Abstract
Heme proteins play essential roles in biology, but little is known about heme transport inside mammalian cells or how heme is inserted into soluble proteins. We recently found that nitric oxide (NO) blocks cells from inserting heme into several proteins, including cytochrome P450s, hemoglobin, NO synthases, and catalase. This finding led us to explore the basis for NO inhibition and to identify cytosolic proteins that may be involved, using inducible NO synthase (iNOS) as a model target. Surprisingly, we found that GAPDH plays a key role. GAPDH was associated with iNOS in cells. Pure GAPDH bound tightly to heme or to iNOS in an NO-sensitive manner. GAPDH knockdown inhibited heme insertion into iNOS and a GAPDH mutant with defective heme binding acted as a dominant negative inhibitor of iNOS heme insertion. Exposing cells to NO either from a chemical donor or by iNOS induction caused GAPDH to become S-nitrosylated at Cys152. Expressing a GAPDH C152S mutant in cells or providing a drug to selectively block GAPDH S-nitrosylation both made heme insertion into iNOS resistant to the NO inhibition. We propose that GAPDH delivers heme to iNOS through a process that is regulated by its S-nitrosylation. Our findings may uncover a fundamental step in intracellular heme trafficking, and reveal a mechanism whereby NO can govern the process.
Heme proteins are essential to life and play key roles in electron transfer, oxidative transformations, free radical scavenging, signal transduction, and oxygen storage and transport (1–4). Among the four major types of heme found in different proteins (A, B, C, and O), heme B (Fe-protoporphyrin IX) is most common (5). The final step of heme biosynthesis occurs in the mitochondria (6). Because free heme is toxic to cells as a result of its potential to generate reactive oxygen species that can cause damage to DNA, proteins, and lipids (3), cells keep intracellular-free heme levels to the minimum (6–8) and heme homeostasis is one of the most highly regulated processes in mammals (9). Heme insertion into apo-proteins is a key posttranslational event that must take place within this context. Surprisingly, almost nothing is known about the mechanisms of intracellular heme transport and heme insertion into soluble apo-protein targets, or how these processes are regulated in mammals (6). This lack of knowledge is particularly true for the many B-type hemeproteins that reside in cells outside the mitochondria, like nitric oxide synthases (NOS), globins, and soluble guanylate cyclase. Because heme needs to travel from mitochondria to reach apo-protein targets in the endoplasmic reticulum or cytosol, intracellular heme transport has long been suspected to involve a protein carrier (6, 10). Over the years, many cytosolic heme binding proteins have been described (11) but none have been shown to function in intracellular heme transport or heme insertion reactions. In fact, the studies to date have not identified any cytosolic heme transport proteins in mammals and only suggest that heme insertion may be a postendoplasmic reticulum process and may involve a chaperone (HSP90) (12–15).
In 1996, we reported that heme insertion into the inducible NOS (iNOS) can be inhibited by NO (16). In that study, the NO was produced naturally by iNOS in cells and was found to act somewhere near or at the point of heme insertion. More recently, we showed that NO can inhibit cellular heme insertion into a broad range of mammalian proteins, including all three NOS enzymes, hemoglobin, catalase, and cytochrome P450 (17). Importantly, the mechanism of NO action did not involve changes in heme availability or effects of NO on cellular energy production or activation of soluble guanylate cyclase (17). The results implied that NO might act on an uncharacterized yet common cellular component or process to generally inhibit heme insertion into proteins. This concept served as a starting point to search for proteins that might drive and regulate intracellular heme insertion reactions in mammals. In our current study, we used iNOS as a model heme protein target, and identify a cytosolic protein that binds heme, functions in heme insertion, and is the focal point for NO regulation.
Results
GAPDH Is a NO-Sensitive Heme Binding Protein.
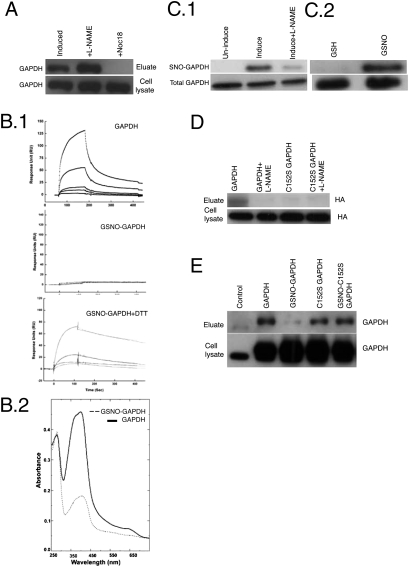
We used hemin-agarose affinity chromatography coupled with proteomic methods (protein 2D gel chromatography and MS) to identify the cytosolic heme binding proteins that are present in macrophage-like RAW 264.7 cells that were cultured with succinyl acetone (SA) to become heme-deficient (18) (Fig. S1). GAPDH was among the proteins we identified by this method (Fig. S1), confirming earlier reports that GAPDH can bind heme (11, 19). However, GAPDH was unusual because its binding to the hemin-agarose became greatly diminished if the cells (Fig. 1A) or the cell cytosols (Fig. S1) were treated with a NO donor (Noc18) or glutathione-NO (GSNO) before chromatography. We explored this relationship further using purified GAPDH. Adding heme to a solution of pure GAPDH formed a stable protein–heme complex that remained intact after passage through a sephadex PD-10 column (heme content typically ranged from 0.4 to 1.2 heme bound/GAPDH tetramer). However, treating GAPDH with GSNO before heme addition diminished its heme-binding property, as determined by size-exclusion chromatography or by surface plasmon resonance (SPR) (Fig. 1 B.1 and B.2). Exposing cells to NO donor commonly results in the S-nitrosylation of GAPDH (20). We therefore tested if DTT, which can reverse protein S-nitrosylation (20) could restore the heme-binding property of NO-treated GAPDH, and found that it could do so (Fig. 1B.1). This finding suggested that NO inhibited GAPDH heme binding by causing a reversible modification of a cysteine thiol.
Fig. 1.
S-nitrosylation of GAPDH regulates its heme binding. (A) Heme binding proteins from RAW264.7 cells induced by LPS and IFN-γ (± NOC-18 or L-NAME) were pulled down with hemin-agarose, eluted, separated by SDS/PAGE, and Western blotted with GAPDH antibody. Initial cell lysates contained equal amounts of GAPDH in all cases. (B.1) Representative sensograms of binding interaction between free heme (0–50 μM) and chip-immobilized human GAPDH, GSNO-treated GAPDH, and GSNO-treated GAPDH + DTT as measured in a Biacore 3000. (B.2) Equal amounts (20 μM) of unmodified or GSNO-treated GAPDH were incubated with 30 μM heme for 45 min at room temperature. The GAPDH-heme complexes were isolated using PD-10 columns and their UV-visible spectra are shown. The ratio of the absorbance at 410–280 nm indicates the heme content of each complex. (C) Biotin switch followed by neutravidin-linked Sepharose pull-downs compare the levels of SNO-GAPDH versus total GAPDH in (1) cell lysates from cytokine-induced RAW264.7 cells that either did or did not (+L-NAME) generate NO, and (2) pure GAPDH treated either with GSH or GSNO. Samples processed by SDS/PAGE and Western blot analysis. (D) Cell lysates from cytokine-induced (NO producing) RAW264.7 cells stably transfected with GAPDH-HA or C152S-GAPDH-HA underwent biotin switch and neutravidin-linked Sepharose pull-downs to compare their levels of S-nitrosylation. The C152S mutation protected GAPDH from S-nitrosylation. Samples processed by SDS/PAGE and Western blot analysis. (E) SA-pretreated RAW264.7 cells stably expressing GAPDH or the C152S mutant were treated with 400 μM GSNO or vehicle for 45 min followed by cell lysis and hemin agarose pull-down. GAPDH levels in the cell lysates and the hemin agarose eluates were probed by SDS/PAGE and Western blot analysis.
GAPDH Heme Binding Is Diminished upon S-nitrosylation at Cys-152.
We used the biotin switch method (21) and confirmed that GAPDH becomes S-nitrosylated (SNO-GAPDH) in cytokine-activated NO-generating cells or when pure GAPDH was treated with an NO donor (Fig. 1 C.1 and C.2). Interestingly, formation of SNO-GAPDH in cells occurred under the same NO treatment conditions that block cellular heme insertion into iNOS or other heme proteins (17). One conserved Cys residue in GAPDH has a low pKa (Cys152), and thus becomes preferentially S-nitrosylated in NO-generating cells or when NO donors are added (22). Indeed, we found that the C152S mutant of GAPDH did not become S-nitrosated when it was expressed in NO-generating cells (Fig. 1D). The C152S GAPDH had normal heme-binding properties despite being catalytically inactive (20), indicating that GAPDH heme binding is independent of its enzymatic function. Significantly, the heme-binding properties of C152S GAPDH were unaffected by treatment with NO donor in both intact cells or when tested in pure form (Fig. 1E and Fig. S2). Together, our findings show that GAPDH heme binding can be regulated by NO through a specific S-nitrosylation at Cys152.
Inducible NOS and GAPDH Interact in Cells.
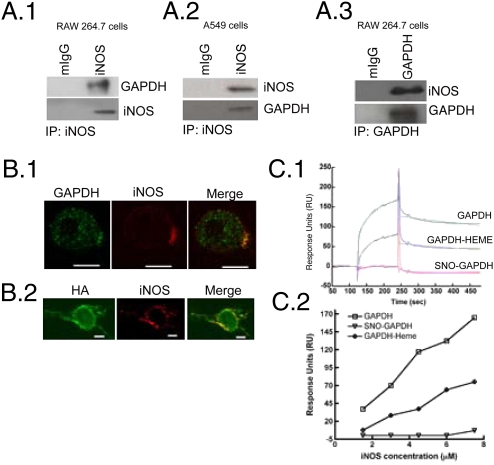
The above properties encouraged us to investigate a role for GAPDH in cellular heme insertion into iNOS. Immunoprecipitation of endogenous iNOS from two different cell types (mouse cytokine-activated RAW264.7 cells or human lung epithelium A549 cells) indicated that GAPDH is associated with iNOS (Fig. 2 A.1 and A.2). Reverse immunoprecipitation of GAPDH from cytokine-activated macrophages also pulled down iNOS, confirming their association (Fig. 2A.3). Blocking cellular NO synthesis with L-NAME increased the extent of iNOS-GAPDH association (Fig. S3). Immunofluorescence images obtained by confocal microscopy showed that GAPDH and iNOS colocalize in cytokine-activated macrophages (Fig. 2B.1). To understand what fraction of GAPDH binds to iNOS, we cotransfected HA-tagged GAPDH and iNOS in HEK cells. At 4 h posttransfection, we found HA-tagged GAPDH and iNOS colocalize, thereby indicating the freshly made GAPDH and iNOS were capable of associating in the cells (Fig. 2B.2).
Fig. 2.
Inducible NOS binds GAPDH. (A) Immunoprecipitation of iNOS from cell lysates of activated (A.1) RAW 264.7 or (A.2) A549 cells shows GAPDH binds iNOS. (A.3) Immunoprecipitation of GAPDH from cell lysates of activated RAW264.7 cells also pulls-down iNOS. (B.1) Confocal image of activated RAW264.7 cells stained for GAPDH (green), iNOS (red), and their merge is shown. (B.2) HEK 293 cells cotransfected with HA-tagged GAPDH and iNOS were stained for HA (green) and iNOS (red). Yellow spots on the merge images show the colocalization of iNOS and GAPDH. (Scale bar, 10 μm.) (C.1) Sensogram of the interaction between immobilized human GAPDH, GAPDH-heme, or SNO-GAPDH with murine iNOSfl protein at 7.5 μM, measured by Biacore 3000. (C.2) Sensogram data comparing the interactions of GAPDH, GAPDH-heme, or SNO-GAPDH with different indicated concentrations of murine iNOSfl is plotted.
We next explored the interaction between purified GAPDH and iNOS. SPR measurements using murine iNOS and immobilized human GAPDH showed they have a relatively strong interaction with estimated dissociation constant (Kd) = 0.6 nM (Fig. 2C and Fig. S4). Binding heme to the GAPDH before measurement only weakly antagonized the interaction, whereas S-nitrosylation of GAPDH abolished it (Fig. 2C). Purified endothelial NOS and neuronal NOS also interacted with GAPDH in similar SPR experiments (Fig. S4). Together, the data show that iNOS and GAPDH form stable complexes both in cells and in vitro, and that S-nitrosylation of GAPDH antagonizes complex formation.
Cellular Heme Insertion into iNOS Involves GAPDH and Is Regulated by S-nitrosylation of GAPDH at Cys152.
The above-mentioned properties of GAPDH provided a unique platform to investigate its role in cellular heme insertion and in NO control of the process. We recently developed a cell culture method (17) that allows study of posttranslational heme insertion into target apo-proteins over a 2-h period (Fig. S5). By focusing on heme incorporation into preformed apo-proteins in cells, incorporating a short time-window for observation, and preventing any new protein biosynthesis during the measurement, our method ensures that any effects on heme insertion we may observe do not result from changes in gene induction or protein expression that could otherwise occur under less-restrictive culture conditions. In this system, GSNO inhibited cellular heme insertion into iNOS in a dose-dependent manner, as judged by a spectroscopic measure of iNOS heme content or by the gain in NO synthesis activity, which strictly requires that heme become inserted into iNOS (Fig. S6).
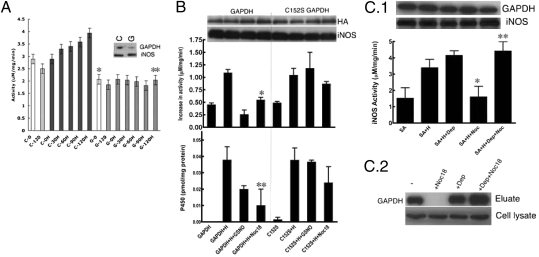
We used our cell culture method along with GAPDH knockdown or transient and stable gene-expression experiments to investigate a role for GAPDH in cellular heme insertion. Treating cells with si-GAPDH caused a significant reduction in GAPDH protein expression (more than 50%) and caused an almost complete inhibition of heme insertion into iNOS, as judged by its eliminating the time-dependent increase in iNOS NO synthesis activity that otherwise occurred during heme insertion (Fig. 3A). The GAPDH knockdown had no effect on the cellular ATP level (Fig. S7). In any case, NO inhibition of cellular heme insertion is known to be insensitive to changes in cellular energy levels (17). Overexpression of HA-tagged GAPDH increased the cell's capacity to insert heme into iNOS (Fig. S8) but did not prevent NO from inhibiting heme insertion into iNOS (Fig. 3B), as judged by measuring iNOS activity or heme content (Fig. 3B). In contrast, overexpressing the HA-tagged C152S GAPDH made heme insertion into iNOS resistant to the NO inhibition (Fig. 3B). These findings support a role for GAPDH in cellular heme insertion into iNOS, and imply that NO inhibits the process by causing a specific S-nitrosylation of GAPDH at Cys152.
Fig. 3.
Expression level and S-nitrosylation of GAPDH controls cellular heme insertion. (A) GAPDH-silenced SA-treated RAW264.7 cells were unable to insert heme (H) into apo-iNOS as measured by the gain in NO synthesis activity achieved over a 2-h (0–120 min) incubation of cells with heme. GAPDH (G) silenced vs. control (C) silenced (n = 3, mean ± SEM; *P = 0.006 vs. C0; **P = 0.008 vs. C120H by two-tailed Student's t test at equal variance). The inset shows the cells contained a reduced amount of GAPDH protein after silencing GAPDH (G) compared with control siRNA (C). Levels of iNOS protein expression were unaffected. (B) Overexpression of C152S GAPDH but not GAPDH makes cellular heme (H) insertion into apo-iNOS resistant to NO donor (400 μM GSNO or 250 μM Noc18) as measured by the gain in NO synthesis activity or P450 spectral measure of iNOS heme content (n = 3, mean ± SEM; *P = 0.024 vs. GAPDH+H; **P = 0.008 vs. GAPDH+H by two-tailed Student's t test at equal variance). Cell levels of GAPDH or iNOS protein were equivalent across the various treatments. (C.1) Addition of 5 nM deprenyl to the cell culture made heme insertion into apo-iNOS resistant to the inhibition by the NO donor Noc18 (n = 3, mean ± SEM; *P = 0.0101 vs. SA+H; and **P = 0.0222 vs. SA+Noc18 by two-tailed paired Student's t test). Protein levels were unaffected by the treatments as judged by Western blots. (C.2) The presence of 5 nM deprenyl in cultures of RAW 264.7 cells made the heme-binding property of cellular GAPDH resistant to Noc18, but did not affect GAPDH expression, as shown by hemin-agarose pull-downs and Western blotting for GAPDH.
We further explored this relationship using the pharmacologic drug deprenyl, which is a small molecule inhibitor that binds tightly to GAPDH and prevents its S-nitrosylation at Cys152 (23). Heme insertion into iNOS became resistant to NO inhibition in cells given deprenyl (Fig. 3C.1). This protection was associated with deprenyl enabling GAPDH to maintain its normal heme binding properties in cells that were treated with the NO donor (Fig. 3C.2). These results strengthen the link between GAPDH and cellular heme insertion into iNOS, and confirm that S-nitrosylation of GAPDH compromises this function.
K227A GAPDH Inhibits Heme Insertion into iNOS.
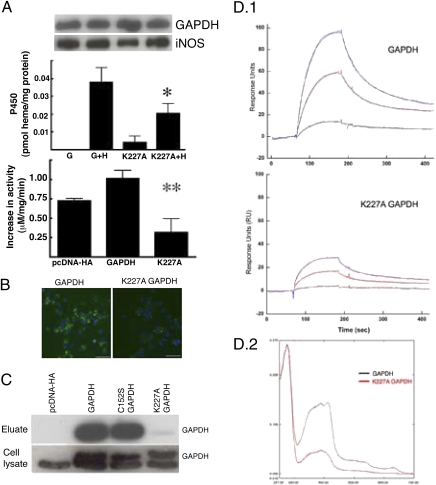
We next assessed whether mutants of GAPDH (i.e., Tyr41F, Lys162A, and Lys227A) that are known to affect some of its nonglycolytic functions (22, 24) would impact cellular heme insertion into iNOS. Overexpression of one mutant (K227A GAPDH) antagonized cellular heme insertion into iNOS by a mechanism that was independent of NO (Fig. 4 A and B). Visualization of heme-containing iNOS using our previously described fluorescent reagent, PIF (25), confirmed decreased levels of heme insertion in the K227A GAPDH overexpressing cells (Fig. 4B). The K227A mutation did not diminish GAPDH enzymatic activity (Fig. S9). We found that the K227A GAPDH had reduced binding to heme-agarose resin and to free heme in solution (Fig. 4 C and D). Thus, K227A GAPDH acted as a dominant negative inhibitor of cellular heme insertion into iNOS and this was associated with a defective heme binding property.
Fig. 4.
K227 GAPDH acts as a dominant negative inhibitor for heme insertion and displays a defective heme binding. (A) Overexpression of K227A GAPDH in SA-treated RAW264.7 cells antagonized heme insertion into apo-iNOS, as measured by iNOS NO synthesis activity and P450 spectral measurement (n = 3, mean ± SEM; *P = 0.048 vs. GAPDH+H; **P = 0.035 vs., GAPDH by two-tailed Student's t test at equal variance). There was similar expression of GAPDH or iNOS protein in the cell lysates as judged from Western blots. (B) PIF fluorescence staining for heme-containing iNOS was obtained during 2 h of heme incorporation in SA-treated, cytokine-induced RAW264.7 cells that were stably overexpressing either GAPDH or K227A GAPDH. A lesser heme incorporation into apo-iNOS is apparent in the K227A GAPDH-expressing cells. (Scale bar, 10 μm.) (C) Hemin agarose pull-downs from HEK cells that were transfected with HA-tagged GAPDH or the C152S and K227A mutants were separated by SDS/PAGE and probed with GAPDH antibody. K227A GAPDH exhibited defective binding to the hemin-agarose. Expression levels of native or HA-tagged GAPDH proteins in the cell lysates were equivalent. (D.1) Representative sensograms of binding interaction between free heme (0–25 μM) and chip-immobilized human GAPDH or K227A GAPDH protein as measured in a Biacore 3000. (D.2) UV-visible spectra recorded after PD-10 column separation of preformed heme complexes of GAPDH or K227A GAPDH show that less heme is retained by the mutant. The ratio of the absorbance at 410–280 nm indicates the heme content of each complex.
Endogenous NO Inhibits iNOS Heme Insertion Through Formation of SNO-GAPDH.
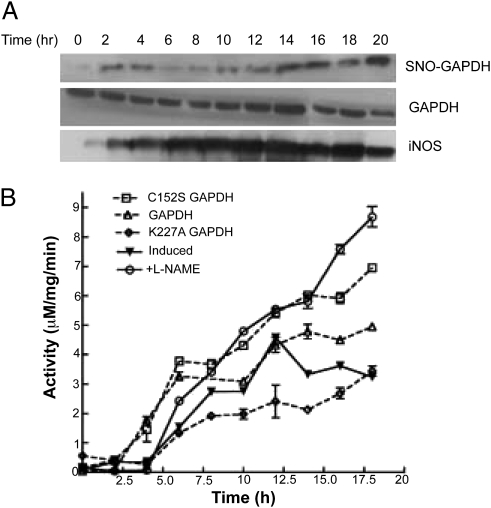
We next tested how endogenous NO formed by iNOS in activated RAW264.7 cells would correlate with SNO-GAPDH buildup and inhibition of iNOS heme insertion. Fig. 5A shows that SNO-GAPDH formation was an early event after the cells were activated with IFN-γ and LPS and appeared coincident with detectable iNOS protein expression. Fig. 5B shows that heme insertion into iNOS became progressively inhibited over time in the activated NO-generating cells, as judged by their iNOS activity reaching a plateau. This effect was prevented if a NOS inhibitor was added to the cells during the activation (L-NAME), as established previously (16). The different extents of iNOS heme insertion were also apparent from the ratio of iNOS dimer–monomer that was present in the cells after 18 h of activation (Fig. S10). In the same system, overexpression of the wild-type or C152S GAPDH proteins caused either a partial or near-complete protection against the effect of endogenous NO on heme insertion into iNOS (Fig. 5B and Fig. S11). In contrast, overexpression of K227A GAPDH further antagonized heme insertion (Fig. 5B). These results indicate that GAPDH and SNO-GAPDH are involved in controlling cellular heme insertion during the endogenous iNOS expression and NO production that occurs following immunostimulation.
Fig. 5.
Effect of endogeneous NO production on iNOS activity is rescued by L-NAME and C152S GAPDH: (A) Activated RAW264.7 cells were harvested every 2 h during the 0- to 20-h time-course and analyzed for SNO-GAPDH content by biotin-switch followed by pull-downs with neutravidin-linked Sepharose. Eluates were probed for GAPDH. Western analyses show the levels of iNOS and GAPDH proteins present in the cell supernatant samples. (B) Inducible NOS activity in the cell supernatant samples collected every 2 h during 0–20 h of postinduction (LPS and IFN-γ treatment) in RAW264.7 cells (± L-NAME), GAPDH, or mutant overexpressing stable cell lines.
GAPDH Enables Heme Transfer to Apo-iNOS in Vitro.
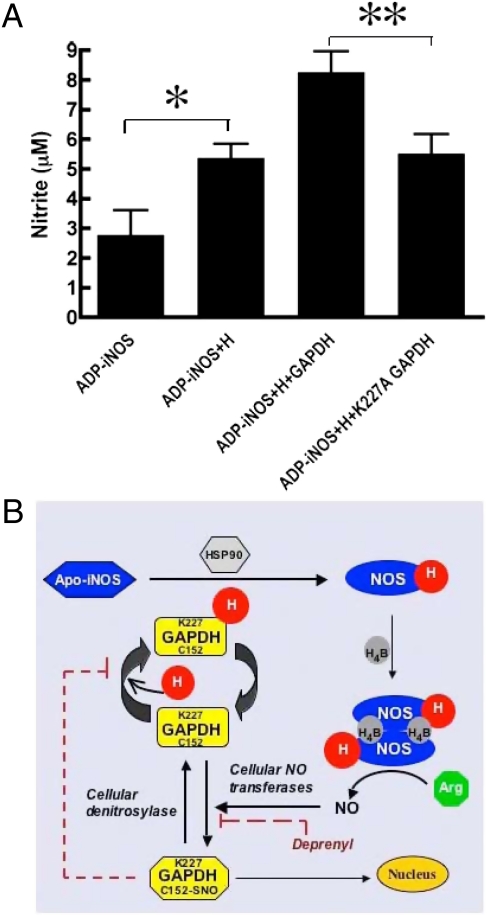
To test if GAPDH can deliver heme to iNOS, we performed in vitro studies using apo-iNOS protein that we isolated from SA-treated activated macrophages using 2′,5′-ADP-Sepharose beads. Incubation of the iNOS beads with a GAPDH-heme complex enabled heme incorporation into iNOS, as judged by the gain in NO synthesis activity (Fig. 6A). The presence of GAPDH facilitated the heme insertion reaction relative to what we observed by adding free heme. The effect of GAPDH was specific, because the K227A GAPDH mutant was unable to increase heme insertion into iNOS in otherwise identical experiments (Fig. 6A). Our in vitro data are consistent with GAPDH enabling heme insertion into iNOS.
Fig. 6.
GAPDH delivers heme to iNOS. (A) Apo-iNOS from cytokine activated SA-treated RAW264.7 cells was pulled down and incubated 45 min with heme (4 mM) or with GAPDH protein (2 mM) plus heme (4 mM). Heme insertion into iNOS was measured by the increase in NO synthesis activity. GAPDH facilitated heme delivery into iNOS but the K227A GAPDH mutant did not (n = 4, mean ± SEM; *P = 0.03; **P = 0.036 by two-tailed Student's t test at equal variance). (B) Model for GAPDH function in heme insertion and its regulation by S-nitrosylation is shown. See text for details.
Discussion
Large gaps exist in our understanding of heme protein assembly in mammals. In particular, there is almost no information on how heme travels from the mitochondria and becomes inserted into soluble heme proteins in cells, or how this process might be regulated (6, 26, 27). We approached this problem by focusing on the NO regulation of heme insertion to drive our discovery process, and by using a cell-culture protocol we developed (17) that only monitors posttranslational events (i.e., events that affect heme insertion into apo-protein that has already accumulated in cells). Our current study of iNOS heme insertion makes two unique and important contributions: (i) We identify GAPDH as the first cytosolic protein whose heme binding property is now tied to a heme insertion reaction taking place in cells, specifically heme insertion into iNOS. (ii) We uncover a mechanism whereby NO inhibits heme insertion into iNOS as a possible feedback mechanism to control inflammation. The mechanism relies on a site-specific S-nitrosylation of GAPDH, and appears to diminish GAPDH function in heme insertion by modifying GAPDH heme binding properties and its protein interactions.
Fig. 6B illustrates a model for cellular heme insertion that is consistent with our findings and describes a unique feedback mechanism for NO to limit assembly of active iNOS in cells: GAPDH binds heme (H) and can interact with apo-iNOS in cells. This process enables heme transfer and insertion into iNOS through a process that may also involve HSP90 (13) and possibly other cellular proteins. The heme-containing iNOS monomer then dimerizes and binds its H4B cofactor to form an active iNOS that can synthesize NO from L-arg (28). The NO produced, possibly through the actions of cellular thiols or NO transferase enzymes (29), causes GAPDH to become S-nitrosylated at Cys152 to form SNO-GAPDH. SNO-GAPDH has altered heme and protein-binding properties and its buildup in cells inhibits heme insertion into iNOS. Importantly, the SNO-GAPDH formation can be pharmacologically blocked with deprenyl (23), and this protects iNOS heme insertion against the NO inhibition. The level of SNO-GAPDH in cells may also become up- or down-regulated through the actions of cellular denitrosylase enzymes like GSNOR or Trx1 (30). Besides impacting cellular heme insertion, SNO-GAPDH is active in other cell functions, including its translocation to the nucleus to control apoptosis (22).
A new function for GAPDH in cellular heme insertion complements its emerging roles in the immune response, apoptosis, and cancer (31–34). Notably, none of these newer functions of GAPDH rely on its inherent dehydrogenase activity. In our case, this was most clearly shown by work with the K227A GAPDH mutant, which acts as a dominant negative inhibitor of iNOS heme insertion despite having normal dehydrogenase activity, and by the C152S mutant, as well as deprenyl, being protective despite their inhibiting GAPDH enzymatic activity (20, 35).
Our results appear to rule out an alternative mechanism for NO action, which would have NO bind directly to the heme during its transport in cells to somehow inhibit the heme insertion process (36). Given that NO has a good propensity to bind to heme, it is truly remarkable that NO impacts cellular heme insertion instead through a mechanism involving protein–S-nitrosylation. This finding underscores the growing importance of SNO protein modifications in a host of cellular processes (29). Because our work links an essential cellular process (heme protein assembly) to NO biology, it stands to impact many facets of human health and disease, including inflammation, hematopoesis, drug metabolism, oxidative stress, asthma, anemia, and the growth of intracellular parasites.
Material and Methods
Cell Culture.
RAW264.7 and HEK293 (ATCC) cells were cultured as described elsewhere (25). Cells were pretreated with 2,4-dioxoheptanoic acid (250 μM) for 2–3 d to make heme deficient. Cells were activated by following a mixture of cytokines, 25 μM LPS, and 10 units/mL of IFN-γ for RAW 264.7 cells or 10 ng/mL TNF-α, 0.5 ng/mL IL-1β, and 5,000 units/mL of human IFN-γ for A549 cells during 18 or 36 h, respectively, at 37 °C and 5% CO2 in a humidified incubator. For time-course experiments, RAW264.7 cells were harvested every 2 h postactivation during the period of 0–20 h followed by immediate washing, scraping, and freezing (−80 °C) of cells to store. Later, all of the samples were analyzed together. Heme insertion studies were done by adding 7.5 μM heme as shown in Fig. S5. Silencing of GAPDH in SA-RAW264.7 cells was done by transfection of 10 nM GAPDH/control siRNA with siPORT transfection reagent (Ambion Inc.). During the course of silencing, medium was supplemented with 1 mM pyruvate and after 36 h of transfection, cells were induced with LPS and IFN-γ. For generation of stable cell lines of GAPDH and mutants, constructs were transfected in RAW264.7 cells using lipofectamine 2000 followed by G418 (800 μM) selection. For reagent details, see SI Matrials and Methods.
Hemin Agarose Pull-Down.
Cell lysates were made using lysis buffer (Tris 10 mM, 0.5 M NaCl, pH = 7.4) followed by centrifugation at 10,000 × g for 25 min at 4 °C. Lysates containing equal amount of proteins were incubated with washed hemin agarose (37) for 1 h at 4 °C. Resin was thoroughly washed with buffer (three times, 15 min each) and then with high-salt buffer (1 M NaCl) followed by addition of SDS containing reducing sample buffer. Samples were analyzed by Western blotting.
Immunofluorescence.
Cells were cultured on glass coverslips and treated as per the experiment followed by staining protocol as described in SI Materials and Methods.
S-nitrosylation Biotin Witch Assay.
The biotin switch assay was performed as mentioned (21) and pull-downs from NeutrAvidin resin (Pierce) were probed by Western blotting.
P450 and iNOS Activity Measurement.
Cell lysates were processed as described elsewhere (16, 25). All of the experiments were repeated at least three times and the average of all is shown in the results.
Protein Purification.
GAPDH and mutant proteins were expressed using GST constructs in Escherichia coli and grown initially in100 mL luria broth for 16 h at 37 °C, followed by inoculation in 2 L of terrific broth. Cells were induced with 1 mM IPTG after the cell density reached to 0.8 to 1.0 OD, and were incubated for another 48 h at room temperature. Cells were harvested and purified using glutathione Sepharose according to the manufacturer's instructions (Amersham Biosciences). After thrombin cleavage (37 °C, 1 h), thrombin was removed with HiTrap Benzamidine FF affinity column (GE Healthcare).
In Vitro Reconstitution.
Apo-iNOS was pulled down from the cell lysates of SA-treated activated RAW264.7 cells using washed ADP-Sepharose for 2 h at 4 °C. Beads were thoroughly washed and resuspended with heme solutions or buffer alone. GAPDH proteins and heme were premixed for 10 min at room temperature before use. Following 45-min incubation at room temperature, beads were pulled down and washed thoroughly to remove any free heme. Beads were then used to measure the iNOS enzyme activity.
Determination of Protein–Protein/Ligand Interaction by SPR.
All SPR experiments were carried out using CM5 sensor chip and BIACORE 3000 instrument (Biacore). For details, see SI Materials and Methods.
UV-Visible Spectra.
The 20 μM of pure protein was mixed with 30 μM of heme in 150 μL of final HES-N buffer and the solution was incubated for 45 min at room temperature. The PD-10 columns (GE Healthcare) were washed with distilled water and equilibrated with HES-N buffer. The solution was allowed to separate on equilibrated PD-10 column and fractions were analyzed using a Hitachi U-3110 spectrophotometer.
Supplementary Material
Acknowledgments
We thank Dr. Satya P. Yadav for maintenance of the Biacore Facility (NIH-SIGRR016789-01A1), Drs. Serpil C. Erzurum and Saurav Misra for helpful discussions and critical reading of the manuscript, and Deborah Durra for excellent technical assistance. This work was supported by National Institute of Health Grants HL076491 and GM51491 (to D.J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008133107/-/DCSupplemental.
References
- 1.Mense SM, Zhang L. Heme: A versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 2006;16:681–692. doi: 10.1038/sj.cr.7310086. [DOI] [PubMed] [Google Scholar]
- 2.Choi AM. Heme oxygenase-1 protects the heart. Circ Res. 2001;89(2):105–107. [PubMed] [Google Scholar]
- 3.Camejo G, et al. Hemin binding and oxidation of lipoproteins in serum: Mechanisms and effect on the interaction of LDL with human macrophages. J Lipid Res. 1998;39:755–766. [PubMed] [Google Scholar]
- 4.Atamna H, Killilea DW, Killilea AN, Ames BN. Heme deficiency may be a factor in the mitochondrial and neuronal decay of aging. Proc Natl Acad Sci USA. 2002;99:14807–14812. doi: 10.1073/pnas.192585799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsiftsoglou AS, Tsamadou AI, Papadopoulou LC. Heme as key regulator of major mammalian cellular functions: Molecular, cellular, and pharmacological aspects. Pharmacol Ther. 2006;111:327–345. doi: 10.1016/j.pharmthera.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 6.Severance S, Hamza I. Trafficking of heme and porphyrins in metazoa. Chem Rev. 2009;109:4596–4616. doi: 10.1021/cr9001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hvidberg V, et al. Identification of the receptor scavenging hemopexin-heme complexes. Blood. 2005;106:2572–2579. doi: 10.1182/blood-2005-03-1185. [DOI] [PubMed] [Google Scholar]
- 8.Kumar S, Bandyopadhyay U. Free heme toxicity and its detoxification systems in human. Toxicol Lett. 2005;157(3):175–188. doi: 10.1016/j.toxlet.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Wu N, Yin L, Hanniman EA, Joshi S, Lazar MA. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbalpha. Genes Dev. 2009;23:2201–2209. doi: 10.1101/gad.1825809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latunde-Dada GO, Simpson RJ, McKie AT. Recent advances in mammalian haem transport. Trends Biochem Sci. 2006;31(3):182–188. doi: 10.1016/j.tibs.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Li X, et al. A novel approach for identifying the heme-binding proteins from mouse tissues. Genomics Proteomics Bioinformatics. 2003;1(1):78–86. doi: 10.1016/S1672-0229(03)01011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nauseef WM, McCormick S, Yi H. Roles of heme insertion and the mannose-6-phosphate receptor in processing of the human myeloid lysosomal enzyme, myeloperoxidase. Blood. 1992;80:2622–2633. [PubMed] [Google Scholar]
- 13.Billecke SS, et al. The role of hsp90 in heme-dependent activation of apo-neuronal nitric-oxide synthase. J Biol Chem. 2004;279:30252–30258. doi: 10.1074/jbc.M403864200. [DOI] [PubMed] [Google Scholar]
- 14.Bender AT, et al. Neuronal nitric-oxide synthase is regulated by the Hsp90-based chaperone system in vivo. J Biol Chem. 1999;274:1472–1478. doi: 10.1074/jbc.274.3.1472. [DOI] [PubMed] [Google Scholar]
- 15.Pratt WB, Morishima Y, Osawa Y. The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J Biol Chem. 2008;283:22885–22889. doi: 10.1074/jbc.R800023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albakri QA, Stuehr DJ. Intracellular assembly of inducible NO synthase is limited by nitric oxide-mediated changes in heme insertion and availability. J Biol Chem. 1996;271:5414–5421. doi: 10.1074/jbc.271.10.5414. [DOI] [PubMed] [Google Scholar]
- 17.Waheed SM, et al. Nitric oxide blocks cellular heme insertion into a broad range of heme proteins. Free Radic Biol Med. 2010;48:1548–1558. doi: 10.1016/j.freeradbiomed.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebert PS, Hess RA, Frykholm BC, Tschudy DP. Succinylacetone, a potent inhibitor of heme biosynthesis: Effect on cell growth, heme content and delta-aminolevulinic acid dehydratase activity of malignant murine erythroleukemia cells. Biochem Biophys Res Commun. 1979;88:1382–1390. doi: 10.1016/0006-291x(79)91133-1. [DOI] [PubMed] [Google Scholar]
- 19.Campanale N, et al. Identification and characterization of heme-interacting proteins in the malaria parasite, Plasmodium falciparum. J Biol Chem. 2003;278:27354–27361. doi: 10.1074/jbc.M303634200. [DOI] [PubMed] [Google Scholar]
- 20.Padgett CM, Whorton AR. S-nitrosoglutathione reversibly inhibits GAPDH by S-nitrosylation. Am J Physiol. 1995;269:C739–C749. doi: 10.1152/ajpcell.1995.269.3.C739. [DOI] [PubMed] [Google Scholar]
- 21.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001(86):pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 22.Hara MR, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 23.Hara MR, et al. Neuroprotection by pharmacologic blockade of the GAPDH death cascade. Proc Natl Acad Sci USA. 2006;103:3887–3889. doi: 10.1073/pnas.0511321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tisdale EJ, Artalejo CR. A GAPDH mutant defective in Src-dependent tyrosine phosphorylation impedes Rab2-mediated events. Traffic. 2007;8:733–741. doi: 10.1111/j.1600-0854.2007.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panda K, et al. Visualizing inducible nitric-oxide synthase in living cells with a heme-binding fluorescent inhibitor. Proc Natl Acad Sci USA. 2005;102:10117–10122. doi: 10.1073/pnas.0408972102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reedy CJ, Gibney BR. Heme protein assemblies. Chem Rev. 2004;104:617–649. doi: 10.1021/cr0206115. [DOI] [PubMed] [Google Scholar]
- 27.Mendel RR, Smith AG, Marquet A, Warren MJ. Metal and cofactor insertion. Nat Prod Rep. 2007;24:963–971. doi: 10.1039/b703112m. [DOI] [PubMed] [Google Scholar]
- 28.Tzeng E, Billiar TR, Robbins PD, Loftus M, Stuehr DJ. Expression of human inducible nitric oxide synthase in a tetrahydrobiopterin (H4B)-deficient cell line: H4B promotes assembly of enzyme subunits into an active dimer. Proc Natl Acad Sci USA. 1995;92:11771–11775. doi: 10.1073/pnas.92.25.11771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: A current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takasaki Y, et al. Glyceraldehyde 3-phosphate dehydrogenase is a novel autoantigen leading autoimmune responses to proliferating cell nuclear antigen multiprotein complexes in lupus patients. Int Immunol. 2004;16:1295–1304. doi: 10.1093/intimm/dxh131. [DOI] [PubMed] [Google Scholar]
- 32.Colell A, Green DR, Ricci JE. Novel roles for GAPDH in cell death and carcinogenesis. Cell Death Differ. 2009;16:1573–1581. doi: 10.1038/cdd.2009.137. [DOI] [PubMed] [Google Scholar]
- 33.Pumain R, Laschet J. A key glycolytic enzyme plays a dual role in GABAergic neurotransmission and in human epilepsy. Crit Rev Neurobiol. 2006;18:197–203. doi: 10.1615/critrevneurobiol.v18.i1-2.200. [DOI] [PubMed] [Google Scholar]
- 34.Hara MR, Snyder SH. Nitric oxide-GAPDH-Siah: A novel cell death cascade. Cell Mol Neurobiol. 2006;26:527–538. doi: 10.1007/s10571-006-9011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlile GW, et al. Reduced apoptosis after nerve growth factor and serum withdrawal: Conversion of tetrameric glyceraldehyde-3-phosphate dehydrogenase to a dimer. Mol Pharmacol. 2000;57(1):2–12. [PubMed] [Google Scholar]
- 36.Cooper CE. Nitric oxide and iron proteins. Biochim Biophys Acta. 1999;1411:290–309. doi: 10.1016/s0005-2728(99)00021-3. [DOI] [PubMed] [Google Scholar]
- 37.Tsutsui K, Mueller GC. Affinity chromatography of heme-binding proteins: An improved method for the synthesis of hemin-agarose. Anal Biochem. 1982;121:244–250. doi: 10.1016/0003-2697(82)90475-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.