Abstract
Cancer-testis (CT) antigens/genes show restricted expression in normal tissues but widespread expression in many tumour types. This, coupled with their ability to induce cytotoxic T-lymphocyte responses, makes them attractive vaccine candidates. Following our identification of PASD1, we have used RT-PCR to analyse the mRNA expression profile of a large panel of CT genes in cell lines derived from haematological malignancies, and have studied Sp17 protein expression in the same cell lines and diffuse large B-cell lymphoma (DLBCL) biopsies. Our extensive analysis revealed multiple CT transcripts exhibiting widespread expression across cell lines derived from 21 B- and 4 T-cell malignancies. The broadest mRNA expression profiles were observed for the following eight CT genes: Sp17 (25/25), PRAME (25/25), CSAGE (24/25), PASD1 (22/25), CAGE/DDX53 (19/25), CTAGE1 (19/25), HAGE/DDX43 (16/25) and PLU-1/JARID1B (15/25). Cell lines derived from more aggressive lymphoma subtypes generally expressed CT transcripts at higher frequency. Sp17 protein was detected in a number of cell lines and in six of eleven (54.5%) DLBCL biopsies. Analysis of Sp17 protein expression, by immunohistochemistry and Western blotting, broadens the scope of this CT antigen as a potentially relevant clinical target in haematological malignancies. Further studies of protein expression are now needed to validate these antigens as vaccine candidates.
Keywords: human, haematological cancer, CT antigens, RT-PCR, immunohistochemistry, Western blotting
Introduction
Despite modern chemotherapeutic agents and monoclonal antibody therapy, the prognosis for many patients with haematological malignancies, particularly those with aggressive diseases such as diffuse large B-cell lymphoma (DLBCL), mantle cell lymphoma (MCL) and multiple myeloma (MM), remains poor (1-4). There is, however, increasing evidence that the immune system may play an important role in lymphoma patients, particularly in those with indolent lymphomas such as follicular lymphoma (FL) where spontaneous remission can occur (5), and gene expression profiling studies have identified microenvironmental signatures rather than tumour profiles as predicting patient survival (6). Furthermore, animal models and clinical trials have demonstrated that vaccines containing idiotypic proteins can induce a specific immune response in both lymphoma and myeloma patients (7-9). However, the production of these patient-specific vaccines may require considerable effort and thus a relatively high cost.
The development of immunotherapeutic options for the treatment of cancers requires the identification and characterisation of antigenic tumour proteins. Optimal immunotherapeutic target proteins should be restricted to malignant cells and be expressed in a wide range of tumour types where current therapy is ineffective. Ideal antigens should be crucial for tumour cell survival to prevent selection favouring loss of antigen expression and subsequent immune evasion. Cancer-testis (CT) antigens, encoded by CT genes, which show restricted normal tissue distribution but a widespread neoplastic expression, thus represent attractive targets for the development of generic cancer vaccines (10-12).
Normal tissue mRNA expression patterns are used to categorise CT genes. Scanlan et al. (11) classified them as: (i) testis-restricted, (ii) tissue-restricted to two or fewer non-gametogenic tissues, (iii) differentially expressed in 3-6/13 non-gametogenic tissues and (iv) ubiquitously expressed, whereas a more recent study (13) has described (i) testis-restricted, (ii) testis/brain-restricted and (iii) testis-selective groups. Testis-restricted CT antigens whose normal tissue protein expression is as restricted as the mRNA obviously represent the ultimate candidates for therapy. However, as published data suggest that many CT antigens have heterogeneous expression in tumours, it is likely that multiple antigens will be required in vaccine preparations (14). This should also minimize the risk of escape variants lacking individual CT gene expression in response to targeted therapy.
We previously utilised the serological recombinant cDNA expression cloning (SEREX) technique to identify lymphoma-associated antigens that might represent new diagnostic and/or prognostic markers and targets for immunotherapy. We identified the PAS domain containing 1 (PASD1) molecule, which has a CT gene expression profile, is expressed in DLBCL-derived cell lines, and induces humoral immune responses in DLBCL and acute myeloid leukaemia patients (15-17) and cellular immune responses in DLBCL patients (18). Our monoclonal antibodies to the PASD1 protein have confirmed its restricted normal tissue distribution and expression in a range of lymphomas and MMs (19, 20). These studies confirm that PASD1 is heterogeneously expressed both within and between individual cases and that it represents a potential candidate to be utilised in a multi-antigen vaccine.
While some tumours such as melanoma and non-small cell lung cancer are high CT gene expressors [expressing >50% of CT antigens/genes examined at a frequency >20%; (11)], previous reports indicated that CT genes, although expressed in T-cell lymphomas, are rarely expressed in B-cell lymphomas (11, 21, 22). However, in contrast to these findings, our analyses of PASD1 protein identified a relatively frequent expression pattern of this protein in B-cell lymphomas (19). As a result, we decided to investigate the mRNA expression in cell lines derived from haematological malignancies of additional CT genes/antigens, including those that were less widely studied. Our aims were to identify other CT genes that might also exhibit frequent expression in haematological malignancies, to compare their expression to that of PASD1, and to identify potential candidates for combination within lymphoma vaccines. We have assessed the expression of 32 CT gene transcripts in 25 neoplastic cell lines (21 B-cell derived and 4 T-cell derived) by RT-PCR. This extensive panel incorporated some of the more traditionally investigated CT transcripts e.g. NY-ESO-1, SSX2 and MAGE-A1 as well as a number of the lesser-studied CT genes that do not map to the X chromosome e.g. LIPI and PLU-1/JARID1B. We report our identification of nine frequently expressed CT transcripts that are derived from eight CT genes: PRAME and Sp17 (25/25), CSAGE (24/25), PASD1_v2 (22/25), PASD1_v1 (21/25), CAGE/DDX53 (19/25), CTAGE1 (19/25), HAGE/DDX43 (16/25) and PLU-1/JARID1B (15/25). Furthermore, we demonstrate that the protein expression profile of Sp17 suggests that this antigen could also be a vaccine candidate in B-cell non-Hodgkin lymphomas (NHLs).
Results
Expression of CT genes in DLBCL cell lines
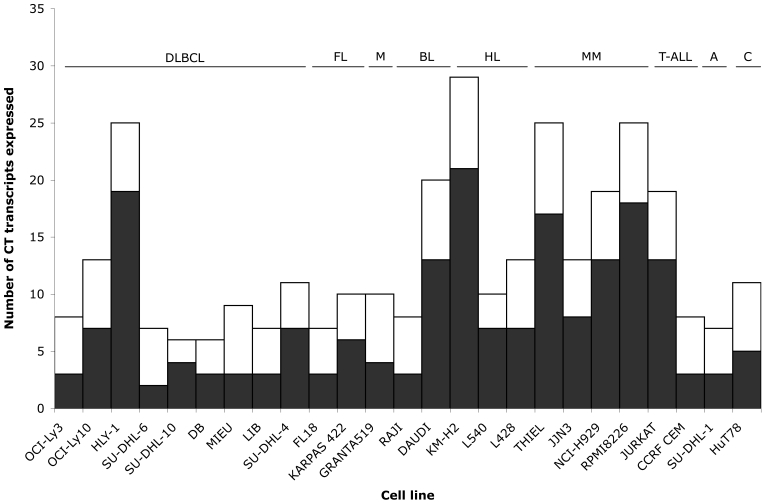
Messenger RNA expression of 32 CT transcripts was assessed by RT-PCR in 9 DLBCL-derived cell lines, three of which exhibit an activated B-cell (ABC)-like gene expression profile associated with a poor prognosis (23, 24). Representative results are illustrated in Figure 1 whilst all results are tabulated in Table 1. All 9 DLBCL cell lines expressed six or more CT transcripts. The most frequently expressed were CSAGE, Sp17 and PRAME (9/9, 100%), followed by PASD1_v1 and CAGE/DDX53 (7/9, 78%), PASD1_v2 and CTAGE1 (6/9, 67%), and PLU-1/JARID1B (5/9, 56%). Overall, we observed that CT genes were more frequently expressed in ABC-DLBCL cell lines (OCI-Ly3, OCI-Ly10, HLY-1) than in non-ABC/germinal centre (GC)-like DLBCL cell lines (SU-DHL-6, SU-DHL-10, DB, MIEU, LIB, SU-DHL-4) (Figure 2). The ABC lines also tended to express more CT genes that localize to chromosome X (CT-X) e.g. ratio of CT-X to non CT-X genes was 7:6 for OCI-Ly10 and 19:6 HLY-1 (both ABC) whereas in the remaining DLBCL cell lines there was no obvious difference in expression levels between CT-X and non CT-X genes e.g. DB 3:3, LIB 3:4 (Figure 3). However, the number of CT genes and cell lines are too small to determine whether these observations have any statistical significance. Based on the definition that medium CT expressors are those that express 30-50% of CT genes at a frequency >20% (11), DLBCL cell lines are, on average, medium CT gene expressors.
Figure 1.
RT-PCR data showing high frequency of expression of non CT-X transcripts CTAGE1, HAGE/DDX43, CAGE/DDX53, Sp17 and PLU-1/JARID1B CT transcripts as opposed to low frequency of expression of CT-X transcripts GAGE1/2 and SSX2 in cell lines derived from haematological malignancies. Although the results were not assessed quantitatively, differential expression of transcripts between cell lines can clearly be seen.
Table 1.
Expression of CT transcripts in cell lines derived from DLBCL as assessed by RT-PCR.
Figure 2.
Graphical representation of the total number of CT transcripts expressed in individual cell lines.
Figure 3.
Graphical representation of CT-X (black) and non CT-X (white) transcripts expressed in individual cell lines.
Expression of CT genes in other B- and T-cell malignancies
The expression of CT gene transcripts was also assessed by RT-PCR in 12 cell lines derived from B-cell malignancies other than DLBCL, including those derived from FL (n = 2), MCL (n = 1), Burkitt lymphoma (BL) (n = 2), Hodgkin lymphoma (HL) (n = 3) and MM (n = 4). Representative results are shown in Figure 1 whilst all results are tabulated in Table 2. FL18, the FL-derived cell line, expressed the lowest number of transcripts (7/32) whilst KM-H2 (HL) expressed the most (29/32) (Figure 2), although on average, it was the MM lines that expressed the highest number of CT transcripts (21/32). The most frequently expressed antigens in B-cell malignancies were Sp17, PASD1_v2, CSAGE and PRAME (each 12/12, 100%) followed by PASD1_v1 (11/12, 91%), CAGE/DDX53 and CTAGE1 (both 10/12, 83%) and HAGE/DDX43 (9/12, 75%). We observed that the HL and MM lines not only expressed more CT-X transcripts than did the other lines, but these were more frequently expressed than non CT-X transcripts (average CT-X:non CT-X ratios were 11:5 for HL and 14:6 for MM) (Figure 3). These results indicate that cell lines derived from non-DLBCL B-cell malignancies are, on average, high expressors of CT genes, although this is due predominantly to the high levels of transcripts found in the HL and MM lines. The most frequently expressed CT genes in B-cell derived malignancies were also most frequently expressed in T-cell lines, e.g. Sp17, PRAME, PASD1_v2 (4/4, 100%); HAGE/DDX43, PLU-1/JARID1B, CSAGE, PASD1_v1, CTAGE1 (all 3/4, 75%); and CAGE/DDX53 (2/4, 50%). SU-DHL-1, the anaplastic large cell lymphoma (ALCL) derived cell line, expressed the lowest number of transcripts (7/32). Although the number of T-cell lines tested is small, these results suggest they are at least medium expressors of CT genes.
Table 2.
Expression of CT transcripts in cell lines derived from non-DLBCL B-cell and T-cell malignancies as assessed by RT-PCR.
Expression of CT transcripts in normal tissues
The normal tissue mRNA expression of PASD1 has previously been reported and shown to be testis-restricted (16). The mRNA expression of the remaining seven CT transcripts with the broadest cell line expression was assessed by RT-PCR using a panel of sixteen normal human tissue cDNAs. PRAME and CAGE/DDX53 were testis-restricted, CTAGE1 and Sp17 were detected in 6/13 and 10/13 non-gametogenic tissues respectively, whilst CSAGE, HAGE/DDX43 and PLU-1/JARID1B were found to be ubiquitously expressed (Supplementary Figure 1).
Immunohistochemistry and Western blotting
Protein expression is more relevant than mRNA expression in terms of selecting targets for lymphoma immunotherapy, yet there are still relatively few monoclonal antibodies available to study CT protein expression. We have already reported PASD1 protein expression in B- and T-cell NHL (19), and Sp17 is one of the very few antigens that we have shown here to be widely expressed at the mRNA level in our cell lines to which there is a fully-validated, well-characterised monoclonal antibody available (25). Therefore, we selected this antigen for further study of its protein expression.
Sp17 protein expression was initially investigated by immunohistochemistry (IHC) on tissue microarrays (TMAs) of formalin-fixed paraffin-embedded cell pellets derived from those haematological malignancies used in the RT-PCR studies, with the exception of Karpas422, L540 and Hut78. Tissue sections from normal testis and tonsil were included as positive and negative controls respectively. The anti-Sp17 antibody detected protein in the cytoplasm of the spermatogonia (weak staining), and in the cytoplasm and nuclei of the primary spermatocytes and the spermatozoa (strong staining) of normal testis; no Sp17 protein expression was seen in normal tonsil tissue (Figure 4A). IHC on biopsies from DLBCL patients identified scattered positive cells, exhibiting predominantly nuclear Sp17 expression, in six of eleven cases (54.5%) (Figure 4A). Sp17 mRNA was abundant and showed widespread expression in the cell lines, yet surprisingly Sp17 protein was detected in only 4/22 (18%) cell lines tested by IHC (L428, Jurkat, SU-DHL-10 and MIEU; data not shown). To investigate whether Sp17 protein was present at low levels that were undetectable by IHC techniques, whole cell lysates prepared from 23/25 cell lines used in the RT-PCR studies, together with a selection of commercially-available quantified normal human tissue lysates, were Western blotted with the anti-Sp17 antibody. A protein of approximately 25 kDa, consistent with the 24.5 kDa protein previously reported (25), was detected with the anti-Sp17 antibody in normal testis and all cell lines tested (Figure 4B). Extremely low levels of protein were also observed in normal tonsil and ovary tissues following a prolonged exposure to film (Supplementary Figure 2).
Figure 4.
Sp17 protein expression. (A) Anti-Sp17 antibody labels the cytoplasm of spermatogonia (weakly), and both cytoplasm and nuclei of spermatocytes and spermatozoa of normal testis. The intensity of the labelling increases as the cells increase in maturity. No staining was observed in normal tonsil sections. Sp17 expression was observed in scattered cells in DLBCL biopsies, DLBCL002, DLBCL005 and DLBCL003. A negative case is also illustrated (DLBCL013). (B) Western blotting showed expression of Sp17 protein (upper panel) in neoplastic B- and T-cell derived cell lines and testis. Heterogeneity of expression can also be observed between cell lines since the β-actin loading control (lower panel) suggests equal amounts of proteins were loaded. No Sp17 protein was detected in normal tonsil lysate. Samples: M, marker; To, normal tonsil lysate; Te, normal testis lysate; -, empty lane; DLBCL, lysates from DLBCL-derived cell lines; FL, lysates from follicular lymphoma-derived cell lines; BL, lysates from Burkitt's lymphoma-derived cell lines; HL, lysates from Hodgkin lymphoma-derived cell lines; MM, lysates from multiple myeloma-derived cell lines; T-ALL, lysates from T-acute lymphoblastic leukaemia/lymphoma-derived cell lines; A, lysate from anaplastic large cell lymphoma-derived cell line.
Discussion
An early study investigating the potential of CT genes as vaccines for NHLs studied the expression of eight genes (MAGEA3, MAGEA4, MAGEC1, SSX2, SSX1, SSX4, SCP1 and HOM-TES-85/LUZP4) in a variety of NHL subtypes (22). While SCP1, and to some degree SSX1 and CT-7, were proposed as candidates for lymphoma vaccine development, the authors reported that it was only the majority of T-cell lymphomas and a small proportion of DLBCLs that expressed at least one CT gene. They proposed that the identification of additional antigens exhibiting frequent expression in lymphoma was desirable.
A follow-up study using the SEREX technique to screen a testis library with serum from 25 lymphoma patients failed to yield any additional lymphoma-associated CT antigens, leading the authors to conclude that novel strategies would be necessary to identify vaccine targets for a broad spectrum of NHL patients (21). In contrast, other studies have identified good potential CT targets for cutaneous T-cell lymphomas (26) and for MMs (27-31). To date however, there have been very few CT genes reported to have a broad spectrum of expression in B-cell NHLs.
Monoclonal antibodies to PASD1, a novel CT antigen that elicits humoral responses in DLBCL (16, 17) and AML (15) patients and cellular responses in DLBCL patients (18), showed widespread expression of the PASD1 protein in a number of haematological malignancies (19, 20), which suggested that other CT antigens/genes were worthy of investigation. This report clearly shows that a more extensive study of CT genes, including the less-commonly studied non CT-X genes, may provide a new insight into prospective targets for the immunotherapeutic treatment of NHLs. While many CT genes are indeed, as previously reported, infrequently expressed in B-cell-derived NHLs, others exhibit the broad expression pattern that is desirable in potential vaccine candidates. Nine CT transcripts (from eight genes) (HAGE/DDX43, PLU-1/JARID1B, CTAGE1, CAGE/DDX53, CSAGE, Sp17, PRAME and PASD1) are expressed in over 55% of the B-cell lines (and over 60% of all lines tested). Of particular interest is the expression of all nine of these frequently expressed CT transcripts in the MCL cell line, Granta519, as this malignancy has such a poor prognosis. Given that HAGE/DDX43, PLU-1/JARID1B, CTAGE1, CAGE/DDX53, CSAGE, Sp17 and PRAME have very similar mRNA expression profiles to PASD1, and that PASD1 protein is expressed in a number of haematological malignancies (19, 20), these other CT genes may also be valid candidates for therapy of B- and T-cell derived NHLs.
The high frequency of CT gene expression in lymphoma cell lines could potentially reflect unrestricted normal tissue expression or expression in lymphoid tissues. The normal tissue expression patterns of six of the eight genes discussed here have been analysed by 35-cycle RT-PCR in 16 normal tissues including leukocytes, spleen and thymus (11). Of these, CTAGE1 and CSAGE were testis-restricted; HAGE/DDX43 was tissue-restricted (expressed in testis and pancreas); CAGE/DDX53 was differentially expressed (testis, leukocytes, brain and pancreas); while PLU-1/JARID1B and Sp17 were ubiquitously expressed. Our own RT-PCR data on seven transcripts, using the same human tissue cDNA panel as Scanlan et al. (11), showed CAGE/DDX53 and PRAME to be testis-restricted; CTAGE1 to be differentially-expressed (6 non-gametogenic tissues), while Sp17, PLU-1/JARID1B, CSAGE and HAGE/DDX43 were ubiquitously expressed. The normal tissue mRNA expression of six of the eight genes has also been reported by Hofmann et al. (13): CAGE/DDX53 was testis-restricted; CTAGE1 and PASD1 were testis/brain-restricted, while HAGE/DDX43, PLU-1/JARID1B and Sp17 were testis-selective, although HAGE/DDX43 was not as widely expressed as PLU-1/JARID1B and Sp17. We have also previously reported that PASD1 is not expressed in normal lymphocytes (16, 19). The PRAME results in this study are supported by BioGPS microarray data, and a recent paper reported the expression of PRAME in only 1/17 non-gametogenic tissues by real-time PCR (32). In addition, a study using the same PCR primers as in our study reported no PRAME expression in normal bone marrow (n = 11) or PBMCs (n = 17) (33). Therefore, using restricted mRNA expression in normal tissues as the basis for selecting therapeutic candidates from the eight genes highlighted here, the Scanlan et al. study (11) would predict CSAGE, CTAGE1 and possibly HAGE/DDX43; the Hofmann et al. study (13) would predict CAGE/DDX53, CTAGE1 and PASD1, whereas our data identify CAGE/DDX53 and PRAME as prime targets, as well as PASD1 (16). Data from all three studies of mRNA expression indicate that PLU-1/JARID1B and Sp17 may not be ideal therapeutic candidates. Comparing just these three sets of data serves to highlight the conflicting results that exist in published databases and the literature. Indeed, on the basis of the normal tissue data available in Oncomine (34), PLU-1/JARID1B would perhaps appear to be a good candidate for therapy. A comparison summary of the normal tissue expression of these eight genes is given in Supplementary Table 1. These conflicting results may arise because of differences in technique sensitivity, sample preparation, or variation between equipment used, to list but a few reasons.
However, the relevance of mRNA expression in prioritising the best targets for immunotherapy is unclear as we have shown that whilst the Sp17 transcript is ubiquitously expressed in normal tissues, the same is not true for the protein, although the same source material was not used for both assays. In addition, HAGE/DDX43 protein has been reported to be absent by IHC in 9/12 normal tissues where HAGE/DDX43 mRNA had been detected (35). Whilst some of the transcripts here may not fulfil the definition of a CT gene, they may be CT proteins or antigens. Therefore, it is vital to study the protein distribution of CT genes with well-validated reagents in order to determine relevant therapeutic targets.
Interestingly, of the nine frequently-expressed transcripts, only CSAGE and PASD1 localize to chromosome X. Chromosome X, and Xq28 in particular, appears to be a hotspot for CT antigens/genes, especially those belonging to gene families, e.g. MAGEA, MAGEB, GAGE, and SSX. Indeed, localization to chromosome X was almost considered a defining factor in the early studies of CT genes (36, 37), and this, together with their expression in carcinomas, may be a reason why these CT-X transcripts have historically been more intensively studied.
At least two subtypes of DLBCLs have been identified by expression profiling: good prognosis GC-derived and poor-prognosis non-GC (NGC)-derived that includes the ABC-subtype (23, 38). The ABC DLBCL lines in this study express more CT transcripts than the other DLBCL lines (on average 15 versus 7), whilst cell lines derived from FL, an indolent lymphoma, expresses only seven transcripts. This is consistent with previous reports of increased CT antigen expression being associated with more aggressive diseases, or advanced stages of disease (39, 40). In addition, there are reports of higher levels of CT-X expression correlating to poorer prognosis (31). In addition to our findings of widespread PRAME expression across DLBCL-derived cell lines, a recent microarray study has shown that DLBCL cases with resistance to anthracycline-containing chemotherapy regimens showed high expression of PRAME, and that PRAME-positive patients had significantly worse progression-free survival than PRAME-negative patients (41). However, the small number of cell lines used here does not enable any statistically significant conclusions to be drawn regarding any correlation between CT expression and DLBCL subtype.
Although there are a great many studies of CT gene expression patterns, relatively few CT antigens/genes have been studied at the protein level using fully-validated reagents. We have already characterized PASD1 protein expression in haematological malignancies, including DLBCL and MM (19, 20) and now have also shown it can induce a cellular immune response (18). Since the anti-Sp17 antibody has been fully-validated (25) and the mRNA was abundant and widespread in our study, we selected this antigen for further characterisation at the level of protein expression. Sp17 was first demonstrated to be a CT antigen in MM (42) and has since been shown to induce a cellular immune response and has been validated as a target for immunotherapy of this disease (43) and ovarian carcinoma (44). Although Sp17 protein expression was detected in only 4/22 cell lines by IHC techniques, Western blotting showed expression in all 23 cell lines derived from haematological malignancies that were tested, including those that were originally negative by IHC. The Jurkat cell line expresses relatively high levels of Sp17 protein, which may be why this cell line was positive by both Western blotting and IHC. However, the level of Sp17 expression in SU-DHL-10 was comparable to other IHC-negative cell lines by Western blotting. Discrepancies between IHC and Western blotting may also be caused by a number of technical issues such as fixation of the tissues or the effectiveness of antigen retrieval. As the level of expression in the cell line panel was largely comparable to that in the MM cell line Thiel, these data suggest that Sp17 represents a promising vaccine target in haematological malignancies other than MM. Sp17 protein was also detected in a number of DLBCL biopsies and, although the staining is heterogeneous, something that is characteristic of CT antigens, high levels of expression are not necessarily required for an effective immune response (35). Importantly we also show that the expression of Sp17 mRNA in normal tissues is not indicative of protein expression, which suggests that the expression of Sp17 in lymphomas may be regulated post-transcriptionally, perhaps by mechanisms enabling Sp17 translation (e.g. microRNAs) or increasing protein stability.
CT antigens are considered to be promising candidates for cancer immunotherapy with minimal autoimmune side effects because of their widespread expression in tumours but restricted normal tissue distribution. Importantly, the blood-testis barrier means that testis is considered to represent an immune-privileged site. Some of the CT antigens discovered in recent years are now being included in clinical trials [e.g. NY-ESO-1 (45-47), MAGEA3 (48, 49) and Sp17 (50)] with mixed success. Although CT antigens have been studied extensively in solid tumours, more recently, CT antigens have also been shown to induce humoral (15, 16, 42, 51-54) and cellular (18, 43, 55-60) immune responses in haematological malignancies, suggesting that CT vaccines would be a viable option for these diseases.
Polyvalent vaccines serve two purposes: to increase the number of patients who will benefit from a vaccination strategy, and to reduce the risk of tumour escape from immune surveillance. The heterogeneity of CT antigen expression that has been described both within and between malignancies (61, 62) also suggests that a combination of antigens in a vaccine would be beneficial. From our studies, a vaccine combining two or more of the eight frequently expressed CT genes has the potential to target a broad range of haematological malignancies.
In conclusion, we have identified a panel of eight CT genes (nine transcripts) showing frequent expression in cell lines derived from B- and T-cell malignancies. Of these, six are already known to be immunogenic in patients (PASD1, Sp17, CAGE/DDX53, HAGE/DDX43, CTAGE1 and PRAME). We have also demonstrated the expression of Sp17 at the protein level in cell lines derived from T- and B-cell malignancies, which expands the range of malignancies that may be therapeutic targets for this antigen. Sp17 protein expression was observed in 54.5% of the DLBCL biopsies examined by immunohistochemistry, indicating that Sp17 is indeed expressed at the protein level in patients, a prerequisite for a therapeutic vaccine. Although some CT genes are reported to have widespread normal tissue mRNA expression, this does not preclude a restricted protein expression. Until protein expression profiles are determined using fully-validated reagents, a conclusion on a CT gene's worthiness as a therapeutic target cannot be drawn. Therefore, further research is now required to investigate the protein expression of the remaining six CT genes highlighted by this study, to investigate their ability to induce cellular immune responses, and to determine which antigens might contribute to the disease process before deciding on the optimum combination of CT antigens for B- and T-cell NHL vaccine development.
Abbreviations
- ABC
activated B-cell
- CT
cancer-testis
- CT-X
CT genes localising to the X chromosome
- DLBCL
diffuse large B-cell lymphoma
- HL
Hodgkin lymphoma
- IHC
immunohistochemistry
- MM
multiple myeloma
- NHL
non-Hodgkin lymphoma
Acknowledgements
This work was funded by Leukaemia & Lymphoma Research.
References
- 1.Ng AK. Diffuse large B-cell lymphoma. Semin Radiat Oncol. 2007;17:169–175. doi: 10.1016/j.semradonc.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Weigert O, Unterhalt M, Hiddemann W, Dreyling M. Current management of mantle cell lymphoma. Drugs. 2007;67:1689–1702. doi: 10.2165/00003495-200767120-00004. [DOI] [PubMed] [Google Scholar]
- 3.Richardson PG, Mitsiades C, Schlossman R, Munshi N, Anderson K. New drugs for myeloma. Oncologist. 2007;12:664–689. doi: 10.1634/theoncologist.12-6-664. [DOI] [PubMed] [Google Scholar]
- 4.Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD. The World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues. Report of the Clinical Advisory Committee meeting, Airlie House, Virginia, November, 1997. Ann Oncol. 1999;10:1419–1432. doi: 10.1023/a:1008375931236. [DOI] [PubMed] [Google Scholar]
- 5.Horning SJ, Rosenberg SA. The natural history of initially untreated low-grade non-Hodgkin's lymphomas. N Engl J Med. 1984;311:1471–1475. doi: 10.1056/NEJM198412063112303. [DOI] [PubMed] [Google Scholar]
- 6.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, Fisher RI, Braziel RM, Rimsza LM, Grogan TM, Miller TP, LeBlanc M, Greiner TC, Weisenburger DD, Lynch JC, Vose J, Armitage JO, Smeland EB, Kvaloy S, Holte H, Delabie J, Connors JM, Lansdorp PM, Ouyang Q, Lister TA, Davies AJ, Norton AJ, Muller-Hermelink HK, Ott G, Campo E, Montserrat E, Wilson WH, Jaffe ES, Simon R, Yang L, Powell J, Zhao H, Goldschmidt N, Chiorazzi M, Staudt LM. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 7.Veelken H, Osterroth F. Vaccination strategies in the treatment of lymphomas. Oncology. 2002;62:187–200. doi: 10.1159/000059565. [DOI] [PubMed] [Google Scholar]
- 8.Ruffini PA, Neelapu SS, Kwak LW, Biragyn A. Idiotypic vaccination for B-cell malignancies as a model for therapeutic cancer vaccines: from prototype protein to second generation vaccines. Haematologica. 2002;87:989–1001. [PubMed] [Google Scholar]
- 9.Bogen B, Ruffini PA, Corthay A, Fredriksen AB, Frøyland M, Lundin K, Røsjø E, Thompson K, Massaia M. Idiotype-specific immunotherapy in multiple myeloma: suggestions for future directions of research. Haematologica. 2006;91:941–948. [PubMed] [Google Scholar]
- 10.Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 11.Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 2004;4:1. http://www.cancerimmunity.org/v4p1/031220.htm [PubMed] [Google Scholar]
- 12.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann O, Caballero OL, Stevenson BJ, Chen YT, Cohen T, Chua R, Maher CA, Panji S, Schaefer U, Kruger A, Lehvaslaiho M, Carninci P, Hayashizaki Y, Jongeneel CV, Simpson AJ, Old LJ, Hide W. Genome-wide analysis of cancer/testis gene expression. Proc Natl Acad Sci U S A. 2008;105:20422–20427. doi: 10.1073/pnas.0810777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juretic A, Spagnoli GC, Schultz-Thater E, Sarcevic B. Cancer/testis tumour-associated antigens: immunohistochemical detection with monoclonal antibodies. Lancet Oncol. 2003;4:104–109. doi: 10.1016/s1470-2045(03)00982-3. [DOI] [PubMed] [Google Scholar]
- 15.Guinn BA, Bland EA, Lodi U, Liggins AP, Tobal K, Petters S, Wells JW, Banham AH, Mufti GJ. Humoral detection of leukaemia-associated antigens in presentation acute myeloid leukaemia. Biochem Biophys Res Commun. 2005;335:1293–1304. doi: 10.1016/j.bbrc.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 16.Liggins AP, Brown PJ, Asker K, Pulford K, Banham AH. A novel diffuse large B-cell lymphoma-associated cancer testis antigen encoding a PAS domain protein. Br J Cancer. 2004;91:141–149. doi: 10.1038/sj.bjc.6601875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liggins AP, Guinn BA, Hatton CS, Pulford K, Banham AH. Serologic detection of diffuse large B-cell lymphoma-associated antigens. Int J Cancer. 2004;110:563–569. doi: 10.1002/ijc.20170. [DOI] [PubMed] [Google Scholar]
- 18.Ait-Tahar K, Liggins AP, Collins GP, Campbell A, Barnardo M, Lawrie C, Moir D, Hatton C, Banham AH, Pulford K. Cytolytic T-cell response to the PASD1 cancer testis antigen in patients with diffuse large B-cell lymphoma. Br J Haematol. 2009;146:396–407. doi: 10.1111/j.1365-2141.2009.07761.x. [DOI] [PubMed] [Google Scholar]
- 19.Cooper CD, Liggins AP, Ait-Tahar K, Roncador G, Banham AH, Pulford K. PASD1, a DLBCL-associated cancer testis antigen and candidate for lymphoma immunotherapy. Leukemia. 2006;20:2172–2174. doi: 10.1038/sj.leu.2404424. [DOI] [PubMed] [Google Scholar]
- 20.Sahota SS, Goonewardena CM, Cooper CD, Liggins AP, Ait-Tahar K, Zojer N, Stevenson FK, Banham AH, Pulford K. PASD1 is a potential multiple myeloma-associated antigen. Blood. 2006;108:3953–3955. doi: 10.1182/blood-2006-04-014621. [DOI] [PubMed] [Google Scholar]
- 21.Huang S, Preuss KD, Xie X, Regitz E, Pfreundschuh M. Analysis of the antibody repertoire of lymphoma patients. Cancer Immunol Immunother. 2002;51:655–662. doi: 10.1007/s00262-002-0320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie X, Wacker HH, Huang S, Regitz E, Preuss KD, Romeike B, Parwaresch R, Tiemann M, Pfreundschuh M. Differential expression of cancer testis genes in histological subtypes of non-Hodgkin's lymphomas. Clin Cancer Res. 2003;9:167–173. [PubMed] [Google Scholar]
- 23.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 24.Brown PJ, Ashe SL, Leich E, Burek C, Barrans S, Fenton JA, Jack AS, Pulford K, Rosenwald A, Banham AH. Potentially oncogenic B-cell activation-induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL. Blood. 2008;111:2816–2824. doi: 10.1182/blood-2007-09-115113. [DOI] [PubMed] [Google Scholar]
- 25.Straughn JM Jr, Shaw DR, Guerrero A, Bhoola SM, Racelis A, Wang Z, Chiriva-Internati M, Grizzle WE, Alvarez RD, Lim SH, Strong TV. Expression of sperm protein 17 (Sp17) in ovarian cancer. Int J Cancer. 2004;108:805–811. doi: 10.1002/ijc.11617. [DOI] [PubMed] [Google Scholar]
- 26.Häffner AC, Tassis A, Zepter K, Storz M, Tureci O, Burg G, Nestle FO. Expression of cancer/testis antigens in cutaneous T cell lymphomas. Int J Cancer. 2002;97:668–670. doi: 10.1002/ijc.1643. [DOI] [PubMed] [Google Scholar]
- 27.Dhodapkar MV, Osman K, Teruya-Feldstein J, Filippa D, Hedvat CV, Iversen K, Kolb D, Geller MD, Hassoun H, Kewalramani T, Comenzo RL, Coplan K, Chen YT, Jungbluth AA. Expression of cancer/testis (CT) antigens MAGE-A1, MAGE-A3, MAGE-A4, CT-7, and NY-ESO-1 in malignant gammopathies is heterogeneous and correlates with site, stage and risk status of disease. Cancer Immun. 2003;3:9. http://www.cancerimmunity.org/v3p9/030709.htm [PubMed] [Google Scholar]
- 28.Jungbluth AA, Ely S, DiLiberto M, Niesvizky R, Williamson B, Frosina D, Chen YT, Bhardwaj N, Chen-Kiang S, Old LJ, Cho HJ. The cancer-testis antigens CT7 (MAGE-C1) and MAGE-A3/6 are commonly expressed in multiple myeloma and correlate with plasma-cell proliferation. Blood. 2005;106:167–174. doi: 10.1182/blood-2004-12-4931. [DOI] [PubMed] [Google Scholar]
- 29.Taylor BJ, Reiman T, Pittman JA, Keats JJ, de Bruijn DR, Mant MJ, Belch AR, Pilarski LM. SSX cancer testis antigens are expressed in most multiple myeloma patients: co-expression of SSX1, 2, 4, and 5 correlates with adverse prognosis and high frequencies of SSX-positive PCs. J Immunother. 2005;28:564–575. doi: 10.1097/01.cji.0000175685.36239.e5. [DOI] [PubMed] [Google Scholar]
- 30.Atanackovic D, Arfsten J, Cao Y, Gnjatic S, Schnieders F, Bartels K, Schilling G, Faltz C, Wolschke C, Dierlamm J, Ritter G, Eiermann T, Hossfeld DK, Zander AR, Jungbluth AA, Old LJ, Bokemeyer C, Kröger N. Cancer-testis antigens are commonly expressed in multiple myeloma and induce systemic immunity following allogeneic stem cell transplantation. Blood. 2007;109:1103–1112. doi: 10.1182/blood-2006-04-014480. [DOI] [PubMed] [Google Scholar]
- 31.Condomines M, Hose D, Raynaud P, Hundemer M, De Vos J, Baudard M, Moehler T, Pantesco V, Moos M, Schved JF, Rossi JF, Rème T, Goldschmidt H, Klein B. Cancer/testis genes in multiple myeloma: expression patterns and prognosis value determined by microarray analysis. J Immunol. 2007;178:3307–3315. doi: 10.4049/jimmunol.178.5.3307. [DOI] [PubMed] [Google Scholar]
- 32.Luetkens T, Schafhausen P, Uhlich F, Stasche T, Akbulak R, Bartels BM, Hildebrandt Y, Gontarewicz A, Kobold S, Meyer S, Gordic M, Bartels K, Lajmi N, Cao Y, Kröger N, Bokemeyer C, Brümmendorf TH, Atanackovic D. Expression, epigenetic regulation, and humoral immunogenicity of cancer-testis antigens in chronic myeloid leukemia. Leuk Res. 2010 doi: 10.1016/j.leukres.2010.03.039. in press. [DOI] [PubMed] [Google Scholar]
- 33.van Baren N, Chambost H, Ferrant A, Michaux L, Ikeda H, Millard I, Olive D, Boon T, Coulie PG. PRAME, a gene encoding an antigen recognized on a human melanoma by cytolytic T cells, is expressed in acute leukaemia cells. Br J Haematol. 1998;102:1376–1379. doi: 10.1046/j.1365-2141.1998.00982.x. [DOI] [PubMed] [Google Scholar]
- 34.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathieu MG, Linley AJ, Reeder SP, Badoual C, Tartour E, Rees RC, McArdle SE. HAGE, a cancer/testis antigen expressed at the protein level in a variety of cancers. Cancer Immun. 2010;10:2. http://www.cancerimmunity.org/v10p2/091211.htm [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YT, Güre AO, Tsang S, Stockert E, Jager E, Knuth A, Old LJ. Identification of multiple cancer/testis antigens by allogeneic antibody screening of a melanoma cell line library. Proc Natl Acad Sci U S A. 1998;95:6919–6923. doi: 10.1073/pnas.95.12.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Old LJ. Cancer/Testis (CT) antigens - a new link between gametogenesis and cancer. Cancer Immun. 2001;1:1. http://www.cancerimmunity.org/v1p1/010304.htm [PubMed] [Google Scholar]
- 38.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Müller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini B, Banham AH, Rosenwald A, Staudt LM, Connors JM, Armitage JO, Chan WC. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 39.Brasseur F, Rimoldi D, Liénard D, Lethe B, Carrel S, Arienti F, Suter L, Vanwijck R, Bourlond A, Humblet Y, Vacca A, Conese M, Lahaye T, Degiovanni G, Deraemaecker R, Beauduin M, Sastre X, Salamon E, Dréno B, Jäger E, Knuth A, Chevreau C, Suciu S, Lachapelle JM, Pouillart P, Parmiani G, Lejeune F, Cerottini JC, Boon T, Marchand M. Expression of MAGE genes in primary and metastatic cutaneous melanoma. Int J Cancer. 1995;63:375–380. doi: 10.1002/ijc.2910630313. [DOI] [PubMed] [Google Scholar]
- 40.Kurashige T, Noguchi Y, Saika T, Ono T, Nagata Y, Jungbluth A, Ritter G, Chen YT, Stockert E, Tsushima T, Kumon H, Old LJ, Nakayama E. Ny-ESO-1 expression and immunogenicity associated with transitional cell carcinoma: correlation with tumor grade. Cancer Res. 2001;61:4671–4674. [PubMed] [Google Scholar]
- 41.Kawano R, Karube K, Kikuchi M, Takeshita M, Tamura K, Uike N, Eto T, Ohshima K, Suzumiya J. Oncogene associated cDNA microarray analysis shows PRAME gene expression is a marker for response to anthracycline containing chemotherapy in patients with diffuse large B-cell lymphoma. J Clin Exp Hematopathol. 2009;49:1–7. doi: 10.3960/jslrt.49.1. [DOI] [PubMed] [Google Scholar]
- 42.Lim SH, Wang Z, Chiriva-Internati M, Xue Y. Sperm protein 17 is a novel cancer-testis antigen in multiple myeloma. Blood. 2001;97:1508–1510. doi: 10.1182/blood.v97.5.1508. [DOI] [PubMed] [Google Scholar]
- 43.Chiriva-Internati M, Wang Z, Salati E, Bumm K, Barlogie B, Lim SH. Sperm protein 17 (Sp17) is a suitable target for immunotherapy of multiple myeloma. Blood. 2002;100:961–965. doi: 10.1182/blood-2002-02-0408. [DOI] [PubMed] [Google Scholar]
- 44.Chiriva-Internati M, Wang Z, Salati E, Timmins P, Lim SH. Tumor vaccine for ovarian carcinoma targeting sperm protein 17. Cancer. 2002;94:2447–2453. doi: 10.1002/cncr.10506. [DOI] [PubMed] [Google Scholar]
- 45.Old LJ. Cancer vaccines: an overview. Cancer Immun. 2008;8(Suppl 1):1. http://www.cancerimmunity.org/v8suppl1p1/080104.htm [PubMed] [Google Scholar]
- 46.Jäger E, Gnjatic S, Nagata Y, Stockert E, Jäger D, Karbach J, Neumann A, Rieckenberg J, Chen YT, Ritter G, Hoffman E, Arand M, Old LJ, Knuth A. Induction of primary NY-ESO-1 immunity: CD8+ T lymphocyte and antibody responses in peptide-vaccinated patients with NY-ESO-1+ cancers. Proc Natl Acad Sci U S A. 2000;97:12198–12203. doi: 10.1073/pnas.220413497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jäger E, Stockert E, Zidianakis Z, Chen YT, Karbach J, Jäger D, Arand M, Ritter G, Old LJ, Knuth A. Humoral immune responses of cancer patients against "Cancer-Testis" antigen NY-ESO-1: correlation with clinical events. Int J Cancer. 1999;84:506–510. doi: 10.1002/(sici)1097-0215(19991022)84:5<506::aid-ijc10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 48.Sadanaga N, Nagashima H, Mashino K, Tahara K, Yamaguchi H, Ohta M, Fujie T, Tanaka F, Inoue H, Takesako K, Akiyoshi T, Mori M. Dendritic cell vaccination with MAGE peptide is a novel therapeutic approach for gastrointestinal carcinomas. Clin Cancer Res. 2001;7:2277–2284. [PubMed] [Google Scholar]
- 49.Marchand M, van Baren N, Weynants P, Brichard V, Dréno B, Tessier MH, Rankin E, Parmiani G, Arienti F, Humblet Y, Bourlond A, Vanwijck R, Liénard D, Beauduin M, Dietrich PY, Russo V, Kerger J, Masucci G, Jäger E, De Greve J, Atzpodien J, Brasseur F, Coulie PG, van der Bruggen P, Boon T. Tumor regressions observed in patients with metastatic melanoma treated with an antigenic peptide encoded by gene MAGE-3 and presented by HLA-A1. Int J Cancer. 1999;80:219–230. doi: 10.1002/(sici)1097-0215(19990118)80:2<219::aid-ijc10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 50.Dadabayev AR, Wang Z, Zhang Y, Zhang J, Robinson WR, Lim SH. Cancer immunotherapy targeting Sp17: when should the laboratory findings be translated to the clinics? Am J Hematol. 2005;80:6–11. doi: 10.1002/ajh.20415. [DOI] [PubMed] [Google Scholar]
- 51.Curioni-Fontecedro A, Knights AJ, Tinguely M, Nuber N, Schneider C, Thomson CW, von Boehmer L, Bossart W, Pahlich S, Gehring H, Moch H, Renner C, Knuth A, Zippelius A. MAGE-C1/CT7 is the dominant cancer-testis antigen targeted by humoral immune responses in patients with multiple myeloma. Leukemia. 2008;22:1646–1648. doi: 10.1038/leu.2008.43. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z, Zhang Y, Mandal A, Zhang J, Giles FJ, Herr JC, Lim SH. The spermatozoa protein, SLLP1, is a novel cancer-testis antigen in hematologic malignancies. Clin Cancer Res. 2004;10:6544–6550. doi: 10.1158/1078-0432.CCR-04-0911. [DOI] [PubMed] [Google Scholar]
- 53.Wang Z, Zhang Y, Liu H, Salati E, Chiriva-Internati M, Lim SH. Gene expression and immunologic consequence of SPAN-Xb in myeloma and other hematologic malignancies. Blood. 2003;101:955–960. doi: 10.1182/blood-2002-06-1930. [DOI] [PubMed] [Google Scholar]
- 54.Adams SP, Sahota SS, Mijovic A, Czepulkowski B, Padua RA, Mufti GJ, Guinn BA. Frequent expression of HAGE in presentation chronic myeloid leukaemias. Leukemia. 2002;16:2238–2242. doi: 10.1038/sj.leu.2402732. [DOI] [PubMed] [Google Scholar]
- 55.Lendvai N, Gnjatic S, Ritter E, Mangone M, Austin W, Reyner K, Jayabalan D, Niesvizky R, Jagannath S, Bhardwaj N, Chen-Kiang S, Old LJ, Cho HJ. Cellular immune responses against CT7 (MAGE-C1) and humoral responses against other cancer-testis antigens in multiple myeloma patients. Cancer Immun. 2010;10:4. http://www.cancerimmunity.org/v10p4/091213.htm [PMC free article] [PubMed] [Google Scholar]
- 56.Goodyear OC, Pratt G, McLarnon A, Cook M, Piper K, Moss P. Differential pattern of CD4+ and CD8+ T-cell immunity to MAGE-A1/A2/A3 in patients with monoclonal gammopathy of undetermined significance (MGUS) and multiple myeloma. Blood. 2008;112:3362–3372. doi: 10.1182/blood-2008-04-149393. [DOI] [PubMed] [Google Scholar]
- 57.Quintarelli C, Dotti G, De Angelis B, Hoyos V, Mims M, Luciano L, Heslop HE, Rooney CM, Pane F, Savoldo B. Cytotoxic T lymphocytes directed to the preferentially expressed antigen of melanoma (PRAME) target chronic myeloid leukemia. Blood. 2008;112:1876–1885. doi: 10.1182/blood-2008-04-150045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frank C, Hundemer M, Ho AD, Goldschmidt H, Witzens-Harig M. Cellular immune responses against the cancer-testis antigen SPAN-XB in healthy donors and patients with multiple myeloma. Leuk Lymphoma. 2008;49:779–785. doi: 10.1080/10428190801911688. [DOI] [PubMed] [Google Scholar]
- 59.van Rhee F, Szmania SM, Zhan F, Gupta SK, Pomtree M, Lin P, Batchu RB, Moreno A, Spagnoli G, Shaughnessy J, Tricot G. NY-ESO-1 is highly expressed in poor-prognosis multiple myeloma and induces spontaneous humoral and cellular immune responses. Blood. 2005;105:3939–3944. doi: 10.1182/blood-2004-09-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodyear O, Piper K, Khan N, Starczynski J, Mahendra P, Pratt G, Moss P. CD8+ T cells specific for cancer germline gene antigens are found in many patients with multiple myeloma, and their frequency correlates with disease burden. Blood. 2005;106:4217–4224. doi: 10.1182/blood-2005-02-0563. [DOI] [PubMed] [Google Scholar]
- 61.Jungbluth AA, Chen YT, Stockert E, Busam KJ, Kolb D, Iversen K, Coplan K, Williamson B, Altorki N, Old LJ. Immunohistochemical analysis of NY-ESO-1 antigen expression in normal and malignant human tissues. Int J Cancer. 2001;92:856–860. doi: 10.1002/ijc.1282. [DOI] [PubMed] [Google Scholar]
- 62.Jungbluth AA, Stockert E, Chen YT, Kolb D, Iversen K, Coplan K, Williamson B, Altorki N, Busam KJ, Old LJ. Monoclonal antibody MA454 reveals a heterogeneous expression pattern of MAGE-1 antigen in formalin-fixed paraffin embedded lung tumours. Br J Cancer. 2000;83:493–497. doi: 10.1054/bjoc.2000.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pulford K, Banham AH, Lyne L, Jones M, Ippolito GC, Liu H, Tucker PW, Roncador G, Lucas E, Ashe S, Stockwin L, Walewska R, Karran L, Gascoyne RD, Mason DY, Dyer MJ. The BCL11AXL transcription factor: its distribution in normal and malignant tissues and use as a marker for plasmacytoid dendritic cells. Leukemia. 2006;20:1439–1441. doi: 10.1038/sj.leu.2404260. [DOI] [PubMed] [Google Scholar]
- 64.Sambrook J, Russell DW. New York (NY): Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning: A Laboratory Approach. 3rd ed. [Google Scholar]
- 65.van Baren N, Brasseur F, Godelaine D, Hames G, Ferrant A, Lehmann F, André M, Ravoet C, Doyen C, Spagnoli GC, Bakkus M, Thielemans K, Boon T. Genes encoding tumor-specific antigens are expressed in human myeloma cells. Blood. 1999;94:1156–1164. [PubMed] [Google Scholar]
- 66.Yuasa T, Okamoto K, Kawakami T, Mishina M, Ogawa O, Okada Y. Expression patterns of cancer testis antigens in testicular germ cell tumors and adjacent testicular tissue. J Urol. 2001;165:1790–1794. [PubMed] [Google Scholar]
- 67.Güre AO, Wei IJ, Old LJ, Chen YT. The SSX gene family: characterization of 9 complete genes. Int J Cancer. 2002;101:448–453. doi: 10.1002/ijc.10634. [DOI] [PubMed] [Google Scholar]
- 68.Chen YT, Scanlan MJ, Sahin U, Türeci O, Gure AO, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old LJ. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci U S A. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scanlan MJ, Altorki NK, Gure AO, Williamson B, Jungbluth A, Chen YT, Old LJ. Expression of cancer-testis antigens in lung cancer: definition of bromodomain testis-specific gene (BRDT) as a new CT gene, CT9. Cancer Lett. 2000;150:155–164. doi: 10.1016/s0304-3835(99)00385-7. [DOI] [PubMed] [Google Scholar]
- 70.Güre AO, Stockert E, Arden KC, Boyer AD, Viars CS, Scanlan MJ, Old LJ, Chen YT. CT10: a new cancer-testis (CT) antigen homologous to CT7 and the MAGE family, identified by representational-difference analysis. Int J Cancer. 2000;85:726–732. doi: 10.1002/(sici)1097-0215(20000301)85:5<726::aid-ijc21>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 71.Liu XF, Helman LJ, Yeung C, Bera TK, Lee B, Pastan I. XAGE-1, a new gene that is frequently expressed in Ewing's sarcoma. Cancer Res. 2000;60:4752–4755. [PubMed] [Google Scholar]
- 72.Martelange V, De Smet C, De Plaen E, Lurquin C, Boon T. Identification on a human sarcoma of two new genes with tumor-specific expression. Cancer Res. 2000;60:3848–3855. [PubMed] [Google Scholar]
- 73.Scanlan MJ, Gordon CM, Williamson B, Lee SY, Chen YT, Stockert E, Jungbluth A, Ritter G, Jäger D, Jäger E, Knuth A, Old LJ. Identification of cancer/testis genes by database mining and mRNA expression analysis. Int J Cancer. 2002;98:485–492. doi: 10.1002/ijc.10276. [DOI] [PubMed] [Google Scholar]
- 74.Eichmuller S, Usener D, Dummer R, Stein A, Thiel D, Schadendorf D. Serological detection of cutaneous T-cell lymphoma-associated antigens. Proc Natl Acad Sci U S A. 2001;98:629–634. doi: 10.1073/pnas.021386498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin C, Mak S, Meitner PA, Wolf JM, Bluman EM, Block JA, Terek RM. Cancer/testis antigen CSAGE is concurrently expressed with MAGE in chondrosarcoma. Gene. 2002;285:269–278. doi: 10.1016/s0378-1119(02)00395-5. [DOI] [PubMed] [Google Scholar]
- 76.Cho B, Lim Y, Lee DY, Park SY, Lee H, Kim WH, Yang H, Bang YJ, Jeoung DI. Identification and characterization of a novel cancer/testis antigen gene CAGE. Biochem Biophys Res Commun. 2002;292:715–726. doi: 10.1006/bbrc.2002.6701. [DOI] [PubMed] [Google Scholar]
- 77.Sahin U, Koslowski M, Türeci O, Eberle T, Zwick C, Romeike B, Moringlane JR, Schwechheimer K, Feiden W, Pfreundschuh M. Expression of cancer testis genes in human brain tumors. Clin Cancer Res. 2000;6:3916–3922. [PubMed] [Google Scholar]
- 78.Barrett A, Madsen B, Copier J, Lu PJ, Cooper L, Scibetta AG, Burchell J, Taylor-Papadimitriou J. PLU-1 nuclear protein, which is upregulated in breast cancer, shows restricted expression in normal human adult tissues: a new cancer/testis antigen? Int J Cancer. 2002;101:581–588. doi: 10.1002/ijc.10644. [DOI] [PubMed] [Google Scholar]
- 79.Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, Zlotnik A. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7:67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- 80.Ge X, Yamamoto S, Tsutsumi S, Midorikawa Y, Ihara S, Wang SM, Aburatani H. Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics. 2005;86:127–141. doi: 10.1016/j.ygeno.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 81.Shyamsundar R, Kim YH, Higgins JP, Montgomery K, Jorden M, Sethuraman A, van de Rijn M, Botstein D, Brown PO, Pollack JR. A DNA microarray survey of gene expression in normal human tissues. Genome Biol. 2005;6:R22. doi: 10.1186/gb-2005-6-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hsiao LL, Dangond F, Yoshida T, Hong R, Jensen RV, Misra J, Dillon W, Lee KF, Clark KE, Haverty P, Weng Z, Mutter GL, Frosch MP, Macdonald ME, Milford EL, Crum CP, Bueno R, Pratt RE, Mahadevappa M, Warrington JA, Stephanopoulos G, Stephanopoulos G, Gullans SR. A compendium of gene expression in normal human tissues. Physiol Genomics. 2001;7:97–104. doi: 10.1152/physiolgenomics.00040.2001. [DOI] [PubMed] [Google Scholar]
Materials and methods
Cell lines and culture conditions
The OCI-Ly3, OCI-Ly10 (ABC-derived), SU-DHL-6, SU-DHL-10 and DB (GC-derived) DLBCL cell lines were a kind gift from Dr. Eric Davis, Bethesda USA, and the MIEU (GC-derived), HLY-1 (ABC-derived) and LIB (non-ABC-derived) (24) DLBCL-derived cell lines were generously provided by Dr. Talal Al Saati, Toulouse, France. All other cell lines were obtained as previously reported (63). Cells were maintained in RPMI 1640 media supplemented with 10% FCS, 2 mM glutamine and antibiotics [streptomycin (50 µg/ml) and penicillin (50 U/ml)] at 37˚C and 5% CO2. Cells were washed twice in RNase-free PBS prior to RNA extraction.
RT-PCR
Messenger RNA from cell lines was extracted using µMACS mRNA Isolation kits (Miltenyi Biotec, Germany), treated with DNaseI (Ambion, Huntingdon, UK) to ensure complete removal of genomic DNA, and then quantified. Total RNA from human testis (Clontech-Takara Bio Europe, France) was resuspended at 1 µg/µl. Complementary DNA (cDNA) was reverse transcribed at 42˚C for 50 minutes from 100 ng cell line mRNA or 200 ng testis total RNA in a 25 µl reaction containing 200 U Superscript II RNase H- reverse transcriptase, 1x First Strand Buffer, 4 mM DTT, 250 µM dNTPs (Invitrogen, Paisley, UK), 1 µl RNasin® ribonuclease inhibitor and 100 ng random hexamers (both Promega, Madison, WI, USA). Complementary DNA synthesis reactions lacking the reverse transcriptase were also prepared for use in PCR to confirm amplified products did not arise from genomic DNA, which was particularly important where primers did not span introns, or for transcripts that did not contain any introns. Two normal tissue cDNA panels were also purchased from Takara BioEurope, France (Human MTC Panels I and II). The integrity of cDNA templates was assessed using gene-specific primers to β-actin. PCR reactions (25 µl) contained 2 µl cell line cDNA, 2 µl human testis cDNA or 2.5 µl normal tissue cDNA, 200 µM each dNTP, 10 µM each primer, 1x PCR buffer and 1x Advantage 2 Polymerase mix (Clontech-Takara Bio Europe, France). Gene-specific primers and annealing temperatures are given in Table 3. Cycling parameters were: 94˚C 5 minutes (initial denaturation), then 35 cycles of 94˚C 30 seconds, appropriate annealing temperature 30 seconds, 72˚C up to 3 minutes. Sequencing of selected PCR products generated by each primer pair confirmed their identity.
Table 3.
Primer sequences and annealing temperatures used in RT-PCR analyses of CT transcripts.
Immunohistochemistry
Paraffin-embedded normal tissues and DLBCL biopsies were obtained from the Histopathology Department and the ENT Department, John Radcliffe Hospital, Oxford, U.K. All tissues were used only after informed consent had been obtained and the study was performed under local ethical committee approval from the Oxford Research Ethics Committee. Paraffin embedded cells, tissue sections and tissue microarrays (TMAs) were dewaxed, rehydrated and then subjected to microwave antigen retrieval in 50 mM Tris/2 mM EDTA (pH 9) for 3 minutes (TMAs were baked at 65˚C for 10 minutes prior to dewaxing). Sections were incubated with the anti-Sp17 monoclonal antibody (S.H. Lim, Texas) at a dilution of 1:500 and labelling was detected using the DAKO REAL™ EnVision™ Detection System (DakoCytomation, Glostrup, Denmark).
Western blotting
Whole cell lysates were prepared by resuspending 1 x 107 cells or four 8 µm frozen tissue sections in 300 µl SDS loading buffer (64). Quantified normal human tissue lysates were purchased from Abcam (Cambridge, UK). Samples were resolved by SDS-PAGE then transferred to Hybond ECL nitrocellulose membrane. Membranes were blocked in 5% dried milk in TBS overnight, incubated with anti-Sp17 antibody at a 1:2000 dilution, and antigen/antibody complexes were detected using ECL (GE Healthcare, Amersham, UK).
Supplemental data
Download from http://www.cancerimmunity.org/v10p8/100708_suppl_fig1.pdf (480 KB PDF file).
Download from http://www.cancerimmunity.org/v10p8/100708_suppl_fig2.pdf (283 KB PDF file).
Download from http://www.cancerimmunity.org/v10p8/100708_suppl_tab1.pdf (119 KB PDF file).
Download from http://www.cancerimmunity.org/v10p8/100708_suppl_data.pdf (1.4 MB PDF file).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Download from http://www.cancerimmunity.org/v10p8/100708_suppl_fig1.pdf (480 KB PDF file).
Download from http://www.cancerimmunity.org/v10p8/100708_suppl_fig2.pdf (283 KB PDF file).
Download from http://www.cancerimmunity.org/v10p8/100708_suppl_tab1.pdf (119 KB PDF file).
Download from http://www.cancerimmunity.org/v10p8/100708_suppl_data.pdf (1.4 MB PDF file).