Abstract
Although metazoan body plans are remarkably diverse, the structure and function of many embryonic regulatory genes are conserved because large changes would be detrimental to development. However, the fushi tarazu (ftz) gene has changed dramatically during arthropod evolution from Hox-like to a pair-rule segmentation gene in Drosophila. Changes in both expression and protein sequence contributed to this new function: ftz expression switched from Hox-like to stripes and changes in Ftz cofactor interaction motifs led to loss of homeotic and gain of segmentation potential. Here, we reconstructed ftz changes in a rigorous phylogenetic context. We found that ftz did not simply switch from Hox-like to segmentation function; rather, ftz is remarkably labile, having undergone multiple changes in sequence and expression. The segmentation LXXLL motif was stably acquired in holometabolous insects after the appearance of striped expression in early insect lineages. The homeotic YPWM motif independently degenerated multiple times. These “degen-YPWMs” showed varying degrees of homeotic potential when expressed in Drosophila, suggesting variable loss of Hox function in different arthropods. Finally, the intensity of ftz Hox-like expression decreased to marginal levels in some crustaceans. We propose that decreased expression levels permitted ftz variants to arise and persist in populations without disadvantaging organismal development. This process, in turn, allowed evolutionary transitions in protein function, as weakly expressed “hopeful gene variants” were coopted into alternative developmental pathways. Our findings show that variation of a pleiotropic transcription factor is more extensive than previously imagined, suggesting that evolutionary plasticity may be widespread among regulatory genes.
Keywords: molecular evolution, protein module, cis-regulatory module
Developmental regulatory genes encode transcription factors that participate in evolutionarily conserved gene regulatory networks (GRNs) crucial for regional specification and patterning during embryonic development (1–5). This “toolkit” of regulatory genes controls the development of diverse animals with different types of body plans (6). Furthermore, these genes are pleiotropic, being reused within individual animal lineages in different combinations and at different developmental stages (7). These findings raise two related issues. (i) How do regulatory genes change during evolution to direct the development of diverse animals? (ii) How can these changes be tolerated during development, as they are expected to be highly detrimental, reminiscent of Goldschmidt's “hopeful monster” (8)? The modularity of toolkit genes provides a partial answer to these questions, as it allows for changes in both expression and function in only specific tissues or at specific developmental times (9). Thus, although coding regions may be similar in diverse taxa, their differential expression resulting from changes in modular cis-regulatory elements (CREs) contributes to morphological diversity throughout Metazoa (10, 11). However, this modularity also applies to protein-coding regions, such that changes in coding regions that affect distinct functions of a particular protein also contribute to morphological evolution. These changes may result in gain or loss of cofactor interaction motifs, posttranslational modifications, DNA binding specificity, or other functions (9, 12–20).
One scenario for changes in developmental networks is gene duplication followed by divergence (21, 22). The Hox genes, which pattern the body plans of most metazoans, provide a prime example of this (2, 6, 23, 24). Duplication events that generated Hox clusters in early Bilateria (25) provided opportunities for genes to diverge, partitioning existing functions (subfunctionalization) or acquiring new functions (neofunctionalization) (22). A dramatic example of neofunctionalization is the Hox gene fushi tarazu (ftz), which switched function from an ancestral Hox gene to a pair-rule segmentation gene, originally identified in Drosophila melanogaster (13, 26, 27). Initial changes in ftz were likely enabled by the relaxation of constraints because of overlap in expression and function between ftz and Antp or Scr. This theory is supported by the finding that Ftz from several insects showed Antp-like functional specificity when expressed in Drosophila (13) and sequence comparisons that suggest ftz and Antp are closely related (25).
We previously showed that changes in two cofactor interaction motifs in Ftz switched its regulatory specificity from a canonical homeotic protein to a segmentation protein, found in Drosophila: (i) an LXXLL motif in Dm-Ftz confers strong interaction with the orphan nuclear receptor Ftz-F1 and is required for segmentation function (28–30); (ii) the YPWM motif, present in most Hox proteins, is degenerate in Dm-Ftz. The YPWM motif is required for homeotic function by virtue of interaction with cofactor Extradenticle (Exd), a TALE family homeodomain protein (31–35). These two protein changes resulted in gain of segmentation potential and loss of homeotic potential, specializing Dm-Ftz for segmentation. Ftz proteins that include an intact YPWM motif, such as grasshopper Sg-Ftz and beetle Tc-Ftz, have homeotic potential when expressed in Drosophila, and addition of a YPWM motif to Dm-Ftz restored ancestral homeotic function (14). In addition to these protein changes, ftz expression changed during arthropod evolution from a Hox-like domain in an arthropod ancestor (25, 36–38) to seven pair-rule stripes, seen in modern day drosophilids (39, 40). Striped expression was also observed in the basal insect Thermobia (41) and two other holometabolous insects, the beetle Tribolium casteneum and the honey bee Apis mellifera (42, 43), but stripes were absent in a grasshopper Schistocerca gregaria (44). This finding suggests that striped expression was either gained twice in arthropods, in a basal insect lineage and during early radiations of holometabolous insects, or was gained once in basal insects and lost in orthopteran lineages.
Here, we address the question: When and in what order did the changes in ftz expression and function occur during arthropod evolution? We find that the LXXLL motif was stably acquired at the base of the holometabolous insects but the YPWM degenerated in sequence and function multiple times independently in various arthropod lineages. Although strong ftz Hox-like expression is likely ancestral, it has decreased to marginal levels in a crustacean, the brine shrimp Artemia, where Ftz lacks an LXXLL and carries a degenerate YPWM motif. We suggest a mechanism that incorporates both cis-regulatory and coding changes to explain how large variations in an embryonic transcription factor can be tolerated during evolution.
Results
ftz Gene Diversity in the Arthropod Tree of Life.
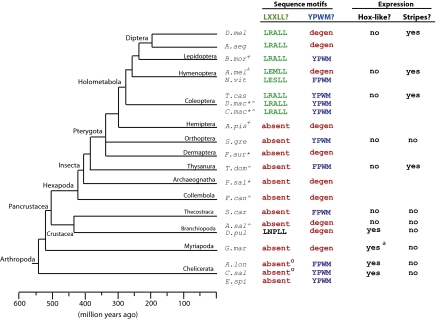
To identify when during arthropod evolution Ftz segmentation and homeotic cofactor interaction motifs were gained and lost, ftz orthologs were isolated and sequenced from organisms at representative points along the phylogenetic path from the base of Arthropoda to D. melanogaster, spanning ∼550 million years of geological time (45). These data were combined with published ftz genes and reconstructed sequence information from ongoing genome projects (Figs. 1, and 2, and Fig. S1). Full-length ftz cDNAs were isolated from embryonic RNA of organisms that could be cultured: beetles Callosobruchus maculatus (Cm) and Dermestes maculatus (Dmac), thysanuran Thermobia domestica (Td), collembolan Folsomia candida (Fc), and brine shrimp Artemia salina (As). For the dermapteran Forficula auricularia (Fa) and archaeognathan Pedetontus saltator (Ps), putative full-length ftz coding regions were isolated from genomic DNA. Although the Ftz homeodomain is similar to that of other Hox proteins, the characteristic nine residues at the amino terminal end of the homeodomain [KR(T/S)RQTYTR] distinguish Ftz from other Hox paralog groups. Thermobia, Folsomia, and Artemia partial ftz homeobox, and 3′ fragments had been previously identified (41, 47). We used these sequences to design ftz-specific primers to isolate full-length sequence. Because there was no ftz sequence data in the literature for Callosobruchus, Dermestes, Forficula, or Pedetontus, partial homeobox sequence was isolated by degenerate PCR, using primers specific for the Ftz homeodomain N-terminal arm and another highly conserved region of the homeodomain (QIKIWFQN). Once ftz was positively identified, sequence up- and downstream of the homeobox was isolated using 5′ and 3′ RACE (rapid amplification of cDNA ends) or modified, gene-specific AFLP (amplified fragment-length polymorphism) and genomic walking (Materials and Methods). In combination with ftz genes assembled from available genomes, we report nine previously unrecorded Ftz sequences: Bm-Ftz (447 amino acids), Am-Ftz (713 amino acids), Cm-Ftz (368 amino acids), Dmac-Ftz (377 amino acids), Td-Ftz (369 amino acids), Fa-Ftz (191 amino acids), Ps-Ftz (134 amino acids), Fc-Ftz (161 amino acids), and As-Ftz (201 amino acids) (Fig. 2 and Fig. S1). Adding these 9 previously unrecorded sequences to the 11 previously described yields 20 full-length arthropod ftz gene sequences available for analysis (Fig. 1).
Fig. 1.
The ftz genes from diverse arthropods display remarkable flexibility. Cladogram of major arthropod taxa is shown with divergence timeline below. The presence of cofactor interaction motifs (LXXLL motif, green; YPWM motif, blue; absent, red) and observed expression patterns (stripes; Hox-like) are indicated. +ftz assembled from genome project contigs [Sources: B. mor (69); A. mel, Honey bee Genome Sequencing Consortium; N. vit, Nas_1.0, 2007; A. pis, BCM-HGSC]; ^ftz sequence isolated in this study by RACE; *ftz sequence isolated in this study by modified AFLP from gDNA; aStriped expression seen only after segments formed (38); osequence not full length. Other sequences: (Sg) (44); (Dp) (36); (Sc) (46); (La) (38); (Gm) (37); (Cs) (70). Partial ftz sequences: AAS17755 (Td), AAK51915 (Fc), CAA49684 (As) (36), AAF63162 (Al), CAI91292 (Cs).
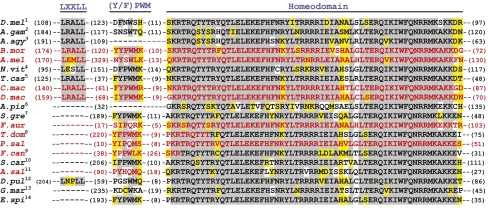
Fig. 2.
Ftz orthologs have similar homeodomain sequences but vary in their cofactor interaction motifs and protein lengths. Residues in the segmentation LXXLL, homeotic YPWM, and homeodomain are shown as identical (gray) or similar (yellow) to the most common amino acid at that position in Ftz. All Ftz proteins share a characteristic nine amino acid N-terminal homeodomain arm (KRT/sRQT/sYT/sR/k). Sequences in red were isolated in this study. Other sequences: National Center for Biotechnology Information accession numbers: 1, NP_477498; 2, NT_078265; 3, CH477233; 4, XP_001603670; 5, NP_001034539; 6, NW_001923321; 7, CAA52160; 8, AAS17755; 9, AAK51915; 10, AAM50460; 11, CAA49684; 12, ABQ22961; 13, CAJ56096; 14, ABD46730.
Arthropod Ftz orthologs differ greatly in size and composition (Fig. 2 and Table S1). The putative Ps-Ftz and Fa-Ftz sequences have very short coding regions upstream of the homeodomain (<30 amino acids), and As-Ftz and Fc-Ftz have slightly longer protein sequences upstream of the homeodomain (∼100 amino acids). Interestingly, Ftz sequences that have an LXXLL motif are much larger (Fig. 2). Although we cannot positively confirm the coding sequences of Ps-Ftz and Fa-Ftz because embryonic RNA is not available, we have several reasons to believe these sequences are full-length. First, there are splice donor (GT) and splice acceptor (AG) sites flanking small introns directly upstream of the homeobox, which are comparable in size to other ftz introns (Table S1). Second, there are no other ORFs with a splice donor site ∼800-bp upstream of the homeodomain. Third, there are several possible transcription initiator and TATA-consensus sequences upstream of the translation start site. Finally, sequence from the aphid genome shows that the predicted ftz gene in this organism does not encode an LXXLL or YPWM motif, and has very little coding region upstream of the homeodomain (32 amino acids; Aphid Genome Project).
LXXLL Was Stably Acquired at the Base of Holometabola.
The LXXLL motif in Dm-Ftz is necessary for segmentation function and mediates interaction with the cofactor Ftz-F1 (29, 30, 48). Ftz from the flour beetle T. castaneum (Tc-Ftz) contains an LXXLL motif and displayed segmentation potential when expressed in Drosophila (13). We found that Ftz orthologs from Callosobruchus and Dermestes, long- and intermediate-germ beetles, encode proteins very similar to Tc-Ftz, including LRALL sequences and similar flanking amino acids. Ftz sequences assembled from the genomes of the silkworm Bombyx mori (Bm-Ftz), honey bee A. mellifera (Am-Ftz), and mosquitoes Aedes agypti (Aa-Ftz) and Anopheles gambiae (Ag-Ftz) all include LXXLL motifs. Interestingly, most of these Ftz proteins share an LRALL sequence. Although the importance of the “RA” in Ftz has not been studied, Am-Ftz and Nv-Ftz (wasp) have EM and ES substituted at these positions. This finding suggests the internal residues (XX) are somewhat flexible, but the three leucine residues required for interaction with Ftz-F1 (14) are constrained. Whereas all Ftz proteins isolated to date from holometabolous insects harbor LXXLL motifs (Fig. 1, green), no other insect ftz encodes this motif: Sg-Ftz, Ap-Ftz, Fa-Ftz, Td-Ftz, Ps-Ftz, Fc-Ftz, and As-Ftz all lack LXXLL sequences. A Ftz LXXLL motif (LNPLL) appears in one other arthropod, the crustacean Daphnia pulex (Dp-Ftz). However, although functional experiments will be interesting in the future, as proposed by Papillon and Telford (36), this motif is probably not functional in Daphnia, as it is unlikely to participate in segmentation, particularly in light of the Hox-like expression of Dp-ftz. Together, these findings suggest that the segmentation LXXLL motif was acquired once at the stem of the holometabolous clade and that it has been stably retained in this lineage.
YPWM Motif “Flickers” in Arthropod Phylogeny.
Although the homeodomain is sufficient for binding DNA, a (Y/F)PWM sequence (referred to throughout as “YPWM motif”) found at variable distances upstream of the homeodomain in most Hox proteins is crucial for cooperative binding to Exd/Pbx (33, 49–51) and biological specificity in vivo (35, 52). The YPWM motif is found in diverse Antp and Ubx proteins (Fig. S2) and is considered the ancestral condition for Ftz, represented by a chelicerate (mite, Al-Ftz) (25) and Onychophora (53), an arthropod outgroup. Consensus YPWM motifs are also found in Ftz in both holometabolous (beetles Tc-Ftz, Cm-Ftz, Dmac-Ftz) and other insects (grasshopper Sg-Ftz, firebrat Td-Ftz). However, a degenerate motif (FNWS), with decreased Exd-binding ability and homeotic potential, is found in Dm-Ftz (14). We found degenerate motifs (degen-YPWMs) in several other Ftz sequences, including a YPPWLK in Fc-Ftz, a YHQM in As-Ftz, an IPQM in Ps-Ftz, and an IPQRK in Fa-Ftz (Fig. 2 and Fig. S1). These sequences all resemble YPWM, and are considered degenerate rather than completely lost. Additionally, degenerations appear to have occurred independently, as each motif has a different sequence. Dollo parsimony, which allows only losses after one initial gain, indicates that the motif degenerated eight times (Fig. 1: Diplopoda, Branchiopoda, Collembola, Archaeognatha, Dermaptera, Hemiptera, Hymenoptera, Diptera). Alternatively, a strict parsimony analysis, which minimizes the number of total evolutionary events regardless of direction, suggests five losses (Diplopoda, Dermaptera, Hemiptera, Hymenoptera, Diptera) and two gains (Thecostraca, Insecta). We favor the Dollo parsimony analysis, suggesting that this motif independently degenerated multiple times for several reasons. First, in each case the specific sequence change is different, sometimes involving changes in amino acid sequence (e.g., FNWS or IPQM), other times involving insertions and amino acid substitutions (e.g., YPPWLK). Second, within multiple taxa, closely related species “flicker” (54) with respect to YPWM. For example, within Hymenoptera, honey bee Ftz (Am-Ftz) has a degenerate YPWM but wasp Ftz (Nv-Ftz) retains a consensus YPWM; within crustaceans, brine shrimp Ftz (As-Ftz) YPWM is degenerate but barnacle Ftz (Sc-Ftz) retains YPWM (46). Third, some losses (e.g., dipterans) may be secondary, occurring after addition of LXXLL, and presumed gain of segmentation function. In sum, whereas the LXXLL motif of Ftz has established itself at the base of the holometabolous insects, the YPWM motif in Ftz proteins shows a complex evolutionary history with a flickering pattern in arthropod phylogeny, suggesting that it has been independently lost in multiple lineages.
Degen-YPWMs Vary in Homeotic Potential.
The lability of the YPWM motif through phylogeny reflects surprising evolutionary flexibility in this homeotic cofactor interaction motif. In contrast to what is observed for Ftz, the homeodomains and YPWM motifs encoded by neighboring Hox genes are highly conserved. Comparison of Antp, Scr, and Ubx from five divergent taxa (D. mel, A. mel, T. cas, A. pis, D. pul) revealed only one amino acid change in a YPWM motif of the 15 proteins, and virtually identical homeodomains among orthologous proteins (Fig. S2). Thus, the changes in Ftz YPWM, as well as the divergence seen within the homeodomains (Fig. 2), are specific to this protein and not a general feature of Hox proteins. The YPWM motif is required for interaction with a hydrophobic pocket on the surface of the Exd homeodomain and, based upon the mode of action of YPWM in mediating interaction with Exd (33, 49–51), the observed deviations from YPWM reported here are all expected to result in loss of interaction with Exd. We therefore asked whether these degen-YPWMs retain homeotic potential in vivo. We previously showed that ectopic expression of Dm-Ftz in imaginal discs did not cause a homeotic transformation, but rather resulted in antennal truncation because of cell death. In contrast, more homeotic-like ftz genes, such as Tc-ftz, resulted in Antp-like transformations of antennae to legs accompanied by activation of Antp-target genes (13). Additionally, replacement of FNWS in Dm-Ftz with YPWM conferred homeotic function to Dm-Ftz (14). Here, we used a similar strategy to assess the activity of degen-YPWMs from Ftz in other taxa. The homeotic potential of DmFtz-FNWS (Drosophila degenerate motif), DmFtz-YPPWLK (Folsomia degenerate motif), and DmFtz-YHQM (Artemia degenerate motif) were compared with that of a protein that completely lacked a functional motif, DmFtz-AAAA. All mutations were made in a Dm-Ftz background that included a mutation of LRALL to LRAAA because homeotic effects were found to be stronger when the LXXLL motif was inactivated (14). Additionally, the degen-YPWMs tested in this experiment were derived from Ftz proteins lacking LXXLL motifs (Fig. 2).
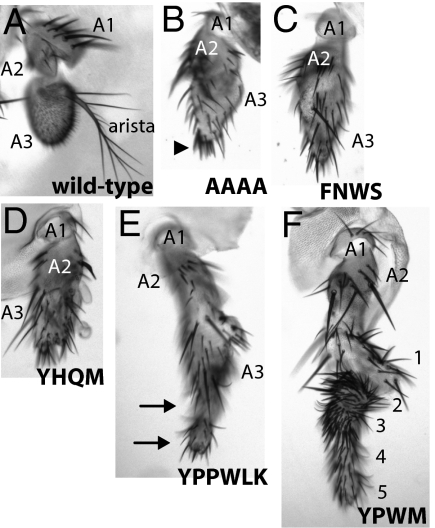
Multiple independent transformant lines were established for each construct and modified Ftz proteins were expressed in developing imaginal discs with a Dll-GAL4 driver (Figs. 3 and Fig. S3). Transgenic flies expressing UAS-lacZ (negative control) had wild-type antennae with three antennal segments (A1–A3) and aristae, demonstrating that phenotypes seen with ftz transgenes were specific and not caused by the GAL4 driver (Fig. 3A). Expression of DmFtz-AAAA resulted in antennae with normal A1 and A2 segments, but a malformed A3 segment with extra bristles and a truncated arista (Fig. 3B, arrowhead). Expression of DmFtz-FNWS (Fig. 3C) (14) and DmFtz-YHQM (Fig. 3D) caused phenotypes similar to DmFtz-AAAA, suggesting neither the Drosophila FNWS nor the Artemia YHQM conferred any further homeotic potential to Dm-Ftz. In contrast, the YPPWLK motif (DmFtz-YPPWLK) conferred some homeotic potential (Fig. 3E), but the transformation was not as strong as that induced by DmFtz-YPWM (Fig. 3F): DmFtz-YPWM transformed antennae to complete legs with five distinguishable segments (Fig. 3F) but DmFtz-YPPWLK animals showed only two distal leg segments (Fig. 3E, arrows) and a malformed A3 segment. Together, these results suggest that the YPWM motif has functionally degenerated independently multiple times and that it has lost function to different extents in different lineages.
Fig. 3.
Degenerate YPWM motifs retain varying degrees of homeotic potential. The ftz transgenes carrying examples of natural variation in YPWM motifs were expressed in developing imaginal discs with the Dll-Gal4 driver. (A) Control, expression of UAS-lacZ did not cause homeotic transformation of antennae. (B) DmFtz-AAAA animals showed normal A1 and A2, but abnormal A3 segments with bristles (arrowhead) and no aristae. (C) DmFtz-FNWS and (D) DmFtz-YHQM effects were similar to DmFtz-AAAA. (E) DmFtz-YPPWLK caused transformation of aristae into partial legs with two segments (arrows) and malformed A3 segments. (F) DmFtz-YPWM caused complete transformation of aristae to legs with five segments.
Loss of Hox-Like Expression in Crustaceans.
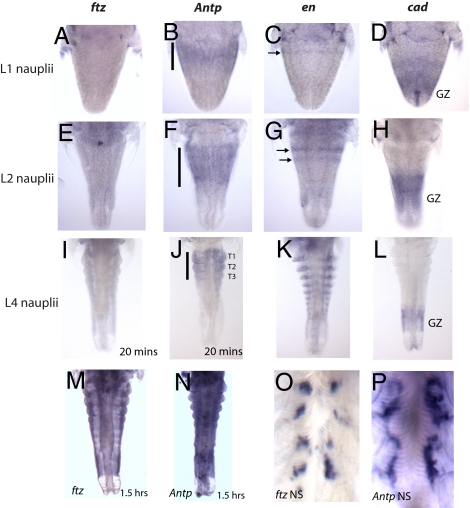
In addition to changes in sequence, ftz expression has changed during the radiation of arthropods from Hox-like to stripes. Our sequence of Td-Ftz suggests that the LXXLL motif was acquired after ftz was expressed in a striped pattern (Fig. 1). To investigate how a ftz gene encoding neither an LXXLL nor YPWM motif is expressed, we examined ftz expression in the brine shrimp Artemia. Artemia naupliar development is similar to that of short-germ insects, in that upon hatching the antennular, antennal, and mandibular segments are present, but the remaining segments are added on sequentially from the growth zone during postembryonic development. As shown in Fig. 4, As-ftz is not expressed in nauplii in a pattern seen for ftz in other species. The expression of ftz was undetectable in L1 and L2 nauplii (Fig. 4 A, E, I) and later, ftz was weakly expressed throughout the trunk of the L4 nauplius (Fig. 4I). This pattern was better visualized when Artemia were overstained (Fig. 4 M and N). Quantitative RT-PCR with L1 and L4 nauplii confirmed ftz expression in the trunk was not background or a staining artifact, and strong ftz expression in the nervous system later in development (Fig. 4O) confirmed that the weak expression observed was not the result of technical problems with the probe. In other taxa, ftz expression has been observed in the growth zone, in stripes, or in a Hox-like pattern in segment primordia (Fig. 1). Although we were able to detect expression in these regions using various probes [growth zone: cad (55) (Fig. 4 D, H, L); stripes: en (56) (Fig. 4 C, G, K); Hox: Antp (57)(Fig. 4 B, F, J)], none of these patterns were seen for ftz in Artemia (Fig. 4 A, E, I). Together, the expression and quantitative data suggest that ftz has lost Hox-like expression and potential to participate in homeosis in Artemia nauplii.
Fig. 4.
Fading Hox-like ftz expression in the crustacean Artemia salina. Expression patterns determined by in situ hybridization. (A, E, I, M, O) As-ftz; (B, F, J, M, P) As-Antp; (C, G, K) As-en; (D, H, L) As-cad in L1 (A–D), L2 (E–H), or L4 (I–N) nauplii. As-ftz was not detected in L1 or L2 and was weakly expressed in L4 nauplii. Antp was Hox-like in thoracic primordia (bar). (M and N) Overstaining highlights relative weakness of As-ftz compared with As-Antp; engrailed was detected in the posterior of every segment (C, G, K, arrows); caudal localized to the growth zone (D, H, L). Both As-ftz (O) and As-Antp (P) were detected later in the nervous system.
Discussion
Embryonic regulatory genes are remarkably conserved across broadly divergent taxa. Their structure and functional specificity are generally thought to be highly constrained, yet Hox genes are master regulatory genes that pattern body plans of diverse types of animals (2, 5, 23). This paradox has raised questions as to how Hox GRNs have evolved. One answer to this is provided by the cis-regulatory hypothesis, which posits that changes in the expression of either the Hox genes themselves, or CREs of Hox gene targets have changed during evolution (6, 11). For example, the loss of limbs in the ancestor of snakes is thought to be the result of a shift in Hox expression (58), and shifts in the borders of Ubx/Abd-A expression correlate with changes in appendage morphology in myriapods and crustaceans (59). Ubx regulates the development of both the butterfly hind-wing and fruit fly haltere, two structures with very different morphology, as a result of evolutionary changes in Ubx-responsive target genes (60). In mammals, where duplications have led to four paralogous Hox complexes, exchange of the protein coding regions of the paralogs Hox A3 and D3 revealed functional equivalence; differential gene function in vivo results from differences in gene expression (61). However, evolution does not work by one mechanism alone and changes in the coding regions of Hox genes have also been correlated with morphological diversification. For example, changes in Ubx protein led to the acquisition of a repression domain in insects that contributed to differences in limb number between crustaceans and hexapods (15, 16, 62), and changes in Hox-A11 altered its regulatory specificity such that it regulates prolactin production, critical for pregnancy, specifically in eutherian mammals (63). More dramatic perhaps than these are the changes in Hox3 and ftz in arthropods, which have escaped the rules of colinearity and taken on new roles during embryogenesis in different taxa. Duplication of Hox3 in flies generated the zen and bcd genes, which have novel functions because of shifts in expression patterns and changes in protein sequence (64). Bcd switched DNA-binding specificity because of a single amino acid change in the homeodomain (65), and acquired RNA-binding ability (reviewed in ref. 19).
Here we initiated a phylogenetically structured analysis to reconstruct the sequence of events leading to the switch in Ftz function. Because Hox genes are thought to be so highly constrained, we began with an assumption that a minimum number of changes (three total: switch to pair-rule stripes, YPWM degeneration, LXXLL acquisition) would be sufficient to describe the evolutionary trajectory of ftz. Thus, our initial goal for the present study was to map the switch points for each of these changes with the expectation that each would map to a distinct branch. Contrary to this expectation, we found that ftz has varied multiple times in both coding sequence and expression pattern (Fig. 1). (i) Expression of ftz changed at least three times during arthropod evolution: loss of Hox-like expression, gain of striped expression, and secondary loss of striped expression. (ii) The homeotic YPWM motif degenerated independently at least eight times. (iii) The LXXLL motif was stably acquired in a single “switch” at the base of the holometabolous insects. This acquisition appears be under functional constraint in holometabolous insects, as an LXXLL motif is found in Ftz throughout this taxon. The gain of a striped expression pattern in early hexapod lineages, represented by Td-ftz (41), preceded the stable gain of the segmentation LXXLL motif. This “snapshot” of molecular evolution in progress revealed a surprisingly dynamic pattern of changes in a transcription factor whose pleiotropic roles during embryonic development would be expected to restrict functional changes. We suggest that deep phylogentic sampling, such as that carried out here, will reveal similar variation in expression and function of other regulatory genes, exemplified by variations in Ubx protein domains from different taxa (see above) and loss of Abd-A expression in Artemia (66). These changes in protein motifs and expression beg for a mechanistic explanation as loss- and gain-of-function changes in Hox proteins are deleterious and ectopic expression of transcription factors usually results in lethality, even in the unchallenging environment of a laboratory.
Model for Regulatory Transcription Factor Flexibility.
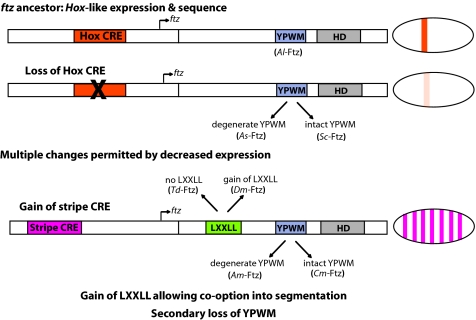
How could changes in ftz be so pervasive in nature? We propose that cis-regulatory changes that altered ftz expression were permissive for changes in protein function, enabling flexibility and variation (Fig. 5). Decreased Hox expression, seen in extant crustaceans (Fig. 4), presumably because of mutation in a CRE directing Hox-like expression (Hox CRE), removed ftz from homeotic pathways, relieving constraints on its homeotic function and allowing degeneration of the YPWM motif and eventual loss of homeotic potential. We propose that reduced levels of Hox-like expression, seen in at least two crustaceans, represent a transition state that was permissive for additional changes in ftz expression and sequence (Fig. 5): low levels of gene expression provide a platform for changes that impact protein function because their weak expression dampens activity and thus minimizes impact on existing GRNs. Although many protein variants could produce inviable “hopeful monsters” (8) if expressed at higher levels, at subthreshold levels they can provide raw material for cooption of regulatory proteins with unique functions into alternate GRNs. Some “hopeful gene variants” can endure to take on new and essential roles, exemplified by the pair-rule function of Dm-Ftz. A second cis-regulatory change in ftz was the acquisition of a striped expression pattern (Stripe CRE). This pattern arose earlier but was stabilized in holometabolous insects where acquisition of an LXXLL motif conferred interaction with the cofactor Ftz-F1, generating a Ftz able to regulate whole new sets of downstream target genes (28, 30). We suggest that maintenance of stripes in this lineage is in turn explained by the regulatory switch in Ftz (LXXLL acquisition), as interaction with Ftz-F1 allowed for ftz autoregulation (67), thus reinforcing striped expression.
Fig. 5.
The modularity of ftz CREs and protein motifs allows for extensive variation in ftz throughout arthropods. Ancestrally ftz was expressed in a Hox-like pattern because of a “Hox CRE.” CRE mutation weakened ftz Hox-like expression. Low expression levels enabled additional protein changes without deleterious consequence. The YPWM motif degenerated and lost homeotic function in multiple lineages. A CRE directing striped expression was gained and ftz was coopted into segmentation GRNs when the LXXLL motif was acquired, providing an interaction with the cofactor Ftz-F1.
Materials and Methods
Arthropod Sources and Care.
A. salina were obtained as dehydrated cysts from Carolina Biological and rehydrated in 3% salt water. Once hatched, they were maintained in a salt water solution containing an air source and fed a dilute yeast solution. Thermobia domestica were raised at 35 °C in a humid incubator, and fed oatmeal and hermit crab food. Folsomia candida were kept in Petri dishes containing charcoal/plaster of Paris and fed dry yeast. P. saltator and F. auricularia were captured in the field, preserved in >95% EtOH, and stored at −80 °C before isolation of genomic DNA.
Isolation of ftz Sequence by RLM-RACE and Modified AFLP.
RNA was extracted from 0 to 4 d Artemia nauplii, 0 to 4 d Folsomia eggs, and 0 to 9 d Thermobia eggs using the TRIzol reagent (Invitrogen) and Qiagen RNA extraction kit. Full-length ftz cDNAs were obtained by 5′ and 3′ RLM-RACE (Ambion) and PCR, using primers designed to previously identified partial ftz homeobox regions [National Center for Biotechnology Information (NCBI) accession numbers: X70079, AF361331, AY456923]. Genomic DNA was extracted from Pedetontus and Forficula using standard Drosophila protocols. Additional sequence was obtained by modified, gene-specific AFLP (68) and genomic-walking. Primer sequences are available by request.
Artemia Expression Analysis.
Artemia nauplii were fixed in 4% paraformaldehyde for 2 h at room temperature, and taken through a series of PBS/MeOH rinses: 75, 50, and 25%. After four additional washes in 100% MeOH, fixed nauplii were stored at −20 °C. Digoxygenin-labeled probes were made with T7/T3 polymerase [NCBI references: Antp: AF435786 (57); en: X70939 (56); cad: AJ567452]. Expression was examined in Artemia using protocols established by others (56). Nauplii were mounted in 90% glycerol and viewed with Leica DMRB microscopy.
Transgenic Drosophila.
Mutations to alter the FNWS in Dm-Ftz were generated by site-directed mutagenesis, as previously described (14). Multiple independent transformant lines were generated by Rainbow Transgenic Flies. Phenotypes shown were observed in at least five independent transgenic lines for each construct, and only one phenotype—that shown—was observed for each transgene. The levels of expression of the transgenes shown (Fig. S3) were similar, as determined by RT-PCR using cDNA generated from L1 larvae.
Supplementary Material
Acknowledgments
We thank Lawrence Blumer (Morehouse College, Atlanta, GA) for providing Callosobruchus, George Keeney (Ohio State University, Columbus, OH) for providing Thermobia and Folsomia, David Stern (Princeton University, Princeton, NJ) for sharing aphid sequences before publication, Kawther Abdilleh and Cristian Castillo-Davis for advice, and Alexa Bely, Eric Haag, and Bill Jeffery for helpful discussions and comments on the manuscript. J.W.S. was supported by the Maryland Agricultural Experiment Station. This work was supported by National Science Foundation Grant IBN0641717.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. HQ287864–HQ287870).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010746107/-/DCSupplemental.
References
- 1.Gehring WJ, Affolter M, Bürglin T. Homeodomain proteins. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 2.Gehring WJ, Kloter U, Suga H. Evolution of the Hox gene complex from an evolutionary ground state. Curr Top Dev Biol. 2009;88:35–61. doi: 10.1016/S0070-2153(09)88002-2. [DOI] [PubMed] [Google Scholar]
- 3.Frigerio G, Burri M, Bopp D, Baumgartner S, Noll M. Structure of the segmentation gene paired and the Drosophila PRD gene set as part of a gene network. Cell. 1986;47:735–746. doi: 10.1016/0092-8674(86)90516-7. [DOI] [PubMed] [Google Scholar]
- 4.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 5.Akam M. Hox genes and the evolution of diverse body plans. Philos Trans R Soc Lond B Biol Sci. 1995;349:313–319. doi: 10.1098/rstb.1995.0119. [DOI] [PubMed] [Google Scholar]
- 6.Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. 2nd Ed. Oxford: Blackwell Science Ltd.; 2005. [Google Scholar]
- 7.Stern DL, Orgogozo V. The loci of evolution: How predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldshmidt R. The Material Basis of Evolution. New Haven: Yale University Press; 1940. [Google Scholar]
- 9.Schlosser G, Wagner GP. Modularity in Development and Evolution. Chicago: University of Chicago Press; 2004. [Google Scholar]
- 10.Prud'homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 12.Berry M, Gehring W. Phosphorylation status of the SCR homeodomain determines its functional activity: Essential role for protein phosphatase 2A,B'. EMBO J. 2000;19:2946–2957. doi: 10.1093/emboj/19.12.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Löhr U, Yussa M, Pick L. Drosophila fushi tarazu. A gene on the border of homeotic function. Curr Biol. 2001;11:1403–1412. doi: 10.1016/s0960-9822(01)00443-2. [DOI] [PubMed] [Google Scholar]
- 14.Löhr U, Pick L. Cofactor-interaction motifs and the cooption of a homeotic Hox protein into the segmentation pathway of Drosophila melanogaster. Curr Biol. 2005;15:643–649. doi: 10.1016/j.cub.2005.02.048. [DOI] [PubMed] [Google Scholar]
- 15.Ronshaugen M, McGinnis N, McGinnis W. Hox protein mutation and macroevolution of the insect body plan. Nature. 2002;415:914–917. doi: 10.1038/nature716. [DOI] [PubMed] [Google Scholar]
- 16.Galant R, Carroll SB. Evolution of a transcriptional repression domain in an insect Hox protein. Nature. 2002;415:910–913. doi: 10.1038/nature717. [DOI] [PubMed] [Google Scholar]
- 17.Lynch VJ, Wagner GP. Resurrecting the role of transcription factor change in developmental evolution. Evolution. 2008;62:2131–2154. doi: 10.1111/j.1558-5646.2008.00440.x. [DOI] [PubMed] [Google Scholar]
- 18.Mann RS, Carroll SB. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev. 2002;12:592–600. doi: 10.1016/s0959-437x(02)00344-1. [DOI] [PubMed] [Google Scholar]
- 19.Hsia CC, McGinnis W. Evolution of transcription factor function. Curr Opin Genet Dev. 2003;13:199–206. doi: 10.1016/s0959-437x(03)00017-0. [DOI] [PubMed] [Google Scholar]
- 20.Hoekstra HE, Coyne JA. The locus of evolution: Evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 21.Ohno S. Evolution by Gene Duplication. Berlin: Springer-Verlag; 1970. [Google Scholar]
- 22.Force A, et al. The origin of subfunctions and modular gene regulation. Genetics. 2005;170:433–446. doi: 10.1534/genetics.104.027607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 24.Wagner GP, Amemiya C, Ruddle F. Hox cluster duplications and the opportunity for evolutionary novelties. Proc Natl Acad Sci USA. 2003;100:14603–14606. doi: 10.1073/pnas.2536656100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telford MJ. Evidence for the derivation of the Drosophila fushi tarazu gene from a Hox gene orthologous to lophotrochozoan Lox5. Curr Biol. 2000;10:349–352. doi: 10.1016/s0960-9822(00)00387-0. [DOI] [PubMed] [Google Scholar]
- 26.Gibson G. Evolution: Hox genes and the cellared wine principle. Curr Biol. 2000;10:R452–R455. doi: 10.1016/s0960-9822(00)00531-5. [DOI] [PubMed] [Google Scholar]
- 27.Alonso CR, Maxton-Kuechenmeister J, Akam M. Evolution of Ftz protein function in insects. Curr Biol. 2001;11:1473–1478. doi: 10.1016/s0960-9822(01)00425-0. [DOI] [PubMed] [Google Scholar]
- 28.Yu Y, et al. The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature. 1997;385:552–555. doi: 10.1038/385552a0. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz CJE, et al. FTZ-Factor1 and Fushi tarazu interact via conserved nuclear receptor and coactivator motifs. EMBO J. 2001;20:510–519. doi: 10.1093/emboj/20.3.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yussa M, Löhr U, Su K, Pick L. The nuclear receptor Ftz-F1 and homeodomain protein Ftz interact through evolutionarily conserved protein domains. Mech Dev. 2001;107:39–53. doi: 10.1016/s0925-4773(01)00448-8. [DOI] [PubMed] [Google Scholar]
- 31.Bürglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK. Structure of a DNA-bound ultrabithorax-extradenticle homeodomain complex. Nature. 1999;397:714–719. doi: 10.1038/17833. [DOI] [PubMed] [Google Scholar]
- 33.Johnson FB, Parker E, Krasnow MA. Extradenticle protein is a selective cofactor for the Drosophila homeotics: Role of the homeodomain and YPWM amino acid motif in the interaction. Proc Natl Acad Sci USA. 1995;92:739–743. doi: 10.1073/pnas.92.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mann RS, Chan S-K. Extra specificity from extradenticle: The partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 35.Zhao JJ, Lazzarini RA, Pick L. Functional dissection of the mouse Hox-a5 gene. EMBO J. 1996;15:1313–1322. [PMC free article] [PubMed] [Google Scholar]
- 36.Papillon D, Telford MJ. Evolution of Hox3 and ftz in arthropods: Insights from the crustacean Daphnia pulex. Dev Genes Evol. 2007;217:315–322. doi: 10.1007/s00427-007-0141-8. [DOI] [PubMed] [Google Scholar]
- 37.Janssen R, Damen WG. The ten Hox genes of the millipede Glomeris marginata. Dev Genes Evol. 2006;216:451–465. doi: 10.1007/s00427-006-0092-5. [DOI] [PubMed] [Google Scholar]
- 38.Hughes CL, Kaufman TC. Exploring the myriapod body plan: Expression patterns of the ten Hox genes in a centipede. Development. 2002;129:1225–1238. doi: 10.1242/dev.129.5.1225. [DOI] [PubMed] [Google Scholar]
- 39.Hafen E, Kuroiwa A, Gehring WJ. Spatial distribution of transcripts from the segmentation gene fushi tarazu during Drosophila embryonic development. Cell. 1984;37:833–841. doi: 10.1016/0092-8674(84)90418-5. [DOI] [PubMed] [Google Scholar]
- 40.Maier D, Preiss A, Powell JR. Regulation of the segmentation gene fushi tarazu has been functionally conserved in Drosophila. EMBO J. 1990;9:3957–3966. doi: 10.1002/j.1460-2075.1990.tb07616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes CL, Liu PZ, Kaufman TC. Expression patterns of the rogue Hox genes Hox3/zen and fushi tarazu in the apterygote insect Thermobia domestica. Evol Dev. 2004;6:393–401. doi: 10.1111/j.1525-142X.2004.04048.x. [DOI] [PubMed] [Google Scholar]
- 42.Brown SJ, Hilgenfeld RB, Denell RE. The beetle Tribolium castaneum has a fushi tarazu homolog expressed in stripes during segmentation. Proc Natl Acad Sci USA. 1994;91:12922–12926. doi: 10.1073/pnas.91.26.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dearden PK, et al. Patterns of conservation and change in honey bee developmental genes. Genome Res. 2006;16:1376–1384. doi: 10.1101/gr.5108606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dawes R, Dawson I, Falciani F, Tear G, Akam M. Dax, a locust Hox gene related to fushi-tarazu but showing no pair-rule expression. Development. 1994;120:1561–1572. doi: 10.1242/dev.120.6.1561. [DOI] [PubMed] [Google Scholar]
- 45.Regier JC, et al. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature. 2010;463:1079–1083. doi: 10.1038/nature08742. [DOI] [PubMed] [Google Scholar]
- 46.Mouchel-Vielh E, Blin M, Rigolot C, Deutsch JS. Expression of a homologue of the fushi tarazu (ftz) gene in a cirripede crustacean. Evol Dev. 2002;4:76–85. doi: 10.1046/j.1525-142x.2002.01063.x. [DOI] [PubMed] [Google Scholar]
- 47.Averof M, Akam M. HOM/Hox genes of Artemia: Implications for the origin of insect and crustacean body plans. Curr Biol. 1993;3:73–78. doi: 10.1016/0960-9822(93)90158-k. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki T, Kawasaki H, Yu RT, Ueda H, Umesono K. Segmentation gene product Fushi tarazu is an LXXLL motif-dependent coactivator for orphan receptor FTZ-F1. Proc Natl Acad Sci USA. 2002;98:12403–12408. doi: 10.1073/pnas.221552998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang C-P, et al. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 50.Phelan ML, Rambaldi I, Featherstone MS. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol Cell Biol. 1995;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neuteboom ST, Peltenburg LT, van Dijk MA, Murre C. The hexapeptide LFPWMR in Hoxb-8 is required for cooperative DNA binding with Pbx1 and Pbx2 proteins. Proc Natl Acad Sci USA. 1995;92:9166–9170. doi: 10.1073/pnas.92.20.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tour E, Hittinger CT, McGinnis W. Evolutionarily conserved domains required for activation and repression functions of the Drosophila Hox protein Ultrabithorax. Development. 2005;132:5271–5281. doi: 10.1242/dev.02138. [DOI] [PubMed] [Google Scholar]
- 53.Grenier JK, Garber TL, Warren R, Whitington PM, Carroll S. Evolution of the entire arthropod Hox gene set predated the origin and radiation of the onychophoran/arthropod clade. Curr Biol. 1997;7:547–553. doi: 10.1016/s0960-9822(06)00253-3. [DOI] [PubMed] [Google Scholar]
- 54.Marshall CR, Raff EC, Raff RA. Dollo's law and the death and resurrection of genes. Proc Natl Acad Sci USA. 1994;91:12283–12287. doi: 10.1073/pnas.91.25.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Copf T, Schröder R, Averof M. Ancestral role of caudal genes in axis elongation and segmentation. Proc Natl Acad Sci USA. 2004;101:17711–17715. doi: 10.1073/pnas.0407327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manzanares M, Marco R, Garesse R. Genomic organization and developmental pattern of expression of the engrailed gene from the brine shrimp Artemia. Development. 1993;118:1209–1219. doi: 10.1242/dev.118.4.1209. [DOI] [PubMed] [Google Scholar]
- 57.Averof M, Akam M. Hox genes and the diversification of insect and crustacean body plans. Nature. 1995;376:420–423. doi: 10.1038/376420a0. [DOI] [PubMed] [Google Scholar]
- 58.Cohn MJ, Tickle C. Developmental basis of limblessness and axial patterning in snakes. Nature. 1999;399:474–479. doi: 10.1038/20944. [DOI] [PubMed] [Google Scholar]
- 59.Abzhanov A, Kaufman TC. Crustacean (malacostracan) Hox genes and the evolution of the arthropod trunk. Development. 2000;127:2239–2249. doi: 10.1242/dev.127.11.2239. [DOI] [PubMed] [Google Scholar]
- 60.Weatherbee SD, et al. Ultrabithorax function in butterfly wings and the evolution of insect wing patterns. Curr Biol. 1999;9:109–115. doi: 10.1016/s0960-9822(99)80064-5. [DOI] [PubMed] [Google Scholar]
- 61.Greer JM, Puetz J, Thomas KR, Capecchi MR. Maintenance of functional equivalence during paralogous Hox gene evolution. Nature. 2000;403:661–665. doi: 10.1038/35001077. [DOI] [PubMed] [Google Scholar]
- 62.Grenier JK, Carroll SB. Functional evolution of the Ultrabithorax protein. Proc Natl Acad Sci USA. 2000;97:704–709. doi: 10.1073/pnas.97.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lynch VJ, et al. Adaptive changes in the transcription factor HoxA-11 are essential for the evolution of pregnancy in mammals. Proc Natl Acad Sci USA. 2008;105:14928–14933. doi: 10.1073/pnas.0802355105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt-Ott U, Wimmer EA. Starting the segmentation gene cascade in insects. In: Wagner GP, editor. Modularity in Development and Evolution. Chicago: University of Chicago Press; 2004. pp. 395–412. [Google Scholar]
- 65.Hanes SD, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 66.Hsia CC, Paré AC, Hannon M, Ronshaugen M, McGinnis W. Silencing of an abdominal Hox gene during early development is correlated with limb development in a crustacean trunk. Evol Dev. 2010;12:131–143. doi: 10.1111/j.1525-142X.2010.00399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hiromi Y, Gehring WJ. Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell. 1987;50:963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- 68.Biedler J, et al. Transposable element (TE) display and rapid detection of TE insertion polymorphism in the Anopheles gambiae species complex. Insect Mol Biol. 2003;12:211–216. doi: 10.1046/j.1365-2583.2003.00403.x. [DOI] [PubMed] [Google Scholar]
- 69.Mita K, et al. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004;11:27–35. doi: 10.1093/dnares/11.1.27. [DOI] [PubMed] [Google Scholar]
- 70.Damen WG, Janssen R, Prpic NM. Pair rule gene orthologs in spider segmentation. Evol Dev. 2005;7:618–628. doi: 10.1111/j.1525-142X.2005.05065.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.