetochores at metaphase.
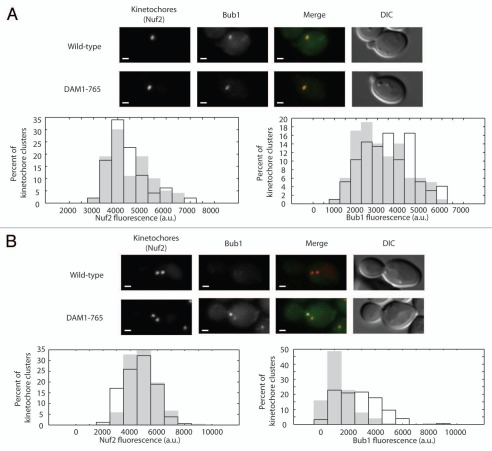
Bub1 has been proposed to localize to laterally attached kinetochores, although this has not been demonstrated directly in yeast.6 To determine whether Bub1 is enriched on DAM1-765 kinetochores, we measured the amount of Bub1 present at kinetochores in small budded cells prior to kinetochore separation and in metaphase spindles with well-separated kinetochore clusters. Prior to kinetochore separation, the amount of Bub1 on DAM1-765 kinetochore clusters (3,500 ± 100 arbitrary units (a.u); mean ± SEM, N = 97) was similar to wild-type (3,200 ± 100 a.u., N = 100) (Fig. 3A). At metaphase, however, the amount of Bub1 per kinetochore cluster was significantly higher (p < 0.0001) in DAM1-765 cells (2,800 ± 100 a.u., N = 158) compared to wild type cells (1,400 ± 100 a.u.; N = 260) (Fig. 3B). Roughly 60% of DAM1-765 cells exhibited Bub1 localization on both kinetochore clusters, while 35% showed Bub1 enrichment predominantly on one of the two kinetochore clusters. An asymmetrical kinetochore distribution has been described previously for human Bub1.19 The checkpoint protein Bub3 also showed increased localization to DAM1-765 kinetochores, however Bub3 fluorescence was weaker than Bub1 (data not shown). Localization of Bub1 to DAM1-765 kinetochores suggests that Bub1 recruitment to kinetochores may be correlated with the absence of end-on attachments.
Figure 3.
Bub1 localizes to DAM1-765 metaphase kinetochores. Bub1 fluorescence at kinetochores was analyzed as described in Materials and Methods for (A) small-budded cells and (B) metaphase spindles. In merged images, Nuf2-mCherry is in red and Bub1-GFP is in green. Bars are 1 µm. Histograms of total kinetochore (Nuf2) and Bub1 fluorescence for wild-type (solid grey, MSY202-8C) and DAM1-765 (black outline, MSY201-4C) kinetochore clusters are plotted.
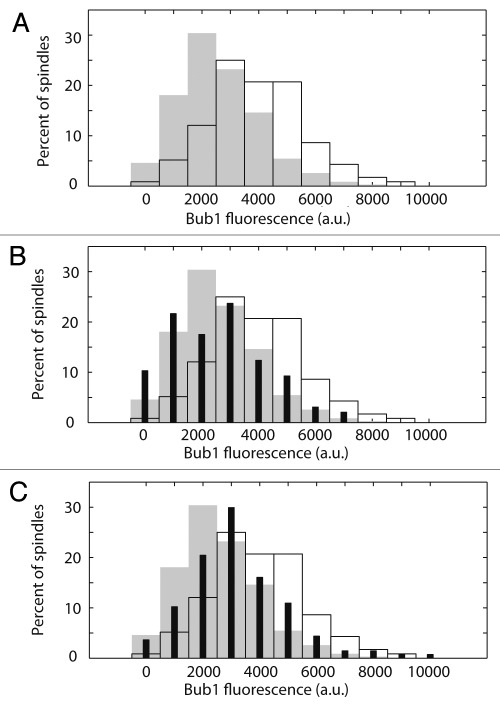
The mechanism by which Bub1 detects lateral attachments is unclear, however one possibility is that Bub1 monitors interactions between kinetochores and microtubule plus-end associated proteins. Because Bik1 and Kip3 do not colocalize with kinetochores in the DAM1-765 mutant, we hypothesized that loss of an interaction between Bik1 or Kip3 and the kinetochore could be responsible for Bub1 recruitment. We therefore examined whether deletion of Bik1 or Kip3 is sufficient to recruit Bub1 to kinetochores even in cells with the wild-type DAM1 allele. We measured the total amount of Bub1-GFP fluorescence on metaphase spindles in strains expressing the spindle pole body marker, Spc110-mCherry. Consistent with the above results, the total amount of Bub1 on DAM1-765 spindles (4,000 ± 200 a.u; mean ± SEM, N = 116) was significantly higher (p < 0.0001) than on wild-type spindles (2,500 ± 100 a.u, N = 349) (Fig. 4A). Neither bik1Δ (2,600 ± 200 a.u, N = 97, Fig. 4B) nor kip3Δ (3,200 ± 200 a.u, N = 137, Fig. 4C) elevated Bub1 fluorescence to DAM1-765 levels. Thus, disrupting the interaction of either Bik1 or Kip3 with kinetochores is not sufficient to recruit Bub1 to the extent seen in DAM1-765 cells.
Figure 4.
Deletion of BIK1 or KIP3 does not reproduce DAM1-765 levels of Bub1 recruitment to kinetochores. Total Bub1 fluorescence on metaphase spindles was analyzed as described in Materials and Methods. (A) Histograms of total Bub1 fluorescence for wild-type (solid grey, MSY319-1D) and DAM1-765 (black outline, MSY318-7A) spindles are plotted. the DAM1-765 histogram is shifted toward higher Bub1 fluorescence values relative to wild-type. (B) Histograms of total Bub1 fluorescence for bik1Δ (solid black bars, MSY331-2D), wild-type (solid grey bars) and DAM1-765 (black outline) spindles are plotted. The bik1Δ histogram overlaps with wild-type. (C) Histograms of total Bub1 fluorescence for kip3Δ (solid black bars, MSY317-10C), wild-type (solid grey bars) and DAM1-765 (black outline) spindles are plotted. The kip3Δ histogram is shifted relative to wild-type, but does not reproduce the high levels of Bub1 fluorescence measured in DAM1-765 spindles.
DAM1-765 cells exhibit a slight cell cycle delay.
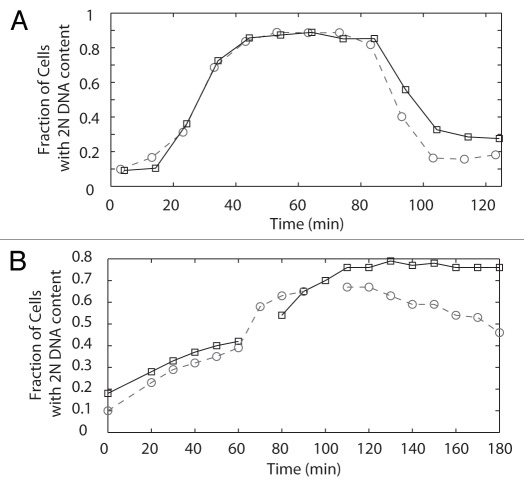
Previously we showed that DAM1-765 cells are not temperature-sensitive and progress through the cell cycle at the same rate as wild-type cells.27 Because checkpoint protein localization to kinetochores is associated with an activated spindle checkpoint, we decided to re-examine the cell cycle in DAM1-765. We found that DAM1-765 cells grow at 30°C with a doubling time (92 ± 1 min; mean ± SEM) that is 7% longer than wild-type cells (86 ± 1 min). When monitored in synchronous cultures, the delay was detectable but less than the 10 min interval between time points (Fig. 5A). Thus, the absence of stable end-on attachments in DAM1-765, although recognized by the Bub1 protein, has little impact on the rate of cell cycle progression.
Figure 5.
(A) Flow cytometry of DAM1-765 cells reveals a slight cell cycle delay. Cells were synchronized with alpha factor, released at 30°C and re-arrested with alpha factor in the following G1. Samples were fixed for flow cytometry every 10 minutes. The fraction of cells with a 2N DNA content at each time point was determined as described in Materials and Methods. The graph represents the average of two experiments, with the strains aligned according to when 50% of the cells reached a 2N DNA content. Wild-type, dashed line, circles (MSY216-1A); DAM1-765, solid line, squares (MSY165-19B). (B) DAM1-765 cells delay in response to lack of tension caused by mcd1-1 and the delay is Ipl1-dependent. Cells were synchronized at 25°C with alpha factor, released at 37°C and re-arrested with alpha factor in the following G1. Samples were fixed for flow cytometry every 10 minutes. DAM1-765 mcd1-1, solid line, squares (MSY289-32D), DAM1-765 mcd1-1 ipl1-321 dashed line, circles (MSY312-12B).
DAM1-765 cells do not exhibit increased chromosome loss.
The slight cell cycle delay in DAM1-765 cells prompted us to examine whether the lack of stable end-on attachments causes cells to missegregate chromosomes at a higher rate. We measured the frequency with which diploid cells lost one copy of Chromosome III and found the frequencies for wild-type (2.2 × 10−6) and DAM1-765 (2.7 × 10−6) to be similar. Consistent with our previous results that DAM1-765 cells biorient their chromosomes as well as wild-type cells,27 we find that DAM1-765 cells do not exhibit an elevated rate of chromosome loss. Additionally, DAM1-765 cells were viable even after deletion of the checkpoint protein genes BUB1, MAD2 or MAD1 (data not shown; reviewed in ref. 27), demonstrating that DAM1-765 cells are not relying on the spindle checkpoint for survival.
The lateral attachments in DAM1-765 cells satisfy the tension checkpoint.
In wild-type cells, the lack of tension caused by defects in biorientation results in a cell cycle delay that depends on the chromosomal passengers. The lack of a significant delay in DAM1-765 cells suggests that either the laterally attached kinetochores support enough tension to satisfy the tension-dependent checkpoint or the cells do not have a functional tension checkpoint. To test whether the tension checkpoint operates properly even in the presence of lateral attachments, we examined the ability of DAM1-765 cells to delay the cell cycle in response to a loss of tension in a cohesin mutant (mcd1-1). At the restrictive temperature, cohesin is inactivated and this causes a cell cycle delay dependent on the chromosomal passengers.7,8 We found that the tension checkpoint was still active in the presence of DAM1-765; DAM1-765 mcd1-1 cells delayed with large-budded cells and a 2N DNA content (Fig. 5B). This delay was Ipl1-dependent, as a DAM1-765 mcd1-1 ipl1-321 triple mutant showed a reduced delay (Fig. 5B).
Previously, we showed that the chromosomal passengers Ipl1, Sli15 and Bir1 are not localized to metaphase kinetochores in wild-type cells but that kinetochore localization of Bir1 is enhanced in the absence of tension.13 To determine whether lateral attachments might retain chromosomal passengers at metaphase kinetochores, we examined whether Bir1 was localized to DAM1-765 kinetochores. We compared the fluorescence distribution of Bir1 at metaphase to that of the kinetochore component Nuf2. Similarly to wild-type cells, Bir1 was localized along the spindle between DAM1-765 kinetochores (Fig. 6A; reviewed in ref. 13). This is in contrast to an mcd1-1 cohesin mutant, in which kinetochores are also positioned close to the spindle poles, but not under tension, and Bir1 is strongly localized to kinetochores.13 Together these results indicate that the DAM1-765 cells possess a functional tension checkpoint and that laterally attached kinetochores are able to satisfy the checkpoint.
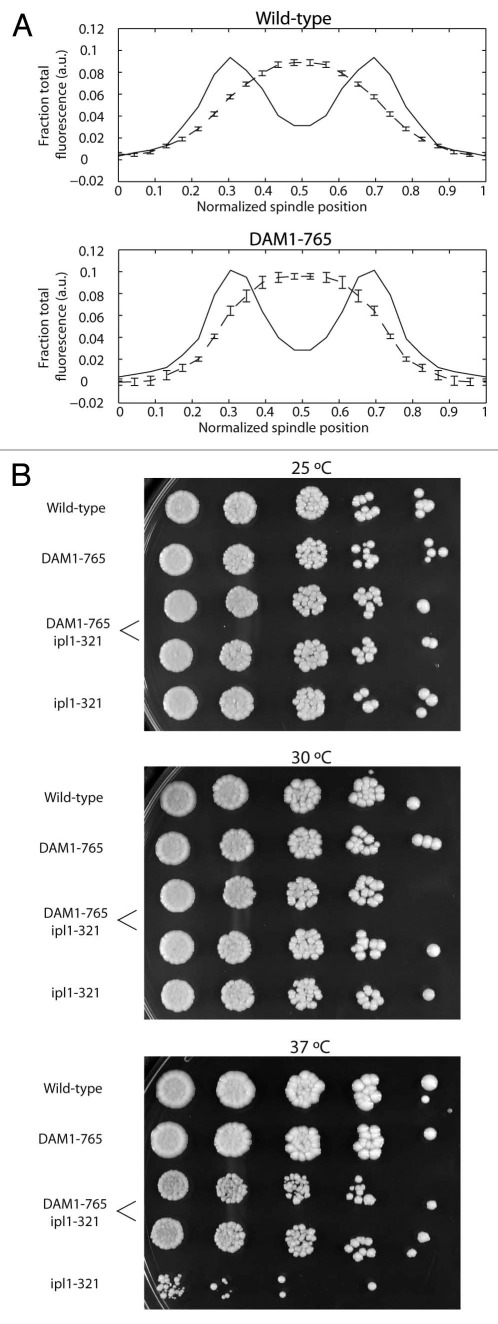
Figure 6.
(A) Bir1 localizes between metaphase kinetochores in both DAM1-765 and wild-type cells. Wild-type and DAM1-765 strains carrying Nuf2-CFp and Bir1-Venus (MSY279-6C and MSY279-7B) were imaged and fluorescence distributions analyzed as described in Materials and Methods. The solid lines represent average Nuf2 (kinetochore) fluorescence and the dashed lines represent average Bir1 fluorescence. Error bars are the standard error of the mean. (B) DAM1-765 rescues ipl1-321 temperature-sensitivity. The indicated yeast strains were grown in liquid culture at 25°C to mid-log phase (MSY216-1A, MSY165-19B, PWY261-2B, pWY261-3D, SFY233-2D). Cells were diluted to a density equal to approximately 5 Klett units. 5-fold serial dilutions were spotted (3 µl per spot) onto YPD plates and grown at the indicated temperatures for 2–3 days.
DAM1-765 reduces the temperature sensitivity and chromosome missegregation of an ipl1-321 mutant.
Interestingly, lateral attachments in DAM1-765 not only satisfy the tension checkpoint, but also appear to reduce dependence on Ipl1 activity. Cells carrying both the DAM1-765 allele and a temperature-sensitive ipl1-321 allele were able to grow at higher temperatures than cells with the ipl1-321 allele alone (Fig. 6B). Cells carrying the ipl1-321 allele exhibit chromosome missegregation at the non-permissive temperature.13,17,41 We found that 25% of ipl1-321 cells (N = 106) incorrectly segregate a pair of fluorescently tagged centromeres to the same spindle pole after 60 min at the non-permissive temperature.13 Combining the ipl1-321 allele with DAM1-765 reduced the frequency of missegregation to 8% (N = 100), only slightly higher than wild-type (3%, N = 104) or DAM1-765 alone (3%, N = 99). This suggests that biorientation in DAM1-765 shows a reduced requirement for Ipl1-dependent corrective activity.
Discussion
Kinetochore attachment to the ends of kinetochore microtubules has been observed in several organisms and is believed to be essential for proper chromosome segregation. In our previous work, we characterized a yeast kinetochore mutation, DAM1-765, that disrupts the stable interaction between kinetochores and the ends of microtubules without impairing the cell's ability to properly biorient and segregate its chromosomes.27 Here we provide further evidence that the kinetochores in DAM1-765 cells not only contain all of the examined kinetochore components, but also exhibit changes in the organization of microtubule-binding components that are consistent with a disruption of the attachments between kinetochores and microtubule plus ends.
Despite localization of microtubule plus-end associated proteins such as Stu2, Bik1, Cin8 and Kip3 to microtubules, microtubule lengths in DAM1-765 cells are misregulated. This suggests that the examined microtubule regulators are not sufficient for proper microtubule length control in the absence of end-on kinetochore attachments. Indeed, members of the Dam1 and Ndc80 kinetochore complexes have been shown to exert direct effects on plus-end behavior, and a mutation in Ndc10 also affects microtubule dynamics.34,42,43 Additionally, both the kinetochore and Stu2 have been shown to alter microtubule dynamics independently of one another, suggesting kinetochores and plus-end binding proteins cooperate to properly regulate microtubule length.34
Despite their laterally attached kinetochores, DAM1765 cells delay the cell cycle in response to a lack of tension. Furthermore, although the kinetochores in DAM1-765 cells are not stably associated with the ends of microtubules, Bir1 is not localized to metaphase kinetochores. Therefore, the mechanisms underlying proper Bir1 localization and the Ipl1-dependent tension checkpoint do not depend on formation of stable end-on attachments. The absence of a significant cell cycle delay in DAM1-765 cells during normal growth suggests that biorientation and tension are sufficient to prevent detachment of laterally attached kinetochores by the Ipl1 kinase. In fact, the DAM1-765 mutant displays a reduced requirement for Ipl1 activity, suggesting that the DAM1-765 kinetochores may be more likely to achieve biorientation on the first attempt than wild-type kinetochores. Alternatively, the DAM1-765 kinetochore attachments may be less stable prior to proper biorientation and tension-dependent stabilization. Whereas mono-oriented end-on attachments must be detached by Ipl1, mono-oriented DAM1-765 kinetochores may be more likely to detach on their own, thus reducing the requirement for Ipl1 activity.
Bub1 localization to kinetochores in DAM1-765 cells supports a role for yeast Bub1 in monitoring lateral attachments. Consistent with previous results, Bub1 recruitment to kinetochores is not sufficient to cause a metaphase arrest,6 supporting the idea that a full checkpoint arrest may require the presence of unattached kinetochores, which have failed to attach to the spindle or had their incorrect attachments severed by the corrective activity of Ipl1. This is consistent with recent work showing that a cell cycle arrest can be bypassed by preventing detachment of incorrect attachments.44 Because each budding yeast kinetochore attaches to a single microtubule, arresting the cell cycle solely in response to unattached kinetochores can be effective when coupled with a tension-dependent detachment mechanism. It will be important to determine how the expanded repertoire of spindle checkpoint components monitors kinetochore-microtubule attachment in higher eukaryotes, where kinetochores attach to multiple microtubules.
Materials and Methods
Plasmids and strains.
All strains were derived from W303 and are listed in Table 1. All fluorescent tags were introduced by PCR at the 3′ end of the target genes with plasmids from the Yeast Resource Center as described (depts.washington.edu/yeastrc “Plasmids and Protocols”, reviewed in ref. 45).
Table 1.
Yeast strains
| Strain | Genotype | Reference |
| EMY84-6C | MATa lys2Δ::HIS3 | 51 |
| MSY58 | MATa ade3Δ BIK1-GFP::TRP1 DAM1-765-13xmyc::KanMX lys2Δ::HIS3 NUF2-mCherry::HphMX URA3 | 27 |
| MSY165-19B | MATa ade3Δ DAM1-765-13xmyc::KanMX | |
| MSY197-3B | Matα ade3Δ lys2Δ::HIS3 DAM1-765-myc::KanMX MAD1-GFP::HIS3MX6 NUF2-Cherry::hphMX URA | |
| MSY198-11D | Mata ade3Δ lys2Δ::HIS3 DAM1-myc::KanMX MAD1-GFP::HIS3MX6 NUF2-Cherry::hphMX | |
| MSY201-4C | MATa ade3Δ BUB1-GFP::HIS3MX6 DAM1-765-13xmyc::KanMX NUF2-mCherry::HphMX | |
| MSY202-8C | MATa ade3Δ BUB1-GFP::HIS3MX6 DAM1-myc::KanMX NUF2-mCherry::HphMX | |
| MSY216-1A | MATa ade3Δ DAM1-13xmyc::KanMX | |
| MSY255-15C | MATa ade3Δ cyh2r DAM1-765-13xmyc::KanMX MTW1-GFP::HIS3MX6 NUF2-mCherry::HphMX | |
| MSY255-1C | MATa ade3Δ cyh2r MTW1-GFP::HIS3MX6 NUF2-mCherry::HphMX | |
| MSY263-23B | MATa ade3Δ KIP3-GFP::HIS3MX6 NUF2-mCherry::HphMX | |
| MSY263-46D | MATa ade3Δ DAM1-765-13xmyc::KanMX KIP3-GFP::HIS3MX6 NUF2-mCherry::HphMX | |
| MSY267-2A | MATa ade3Δ NUF2-mCherry::HphMX SPC105-GFP::HIS3MX6 | |
| MSY267-7D | MATa ade3Δ DAM1-765-13xmyc::KanMX NUF2-mCherry::HphMX SPC105-GFP::HIS3MX6 | |
| MSY279-6C | MATa ade3Δ BIR1-Venus::KanMX NUF2-CFP::HphMX | |
| MSY279-7B | MATa ade3Δ BIR1-Venus::KanMX DAM1-765-13xmyc::KanMX NUF2-CFP::HphMX | |
| MSY298-32D | MATa DAM1-765-13xmyc::KanMX mcd1-1 PDS1-3xHA::URA3 | |
| MSY304-18C | MATa ade3Δ BIM1-GFP::NatMX DAM1-13xmyc::KanMX NUF2-mCherry::HphMX | |
| MSY305-14C | MATa ade3Δ CIN8-GFP::HIS3MX6 DAM1-765-13xmyc::KanMX NUF2-mCherry::HphMX | |
| MSY306-5C | MATa ade3Δ CIN8-GFP::HIS3MX6 DAM1-13xmyc::KanMX NUF2-mCherry::HphMX | |
| MSY307-16C | MATa ade3Δ NUF2-mCherry::HphMX STU2-GFP::HIS3MX6 | |
| MSY307-18D | MATa ade3Δ cyh2r DAM1-765-13xmyc::KanMX NUF2-mCherry::HphMX STU2-GFP::HIS3MX6 | |
| MSY312-12B | MATa DAM1-765-13xmyc::KanMX his3-11::pCUP1-GFP12lacI12::HIS3 ipl1-321 mcd1-1 PDS1-3xHA::URA3 trp1-1::URA3::lacO::TRP1 | |
| MSY317-10C | MATa ade3Δ BUB1-GFP::HIS3MX6 kip3Δ::HIS3 SPC110-mCherry::HphMX | |
| MSY318-7A | MATa ade3Δ BUB1-GFP::HIS3MX6 DAM1-765-13xmyc::KanMX SPC110-mCherry::HphMX URA3 | |
| MSY319-1D | MATa ade3Δ BUB1-GFP::HIS3MX6 SPC110-mCherry::HphMX | |
| MSY331-2D | MATa ade3Δ bik1Δ::NatMX BUB1-GFP::HIS3MX6 cyh2r SPC110-mCherry::HphMX | |
| MSY346-1C | MATa ade3Δ mad2Δ::NatMX cyh2r | |
| MSY351-1B | MATa ade3Δ DAM1-765-13xmyc::KanMX mad2Δ::NatMX | |
| MSY351-3D | MATα ade3Δ DAM1-765-13xmyc::KanMX mad2Δ::NatMX cyh2r | |
| PWY247-30C | MATa Chr8::CEN-lacO::TRP1 his3-11::pCUP1-GFP12-lacI12::HIS3 SPC110-mCherry::HphMX | 13 |
| PWY261-2B | MATa DAM1-765-13xmyc::KanMX ipl1-321 | |
| PWY261-3D | MATα DAM1-765-13xmyc:KanMX ipl1-321 | |
| PWY266-8B | MATα ade3Δ BIK1-GFP::TRP1 lys2Δ::HIS3 NUF2-mCherry::HphMX | |
| PWY277-44C | MATa Chr8::CEN-lacO::TRP1 DAM1-765-13xmyc::KanMX his3-11::pCUP1-GFP12-lacI12::HIS3 SPC110-mCherry::hphMX | |
| PWY277-3A | MATa Chr8::CEN-lacO::TRP1 DAM1-765-13xmyc::KanMX his3-11::pCUP1-GFP12-lacI12::HIS3 ipl1-321 SPC110-mCherry::hphMX | |
| PWY286-23A | MATa Chr8::CEN-lacO::TRP1 his3-11::pCUP1-GFP12-lacI12::HIS3 ipl1-321 SPC110-mCherry::hphMX | 13 |
| SFY145 | MATa ade3Δ DAM1-765-13xmyc::KanMX lys2Δ::HIS3 URA3 | |
| SFY233-2D | MATa ipl1-321 | 13 |
| W303 | ade2-1oc can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 |
Fluorescence microscopy.
Live cells were mounted for microscopy as described.46 Images of cells with fluorescent tags were taken with a 100x, 1.35 numerical aperture oil objective lens, an Olympus IX-70 inverted fluorescence microscope and a CoolSnap HQ camera (Photometrics) managed by softWorX software (Applied Precision). Exposures were typically 0.4–0.5 s at a single focal plane, with 1 × 1 binning, except as otherwise noted.
Metaphase distribution of spindle components.
Spindles were identified by the kinetochore marker, Nuf2, using the cutup.m program as previously described13 or a collection of integrated Matlab scripts developed for the analysis of fluorescent images (Fluorcal) based on Fretscal.47 Fluorcal is available upon request. Briefly, 12-bit softWoRx data were converted to 8- and 16-bit TIFF images using R3DConverter as previously described.13 The Fluorcal program allows the user to specify criteria that are used to identify peaks of fluorescence as areas of interest (AOIs) in the 16-bit TIFF images. These criteria were chosen such that AOIs were identified solely in the channel representing kinetochore (Nuf2) fluorescence, the signal to background was ≥1.25, the full width at half maximum (FWHM) of the Gaussian fit was ≤5.6 for wild-type and ≤5.3 for DAM1-765, which has tighter kinetochore clusters. The minimum distance between AOIs was 8 pixels (0.51 µm), the size of the AOI was 7 pixels × 7 pixels and the background fluorescence was calculated from 2 concentric rings beginning 2 pixels away from the AOI. Pairs of AOIs within 25 pixels (1.6 µm) of each other were identified and visually screened to ensure that each pair represented an in-focus mitotic spindle. The fluorescence distributions were then analyzed using calc.m as previously described,13 with a pushout factor of 0.75. Metaphase kinetochore cluster separations were 14–17 pixels (0.9–1.1 µm) for wild-type and 12–16 pixels (0.8–1.0 µm) for DAM1-765. Spindle profiles were normalized to a constant length. Half-spindle profiles were averaged and then mirrored to produce a symmetric plot of the average profile across the whole spindle.
Metaphase Bir1 distribution was analyzed as described above with longer (1 s) exposures and 2 × 2 binning due to the relatively low levels of fluorescence. As a result, metaphase kinetochore cluster separations were 7–8.5 pixels (0.9–1.1 µm) for wild-type and 6–8 pixels (0.8–1.0 µm) for DAM1-765.
Nocodazole treatment.
Cells were grown to mid-log phase in YPD liquid medium48 containing 75 µg/ml adenine, resuspended at a density of 25 klett units and incubated at 30°C with 15 µg/ml nocodazole for 2 hr and 15 min.
Quantitative analysis of Bub1 fluorescence.
For measurements of Bub1 fluorescence prior to kinetochore separation, Fluorcal was used to identify AOIs based on the kinetochore marker, Nuf2, with the following criteria. The signal to background for each AOI was ≥1.25, the FWHM of the Gaussian fit was ≤13.0, the size of the AOI was 11 pixels × 11 pixels and the background fluorescence was calculated from 2 concentric rings 2 pixels away from the AOI. Images were visually screened to identify small-budded cells, in which kinetochores have not yet separated into two clusters. For each AOI, the amount of fluorescence was measured in both color channels and background was subtracted.
For the quantification of Bub1 at metaphase kinetochores, spindles were identified by the kinetochore marker, Nuf2, using the Fluorcal program described above with the following modifications: the signal to background for each AOI was ≥1.2, the FWHM of the Gaussian fit was ≤6.0, the size of the AOI was 9 pixels – 9 pixels and the background fluorescence was calculated from a concentric ring 2 pixels away from the AOI. The trend was similar, regardless of kinetochore separation, so kinetochore separations of 12–17 pixels (0.8–1.1 µm) were analyzed. For each spindle, the amount of fluorescence was measured for each AOI in both color channels and background was subtracted.
For the quantification of Bub1 across the entire metaphase spindle, spindles were identified by the spindle pole body marker, Spc110, using the Fluorcal program with the following modifications. The signal to background for each AOI was ≥1.5, the FWHM of the Gaussian fit was ≤4.7, the minimum distance between AOIs was 12 pixels (0.76 µm), the size of the AOI was 5 pixels × 5 pixels and the background fluorescence was calculated from concentric rings 3–4 pixels away from the AOI. Spindles were identified and Bub1 fluorescence analyzed as described above for the metaphase distribution of spindle components, with a pushout factor of 0.3. Total fluorescence was calculated using summation.pl as previously described.13
Flow cytometry.
Cells were synchronized in G1 with alpha factor at 30°C (25°C for mcd1-1 strains) and released (at 37°C for mcd1-1 strains). DNA content was monitored by flow cytometry to compare progression through the cell cycle. To facilitate comparison of the metaphase to anaphase transition, alpha factor was added back after release into the cell cycle to re-arrest cells in the following G1. Cells were fixed with two volumes of 100% ethanol at 22°C overnight and stained with propidium iodide. Fluorescence was analyzed using a Becton Dickinson FACSCAN flow cytometer and data were collected with Cell Quest software. The fraction of cells with 2N DNA content was determined using Flowjo software. The fraction of 2N cells represents the number of cells in the 2N peak divided by the total number of cells in the 1N and 2N peaks.
Chromosome loss assays.
The rates at which diploid cells lost a tagged Chromosome III were measured as described previously.49 Nineteen single colonies were picked after growth for 2 days on YPD at 25°C and each colony was suspended in 1 ml of distilled H2O. Suspensions were sonicated and 100 µl of each suspension was plated onto 19 separate YPD plates containing 2 µg/ml cycloheximide. Equal volumes from each of the 19 suspensions were pooled and the pooled sample was serially diluted on YPD plates to determine the average number of colony forming units per suspension. After incubation at 30°C for 4 days, cycloheximide-resistant colonies were tested for mating ability, to differentiate between chromosome loss and recombination events. The rate of chromosome loss was calculated by the method of the median.50 The median number of mating colonies per suspension was used to determine the median number of chromosome loss events per original colony. This number was divided by the average number of colony forming units per suspension to obtain a frequency of chromosome loss per cell division.
Acknowledgements
We thank Sue Biggins for helpful advice and critical reading of the manuscript. This work was funded by NIGMS RO1-GM40506 to T.N.D. E.G.M. was supported by NCRR P41-RR011823. M.M.S. and M.M.W. were supported by National Institute of General Medical Sciences grant T32-GM07270.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12907
References
- 1.Campbell MS, Chan GK, Yen TJ. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci. 2001;114:953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- 2.Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott RJ, Cairo LV, Van de Vosse DW, Wozniak RW. The nuclear export factor Xpo1p targets Mad1p to kinetochores in yeast. J Cell Biol. 2009;184:21–29. doi: 10.1083/jcb.200804098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillett ES, Espelin CW, Sorger PK. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stern BM, Murray AW. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr Biol. 2001;11:1462–1467. doi: 10.1016/s0960-9822(01)00451-1. [DOI] [PubMed] [Google Scholar]
- 9.Sandall S, Severin F, McLeod IX, Yates JR, 3rd, Oegema K, Hyman A, et al. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 2006;127:1179–1191. doi: 10.1016/j.cell.2006.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakajima Y, Tyers RG, Wong CC, Yates JR, 3rd, Drubin DG, Barnes G. Nbl1p: a Borealin/Dasra/CSC-1-like protein essential for Aurora/Ipl1 complex function and integrity in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:1772–1784. doi: 10.1091/mbc.E08-10-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeyaprakash AA, Klein UR, Lindner D, Ebert J, Nigg EA, Conti E. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007;131:271–285. doi: 10.1016/j.cell.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 12.Kang J, Cheeseman IM, Kallstrom G, Velmurugan S, Barnes G, Chan CS. Functional cooperation of Dam1, Ipl1 and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J Cell Biol. 2001;155:763–774. doi: 10.1083/jcb.200105029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimogawa MM, Widlund PO, Riffle M, Ess M, Davis TN. Bir1 is required for the tension checkpoint. Mol Biol Cell. 2009;20:915–923. doi: 10.1091/mbc.E08-07-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JH, Kang JS, Chan CS. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1381–1394. doi: 10.1083/jcb.145.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewar H, Tanaka K, Nasmyth K, Tanaka TU. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- 16.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome biorientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 18.Skoufias DA, Andreassen PR, Lacroix FB, Wilson L, Margolis RL. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochoretension checkpoints. Proc Natl Acad Sci USA. 2001;98:4492–4497. doi: 10.1073/pnas.081076898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor SS, Hussein D, Wang Y, Elderkin S, Morrow CJ. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J Cell Sci. 2001;114:4385–4395. doi: 10.1242/jcs.114.24.4385. [DOI] [PubMed] [Google Scholar]
- 20.Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- 21.Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basu J, Bousbaa H, Logarinho E, Li Z, Williams BC, Lopes C, et al. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jablonski SA, Chan GK, Cooke CA, Earnshaw WC, Yen TJ. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 1998;107:386–396. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- 24.Meraldi P, Sorger PK. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 2005;24:1621–1633. doi: 10.1038/sj.emboj.7600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logarinho E, Resende T, Torres C, Bousbaa H. The human spindle assembly checkpoint protein Bub3 is required for the establishment of efficient kinetochore-microtubule attachments. Mol Biol Cell. 2008;19:1798–1813. doi: 10.1091/mbc.E07-07-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 27.Shimogawa MM, Graczyk B, Gardner MK, Francis SE, White EA, Ess M, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 30.Gardner MK, Bouck DC, Paliulis LV, Meehl JB, O'Toole ET, Haase J, et al. Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 2008;135:894–906. doi: 10.1016/j.cell.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Breugel M, Drechsel D, Hyman A. Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J Cell Biol. 2003;161:359–369. doi: 10.1083/jcb.200211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolyniak MJ, Blake-Hodek K, Kosco K, Hwang E, You L, Huffaker TC. The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol Biol Cell. 2006;17:2789–2798. doi: 10.1091/mbc.E05-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosco KA, Pearson CG, Maddox PS, Wang PJ, Adams IR, Salmon ED, et al. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol Biol Cell. 2001;12:2870–2880. doi: 10.1091/mbc.12.9.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson CG, Maddox PS, Zarzar TR, Salmon ED, Bloom K. Yeast kinetochores do not stabilize Stu2pdependent spindle microtubule dynamics. Mol Biol Cell. 2003;14:4181–4195. doi: 10.1091/mbc.E03-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta M, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- 36.Wargacki M, Tay J, Muller E, Asbury C, Davis T. Kip3, the yeast kinesin-8, is required for clustering of kinetochores at metaphase. Cell Cycle. 2010;9:2581–2588. doi: 10.4161/cc.9.13.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blake-Hodek KA, Cassimeris L, Huffaker TC. Regulation of microtubule dynamics by Bim1 and Bik1, the budding yeast members of the EB1 and CLIP-170 families of plus-end tracking proteins. Mol Biol Cell. 21:2013–2023. doi: 10.1091/mbc.E10-02-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin H, de Carvalho P, Kho D, Tai CY, Pierre P, Fink GR, et al. Polyploids require Bik1 for kinetochoremicrotubule attachment. J Cell Biol. 2001;155:1173–1184. doi: 10.1083/jcb.200108119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tytell JD, Sorger PK. Analysis of kinesin motor function at budding yeast kinetochores. J Cell Biol. 2006;172:861–874. doi: 10.1083/jcb.200509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma L, McQueen J, Cuschieri L, Vogel J, Measday V. Spc24 and Stu2 promote spindle integrity when DNA replication is stalled. Mol Biol Cell. 2007;18:2805–2816. doi: 10.1091/mbc.E06-09-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franck AD, Powers AF, Gestaut DR, Gonen T, Davis TN, Asbury CL. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–837. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z, Kenny AE, Brito DA, Rieder CL. Cells satisfy the mitotic checkpoint in Taxol, and do so faster in concentrations that stabilize syntelic attachments. J Cell Biol. 2009;186:675–684. doi: 10.1083/jcb.200906150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 46.Muller EG, Snydsman BE, Novik I, Hailey DW, Gestaut DR, Niemann CA, et al. The organization of the core proteins of the yeast spindle pole body. Mol Biol Cell. 2005;16:3341–3352. doi: 10.1091/mbc.E05-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kollman JM, Zelter A, Muller EG, Fox B, Rice LM, Davis TN, et al. The structure of the gamma-tubulin small complex: implications of its architecture and flexibility for microtubule nucleation. Mol Biol Cell. 2008;19:207–215. doi: 10.1091/mbc.E07-09-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherman F, Fink GR, Hicks JB. Cold Spring Harbor Laboratory: Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 49.Widlund PO, Lyssand JS, Anderson S, Niessen S, Yates JR, 3rd, Davis TN. Phosphorylation of the chromosomal passenger protein Bir1 is required for localization of Ndc10 to the spindle during anaphase and full spindle elongation. Mol Biol Cell. 2006;17:1065–1074. doi: 10.1091/mbc.E05-07-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–284. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen T, Vinh DB, Crawford DK, Davis TN. A genetic analysis of interactions with Spc110p reveals distinct functions of Spc97p and Spc98p, components of the yeast gamma-tubulin complex. Mol Biol Cell. 1998;9:2201–2216. doi: 10.1091/mbc.9.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]