The liver executes a multitude of functions in human physiology such as detoxification or hormone synthesis. Consequently, hepatic dysfunction, be it inborn or acquired, can lead to diverse pathological manifestations as for instance argininemia or hypercholesterolemia. To analyze liver function at the cellular scale and to investigate underlying genotype–phenotype relationships, a growing number of high-throughput data has been generated in recent years. However, mechanistic computational models are equally important in order to interpret and contextualize this data and to obtain a holistic understanding of human liver functionality per se.
In a first attempt to capture human metabolism in a rigorous quantitative sense, two generic stoichiometric models at genome scale have been developed recently (Duarte et al, 2007; Ma et al, 2007). They correlate pathway information with gene annotation thereby describing basic metabolic biochemistry. These models successfully identified selected drug targets, whereas investigation of tissue-specific metabolism was only possible by additionally considering gene-expression measurements, which revealed flux activity of disease-related enzymes in different organs (Shlomi et al, 2008). This approach, however, cannot compensate for the lacking tissue-specific metabolic networks, which are essential to analyze modes of drug action by probing strategies of structural network intervention in silico.
Now two research groups have overcome this limitation and independently developed genome-scale stoichiometric models of human liver metabolism, thus allowing in-depth mechanistic insights into hepatic functioning (Gille et al, 2010; Jerby et al, 2010). Interestingly, the approaches for network reconstruction chosen by the respective groups are fairly different even though both have started from the same generic metabolic models. Gille et al performed a large-scale analysis of bibliomic data and subsequently reconciled the network named HepatoNet1 by hand. In comparison, Jerby et al used a model-building algorithm (MBA) for network reconstruction, which has a broad applicability to generic models in general. Regarding the resulting compartmentalization, both models consider cytosol, mitochondria, lysosome, endoplasmatic reticulum and nucleus, but peroxisome, bile canaliculus and sinusoidal space are unique to HepatoNet1, whereas the MBA-based model includes a generic extracellular space. This divergence in overall model structure is also reflected in the total number of network reactions, which is higher for HepatoNet1 (2539 versus 1827 reactions), albeit a direct comparison of both models is difficult due to different exchange reactions and compound names. In this respect, development of a consensus network such as previously undertaken for yeast (Herrgard et al, 2008) will be a rewarding endeavor for the future.
Both models were subsequently tested for an accurate description of both gluconeogenesis and detoxification of ammonia, two central functions of liver metabolism. In addition, Gille et al show the accuracy of their model by validating the fulfillment of 442 metabolic tasks of various degrees of complexity. These can be considered as hepatic equivalents to simpler cellular objective functions identified in microbial cells (Schuetz et al, 2007). Jerby et al implicitly account for similar information, though presumably to a lesser extent, by structural inclusion of tissue-specific ‘omics' data and additionally evaluate the validity of their model by predicting hepatic flux distributions and metabolic biomarkers. Finally, both groups compare the predictive performance of their hepatic models to the earlier generic models of human metabolism and demonstrate a considerable improvement in both cases. The newly developed models therefore represent promising platforms for further mechanistic studies of liver function.
With human metabolic models coming of age, medical, diagnostic and pharmaceutical applications are now becoming feasible. Network models offer a structural scaffold onto which high-throughput data sets can be mapped in order to analyze experimental findings mechanistically. Hence, the impact of structural network modifications and interventions by genetic variation or drug administration can be investigated by simulating the resulting effect on metabolic states. Potential diagnostic applications are, for instance, model-based identification of specific biomarkers with altered levels in biofluids (Shlomi et al, 2009) or mapping of disease networks to human metabolism (Lee et al, 2008).
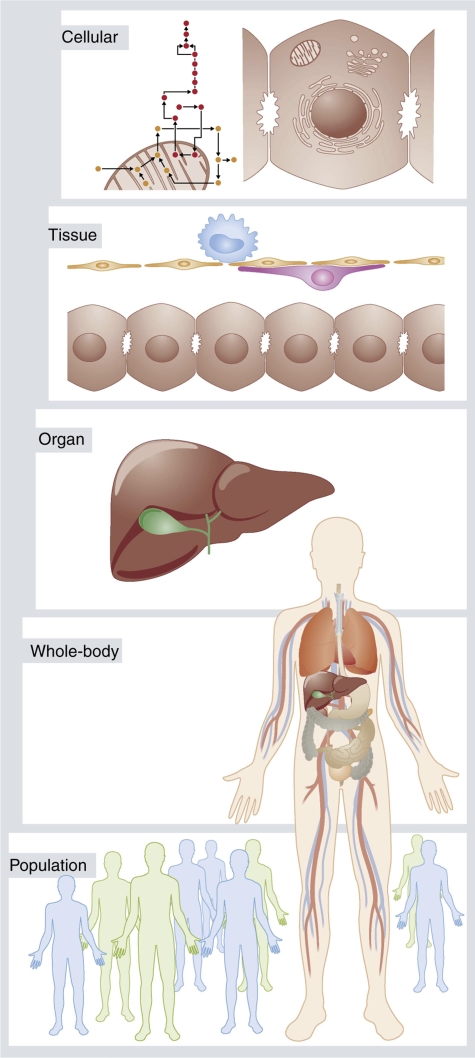
Hepatic metabolism closely interacts with whole-body physiology. Hence, the next step clearly requires moving from the cellular level to the organ scale, for example by consideration of hepatic zonation, and ultimately expansion to the organism level (Figure 1). With such fully integrated models, it will be possible to analyze hepatic metabolism in a clinical, patient-level context and, for instance, simulate the impact of metabolic activity on pharmacokinetic drug distribution or, contrariwise, investigate changes in biomarker levels in response to external stimuli (de Graaf et al, 2009).
Figure 1.
A multiscale representation of human physiology. Genome-scale models of human liver metabolism can be incorporated in organ models and may then be analyzed at a whole-body context. Specific phenotypes observed in population-wide studies (marked in blue and green) can thus be assigned mechanistically to a particular genotype.
Population-wide genotype information regarding metabolic activity has already been used in whole-body models (Willmann et al, 2009). There is reasonable hope that, in future, such computer-based models will greatly help to mechanistically explain pathophysiological states and support the development of targeted therapeutic strategies. With the genome-scale representations of hepatic metabolism by Gille et al and Jerby et al at hand, detailed models will become available, assigning a specific genetic background to an observed phenotype. Ultimately, such multiscale models will help to explain the complex interactions of the various networks in the human body from the cellular level up to the organism scale in an integrative way, thereby creating a comprehensive picture of whole-body physiology in its entirety.
Footnotes
The author declares that he has no conflict of interest.
References
- de Graaf AA, Freidig AP, De Roos B, Jamshidi N, Heinemann M, Rullmann JA, Hall KD, Adiels M, van Ommen B (2009) Nutritional systems biology modeling: from molecular mechanisms to physiology. PLoS Comput Biol 5: e1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BO (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA 104: 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille C, Bölling C, Hoppe A, Bulik S, Hoffmann S, Hübner K, Karlstädt A, Ganeshan R, König M, Rothers K, Weidlich M, Behre J, Holzhütter HG (2010) HepatoNet1: a comprehensive metabolic reconstruction of the human hepatocyte for the analysis of liver physiology. Mol Syst Biol 6: 411 10.1038/msb201062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrgard MJ, Swainston N, Dobson P, Dunn WB, Arga KY, Arvas M, Bluthgen N, Borger S, Costenoble R, Heinemann M, Hucka M, Le Novere N, Li P, Liebermeister W, Mo ML, Oliveira AP, Petranovic D, Pettifer S, Simeonidis E, Smallbone K et al. (2008) A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol 26: 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerby L, Shlomi T, Ruppin E (2010) Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol Syst Biol 6: 401 10.1038/msb201056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DS, Park J, Kay KA, Christakis NA, Oltvai ZN, Barabási AL (2008) The implications of human metabolic network topology for disease comorbidity. Proc Natl Acad Sci USA 105: 9880–9885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Sorokin A, Mazein A, Selkov A, Selkov E, Demin O, Goryanin I (2007) The Edinburgh human metabolic network reconstruction and its functional analysis. Mol Syst Biol 3: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz R, Kuepfer L, Sauer U (2007) Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol Syst Biol 3: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E (2008) Network-based prediction of human tissue-specific metabolism. Nat Biotechnol 26: 1003–1010 [DOI] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Ruppin E (2009) Predicting metabolic biomarkers of human inborn errors of metabolism. Mol Syst Biol 5: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann S, Edginton AN, Coboeken K, Ahr G, Lippert J (2009) Risk to the breast-fed neonate from codeine treatment to the mother: a quantitative mechanistic modeling study. Clin Pharmacol Ther 86: 634–643 [DOI] [PubMed] [Google Scholar]