An actin-regulated importin α/β-dependent extended bipartite NLS directs nuclear import of MRTF-A
The transcriptional coactivator MRTF-A/Mal binds G-actin, which sequesters it in the cytoplasm. In this study, Treisman and colleagues identify an unusual bipartite nuclear localisation signal in MRTF-A, and show that importin α/β-mediated import through this NLS is directly inhibited by G-actin binding.
Keywords: actin, importin, MRTF, NLS, SRF
Abstract
Myocardin-related transcription factors (MRTFs) are actin-regulated transcriptional coactivators, which bind G-actin through their N-terminal RPEL domains. In response to signal-induced actin polymerisation and concomitant G-actin depletion, MRTFs accumulate in the nucleus and activate target gene transcription through their partner protein SRF. Nuclear accumulation of MRTFs in response to signal is inhibited by increased G-actin level. Here, we study the mechanism by which MRTF-A enters the nucleus. We show that MRTF-A contains an unusually long bipartite nuclear localisation signal (NLS), comprising two basic elements separated by 30 residues, embedded within the RPEL domain. Using siRNA-mediated protein depletion in vivo, and nuclear import assays in vitro, we show that the MRTF-A extended bipartite NLS uses the importin (Imp)α/β-dependent import pathway, and that import is inhibited by G-actin. Interaction of the NLS with the Impα–Impβ heterodimer requires both NLS basic elements, and is dependent on the Impα major and minor binding pockets. Binding of the Impα–Impβ heterodimer to the intact MRTF-A RPEL domain occurs competitively with G-actin. Thus, MRTF-A contains an actin-sensitive nuclear import signal.
Introduction
Regulated nuclear entry and export has a critical role in controlling the activity of the MRTF transcriptional coactivators. These new G-actin-binding proteins regulate a subset of genes controlled by serum response factor (SRF), linking their transcription to changes in actin dynamics (Sotiropoulos et al, 1999; Cen et al, 2003; Miralles et al, 2003). In resting cells, MRTF-A (also known as MAL and MKL1) rapidly shuttles in and out of the nucleus, its cytoplasmic location being promoted by interaction of G-actin with its N-terminal RPEL domain (Miralles et al, 2003; Posern et al, 2004; Vartiainen et al, 2007; Guettler et al, 2008). Serum-induced RhoA activation promotes F-actin assembly, resulting in the depletion of G-actin pool and reduced actin–MRTF-A interaction, which inhibits actin-dependent MRTF-A export, thereby promoting its nuclear accumulation (Vartiainen et al, 2007). Interestingly, although MRTF-A nuclear import occurs constitutively in fibroblasts and its rate does not increase upon G-actin depletion, it is inhibited upon overexpression of G-actin (Vartiainen et al, 2007). Nuclear import of MRTF-A requires a putative import signal (B2) within the RPEL domain (Miralles et al, 2003; Vartiainen et al, 2007), but the nature of this signal and its functional relationship with actin binding has not been investigated.
Although proteins smaller than ∼40 kDa in size can traverse the NPC by passive diffusion, nuclear accumulation of larger proteins is energy-dependent and is mediated by specialised import receptors, the majority of which belong to the Impβ superfamily (Görlich and Kutay, 1999; Fried and Kutay, 2003; Poon and Jans, 2005; Stewart, 2007). These receptors, which continuously transit between the cytoplasm and the nucleus, interact with import signals in the cargo protein, either directly or through adaptor proteins such as Impα (Görlich and Kutay, 1999; Pemberton and Paschal, 2005; Poon and Jans, 2005; Stewart, 2007). Nuclear localisation signals (NLS) can be operationally defined as required for nuclear import in their natural context, functional when attached to an heterologous protein, and sufficient for effective interaction with the import receptors or receptor complexes (Damelin et al, 2002; Lange et al, 2007). Classical NLS sequences consist either of a single basic element, as in SV40 large T, or are bipartite, comprising two basic elements separated by a 10–12 residue linker, as in nucleoplasmin (Dingwall and Laskey, 1991; Robbins et al, 1991). Recent studies have identified atypical bipartite NLS that contain extended linkers (see Corredor et al, 2010; Lange et al, 2010). Short NLS sequences engage only one of the two binding pockets of the adaptor Impα, whereas bipartite ones interact simultaneously with both (Fontes et al, 2000, 2003; Kosugi et al, 2009). Impα in turn associates with Impβ through its Impβ-binding (IBB) domain to assemble a complex competent for import (Gorlich et al, 1995; Lange et al, 2007). At least some atypical bipartite NLS also interact with the classical Impα/β-dependent import machinery (Lange et al, 2010).
In this study, we demonstrate that the RPEL domain of MRTF-A contains an unusually extended bipartite NLS with a 30-residue linker, and that the integrity of both its basic elements is required for serum-induced MRTF-A nuclear accumulation. Using siRNA approaches, in vitro nuclear import assays and protein interaction experiments, we demonstrate that MRTF-A uses the Impα–Impβ complex to enter the nucleus, and that actin both inhibits nuclear import in vitro and competes with the Impα–Impβ complex for binding to the extended NLS. The MRTF-A thus contains an actin-sensitive nuclear import signal.
Results
The RPEL domain contains two basic clusters required for MRTF-A nuclear import
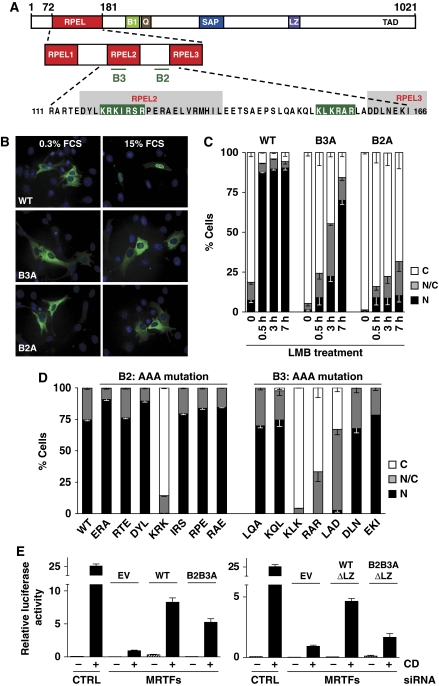
The MRTF-A RPEL domain, together with the 72 residues N-terminal to it, are sufficient to confer actin-regulated nucleocytoplasmic shuttling to heterologous proteins, such as GFP or pyruvate kinase (Miralles et al, 2003; Vartiainen et al, 2007; Guettler et al, 2008). We previously demonstrated that a short basic sequence between RPEL2 and RPEL3, termed B2, is necessary for MRTF-A nuclear import (Miralles et al, 2003; Vartiainen et al, 2007). As our preliminary experiments suggested that this sequence does not function as an effective NLS element (see below), we sought to identify other sequences required for MRTF-A nuclear import. Inspection of the RPEL domain sequence revealed an additional short basic element, 119KRKIRSR125, embedded within RPEL2 sequences mediating actin contact (Mouilleron et al, 2008), to which we will refer as B3 (Figure 1A).
Figure 1.
The MRTF-A RPEL domain contains two basic elements required for nuclear import. (A) Domain organisation of MRTF-A. RPEL, RPEL domain; B1, basic region 1; Q, Q-rich region; SAP (SAP, SAF-AIB, Acinus, Pias) domain; LZ, leucine zipper motif; TAD, transcription activation domain. The RPEL domain and motifs (pfam PF02755) are shown in red and the basic boxes are indicated with bars (B2 and B3). Bottom: sequence of the RPEL domain fragment containing the two basic boxes. (B) Mutation of B2 or B3 blocks serum-induced MRTF-A nuclear accumulation. Cells were transiently transfected with wild type or mutants of MRTF-A–GFP, starved for 24 h and stimulated with 15% serum for 30 min. The localisation of the proteins in NIH 3T3 cells was visualised by fluorescence microscopy. (C) The B2 and B3 mutations block MRTF-A import. Localisation of MRTF-A proteins after leptomycin B treatment (30 nM) was assessed as in panel B. C, cytoplasmic; N/C, pancellular; N, nuclear (⩾100 cells per condition). (D) Alanine-scanning mutagenesis of the B2 and B3 regions. Wild-type MRTF-A–GFP or the mutants in three consecutive residues were analysed for localisation after serum stimulation as in panel C. (E) The extended NLS is required for full reporter activation. B16F2 cells were treated with MRTF-A/MRTF-B siRNA and transfected with expression plasmids for MRTF-A, either containing or lacking the leucine zipper sequence, with MRTF activation by Cytochalasin D treatment (2 μM), as indicated. Error bars represent s.e.m. values (n=4).
An alanine substitution mutant of the three consecutive B3 basic residues 119KRK121 (B3A hereafter) localised to the cytoplasm in resting cells, and failed to accumulate in the nucleus on serum stimulation, as did B2A, an alanine substitution mutation of B2 residues 152KLK154 (Figure 1B). To confirm that B3 is involved in nuclear import, we tested whether the B3A mutation impaired nuclear accumulation of MRTF-A after treatment with leptomycin B (LMB), which blocks nucleocytoplasmic shuttling in resting cells by inactivating exportin 1/Crm1. The B3A mutant exhibited more pronounced cytoplasmic localisation in untreated cells, and, although wild-type MRTF-A became entirely nuclear after a 30-min LMB treatment, the B3A mutant showed little change from untreated cells (Figure 1C). Unlike the B2A mutant, however, which showed very little nuclear accumulation even over a 7-h LMB treatment, B3A did accumulate in the nucleus at later time points (Figure 1C), suggesting either that the B2 element can act as a weak autonomous NLS, or that the alanine substitution does not completely inactivate this element (see below). Taken together, the data identify the B3 element as an additional basic sequence required for efficient MRTF-A nuclear import.
We carried out alanine-scanning mutagenesis through the regions including the B3 and B2 elements to define them more rigorously. Within B3, only mutation of the basic triplet prevented MRTF-A nuclear accumulation in serum-stimulated cells, whereas within the B2 region, mutations in three successive triplets (152KLKRARLAD160), all located within the RPEL2–RPEL3 linker sequence, impaired serum-induced nuclear accumulation (Figure 1D). In contrast, mutations at neighbouring residues implicated in actin contacts led to increased MRTF-A nuclear accumulation in resting cells (Supplementary Figure S1), consistent with our previous results (Mouilleron et al, 2008).
To test the significance of the NLS for MRTF-A function, we used siRNA treatment to deplete cells of endogenous MRTFs. Expression of an siRNA-resistant form of MRTF-A rescued both basal and serum-induced transcription in cells, and this was significantly reduced by mutation of the NLS (Figure 1E). The effect of the NLS mutation was more pronounced in the context of MRTF-A lacking its leucine zipper, suggesting that at least part of the residual activity is due to import with residual MRTF-A.
The basic elements constitute an extended bipartite NLS
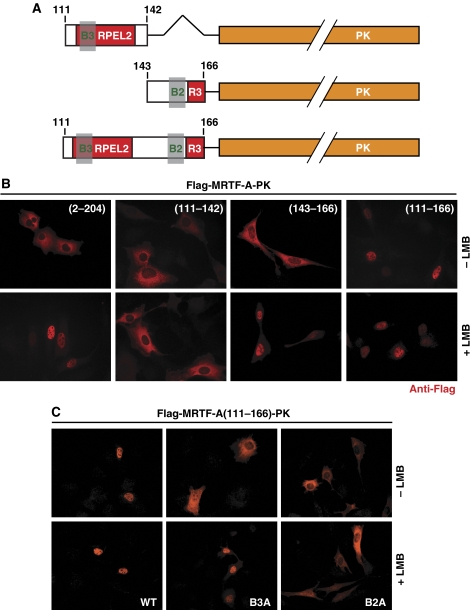
As independent mutation of either the B2 or B3 sequences effectively abolished MRTF-A nuclear import, we speculated that together the two elements might constitute a bipartite nuclear import signal. To test whether this is the case, we assessed the subcellular localisation of fusion proteins in which peptide sequences encompassing B2, B3 or both elements were joined to the normally cytoplasmic pyruvate kinase protein (Figure 2A). Fusion proteins containing either B2 or B3 alone were predominantly cytoplasmic (Figure 2B). In contrast, the fusion protein containing the entire putative NLS sequence (111/166–PK) localised exclusively to the nucleus, and this was dependent on the integrity of the two basic elements, as seen with MRTF-A itself (Figure 2C). Actin overexpression did not cause relocalisation of 111/166–PK to the cytoplasm (data not shown), consistent with the observation that the entire RPEL domain is required for correct MRTF-A regulation (Miralles et al, 2003; Vartiainen et al, 2007; Guettler et al, 2008).
Figure 2.
The B2 and B3 elements constitute an extended bipartite NLS sufficient for nuclear import of a heterologous protein. NIH 3T3 cells were transfected with the PK fusion proteins and analysed by immunofluorescence microscopy 24 h later with or without a 30-min treatment with 30 nM leptomycin B (LMB). (A) Schematic representation of pyruvate kinase (PK) fusion proteins. (B) The B2 and B3 elements constitute a bipartite NLS, and the B2 element possesses weak autonomous NLS activity. (C) Nuclear accumulation of MRTF-A(111–166)–PK requires intact B2 and B3 elements.
The basic elements of some bipartite NLS exhibit some ability to function as weak monopartite NLS, which may be revealed when nuclear export is inhibited (Kosugi et al, 2009; Corredor et al, 2010). To test whether this is also the case with the MRTF-A extended NLS, we evaluated the behaviour of the peptide–PK fusion proteins in cells treated with LMB. In this assay, the B2–PK fusion protein displayed LMB-dependent nuclear accumulation, indicating that this element does possess autonomous NLS activity in this context (Figure 2B). In contrast, the B3 element did not display autonomous NLS activity either alone or in the context of the B2+B3 peptide with the B2 element mutated to alanine (Figure 2C). Taken together these results identify the B2 and B3 elements as parts of an unusually extended bipartite nuclear import sequence.
Impβ1 activity is required for MRTF-A nuclear import in vivo
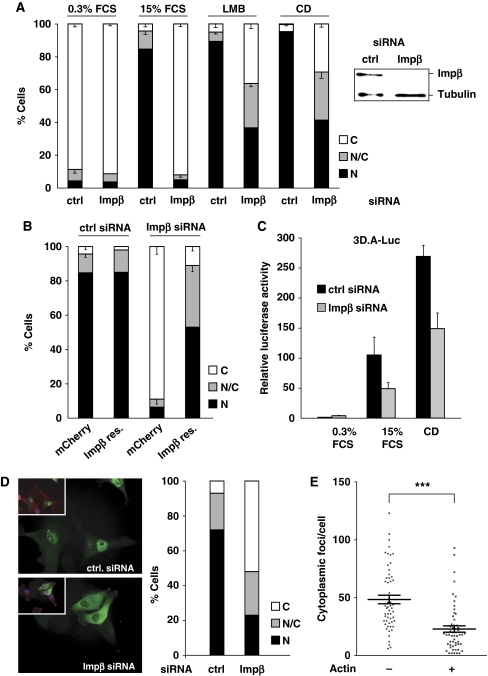
We next sought to identify the import machinery involved in MRTF-A nuclear import. As Impβ has been shown to function as a nuclear import receptor for proteins containing a conventional bipartite NLS, we tested its involvement in MRTF-A nuclear import by use of siRNA to silence its expression. Knockdown of Impβ in NIH 3T3 cells stably expressing MRTF-A–GFP resulted in the substantial inhibition of LMB -induced MRTF-A nuclear accumulation (Figure 3A). Similar results were obtained when MRTF-A nuclear accumulation was induced by cytochalasin D, which interferes with actin–MRTF-A interaction, and thereby blocks actin-mediated MRTF-A export (Vartiainen et al, 2007). The effect of Impβ depletion on serum-stimulated nuclear accumulation was more marked (Figure 3A). Probably, this reflects the fact that serum stimulation, unlike CD or LMB treatment, does not completely block nuclear export (Vartiainen et al, 2007), although we cannot exclude the possibility that basal import involves additional Impβ-independent import signal(s) active only in unstimulated conditions. Expression of an siRNA-resistant Impβ derivative restored nuclear accumulation of MRTF-A in response to serum stimulation, indicating that the effect was specific (Figure 3B). Depletion of Impβ also impaired serum- or cytochalasin D-induced activation of an SRF reporter gene (Figure 3C). In MDA-MB-231 cells, in which MRTF-A is nuclear even under resting conditions (Medjkane et al, 2009), Impβ depletion restored its cytoplasmic localisation (Figure 3D).
Figure 3.
Importin β activity is required for MRTF-A nuclear accumulation. (A) Impβ was silenced in R332 cells (NIH 3T3 cells stably expressing MRTF-A–GFP) using RNA interference, and the localisation of the protein was analysed under different conditions. FCS, fetal calf serum; CD, Cytochalasin D (2 μM); LMB, leptomycin B. (⩽100 cells per point, n=3 independent experiments, error bars indicate s.e.m. values). Right: western blotting showing Impβ depletion (B) Similar as in panel A, serum stimulation. The Impβ knockdown was rescued by transient transfection of an siRNA-resistant form of Impβ–mCherry. mCherry empty plasmid was used as control. (C) Inhibition of SRF reporter 3D.A-Luc activation after Impβ depletion. Three independent experiments were performed; error bars represent s.e.m. values. (D) The localisation of transiently transfected MRTF-A–GFP in resting MDA-MB-231 cells. Phalloidin staining in red. Right: quantification. (E) Impβ–MRTF-A interaction detected by proximity ligation assay, using anti-impβ and anti-MRTF-A antibodies, is reduced in cells transfected with a β-actin expression plasmid, identified by coexpressed GFP marker. PLA was scored as cytoplasmic foci per cell (error bar indicates s.e.m. values; n=55; ***P<0.001, unpaired Mann–Whitney test).
To confirm directly that Impβ interacts with MRTF-A in vivo, we used an in situ proximity ligation assay, in which a subset of target protein interactions are visualised in fixed cells as discrete foci generated by localised fluorescent rolling-circle amplification reactions dependent on the close proximity of antibodies against the target proteins (Soderberg et al, 2006). In this assay, MRTF-A–Impβ interactions were detectable as distinct cytoplasmic foci, number of which was substantially reduced on overexpression of β-actin (Figure 3E), which impairs nuclear import of MRTF-A (Vartiainen et al, 2007).
These results show that Impβ is involved in MRTF-A nuclear import, but give no insight into the potential role of Impα proteins, which function in many cases as heterodimers with Impβ. Although siRNAs targeting individual Impα genes were also tested for the ability to affect MRTF-A nuclear accumulation, in no case did we observe significant inhibition of MRTF-A nuclear import (data not shown). Given the in vitro data described below, this probably reflects functional redundancy among Impα proteins (Goldfarb et al, 2004; see Discussion section).
Impα and impβ are both required for MRTF-A NLS function in vitro
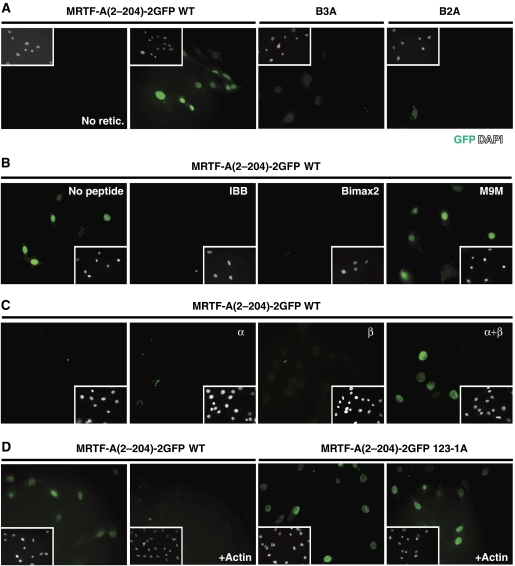
To investigate MRTF-A nuclear import directly, and to gain insight into whether it requires Impα, we developed an in vitro nuclear import assay. Semi-permeabilised cells were incubated with recombinant GFP-tagged MRTF-A derivatives, together with reticulocyte lysate as a source of import factors, and an energy source. MRTF-A(2–204)–2GFP, which exhibits similar regulation to intact MRTF-A in vivo (Guettler et al, 2008), was imported into nuclei in this assay; import was dependent on soluble import factors, as it was inhibited by RanQ69L, which mimics RanGTP, dissociating importins from their cargoes (data not shown). Mutation of basic elements B2 and B3 also prevented nuclear import of MRTF-A(2–204)–2GFP (Figure 4A). The in vitro assay thus faithfully reproduces MRTF-A import.
Figure 4.
Analysis of the MRTF-A nuclear import in vitro. (A) The in vitro nuclear import assay. NIH 3T3 cells were semi-permeabilised with digitonin and nuclear import of recombinant MRTF-A(2–204)–2GFP or mutant derivatives was analysed. Where indicated, reticulocyte lysate was omitted. Addition of 5 μM RanQ69L blocked import (data not shown). (B) Peptide competition analysis. Impα−Impβ is required for MRTF-A(2–204)–2GFP import. Competitor peptides (10 μM) were added to import reactions as indicated: IBB, Impβ-binding domain peptide that blocks Impα–Impβ interactions; bimax2, high affinity Impα-binding peptide; M9 M, Impβ2-specific NLS peptide. (C) Impα–Impβ heterodimers are sufficient for import in vitro. Import assays were carried out with purified cofactors, including Impα, Impβ or both as indicated. (D) Interaction of actin with the RPEL domain inhibits nuclear import. Import reactions were performed with wild-type MRTF-A(2–204)–2GFP or its 123-1A mutant derivative that cannot bind actin (Vartiainen et al, 2007). Import reactions contained latrunculin B–actin (2 μM) where indicated.
Impβ functions together with Impα for import of many of its cargoes (Lange et al, 2007). Consistent with the involvement of Impα, nuclear import of MRTF-A(2–204)–2GFP was inhibited by the bimax2 peptide, a high-affinity Impα ligand derived from iterative mutational analysis of the SV40 large T NLS (Kosugi et al, 2008), and was also blocked by an Impα IBB peptide, which disrupts Impα–Impβ interactions (Figure 4B). In contrast, import was not affected by the M9M peptide, an Impβ2-specific competitor peptide derived from the transportin PY-type NLS (Cansizoglu et al, 2007). These results strongly suggest that Impα–Impβ heterodimers are required for activity of the MRTF-A NLS. To test whether they are sufficient, we used a reconstituted import system containing recombinant NTF2, RanGDP, RanGAP, RanBP1 and importins. In this assay, the combination of Impα3 and Impβ was sufficient for import, but neither protein functioned alone (Figure 4C).
Actin overexpression inhibits MRTF-A nuclear accumulation in vivo, whereas RPEL domain mutants that cannot bind actin in vitro exhibit constitutive nuclear localisation in vivo and are insensitive to actin overexpression (Miralles et al, 2003; Vartiainen et al, 2007; Guettler et al, 2008). Consistent with these observations, addition of actin (as nonpolymerizable latrunculin B (LatB)–actin) to the in vitro assay prevented nuclear import of MRTF-A(2–204)–2GFP (Figure 4D). A mutant version of the MRTF-A(2–204)–2GFP protein containing a mutation that abrogates actin binding (Vartiainen et al, 2007; Guettler et al, 2008) was efficiently imported into nuclei in vitro, but was insensitive to LatB–actin addition (Figure 4D). Together, these data show that actin directly inhibits nuclear import of MRTF-A, and this requires its interaction with the RPEL domain.
Actin inhibits interaction of the MRTF-A NLS with Impα–Impβ in vitro
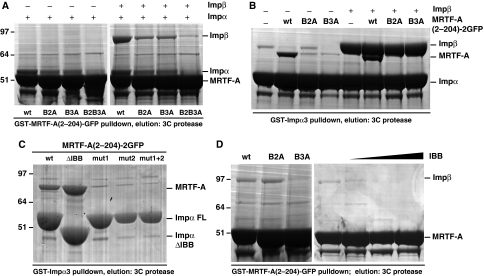
To study the interaction of the Impα−Impβ heterodimer with the MRTF-A NLS in more detail, we analysed their interaction in vitro. In glutathione S-transferase (GST) pulldown experiments, GST–MRTF-A(2–204)–GFP recruited recombinant Impα3 and the Impα−Impβ heterodimer (Figure 5A). Recovery was significantly impaired by mutation of the B2 or B3 basic elements of the NLS, and effectively abolished by their simultaneous mutation (Figure 5A). Similar results were obtained when GST–Impα3 was used as the affinity matrix, although the effects of the B2 and B3 mutations were more marked, probably reflecting limiting concentration of MRTF-A(2–204)–2GFP (Figure 5B). Classical monopartite NLS elements interact with the Impα major binding pocket, whereas bipartite NLS C- and N-terminal basic elements interact with the major and minor binding pockets, respectively (Conti et al, 1998; Conti and Kuriyan, 2000). In pulldown experiments using recombinant Impα3 derivatives, effective interaction with the MRTF-A extended NLS was dependent on the integrity of both binding pockets, and increased upon removal of the N-terminal Impα IBB domain (Figure 5C). Peptide array experiments confirmed that residues involved in Impα interaction encompassed the B2 element identified in the alanine scanning experiments, including residues overlapping the N-terminal helix of RPEL3 (Supplementary Figure S2; see Discussion section). Taken together with the functional data, these results show that MRTF-A nuclear import requires interaction of the Impα−Impβ heterodimer with the basic elements of the extended NLS.
Figure 5.
The MRTF-A extended bipartite NLS binds Impα3–Impβ heterodimers. (A) Impα3 and Impα3–Impβ interact with B2 and B3. GST–MRTF-A(2–204)–GFP derivatives were used for pulldown of recombinant Impα3 (left) and Impα3–Impβ (right). Bound proteins were freed from the matrix using 3C protease and analysed by SDS–PAGE. (B) Assays were performed as in panel A, but using GST–Impα3 as affinity matrix. (C) Interaction of Impα with the MRTF-A extended NLS requires both its major and minor binding pockets. Assays were performed with GST–Impα3 (mut1 and mut2 correspond to mutations in major and minor binding pockets, respectively). (D) Impβ interacts with the B3 element. Left, MRTF-A(2–204)–GFP derivatives were used for pulldown of recombinant Impβ as in panel A. Right, pulldown experiments using wild-type MRTF-A(2–204)–GFP were performed in the presence of increasing concentrations of IBB peptide (0.1–10 μM).
Impβ could be also recovered directly, although with low efficiency, in pulldown assays with GST–MRTF-A(2–204)–GFP, and this interaction was dependent on the B3 element but not on B2 (Figure 5D). The Impβ–B3 interaction was competed by the IBB peptide, indicating that the Impβ sequences involved overlap with those mediating dimerisation with Impα (Figure 5D). The Impβ–B3 interaction cannot be sufficient for import, which also requires B2 (see Discussion section).
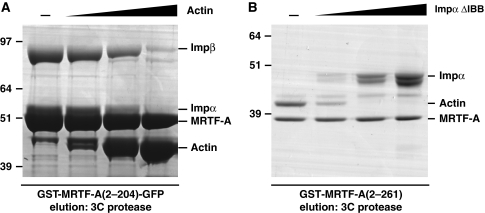
Actin binding inhibits MRTF-A nuclear import both in vitro and in vivo, and our data demonstrate that the basic importin-binding elements within the RPEL domain are intimately associated with its actin-binding sites: B3 is occluded by actin contacts with RPEL2 (Mouilleron et al, 2008), whereas B2 abuts RPEL3. We tested whether the inhibitory effect of actin on MRTF-A nuclear import reflected simple competition for importin binding by titrating LatB–actin into Impα–Impβ pulldown assays with GST–MRTF-A(2–204)–GFP. As LatB–actin input was increased, recovery of Impα–Impβ was correspondingly reduced (Figure 6A). Similarly, titration of recombinant Impα3 ΔIBB into reactions containing MRTF-A(2–261) and LatB–actin reduced recovery of actin (Figure 6B). Thus, actin and Impα–Impβ compete for interaction with the MRTF-A RPEL domain.
Figure 6.
Binding of the extended bipartite NLS to Impα3–Impβ heterodimers is actin sensitive. (A) GST–MRTF-A(2–204)–GFP was used for pulldown of Impα3–Impβ in the presence of increasing amounts of Latrunculin B–actin (0.25–10 μM). (B) GST–MRTF-A(2–261) was used for pulldown of actin in the presence of increasing amounts of Impα–ΔIBB.
Discussion
In this study, we studied the nuclear import mechanism of MRTF-A. We observed that the RPEL domain contains an extended bipartite nuclear localisation signal comprising two basic elements separated by a 30-residue linker. Both basic elements are required for effective MRTF-A nuclear import, and the extended NLS can confer import on a heterologous protein. The nuclear import of MRTF-A is dependent on the classical nuclear import pathway, using Impβ and the adaptor protein Impα, which interact with the NLS basic elements. Our findings support a recent proposal that the definition of a bipartite classical NLS should be relaxed to include linkers 20–30 residues in length (Lange et al, 2010). Such extended linkers have been identified in the Impα–Impβ-dependent import of yeast Ty1 integrase and Rrp4 (Lange et al, 2010) and BIV-1 Rev (Corredor et al, 2010), whereas widely separated basic elements are also necessary for import of Smad4 (Xiao et al, 2003), topoisomerase II (Kim et al, 2002) and cFLIP-L (Katayama et al, 2010). The MRTF-A NLS basic elements are intimately associated with its actin-binding RPEL motifs, and actin binds competitively with the Impα−Impβ heterodimer, directly inhibiting MRTF-A nuclear import. The MRTF-A extended NLS therefore represents an actin-sensitive nuclear import signal. The possibility remains, however, that MRTF-A contains Impα–Impβ-independent NLS not detected in our experiments.
The Impα−Impβ pathway, which may be responsible for import of 50% of all nuclear proteins in yeast, has been extensively characterised (Goldfarb et al, 2004; Lange et al, 2007). In this study, we have demonstrated that human Impα3–Impβ can mediate MRTF-A import, but at least six genes in the human genome encode for Impα proteins (Goldfarb et al, 2004; Mason et al, 2009), and these exhibit a broad functional redundancy (Nadler et al, 1997; Kohler et al, 1999, 2002; Goldfarb et al, 2004; Quensel et al, 2004; Yasuhara et al, 2007). For example, hypoxia-inducible factors (HIFs) were observed to bind Impα1, 3, 5 and 7 in vitro (Depping et al, 2008), whereas nuclear factor kappa B (NF-κB) nuclear import relies mostly on closely related Impα3 and Impα4 (Fagerlund et al, 2005). We think it is likely that Impα3 functions redundantly in MRTF-A import with other Impα family members, as we were unable to see any significant defect in serum-induced MRTF-A nuclear accumulation on siRNA-induced depletion of individual family members, including Impα3. As we have also observed partial relocalisation of myocardin from the nucleus to the cytoplasm on Impβ silencing (MK Vartiainen, unpublished data), we speculate that import of all three myocardin-family members may be dependent on the classical Impα−Impβ pathway.
Our biochemical studies indicate that Impα−Impβ heterodimers mediate MRTF-A import in vitro, and that direct interaction between Impα and both the B3 and B2 basic elements is required for Impα−Impβ recruitment. The NLS-binding domain of Impα is formed from 10 armadillo (ARM) repeats, and contains two binding pockets formed by repeats 2–4 and 7–9, known as the major and minor binding sites, respectively (Conti et al, 1998; Conti and Kuriyan, 2000). Monopartite NLS elements, such as that from SV40 large T, can interact with either the major or minor sites, whereas the N- and C-terminal basic elements of a conventional bipartite NLS interact with the minor and major sites respectively, with the 10–12 residue matching the distance between them and making few if any specific contacts (Conti and Kuriyan, 2000; Fontes et al, 2000, 2003). The extended bipartite MRTF-A NLS also uses the two Impα-binding pockets, and its longer linker sequence may therefore be looped out from the complex. We observed that the B2 element of the extended NLS, but not the B3 element, retains a weak ability to act autonomously, and suggest that it interacts with the major Impα-binding pocket. We propose that the functionally effective complex formation with Impα is dependent on the additional interaction between the NLS B3 element and the Impα minor binding pocket. These observations, which are consistent with other recent observations (Kosugi et al, 2009; Corredor et al, 2010), begin to blur the distinction between bipartite and classical monopartite NLS elements.
Our results indicate that the B2 element of the extended NLS also encompasses acidic residues C-terminal to the basic residues implicated in pocket binding, and peptide array experiments indicate that these residues are required for Impα binding. Previous studies have implicated residues outside the basic elements in recognition of both conventional and bipartite NLS by Impα (Kosugi et al, 2008, 2009). We also observed that Impβ alone exhibits affinity for the B3 basic element, and that the surface involved in this interaction overlaps with that which interacts with Impα. This interaction cannot be sufficient for nuclear import, however, which also requires the B2 element. Perhaps Impβ–B3 interaction facilitates assembly of Impα–Impβ–MRTF-A complex. More detailed biochemical analysis of the MRTF-A NLS interaction with the two importins, including structural studies, is underway to address these issues.
The activity of nuclear import and nuclear export signals can be controlled through regulation of their interactions with importins and exportins, respectively (Kaffman and O'Shea, 1999; Poon and Jans, 2005). Such regulation can occur through intramolecular masking of the import signal, frequently by phosphorylation at sites within or near the signal itself, as in the case of yeast Pho4 at Ser152 (Kaffman et al, 1998), or by occlusion of the signal through interaction of the potential cargo with other proteins, as in the case of IκB–NFκB interaction (Beg et al, 1992). Regulation of MRTF-A nuclear import is an example of the latter mechanism, as Impα–Impβ competes with G-actin for access to the NLS, which is located within the MRTF-A actin-binding RPEL domain. This competition is probably direct, as the B3 basic element is embedded within sequences physically masked by RPEL2-actin interactions, whereas the B2 basic element is in close apposition to RPEL3. It remains unclear whether the utilisation of the extended bipartite NLS is functionally significant in this context, or whether it has been selected because the shorter NLS elements cannot be successfully embedded within functional G-actin-binding sites. The RPEL domain contains multiple actin-binding sites, however, and it is tempting to speculate that the widely separated basic elements allow the activity of the NLS to be fine-tuned according to the stoichiometry of actin binding. This possibility, as well as the mechanism by which actin binding promotes MRTF nuclear export, will be interesting subjects for future studies.
Materials and methods
Plasmids
Sequences containing parts of the putative NLS of mouse MRTF-A were inserted into a pEF-Flag–PK vector for mammalian expression described previously (Sotiropoulos et al, 1999; Miralles et al, 2003). The B2A and B3A mutations used in the study are alanine substitutions of 152KLK154 and 119KRK121 within the B2 and B3 elements. The MRTF-A–GFP fusion protein has been described previously (Vartiainen et al, 2007). Dimerisation-defective MRTF-A ΔLZ lacked residues 612–652 (Miralles et al, 2003). In siRNA-resistant MRTF-A, 5′-518TGGAGCTGGTGGAGAAGAA536-3′ within RPEL 3 was changed to 5′-TGGAACTAGTAGAAAAGAA-3′. The MRTF-A 123-1A mutation has been described earlier (Vartiainen et al, 2007).
For bacterial expression of GST-tagged fusions, MRTF-A(2–204) linked to one or two GFP molecules, human Impα3 and Impβ sequences were expressed using a pET-41a(+) derivative as described previously (Vartiainen et al, 2007; Mouilleron et al, 2008). Impα3–ΔIBB lacks residues 1–55. The Impα3 major and minor pocket mutants contain two alanine substitutions at W179/N183 and W390/N394, respectively.
Mutations were introduced with quick-change exchange amino-acids method and the full sequences were verified by sequencing.
Proteins and peptides
The GST fusion proteins were purified from 2-l culture using 2 ml of glutathione–Sepharose. After elution, the GST moiety was cleaved off with 3C protease (20 μg) at 4°C overnight in 4 ml of buffer (50 mM Tris (pH 7.5), 100 mM NaCl, 1 mM DTT and protease inhibitors). Proteins were further purified using ion-exchange chromatography, followed by dialysis into binding buffer (50 mM Tris pH (7.5), 50 mM NaCl and 5 mM MgCl2) containing 2 mM DTT and protease inhibitors. LatB–actin was prepared as described previously (Morton et al, 2000; Hertzog et al, 2004; Mouilleron et al, 2008).
Peptides used in this study were synthesised and purified by the LRI Peptide Chemistry Laboratory. Peptide sequences were as follows:
IBB: AARLHRFKNKGKDSTEMRRRRIEVNVELRKAKKDDQMLKRRNVS
M9M: GGSYNDFGNYNNQSSNFGPMKGGNFGGRFEPYANPTKR
bimax2: RRRRRRKRKREWDDDDDPPKKRRRLD
In vitro import assays
The NIH 3T3 cells were grown on coverslips in 10% FCS DMEM until they reached 60–70% confluency. Semi-permeabilisation was performed in import buffer (250 mM sucrose, 20 mM HEPES (pH 7.3), 110 mM KOAc, 5 mM MgOAc, 0.5 mM EGTA, 2 mM DTT and protease inhibitors) with 20 μg/ml digitonin for 5–10 min. After washing with import buffer without detergent, the cells were incubated with 50 μl import mix (60% reticulocyte lysate dialysed against import buffer, 10 mM creatine phosphate, 0.5 mM ATP, 0.5 mM GTP and 20 U/ml creatine kinase in import buffer) containing 1 μM test proteins for 10 min at room temperature. For assays with purified components, the reticulocyte lysate was replaced with import buffer containing 0.8 μM NTF2, 4 μM RanGDP, 0.05 μM RanBP1, 0.4 μM RanGAP, with 1 μM Impα and 2 μM Impβ (final concentrations) as required. To stop the reaction, the cells were rinsed briefly with PBS and fixed with formaldehyde; nuclei were stained with DAPI. Competitor peptides were included at 10 μM and LatB–actin at 2 μM.
RNAi analysis
For the NLS studies, 70% confluent cells were transfected with 100 ng of plasmid (pyruvate kinase fusions, MRTF-A–GFP and their derivatives) using LipofectamineTM 2000 (Invitrogen) according to the manufacturers instructions. After transfection, cells were maintained in 0.3% FCS DMEM for 24 h and subsequently treated with 30 nM LMB, 0.5 μM LatB or 15% FCS for 30 min, unless stated otherwise. When required (PK fusions), the staining was performed with anti-Flag antibody (F7425, Sigma). The localisation of MRTF-A was scored as predominantly nuclear, predominantly cytoplasmic or pancellular in ⩾100 cells.
For siRNA transfections, B16F2 cells, NIH 3T3 cells or R332 cells (Vartiainen et al, 2007) were plated onto 24-well tissue culture plates at a density of 40 000 cells per well. The following day, cells were transfected with siRNA (mouse Impβ: 5′-CAACUGAAACCAUUAGUCA-3′ from Sigma Genosys; negative control: AllStars negative control from Qiagen; MRTF-A/MRTF-B, 5′-UGGAGCUGGUGGAGAAGAA-3′ (Medjkane et al, 2009)). On day 4, cells were transfected with DNA constructs when required (SRF reporter: 3D.A–Luc; control, pTK–renilla, mCherry, mCherry–Impβ, Flag–MRTF-A (wild type or NLS mutants)) as described previously (Vartiainen et al, 2007) and cultured in 0.3% FCS DMEM overnight. On the last day, cells were subjected to appropriate treatment and processed for western blotting, microscopy or luciferase assay as described earlier (Vartiainen et al, 2007). When using R332 cells, tetracycline (1 μg/ml) was added to the growth medium to induce the expression of MRTF-A–GFP.
Protein interaction assays
For GST pulldown analyis, glutathione–Sepharose was saturated with GST–MRTF-A(2–204)–GFP, GST–MRTF-A(2–261) or GST–Impα3 derivatives from bacterial lysates. After extensive washing, the beads were incubated with purified recombinant importins or MRTF-A(2–204)–2GFP derivatives for 3 h at 4°C in binding buffer (50 mM Tris (pH 7.5), 50 mM NaCl and 5 mM MgCl2). The resin was washed four times with binding buffer, after which 3C protease was used to cut MRTF-A or Impα3 derivatives off the GST. The resin was spun down and the supernatant was subjected to 4-12% SDS–PAGE. Gels were stained with Coomassie Brilliant Blue.
For proximity ligation analysis of protein interactions in cells (Soderberg et al, 2006), we used the Duolink II proprietary system according to the manufacturer's protocol, using antibodies against MRTF-A (C19, Santa Cruz Biotechnology) and Impβ (2811, Abcam).
Supplementary Material
Acknowledgments
We thank Dr R Depping for providing Impα plasmids; B Schmierer, M Fornerod, T Güttler and D Gohrlich for sharing reagents; N O'Reilly and the peptide synthesis lab for peptide synthesis; M Howell for proximity ligation assay data analysis; and S Guettler, C Hill, M Way and members of the Transcription Laboratory for stimulating discussions and helpful comments. This study was funded by Cancer Research UK. MV was the recipient of a long-term Fellowship from EMBO during the early stages of this project. The part of this study in MV's laboratory is funded by the Academy of Finland and the Finnish Cancer Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Beg AA, Ruben SM, Scheinman RI, Haskill S, Rosen CA, Baldwin AS Jr (1992) I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: a mechanism for cytoplasmic retention. Genes Dev 6: 1899–1913 [DOI] [PubMed] [Google Scholar]
- Cansizoglu AE, Lee BJ, Zhang ZC, Fontoura BM, Chook YM (2007) Structure-based design of a pathway-specific nuclear import inhibitor. Nat Struct Mol Biol 14: 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen B, Selvaraj A, Burgess RC, Hitzler JK, Ma Z, Morris SW, Prywes R (2003) Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol Cell Biol 23: 6597–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Kuriyan J (2000) Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure 8: 329–338 [DOI] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94: 193–204 [DOI] [PubMed] [Google Scholar]
- Corredor AG, St-Louis MC, Archambault D (2010) Molecular and biological aspects of the bovine immunodeficiency virus. Curr HIV Res 8: 2–13 [DOI] [PubMed] [Google Scholar]
- Damelin M, Silver PA, Corbett AH (2002) Nuclear protein transport. Methods Enzymol 351: 587–607 [DOI] [PubMed] [Google Scholar]
- Depping R, Steinhoff A, Schindler SG, Friedrich B, Fagerlund R, Metzen E, Hartmann E, Kohler M (2008) Nuclear translocation of hypoxia-inducible factors (HIFs): involvement of the classical importin-alpha/beta pathway. Biochim Biophys Acta 1783: 394–404 [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA (1991) Nuclear targeting sequences—a consensus? Trends Biochem Sci 16: 478–481 [DOI] [PubMed] [Google Scholar]
- Fagerlund R, Kinnunen L, Kohler M, Julkunen I, Melen K (2005) NF-{kappa}B is transported into the nucleus by importin {alpha}3 and importin {alpha}4. J Biol Chem 280: 15942–15951 [DOI] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Jans D, Brinkworth RI, Kobe B (2003) Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha. J Biol Chem 278: 27981–27987 [DOI] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Kobe B (2000) Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J Mol Biol 297: 1183–1194 [DOI] [PubMed] [Google Scholar]
- Fried H, Kutay U (2003) Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci 60: 1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA (2004) Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol 14: 505–514 [DOI] [PubMed] [Google Scholar]
- Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey RA, Hartmann E, Prehn S (1995) Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol 5: 383–392 [DOI] [PubMed] [Google Scholar]
- Görlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Guettler S, Vartiainen MK, Miralles F, Larijani B, Treisman R (2008) RPEL motifs link the serum response factor cofactor MAL but not myocardin to Rho signaling via actin binding. Mol Cell Biol 28: 732–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog M, van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, Didelot G, Preat T, Knossow M, Guittet E, Carlier MF (2004) The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell 117: 611–623 [DOI] [PubMed] [Google Scholar]
- Kaffman A, O'Shea EK (1999) Regulation of nuclear localization: a key to a door. Annu Rev Cell Dev Biol 15: 291–339 [DOI] [PubMed] [Google Scholar]
- Kaffman A, Rank NM, O'Shea EK (1998) Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev 12: 2673–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama R, Ishioka T, Takada S, Takada R, Fujita N, Tsuruo T, Naito M (2010) Modulation of Wnt signaling by the nuclear localization of cellular FLIP-L. J Cell Sci 123(Part 1): 23–28 [DOI] [PubMed] [Google Scholar]
- Kim KH, Kanbe T, Akashi T, Mizuguchi I, Kikuchi A (2002) Identification of a single nuclear localization signal in the C-terminal domain of an Aspergillus DNA topoisomerase II. Mol Genet Genomics 268: 287–297 [DOI] [PubMed] [Google Scholar]
- Kohler M, Fiebeler A, Hartwig M, Thiel S, Prehn S, Kettritz R, Luft FC, Hartmann E (2002) Differential expression of classical nuclear transport factors during cellular proliferation and differentiation. Cell Physiol Biochem 12: 335–344 [DOI] [PubMed] [Google Scholar]
- Kohler M, Speck C, Christiansen M, Bischoff FR, Prehn S, Haller H, Gorlich D, Hartmann E (1999) Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol Cell Biol 19: 7782–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Entani T, Takayama S, Tomita M, Yanagawa H (2008) Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chem Biol 15: 940–949 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H (2009) Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem 284: 478–485 [DOI] [PubMed] [Google Scholar]
- Lange A, McLane LM, Mills RE, Devine SE, Corbett AH (2010) Expanding the definition of the classical bipartite nuclear localization signal. Traffic 11: 311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH (2007) Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem 282: 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason DA, Stage DE, Goldfarb DS (2009) Evolution of the metazoan-specific importin alpha gene family. J Mol Evol 68: 351–365 [DOI] [PubMed] [Google Scholar]
- Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R (2009) Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol 11: 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R (2003) Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113: 329–342 [DOI] [PubMed] [Google Scholar]
- Morton WM, Ayscough KR, McLaughlin PJ (2000) Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat Cell Biol 2: 376–378 [DOI] [PubMed] [Google Scholar]
- Mouilleron S, Guettler S, Langer CA, Treisman R, McDonald NQ (2008) Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL. EMBO J 27: 3198–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler SG, Tritschler D, Haffar OK, Blake J, Bruce AG, Cleaveland JS (1997) Differential expression and sequence-specific interaction of karyopherin alpha with nuclear localization sequences. J Biol Chem 272: 4310–4315 [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM (2005) Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6: 187–198 [DOI] [PubMed] [Google Scholar]
- Poon IKH, Jans DA (2005) Regulation of nuclear transport: central role in development and transformation? Traffic 6: 173–186 [DOI] [PubMed] [Google Scholar]
- Posern G, Miralles F, Guettler S, Treisman R (2004) Mutant actins that stabilise F-actin use distinct mechanisms to activate the SRF coactivator MAL. EMBO J 23: 3973–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quensel C, Friedrich B, Sommer T, Hartmann E, Kohler M (2004) In vivo analysis of importin alpha proteins reveals cellular proliferation inhibition and substrate specificity. Mol Cell Biol 24: 10246–10255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C (1991) Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64: 615–623 [DOI] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3: 995–1000 [DOI] [PubMed] [Google Scholar]
- Sotiropoulos A, Gineitis D, Copeland J, Treisman R (1999) Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 98: 159–169 [DOI] [PubMed] [Google Scholar]
- Stewart M (2007) Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol 8: 195–208 [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R (2007) Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316: 1749–1752 [DOI] [PubMed] [Google Scholar]
- Xiao Z, Latek R, Lodish HF (2003) An extended bipartite nuclear localization signal in Smad4 is required for its nuclear import and transcriptional activity. Oncogene 22: 1057–1069 [DOI] [PubMed] [Google Scholar]
- Yasuhara N, Shibazaki N, Tanaka S, Nagai M, Kamikawa Y, Oe S, Asally M, Kamachi Y, Kondoh H, Yoneda Y (2007) Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nat Cell Biol 9: 72–79 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.