SUMMARY
Mouse iNKT cell receptors (iNKT TCRs) use a single Vα14-Jα18 sequence and Vβs that are almost always Vβ8.2, Vβ7 or Vβ2, although the basis of this differential usage is unclear. We show that the Vβ bias occurs as a consequence of the CDR2β loops determining the affinity of the iNKT TCR for CD1d/glycolipids, thus controlling positive selection. Within a conserved iNKT-TCR-CD1d docking framework, these inherent Vβ-CD1d affinities are further modulated by the hypervariable CDR3β loop, thereby defining a functional interplay between the two iNKT TCR CDRβ loops. These Vβ biases reveal a broadly hierarchical response in which Vβ8.2 > Vβ7 > Vβ2 in the recognition of diverse CD1d ligands. This restriction of the iNKT TCR Vβ repertoire during thymic selection paradoxically ensures that each peripheral iNKT cell recognizes a similar spectrum of antigens.
INTRODUCTION
T lymphocyte recognition of antigen via their somatically rearranged αβ TCRs is central to the proper functioning of the immune system. The molecular basis for the recognition of antigenic peptides bound to major histocompatibility complex (MHC) molecules by αβ TCRs has been characterized. Although some variability of the TCR docking modes onto the pMHC surfaces has been observed, a rough docking mode between the TCR-pMHC appears preserved, in which the Vα and Vβ domains are positioned over the α2-helix and α1-helix, respectively (Rudolph et al., 2006). Within this consensus footprint, the individual contributions of the germline-encoded and non-germline encoded CDR loops to the interaction can vary greatly (Gras et al., 2008). Nevertheless, recent findings suggest that such a mode of recognition might be the result of structural constraints within the TCR and MHC molecules selected through co-evolution of genes encoding for these molecules (Feng et al., 2007; Garcia et al., 2009; Marrack et al., 2008).
A sizeable portion of αβ TCR+ T cells does not recognize peptide antigens presented by conventional polymorphic MHC I/MHC II molecules. αβ TCRs have been described that recognize lipid or glycolipid antigens presented by members of the monomorphic lipid-binding family of molecules, CD1 (Brigl and Brenner, 2004). The most extensively studied lipid-reactive T cells are natural killer T (NKT) cells, which recognize a number of glycolipid antigens in association with CD1d. Two broad classes of NKT cells have been defined on the basis of TCR expression and antigen reactivity (Godfrey et al., 2004). Most studies of these cells focus on type I, or iNKT cells, which are the most prevalent NKT cells in mice (Matsuda et al., 2008). These iNKT cells express a TCR that is the product of a canonical rearrangement between the Vα14 gene segment (Vα24 in human) and the Jα18 gene segment, with a CDR3α region invariant at the amino acid level (Koseki et al., 1990; Lantz and Bendelac, 1994). This Vα14 invariant chain is co-expressed with a limited set of Vβ chains, predominantly Vβ8.2, Vβ7 and Vβ2 in mice and Vβ11 in humans (Dellabona et al., 1994; Koseki et al., 1990; Lantz and Bendelac, 1994; Porcelli et al., 1993). However, unlike their TCRα chain, these Vβ chains are highly diverse in both their CDR3 composition and association with Jβ segments (Apostolou et al., 2000; Behar et al., 1999; Lantz and Bendelac, 1994; Matsuda et al., 2001) and the basis for this diversity is unclear. iNKT cells expressing these TCRs can recognize several microbe-derived glycosphingolipid (Kinjo et al., 2005; Mattner et al., 2005) and diacylglycerol antigens (Kinjo et al., 2006), including the prototypical glycosphingolipid antigen α-galactosylceramide (αGC) (Burdin et al., 1998; Kawano et al., 1997), and are best identified using the CD1d tetramer loaded with this antigen (Benlagha et al., 2000; Matsuda et al., 2000).
Upon TCR engagement, iNKT cells rapidly release large amounts of cytokines and chemokines that can enhance or suppress immune responses. As such, iNKT cells are often considered an important link between the innate and adaptive immune systems, and have been shown to influence a broad range of diseases, including cancer, autoimmunity, allergy and infection (Bendelac et al., 2006; Kronenberg, 2005). Identification of the molecular features of iNKT TCR receptor-antigen recognition and the mechanisms responsible for the formation of the iNKT cell repertoire are of fundamental importance to our understanding of the biology of these cells.
Recent crystallographic and mutational analyses have shown how the iNKT TCR recognizes glycolipid/CD1d complexes. The crystal structure of iNKT TCRs, unliganded (Gadola et al., 2006; Kjer-Nielsen et al., 2006; Zajonc et al., 2008), and in complex with αGC/CD1d revealed a unique docking strategy that differs from most TCR/MHC/peptide interactions (Borg et al., 2007; Pellicci et al., Submitted). Namely, the iNKT TCR docked at the extreme end of, and parallel to, the αGC/CD1d complex. Here, the CDR1α and CDR3α loops of the invariant TCRα chain dominated the interaction with the antigen and αGC/CD1d respectively, while the role of the TCRβ chain was restricted to the CDR2β loop interacting with CD1d. CDR3β, the only “non-germline encoded” region of the iNKT TCR, does not make contact with the antigen but instead is positioned over the α2 helix of CD1d. Thus, recognition of αGC/CD1d by the iNKT TCR seems to be entirely mediated by the germline-encoded surface of the iNKT TCR.
Furthermore, extensive mutational analyses of both mouse and human iNKT TCRs (Scott Browne et al., 2007; Wun et al., 2008) revealed that an energetic ‘hot-spot’ was formed by residues within the CDR1α, CDR3α and CDR2β loops of the NKT TCR that were critical for the recognition of the αGC/CD1d complex and provided a snapshot into the basis of iNKT cell recognition.
The mechanisms that drive the Vβ bias of in the iNKT cell repertoire remain unclear. Moreover, it is not known whether different TCRβ chains might allow the iNKT TCR to change its specificity for different antigens. Several scenarios, including possible pairing preferences with the invariant NKT TCRα chain (Degermann et al., 1999; Gui et al., 2001) and/or influence of thymic selection during development (Schumann et al., 2006; Wei et al., 2006), have been proposed to explain the Vβ bias. Here, we demonstrate that specific sequences within the CDR2β and CDR3β loops are responsible for shaping the iNKT cell repertoire in vivo, and do so by constraining iNKT TCR interaction with antigen(s)/CD1d complexes.
RESULTS
Vβ chains confer differential affinity for CD1d
The TCRβ repertoire of mouse iNKT cells is biased, with preferential usage of Vβ8, Vβ7 and Vβ2 segments. Such a bias has been proposed to result, at least partly, from poor pairing between the Vα14 invariant chain and certain Vβ chains (Gui et al., 2001). To rigorously test whether pairing constraints can contribute to the observed bias in the repertoire, we co-expressed the Vα14 invariant chain with each of the Vβ chains in an αβ− TCR hybridoma (White et al., 1993). We first transduced the hybridoma with an MSCV-based retrovirus encoding the Vα14 invariant chain and GFP as a reporter. Transduced cells were selected on the basis of GFP expression and do not express any TCR on their cell surfaces (data not shown). Next, we cloned each of the 20 different mouse Vβ chains in the context of the same CDR3β rearrangement (i.e. the CDR3β and Jβ) from the DO-11.10 TCRβ chain into another MSCV-based retroviral plasmid and transduced the Vα14-expressing hybridoma separately with each of the TCRβ chains. Transduced hybridomas were sorted for cell surface expression of the TCR and tested for their interaction with the αGC/CD1d tetramer. As seen in Fig. 1A, under these conditions, all of the individual TCRβ chains paired with the Vα14 chain and all the hybridomas expressed TCR molecules on their cell surface. Some variability in TCR expression level between the various Vβ chains was observed that might be related to the efficiency of transduction between experiments. Nevertheless, these results suggested that all Vβ chains can pair with the Vα14 invariant chain.
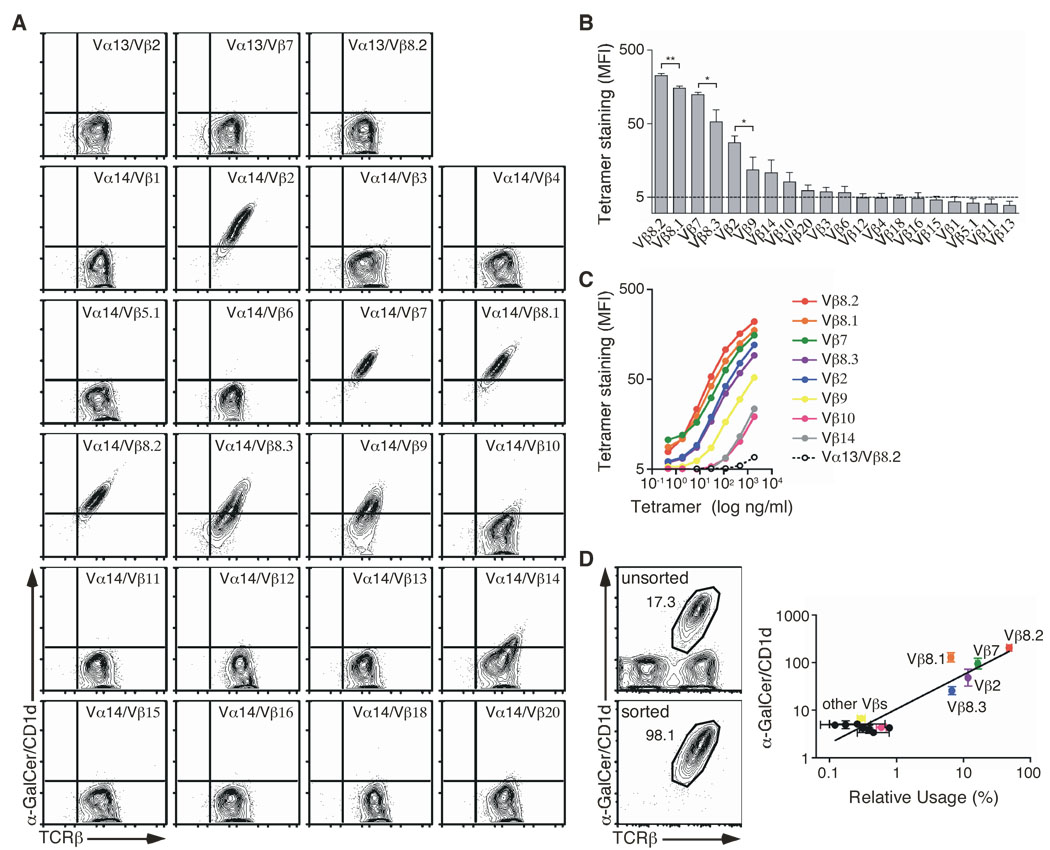
Figure 1.
Vβ chains confer differential affinity for CD1d. (A) Staining of hybridomas expressing the Vα14i TCRα chain, or Vα13-Jα18 TCRα chain (negative controls), paired with the indicated Vβ chains in the context of a unique CDR3β. (B) The MFI of αGC/CD1d tetramer staining for each hybridoma was determined for a narrow TCR gate. Data represent the mean + s.e.m. of three independent experiments. (C) The indicated hybridomas were stained with increasing concentrations of αGC/CD1d tetramer. The MFI of tetramer staining was determined for a narrow TCR gate. The data represent the mean of two independent experiments. (D) Ex vivo sorted NKT cells were stained with specific anti-Vβ antibodies. Data represent a plot of the relative Vβ usage (x axis) against the αGC/CD1d tetramer MFI of the appropriate Vβ-expressing hybridomas as determined in (B) (y axis). Data represent the mean ± s.e.m. of two independent experiments.
TCRs containing Vβ8.2, Vβ8.1, Vβ8.3, Vβ7, Vβ2 stained intensely with αGC/CD1d tetramer, while Vβ9, Vβ10 and Vβ14-containing TCRs bound the αGC/CD1d tetramer weakly. TCRs containing the other Vβ chains did not stain above background (Fig. 1A). Comparison of tetramer staining intensity relative to TCR expression and staining experiments using a wide range of αGC/CD1d tetramer concentrations revealed different relative avidities for the αGC/CD1d complex as a function of the Vβ chain usage, with the hierarchy Vβ8.2 > Vβ8.1 ≥ Vβ7 > Vβ8.3 ≥ Vβ2 > Vβ9 ≥ Vβ14 ≥ Vβ10 (Fig. 1B, C).
Because CD4 expression by the original TCRαβ- parent hybridoma is heterogeneous, we generated two sets of hybridomas expressing the same panel of TCRs with or without CD4 expression. The Vβ hierarchy of the relative avidity of the various TCRs for the αGC/CD1d complex remained the same irrespective of CD4 expression on the surface of the hybridoma, with the exception of the Vβ14-containing TCR (Supplementary Fig. 1A). Transduction of the double-negative (4- CD8-) hybridoma DN32D3 (Lantz and Bendelac, 1994) with a mouse CD4-encoding retrovirus also did not alter the binding of the TCR to the αGC/CD1d complex (Supplementary Fig. 1B) suggesting that CD4 only plays a minor role, if any, in modulating the affinity of the iNKT TCR for the αGC/CD1d complex.
To analyze the natural Vβ chain repertoire expressed by the iNKT cell population, we sorted αGC/CD1d tetramer+ TCRβ+ cells from the thymus of C57BL/6 mice and expanded the cells in the presence of IL-15, as previously described (Matsuda et al., 2002). After four days of culture, the cells were stained with a panel of anti-Vβ mAbs and the repertoire analyzed by flow cytometry. This strategy allows for the analysis of a large number of pure iNKT cells (Fig. 1D) and prevents any potential competition for binding that might occur during co-staining with the αGC/CD1d tetramer and certain anti-Vβ mAbs (data not shown). Interestingly, the prevalence of the various Vβ chains analyzed in the ex vivo iNKT cell repertoire correlated positively with the relative apparent avidity for the αGC/CD1d tetramer conferred to the TCR by the usage of these various Vβ chains. For example, an iNKT TCR containing Vβ8.2 binds αGC/CD1d tetramer the best and Vβ8.2 was the Vβ most frequently used by iNKT cells. Conversely, iNKT TCRs including Vβ9 bind the tetramer poorly and this Vβ is rarely used by iNKT cells.
Together, these data demonstrate that the iNKT TCRα chain can pair with all Vβ segments. The results also show that the differential Vβ usage in the iNKT TCR affects its apparent affinity for αGC/CD1d. Furthermore, the hierarchy with which Vβs confer ligand-affinity on the iNKT TCRs correlates with the frequency with which these Vβs are used in iNKT TCRs in vivo. This suggests that Vβs might affect recognition of αGC/CD1d and endogenous positively selecting ligand(s) similarly.
Clustering of Vβ-containing iNKT TCRs according to antigen stimulation
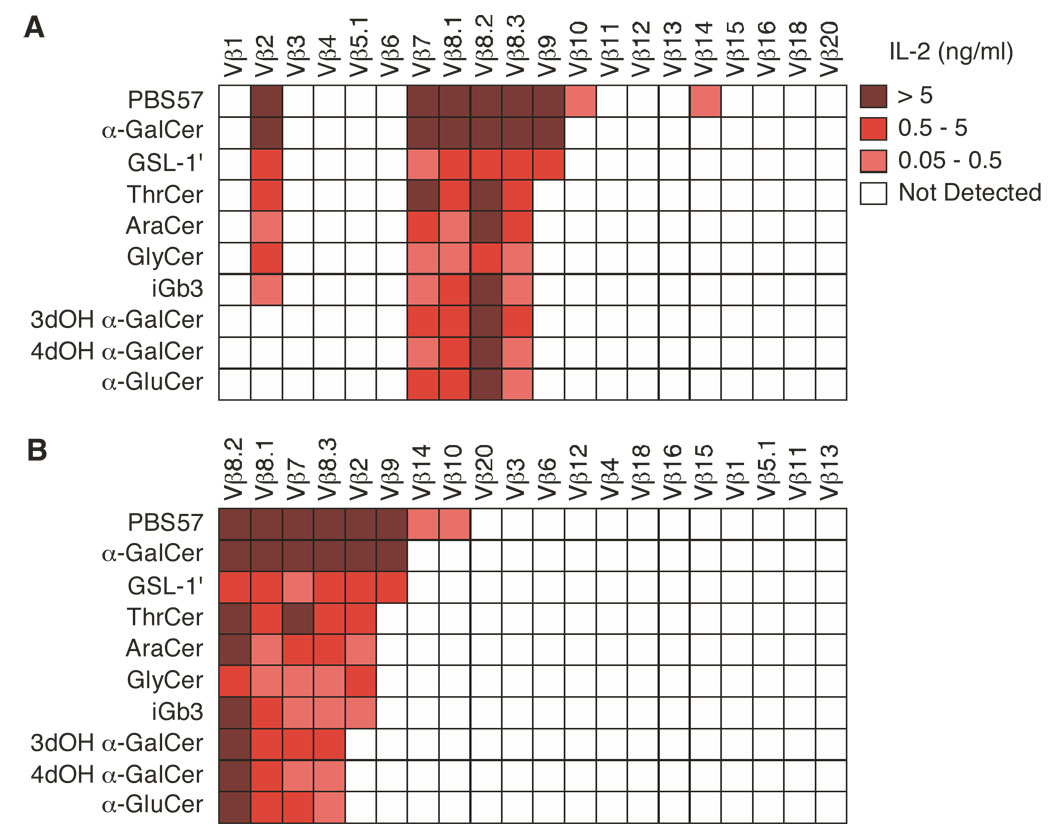
Because different Vβ usage in the iNKT TCR has been suggested to influence antigen specificity, we next tested how our panel of hybridomas responded to 10 structurally different antigens known to stimulate iNKT cells when presented by CD1d molecules, including the self glycosphingolipid iGb3 (Zhou et al., 2004), the microbial α-glycosphingolipid (GSL-1’) (Kinjo et al., 2005; Mattner et al., 2005) and several nonglycosidic compounds (Silk et al., 2008). Hybridomas expressing Vβ8.2, Vβ8.1, Vβ7 and Vβ8.3- containing iNKT TCRs responded to all the antigens tested (Fig. 2A), while hybridomas expressing Vβ2 responded only to PBS57, αGC, GSL-1’, ThrCer, Ara-Cer, GlyCer and iGb3, but not to 3’- and 4’ hydroxyl variants of αGC (3dOH αGC and 4dOH αGC0) nor to α-glucosylceramide (α-GluCer0). Vβ9 expressing hybridomas responded to a limited number of antigens (PBS57, αGC and GSL-1’), while the response of Vβ10- and Vβ14-containing iNKT TCRs was further restricted to the synthetic analog PBS57 (Liu et al., 2006) only (Fig. 2A). Hybridomas expressing the other Vβ chains did not respond to any antigen used in this study, although they were equally responsive to anti-CD3 stimulation (data not shown). These results indicate that the response of the iNKT cell hybridomas is not randomly distributed, but instead varies as a function of the Vβ usage. By rearranging the strength of the response versus the number of hybridomas stimulated by a given antigen, we found that Vβ segment usage correlated with the relative “potency” of the stimulating antigen (Fig. 2B). While hybridomas expressing iNKT TCRs with a relatively high affinity for αGC/CD1d, such as those containing Vβ8s and Vβ7 chains, responded to all antigens, hybridomas with iNKT TCRs of apparently lower affinity for αGC/CD1d (Fig. 1B, C) responded only to the most potent antigens. These results suggest that the relative apparent avidity of the iNKT TCR for a given antigen/CD1d complex can vary as a function of the Vβ chain used.
Figure 2.
Clustering of Vβ-containing iNKT TCRs according to antigen stimulation. (A, B) Enzyme-linked immunosorbent assay of IL-2 production by hybridomas expressing iNKT TCRs containing Vβ1 to Vβ20 TCRβ chains, stimulated with mCD1d-expressing A20 cells in the presence of PBS57 (1 µg/ml), αGC (1 µg/ml), GSL-1’ (1 µg/ml), ThrCer (0.2 µg/ml), AraCer (0.2 µg/ml), GlyCer (0.2 µg/ml), iGb3 (10 µg/ml), 3dOH αGC (1 µg/ml), 4dOH αGC (1 µg/ml) and α-GluCer (1 µg/ml). (B) Rearrangement of the strength of the response versus the number of hybridomas stimulated. Data represent the mean of two to four independent experiments.
Mutational analysis of Vβ8.2, Vβ7 or Vβ2-containing iNKT TCRs
Both crystallographic and mutational studies have revealed a very similar footprint for human Vβ11- and mouse Vβ8.2- containing iNKT TCRs on the αGC/CD1d complex (Borg et al., 2007; Scott Browne et al., 2007; Wun et al., 2008). The binding surface between the iNKT TCR and αGC/CD1d complex is composed entirely of germline-encoded residues within CDR1α, CDR3α and CDR2β loops. To test whether the underlying energetic basis of the iNKT TCR αGC/CD1d complex interaction might change as a function of the Vβ chain in the TCR, we compared the role of individual CDR1α, CDR2α and CDR3α residues in the Vα14 invariant chain when paired with wildtype Vβ8.2, Vβ7 or Vβ2 chains. Our results demonstrated that the same residues within the CDR1α (P28α, N31α), and CDR3α (D94α, R95α, G96α, S97α, L99α) loops were important for the recognition of the αGC/CD1d complex, irrespective of the Vβ chain associated with the Vα14 invariant chain (Fig. 3A, B, C). These results suggest that the docking of the Vα14 domain within Vβ8.2, Vβ7, and Vβ2-containing iNKT TCRs on the αGC/CD1d-glycolipid complex is similar, a conclusion that is supported by the Vβ8.2- and Vβ7- containing iNKT TCRs in complex with αGC presented by mouse CD1d molecules (Pellicci et al., Submitted).
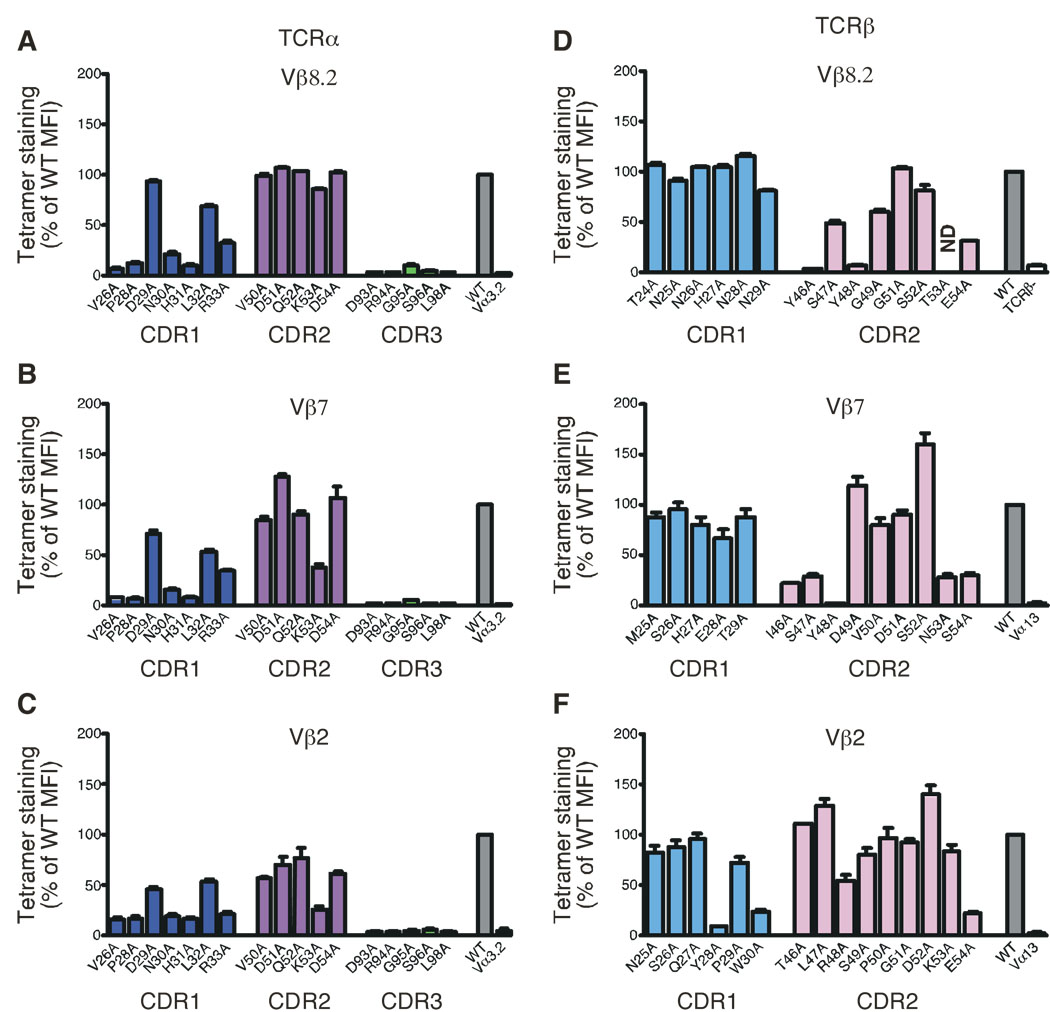
Figure 3.
Mutational analysis of Vβ8.2, Vβ7 and Vβ2-containing iNKT TCRs. Staining of hybridomas expressing mutant versions of the Vα14i TCRα chain associated with the wild-type (A) Vβ8.2, (B) Vβ7 or (C) Vβ2 TCRβ chains. Staining of hybridomas expressing alanine substitutions of (D) Vβ8.2, (E) Vβ7 or (F) Vβ2 associated with the wild-type Vα14i TCRα chain. Dark blue, CDR1α; magenta, CDR2α; green, CDR3α, light blue, CDR1β and pink, CDR2β. WT, unsubstituted Vα14i TCRα chain paired with unsubstituted Vβ8.2, Vβ7 or Vβ2 TCRβ chains (wild-type controls). Vα3.2, Vα14i TCRα chain in which the CDR1α region is swapped for the Vα3.2 CDR1α region, and paired with the appropriate TCRβ chains (negative controls for TCRα substitutions). Vα13, Vα13-Jα18 TCRα chain paired with the appropriate TCRβ chains (negative controls for the TCRβ substitutions). ND, not done. The MFI of tetramer staining for each mutant was determined for a narrow TCR gate and normalized to wild-type MFI (set as 100%). Data represent the mean + s.e.m. of three independent experiments. Analysis of the Vβ8.2 mutants, with the exception of Y46A and E54A mutants, has been published previously in (Scott Browne et al., 2007) and is shown for comparison.
Our previous mutational analysis of a Vβ8.2-containing iNKT TCR identified several CDR2β residues (Y46β, Y48β and to some extent E54β) required for the recognition of the antigen/CD1d complex (Scott Browne et al., 2007). Alanine substitutions of these residues revealed that while only a few contacts are mediated via the Vβ domain, they are absolutely critical to the interaction. These residues are identical, or similar, between human Vβ11 and mouse Vβ8s (Supplementary Fig. 2). However, the CDR1/2β regions of mouse Vβ2 and Vβ7 are unexpectedly dissimilar from those of Vβ8s. To understand further the molecular basis for the restricted TCRβ repertoire, we mutated individual residues in the CDR1β and CDR2β loops to alanine in Vβ7 and Vβ2 chains and expressed each mutant with the wild-type Vα14 partner in TCR-deficient hybridomas. We then analyzed the influence of each substitution on the recognition of CD1d tetramers loaded with αGC (Fig. 3 D, E, F). For the Vβ7 chain, alanine substitutions in CDR1β did not substantially affect αGC/CD1d tetramer binding. By contrast, tetramer binding was completely abrogated for the βY48A substitution and reduced by about 70% for βI46A, βS47A, βN53A and βS54A mutations. By contrast, βD49A and βS52A substitutions enhanced tetramer binding (Fig. 3E). In the Vβ2 TCRβ CDR2 region, βR48A and βE54A substitutions reduced the tetramer binding by 50% and 80% respectively, whereas other substitutions in CDR2β did not affect, or even enhanced, αGC/CD1d tetramer binding (Fig. 3F). CDR1β alanine mutants bound the αGC/CD1d tetramer equivalently to their wild-type counterpart, with the exception of βY28A that almost completely abrogated tetramer binding, and βW30A that reduced the binding by 70% (Fig. 3F), which suggest a potential involvement of the CDR1β loop in mediating recognition with CD1d.
The fact that similar residues, located at the same positions, between Vβ8.2 and Vβ7 (CDR2β 46, 48 and 54) are required for recognition of the αGC/CD1d complex suggested that Vβ8.2 and Vβ7 make similar contacts on CD1d-glycolipid complexes, consistent with recent crystallographic data (Pellicci et al., Submitted). Curiously, while Vβ2 also requires positions R48β and E54β to bind α-GalCer/CD1d complexes, there is an increased effect of CDR1β mutations compared to Vβ8.2 and Vβ7, suggesting that Vβ2 might dock slightly differently on CD1d.
CDR2β dictates iNKT TCR interactions with mCD1d + foreign or self ligands
The in vitro experiments described above highlighted the importance of CDR2β residues in αGC/CD1d recognition, raising the possibility that the Vβ bias of the iNKT cell repertoire might depend on the ability of particular CDR2β residues to make CD1d contacts in the context of the interaction determined by the canonical TCRα chain. To assess directly whether the CDR2β loop influences the development of iNKT cells, we produced TCRβ “retrogenic” mice using the wildtype Vβ8.2 and three Vβ8.2 mutants, βY46A, βY48A and βE54A. Bone marrow cells from 5-FU treated TCR Cβ/Cδ-deficient mice were transduced using retroviral constructs encoding one of the Vβ8.2 chains and transferred to sub-lethally irradiated RAG-deficient mice. Four to six weeks post-reconstitution, chimeric mice were sacrificed and analyzed for the presence of iNKT cells in the thymus by αGC/CD1d tetramer staining. While the wild-type Vβ8.2 TCRβ chain and βE54A mutant chains restored T cell development (Scott Browne et al., 2009) as well as a sizeable iNKT cell population, iNKT cells were undetectable in the chimeric mice expressing the βY46A and βY48A mutant chains (Supplementary Fig. 3). Comparable results were obtained with conventional TCR transgenic animals generated using the wildtype Vβ8.2 and the βY48A chains (data not shown). These results suggest that the same contacts between the CDR2β region of the iNKT TCR are needed for engagement of natural ligand(s)/mCD1d complex(es) and for positive selection of iNKT cells.
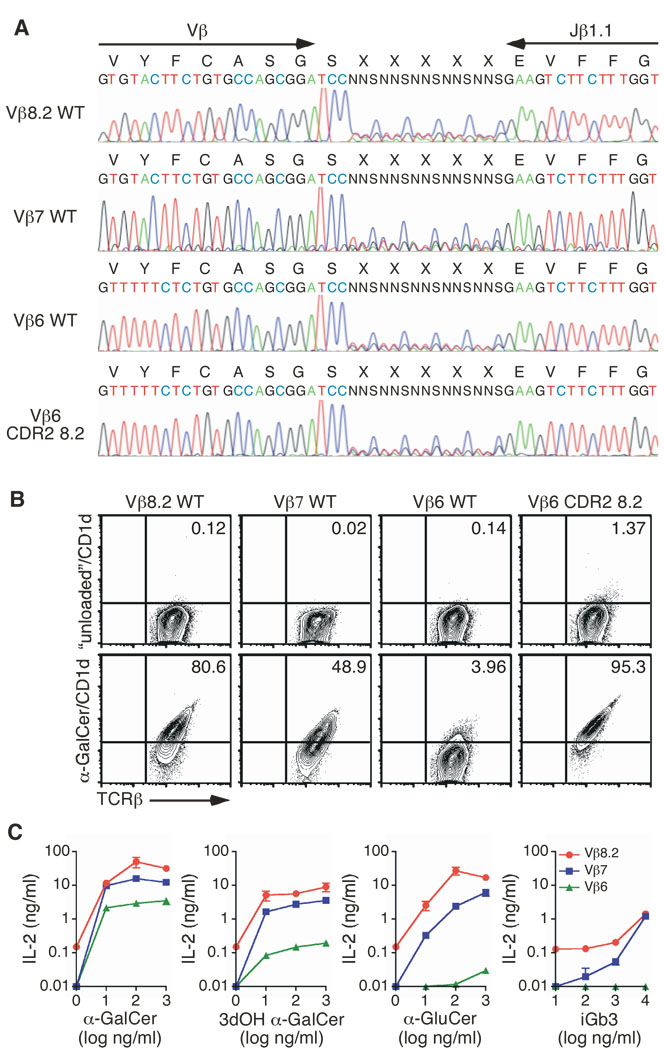
When paired with the invariant Vα14 chain, some Vβ chains, such as Vβ6, do not bind the αGC/CD1d tetramer (Fig. 1) nor do they respond to any of the glycolipid antigens presented by CD1d that we tested (Fig. 2). We hypothesized that specific CDR2β residues within these Vβ chains might prevent interaction with mCD1d or that the residues that favor the interaction might be absent. To test this possibility directly, we decided to swap the entire CDR2β region of Vβ8.2 (including 2 frame-work residues T53β and E54β) into the Vβ6 TCR chain (Fig. 4A). The Vβ6 chain was chosen because 1) it is relatively similar to Vβ8.2 (44% identity; 77% similarity); 2) the Y46β residue is already present in Vβ6 and 3) Vα14i/Vβ6 TCRs with the CDR3β DO-11.10 do not interact with αGC/CD1d tetramers nor do they recognize any of the 10 different iNKT cell antigens used in this study (Fig. 1, Fig. 2 and Fig. 4B).
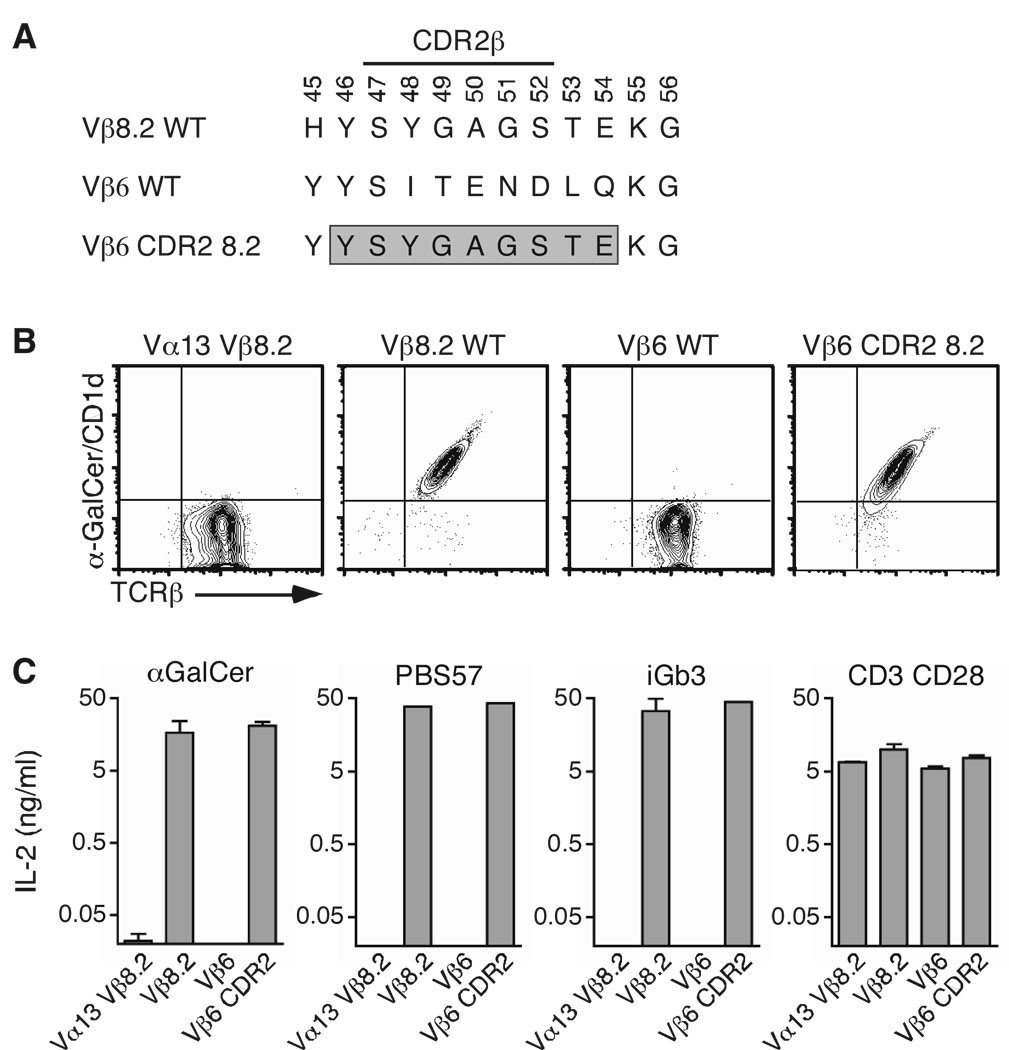
Figure 4.
CDR2β swapping restores CD1d-glycolipid recognition. (A) The Vβ6 CDR2β was substituted with that of Vβ8.2, from positions 46 to 54 (boxed sequence). (B) Staining of hybridomas expressing the indicated TCRβ chain associated with the Vα14i TCRα chain. (C) Enzyme-linked immunosorbent assay of IL-2 production by hybridomas stimulated with mCD1d-expressing A20 cells in the presence of αGC (1 µg/ml), PBS57 (1 µg/ml) or iGb3 (10 µg/ml), or plate-bound anti-CD3 (5 µg/ml) and anti-CD28 (2 µg/ml) antibodies. Data represent the mean + s.e.m. of three independent experiments.
The CDR2-modified Vβ6 chain was expressed in the TCRαβ− hybridoma with the Vα14 invariant chain and TCR-expressing hybridomas were sorted by flow cytometry. The cells were then stained with αGC/CD1d tetramers. Strikingly, swapping the Vβ6 CDR2 sequence with that of Vβ8.2 was sufficient to allow the TCR to interact with the αGC/CD1d complex (Fig. 4B). In addition, hybridomas expressing Vβ8.2- and Vβ6-modified iNKT TCRs responded similarly to APCs presenting αGC, PBS57 and iGb3 (Fig. 4C). We wanted to extend these findings to another Vβ8.2-related Vβ chain. The Vβ14 CDR2β loop encodes for two of the three residues (βY46 and βE54) that are important for the binding of Vβ8.2- and Vβ7-containing TCRs to the antigen/CD1d complex (Fig 3). However, an isoleucine residue is found at position 48 in place of the tyrosine residue found in Vβ8.2 and Vβ7 CDR2 loops (supplementary Fig 2). Therefore, we decided to replace the I48 residue by a tyrosine into this Vβ. The mutant TCRβ chain was expressed with the invariant Vα14 chain into the 5KC hybridoma; TCR-expressing cells were sorted and stained with the αGC/CD1d tetramer. The single introduction of a Y residue at position 48 of the CDR2β of the Vβ14 chain was enough to increase the MFI of αGC/CD1d tetramer binding to this TCR by more than 4 fold compared to the binding to the wildtype Vβ14-containing TCR (supplementary figure 4). Altogether, these results demonstrate that residues within the CDR2β loop are necessary and sufficient to dictate iNKT TCR reactivity with αGC/CD1d complexes.
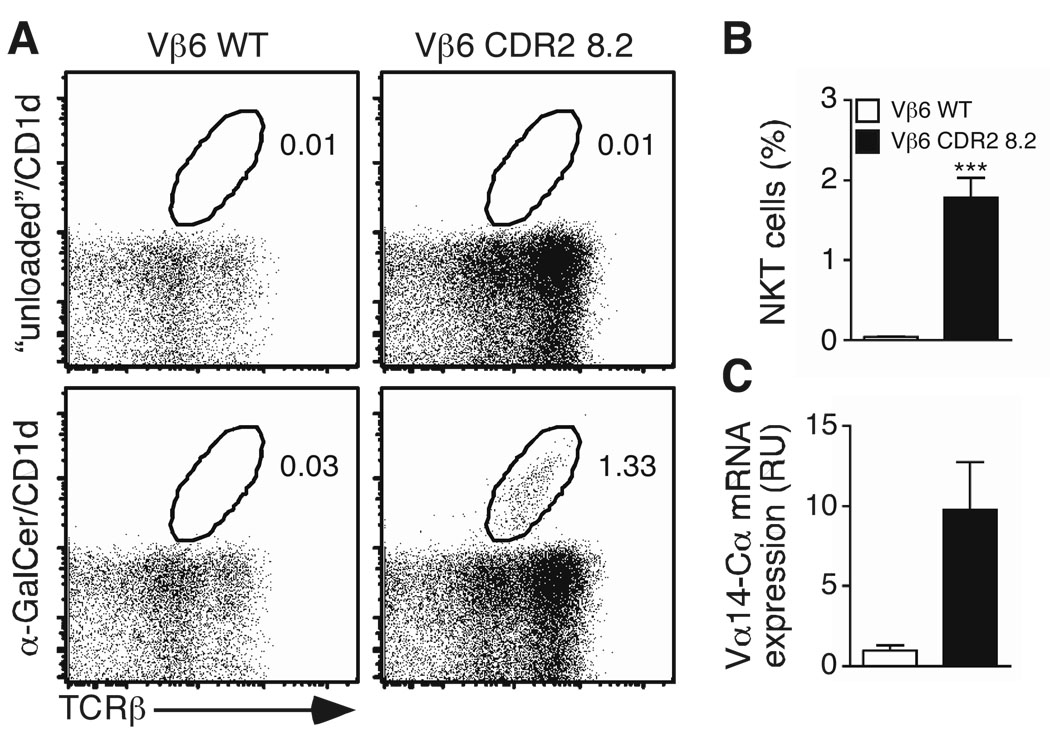
Next, we tested whether CDR2β residues also influence the positive selection of iNKT cells. TCRβ “retrogenic” mice were produced using the wildtype Vβ6 and the CDR2-modified Vβ6 mutant. Bone marrow cells from 5-FU treated TCR Cβ-deficient mice were transduced with either of the two retroviral constructs and transferred into lethally irradiated CD45.1 congenic mice. Four to six weeks post-reconstitution, chimeric mice were sacrificed and analyzed for the presence of iNKT cells in the thymus. iNKT cells were not detectable in chimeric mice expressing the wild-type Vβ6 chain (Fig. 5A). By contrast, mice expressing the CDR2β-modified Vβ6 chain developed αGC/CD1d tetramer+ cells (Fig. 5A, B). To formally exclude that the apparent absence of αGC/CD1d tetramer+ cells in Vβ6-expressing retrogenic mice might be due to the inability of the tetramer to interact with this particular TCR (Fig. 4), we also measured by quantitative PCR the amount of Vα14-Cα mRNA found within sorted CD45.1− CD8− GFP+ TCRβ+ cells from the thymus of the retrogenic mice (Fig. 5C). The results demonstrated that Vα14-Cα mRNA was only significantly detected within retrogenic T cells expressing the CDR2-modified Vβ6 chain but not the wildtype Vβ6 chain.
Figure 5.
CDR2β swapping restores iNKT-cell development in vivo. TCRβ-deficient donor bone marrow cells were infected with retroviruses encoding the indicated TCRβ chain and eGFP as a reporter. Cells were injected i.v. into sub-lethally irradiated CD45.1 congenic recipient mice. (A, B) Thymocytes were stained 5 weeks post-reconstitution. Cells were gated on eGFP+ B220- CD8- F4/80− Gr-1- cells and presented plots are representative of two independent experiments (3 mice/group) (A). Percentage of αGC/CD1d tetramer+/TCRβ+ cells in the thymus of TCRβ retrogenic mice (B) Data shown are the mean percentage + s.e.m. of 6 mice/group. Statistical significance (p < 0.001) was determined using unpaired Student’s t test. (C) Thymocytes from TCRβ retrogenic mice were depleted of cells expressing CD8α and CD45.1 and sorted for eGFP, TCRβ and CD4 expression and total RNA was prepared. Amounts of Vα14-Cα transcripts were analyzed by quantitative PCR. Normalization of the samples was relative to the quantity of Cα transcripts. Data are representative of two experiments.
Altogether, these results demonstrate that structural constraints imposed by the amino acid composition of the CDR2β loop play a critical role in the TCRβ bias of the selected iNKT cell repertoire in vivo.
CDR3β modulates iNKT TCR affinity
Comparative analysis of TCRβ chains in the context of the DO-11.10 CDR3β revealed that most Vβ chains, including Vβ6, did not result in glycolipid/CD1d complex interaction (Fig. 1) and, at least for Vβ6, did not drive the development of iNKT cells in vivo (Fig. 5A). However, Vβ6-expressing iNKT cells exist in vivo, albeit at a low frequency, and Vβ6-expressing iNKT cell hybridomas have been described previously (Behar et al., 1999). One possibility that might reconcile these apparently contradictory results may lie within the CDR3β amino acid composition of the various Vβ6+ iNKT TCRs studied. We previously reported that CDR3β amino-acid composition can significantly modulate the overall affinity of the iNKT TCR for the antigen/CD1d complex (Scott Browne et al., 2007). Consequently, one might anticipate that the amino acid diversity tolerated within the CDR3β loop might vary depending upon the Vβ used. To formally test this hypothesis in a controlled system, we used retroviruses to generate four CDR3β libraries encoding the wildtype Vβ8.2 chain, the wildtype Vβ7 chain, the wildtype Vβ6 chain or the Vβ6 chain with its CDR2β replaced by the Vβ8.2 CDR2β sequence. Five positions at the tip of the CDR3β loops were randomized for each of the libraries (Fig. 6A), and were estimated to encode between 6,000 and 17,000 different sequences (data not shown). Retroviruses were prepared and used to transduce the Vα14i-expressing TCRβ-negative hybridoma. Transduced cells were sorted for TCRβ expression and stained with the αGC/CD1d tetramer. Over 80% of the CDR3β sequences in the context of wildtype Vβ8.2–expressing TCRs interacted with the tetramer (Fig. 6B), suggesting that most of these CDR3β sequences are compatible with αGC/CD1d recognition, in agreement with previous results (Lantz and Bendelac, 1994; Matsuda et al., 2001).
Figure 6.
CDR3β modulates iNKT TCR affinity. (A) Sequence of the wild-type Vβ8.2, wild-type Vβ7, wild-type Vβ6 WT and CDR2-modified Vβ6 CDR3β random constructs. (B) Staining of hybridomas expressing the indicated CDR3β random construct associated with the Vα14i TCRα chain. The percentage of αGC/CD1d tetramer+ cells is shown. Plots are gated on live eGFP+ cells and are representative of four independent experiments. (C) Enzyme-linked immunosorbent assay of IL-2 production by hybridomas stimulated with the indicated antigen in the presence of mCD1d-expressing A20 cells. One representative experiment out of two is shown.
Interestingly, only 50% of the TCRs expressing Vβ7 and 4% of the TCRs expressing the wild-type Vβ6 libraries interacted with the αGC/CD1d tetramer (Fig. 6B). Remarkably, more than 90% of the TCRs expressing the modified Vβ6 bound the tetramer. Altogether, these results demonstrate that the CDR3β amino acid composition diversity that is tolerated by the iNKT TCR and still preserves αGC/CD1d tetramer recognition varies with Vβ usage. However, only a few wildtype Vβ6-containing iNKT TCRs appeared compatible with αGC/CD1d recognition. To determine the nature of the CDR3β responsible for this reactivity, αGC/CD1d tetramer positive and negative cells from the Vβ6+ TCR library were sorted and mRNA extracted from each population. After cDNA synthesis, the Vβ6 TCRs were amplified by PCR using appropriate primers, cloned into the retroviral vector and sequenced. In addition, each Vβ6 TCR was expressed separately with the invariant Vα14 chain into the 5KC hybridoma and stained with the αGC/CD1d tetramer. As shown in supplementary Table I, a unique combination of CDR3β sequences was found within the tetramer+ population, while the sequences found within the tetramer− population were more diverse. Analysis of the CDR3β sequences revealed the I/LXXPL/I motif (where X represents any amino acid, and slashes separate alternative amino acids that may occupy a given position), which in the context of Vβ6-Jβ1.1 rearrangement with a fixed CDR3 length, appears to be required for high binding to the αGC/CD1d tetramer. These results support the hypothesis that only few wildtype Vβ6-containing iNKT TCRs, with specific CDR3β sequences, are in fact compatible with αGC/CD1d recognition. In striking contrast, when the Vβ6 chain had a modified CDR2β region, greater than 90% of the CDR3β sequences within the retroviral library were tolerated and interacted with the αGC/CD1d tetramer (Fig. 6B). Thus, the extent of CDR3β sequence diversity tolerated by iNKT TCRs while maintaining αGC/CD1d tetramer recognition is a function of the Vβ used by the TCR and, more specifically, is controlled by the particular CDR2β amino acid composition of this Vβ chain.
To test if the Vβ hierarchy previously established using Vβ chains with a fixed CDR3β region (Fig. 1), could be extended to include a diverse collection of CDR3βs, the ability of hybridomas from the various TCR libraries to respond to other iNKT cell antigens was assessed (Fig. 6C). In all cases, the Vβ hierarchy remained the same in the context of TCRs with diverse CDR3βs, with Vβ8.2 expressing cells responding better than Vβ7 and Vβ6 expressing cells to all glycolipids tested. These data demonstrate that our previous results with all Vβs in the context of the fixed CDR3β of DO-11.10 are reproducible even in the context of a diverse unselected repertoire.
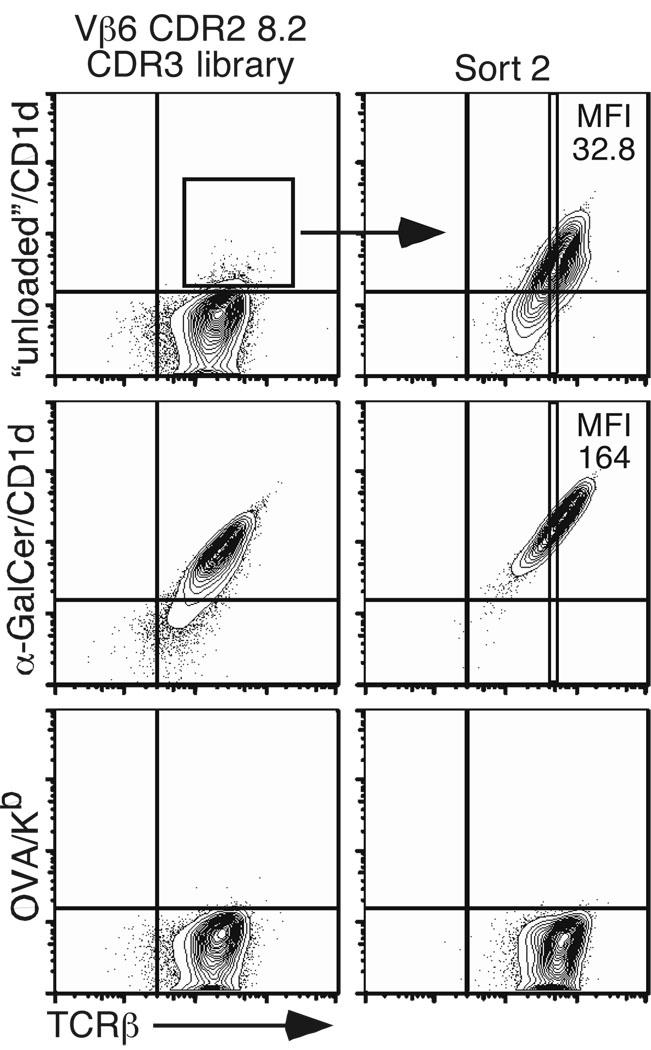
Finally, based on the above results, one might predict that by selecting adequate CDR2β and CDR3β residues, the affinity of the iNKT TCR for the antigenic/CD1d complex might be improved. In fact, while controlling for the specificity of the αGC/CD1d tetramer staining, we realized that a small but reproducible percentage of TCR-expressing hybridomas derived from the CDR2β-modified Vβ6 chain TCR library interacted with the control CD1d tetramer, which is not “loaded” with any exogenous antigen (Fig. 6B). These cells were sorted twice and subsequently re-tested for reactivity with the “unloaded” CD1d tetramer. Although no external antigen was added in the preparation of the tetramer reagent, it is likely that “natural” lipid(s) derived from the 293 cells used to produce CD1d monomers, are in fact loaded within the groove of CD1d molecules (Yuan et al., 2009). As seen in Fig. 7, a homogeneous population of hybridomas expressing TCRs capable of interacting with the “unloaded” mouse CD1d tetramer could clearly be defined. These results demonstrate that the affinity of the iNKT TCR for the CD1d-antigenic complex can be improved upon by optimizing the CDR2 and CDR3 loops of the Vβ chain. Furthermore, at an identical level of TCR expression, the MFI of αGC/CD1d tetramer staining is higher compared with the MFI of “unloaded” tetramer staining (Fig. 7). These results suggest that the affinity of TCRs selected on the basis of the recognition of an unknown self-antigen presented by mouse CD1d molecules can be further improved by the addition of an iNKT cell antigen such as αGC.
Figure 7.
Optimal CDR2β and CDR3β composition improves iNKT TCR affinity for CD1d. “Unloaded”/CD1d tetramer+ hybridomas derived from the CDR2-modified Vβ6 chain TCR library were sorted twice and stained with the indicated tetramers. Plots are gated on live eGFP+ cells and are representative of three independent experiments.
DISCUSSION
Identification of the structural features of antigen recognition by iNKT cells is critical to understanding this lymphocyte population and to the design of effective ligands aimed at exploiting iNKT cell functions. iNKT cells express a semi-invariant TCR, composed of Vα24-Jα18 segments paired with Vβ11 in humans and Vα14-Jα18 segments paired with a limited set of Vβ chains in mice (Vβ8.2, Vβ7 and Vβ2). The reasons for this Vβ usage bias have remained unclear. We found no evidence that certain Vβ chains are excluded because they cannot pair with the Vα14 invariant chain, as the hybridoma subclones we generated with each of 20 different Vβs all expressed similar levels of TCR and appeared stable. Notably, only 8 out of the 20 different Vβ segments, when associated with the Vα14 invariant chain, produced a TCR that, at identical level of expression, interacted with the αGC/CD1d tetramer. The relative avidity of these TCRs for the αGC/CD1d complex, as measured by titration of the tetramer, varied greatly. These results demonstrate that the Vβ composition of the iNKT TCR affects its affinity for the αGC/CD1d complex and are in good agreement with previous studies showing that the TCRβ chain influences the avidity of αGC/CD1d binding, with Vβ8.2 conferring higher avidity than Vβ7 and than Vβ2 (Schumann et al., 2003; Stanic et al., 2003).
Interestingly, the relative avidities conferred by the different Vβ correlated positively with the frequency with which the Vβ is used in the natural repertoire of iNKT cells. These results suggest the possibility that a certain affinity threshold of the iNKT TCR for the positively selecting ligand/CD1d complex might be required for positive selection of iNKT cells. In this scenario, Vβ9, Vβ10 and Vβ14-containing TCRs might not have sufficient affinity for the self-ligand(s)/CD1d complexes that positively select iNKT cells. One potential problem with such an interpretation is that the nature of the positively selecting ligand(s) remains uncertain and therefore the Vβ hierarchy observed for reactivity to the αGC/CD1d complex might not translate to other antigen/CD1d complexes. The data presented here suggest that, in general, iNKT TCRs react with different antigens with a defined hierarchy, reacting best with CD1d bound to αGC and PBS57, less well with, for example, CD1d bound to iGb3 and undetectably with CD1d bound to self ligand(s). Again, in general, the affinity of iNKT TCRs for a particular ligand, αGC/CD1d for example, is controlled by CDR2β and CDR3β, such that iNKT TCRs that contain Vβ8.2 usually react more strongly that those that contain Vβ2 and even more strongly than those containing Vβ6. CDR3β sequences modulate this phenomenon, however, so that a few, rare, CDR3β sequences can compensate the inadequacies of Vβ6 CDR2β, and allow recognition. Conversely, a few, rare, CDR3β sequences can interfere with the otherwise excellent recognition properties of iNKT TCRs containing Vβ8.2. Thus our data suggest that, in general, iNKT TCRs recognize CD1d bearing different ligands with the same hierarchy of affinities, an idea supported by crystallographic data showing that the glycolipid ligand is engaged only by the invariant iNKT TCR α chain (Borg et al., 2007; Pellicci et al., Submitted). If this is true for all ligands, the iNKT TCR β chain can affect the affinity of the iNKT TCR by affecting its ability to bind CD1d, but can only affect the specificity of the iNKT TCR if it affects the orientation of the TCR α chain (Pellicci et al., Submitted), or if the glycolipid ligand affects the configuration of the CD1d protein itself.
Analysis of Vβ usage by iNKT cells in vivo under conditions where CD1d was under-expressed revealed an increased frequency of Vβ7+ iNKT cells while the proportion of Vβ8.2+ iNKT cells remained constant (Schumann et al., 2006; Wei et al., 2006). These results were interpreted as a reflection of the preferential positive selection of Vβ7+ iNKT cells at suboptimal endogenous ligand concentration and suggested that the hierarchy of Vβ usage for the endogenous positively selecting ligand(s) is Vβ7>Vβ8.2>Vβ2. A similar Vβ hierarchy was found for the response to the self-glycosphingolipid iGb3, and it was argued that only iNKT cell TCRs composed of Vβ8.2, Vβ7 or Vβ2 were suitable for positive selection by the iGb3/CD1d complex (Schumann et al., 2006; Wei et al., 2006). In our in vitro experiments, we did not find a Vβ7 bias in response to any of the glycolipids that we tested, including iGb3. The reasons for the discrepancy between the in vivo and in vitro results are currently unclear. One possibility is that particular CDR3β sequences are used by Vβ7+ iNKT cells for the recognition of iGb3 in vivo in a way that our CDR3β library would not reveal, perhaps because of the fixed CDR3β length and/or Jβ that we used. On the other hand, the precise nature of the ligand(s) involved in positively selecting the iNKT cell repertoire remains unknown and the role of iGb3 in this process is currently controversial (Christiansen et al., 2008; Li et al., 2008; Li et al., 2009; Porubsky et al., 2007; Speak et al., 2007). Thus, the panel of antigens that we tested in vitro might not be representative of the ligand(s) involved in positive selection. Although such a possibility cannot be formally excluded, it would imply that recognition of the positively selecting “self” by the iNKT TCR is somehow different from the recognition of αGC and the other glycolipids tested. Unfortunately, we cannot further test this hypothesis with our current TCR mutants because the hybridomas do not show any significant autoreactive response in the presence of thymocytes or bone-marrow-derived dendritic cells (data not shown). Nevertheless, we agree with the earlier study suggesting that the iNKT TCR repertoire directed at glycolipid/CD1d complexes could indeed be potentially larger than the actual natural repertoire of iNKT cells (Wei et al., 2006). It is possible that the overall affinity of the iNKT TCR for the glycolipid(s)/CD1d complex responsible for the positive selection of the cells, rather than the specific nature of a particular ligand, might be responsible for this restriction in repertoire diversity.
Mutational analysis of three mouse iNKT TCRs with the invariant Vα14 chain associated with Vβ8.2, Vβ7 or Vβ2 demonstrated that the “energetic hot-spot” of the TCR on the αGC/CD1d complex remains largely similar, regardless of the Vβ chain used. These results are substantiated by the recent analysis of the crystal structures of two mouse iNKT TCRs, containing either Vβ8.2 or Vβ7, in complex with αGC/CD1d (Pellicci et al., Submitted). In both cases, the invariant TCRα chain dominates the interaction with both the glycolipid and mouse CD1d, while the role of the TCRβ chain is mostly restricted to the CDR2β loop interacting with the α1 helix of CD1d. Importantly, the CDR2β residues that mediate these interactions with the α1 helix of CD1d, namely Y46 and Y48, are conserved between human Vβ11 and mouse Vβ8.2. These results suggest that the amino acid sequence of the CDR2β loop is likely to be critically important in determining which Vβ chain, when associated with Vα14, can interact with CD1d molecules. In agreement with this, analysis of mice transgenic for TCRβ chains usually not found in the iNKT cell repertoire demonstrated that they do not support positive selection of iNKT cells in vivo (Dao et al., 2004; Ohteki and MacDonald, 1996). Furthermore, the crystal structure of the Vβ7+ iNKT TCR indicates that the Y48 residue, conserved between Vβ8.2 and Vβ7, makes similar contacts with CD1d. Because the Vβ7 chain leans more towards the CD1d molecule than the Vβ8.2 chain, further contacts mediated by the CDR2β S54 and the CDR1β E28 residues with the CD1d molecules tend to compensate for the absence of the Y46 residue in the CDR2β loop of Vβ7 (Pellicci et al., Submitted). Our alanine scan analysis revealed the involvement of two CDR1β residues (Y30 and W32) in Vβ2-containing iNKT TCR for the binding to the αGC/CD1d complex. These results suggest that, like Vβ7 (Pellicci et al., Submitted), the relative juxta-positioning of the Vβ2 and Vα14 domains might position the Vβ2 chain closer to the CD1d molecule, allowing for a greater contribution of the Vβ2 CDR1β loop to interact with CD1d. Moreover, aromatic residues are known to participate in protein-protein interactions and the bulky aromatics of Y30β and W32β are likely to assist in bridging any potential gap between the iNKT TCR and CD1d.
By simply grafting the CDR2β region (and 2 adjacent frame-work residues) of Vβ8.2 into the Vβ6 chain, we were able to impart mouse CD1d reactivity onto a TCR that otherwise did not interact with mouse CD1d. This Vβ6-modified chain, when associated with the invariant Vα14 chain, responded to all antigens tested and permitted selection of iNKT cells in vivo, suggesting that the introduced modifications probably altered the recognition of CD1d molecules per se and not recognition of a specific antigen/CD1d complex. To our knowledge, this is the first evidence of CDR grafting successfully transferring reactivity for an αβ TCR. Graft of a CDR3δ loop into a naïve αβ TCR also reconstituted reactivity to other non classical class I molecules, T10 and T22 (Adams et al., 2008). These results highlight the evolutionary pressure in focusing the binding energetics on specific CDR loops for the recognition of non-conventional MHC-like molecules.
We propose that structural constraints within the CDR2β loop for interaction with mouse CD1d play a major role in biasing selection and thus the iNKT cell repertoire. It is interesting to note that the same CDR2β residues have been proposed to be important for the generic recognition of MHC molecules by Vβ8.2-containing αβ TCRs (Marrack et al., 2008), although the site of Vβ8.2-MHC recognition is markedly different from that of Vβ8.2-CD1d. It is likely that the entire composition of the CDR2β loop, rather than the simple presence of the Y46 and Y48 residues, is important for mediating proper contacts with CD1d. This is perhaps best illustrated by the effect of CDR2β mutations within residues that do not contact CD1d on the crystal structure (Pellicci et al., Submitted) that nevertheless influence the recognition indirectly (Scott Browne et al., 2007). Similarly, while the I48Y mutation in the CDR2β loop of the Vβ14 chain positively improved the binding to the αGC/CD1d tetramer compared to the wildtype Vβ14 chain, it remained lower compared to the Vβ8.2-containing TCR. These results might explain why other human and mouse Vβ chains (Arden et al., 1995), which also contain various combinations of these two residues but in the context of other frame-work residues, are poorly represented in the iNKT cell repertoire. In addition to sequence differences, slight changes in the angle between the Vα14 and the various Vβ chains are likely to also play some role in selecting the iNKT cell repertoire by influencing the positioning of the CDR2β and CDR1β loops over the CD1d molecules.
With the help of retroviral libraries coding for various Vβ chains randomized at five CDR3β positions, we were able to create an unselected iNKT cell repertoire in vitro. TCRs expressed by αGC-reactive iNKT cells are known to have polyclonal CDR3β sequences and it has been suggested that such diversity might allow for discrimination between different glycolipid antigens loaded on CD1d molecules (Godfrey et al., 2005; Kinjo et al., 2005). We recently showed that despite the CDR3β diversity, individual iNKT TCRs reacted similarly to many glycolipid/CD1d complexes and that the role of the CDR3β region is to modulate the overall affinity of the TCR (Scott Browne et al., 2007). Our present results confirm and extend these findings. First, the extent of CDR3β sequence diversity tolerated by iNKT TCRs while maintaining αGC/CD1d tetramer recognition is a function of the Vβ used by the TCR and, more specifically, is controlled by the unique CDR2β amino acid composition of this Vβ chain. TCRs with Vβ chains that have CDR2β loops that allow appropriate interactions with CD1d probably reach sufficient affinity for the positively selecting ligand(s)/CD1d complex so that little to no energy is required from the CDR3β loop. As a consequence, most CDR3β loops, but not all, are tolerated and allow recognition of the antigen/CD1d complex. Some CDR3β loops may impede, rather than enhance, antigen recognition, as demonstrated in an earlier study of human iNKT TCR binding (Kjer-Nielsen et al., 2006). Overall, when a Vβ ideally suited to interact with CD1d is used, most CDR3β sequences may be positively selected in the iNKT cell repertoire. By contrast, for Vβ chains with a CDR2β loop that contributes to fewer interactions with CD1d, specific CDR3β sequences are necessary to compensate for the lack of energy provided to the interaction. This is perhaps best exemplified by the selection of a particular CDR3β motif that allows some Vβ6 TCRs to recognize CD1d + ligands. Thus, relatively few TCRs using these particular Vβs (such as Vβ6) would then be expected in the natural iNKT cell repertoire, which is consistent with our observations. These results suggest a functional collaboration between the CDR2β and CDR3β loops, where the iNKT cell repertoire might be represented as a “sliding scale” with the affinity of the TCR determined by a hierarchy of Vβ chains and modulated by the CDR3β composition for this particular Vβ chain. Hence, we propose that the inherent CD1d reactivity of a given Vβ chain determines its relative position in the Vβ hierarchy while the CDR3β sequence defines the placement of an individual TCR within the overlapping hierarchy.
What then, are the functional consequences of restricting the iNKT cell repertoire to a limited number of Vβ domains? For “conventional” T cells that recognize highly polymorphic MHC molecules, developing a broad Vα and Vβ repertoire of TCRs allows the immune system to respond to the array of potential foreign peptides. Thus, restricting the TCRβ diversity among the iNKT cell population would potentially exclude cells capable of recognizing some glycolipid antigens. Indeed, we and others (Wei et al., 2006) have observed that some TCRs capable of αGC/CD1d recognition are excluded from the iNKT cell repertoire. One might expect, a priori, that a less diverse iNKT cell population would impair the host’s ability to recognize foreign glycolipid antigens. Alternatively, we propose that restricting the TCRβ repertoire in fact promotes a “functionally diverse” iNKT cell population. Using mutational analyses, supported by crystallographic studies (Borg et al., 2007; Pellicci et al., Submitted), we observed that the iNKT TCR uses primarily germline-encoded residues to recognize CD1d presenting diverse glycolipids (Borg et al., 2007; Scott Browne et al., 2007). Based on these data, we proposed that the iNKT TCR functions as a pattern recognition receptor, where diverse antigens are recognized using a conserved TCR recognition strategy (Scott Browne et al., 2007). The three NKT TCR-CD1d structures currently available are broadly consistent with this proposition. While thymic selection restricts the iNKT population to only a few Vβ, the majority of cells in this repertoire is capable of recognizing a wide array of potential antigens. Using this mechanism during development excludes iNKT cells, which may only recognize the highest affinity antigens. Thus, seemingly restricting the iNKT cell repertoire promotes and magnifies efficient recognition of many potential pathogens.
EXPERIMENTAL PROCEDURES
Reagents
α-galactosylceramide (αGC) and isoglobotrihexosylceramide (iGb3) were purchased from Alexis Biochemicals. PBS57 was kindly provided by Dr. Paul Savage (Brigham Young University, Provo, Utah), while the GSL-1’ antigen was obtained from the National Institutes of Health core facility. 3dOH αGC, 4dOH αGC and α-glucosylceramide (α-GluCer) compounds were synthesized by Dr. Amy Howell (University of Connecticut, Storrs, Connecticut) and have been described previously (Wun et al., 2008). Nonglycosidic compounds have been described previously (Silk et al., 2008).
TCRαβ constructs and retroviral plasmids
Wild-type and mutant TCRα chains were generated as previously described (Scott Browne et al., 2007). Wild-type and mutant TCRβ chains were constructed by PCR from C57BL/6 thymus-derived cDNA, or alternatively from plasmid templates, with overlapping primers and cloned into engineered restriction sites. TCRβ constructs were cloned into mouse stem cell virus-based plasmids with an internal ribosome entry site (IRES) plus sequence encoding for the human nerve growth factor receptor as a reporter (kindly provided by Dr. Steve Reiner, University of Pennsylvania).
Cell lines and retroviral packaging
Wild-type or mutant TCRβ constructs were expressed together with wild-type or mutant Vα14i TCRα constructs by retroviral transduction of 5KC-78.3.20, a hybridoma selected for loss of both TCRα and TCRβ chains (White et al., 1993). Retroviral plasmids were transfected into Phoenix cells together with the pCLEco accessory plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s specifications. Retrovirus-containing supernatants were harvested 48 h after transfection, centrifuged and filtered to remove debris. Hybridomas were ‘spin-infected’ at 3,300 × g for 90 min at 37 °C in retrovirus-containing supernatants supplemented with polybrene (8 µg/ml). Hybridomas were then sorted on a MoFlo cell sorter (Dakocytomation) on the basis of retroviral reporter and TCRβ expression.
Tetramer staining
Biotinylated recombinant mouse CD1d protein, provided by the National Institutes of Health core facility, was incubated overnight with αGC in a solution of 0.05% (vol/vol) Tween 20 in PBS, followed by the addition of phycoerythrin-conjugated streptavidin. Alternatively, the “unloaded” tetramer was prepared identically without the addition of the antigen αGC. Hybridomas were stained for 45 min at 25°C with the indicated tetramer plus antibody to TCRβ (anti-TCRβ; H57-597; eBiosciences), and cells were analysed on a FACSCalibur or FACScan flow cytometer (BD Biosciences). Data were analysed with FlowJo software (Treestar).
Hybridoma stimulation
5 ×104 hybridomas were cultured for 20 h with 5 ×104 mCD1d-transfected A20 cells, plus the indicated antigens, in complete RPMI medium containing 10% (vol/vol) FCS. Hybridoma responses were measured by an IL-2 enzyme-linked immunosorbent assay according to standard protocols.
Ex vivo NKT cells analysis
Thymocytes from C57BL/6 mice were subjected to depletion by magnetic-activated cell separation with anti-CD8α beads (Miltenyi Biotec) and cultured for 48 h in RPMI medium containing 10% (vol/vol) FCS and IL-15 (50 ng/ml, R&D Systems). Cells were stained for 45 min at 25 °C with mouse CD1d tetramer loaded with α-GalCer plus antibody to TCRβ. αGC/CD1d tetramer+ TCRβ+ cells were sorted using a MoFlo cell sorter and then cultured for another 4 days in complete RPMI medium containing 10% (vol/vol) FCS and IL-15 (50 ng/ml). Cells were stained with mouse CD1d tetramer loaded with αGC plus antibody to TCRβ to control for purity, or with antibodies to Vβ2, Vβ3, Vβ4, Vβ6, Vβ7, Vβ8.1, Vβ8.2, Vβ8.3, Vβ9, Vβ10, Vβ12, Vβ14 antibodies. Vβ8.1 relative usage was determined by differential staining between antibodies to Vβ8.1/8.2 (clone KJ16) and to Vβ8.2 (clone F23.2).
TCRβ ‘retrogenic’ mice
TCRβ constructs were cloned into mouse stem cell virus-based plasmids followed by an IRES and sequence encoding green fluorescent protein (eGFP) as a reporter. Retroviruses were produced as described above. Bone marrow cells from 5 Fluorouracyl-treated TCRβ/δ-deficient or TCRβ-deficient donor mice (0.15 mg/g weight) were harvested and cultured for 4 days in DMEM conditioned medium containing IL-3, IL-6 and SCF. Cells were ‘spin-infected’ on day 1, 2 and 3 after collection and injected on day 4 into sub-lethally irradiated CD45.1 congenic recipient mice (900 rads), or alternatively RAG-deficient mice (400 rads). Five to six weeks post-reconstitution, chimeric mice were sacrificed and thymocytes were stained for 45 min at 4 °C with phycoerythrin-conjugated αGC/CD1d tetramer, PerCP-conjugated anti-TCRβ mAb, allophycocyanin-conjugated anti-CD45.1 mAb, and Pacific Blue-conjugated anti-B220, anti-CD8α, anti-F4/80 and anti-Gr1 mAbs. Cells were acquired on a LSRII flow cytometer (BD Biosciences) and data were analysed with FlowJo software.
Quantitative RT-PCR
Thymocytes from TCRβ retrogenic mice were stained with biotin-conjugated antibodies to CD8α and CD45.1 and were subjected to depletion by magnetic-activated cell separation with anti-biotin beads (Miltenyi Biotec), and then stained with antibodies to CD4 and TCRβ. eGFP+ TCRβ+ CD4+ cells were sorted using a MoFlo cell sorter and total RNA was prepared using TRIzol solution (Invitrogen) according to the manufacturer’s protocol. Reverse transcription was carried out by using the SuperScript III kit (Invitrogen) and the amount of amplicon generated was monitored using a DNA engine Opticon 2 apparatus (Bio-Rad) with gene specific primers and probes and the Platinum Quantitative PCR SuperMix UDG (Invitrogen). Primers and probes have been described previously (Gapin et al., 2001).
CDR3β libraries
Wild-type Vβ8.2, wild-type Vβ7, wild-type Vβ6 and CDR2-modified Vβ6 CDR3β random constructs were generated by PCR using forward primers specific of each Vβ chain and the reverse primer 5' - TCT CAG ATC TTC TAC AAC TGT GAG TCT GGT TCC TTT ACC AAA GAA GAC TTC SNN SNN SNN SNN SNN GGA TCC GCT GGC ACA - 3' (Integrated DNA Technologies). Full-length TCRβ constructs were obtained by overlapping PCR and cloned into engineered restriction sites in the pMSCVpuro vector (Clontech). We estimated the size of the libraries to be 8,000, 6,000, 6,500 and 17,000 sequences for wild-type Vβ8.2, wild-type Vβ7, wild-type Vβ6 and CDR2-modified Vβ6 respectively. Retroviruses were packaged as described above and used to transduce the TCR-negative hybridoma 5KC-78.3.20, together with the Vα14i TCRα chain, at a low infection rate (< 1%) to minimize multiple retroviruses entering a single cell. TCR positive cells were enriched by addition of puromycin (1 µg/ml) into the culture media and hybridomas were subsequently sorted for TCR expression.
Supplementary Material
CD4 expression on T cell hybridomas does not affect TCR affinity. (A) Staining of CD4+ and CD4− hybridomas expressing the Vα14i TCRα chain and the indicated TCRβ chain. The MFI of αGC/CD1d tetramer staining for each hybridoma was determined for a narrow TCR gate. Data represent the mean + s.e.m. of two independent experiments. (B) The NKT hybridoma DN32.D3 was transduced with retroviruses encoding the mouse CD4 molecule. (B, left panel) Dot plots are gated on live cells and are representative of two independent experiments. (B, right panel) Hybridomas were stained with increasing concentrations of αGC/CD1d tetramer. The MFI of tetramer staining was determined for a narrow TCR gate.
Sequence alignment of human and mouse CDR1β and CDR2β loops that allow for CD1d-glycolipid recognition. Conserved residues in CDR2β are highlighted.
TCRβδ-deficient donor bone marrow cells were infected with retroviruses encoding the indicated TCRβ chain and eGFP as a reporter. Cells were injected i.v. into sub-lethally irradiated RAG-deficient recipient mice and thymic cells were stained 5 weeks post-reconstitution. Plots are gated on eGFP+ B220- CD8- and are representative of two independent experiments (3 mice/group).
Mutation in the CDR2β loop of Vβ14 increases iNKT TCR affinity. Vβ14 and Vβ14 I48Y TCRβ chains were expressed with the invariant Vα14 chain in the 5KC hybridoma. (A) Staining of hybridomas expressing the indicated TCR with αGC/CD1d tetramer and anti-TCRβ mAbs. (B) The MFI of tetramer staining was determined for a narrow TCR gate. Data represent the mean + s.e.m. of two independent experiments.
CDR3β sequences of Vβ6-containing iNKT TCRs that bind (or not) to the αGC/CD1d tetramer. αGC/CD1d tetramer+ and tetramer− cells from the Vβ6 CDR3β library (see Fig 6) were sorted, mRNA extracted and cDNA synthesized. Vβ6 chains were amplified using appropriate Vβ6-Jβ1.1 primers and cloned in frame with the Cβ into a retroviral construct with the puromycin resistance reporter gene. Individual colonies were picked out and sequenced. In addition, retroviruses were produced for each individual clone and used to transduce the Vα14-expressing 5KC hybridoma. Transduced hybridomas were selected by the addition of puromycin in the media for 72h. The hybridomas were subsequently stained with anti-TCRβ and αGC/CD1d tetramer. The MFI of αGC/CD1d tetramer staining was determined for a narrow TCR gate and is indicated in parenthesis. Sequences were ordered as a function of the αGC/CD1d tetramer MFI and separated in three groups. Sequences with αGC/CD1d tetramer MFI two fold over background (MFI = 5) were considered to marginally bind the tetramer, while sequences with αGC/CD1d tetramer MFI that were three fold or more over background were considered positive. Data represent the mean of two independent experiments. Recurrent amino acids at each position are highlighted in red.
ACKNOWLEGMENTS
We thank J. Cambier and R. Torres for thoughtful discussion and critical comments on the manuscript. We also thank NJH and UCD flow cytometry facilities for assistance with cell sorting and the NIH core facility for CD1d tetramers and the GSL-1´ compound. Supported by National Institute of Health (AI057485 to L. G., AI18785 to P. M. and AI057519 to A. H.); the Howard Hughes Medical Institute (P. M.); the Australian Research Council Federation Fellowship (to J. R.); the National Health and Medical Research Council (NHMRC) Research Fellowship to D. I. G., NHMRC Program grants to D. I. G., J. M. and the Cancer Council of Victoria grant (to J. M. and J. R.).
Footnotes
AUTHOR CONTRIBUTIONS
T. M., J. P. S.-B., J. L. M., M. H. Y. and L. G. designed, did the experimental work, analyzed the experiments and prepared the manuscript. V. C., S. K. R., M. T. and A. R. H. provided reagents. D. G. P., O. P., L. K.-N., J. M., D. G. and J. R. provided intellectual insight and crystallographic data. T. M., J. P. S.-B., P.M. and L.G. devised the project. T. M., J. P. S.-B., D. I. G., J. R., P. M. and L.G wrote the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare no financial conflict of interest.
REFERENCES
- Adams EJ, Strop P, Shin S, Chien YH, Garcia KC. An autonomous CDR3™ is sufficient for recognition of the nonclassical MHC class I molecules T10 and T22 by ©™ T cells. Nat Immunol. 2008;9:777–784. doi: 10.1038/ni.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou I, Cumano A, Gachelin G, Kourilsky P. Evidence for two subgroups of CD4−CD8− NKT cells with distinct TCRαβ repertoires and differential distribution in lymphoid tissues. J Immunol. 2000;165:2481–2490. doi: 10.4049/jimmunol.165.5.2481. [DOI] [PubMed] [Google Scholar]
- Arden B, Clark SP, Kabelitz D, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- Behar SM, Podrebarac TA, Roy CJ, Wang CR, Brenner MB. Diverse TCRs recognize murine CD1. J Immunol. 1999;162:161–167. [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L. The Biology of NKT Cells. Annu Rev Immunol. 2006 doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- Burdin N, Brossay L, Koezuka Y, Smiley ST, Grusby MJ, Gui M, Taniguchi M, Hayakawa K, Kronenberg M. Selective ability of mouse CD1 to present glycolipids: α-galactosylceramide specifically stimulates Vα14+ NK T lymphocytes. J Immunol. 1998;161:3271–3281. [PubMed] [Google Scholar]
- Christiansen D, Milland J, Mouhtouris E, Vaughan H, Pellicci DG, McConville MJ, Godfrey DI, Sandrin MS. Humans lack iGb3 due to the absence of functional iGb3-synthase: implications for NKT cell development and transplantation. PLoS Biol. 2008;6:e172. doi: 10.1371/journal.pbio.0060172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao T, Guo D, Ploss A, Stolzer A, Saylor C, Boursalian TE, Im JS, Sant'Angelo DB. Development of CD1d-restricted NKT cells in the mouse thymus. Eur J Immunol. 2004;34:3542–3552. doi: 10.1002/eji.200425546. [DOI] [PubMed] [Google Scholar]
- Degermann S, Sollami G, Karjalainen K. Impaired NK1.1 T cell development in mice transgenic for a T cell receptor β chain lacking the large, solvent-exposed Cβ FG loop. J Exp Med. 1999;190:1357–1362. doi: 10.1084/jem.190.9.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellabona P, Padovan E, Casorati G, Brockhaus M, Lanzavecchia A. An invariant Vα24-JαQ/Vβ11 T cell receptor is expressed in all individuals by clonally expanded CD4−8− T cells. J Exp Med. 1994;180:1171–1176. doi: 10.1084/jem.180.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction 'codon'. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- Gadola SD, Koch M, Marles-Wright J, Lissin NM, Shepherd D, Matulis G, Harlos K, Villiger PM, Stuart DI, Jakobsen BK, et al. Structure and binding kinetics of three different human CD1d-α-galactosylceramide-specific T cell receptors. J Exp Med. 2006;203:699–710. doi: 10.1084/jem.20052369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what's in a name? Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, McCluskey J, Rossjohn J. CD1d antigen presentation: treats for NKT cells. Nat Immunol. 2005;6:754–756. doi: 10.1038/ni0805-754. [DOI] [PubMed] [Google Scholar]
- Gras S, Kjer-Nielsen L, Burrows SR, McCluskey J, Rossjohn J. T-cell receptor bias and immunity. Curr Opin Immunol. 2008;20:119–125. doi: 10.1016/j.coi.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Gui M, Li J, Wen LJ, Hardy RR, Hayakawa K. TCRβ chain influences but does not solely control autoreactivity of Vα14Jα281 T cells. J Immunol. 2001;167:6239–6246. doi: 10.4049/jimmunol.167.11.6239. [DOI] [PubMed] [Google Scholar]
- Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, et al. CD1d-restricted and TCR-mediated activation of Vα14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong CH, Kronenberg M. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Borg NA, Pellicci DG, Beddoe T, Kostenko L, Clements CS, Williamson NA, Smyth MJ, Besra GS, Reid HH, et al. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J Exp Med. 2006;203:661–673. doi: 10.1084/jem.20051777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseki H, Imai K, Nakayama F, Sado T, Moriwaki K, Taniguchi M. Homogenous junctional sequence of the V14+ T-cell antigen receptor α chain expanded in unprimed mice. Proc Natl Acad Sci U S A. 1990;87:5248–5252. doi: 10.1073/pnas.87.14.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg M. Toward an Understanding of NKT Cell Biology: Progress and Paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- Lantz O, Bendelac A. An invariant T cell receptor α chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4−8− T cells in mice and humans. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Teneberg S, Thapa P, Bendelac A, Levery SB, Zhou D. Sensitive detection of isoglobo and globo series tetraglycosylceramides in human thymus by ion trap mass spectrometry. Glycobiology. 2008;18:158–165. doi: 10.1093/glycob/cwm129. [DOI] [PubMed] [Google Scholar]
- Li Y, Thapa P, Hawke D, Kondo Y, Furukawa K, Furukawa K, Hsu FF, Adlercreutz D, Weadge J, Palcic M, et al. Immunologic glycosphingolipidomics and NKT cell development in mouse thymus. J Proteome Res. 2009 doi: 10.1021/pr801040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Goff RD, Zhou D, Mattner J, Sullivan BA, Khurana A, Cantu C, 3rd, Ravkov EV, Ibegbu CC, Altman JD, et al. A modified α-galactosyl ceramide for staining and stimulating natural killer T cells. J Immunol Methods. 2006;312:34–39. doi: 10.1016/j.jim.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda JL, Gapin L, Fazilleau N, Warren K, Naidenko OV, Kronenberg M. Natural killer T cells reactive to a single glycolipid exhibit a highly diverse T cell receptor β repertoire and small clone size. Proc Natl Acad Sci U S A. 2001;98:12636–12641. doi: 10.1073/pnas.221445298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda JL, Gapin L, Sidobre S, Kieper WC, Tan JT, Ceredig R, Surh CD, Kronenberg M. Homeostasis of Vα 14i NKT cells. Nat Immunol. 2002;3:966–974. doi: 10.1038/ni837. [DOI] [PubMed] [Google Scholar]
- Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the 'Swiss-Army knife' of the immune system. Curr Opin Immunol. 2008 doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, Kronenberg M. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, 3rd, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- Ohteki T, MacDonald HR. Stringent Vβ requirement for the development of NK1.1+ T cell receptor-α/β+ cells in mouse liver. J Exp Med. 1996;183:1277–1282. doi: 10.1084/jem.183.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicci DG, Patel O, Kjer-Nielsen L, Pang S-S, Kyparissoudis K, Sullivan LC, Brooks AG, Reid HH, Smyth MJ, Mallevaey T, et al. Differential Vβ8.2 and Vβ7-mediated NKT T-cell receptor recognition of CD1d-α-galactosylceramide. (Submitted) [Google Scholar]
- Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4−8− αβ T cells demonstrates preferential use of several Vβ genes and an invariant TCR α chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porubsky S, Speak AO, Luckow B, Cerundolo V, Platt FM, Grone H-F. Normal development and function of invariant natural killer T cells in mice with isoglobotrihexosylceramide (iGb3) deficiency. Proc Natl Acad Sci U S A. 2007;104:5977–5982. doi: 10.1073/pnas.0611139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- Schumann J, Mycko MP, Dellabona P, Casorati G, MacDonald HR. Cutting edge: influence of the TCR Vβ domain on the selection of semi-invariant NKT cells by endogenous ligands. J Immunol. 2006;176:2064–2068. doi: 10.4049/jimmunol.176.4.2064. [DOI] [PubMed] [Google Scholar]
- Schumann J, Voyle RB, Wei BY, MacDonald HR. Cutting Edge: Influence of the TCR Vβ Domain on the Avidity of CD1d:α-Galactosylceramide Binding by Invariant Vα14 NKT Cells. J Immunol. 2003;170:5815–5819. doi: 10.4049/jimmunol.170.12.5815. [DOI] [PubMed] [Google Scholar]
- Scott Browne J, Matsuda JL, Mallevaey T, Borg NA, White J, McCluskey J, Rossjohn J, Kappler J, Marrack P, Gapin L. Germline-encoded recognition of diverse glycolipids by NKT cells. Nat Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- Scott Browne J, White J, Kappler J, Gapin L, Marrack P. αβ TCR recognition of MHC is germline encoded. Nature. In press. [Google Scholar]
- Silk JD, Salio M, Reddy BG, Shepherd D, Gileadi U, Brown J, Masri SH, Polzella P, Ritter G, Besra GS, et al. Cutting edge: nonglycosidic CD1d lipid ligands activate human and murine invariant NKT cells. J Immunol. 2008;180:6452–6456. doi: 10.4049/jimmunol.180.10.6452. [DOI] [PubMed] [Google Scholar]
- Speak AO, Salio M, Neville DCA, Fontaine J, Priestman DA, Platt N, Heare T, Butters TD, Dwek RA, Trottein F, et al. Implications for CD1d-restricted natural killer-like T cell ligands by the restricted presence of isoglobotrihexosylceramide in mammals. Proc Natl Acad Sci U S A. 2007;104:5971–5976. doi: 10.1073/pnas.0607285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanic AK, Shashidharamurthy R, Bezbradica JS, Matsuki N, Yoshimura Y, Miyake S, Choi EY, Schell TD, Van Kaer L, Tevethia SS, et al. Another view of T cell antigen recognition: cooperative engagement of glycolipid antigens by Vα14Jα18 natural T (iNKT) cell receptor. J Immunol. 2003;171:4539–4551. doi: 10.4049/jimmunol.171.9.4539. [DOI] [PubMed] [Google Scholar]
- Wei DG, Curran SA, Savage PB, Teyton L, Bendelac A. Mechanisms imposing the Vβ bias of Vα14 natural killer T cells and consequences for microbial glycolipid recognition. J Exp Med. 2006;203:1197–1207. doi: 10.1084/jem.20060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Pullen A, Choi K, Marrack P, Kappler JW. Antigen recognition properties of mutant Vβ3+ T cell receptors are consistent with an immunoglobulin-like structure for the receptor. J Exp Med. 1993;177:119–125. doi: 10.1084/jem.177.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wun KS, Borg NA, Kjer-Nielsen L, Beddoe T, Koh R, Richardson SK, Thakur M, Howell AR, Scott-Browne JP, Gapin L, et al. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J Exp Med. 2008;205:939–949. doi: 10.1084/jem.20072141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Kang SJ, Evans JE, Cresswell P. Natural lipid ligands associated with human CD1d targeted to different subcellular compartments. J Immunol. 2009;182:4784–4791. doi: 10.4049/jimmunol.0803981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, Savage PB, Bendelac A, Wilson IA, Teyton L. Crystal structures of mouse CD1d-iGb3 complex and its cognate Vα14 T cell receptor suggest a model for dual recognition of foreign and self glycolipids. J Mol Biol. 2008;377:1104–1116. doi: 10.1016/j.jmb.2008.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Mattner J, Cantu C, 3rd, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CD4 expression on T cell hybridomas does not affect TCR affinity. (A) Staining of CD4+ and CD4− hybridomas expressing the Vα14i TCRα chain and the indicated TCRβ chain. The MFI of αGC/CD1d tetramer staining for each hybridoma was determined for a narrow TCR gate. Data represent the mean + s.e.m. of two independent experiments. (B) The NKT hybridoma DN32.D3 was transduced with retroviruses encoding the mouse CD4 molecule. (B, left panel) Dot plots are gated on live cells and are representative of two independent experiments. (B, right panel) Hybridomas were stained with increasing concentrations of αGC/CD1d tetramer. The MFI of tetramer staining was determined for a narrow TCR gate.
Sequence alignment of human and mouse CDR1β and CDR2β loops that allow for CD1d-glycolipid recognition. Conserved residues in CDR2β are highlighted.
TCRβδ-deficient donor bone marrow cells were infected with retroviruses encoding the indicated TCRβ chain and eGFP as a reporter. Cells were injected i.v. into sub-lethally irradiated RAG-deficient recipient mice and thymic cells were stained 5 weeks post-reconstitution. Plots are gated on eGFP+ B220- CD8- and are representative of two independent experiments (3 mice/group).
Mutation in the CDR2β loop of Vβ14 increases iNKT TCR affinity. Vβ14 and Vβ14 I48Y TCRβ chains were expressed with the invariant Vα14 chain in the 5KC hybridoma. (A) Staining of hybridomas expressing the indicated TCR with αGC/CD1d tetramer and anti-TCRβ mAbs. (B) The MFI of tetramer staining was determined for a narrow TCR gate. Data represent the mean + s.e.m. of two independent experiments.
CDR3β sequences of Vβ6-containing iNKT TCRs that bind (or not) to the αGC/CD1d tetramer. αGC/CD1d tetramer+ and tetramer− cells from the Vβ6 CDR3β library (see Fig 6) were sorted, mRNA extracted and cDNA synthesized. Vβ6 chains were amplified using appropriate Vβ6-Jβ1.1 primers and cloned in frame with the Cβ into a retroviral construct with the puromycin resistance reporter gene. Individual colonies were picked out and sequenced. In addition, retroviruses were produced for each individual clone and used to transduce the Vα14-expressing 5KC hybridoma. Transduced hybridomas were selected by the addition of puromycin in the media for 72h. The hybridomas were subsequently stained with anti-TCRβ and αGC/CD1d tetramer. The MFI of αGC/CD1d tetramer staining was determined for a narrow TCR gate and is indicated in parenthesis. Sequences were ordered as a function of the αGC/CD1d tetramer MFI and separated in three groups. Sequences with αGC/CD1d tetramer MFI two fold over background (MFI = 5) were considered to marginally bind the tetramer, while sequences with αGC/CD1d tetramer MFI that were three fold or more over background were considered positive. Data represent the mean of two independent experiments. Recurrent amino acids at each position are highlighted in red.