Abstract
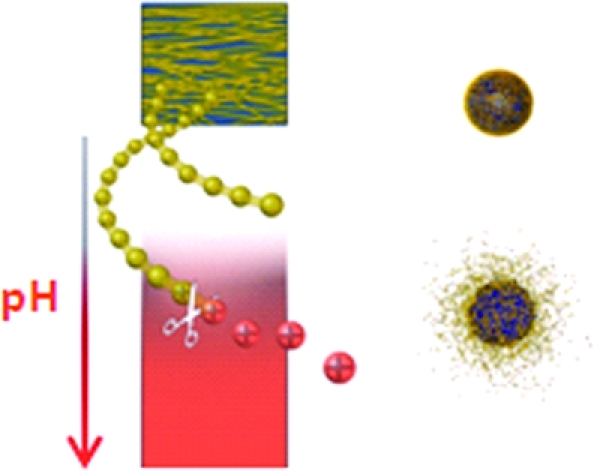
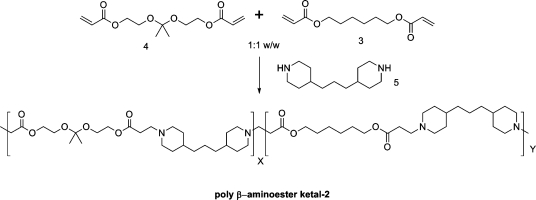
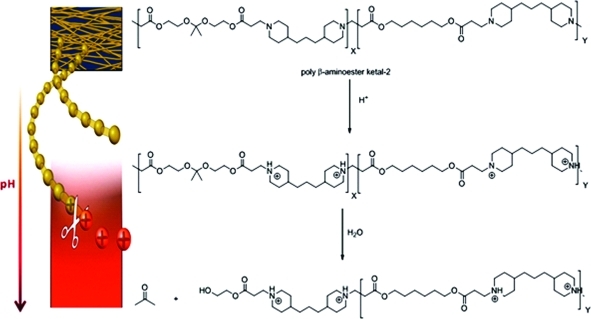
Logic gate nanoparticles, where two chemical transformations take place one after the other, were successfully formulated from a newly synthesized random co-polymer. This polymer, poly([2,2′-(propane-2,2-diylbis(oxy))bis(ethane-2,1-diyl) diacrylate ]-co-[hexane-1,6-diyl diacrylate]-4,4′ trimethylene dipiperidine), (poly-β-aminoester ketal-2) contains two pH responsive moieties within its backbone. As nanoparticles they function akin to an AND logic gate. The β-aminoester backbone moiety provides a pH triggered solubility switch, only when this switch is “ON” does the ketal moiety also turn “ON” to undergo rapid acid catalyzed hydrolysis. These AND logic gate polymeric nanoparticles were prepared using an oil in water emulsion method. Their degradation in the pH range of 7.4−5 was monitored by dynamic light scattering and showed excellent stability at pH 7.4 and rapid degradation at pH 5. Our results indicate that the prepared logic gate nanoparticles may prove valuable in delivering therapeutics and diagnostics to cells and diseased tissue.
Keywords: logic gate, nanoparticles, burst release, dual pH response polymers, poly-β-aminoester ketal
Nanoscale carriers capable of a controlled and rapid triggered response to physiological events such as changes in extracellular pH, temperature and reactive oxygen species are particularly useful in the delivery of therapeutics and diagnostics to diseased cells and tissue.1−8 Such carriers can maximize therapeutic efficacy and minimize undesirable side effects by decreasing their dosage.(9) In addition, encapsulation of therapeutics and imaging agents protects them from harsh in vivo conditions that lead to proteolytic degradation,(10) sequestration, and renal clearance. Encapsulation within biodegradable polymers is one strategy by which bioactives are delivered to diseased cells via endosome to cytosol release.11−13 Cytosolic delivery is particularly challenging and is a major hurdle for effective therapeutic delivery.14,15 Burst-degrading drug delivery systems hold promise in achieving increased cytosolic release through elevated osmotic pressure within the endosomes.16,17 To this effect, hydrogels utilizing ketal cross-links were developed and are promising; however, their payloads are limited to large water-soluble macromolecules and the catalytic nature of their degradation leads to significant degradation at physiological pH over time.(18) Similarly hydrophobic polyketals are also promising classes of polymeric biomaterials as they can encapsulate both hydrophobic and hydrophilic payloads, but as nanoparticles they no longer undergo rapid acid catalyzed hydrolysis unless fully hydrated.(19)
Formulation of nanoparticles from polymers requires them to have a hydrophobic character. This, however dramatically slows down their hydrolysis degradation kinetics. Here degradation would occur slowly by a surface erosion mechanism.19−22 This leads to a catch 22 situation, and we hypothesized that formulating nanoparticles from polymers with a hydrophilic pH switch can both ensure the stability of the nanoparticles in physiological pH and still achieve the desired rapid catalytic degradation in acidic conditions.
We hypothesized that developing a system that has two or more pH response mechanisms to a single triggering event would more finely tune the response to pH stimuli. Among pH sensitive polymers which are able to switch from hydrophobic to hydrophilic, poly-β-amino esters, (PbAE), are widely studied because of their excellent tunability23−27 and ease of preparation.26,28 Hydrophobic nanoparticles made of PbAE undergo protonation of the amine backbone upon decreasing the pH, leading to immediate dissolution in aqueous solutions.29−32
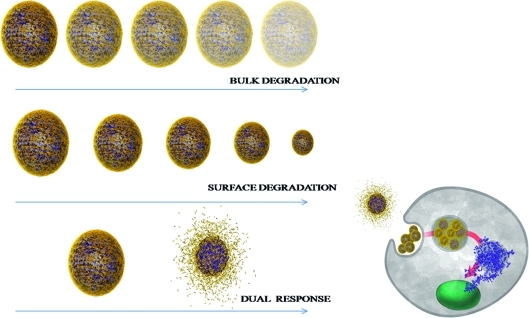
In the present work we developed dual pH responsive random co-polymer poly([2,2′-(propane-2,2-diylbis(oxy))bis(ethane-2,1-diyl) diacrylate]-co-[hexane-1,6-diyl diacrylate]-4,4′-trimethylene dipiperidine) (poly-β-aminoester ketal-2) that utilizes both mechanisms described in the previous paragraphs (1) to undergo degradation. In response to a single triggering event of a decrease in pH, the amine backbone undergoes a sharp hydrophobic−hydrophilic switch. This leads to an increase in uptake of water (bulk dissolution) and hence an increase in ketal hydrolysis (surface degradation). We reasoned that the second degradation step should proceed by both surface and bulk erosion simultaneously. The degradation and release profile of this newly developed system has promise in exhibiting increased cytosolic release (1).(33)
Scheme 1. Synthesis of random co-poly-β-aminoester ketal-2.
Figure 1.
Different release profiles from nanoparticles result in different cellular response.
Nanoparticles formulated using the poly-β-aminoester ketal-2 become hydrophilic at mildly acidic pH 6.5−5.0 and in turn lead to accelerated hydrolysis of the ketal moieties. The pronounced effect of a hydrophilic−hydrophobic balance is evidenced by the fact that our degradation times are significantly faster than that obtained for other hydrophobic polyketals.21,22 Furthermore, our dual pH response design showed better stability at physiological pH (7.4) than other hydrophilic polyketals,(34) while maintaining the desired rapid degradation at acidic pH.
Results and Discussion
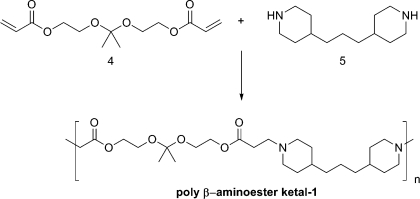
To test our hypothesis of creating a dual pH response system we initially synthesized poly-β-aminoester ketal-1 shown in 2.
Scheme 2. Synthesis of poly-β-aminoester ketal-1.
Nanoparticles formulated from poly-β-aminoester ketal-1 were found to dissolve rapidly at pH 7.4 owing to the increased charge density of the polymer backbone. While the pka of the polymer is expected to be similar to the ones calculated in the literature,35,36 the solubility switch of the polymer depends on a hydrophilic−hydrophobic balance between the hydrophilic protonated amines and hydrophobic alkyl backbone. We hypothesized that the presence of the ketal group on every monomer unit makes the backbone more hydrophilic and results in a more rapidly soluble nanoparticle at pH 7.4. To improve on our original design we synthesized poly-β-aminoester ketal-2 in which we incorporated a more hydrophobic spacer in order to increase the hydrophobicity of the backbone and thereby improve the stability of the nanoparticles at pH 7.4.
Poly-β-aminoester ketal-2 was synthesized as shown in 1via Michael-type addition of bis(secondary amine) monomers to diacrylate ester and diacrylate ester ketal monomers in a 2:1:1 mixture. The resulting poly-β-aminoester ketal-2 was then formulated using emulsion techniques into logic gate nanoparticles. TEM (transmission electron microscope) images of the nanoparticles formulated (Supporting Information, Figure 5) show diameters of 100−150 nm while their average hydrodynamic radius was ∼300 nm by dynamic light scattering, DLS (Supporting Information, Figure 6).
Influence of pH on Nanoparticles Size
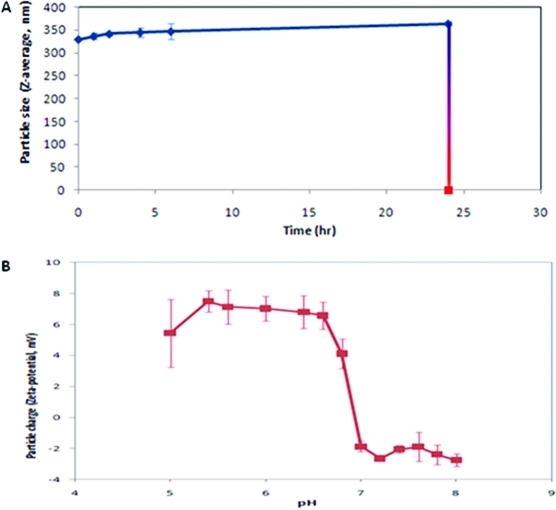
The nanoparticles were monitored for 24 h by DLS, using a Zetasizer−ZS (Malvern, U.K). DLS measures hydrodynamic diameter and charge of particles. 2 shows that the nanoparticles remain stable at pH 7.4 over a period of 24 h without a significant change in size for the first 4 h at pH 7.4 (p > 0.05). Subsequently we note a slight but significant increase between 6 and 24 h (p = 0.044 and 0.002, respectively). This increase in diameter reflects the hydration process of the polymeric nanoparticle due to partial protonation of the amino groups (pKa ≈ 6.7)35,36 at this pH (also see 2). Changing the pH to 5 (pH of many cellular subcompartments) caused a sudden or burst degradation of the nanoparticles. The drop we see in particle size is very dramatic compared to the nanoparticle systems previously published in the literature.21,22
Figure 2.
(A) Influence of pH on particle size (Z-average) and (B) particle charge (zeta-potential) of poly-β-aminoester ketal-2 nanoparticles.
Influence of pH on Nanoparticle Charge
We also measured the zeta potential of the nanoparticles at different pH (2) to detect the protonation of these polymeric nanoparticles with decrease in pH. Zeta potential measurements using DLS gives us the charge on these dual response nanoparticles as a function of pH and thus can help elucidate the degradation mechanism. Poly-β-aminoester ketal-2 nanoparticles were prepared as described and washed using water. A 100 μL suspension of nanoparticles was dispersed in different phosphate buffers with different pH values. We note that the charge on the particle increases with decrease in the pH confirming that the amines on the polymer backbone are progressively protonated and the degree of protonation increases dramatically around pH 7−6 which corresponds to the pKa of the backbone.
Polymer Degradation Studies by GPC and NMR
Polymer degradation studies were performed in order to observe the degradation products and molecular weights of the fragments. NMR studies were carried out by dissolving the polymer in pH 5 phosphate buffer and recording spectra at various time intervals. We found that acetone peaks appeared immediately and grew until about 2 h. We continued to record NMR spectra (Supporting Information, Figure 7) for a two week period and observed no further degradation of the backbone. A 100 mg portion of the polymer was also separately incubated at 37 °C in pH 5 phosphate buffer. Samples were withdrawn at various time points, lyophilized, and analyzed via GPC. The GPC traces show that the polymer fragments into smaller fragments at this pH (Supporting Information, Figure 8). On the basis of these findings, this new hydrophobic polymeric nanoparticle is stable at pH 7.4; however, upon decreasing the pH the tertiary amines along the polymer backbone become protonated and the polymer becomes more hydrophilic. This results in an increased uptake of water followed by acid catalyzed hydrolysis of the ketal groups along the polymeric backbone (3). The ease of degradation of these polymeric nanoparticles is a significant advantage over other systems especially in gene delivery applications as it could minimize the cytotoxicity of the carrier in contrast to other more toxic polyamine systems.(37)
Scheme 3. Mechanism of degradation of the polymer backbone of poly-β-aminoester ketal-2.
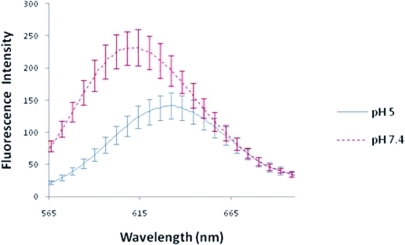
Influence of pH on Nile Red Release from Poly-β-aminoester Ketal-2 Nanoparticles (3)
Figure 3.
Significant red and hypochromic shifts (p < 0.001 and p = 0.010, respectively) were observed in the fluorescence absorption spectra of Nile Red poly-β-aminoester ketal-1 nanoparticle suspensions upon changing the pH from 7.4 to 5. These shifts indicate the presence of Nile Red in a hydrophobic environment such as within the nanoparticles which suddenly changes upon decreasing the pH to a more hydrophilic environment.
Nile red, a nonpolar probe, fluorescent in hydrophobic environments was encapsulated in our dual response nanoparticles in order to investigate the ability of these dual response nanoparticles to release a small hydrophobic molecule, as a model pharmaceutical.
Nanoparticles containing Nile Red were prepared and the fluorescence of Nile Red poly-β-aminoester ketal-2 nanoparticles was measured at pH 7.4 and 5 (3). Upon changing the pH from 7.4 to 5 we observed a decrease in fluorescence intensity coupled with a red shift in the fluorescence peak. These results are indicative of Nile Red release immediately upon decreasing the pH to 5.
Cytoxicity and Cellular Internalization Studies of the Poly-β-aminoester Ketal-2 Nanoparticles
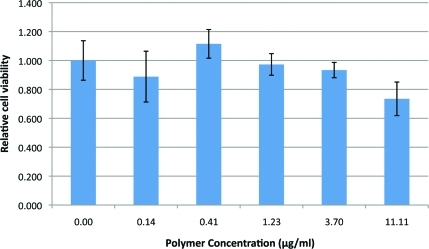
We evaluated the cytotoxicity of poly-β-aminoester ketal-2 nanoparticles in cells by a MTT assay. RAW 264.7 cells were incubated with various amounts of nanoparticles for 20 h. 4 illustrates the comparison of cytotoxicity between cells treated with increasing concentrations of poly-β-aminoester ketal-2 with respect to polymer and control cells. There was no significant cytotoxicity observed until we reached high concentrations of polymer (beyond 11.1 μg/mL).
Figure 4.
Cytotoxicity of poly-β-aminoester ketal-2 nanoparticles at different concentrations in RAW 264.7 macrophage cells.
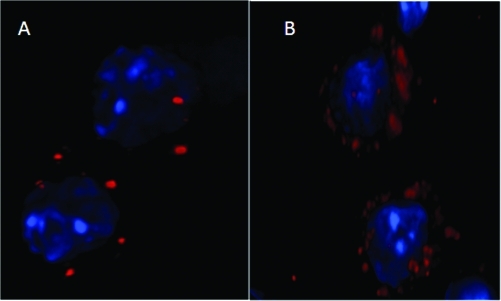
Finally, the uptake of our nanoparticles by RAW 264.7 macrophages was evaluated by fluorescence microscopy. We observed more diffuse fluorescence from our dual pH responsive polymeric nanoparticles encapsulating labeled BSA, compared with poly-lactic-co-glycolic acid (PLGA) encapsulated fluorescent BSA where fluorescence was more punctuate (5). We reasoned from these images that the uptake of poly-β-aminoester ketal-2 nanoparticles and release of fluorescent BSA is enhanced when compared to PLGA nanoparticles (5).
Figure 5.
Uptake of nanoparticles and loaded with fluorescent BSA by macrophage cells. Raw 264.7 macrophage cells were treated for 2 h with poly-β-aminoester ketal-2 or PLGA nanoparticles containing BSA-Alexa Fluor 594 (red) and stained with DAPI (blue).
Conclusions
We have shown that two pH response moieties, a pH solubility switch and a pH labile group can be incorporated into the backbone of polymers which can then be formulated into dual responsive nanoparticles encapsulating small hydrophobic molecules and larger protein payloads. Our dual pH response moieties function like a logic gate to fine-tune the degradation of and release from these nanoparticles. The nanoparticle formulations were stable for 24 h in healthy physiological pH, and upon reducing the pH to endosomal levels, pH 5, these dual responsive nanoparticles underwent a rapid and dramatic degradation followed by concomitant release of their payloads. We have shown that their degradation mechanism begins with the protonation of the tertiary amines along the backbone that then switch the polymeric nanoparticle from hydrophobic to hydrophilic. The increased uptake of water under acidic conditions now allows the ketal groups to rapidly hydrolyze to give a bulk degradation profile. In acidic pH, these polymeric nanoparticles degrade into innocuous fragments, namely diols and acetone. Furthermore, the degradation byproduct, acetone, is anti-inflammatory, alleviating concerns of an inflammatory response of acidic byproduct typical of traditional polyester biomaterials. Such rapid hydrolysis is known to increase the osmotic pressure inside subcellular compartments leading to cytoplasmic release of encapsulated payloads. This system seems to be a promising vehicle for the administration of hydrophilic and hydrophobic payloads into target areas of the human body. Continued efforts in designing optimal bioresponsive polymers would improve limitations in drug, diagnostic, and biopharmaceutical delivery. We are currently applying these novel systems to sense and image early stages of metabolic diseases and inflammation.
Materials and Methods
Materials
4,4′-Trimethylenedipiperidine was purchased from Aldrich Chemical Co. (Milwaukee, WI). Triethylamine (TEA), potassium hydrogen phosphate (K2HPO4) anhydrous, potassium dihydrogen phosphate (KH2PO4), and 1,6-hexanediol diacrylate were purchased from Alfa Aesar Organics (Ward Hill, MA). Dichloromethane (DCM, methylene chloride) was purchased from Fisher Scientific (Hampton, NH). Poly(vinyl alcohol) (PVA) (MW 30−70 k) was purchased from Sigma Chemical Co. (St. Louis, MO). All reagents were used without further purification unless otherwise stated.
Synthesis of Poly-β-aminoester Ketal-1 and -2 (1)
The polymers were prepared by Michael addition of the corresponding diacrylates with trimethyl dipiperidine. Typically acrylic ketal monomer 4 was synthesized from commercially available reagents as described in the literature.(38) In a vial, diacrylate 3 (5 mmol) and acrylicketal 4 (4.2 mmol) was dissolved in 1 mL of DCM followed by addition of 1 mL of TEA. A Teflon-coated stir bar was added to mix the reactants, and dipiperidine (10 mmol) was added to the vial which was then sealed with a Teflon-lined screw-cap, The reaction was purged with nitrogen gas. The reaction was stirred at room temperature for 4 days. The solvent was then evaporated, and the crude polymer was disolved in 10 mL of DCM. The polymer was purified by precipitating it into 2 × 200 mL hexane to yield 360 mg of the polymer. The polymer was collected and dried under vacuum prior to analysis.
Poly-β-aminoester Ketal-1
1H NMR (600 MHz, CDCl3) δ 4.24−4.14 (m, 4H), 3.68−3.57 (m, 4H), 2.85 (d, J = 10.3 Hz, 4H), 2.65 (t, J = 7.5 Hz, 4H), 2.52 (t, J = 7.5 Hz, 4H), 1.93 (t, J = 9.9 Hz, 4H), 1.63 (d, J = 6.4 Hz, 4H), 1.35 (d, J = 9.2 Hz, 6H), 1.26 (d, J = 4.3 Hz, 2H), 1.17 (s, 10H).
13C NMR (151 MHz, CDCl3) δ 172.77, 100.36, 100.12, 63.92, 58.97, 54.00, 53.94, 36.86, 35.73, 32.48, 32.32, 24.90, 24.01.
Molecular weight: Estimated by size exclusion chromatography against polystyrene standards in DMF/0.01% LiBr with a VWD (variable wavelength detector) at 250 nm. Mw = 33400, Mn = 13300, PDI = 2.52
Poly-β-aminoester Ketal-2
1H NMR (400 MHz, CDCl3) δ 4.20 (m, 1.6H), 4.07 (t, J = 6.6 Hz, 2.3H), 3.64 (m, 1.6H), 2.87 (d, J = 9.8 Hz, 4H), 2.67 (t, J = 6.9 Hz, 4H), 2.52 (dd, J = 16.3, 8.4 Hz, 4H), 1.95 (s, 4H), 1.64 (s, 6H), 1.37 (s, 5H), 1.22−1.19 (br, m, 14H).
13C NMR (126 MHz, chloroform-D) δ 172.87, 172.72, 100.31, 64.42, 63.88, 58.92, 54.03, 53.95, 53.87, 36.81, 35.69, 32.43, 32.27, 28.58, 25.65, 24.86, 23.96.
Molecular weight: Estimated by size exclusion chromatography against polystyrene standards in DMF/0.01% LiBr with a VWD (variable wavelength detector) at 250 nm. Mw = 6300, Mn = 2880, PDI = 2.18
Preparation of Dual-Responsive Nanoparticles
In a vial, 25 mg of the synthesized polymer was dissolved in 2.5 mL of DCM. The dissolved polymer was added to 50 mL of phosphate buffer (pH 8) containing 1% PVA. The mixture was stirred at 1000 rpm for 10 min to prepare an emulsion. Further emulsification was achieved using a high pressure homogenizer (Microfluidic 110PS, USA) at 23000 psi for three cycles. The nanoparticle suspension was stirred at 1000 rpm using a magnetic stirrer to evaporate the DCM. Concentrated mode tangential flow filtration system using 500 kDa Pellicon XL cassettes (Millipore, USA) was used to remove the PVA. The nanoparticle suspension was concentrated to 10 mL and washed two times using phosphate buffer (pH 8).(39) The pH of nanocapsule suspensions was reduced to 7.4 using KH2PO4 (6.5 w/v%). The pH was acidified to 5 using 0.1 N HCl. This nanoparticles solution was used diluted and used to perform the degradation and zeta potential experiments.
Nile Red Containing Poly-β-aminoester Ketal-2 Nanoparticles
For this study nanoparticles were prepared as described earlier and by the addition of 500 μg Nile Red in the DCM used during the single emulsion preparation step. This nanoparticles solution was diluted with buffer and used as such for the performing the degradation experiments using Nile Red.
BSA-AlexaFlour Containing Poly-β-aminoester Ketal-2 Nanoparticles
Nanoparticles were prepared using double emulsion method water/oil/water (W/O/W). Briefly, 5 mg of BSA Alexa fluor 594 was dissolved in 0.5 mL of PBS buffer and emulsified with 5 mL of DCM containing 100 mg of polymer (poly-β-aminoester ketal-2 or PLGA) using magnetic stirring. The primary emulsion was added to 50 mL of 1% PVA in PBS pH 8, and the secondary emulsion was produced. Subsequently further emulsification was achieved similar to before. This nanoparticles solution was diluted and used as such for the fluorescence microscopy.
Cell Toxicity of Nanoparticles
The cytotoxicity of nanoparticles was investigated using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction assay. RAW 264.7 macrophage cells were seeded at a density of 15000 cells/well in a 96-well plate and incubated for 24 h to reach ∼60% confluency. Cells were treated with various amounts of nanoparticles that correspond to different amounts of polymer content in them (0.14−11.4 μg/mL) and incubated for 20 h. To each well was added 10 μL of MTT solution, and the wells were incubated for 3 h. Dimethyl sulfoxide (DMSO, 100 μL) was added to cells to dissolve the resulting formazan crystals. After 20 min of incubation, the absorbance at 570 nm was measured using a microplate reader (Flexstation, Molecular Device Co.). Cell viability was obtained by comparing the absorbance of nanoparticles-treated cells to that of control cells not treated with particles.
Cellular Internalization of Nanoparticles
The uptake of Alexa Fluor-594-BSA-loaded nanoparticles was studied in RAW264.7 cells using a fluorescent microscope. Cells were plated on 12 mm cover slides at a density of 10 000 cells per well for 24 h followed by treatment with poly-βaminoester ketal-2 or PLGA nanoparticles. The payload in the nanaparticles contained a final concentration of approximately 50ug/mL Alexa Fluor-594-BSA. After 2 h at 37 °C, the cells were washed three times with PBS before mounting and staining with DAPI
Acknowledgments
We thank the NIH New Innovator Award (1 DP2 OD006499-01), and the PhRMA Foundation (Research Starter Grant) for funding of this work. We also thank Dr. Eric Schopf for his help with TEM imaging.
Supporting Information Available
NMR and GPC characterization of the polymer and its degradation data. TEM image of nanoparticles. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Farokhzad O. C.; Karp J. M.; Langer R. Nanoparticle−Aptamer Bioconjugates for Cancer Targeting. Expert Opin. Drug Delivery 2006, 3, 311–324. [DOI] [PubMed] [Google Scholar]
- Farokhzad O. C.; Langer R. Impact of Nanotechnology on Drug Delivery. ACS Nano 2009, 3, 16–20. [DOI] [PubMed] [Google Scholar]
- Ferrari M. Cancer Nanotechnology: Opportunities and Challenges. Nat. Rev. Cancer 2005, 5, 161–171. [DOI] [PubMed] [Google Scholar]
- Ganta S.; Devalapally H.; Shahiwala A.; Amiji M. A Review of Stimuli-Responsive Nanocarriers for Drug and Gene Delivery. J. Controlled Release 2008, 126, 187–204. [DOI] [PubMed] [Google Scholar]
- Langer R. New Methods of Drug Delivery. Science 1990, 249, 1527–1533. [DOI] [PubMed] [Google Scholar]
- LaVan D. A.; McGuire T.; Langer R. Small-Scale Systems for in Vivo Drug Delivery. Nat. Biotechnol. 2003, 21, 1184–1191. [DOI] [PubMed] [Google Scholar]
- Whitesides G. M. The “Right” Size in Nanobiotechnology. Nat. Biotechnol. 2003, 21, 1161–1165. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Gu F. X.; Chan J. M.; Wang A. Z.; Langer R. S.; Farokhzad O. C. Nanoparticles in Medicine: Therapeutic Applications and Developments. Clin. Pharm. Ther. 2008, 83, 761–769. [DOI] [PubMed] [Google Scholar]
- Tewes , F., ; Boury , F.;, Benoit , J. Biodegradable Microspheres: Advances in Production Technology. Microencapsulation: Methods and Industrial Applications; ; Benita S., Ed.; Taylor and Francis: New York, 2006; pp 1−41; (Drugs Pharm. Sci. 2006,. 158). [Google Scholar]
- Almutairi A.; Akers W. J.; Berezin M. Y.; Achilefu S.; Frechet J. M. J. Monitoring the Biodegradation of Dendritic Near-Infrared Nanoprobes by in Vivo Fluorescence Imaging. Mol. Pharm. 2008, 5, 1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D. H.: Controlled Release of Bioactive Agents from Lactide/Glycolide Polymers. Biodegradable Polymers as Drug Delivery Systems; Drugs and the Pharmaceutical Sciences, Vol. 45,Chasin M.,Langer R., Eds.; Marcel Dekker:New York,1990; pp 1−42. [Google Scholar]
- Panyam J.; Labhasetwar V. Biodegradable Nanoparticles for Drug and Gene Delivery to Cells and Tissue. Adv. Drug Delivery. Rev. 2003, 55, 329–347. [DOI] [PubMed] [Google Scholar]
- Shenoy D.; Little S.; Langer R.; Amiji M. Poly(ethylene oxide)-Modified Poly(beta-amino ester) Nanoparticles as a pH-Sensitive System for Tumor-Targeted Delivery of Hydrophobic Drugs: Part 2. In Vivo Distribution and Tumor Localization Studies. Pharm. Res. 2005, 22, 2107–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasir J. K.; Labhasetwar V. Biodegradable Nanoparticles for Cytosolic Delivery of Therapeutics. Adv. Drug Delivery. Rev. 2007, 59, 718–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescalchin A.; Detzer A.; Wecke M.; Overhoff M.; Wuensche W.; Sczakiel G. Cellular Uptake and Intracellular Release are Major Obstacles to the Therapeutic Application of siRNA: Novel Options by Phosphorothioate-Stimulated Delivery. Expert Opin. Biol. Ther. 2007, 7, 1531–1538. [DOI] [PubMed] [Google Scholar]
- Sonawane N. D.; Szoka F. C. Jr.; Verkman A. S. Chloride Accumulation and Swelling in Endosomes Enhances DNA Transfer by Polyamine−DNA Polyplexes. J. Biol. Chem. 2003, 278, 44826–44831. [DOI] [PubMed] [Google Scholar]
- Hu Y.; Litwin T.; Nagaraja A. R.; Kwong B.; Katz J.; Watson N.; Irvine D. J. Cytosolic Delivery of Membrane-Impermeable Molecules in Dendritic Cells Using pH-Responsive Core−Shell Nanoparticles. Nano Lett. 2007, 7, 3056–3064. [DOI] [PubMed] [Google Scholar]
- Cohen J. L.; Almutairi A.; Cohen J. A.; Bernstein M.; Brody S. L.; Schuster D. P.; Frechet J. M. Enhanced Cell Penetration of Acid-Degradable Particles Functionalized with Cell-Penetrating Peptides. Bioconjugate Chem. 2008, 19, 876–881. [DOI] [PubMed] [Google Scholar]
- Yang S. C.; Bhide M.; Crispe I. N.; Pierce R. H.; Murthy N. Polyketal Copolymers: A New Acid-Sensitive Delivery Vehicle for Treating Acute Inflammatory Diseases. Bioconjugate Chem. 2008, 19, 1164–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan M. J.; Kasturi S. P.; Yang S. C.; Pulendran B.; Murthy N. The Stimulation of CD8+ T Cells by Dendritic Cells Pulsed with Polyketal Microparticles Containing Ion-Paired Protein Antigen and Poly(inosinic acid)-Poly(cytidylic acid). Biomaterials 2009, 30, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan M. J.; Murthy N. Polyketal Nanoparticles: A New pH-Sensitive Biodegradable Drug Delivery Vehicle. Bioconjugate Chem. 2005, 16, 1340–1342. [DOI] [PubMed] [Google Scholar]
- Paramonov S. E.; Bachelder E. M.; Beaudette T. T.; Standley S. M.; Lee C. C.; Dashe J.; Frechet J. M. J. Fully Acid-Degradable Biocompatible Polyacetal Microparticles for Drug Delivery. Bioconjugate Chem. 2008, 19, 911–919. [DOI] [PubMed] [Google Scholar]
- Akinc A.; Anderson D. G.; Lynn D. M.; Langer R. Synthesis of Poly(beta-amino ester)s Optimized for Highly Effective Gene Delivery. Bioconjugate Chem. 2003, 14, 979–988. [DOI] [PubMed] [Google Scholar]
- Kohane D. S.; Tse J. Y.; Yeo Y.; Padera R.; Shubina M.; Langer R. Biodegradable Polymeric Microspheres and Nanospheres for Drug Delivery in the Peritoneum. J. Biomed. Mater. Res., A 2006, 77, 351–361. [DOI] [PubMed] [Google Scholar]
- Lynn D. M.; Amiji M. M.; Langer R. pH-Responsive Polymer Microspheres: Rapid Release of Encapsulated Material within the Range of Intracellular pH. Angew. Chem., Int. Ed. 2001, 40, 1707–1710. [PubMed] [Google Scholar]
- Lynn D. M.; Anderson D. G.; Putnam D.; Langer R. Accelerated Discovery of Synthetic Transfection Vectors: Parallel Synthesis and Screening of a Degradable Polymer Library. J. Am. Chem. Soc. 2001, 123, 8155–8156. [DOI] [PubMed] [Google Scholar]
- Lynn D. M.; Langer R. Degradable Poly(β-amino esters): Synthesis, Characterization, and Self-Assembly with Plasmid DNA. J. Am. Chem. Soc. 2000, 122, 10761–10768. [Google Scholar]
- Anderson D. G.; Lynn D. M.; Langer R. Semi-automated Synthesis and Screening of a Large Library of Degradable Cationic Polymers for Gene Delivery. Angew. Chem., Int. Ed. 2003, 42, 3153–3158. [DOI] [PubMed] [Google Scholar]
- Hofmann D.; Entrialgo-Castano M.; Kratz K.; Lendlein A. Knowledge-Based Approach towards Hydrolytic Degradation of Polymer-Based Biomaterials. Adv. Mater. 2009, 21, 3237–3245. [DOI] [PubMed] [Google Scholar]
- Smith R. C.; Leung A.; Kim B.-S.; Hammond P. T. Hydrophobic Effects in the Critical Destabilization and Release Dynamics of Degradable Multilayer Films. Chem. Mater. 2009, 21, 1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Fredin N. J.; Janz J. F.; Sun B.; Lynn D. M. Structure/Property Relationships in Erodible Multilayered Films: Influence of Polycation Structure on Erosion Profiles and the Release of Anionic Polyelectrolytes. Langmuir 2006, 22, 239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z.; Song Y.; Engbersen J. F. J.; Lok M. C.; Hennink W. E.; Feijen J. A Versatile Family of Degradable Non-viral Gene Carriers Based on Hyperbranched Poly(ester amine)s. J. Controlled Release 2005, 109, 317–329. [DOI] [PubMed] [Google Scholar]
- Broaders Kyle E.; Cohen Joel A.; Beaudette Tristan T.; Bachelder Eric M.; Frechet Jean M. J. Acetalated Dextran is a Chemically and Biologically Tunable Material for Particulate Immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 5497–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R.; Standley S. M.; Frechet J. M. J. Synthesis and Degradation of pH-Sensitive Linear Poly(amidoamine)s. Macromolecules 2007, 40, 452–457. [Google Scholar]
- Kim M. S.; Hwang S. J.; Han J. K.; Choi E. K.; Park H. J.; Kim J. S.; Lee D. S. pH-Responsive PEG-poly(beta-amino ester) Block Copolymer Micelles with a Sharp Transition. Macromol. Rapid Commun. 2006, 27, 447–451. [Google Scholar]
- Kim M. S.; Lee D. S.; Choi E.-K.; Park H.-J.; Kim J.-S. Modulation of Poly(beta-amino ester) pH-Sensitive Polymers by Molecular Weight Control. Macromol. Res. 2005, 13, 147–151. [Google Scholar]
- Reineke T. M. Poly(glycoamidoamine)s: Cationic Glycopolymers for DNA Delivery. J. Polym. Sci., Part A 2006, 44, 6895–6908. [Google Scholar]
- Heath W. H.; Palmieri F.; Adams J. R.; Long B. K.; Chute J.; Holcombe T. W.; Zieren S.; Truitt M. J.; White J. L.; Willson C. G. Degradable Crosslinkers and Strippable Imaging Materials for Step-and-Flash Imprint Lithography. Macromolecules 2008, 41, 719–726. [Google Scholar]
- Dalwadi G.; Benson H.; Chen Y. Comparison of Diafiltration and Tangential Flow Filtration for Purification of Nanoparticle Suspensions. Pharm. Res. 2005, 22, 2152–2162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.