Abstract
Background
Gene complementation strategies are important in validating the roles of genes in specific phenotypes. Complementation systems in Helicobacter pylori include shuttle vectors, which transform H. pylori at relatively low frequencies, and chromosomally based approaches. Chromosomal complementation strategies are susceptible to polar effects and disruption of other H. pylori genes, leading to unwanted pleiotropic effects.
Materials and methods
A new complementation strategy was developed for H. pylori by utilizing a suicide plasmid vector that contains fragments of an H. pylori intergenic region (hp0203–hp0204), a chloramphenicol acetyltransferase cassette (cat), and a multiple-cloning site. Genes of interest could be cloned into the intergenic plasmid and the genes integrated into H. pylori by homologous recombination into the intergenic chromosomal region without disrupting any annotated H. pylori gene. The complementation system was validated using the gene encoding arginase (rocF).
Results
A rocF mutant unable to hydrolyze or consume L-arginine regained these functions by complementation with the wild-type rocF gene. Complemented strains also had restored arginase protein as determined by Western blot analysis. The complementation system could be successfully applied to multiple H. pylori strains. The intergenic region varied in length and sequence across 17 H. pylori strains, but the flanking-3′ ends of the hp0203 and hp0204 coding regions were highly conserved. Inserting a cat cassette and wild-type rocF into the intergenic region did not alter the ability of strain SS1 to colonize mice.
Conclusions
This complementation strategy should greatly facilitate genetic experiments in H. pylori.
Keywords: Helicobacter pylori, arginase, complementation
Since its discovery in 1982 by Marshall and Warren [1], Helicobacter pylori has been shown to be the causative agent of chronic gastritis, gastric and duodenal ulcers, and mucosa-associated lymphoid tissue (MALT) lymphoma [2,3]. Many infected individuals fail to develop clinical symptoms of disease, although all exhibit histologic evidence of pathology [4,5]. However, the exact mechanisms employed by H. pylori to cause such varied host pathology is unknown.
Research on H. pylori pathogenesis has been partially hampered by insufficient and suboptimal complementation systems. Complementation is the process of introducing a wild-type copy of a gene into a mutant strain disrupted for the same gene, leading to restoration of phenotypes that were lost in the mutant. Current methods used for gene complementation in H. pylori include shuttle plasmids and chromosomally based systems. Shuttle plasmids for H. pylori are hybrids comprised of H. pylori cryptic plasmids and plasmids isolated from Escherichia coli, and they contain antibiotic resistance markers for selection [6–10]. Unfortunately, these shuttle plasmids transform some H. pylori strains at low frequencies and completely fail to transform other H. pylori strains [6,8]. Chromosomally based complementation systems in H. pylori have been used to complement genes such as luxS, recG, rdxA, comH, and ureB, but these systems interrupt important chromosomal segments or can be susceptible to polar effects that may influence expression of other H. pylori genes [11–15]. Therefore, improved genetic complementation tools are needed for confirming the function of H. pylori genes.
This study was undertaken to develop a different complementation system and was validated by complementing the gene encoding H. pylori arginase, rocF. Arginase is an enzyme that hydrolyzes L-arginine to L-ornithine and urea. The urea is then hydrolyzed by urease to produce CO2 and ammonia, the latter serving to neutralize harmful stomach acid for H. pylori [16,17]. Arginase-mediated hydrolysis of arginine consumes the same substrate required for nitric oxide (NO) synthesis [18], an important innate host defense mechanism. The consumption of host L-arginine by H. pylori arginase can lead to dysregulation of the host immune response through inhibition of T-cell proliferation and CD3ζ chain expression on the T-cell surface [19].
Herein, we describe a gene complementation system that utilizes a suicide plasmid vector containing a specific intergenic region (between genes hp0203 and hp0204) from the H. pylori chromosome. These regions were separated into two fragments, IR203 and IR204, flanking a multicloning site. Once introduced into the bacterium by electroporation, these intergenic regions can undergo double cross-over homologous recombination into the bacterial chromosome, thereby inserting any genes cloned within the multicloning site and eliminating extraneous plasmid sequences. IR203 and IR204 regions were initially selected because the region represents one of the largest intergenic regions in the H. pylori genome. Moreover, this region is reasonably well conserved between the sequenced H. pylori strains (26695 and J99) [20,21]. Additionally, the hp0203 and hp0204 hypothetical genes are adjacent in 26695 and J99 strains. The hp0203 and hp0204 genes are transcribed towards each other, greatly minimizing possible polar effects, and avoiding insertion of DNA fragments into putative promoter regions. Finally, no annotated H. pylori genes will be disrupted by this chromosomally based strategy.
By using this plasmid vector system, the arginase mutant was complemented for the first time in H. pylori, and phenotypic effects lost in the rocF mutant H. pylori could be restored via complementation as determined by arginase activity, Western blot, and L-arginine consumption. It was also shown that the complementation system can be used for in vivo experiments without supplying antibiotics to animals. This technology provides a general method for complementing H. pylori mutants.
Materials and Methods
Bacterial Strains and Growth Conditions
All H. pylori strains were grown on Campylobacter blood agar (Becton, Dickinson, & Co., Sparks, MD, USA) containing 10% sheep blood (Quad Five, Ryegate, MT, USA), at 37 °C, 10% CO2, 5% O2, and 85% N2 atmospheric conditions for 48 hours, unless otherwise stated. For animal experiments H. pylori strains were grown overnight in Mueller–Hinton broth plus 2% fetal bovine serum (FBS) without aeration under the same atmospheric conditions as above. E. coli strains were grown on Luria agar or broth at 37 °C overnight. Concentrations of antibiotics used for H. pylori were kanamycin (15 μg/ml) and chloramphenicol (10 μg/ml). Antibiotics for E. coli strains were used at the following concentrations: ampicillin (100 μg/ml); kanamycin (25 μg/ml); and chloramphenicol (20 μg/ml). Complete listings of bacterial strains used in this study are in Table 1.
Table 1.
Description of strains used in this study
| Bacterial Strain | Description | Source |
|---|---|---|
| H. pylori | ||
| 26695 | Wild-type H. pylori strain | [21] |
| 26695rocF::aphA3 (26695rocF mutant) | Contains rocF gene disrupted by kanamycin (aphA3) cassette | [29] |
| 26695-203C04 (IR203::cat204) | 26695 transformed with pIR203C04 (chloramphenicol resistance cat cassette inserted into hp0203–204 intergenic region) | This study |
| rocF26695-203C04 (IR203::cat204) | 26695rocF mutant transformed with pIR203C04 (chloramphenicol resistance cat cassette inserted into hp0203–204 intergenic region) | This study |
| rocF26695-MLB001 (IR203::cat rocF204) | 26695rocF mutant transformed with pMLB001 (rocF/rocF+) | This study |
| rocF26695-MLB002 (IR203::cat rocF204) | 26695rocF mutant transformed with pMLB002 (rocF/rocF+) | This study |
| rocF26695-MLB003 (IR203::cat rocF204) | 26695rocF mutant transformed with pMLB003 (rocF/rocF+) | This study |
| rocF26695-MLB004 (IR203::cat rocF204) | 26695rocF mutant transformed with pMLB004 (rocF/rocF+) | This study |
| SS1 | Wild-type mouse-adapted H. pylori strain | [38] |
| SS1rocF::aphA3 (SS1rocF mutant) | Contains rocF gene disrupted by kanamycin (aphA3) cassette | [29] |
| SS1 IR203C04 | SS1 transformed with chromosomal DNA from 26695-203C04 (chloramphenicol resistance cat cassette inserted into hp0203–204 intergenic region) | This study |
| SS1rocF/rocF+ (IR203::cat rocF204) | SS1 rocF::aphA3 transformed with chromosomal DNA from rocF26695-MLB002 (WT SS1 rocF gene inserted into hp0203–204 intergenic region) | This study |
| 43504 | Wild-type H. pylori strain | ATCC |
| 3401 | Wild-type H. pylori strain | M. Blaser |
| J63 | Wild-type H. pylori low passage clinical isolate | R. Peek |
| J68 | Wild-type H. pylori low passage clinical isolate | R. Peek |
| J75 | Wild-type H. pylori low passage clinical isolate | R. Peek |
| J99 | Wild-type H. pylori strain | [20] |
| J104 | Wild-type H. pylori low passage clinical isolate | R. Peek |
| J166 | Wild-type H. pylori low passage clinical isolate | R. Peek |
| J178 | Wild-type H. pylori low passage clinical isolate | R. Peek |
| J188 | Wild-type H. pylori low passage clinical isolate | R. Peek |
| J254 | Wild-type H. pylori low passage clinical isolate | R. Peek |
| B128A | Wild-type H. pylori low passage clinical isolate | R. Peek |
| B194A | Wild-type H. pylori low passage clinical isolate | R. Peek |
| B223A | Wild-type H. pylori low passage clinical isolate | R. Peek |
| B228A | Wild-type H. pylori low passage clinical isolate | R. Peek |
| E. coli | ||
| DH5α | F−, supE44 ΔlacU169 ϕ80lacZM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | BD Biosciences Clontech |
ATCC, American Type Culture Collection.
Molecular Biology
Plasmid DNA was extracted by either the alkaline lysis method [22] or the column chromatography method (QIAGEN, Valencia, CA, USA). H. pylori chromosomal DNA was isolated by suspending H. pylori in 480 μl lysis buffer (10 mmol/l Tris, 75 mmol/l NaCl, 25 mmol/l ethylenediamine tetra-acetic acid, 1% SDS, pH 7.8) plus proteinase K (300 μg/ml) and incubated at 55 °C overnight, followed by phenol–chloroform and chloroform extractions and then ethanol precipitation. DNA was resuspended in 50 μl 10 mmol/l Tris-HCl (pH 8.5) plus RNAse (10 μg/ml). Restriction enzymes were from Promega (Madison, WI, USA). Ligations were performed using a Rapid DNA Ligation kit (Roche, Indianapolis, IN, USA). DM3-DM6 primers were obtained from the Biopolymer Laboratory at the University of South Alabama (Mobile, AL, USA). All other primers were obtained from Integrated DNA Technologies, Inc. (Coralville, IA, USA). Primer sequences are listed in Table 2.
Table 2.
Description of plasmids and primers used in this study
| Plasmid or primer | Description | Source |
|---|---|---|
| Plasmid | ||
| pBluescript II SK(−) (pBS) | Cloning vector, ampicillinr (3 kb) | Stratagene |
| pIR204 | pBS containing H. pylori IR204 region (3.33 kb) | This study |
| pIR20304 | pIR204 with H. pylori IR203 inserted (3.66 kb) | This study |
| pIR203C04 | pIR20304 with chloramphenicol resistance cassette (cat) inserted (4.5 kb) | This study |
| pIR203K04 | pIR20304 with kanamycin resistance cassette (aphA3) inserted (4.9 kb) | This study |
| pMLB001 | pIR203C04 with H. pylori strain 43504 rocF gene and native promoter inserted (5.6 kb) | This study |
| pMLB002 | pIR203C04 with H. pylori strain SS1 rocF gene and native promoter inserted (5.6 kb) | This study |
| pMLB003 | pIR203C04 with H. pylori strain J63 rocF gene and native promoter inserted (5.6 kb) | This study |
| pMLB004 | pIR203C04 with H. pylori strain 26695 rocF gene and native promoter inserted (5.6 kb) | This study |
| pJH43504 | pBluescript SK containing H. pylori 43504 rocF gene and native promoter (4.1 kb) | Hovey & McGee, unpublished |
| pJHSS1 | pBluescript SK containing H. pylori SS1 rocF gene and native promoter (4.1 kb) | Hovey & McGee, unpublished |
| pJH63 | pBluescript SK containing H. pylori J63 rocF gene and native promoter (4.1 kb) | Hovey & McGee, unpublished |
| pBS-rocF | pBluescript SK containing H. pylori 26695 rocF gene and native promoter (4.1 kb) | [29] |
| pBS-cat | pBS containing 0.8 kb non-polar chloramphenicol acetyltransferase cassette from Campylobacter coli; subcloned from pSKCAT4 into EcoRI site, chloramphenicolr, ampicillinr (3.8 kb) | This study |
| pBS-kan | pBS containing 1.25 kb non-polar kanamycin resistance cassette (aphA3) from Enterococcus faecalis (4.3 kb) | [29] |
| Primers (5′–3′) | Lowercase denotes restriction sites/extra nucleotides to facilitate cloning. | |
| DM3 (IR203-F1) | gcgagctcTAGAGCAAGCCCAAAAAGG | |
| DM4 (IR203-R1) | gctctagaTATAAAAATTAAAGCGATAA | |
| DM5 (IR204-F1) | ccgctcgagCGTTTTTTTGAAGCCAATTCT | |
| DM6 (IR204-R1) | ggggtacccTAAAATTTGGTTTTTGTTC | |
| DM71 (hp0203-F1) | CCCTCCTATGGGGTTTCATT | |
| DM189 (hp0204-F2) | CCGGTGGTGATTAAAAACCTT | |
| DM228 (hp0204-F3) | CAATAGGGGAAATTCATGCG | |
| cat-F2 | AATCCCAGTTTGTCGCAC | |
| cat-F4 | TGCTGCCTTGCTACACAGTT | |
| kan-F2 | GCGCGAGCTGTATGATTTTT | |
| rocF-F25 | cgggatccCATGATTTTAGTAGGATTAGAAGCAGAGT | |
| rocF-R25 | ggggtaccTCAGTAACTCCTTGCAAAAGAGTGCTTC | |
, resistance.
Polymerase Chain Reaction
Polymerase chain reaction (PCR) conditions were set at 30 cycles, with one cycle consisting of 94 °C for 30 seconds, 55–60 °C for 30 seconds, and 72 °C for 30 seconds with Taq DNA polymerase (Promega, Madison, WI, USA). Complemented strains of H. pylori were confirmed with the following oligonucleotides: cat-F2, kan-F2, rocF-F25, rocF-R25, and hp0203-F1 (Table 2). Amplification and sequence analysis of the intergenic plus adjacent regions was accomplished in different H. pylori strains using primers DM71 and DM189 (Table 2).
Southern Hybridization
Chromosomal DNA from H. pylori strains was digested with BamHI and EcoRI overnight. DNA was transferred to Hybond-N+ nylon membrane (Amersham Biosciences, Piscataway, NJ, USA). An ECL Direct Nucleic Acid Labeling and Detection System (Amersham) was used for labeling a 3.0 kb gel-purified pBluescript II SK(−) (pBS) (Stratagene, LaJolla, CA) fragment digested with EcoRV and PstI. Blots were washed stringently and treated according to the manufacturer’s specifications.
Construction of Plasmid Vectors (Table 2)
All plasmids were confirmed by restriction analysis and DNA sequencing (Iowa State University Sequencing Facility, Ames, IA, USA).
pIR204
The IR204 region (Fig. 1A) was PCR-amplified from H. pylori 26695 using primers DM5 and DM6 and digested with XhoI and KpnI, and the 326 bp fragment was cloned into pBS digested with XhoI and KpnI.
Figure 1.
Development of an H. pylori chromosomal complementation system. (A) Wild-type 26695 H. pylori chromosome region chosen for complementation. The intergenic region (IR) was divided into two fragments for cloning purposes, IR203 and IR204. Arrow indicates direction of transcription for the coding regions. The 326 bp IR203 and 332 bp IR204 regions were cloned. The actual intergenic region size in strain 26695 is shorter (632 bp), based on sequence analysis (see Fig. 6 and Table 4). (B) Construction of cloning vectors: pIR20304, pIR203C04, pMLB001, pMLB002, pMLB003, and pMLB004. Arrows in plasmids denote direction of transcription. MCS, multicloning site.
pIR20304
The pIR204 plasmid and PCR-amplified IR203 DNA (primers DM3 and DM4; 332 bp) were digested with SacI and XbaI restriction endonucleases, gel-purified (QIAGEN, Valencia, CA, USA), and ligated to form pIR20304 (Fig. 1B).
pIR203C04 (Intergenic Plasmid)
pIR20304 was digested with HincII, and pBS-cat was digested with EcoRV and SmaI. After agarose gel electrophoresis, pIR20304 (3.66 kb) and a nonpolar cat (chloramphenicol acetyltransferase) cassette (0.84 kb) from pBS-cat (Table 1) were purified and the fragments ligated to form pIR203C04 (Fig. 1B). Restriction sites remaining for cloning additional fragments are: BamHI, SmaI, EcoRV, and PstI.
pIR203K04 (Intergenic Plasmid)
This plasmid is similar to pIR203C04, except a nonpolar kanamycin resistance cassette was cloned. pIR20304 was digested with HincII and pBS-kan (29) was digested with HincII and SmaI. The pIR20304 vector (3.66 kb) and kan cassette (1.25 kb) insert were purified with QIAquick and ligated to form pIR203K04. Restriction sites remaining for cloning are BamHI, ClaI, HindIII, and SmaI.
pMLB001, pMLB002, pMLB003, and pMLB004
Four plasmids, pJH43504, pJHSS1, pJH63, and pBS-rocF (Table 1), contain the rocF gene cloned from H. pylori 43504, SS1, J63, and 26695, respectively. These plasmids were digested with BamHI and HincII and the 1.1 kb rocF fragment was gel-purified and ligated into the BamHI and SmaI sites of pIR203C04 (4.5 kb) to yield pMLB001, pMLB002, pMLB003, and pMLB004, respectively (Fig. 1B).
Transformation of H. pylori and E. coli
H. pylori was transformed with 3 μg plasmid or chromosomal DNA by electroporation (2.5 kV, 50 μF, 200 ohms, time constant = 10 ms). Electroporation typically reduces viable H. pylori counts by about two orders of magnitude. Following electroporation, bacteria were spotted onto Campylobacter blood agar (CBA) lacking antibiotics and grown for 2 days and then removed and placed on selective media. Transformation frequency was determined by dividing the number of transformed colonies per milliliter by total CFU/ml of bacteria postelectroporation (T0). E. coli cells were transformed by the calcium chloride method [22].
Construction of H. pylori rocF/rocF+ Complemented Strains
The rocF mutant of H. pylori strain 26695 was transformed with pMLB001, pMLB002, pMLB003, or pMLB004 by electroporation, and transformants were selected on CBA agar containing chloramphenicol and kanamycin. Chromosomal DNA from transformed colonies was isolated and confirmation of each strain was performed by PCR analysis (see Results section; Fig. 2A–D). Southern blot was also used to eliminate the possibility that a single cross-over event occurred. Other genetically engineered H. pylori strains (Tables 1 and 3) were constructed and confirmed in the same fashion, except where noted. Confirmed strains were frozen at −80 °C in 1 ml Ham’s F12 (Hyclone, Logan, UT, USA) broth plus 20% glycerol. As a control, both wild-type 26695 and the rocF mutant 26695 were transformed with the pIR203C04 plasmid lacking rocF, selected on CBA plus chloramphenicol, and confirmed by PCR, as above.
Figure 2.
PCR confirmation of complementation strains. (A) 26695–203C04 chromosome and primers used in PCR confirmation. Expected product size using hp0203-F1 and cat-F2 primers is ~550 bp. (B) rocF 26695-MLB001 chromosome and primers used for confirmation. Product size between hp0203-F1 and rocF-F25 is ~1.15 kb, hp0203-F1 and cat-F2 product size is ~1.65 kb. This strain also harbors the rocF::aphA3 mutation described in Fig. 2C. (C) rocF mutant (rocF::aphA3) 26695 chromosome, interrupted by a kanamycin cassette. Expected product size is ~650 bp using primers kan-F2 and rocF-R25. rocF′ and ‘rocF denote incomplete open-reading frame. (D) PCR confirmation of 26695 derivative strains, using hp0203-F1 and cat-F2 primers. Black arrows denote primers. MCS, multicloning site.
Table 3.
Transformation frequency of four wild-type H. pylori strains: 26695, SS1, J99, and 43504 using intergenic plasmids versus chromosomal DNA derived from strain 26695 already transformed with intergenic plasmids. Wild-type strains were transformed with pIR203C04, pMLB004, 26695–203C04 chromosomal (chr.) DNA, or rocF26695-MLB004 chromosomal DNA. Data shown are four separate experiments, with duplicates averaged from each experiment
| Strain | DNA | Frequency (Expt 1) | Frequency (Expt 2) | Frequency (Expt 3) | Frequency (Expt 4) |
|---|---|---|---|---|---|
| 26695 | pIR203C04 | 2.2 × 10−4 | 5.2 × 10−6 | 1.6 × 10−6 | 3.8 × 10−6 |
| 26695 | 26695–203C04 chr. | 2.2 × 10−4 | 5.2 × 10−5 | 3.3 × 10−6 | 3.8 × 10−6 |
| 26695 | pMLB004 | 4.3 × 10−6 | 1.8 × 10−7 | 3.6 × 10−8 | 9.6 × 10−7 |
| 26695 | rocF26695-MLB004 chr. | 2.2 × 10−4 | 5.2 × 10−5 | 1.6 × 10−6 | 1.9 × 10−6 |
| SS1 | pIR203C04 | < 10−10 | < 10−10 | < 10−10 | < 10−10 |
| SS1 | 26695-203C04 chr. | 3.8 × 10−6 | 1.2 × 10−8 | 1.0 × 10−7 | 6.0 × 10−9 |
| SS1 | pMLB004 | < 10−10 | < 10−10 | < 10−10 | < 10−10 |
| SS1 | rocF26695-MLB004 chr. | < 10−10 | 4.5 × 10−9 | < 10−10 | < 10−10 |
| J99 | pIR203C04 | < 10−10 | 1.4 × 10−8 | 4.5 × 10−8 | < 10−10 |
| J99 | 26695–203C04 chr. | 2.6 × 10−3 | 2.1 × 10−6 | 9.0 × 10−6 | 3.7 × 10−7 |
| J99 | pMLB004 | < 10−10 | 4.4 × 10−9 | < 10−10 | < 10−10 |
| J99 | rocF26695-MLB004 chr. | 2.6 × 10−3 | 2.1 × 10−6 | 4.5 × 10−6 | < 10−10 |
| 43504 | pIR203C04 | < 10−10 | < 10−10 | < 10−10 | < 10−10 |
| 43504 | 26695-203C04 chr. | 9.5 × 10−4 | 8.7 × 10−8 | 4.1 × 10−6 | 1.2 × 10−5 |
| 43504 | pMLB004 | 5.9 × 10−5 | 4.6 × 10−9 | 8.3 × 10−7 | 5.0 × 10−8 |
| 43504 | rocF26695-MLB004 chr. | 6.4 × 10−5 | 8.7 × 10−8 | 3.3 × 10−6 | 7.5 × 10−7 |
Arginase Activity
Arginase activity of H. pylori and E. coli was determined as described previously [23]. Data are presented as mean specific activity ± standard deviation. All experiments were conducted at least twice with duplicate measurements, with a representative experiment shown.
Western Blot
Bacterial extracts containing arginase were prepared as described previously [23]. Protein (20 μg) was loaded on a 10% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and electrophoresed (125 V, 120 minutes). Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules, CA, USA), incubated with anti-RocF (1: 3000) [23], and anti-rabbit IgG conjugated with alkaline phosphatase (1: 10,000, Sigma, St. Louis, MO, USA). Nitro-blue tetrazolium (100 μg/ml) and 3-indoxyl phosphate (10 μg/ml) were used as detection reagents.
L-Arginine Consumption by H. pylori
Experiments were conducted in RPMI 1640 medium plus 10% FBS. Bacteria were inoculated at 5 × 105 viable CFU and allowed to grow for 48 or 72 hours. The supernatant was retained for quantification of L-arginine concentration by high-performance liquid chromatography electrochemical detection (HPLC-ECD) as previously described [24], using an ESA-CoulArray Model 540 (ESA Inc, Chelmsford, MA, USA) and an 80 × 3.2 mm column with 120 Å pore size. Supernatants were briefly deproteinized by methanol. After centrifugation at 6000 × g for 10 minutes at 4 °C, the supernatant was derivatized with 0.2 mol/l o-phthaldialdehyde containing β-mercaptoethanol. Each sample (50 μl) was injected into the column. The retention time for the L-arginine standard (prepared in methanol) was 10.2 minutes.
Colonization of Mice
Animal experiments were approved by the Institutional Animal Care and Use Committee at the University of South Alabama. Mice (129 Ev/Sv strain, in-bred internally) were orally inoculated with 50 μl (~1 × 109 CFU/ml in 1× phosphate-buffered saline [PBS]) of either wild-type SS1 (n = 8 animals), the rocF mutant SS1 (n = 7 animals), the wild-type carrying the cat cassette within the hp0203–204 intergenic region (SS1 IR203C04; Table 1) (n = 4 animals) or the SS1 rocF/rocF+ (Table 1) carrying the cat cassette plus the wild-type rocF gene inserted into the hp0203–hp0204 intergenic region (n = 5 animals). Mice were given antibiotic-free water and food ad libitum. At 1 week post-infection, animals were humanely euthanized. Stomachs were removed, dissected longitudinally along the greater curvature, and the chyme removed. The body and antrum were separated, excised into multiple slices, weighed, and then homogenized (Ultra-Turrax T25, IKA Works, Inc; 10–15 seconds at a setting of 3) in 1.5 ml sterile PBS. Antrum and body homogenates and dilutions thereof in PBS were plated for viable counts in duplicate on CBA plates containing 5-fluorocytosine (5 μg/ml), vancomycin (10 μg/ml), amphotericin B (5 μg/ml), bacitracin (30 μg/ml), polymyxin B (10 U/ml), and trimethoprim (10 μg/ml) to suppress normal flora. Some plates also contained chloramphenicol (10 μg/ml) for recovery of the SS1 IR203C04 strain. Plates were incubated for 5 days in an atmosphere of 5% O2, 10% CO2, 85% N2. Data are presented as colony-forming units (CFU)/g antrum or body. One-way analysis of variance was used to analyze the colonization data. p < .05 was considered significant.
Alignments and Phylogenetic Analyses
The hp0203–hp0204 intergenic region plus adjacent DNA from the corresponding -3′ ends of each coding region were aligned and a consensus sequence was obtained using CLUSTALW 1.83 (http://www.ebi.ac.uk/clustalw/) and the aligned files were then analyzed in CLUSTALX 1.81 using the neighbor-joining method. The output file (tre format) was opened in TREEVIEW 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/rod.html) and formatted into a slanted cladogram.
GenBank Accession Numbers
The sequence data for the 3′ ends of the coding regions of hp0203 and hp0204 along with the intergenic region in between have been deposited into the GenBank database as accession numbers DQ538310–DQ538326.
Results
We developed a new complementation system for H. pylori using the intergenic plasmid pIR203C04 (see Fig. 1; Materials and Methods) and used the H. pylori arginase gene as a model to validate this system. After the wild-type or arginase mutant 26695 H. pylori derivatives were transformed with the intergenic plasmid derivatives, transformants were confirmed by PCR analysis and tested for phenotypic effects by arginase activity, Western blot, and L-arginine consumption. Furthermore, transformation frequency was examined in 26695, as well as SS1, J99, and 43504 H. pylori strains to determine the application of this complementation method to genetically diverse strains. Finally, the usefulness of the complementation system was examined in vivo using a mouse model.
Confirmation of H. pylori Strains by PCR and Southern Blot
All genetically engineered H. pylori strains in Table 1 were confirmed by PCR (Fig. 2A–D; data not shown). The product size when using the cat-F2 and hp0203-F1 primers was 550 bp for 26695–203C04 and 1.65 kb for the complemented strains rocF26695-MLB001, rocF26695-MLB002, rocF26695-MLB003, and rocF26695-MLB004. Wild-type 26695 and rocF mutant 26695 had no product with these primers, as expected. The product size using the hp0203-F1 and rocF-F25 primers was 1.15 kb for rocF26695-MLB001, rocF26695-MLB002, and rocF26695-MLB003. The rocF gene in rocF26695-MLB004 is in the forward orientation, so the rocF-R25 and hp0203-F1 primers were used, generating a product of 1.5 kb. Wild-type 26695, rocF mutant 26695, 26695–203C04, and rocF26695–203C04 all produced no products with these primers, as expected. The same confirmation strategy was also used for strains SS1, J99, and 43504 (data not shown). In addition, PCR was executed on all strains of H. pylori carrying the original rocF::aphA3 mutation by using the kan-F2 and rocF-R25 primers. The product was 650 bp, demonstrating that the original rocF mutation was still intact; strains without the original rocF disruption showed no product, as expected. Southern blots were also performed using a pBS probe. The positive control plasmid (pBS) hybridized with the probe, but none of the chromosomal DNAs from the H. pylori strains hybridized with the pBS vector probe, confirming that the recombination did not occur as a single cross-over event (data not shown).
Complementation of Arginase Mutant H. pylori: Restoration of Arginase Activity
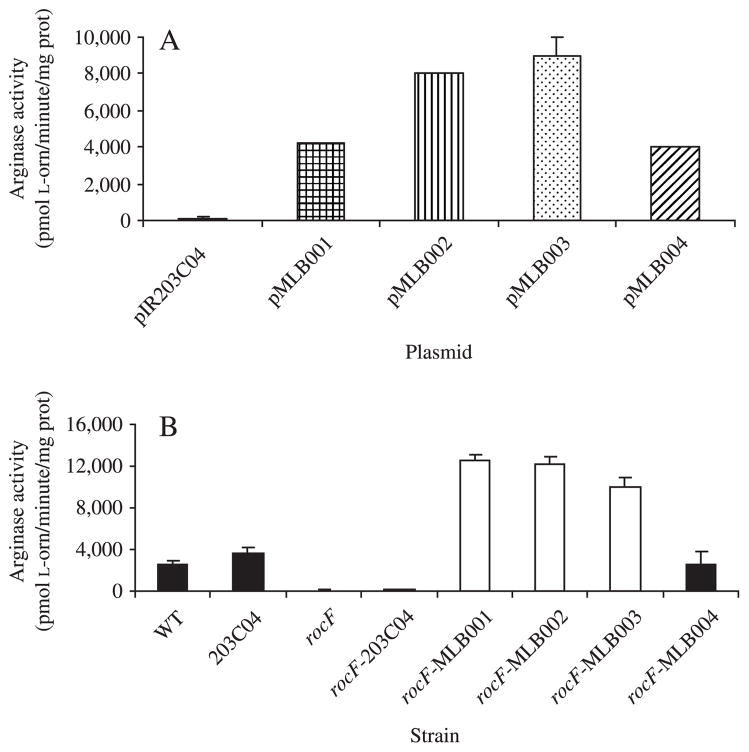
Arginase activity was determined for E. coli strains harboring the intergenic plasmid vectors (pMLB001–004) containing the rocF gene from four different H. pylori strains. High arginase activity occurred in E. coli harboring all four of these plasmids, whereas E. coli carrying the isogenic vector control, pIR203C04, expressed no arginase activity (Fig. 3A). Clear variation in arginase activity was conferred to the E. coli strains by the rocF genes derived from different H. pylori strains (Fig. 3A). In the H. pylori strains, arginase activity determined for the wild-type 26695, 26695-203C04, and the rocF26695-MLB004 complemented strains were not significantly different (Fig. 3B). Also, the rocF mutant and rocF26695-203C04 strains were devoid of arginase activity, indicating that the rocF disruption abolishes production of arginase in these strains. The rocF26695-MLB001, rocF26695-MLB002, and rocF26695-MLB003 strains, which are complemented with wild-type rocF genes from strains 43504, SS1, and J63, respectively, all exhibited activity higher than rocF26695-MLB004 (Fig. 3B). Thus, even when four different rocF alleles were in the same genetic background (either the E. coli model or the H. pylori), arginase activity variation was evident.
Figure 3.
Arginase activity of E. coli expressing H. pylori rocF and of various H. pylori strains. (A) Arginase activity in E. coli carrying pIR203C04 (negative control), pMLB001 (43504 rocF gene), pMLB002 (SS1 rocF gene), pMLB003 (J63 rocF gene), or pMLB004 (26695 rocF gene). (B) Arginase activity in rocF/rocF+ 26695 complemented strains, compared to wild-type and rocF mutant 26695. Strains listed in Table 1. L-orn, L-ornithine; prot, protein.
In the mouse-adapted SS1 H. pylori strain, the rocF mutation also results in abolished arginase activity, whereas complementation of the arginase mutant with the rocF gene from SS1 (SS1 rocF/rocF+ in Table 1) restored arginase activity (data not shown).
Complementation of Arginase Mutant H. pylori: Restoration of Arginase Protein by Western Blot Analysis
Western blot analysis was performed to determine the presence of arginase protein produced in each H. pylori 26695 derivative, as well as DH5α E. coli strains carrying the plasmid derivatives. DH5α E. coli harboring pMLB001, pMLB002, or pMLB003 all had similar RocF levels, while E. coli containing pMLB004 expressed higher levels (Fig. 4A). Arginase protein observed in the wild-type 26695 versus 26695–203C04 was not significantly different, suggesting that the addition of cat cassette and the MCS inserted between the IR203 and IR204 regions did not alter the production of the arginase protein (Fig. 4B). The amount of arginase protein found in rocF26695-MLB004 was also not significantly different from the wild-type 26695, since this strain is complemented with a wild-type copy of 26695 rocF. Both the rocF mutant and the rocF26695-203C04 strains were devoid of arginase protein, as expected (Fig. 4B). The rocF26695-MLB001, rocF26695-MLB002, and rocF26695-MLB003 complemented strains exhibited also had restored expression of the arginase protein (Fig. 4B).
Figure 4.
Arginase production assessed by Western blot analysis. (A) Anti-RocF Western blot analysis of E. coli containing pIR203C04, pMLB001, pMLB002, pMLB003, and pMLB004. Protein (20 μg) was added per well. (B) Anti-RocF Western blot analysis of H. pylori. Protein (20 μg) was added per well. The experiment was repeated twice (independently prepared extracts) with similar results.
Complementation of Arginase Mutant H. pylori: Restoration of L-arginine Consumption
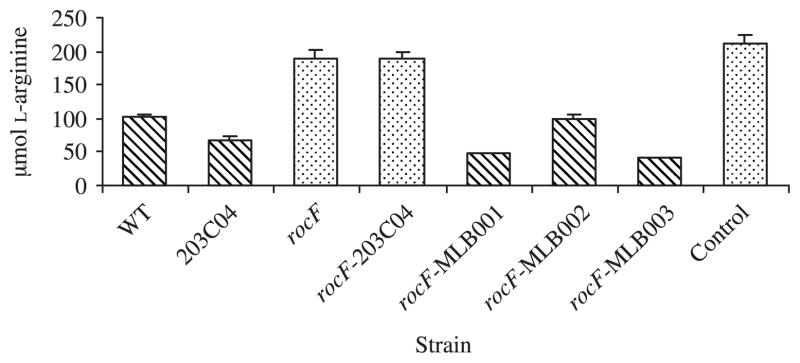
To provide further evidence for complementation of the arginase mutant, L-arginine consumption experiments were conducted. Since H. pylori cannot synthesize its own arginine [21,25–27], wild-type organisms consume L-arginine from the media in which they are grown [25–28]. H. pylori 26695 derivative strains were grown for 48 or 72 hours, and L-arginine consumption was measured (Fig. 5; 48-hour data shown). The rocF mutant strains consumed little to no L-arginine, similar to the uninoculated media; strains with measurable arginase activity (either wild type or complemented with a wild-type rocF) all consumed significant L-arginine in the medium. Strains grown for 72 hours that exhibited arginase activity consumed additional arginine, while strains devoid of arginase activity still did not consume significant arginine (data not shown).
Figure 5.
L-arginine consumption by wild-type and genetically engineered H. pylori strains. H. pylori 26695 strains were inoculated at 5 × 105 CFU/ml in RPMI 1640 medium containing 10% FBS and grown for 48 hours. The culture supernatant was measured for L-arginine by HPLC-ECD as described in Materials and Methods. Control, uninoculated media.
Transformation Frequency in Different H. pylori Strains Using the Intergenic Complementation Plasmid or a Two-Step Transformation Using Chromosomal DNA
To determine whether the complementation system could be used for other H. pylori strains, transformation frequency was compared in wild-type H. pylori strains 26695, SS1, J99, and 43504 transformed with pIR203C04, pMLB004, 26695-203C04 chromosome, or rocF26695-MLB004 chromosome. Data are presented as four independent experiments (Table 3). While the intergenic plasmids transform 26695, transformation was not possible (strain SS1) or observed at low frequencies (strains 43504, J99) in the other strains studied. We therefore examined a two-step transformation, which entails transforming strain 26695 with the intergenic plasmid, isolating the resultant chromosomal DNA and then transforming this DNA into the H. pylori strain of interest. This two-step procedure was successful in all non-26695 strains of H. pylori, but had lower transformation frequencies in strain SS1 than strains 43504 and J99. With strain J99 the two-step transformation was at least two orders of magnitude more successful than transforming with the intergenic plasmids directly into J99 (Table 3). These results suggest that the complementation system may be applied to a broad range of H. pylori strains, including strains (43504, SS1) used in animal studies.
Variation in Intergenic Region in Different H. pylori Strains
To determine whether there is conservation of the hp0203–204 intergenic region in different H. pylori strains, the 3′ ends of the hp0203 and hp0204 coding regions plus the intergenic region were PCR-amplified (using primers DM71 and DM 189 [Table 2]) and directly sequenced from 17 H. pylori strains, including 12 minimally passaged clinical isolates. The results indicated that the 3′ ends of both the hp0203 and hp0204 coding regions were highly conserved across the isolates (Fig. 6A,B). Strikingly, the intergenic region was not conserved in length (Table 4), and there was also substantial sequence heterogeneity (Fig. 6C,D). Remarkably, nucleotide sequences of the hp0203–204 intergenic region were distinct from all strains except strains J75 and B194A. The length of the intergenic region largely correlated with its phylogenetic placement (Table 3 and Fig. 6D). For example, the intergenic region in strains J68, J75, B194A, 3401, and 43504 were similar in length (262–290 bp) and were clustered on the phylogenetic tree (Fig. 6D). Intriguingly, the phylogenetic trees created from the alignments of the 3′ ends of hp0203 and hp0204 coding regions were dissimilar to each other and to the intergenic region (data not shown). The high conservation of the hp0203 and hp0204 3′ ends of the coding regions, however, would potentially allow use of the complementation system in a wide variety of H. pylori strains via the two-step transformation method.
Figure 6.
Variation in the hp0203–hp0204 intergenic region across H. pylori strains. The 3′ ends of the hp0203 and hp0204 coding regions, plus the intergenic region was PCR-amplified and sequenced from 17 H. pylori strains. (A) Multisequence nucleotide alignment of the 3′ end of hp0203 coding region using clustalw. Arrow, extra residue uncovered in the analysis that alters the Hp0203 C-terminal region (lowercase nucleotide). Asterisks depict residues conserved in all 17 H. pylori strains. Underlined, stop codon for hp0203. (B) Multisequence nucleotide alignment of the 3′ end of hp0204 coding region using clustalw. Underlined, stop codon for hp0204. (C) Multisequence nucleotide alignment of the hp0203–204 intergenic region using clustalw. Boxed, repeat elements and palindromes. Underlined, possible stem-loop structures for rho-independent transcriptional termination as determined by m-fold. (D) Bootstrap neighbor-joining phylogenetic tree (slanted cladogram) of the alignment in Fig. 6C using clustalx and treeview. Asterisk denotes that these strains are 100% identical at the nucleotide level over the entire intergenic region.
Table 4.
Variation in the length of the hp0203–hp0203 intergenic region in different H. pylori strains
| Strain | Length of intergenic region (bp) |
|---|---|
| B128A | 262 |
| B194A | 288 |
| B223A | 687 |
| B228A | 681 |
| J63 | 444 |
| J68 | 288 |
| J75 | 288 |
| J99 | 689 |
| J104 | 679 |
| J166 | 708 |
| J178 | 656 |
| J188 | 748 |
| J254 | 719 |
| SS1 | 444 |
| 3401 | 287 |
| 26695 | 632 |
| 43504 | 290 |
Two other observations were made regarding the sequence data obtained on this region. First, the predicted C-terminal end of the Hp0203 protein is nine amino acids longer than reported in the original annotation of strain 26695 [21], owing to an extra G residue between nucleotides 236 and 237 of the entire coding region of hp0203 (Fig. 6A, arrow). This one nucleotide insertion results in a shift in the reading frame that leads to a new stop codon further downstream. The sequenced strain J99 and all strains examined in this study including 26695 have this extra G residue. This updated Hp0203 protein still shares no homology in the database with any other protein, except for the J99 homolog (Jhp_0189).
Second, we noticed several interesting regions in the hp0203–hp0204 intergenic region. The first is a repeat element, which is found in 12 of 17 strains in either one copy (strains SS1, J63 and B128A), two copies (strains B223A, B228A, J99, J104, J254, J178, and 26695 strains), or three copies (strains J188 and J166) (Fig. 6C, boxed). The consensus sequence is TTA(A/C)(T/C)A(G/C)CTATT(G/A). When the sequence is found as TTAATAGCTATTA (as in one of the copies in strains J99, J166, J178, and J188), there is a perfect palindrome (underlined). The second element we identified is TATAACTGACTATTATAATAGTCGGTTATA and forms a near perfect palindrome found in strains B223A, B228A, J99, J104, J254, J188, and J166, with remnants of the element in J178 (Fig. 6C, boxed). The specific roles of these elements are unknown.
The Intergenic Complementation System Can be Used In Vivo
A major drawback to using shuttle plasmids is that antibiotic selection would likely be required for use in animal experiments, leading to potential effects on distributions and levels of normal flora organisms that could affect H. pylori colonization or subsequent immune response. In vitro, the intergenic complementation system did not require antibiotic selection to maintain the chromosomal construct. To ascertain whether insertions into the chromosomal hp0203–hp0204 intergenic region affected the ability of H. pylori to colonize mice, the following experiment was conducted. An SS1 strain containing the cat cassette inserted into the intergenic region (SS1 IR203C04 in Table 1) was constructed by the two-step transformation procedure and compared for its ability to colonize mice versus the isogenic wild-type SS1, the rocF SS1 mutant and the rocF complemented SS1 strain (see Materials and Methods and Table 1). None of the mice were given antibiotics. The results showed that all four strains of H. pylori colonized at similar densities to each other in both the antrum and the body (p > .05) (Table 5). Strains carrying antibiotic resistance prior to infection still retained proper antibiotic resistance profiles after infection (data not shown). We previously reported that the rocF::aphA3 mutant of SS1 colonized Swiss mice at a density of about 1 log lower than wild-type mice [29]. Using a higher starting inoculum, shorter infection time, and different mouse strain than in our previous study, we observed a trend towards a 0.5 log decrease in colonization by the rocF mutant in both the antrum and the body. This result, however, did not reach statistical significance (p > .05). We conclude that the insertion of cat by itself or along with rocF into the hp0203–204 intergenic region does not affect colonization ability, indicating that this chromosomal site can be used for in vivo complementation experiments.
Table 5.
In vivo utilization of the complementation system in H. pylori-infected mice. Animals were inoculated with H. pylori strains and euthanized 1 week later. The body and antrum portions of the stomach were processed and plated for viable counts (see Materials and Methods). There is no significant difference (p > .05) in colonization among the four strains in either the antrum or the body
| Strain (n = number of animals inoculated) | CFU/g antrum | CFU/g body |
|---|---|---|
| WT SS1 (n = 8) | 4.1 × 103 | 1.0 × 104 |
| SS1 rocF::aphA3 (n = 7) | 1.0 × 103 | 6.2 × 103 |
| SS1 rocF/rocF+ (n = 5) | 6.8 × 103 | 2.8 × 104 |
| SS1 IR203C04 (n = 4) | 1.7 × 104 | 1.7 × 104 |
Discussion
Complementation systems are important for assigning and confirming functions of specific genes and for helping to rule out polar effects. Other published methods of gene complementation within the H. pylori chromosome involve homologous recombination into a region critical for virulence, such as urease [12–14]. While this method has been successful in complementing H. pylori mutations, it disrupts the gene coding for the α-subunit of the enzyme urease, ureA. This presents a disadvantage when performing in vivo studies, as urease is required for colonization [30]. Another system interrupts the gene associated with metronidazole resistance, rdxA [15]. Growth of the H. pylori rdxA mutant with metronidazole results in altered protein profiles [31], suggesting that the rdxA mutation can potentially cause unwanted phenotypic effects on the cell. Whether any of these alterations affect H. pylori colonization or host response is unclear. Therefore, we sought to create a system for complementing mutations within a chromosomal region of H. pylori that is likely unassociated with pathogenicity and with minimal chances of pleiotropic and polar effects. The plasmid base, pBluescript, was selected for use because native E. coli plasmids are neither replicated nor stably maintained within H. pylori. The intergenic region within the complementation plasmid pIR203C04 undergoes homologous recombination into the H. pylori hp0203–204 intergenic region. This serves as an attractive alternative to the more traditional shuttle plasmid-mediated gene complementation systems. The system developed here also eliminates copy number effects from shuttle plasmids.
The site chosen for the chromosomal complementation system described here was based on the finding that the hp0203–204 intergenic region is one of the largest in the genome and seemed to be reasonably well conserved based on strains 26695 and J99 [20,21]. The hp0203 and hp0204 coding regions are transcribed in opposite directions towards each other (Fig. 1A). This setting minimizes the possibility of creating polar effects after recombination has occurred, and minimizes risk of inserting DNA fragments into a promoter region. The striking variability in the hp0203–204 intergenic regions across different H. pylori strains, however, suggests that an improved derivative of the system will be required, in which the flanking, highly conserved hp0203 and hp0204 coding regions are utilized. With this proposed modification, the complementation would still be targeted to the hp0203–204 intergenic region. This system is currently under development.
Using the pIR203C04-based chromosomal-targeting complementation plasmid, homologous recombination events occurred not only in strain 26695, but also in 43504 and J99. This occurred despite existence of only ~330 bp of flanking H. pylori DNA in the intergenic plasmid. In contrast, strain SS1 required a two-step transformation. The reason for this is unclear since the SS1 hp0203–204 intergenic region is phylogenetically closer to the 26695 region than strain 43504 is to 26695 (Fig. 6C,D). We speculate that SS1 is more recalcitrant to transformation due to more abundant or distinct restriction–modification systems than found in other strains. Variation in transformation frequencies, when 26695-derived DNA was transformed into heterologous strains, was also probably due to differences in restriction–modification systems, which have been suggested to be important contributors to strain-dependent variations in transformation frequencies [20,21,32]. The variability of the hp0203–204 intergenic region itself could also play a role in transformation frequency differences. To test this idea, the intergenic plasmids derived from different H. pylori strains could be analyzed for transformation frequencies in a single H. pylori strain background.
While the intergenic plasmids were unable to transform strain SS1, transformation using chromosomal DNA from the recombinant 26695-203C04 and rocF26695-MLB004 (via two-step transformation) was achievable in all three heterologous strains studied, suggesting that this complementation system can be adapted to numerous strains. Presumably, the high degree of conservation of the hp0203 and hp0204 coding regions allowed for these recombination events, since all the recombinant strains were shown to be correct by PCR analysis. We recognize that the two-step transformation procedure, while potentially allowing use of the complementation system in broad H. pylori strain backgrounds, has the limitation of generating possible recombination events far away from the hp0203 and hp0204 coding regions, leading to strains that are not truly isogenic.
The complementation system was applied to the arginase mutant of H. pylori. While we cannot completely rule out the possibility for unknown secondary effects in the complemented strains, phenotypes lost in the rocF mutant (arginase activity, arginase protein, arginine consumption) were all restored in the complemented strain. Interestingly, arginase genes from distinct H. pylori strains confer variable arginase activity in the same H. pylori genetic background. For example, the data in Fig. 3B suggest that the rocF genes from strains SS1, 43504, and J63 confer higher arginase activity than rocF from strain 26695, when all rocF genes are in the 26695 rocF mutant strain background. We hypothesize that the sequence variation in the arginase coding region or upstream region plays an important role in the amount of arginase activity observed. This hypothesis is being studied elsewhere (Hovey and McGee, manuscript in preparation). When the 26695 rocF mutant was complemented with a wild-type copy of rocF from 26695 (rocF26695-MLB004), arginase protein and activity were not significantly different from the original wild-type 26695, thereby producing a true complement of the arginase mutant.
Arginase has been shown to be regulated post-translationally by thioredoxin-1 [33], but nothing is yet known about sequence variation of thioredoxin-1 in different H. pylori strains. Recent evidence also suggests that arginase is transcriptionally regulated by the ArsR/S (Hp0165/0166) two-component signal transduction system [34].
Significant genetic polymorphisms exist between different strains of H. pylori [35] (Fig. 6 and Table 4), and horizontal gene transfer is an important contributor to such diversity [36,37]. The high degree of heterogeneity in the hp0203–hp0204 intergenic region suggests there is low selective pressure to maintain conservation of this region. This intergenic region does not appear to play an important role for H. pylori in vivo. Supporting this idea is our finding that insertion of a cat cassette alone or in conjunction with the wild-type arginase gene (rocF) does not alter the ability of H. pylori SS1 to colonize mice (Table 5). Whether the flanking coding regions (hp0203 and hp0204) are important for colonization is unknown, but these regions have a high degree of conservation (Fig. 6A,B), suggesting they have a function that necessitates maintenance of selective pressure on these coding regions. The finding that 16 of 17 strains had distinct hp0203–204 intergenic sequences in our small scale analysis raises the intriguing possibility that this region could be utilized as a novel typing method.
This gene complementation system is a valuable tool that will provide ability to study genes in the native H. pylori background, to investigate the microheterogeneity of H. pylori genes all in the same genetic background, to allow overexpression of genes under the control of a strong promoter, to allow in vivo complementation experiments in the absence of antibiotic selection and to minimize polar and pleiotropic effects. This complementation system should greatly facilitate genetics experiments in H. pylori.
Acknowledgments
This work was supported by Public Health Service grant CA101931 (to D.J.M.) from the National Institutes of Health. We thank Rick Peek (Table 1) for generously providing fresh clinical isolates of H. pylori for this study.
References
- 1.Warren J, Marshall BJ. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1:1273–5. [PubMed] [Google Scholar]
- 2.Cover TL, Blaser MJ. Helicobacter pylori infection, a paradigm for chronic mucosal inflammation: pathogenesis and implications for eradication and prevention. Adv Intern Med. 1996;41:85–117. [PubMed] [Google Scholar]
- 3.Ernst PB, Gold BD. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu Rev Microbiol. 2000;54:615–40. doi: 10.1146/annurev.micro.54.1.615. [DOI] [PubMed] [Google Scholar]
- 4.Annibale B, Negrini R, Caruana P, Lahner E, Grossi C, Bordi C, Delle Fave G. Two-thirds of atrophic body gastritis patients have evidence of Helicobacter pylori infection. Helicobacter. 2001;6:225–33. doi: 10.1046/j.1083-4389.2001.00032.x. [DOI] [PubMed] [Google Scholar]
- 5.Joshi A, Gupta SD, Ahuja V, Sharma MP. Symptom score does not correlate with gastritis grade and Helicobacter pylori infection in non ulcer dyspepsia. Trop Gastroenterol. 2001;22:194–6. [PubMed] [Google Scholar]
- 6.Fischer W, Schwan D, Gerland E, Erlenfeld GE, Odenbreit S, Haas R. A plasmid-based vector system for the cloning and expression of Helicobacter pylori genes encoding outer membrane proteins. Mol General Genet. 1999;262:501–7. doi: 10.1007/s004380051111. [DOI] [PubMed] [Google Scholar]
- 7.Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten SJ, Berg DE, Hoffman PS. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–93. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 8.Heuermann D, Haas R. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol General Genet. 1998;257:519–28. doi: 10.1007/s004380050677. [DOI] [PubMed] [Google Scholar]
- 9.Kleanthous H, Clayton CL, Tabaqchali S. Characterization of a plasmid from Helicobacter pylori encoding a replication protein common to plasmids in Gram-positive bacteria. Mol Microbiol. 1991;5:2377–89. doi: 10.1111/j.1365-2958.1991.tb02084.x. [DOI] [PubMed] [Google Scholar]
- 10.Lee WK, An YS, Kim KH, et al. Construction of a Helicobacter pylori-Escherichia coli shuttle vector for gene transfer in Helicobacter pylori. Appl Environ Microbiol. 1997;63:4866–71. doi: 10.1128/aem.63.12.4866-4871.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton KA, Gilbert JV, Joyce EA, Wanken AE, Thevenot T, Baker P, Plaut A, Wright A. In vivo complementation of ureB restores the ability of Helicobacter pylori to colonize. Infect Immun. 2002;70:771–8. doi: 10.1128/iai.70.2.771-778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsyth MH, Cover TL. Intercellular communication in Helicobacter pylori: luxS is essential for the production of an extracellular signaling molecule. Infect Immun. 2000;68:3193–9. doi: 10.1128/iai.68.6.3193-3199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang J, Tavakoli D, Tschumi A, Aras RA, Blaser MJ. Effect of host species on recG phenotypes in Helicobacter pylori and Escherichia coli. J Bacteriol. 2004;186:7704–13. doi: 10.1128/JB.186.22.7704-7713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loh JT, Forsyth MH, Cover TL. Growth phase regulation of flaA expression in Helicobacter pylori is luxS dependent. Infect Immun. 2004;72:5506–10. doi: 10.1128/IAI.72.9.5506-5510.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smeets LC, Bijlsma JJ, Boomkens SY, Vandenbroucke-Grauls CM, Kusters JG. comH, a novel gene essential for natural transformation of Helicobacter pylori. J Bacteriol. 2000;182:3948–54. doi: 10.1128/jb.182.14.3948-3954.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendz GL, Hazell SL. The urea cycle of Helicobacter pylori. Microbiology. 1996;142:2959–67. doi: 10.1099/13500872-142-10-2959. [DOI] [PubMed] [Google Scholar]
- 17.Sachs G, Weeks DL, Melchers K, Scott DR. The gastric biology of Helicobacter pylori. Annu Rev Physiol. 2003;65:349–69. doi: 10.1146/annurev.physiol.65.092101.142156. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Meininger CJ, Hawker JR, Jr, Haynes TE, Kepka-Lenhart D, Mistry SK, Morris SM, Jr, Wu G. Regulatory role of arginase I and II in nitric oxide, polyamine, and proline syntheses in endothelial cells. Am J Physiol Endocrinol Metab. 2001;280:E75–82. doi: 10.1152/ajpendo.2001.280.1.E75. [DOI] [PubMed] [Google Scholar]
- 19.Zabaleta J, McGee DJ, Zea AH, Hernandez CP, Rodriguez PC, Sierra RA, Correa P, Ochoa AC. Helicobacter pylori arginase inhibits T cell proliferation and reduces the expression of the TCR ζ-chain (CD3ζ) J Immunol. 2004;173:586–93. doi: 10.4049/jimmunol.173.1.586. [DOI] [PubMed] [Google Scholar]
- 20.Alm RA, Ling LS, Moir DT, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–80. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 21.Tomb JF, White O, Kerlavage AR, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–47. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.McGee DJ, Zabaleta J, Viator RJ, Testerman TL, Ochoa AC, Mendz GL. Purification and characterization of Helicobacter pylori arginase, RocF: unique features among the arginase superfamily. Eur J Biochem. 2004;271:1952–62. doi: 10.1111/j.1432-1033.2004.04105.x. [DOI] [PubMed] [Google Scholar]
- 24.Tcherkas YV, Kartsova LA, Krasnova IN. Analysis of amino acids in human serum by isocratic reversed-phase high-performance liquid chromatography with electrochemical detection. J Chromatogr A. 2001;913:303–8. doi: 10.1016/s0021-9673(00)01206-1. [DOI] [PubMed] [Google Scholar]
- 25.Nedenskov P. Nutritional requirements for growth of Helicobacter pylori. Appl Environ Microbiol. 1994;60:3450–3. doi: 10.1128/aem.60.9.3450-3453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds DJ, Penn CW. Characteristics of Helicobacter pylori growth in a defined medium and determination of its amino acid requirements. Microbiology. 1994;140:2649–56. doi: 10.1099/00221287-140-10-2649. [DOI] [PubMed] [Google Scholar]
- 27.Testerman TL, McGee DJ, Mobley HL. Helicobacter pylori growth and urease detection in the chemically defined medium Ham’s F-12 nutrient mixture. J Clin Microbiol. 2001;39:3842–50. doi: 10.1128/JCM.39.11.3842-3850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendz GL, Hazell SL. Amino acid utilization by Helicobacter pylori. Int J Biochem Cell Biol. 1995;27:1085–93. doi: 10.1016/1357-2725(95)00069-2. [DOI] [PubMed] [Google Scholar]
- 29.McGee DJ, Radcliff FJ, Mendz GL, Ferrero RL, Mobley HL. Helicobacter pylori rocF is required for arginase activity and acid protection in vitro but is not essential for colonization of mice or for urease activity. J Bacteriol. 1999;181:7314–22. doi: 10.1128/jb.181.23.7314-7322.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eaton KA, Brooks CL, Morgan DR, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59:2470–5. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAtee CP, Hoffman PS, Berg DE. Identification of differentially regulated proteins in metronidozole resistant Helicobacter pylori by proteome techniques. Proteomics. 2001;1:516–21. doi: 10.1002/1615-9861(200104)1:4<516::AID-PROT516>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 32.Ando T, Xu Q, Torres M, Kusugami K, Israel DA, Blaser MJ. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol Microbiol. 2000;37:1052–65. doi: 10.1046/j.1365-2958.2000.02049.x. [DOI] [PubMed] [Google Scholar]
- 33.McGee DJ, Kumar S, Viator RJ, Bolland JR, Ruiz J, Spadafora D, Testerman TL, Kelly DJ, Pannell LK, Windle HJ. Helicobacter pylori thioredoxin is an arginase chaperone and guardian against oxidative and nitrosative stresses. J Biol Chem. 2006;281:3290–6. doi: 10.1074/jbc.M506139200. [DOI] [PubMed] [Google Scholar]
- 34.Pflock M, Finsterer N, Joseph B, Mollenkopf H, Meyer TF, Beier D. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J Bacteriol. 2006;188:3449–62. doi: 10.1128/JB.188.10.3449-3462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Israel DA, Salama N, Krishna U, Rieger UM, Atherton JC, Falkow S, Peek RM., Jr Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc Natl Acad Sci USA. 2001;98:14625–30. doi: 10.1073/pnas.251551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence JG, Roth JR. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–60. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619–24. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee A, O’Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, Dixon MF. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–97. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]