Abstract
PCR-based gene targeting is a popular method for manipulating yeast genes in their normal chromosomal locations. The manual design of primers, however, can be cumbersome and error-prone. We have developed a straightforward web-based tool that applies user-specified inputs to automate and simplify the task of primer selection for deletion, tagging and/or regulated expression of genes in Schizosaccharomyces pombe. This tool, named PPPP (for Pombe PCR Primer Programs), is available at http://www.sanger.ac.uk/PostGenomics/S_pombe/software/. We also present a searchable Microarray Primer Database to retrieve the sequences and accompanying information for primers and PCR products used to build our in-house Sz. pombe microarrays. This database contains information on both coding and intergenic regions to provide context for the microarray data, and it should be useful also for other applications, such as quantitative PCR. The database can be accessed at http://www.sanger.ac.uk/PostGenomics/S_pombe/microarray/. Copyright © 2006 John Wiley & Sons, Ltd.
Keywords: primer sequence, Sz. pombe, gene deletion, gene expression, gene tagging, microarray, quantitative PCR
Introduction
A major strength of yeast genetics is the ease with which specific genes can be manipulated within the genome, which greatly facilitates functional analyses. Gene targeting takes advantage of homologous recombination between transformed DNA fragments that terminate in short stretches of target sequence and the corresponding genomic sites (Rothstein, 1983; Grimm and Kohli, 1988). The most straightforward approach for gene targeting is to design long primers that include the target sequences, and then carry out PCR with these primers to generate the DNA fragment for transformation (Baudin et al., 1993; Wach et al., 1997). Modular and versatile constructs are available for gene deletion, gene tagging and regulated gene expression in fission yeast (Bähler et al., 1998; Tasto et al., 2001; Sato et al., 2005; Hentges et al., 2005; Van Driessche et al., 2005). These constructs contain different types of markers, tags and promoters; they can be amplified by PCR using a limited number of primers, thus reducing costs and increasing flexibility for analysing gene function. The design of PCR primers involves the selection of appropriate target sequences relative to the gene of interest (typically ∼80 bp) combined with the sequences to amplify the constructs (∼20 bp). Manual primer selection is therefore quite complicated, and any mistakes lead to time loss at best and misleading results at worst. To ease this laborious task, we have developed a web-based tool (Pombe PCR Primer Programs; PPPP) that automatically suggests primer sequences for deletion, tagging and regulatable expression of genes, based on the gene name, length of target sequence and plasmid information that the user specifies. In addition, the tool can design short primers up- or downstream of the target sequences to screen by PCR for correct homologous integration of the transformed fragments. Our group and others now routinely use this tool to facilitate the reliable design of primers for gene targeting.
We also describe a searchable Microarray Primer Database that contains information on primers and PCR products used for our in-house Sz. pombe microarray platform. The intragenic (coding) regions have been used on microarrays for expression profiling studies (Mata et al., 2002; Smith et al., 2002; Chen et al., 2003; Mata and Bähler, 2003; Rodríguez-Gabriel et al., 2003; Gatti et al., 2004; Rustici et al., 2004; Sanders et al., 2004; Watson et al., 2004; Hansen et al., 2005; Harrison et al., 2005; Jenkins et al., 2005; Lee et al., 2005; Mandell et al., 2005; Bachand et al., 2006; Martín et al., 2006; Rodríguez-Gabriel et al., 2006; Sharma et al., 2006; Mata and Bähler, 2006). We have also started to use microarrays covering all intergenic regions for complementary genome-wide studies (e.g. Heichinger et al., 2006). Comprehensive data on intra- and intergenic regions are provided in the Microarray Primer Database to look up particular features of regions of interest represented in the microarray data. Given that primers were selected to cover sequences without cross-hybridization to other genomic sequences and to be located within exon sequences if used for expression profiling (Lyne et al., 2003), the database could also be used for other applications, e.g. to select primers for quantitative PCR.
Implementation
Both PPPP and the Microarray Primer Database are written in Perl. We have designed web interfaces for both web tools using the CGI module of Perl, hosted on an Apache server. Genes can be searched using any of the gene names or systematic identifiers available in GeneDB (http://www.genedb.org/genedb/pombe/; Hertz- Fowler et al., 2004). The outputs of both tools are provided in tabulated HTML format (Figures 1, 2 and 3).
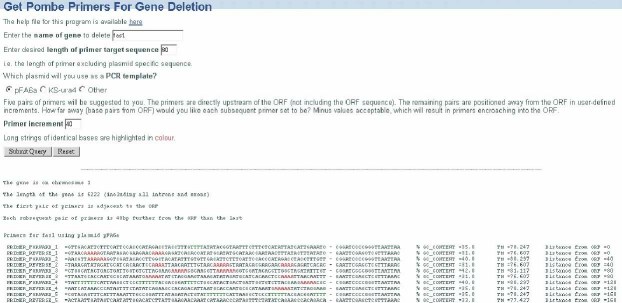
Figure 1.
Screenshot of PPPP output page, using the gene deletion mode with the fas1 gene as an example
Figure 2.
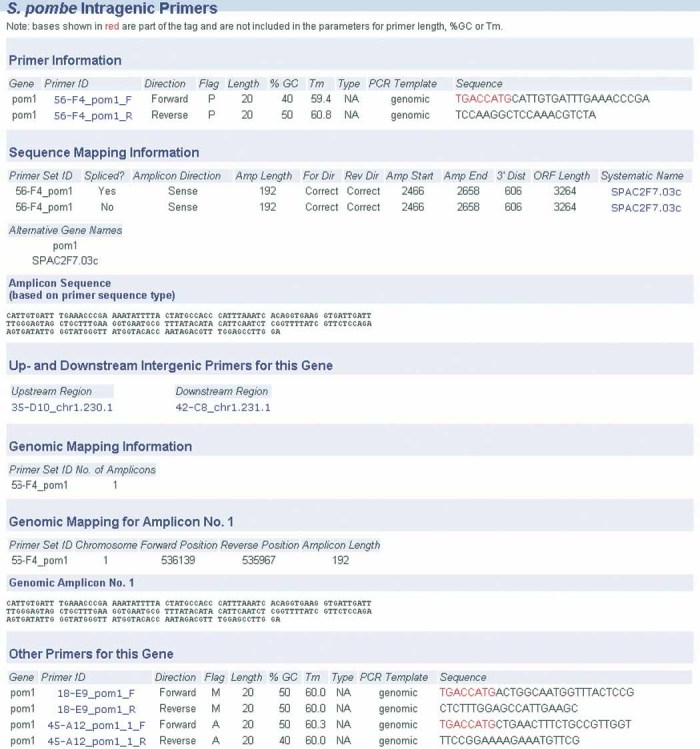
Screenshot of output page from Microarray Primer Database. Full page for intragenic search, using the pom1 gene as an example. The list for other primers at the bottom is only partially shown
Figure 3.

Screenshot of output page from Microarray Primer Database. Full page for intergenic region between pom1 and pmc2 genes. This page has been accessed by clicking on intergenic primer for ‘Downstream Region’ in the page shown in Figure 2
PPPP stores the gene locations and directions, along with gene synonyms, in a Perl disk-based hash (DBM or DataBase Module) file. This file is pre-calculated from data in Sz. pombe GeneDB to speed up the program for primer design without the overhead of creating a relational database. When the program is run, the gene locations are used to extract sequences from the Sz. pombe genome (Wood et al., 2002), from which the primers are designed.
The Microarray Primer Database is implemented as a MySQL relational database running on a UNIX server. The database was created using a Perl-based pipeline with the sequences of the primers used for the intra- and intergenic arrays. Primers are mapped to the genome and, for the intragenic primers, have been checked to map to the correct gene. A set of parameters for each primer is then calculated, including length, GC percentage and melting temperature (Tm) for short primers (Breslauer et al., 1986). Finally, the amplicon sequences are extracted from genomic DNA (or from cDNA sequence if a primer pair flanks intron sequence). Once all of this information has been collected, it is imported into the MySQL database using the Perl DBI module.
The intragenic primers have been designed as described by Lyne et al. (2003). The intergenic primers have been designed using Perl scripts that check EMBL sequence files for regions between any open reading frames (ORFs) and known non-coding RNA genes. Long intergenic regions are divided into sub-sections to increase microarray resolution, as follows. For regions between divergently expressed genes, one section is used for regions up to 1000 bp, whilst regions between 1000 and 4000 bp are divided into two sections. For regions between tandem or convergent genes, one section is used for regions up to 2000 bp, whilst regions between 2000 and 4000 bp are divided into two sections. In all cases, longer regions are divided such that no sub-section is longer than ∼2000 bp. The Perl scripts for the design of intra- and intergenic primers can be found under ‘Other scripts’ on our software page (http://www.sanger.ac.uk/PostGenomics/S_pombe/software/); this page also includes PPPP (this report), YOGY (for integrated orthology and Gene Ontology analyses; Penkett et al., 2006) and a script for initial microarray data processing (Lyne et al., 2003).
Web tool description
Pombe PCR primer programs
The PPPP entry page (http://www.sanger.ac.uk/PostGenomics/S_pombe/software/) provides four separate links to obtain primers for: (a) gene deletion; (b) C-terminal tagging; (c) controlling genes by nmt1 promoter and/or N-terminal tagging; or (d) checking for homologous integration by PCR. For the first three cases, the following information is required as input:
Name of gene to be manipulated.
Length of genomic target sequence (excludes the plasmid-specific sequence; default is 80).
Plasmid to be used as PCR template (refers to plasmids published by Bähler et al., 1998, although primers will work for any compatible plasmids; ‘other’ means no plasmid-specific sequence will be added; for N-terminal tagging, a tag must be specified).
Increment length (see below; default is 40).
Forward and reverse primer sequences are given in the output page (Figure 1). The first set of primers is directly adjacent to the ORF, while the four subsequent primer sets are further away from the ORF at a user-specified distance, depending on the value of the increment used. For example, if the primer increment is set to 40, the second set of primers begins 40 bp before and after the ORF, the third set is placed at 80 bp, etc. A negative increment value results in primers that impinge into the ORF. This allows for a choice of primers with varying positions and base composition. Long strings of identical bases (four nucleotides or more) are highlighted in colour. Additional information is provided on the automatically selected primers as follows:
GC content calculated as percentage of total nucleotide content.
Melting temperature (Tm), as defined for long primers by Sambrook et al. (1989).
Distance of primer from the ORF.
The gene deletion program gives a selection of both forward and reverse primers. For C-terminal tagging, only one forward primer is provided as this is the only option for tagging of full-length proteins. Similarly, for nmt1 regulation/N-terminal tagging, only one reverse primer is provided, which allows manipulation of full-length proteins.
PPPP can also design short PCR primers based on user-defined inputs to check for correct homologous integration of the gene targeting fragments. For this, primers of suitable melting temperature are selected within 1 kb either up- or downstream of the target gene (pointing towards the gene in each case), such that they do not overlap with the target sequence but are close to it. Using these primers in combination with appropriate universal primers within the targeting fragment will result in diagnostic PCR products if the fragment is integrated at the correct genomic location.
Microarray primer database
The entry page of this tool contains input fields for both intra- and intergenic primers (http://www.sanger.ac.uk/PostGenomics/S_pombe/micro-array/). Intragenic primers can be accessed by searching with any valid Sz. pombe GeneDB name including the systematic identifier. If the search identifies more than one set of primers, a list of all primers is provided with basic information on each primer. The database can also be searched using incomplete names with a wild-card option, and again a list of primers is presented if multiple genes represent the incomplete name. The full information pages for the primer sets in these lists can then be obtained by clicking on the hyperlinks associated with the primer names. The search leads directly to the full page if only one primer pair is found. A list of synonyms and systematic identifiers for the gene associated with the primer set is provided on the full page to ensure that the correct gene is selected. Intergenic primers are named by the number of the corresponding intergenic region along the chromosome. Hence, it is easier to access a particular intergenic region by searching for the gene name up- or downstream of the region of interest using the intragenic search; in the intragenic primer output page, the corresponding intergenic regions can then be accessed via hyperlinks to the up- or downstream intergenic region for this gene (see below).
The full page for the intragenic search begins with a table of basic primer information (Figure 2). The primer names include 96-well plate numbers and plate positions used for microarray printing. In addition, the following basic information is included:
Primer direction relative to the gene (forward and reverse primers are given on separate lines).
Flag for information on PCR products (P, present; M, multiple bands; A, absent). A few primers have been designed with an old version of the genome and were either incorrect or better primers have been designed; in these cases, the old primers are marked as A.
Primer length (bp).
GC percentage.
Melting temperature (Tm).
Primer type: indicates if sequence flanked by primers includes introns (cDNA required for PCR template) or not (NA: genomic DNA can be used for PCR template).
PCR template used to amplify the primers for the arrays.
Primer sequence.
Some primers contain an additional universal sequence, highlighted in red, which does not correspond to Sz. pombe genomic sequence. This sequence has been used for second-round PCR amplification with an amino-linked universal primer (Lyne et al., 2003). The universal sequences are not included in the information for primer length, GC content or Tm.
The first table is followed by a table of sequence mapping information for the gene associated with the primer pair (Figure 2). Two lines of data are in this table, one for the spliced and one for the unspliced version of the gene. The mapping data in both lines are identical if both primers are located within the same exon. The following information is provided:
Amplicon direction relative to gene (‘sense’: amplicon strand on array measures transcript in direction of gene; ‘anti-sense’: amplicon strand on array measures reverse transcript relative to gene).
Amplicon length in bp (PCR product).
Direction of forward and reverse primers, indicating whether primers are correctly orientated relative to each other (both ‘correct’; a few primers are wrongly orientated, indicated by ‘wrong’; these will all have correct alternative primers).
Amplicon start position within gene relative to start codon ATG (where A is 1).
Amplicon end position within gene relative to start codon ATG (where A is 1).
Distance (bp) of amplicon end to final base of stop codon.
ORF length (bp).
Systematic name of gene that primers are mapped to with a link to the corresponding page in Sz. pombe Gene DB.
Table with alternative names for this gene as given in Sz. pombe GeneDB.
Formatted version of amplicon sequence excluding any introns.
This gene information is followed by genomic context data (Figure 2). The first table has links to the intergenic regions that are up- and downstream of the mapped gene, which is the easiest way to access intergenic primers of interest. Next, there is a table indicating how many amplicons are possible with the primer pair. Normally this will be 1, but sometimes both primers map to multiple neighbouring positions within the genome that could give rise to multiple amplicons by PCR. Amplicons that would be larger than 4000 bp are not considered.
Next, the following information is shown for each possible amplicon:
Mapped chromosome.
Positions of forward and reverse primers within this chromosome using Sz. pombe GeneDB coordinates.
Amplicon length (bp).
A formatted version of the sequence is then given for each amplicon, including any potential introns (Figure 2). If there are multiple primer sets for the gene, a last table on the output page provides a summary of information for the other primer sets available. The format of this table is the same as for the initial list page.
The full page for the intergenic search also begins with a table of basic primer information (Figure 3). The next table indicates how many amplicons are possible with this primer pair (again, only for amplicons up to a maximum length of 4000 bp). For each amplicon, a summary of genomic mapping information is then shown. This is identical to the table produced for intragenic primers. In addition, it also includes information on the direction of the two genes that flank the intergenic region: ‘tandem’, both genes are in same direction, thus the intergenic region contains a single promoter; ‘convergent’, the 3′ ends of both genes point towards the intergenic region, which therefore contains no promoter; ‘divergent’, the 5′ ends of both genes point towards the intergenic region, which therefore contains two promoters. This is followed by two tables, the first for the upstream gene and the second for the downstream gene. No genes are shown in the tables in cases where the primers are flanked by > 20 000 bp of intergenic sequence. Both tables for flanking genes contain the following information:
Link to the gene in Sz. pombe GeneDB.
Link to the primers available for this gene.
Type of sequence for this gene as specified in Sz. pombe GeneDB (e.g. CDS, tRNA, rRNA).
Direction of the gene relative to the chromosome as specified by Sz. pombe GeneDB.
Start and end positions of the gene within the chromosome using Sz. pombe GeneDB coordinates.
Unspliced length of the ORF (bp).
Spliced length of the ORF (bp).
Shortest distance of primer from end of the closest ORF (a negative value indicates that this primer overlaps with the ORF).
For each possible intergenic amplicon, a formatted version of the amplicon sequence is then presented at the bottom of the page (Figure 3).
Conclusions
PPPP makes primer design for a range of PCR-based gene targeting approaches less painful and more reliable, while the Microarray Primer Database provides searchable information on primers and PCR products used to generate our microarray data and can also help for other applications. We regularly take advantage of these web tools and hope that colleagues of the fission yeast community will find these tools similarly useful for their research.
Acknowledgments
We thank all members of the Bähler laboratory for testing PPPP and the resulting primers, colleagues of the fission yeast community and anonymous reviewers for valuable feedback on PPPP. Roger Pettett helped with installing PPPP on the Sanger Institute website. Rachel Lyne wrote the original microarray primer design scripts. The work in our group is funded by Cancer Research UK (CUK), Grant No. C9546/A6517.
References
- Bachand F, Lackner DH, Bähler J, Silver PA. Autoregulation of ribosome biosynthesis by a translational response in fission yeast. Mol Cell Biol. 2006;26:1731–1742. doi: 10.1128/MCB.26.5.1731-1742.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu J-Q, Longtine MS, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Breslauer KJ, Frank R, Blöcker H, Marky LA. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Toone WM, Mata J, et al. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti L, Chen D, Beretta GL, et al. Global gene expression of fission yeast in response to cisplatin. Cell Mol Life Sci. 2004;61:2253–2263. doi: 10.1007/s00018-004-4218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Kohli J. Observations on integrative transformation in Schizosaccharomyces pombe. Mol Gen Genet. 1988;215:87–93. doi: 10.1007/BF00331308. [DOI] [PubMed] [Google Scholar]
- Hansen KR, Burns G, Mata J, et al. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol Cell Biol. 2005;25:590–601. doi: 10.1128/MCB.25.2.590-601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C, Katayama S, Dhut S, et al. SCFPof1-ubiquitin and its target Zip1 transcription factor mediate cadmium response in fission yeast. EMBO J. 2005;24:599–610. doi: 10.1038/sj.emboj.7600536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bähler J, Nurse P. Genome-wide characterization of fission yeast DNA replication origins (Accepted by EMBO J.) 2006 doi: 10.1038/sj.emboj.7601390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 2005;22:1013–1019. doi: 10.1002/yea.1291. [DOI] [PubMed] [Google Scholar]
- Hertz-Fowler C, Peacock CS, Wood V, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins CC, Mata J, Crane RF, et al. Activation of AP-1-dependent transcription by a truncated translation initiation factor. Eukaryot Cell. 2005;4:1840–1850. doi: 10.1128/EC.4.11.1840-1850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Miklos I, Du H, et al. Impairment of the TFIIH-associated CDK-activating kinase selectively affects cell cycle-regulated gene expression in fission yeast. Mol Biol Cell. 2005;16:2734–2745. doi: 10.1091/mbc.E04-11-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R, Burns G, Mata J, et al. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genom. 2003;4:27. doi: 10.1186/1471-2164-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell JG, Bähler J, Volpe TA, Martienssen RA, Cech TR. Global expression changes resulting from loss of telomeric DNA in fission yeast. Genome Biol. 2005;6:R1. doi: 10.1186/gb-2004-6-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V, Rodríguez-Gabriel MA, McDonald WH, et al. Cip1 and Cip2 are novel RNA-recognition-motif proteins that counteract Csx1 function during oxidative stress. Mol Biol Cell. 2006;17:1176–1183. doi: 10.1091/mbc.E05-09-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bähler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–714. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- Mata J, Bähler J. Correlations between gene expression and gene conservation in fission yeast. Genome Res. 2003;13:2686–2690. doi: 10.1101/gr.1420903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Bähler J. Gene expression during early sexual differentiation in fission yeast: global roles of Ste11p, cell type and pheromone. Proc Natl Acad Sci USA. 2006 doi: 10.1073/pnas.0603403103. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkett CJ, Morris JA, Wood V, Bähler J. YOGY: a web-based, integrated database to retrieve protein orthologs and associated Gene Ontology terms. Nucleic Acids Res. 2006;34:W330–W334. doi: 10.1093/nar/gkl311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Gabriel MA, Burns G, McDonald WH, et al. RNA binding protein Csx1 mediates global control of gene expression in response to oxidative stress. EMBO J. 2003;22:6256–6266. doi: 10.1093/emboj/cdg597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Gabriel MA, Watt S, Bähler J, Russell P. Upf1, an RNA helicase required for nonsense-mediated mRNA decay, modulates the transcriptional response to oxidative stress in fission yeast. Mol Cell Biol. 2006;26:6347–6356. doi: 10.1128/MCB.00286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rustici G, Mata J, Kivinen K, et al. Periodic gene expression program of the fission yeast cell cycle. Nat Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sato M, Dhut S, Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- Sharma N, Watt S, Mehta S, Marguerat S, Bähler J. The fission yeast Rpb4 subunit of RNA polymerase II plays a specialized role in cell separation. Mol Genet Genom. 2006 doi: 10.1007/s00438-006-0161-5. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DA, Toone WM, Chen D, et al. The Srk1 protein kinase is a target for the Sty1 stress-activated MAPK in fission yeast. J Biol Chem. 2002;277:33411–33421. doi: 10.1074/jbc.M204593200. [DOI] [PubMed] [Google Scholar]
- Tasto JJ, Carnahan RH, McDonald WH, Gould KL. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- Van Driessche B, Tafforeau L, Hentges P, Carr AM, Vandenhaute J. Additional vectors for PCR-based gene tagging in Saccharomyces cerevisiae and Schizosaccharomyces pombe using nourseothricin resistance. Yeast. 2005;22:1061–1068. doi: 10.1002/yea.1293. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Watson A, Mata J, Bähler J, Carr A, Humphrey T. Genomic expression responses to ionizing radiation and the regulatory roles of the Rad3-checkpoint and Sty1 stress-response kinases in fission yeast. Mol Biol Cell. 2004;15:851–860. doi: 10.1091/mbc.E03-08-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]