Abstract
Eukaryotes contain an elaborate membrane system, which bounds the cell itself, nuclei, organelles and transient intracellular structures, such as vesicles. The emergence of this system was marked by an expansion of number of structurally distinct classes of lipid-binding domains that could throw light on the early evolution of eukaryotic membranes. The C2 domain is a useful model to understand these events because it is one of the most prevalent eukaryotic lipid-binding domains deployed in diverse functional contexts. Most studies have concentrated on C2 domains prototyped by those in protein kinase C (PKC-C2) isoforms that bind lipid in a calcium-dependent manner. While two other distinct families of C2 domains, namely those in PI3K-C2 and PTEN-C2 are also recognized, a complete picture of evolutionary relationships within the C2 domain superfamily is lacking. We systematically studied this superfamily using sequence-profile searches, phylogenetic and phyletic-pattern analysis and structure-prediction. Consequently, we identified several distinct families of C2 domains including those respectively typified by C2 domains in the Aida (axin interactor, dorsalization associated) proteins, B9 proteins (e.g. Mks1 (Xbx-7), Stumpy (Tza-1) and Tza-2) involved in centrosome migration and ciliogenesis, Dock180/Zizimin proteins which are Rac/CDC42 GDP exchange factors, the EEIG1/Sym-3, EHBP1 and plant RPG/PMI1 proteins involved in endocytotic recycling and organellar positioning and an apicomplexan family. We present evidence that the last eukaryotic common ancestor (LECA) contained at least 10 C2 domains belonging to 6 well-defined families. Further, we suggest that this pre-LECA diversification was linked to emergence of several quintessentially eukaryotic structures, such as membrane repair and vesicular trafficking system, anchoring of the actin and tubulin cytoskeleton to the plasma and vesicular membranes, localization of small GTPases to membranes and lipid-based signal transduction. Subsequent lineage-specific expansions of Zizimin-type C2 domains and functionally linked CDC42/Rac GTPases occurred independently in eukaryotes that evolved active amoeboid motility. While two lipid-binding regions are likely to be shared by majority of C2 domains, the actual constellation of lipid-binding residues (predominantly basic) are distinct in each family potentially reflective of the functional and biochemical diversity of these domains. Importantly, we show that the calcium-dependent membrane interaction is a derived feature limited to the PKC-C2 domains. Our identification of novel C2 domains offers new insights into interaction between both the microtubular and microfilament cytoskeleton and cellular membranes.
Keywords: C2 domain, evolution, remote homology detection, structure and function, membranes, lipid recognition, cytoskeleton, GTPase signaling, vesicular trafficking
1.Introduction
The cell membrane not only serves to contain cellular contents, but also acts as the perimeter for the cytoskeleton, the source of certain signaling molecules and vesicles for intracellular trafficking of proteins and other biomolecules, and as a substrate for cell adhesion (De Matteis and Godi, 2004; Di Paolo and De Camilli, 2006). Eukaryotic cells possess an elaborate membrane system with multiple intracellular membranes, e.g. the nuclear membrane and the organellar membranes, in addition to the plasma membrane that bounds the cell. Membranes of eukaryotic cells are also distinguished by their unique roles in signaling and protein-trafficking. Diverse eukaryotic signaling pathways require specific mechanisms by which soluble proteins localize to the membrane to initiate signaling cascades or transduce signals downstream of membrane receptors (e.g. GTPase signaling). Furthermore, metabolites released by the action of specific enzymes (e.g. phospholipases) on membrane lipids function as intracellular second messengers that play an important role in eukaryotic signaling networks. Similarly, several studies point to the role of the membrane as an anchor for key cytoskeletal components such as the myosins (Patino-Lopez et al., 2010). Localization of proteins to membranes, secretion of extracellular proteins and regulation of receptors via endocytosis requires a complex, multi-protein machinery (the vesicular trafficking machinery) that is also highly developed in eukaryotic cells compared to their bacterial or archaeal counterparts (Raiborg and Stenmark, 2009). In metazoans this process is central to neural function by mediating neurotransmitter release at synaptic junctions (Cremona and De Camilli, 2001). Although these functionally disparate processes are mediated by distinct molecular complexes and interactions, they are all linked by certain common themes. Firstly, different types of membrane phospholipids (e.g. phosphatidylserine, phosphatidylcholine, phosphatidylglycerol, phosphatidylethanolamine, PtdIns(4)P, PtdIns(3)P, PtdIns(4,5)P2, PtdIns(3,5)P2, PtdIns(3,4,5)P3) serve as spatial landmarks on membranes (De Matteis and Godi, 2004; Cho and Stahelin, 2006; Lemmon, 2008). Secondly, these lipids are recognized by proteins by means of an array of distinctive conserved domains (Lemmon, 2008). Thus, specific spatial or temporal interactions between a relatively small set of conserved protein domain superfamilies and distinct lipids facilitate the dynamics of the majority of membrane-related events.
To date several globular protein domains with structurally unrelated folds have been identified to play a central role in recognizing and interacting with specific lipids in eukaryotes. These domains are distinguished in being soluble domains present on the cytoplasmic face of the membrane as opposed to intra-membrane lipid-recognitions domains with TM-helices (e.g. MENTAL and Sterol-sensing domains) (Yabe et al., 2002; Alpy et al., 2005). The globular membrane-recognition domains include the β-barrel domains with the PH-like fold such as the PH, GRAM and myosin tail domains (Gibson et al., 1994; Doerks et al., 2000; Patino-Lopez et al., 2010), the PX, Sec14 and START domains with different α+β folds (Ponting, 1996; Aravind et al., 1999; Iyer et al., 2001), treble clef Zn-fingers such as the FYVE and C1 domains (Azzi et al., 1992; Gaullier et al., 1998), and the C2 and certain versions of discoidin domains with the β-sandwich fold (Nalefski and Falke, 1996; Baumgartner et al., 1998; Lemmon, 2008). While members of the PH-like fold are found in bacteria (e.g. GRAM and PH domains), representatives of most other globular membrane-recognition domains are restricted to eukaryotes. This distinctive phyletic pattern of lipid-recognition or membrane binding domains suggests that understanding their diversification might throw light on the evolution of unique features of membrane organization and dynamics of eukaryotes.
Of these, members of the PH-like fold and C2 domains are prevalent across eukaryotes (Lemmon, 2008) and are combined with a wide range of other domains in the same polypeptide to diverse domains, such as those with enzymatic activities, those related to cytoskeletal interactions and those involved in vesicular trafficking. However, the early evolution of these domains remains poorly understood because not all versions of these domains can be easily identified through straightforward sequence analysis. As part of our program to understand the evolutionary diversification of eukaryote-specific membrane associated functions, we took up the C2 domain as a case for detailed investigation. Currently, three distinct versions of the C2 domain have been recognized, the typical PKC-C2 (PFAM: PF00168) (Ponting and Parker, 1996), PI3K-C2 (PFAM: PF00792) (Vanhaesebroeck et al., 1997) and PTEN-C2 domains (PF10409) (Lee et al., 1999; Leslie et al., 2007). Both PKC-C2 and PTEN-C2 domains are found as a part of a diverse set of domain architectures in eukaryotic membrane-associated proteins, whereas PI3K-C2 domain is always associated with the kinase domain in phosphatidylinositol-3 and phosphatidylinositol-4 kinases (PI3/4Ks). Despite their limited sequence similarity, all C2 domains contain at their core a compact β-sandwich composed of two four-stranded β sheets with highly variable inter-strand regions that might contain one or more α-helices. Most of our current understanding of the membrane-interactions of C2 domains comes from studies on the PKC-C2 versions. The majority of PKC-C2 domains are distinguished by the Ca2+ ion-dependent interactions with membranes. They contain a conserved set of acidic residues that coordinate Ca2+ ions and this allows regulation of membrane recruitment dependent on calcium concentration in the case of conventional PKC isoforms (Verdaguer et al., 1999), rabphilin (Montaville et al., 2007) and cPLA2 (Perisic et al., 1998). Coordination of Ca2+ ions reverses the negative charge of the binding pocket (Murray and Honig, 2002) allowing electrostatic interactions with the acidic phosphatidylserine, the dominant membrane lipid in cells (Lemmon, 2008). Beyond the Ca2+-binding motif, some basic residues at the opposite end of the calcium-binding site have also been shown to be required for membrane penetration by C2 domains (Cho and Stahelin, 2006). While certain PKC-C2 domains have been claimed to participate in protein-protein rather than lipid interactions, it should be noted that the evidence for them not binding any lipid types has not been comprehensively evaluated (Lopez-Lluch et al., 2001; Sklyarova et al., 2002). Recently, an alternative membrane-binding mode has been proposed; this is mediated by a series of basic and aromatic residues present on the face of the β-sandwich between the calcium-coordinating site and the membrane-penetrating basic residues. Although, there have been few studies on the membrane-binding mode of the other C2 domains, electrostatic interactions are believed to be critical for both PI3K-C2 and PTEN-C2 domains in recognizing lipids (Murray and Honig, 2002).
In this study we performed systematic sequence-structure analysis of the C2 domain with the objective of identifying previously uncharacterized versions. We then used the information derived from the newly identified versions to investigate and predict functional mechanisms and possible membrane-binding activities for these domains. We further utilized this in conjunction with phyletic patterns and phylogenetic analysis to infer the ancestral functions and subsequent diversification of C2 domains in course of eukaryotic evolution. We report here identification of several novel versions of the C2 domain and recognize at least three distinct families that have not been previously defined. Our study also found the C2 domains in Dock (dedicator of cytokinesis) to form a family of C2 domains distinct from the rest. The newly identified families provide key functional insights into how certain types of the C2 domain help in anchoring both the microtubule- and microfilament- based cytoskeletal scaffolds and throws new light on previously unrecognized lipid interactions in signaling and organellar movement. Our results also suggest that the initial diversifying radiation of the C2 domain happened between the time of the first eukaryotic common ancestor (FECA) and the last eukaryotic common ancestor (LECA). This radiation appears to have established most of the major families of the C2 domain in parallel with the emergence of the complex membrane-linked functions in eukaryotes. Beyond this point, different families of the C2 domain followed very different trajectories in terms of domain architectural complexity that was largely influenced by the selective forces related to their interactions.
2. Materials and methods
2.1. Homologous Sequence Searching and Sequence Analysis
Iterative profile searches with PSI-BLAST (Altschul et al., 1997) were used to retrieve homologous sequences in the protein non-redundant (NR) database at National Center for Biotechnology Information (NCBI). A profile inclusion expectation (E) value threshold of 0.01, the BLOSUM62 substitution matrix and the gap penalty (existence: 11 and extension: 1) were utilized in the searches. Similarity-based clustering for both classification and culling of nearly identical sequences was performed using the BLASTCLUST program. Multiple sequence alignments were built by Kalign (Lassmann and Sonnhammer, 2005), Muscle (Edgar, 2004), and Promals (Pei and Grishin, 2007) programs, followed by careful manual adjustments based on profile-profile alignment, secondary structure information and structural alignment. The consensus of the alignment were calculated and colored by the Chroma program (Goodstadt and Ponting, 2001). Consensus secondary structures were predicted using the JPred program (Cuff et al., 1998). Multiple alignments of individual families of C2 domains are provided as Figs. S1-S4 (supplementary material), whereas a unified alignment of representatives of all major families is shown in Fig. 1.
Fig. 1.
Multiple sequence and structural alignment for seven families of the C2 domain. The sequences are represented by an abbreviation of species name followed by Genbank GI and PDB ID. The consensus in 65% of the sequences shown below the alignment is derived using the following amino acid classes: a, aromatic (FHWY, black on orange); b, big (EFHIKLMQRWY, green on grey); h, hydrophobic (ACFGHILMTVWY, yellow on dark grey); l, aliphatic (ILV, red on grey); p, polar (CDEHKNQRST, blue on grey); s, small (ACDGNPSTV, cyan on dark grey); t, tiny (AGS, cyan on dark grey). Secondary structures for these C2 families, that were derived from PDB structures or predicted using the Jpred program, are indicated above the alignment of each own family (E in blue, β sheet; H in red, α helix). For each C2 family, conserved or specific residues that are potentially involved in binding membrane lipid are labeled on the top of the alignment and highlighted with the blue background for basic residues, the red background for acidic residues, and the orange background for aromatic residues. Non-conserved or subfamily-specific are indicated in italic. The numbers in bracket are indicative of the excluded residues from sequences. For complete multiple sequence alignments of AIDA-C2, B9-C2, DOCK-C2 and NT-C2 families refer to supplemental Figs. S1 to S4. Species name abbreviations: Ag, Anopheles gambiae; At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Cn, Cryptococcus neoformans; Cr, Chlamydomonas reinhardtii; Dd, Dictyostelium discoideum; Eh, Entamoeba histolytica; Hs, Homo sapiens; Lt, Lachancea thermotolerans; Mb, Monosiga brevicollis; Mi Micromonas sp.; Mt, Medicago truncatula; Nv, Nematostella vectensis; Ot, Ostreococcus tauri; Pi, Phytophthora infestans; Pm, Perkinsus marinus; Ps, Pichia stipitis; Pt, Paramecium tetraurelia; Sp, Strongylocentrotus purpuratus; Ta, Trichoplax adhaerens; Tb, Trypanosoma brucei; Tt, Tetrahymena thermophila; Tv, Trichomonas vaginalis.
2.2. Protein Remote Homology Detection
Profile-profile comparisons were performed using the HHpred program. HHpred utilizes a comparison between HMM constructed using the query via PSI-BLAST search and a preexisting library of HMM compiled from domains found in the PDB database to detect remote relationships (Soding et al., 2005). In parallel, fold recognition was conducted via meta server (Ginalski and Rychlewski, 2003) that combines multiple fold recognition programs including meta-BASIC (Ginalski et al., 2004), ORFeus-2 (Ginalski et al., 2003b), and FFAS03 (Jaroszewski et al., 2005) and further evaluates the modeled three-dimensional structures based on a consensus scoring computed by a 3D-JURY system (Ginalski et al., 2003a). Structure similarity searches were performed using the DaliLite program (Holm et al., 2008) that searches the PDB database with coordinates of a query structure and makes structural alignments.
2.3. Domain Architecture Analysis
For previously characterized domains the PFAM database was used as a guide (Finn et al., 2009). Clustering with BLASTCLUST followed by multiple sequence alignment and further sequence profile searches were used to identify other domains that were not present in the PFAM database. We used the presence of an architecture across multiple species as a guide to eliminate potentially false architecture arising due to errors in gene-prediction.
2.4. Phylogenetic Inference
An unrooted phylogenetic tree for seven versions of the C2 domain was reconstructed using an approximately-maximum-likelihood method implemented in the FastTree 2.1 program under default parameters (Price et al., 2009). In contrast to other phylogenetic analysis programs, FastTree stores sequence profiles of internal nodes. This improves speed considerably and in our bench-marking efforts it provides trees comparable to those generated with the Protml program initiated with starting trees constructed with neighbor-joining or minimal evolution methods. The PhyML program (Guindon and Gascuel, 2003) was also used to determine the maximum likelihood tree using the Jones-Taylor-Thornton (JTT) model for amino acids substitution with a discrete gamma model (eight categories with gamma shape parameter: 5.870). The tree was rendered using MEGA Tree Explorer (Tamura et al., 2007).
2.5. Comparative Structure Modeling and Electrostatic Calculations
The Modeller9v1 program (Sali and Blundell, 1993) was utilized for homology modeling of structures of different C2 domains by using multiple templates chosen based on highest sequence identity to the target sequence and significantly low E value. Since in these low-sequence-identify cases, sequence alignment is the most important factor affecting the quality of the model (Cozzetto and Tramontano, 2005), alignments used in this study have been carefully built and cross-validated on the basis of information from HHpred, meta server, and DaliLite programs, and edited manually using the secondary structure information. Electrostatic potential was calculated using software Adaptive Poisson-Boltzmann Solver (APBS) (Baker et al., 2001) in the VMD environment, which uses the finite element method to solve the non-linear Poisson-Boltzmann equation. The known/modeled structures with their inferred electrostatic surfaces were rendered using the VMD program (Humphrey et al., 1996). It should be noted that these models are necessarily approximate and not substitutes for experimentally determined structures. However, when combined with sequence conservation pattern for a given family/superfamily of proteins they serve as reasonable guides to analyze the functional properties of the domain under consideration.
3. Results and discussion
3.1. Identification of new C2 domains by fold recognition and profile-profile similarity
By combining sequence profile searches with profile-profile comparison and fold recognition methods (see Methods for details) we were able to identify four distinct versions of C2 domains (Fig 1, 2). Both similarity-based clustering and phylogenetic analysis show them to be distinct families that are not closely related to any of the three characterized families of C2 domains (Fig. 3). Three of these families were not previously defined, while the recent solution of the structure of a domain from the Dock protein, when the current article was being prepared, independently showed the 4th of these families to contain a C2 domain (Premkumar et al., 2010). Multiple alignment of these newly detected families with known families of C2 domains shows that despite the low sequence similarity between them they possess a similar structural core comprised of eight primary β-strands and a conserved α-helical segment between strand-6 and strand-7 (Fig. 1). Further, there are several hydrophobic positions corresponding to each of the 8 strands that tend to show reasonably strong conservation across most C2 domains. Also of interest is a well-conserved hydrophobic (usually aromatic) position in the connector between strand-3 and strand-4 that is conserved across most families of the C2 domain. This position along with the conserved helical segment and certain aromatic residues in strands- 2, 4 and 7 (Fig. 1) contribute to a distinctive structural packing and hydrophobic core that differentiates the C2 domain from other known β-sandwich domains. Consistent with this C2 domains never recover other β-sandwich domains in sequence profile searches. Examination of known C2 domain structures shows that the conserved helical segment “blocks” one of the ends of the β-sandwich by lodging into the groove formed by the loops between the strands, thereby leaving only the groove/pocket at the other end available for interactions with lipids (Fig. 4).
Fig. 2.
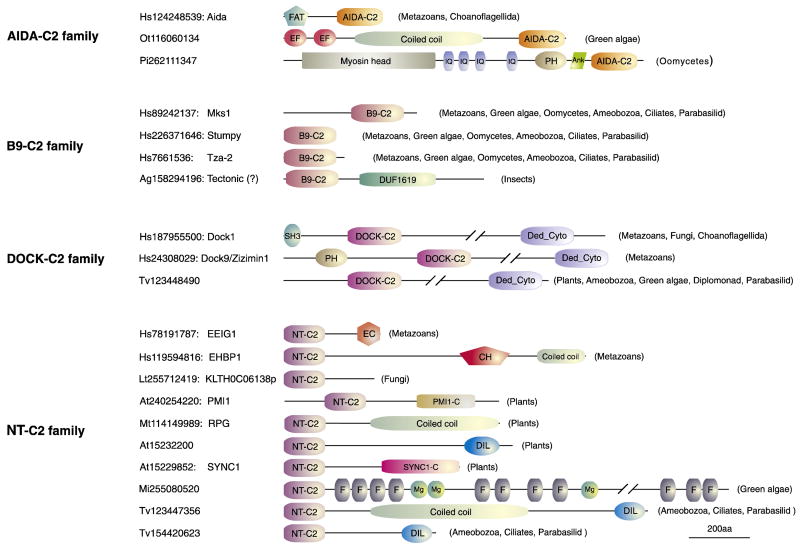
Domain architectures of representatives of the AIDA-C2, B9-C2, DOCK-C2 and NT-C2 domain-containing proteins. The sequences are indicated by species name with Genbank GI. Domains are drawn approximately to scale. Domain abbreviations are as follows: FAT, four-helical up-and-down bundle found in focal adhesion kinase and talin; AIDA-C2, Aida version of the C2 domain; Ank, Ankyrin repeat (PF00023); B9-C2, B9 version of the C2 domain (PF07162); CH, Calponin homology domain (PF00307); Coiled coil, coiled coil region; Ded_Cyto, Dedicator of cytokinesis (PF06920); DIL, DIL domain (PF01843); DOCK-C2, Dock180 version of the C2 domain; EC, EEIG1 C terminal region; EF, Eps15 homology domain or EF Hand (PF00036); Fil, Filamin/ABP280 repeat (PF00630); IQ, IQ calmodulin-binding motif (PF00612); Mg, MgtE family of Mg2+ transporter (PF01769); Myosin head, Myosin head motor domain (PF00063); NT-C2, N-terminal C2 domain (also known as EEIG1 domain in PFAM, PF10358); PH, Pleckstrin homology domain (PF00169); PMI1-C, PMI1 C-terminal region; SH3, Src homology 3 (PF00018); SYNC1-C, SYNC1 C-terminal region. In the case of a subset of insect B9-C2 proteins there has been a systematic mis-annotation of the gene model which has resulted in the B9-C2 gene being fused to an adjacent gene of the Tectonic family (PF07773). Hence this architecture is likely to be artefactual and is indicated with a question mark.
Fig. 3.
Phylogenetic relationship between seven families of the C2 domain. An approximate maximum-likelihood method implemented in the FastTree 2.1 program was used to build the phylogenetic tree for seven versions of the C2 domain. Protein sequences are represented by species names following Genbank GI ID. Main nodes are shown with supporting bootstrap values. Four newly defined families of C2 domain and sub-groups within them are distinguished by different colors. L with a blue circle indicates that those C2 families or subfamilies can be traced to LECA, where question mark (?) with green circle indicates an unresolved evolutionary origination. Species name abbreviations refer to Fig. 1 legend and additional ones are as follows: An, Aspergillus niger; As, Aspergillus terreus; Bm, Brugia malayi; Cc, Coprinopsis cinerea; Ci, Ciona intestinalis; Cm, Cryptosporidium muris; Co, Coccidioides immitis; Cp, Cryptosporidium parvum; Da, Drosophila ananassae; Dr, Danio rerio; Lb, Leishmania braziliensis; Mc, Microsporum canis; Pb, Paracoccidioides brasiliensis; Pg, Pichia guilliermondii; Sc, Saccharomyces cerevisiae; So, Schizosaccharomyces pombe; Ss, Sclerotinia sclerotiorum; St, Strongylocentrotus purpuratus; Ts, Talaromyces stipitatus; Vp, Vanderwaltozyma polyspora; Vv, Vitis vinifera; Zm, Zea mays; Zr, Zygosaccharomyces rouxii.
Fig. 4.
Electrostatic potential surface and cartoon diagrams of structures of seven C2 domains. All structures are depicted in the same orientation. Surface diagrams are colored by electrostatic potential as calculated by APBS in VMD program with positively charged regions (>10 mV) colored in blue and negatively charged regions (<-10 mV) in red. The α helices are shown in purple, β sheets in yellow, and loops in white. Conserved residues that are involved in membrane lipid-binding are highlighted in licorice drawing with basic residues (H, K, R) in blue, acidic residues in red, and aromatic residues (F, W, Y) in orange. The α helix 2 region of B9-C2 and DOCK-C2 domains is not modeled. 1E7U was remodeled for its loop region between beta strand 5 and 6 for which no X-ray data was available.
Most C2 domains also contain two highly conserved hydrophobic residues respectively at the beginning of strand-2 and the end of strand-5 that are exposed on the surface and interact with each other through hydrophobic or aromatic π-π stacking interactions. These residues are part of the constellation of positions implicated in binding PtdIns(4,5)P2, an important regulatory lipid (McLaughlin and Murray, 2005; Guerrero-Valero et al., 2009). In addition, interaction between these two residues on the outer surface of the β sheet conformationally stabilizes its concave curvature that could further allow for ligand-interactions by forming a shallow depression on one side of the β-sandwich. In contrast to these residues, neither the calcium-coordinating nor the basic residues involved in lipid interaction are widely conserved across families. While several biologically interesting properties have been recognized for a subset of the proteins containing the newly detected C2 domains, there has been hardly any mechanistic understanding regarding their roles. Hence we sought to perform a systematic structure-function analysis of C2 domains from these proteins to clarify their potential mode of action. Accordingly, we briefly outline each of the newly defined families with their specific features below.
3.1.1. AIDA-C2 domain is associated with cytoskeletal interaction domains
The axin interaction dorsal-associated (Aida) protein was characterized in zebrafish as a protein that interacts with axin, which is a microtubule-interacting scaffold protein for several distinct signaling proteins in the Wnt cascade (Rui et al., 2007). PFAM annotates the entire Aida protein and its homologues as a domain of unknown function (DUF1855). Our analysis using the SEG program (Wootton and Federhen, 1996) showed that the protein contains two distinct globular domains. Profile-profile comparisons with HHpred and structure similarity searches with DaliLite (using 2QZQ as a query) showed that the C-terminal domain of the Aida protein is a distinct version of the C2 domain (HHpred p= 10-5 corresponding 92% probability, Fig. 1). Accordingly, we named all C2 domains typified by the Aida C-terminal domain as the AIDA-C2 family. A structure similarity search with the N-terminal domain of the Aida recovered the four-helical-up-and-down bundle domain that is typically found in focal adhesion proteins such as the focal adhesion kinase, the ARF GAP Git1, α-catenin and talin (FAT domain) (Hayashi et al., 2002). This domain has been found to be critical for interactions with cytoskeletal proteins in the context of cellular adhesion points. In all proteins the AIDA-C2 domain is found in the C-terminal portion of the protein (Fig. 2). In these proteins too the Aida-C2 domain is combined with diverse domains related to cytoskeletal functions, e.g. EF-hands, coiled coils, IQ calmodulin-binding motifs, ankyrin repeats and myosin head motor domain (Fig. 2). It may also be combined with a second lipid-binding domain the PH domain in certain cases (Fig. 2).
3.1.2. B9-C2 domain is found in ciliary basal body associated proteins
The PFAM database defines the common domain found in three distinct types of proteins, Mks1 (MecKel Syndrome, type 1)/Xbx-7 (X-BoX promoter element regulated family member 7), Stumpy/Tza-1 (ciliary Transition Zone Associate family members 1) and Tza-2 proteins as the B9 domain (PF07162). Mutations in human Mks1 is the cause of the Mechel-Gruber syndrome, a developmental disorder (Kyttala et al., 2006). Disruption of mouse Stumpy results in perinatal hydrocephalus and severe polycystic kidney disease (Town et al., 2008). Studies in both mammal and C. elegans models demonstrated that all the three distinct types of B9 proteins cooperatively localized to the basal body or centrosome of cilia (Kyttala et al., 2006; Dawe et al., 2007; Town et al., 2008; Williams et al., 2008; Bialas et al., 2009). Loss-of-functions mutations in the B2 protein blocked centrosome migration to the apical membrane and resulted in defective formation of cilia (Dawe et al., 2007; Town et al., 2008; Williams et al., 2008; Bialas et al., 2009; Tammachote et al., 2009; Weatherbee et al., 2009). In our sequence searches B9 domains were identified as a specific family of C2 domains. Profile-profile comparisons and fold recognition methods recovered significant hits to previously recognized C2 domains confirming this relationship (meta server Jscores> 55 corresponding to >90% probability match to C2 templates, and HHpred server p<10-4 for best hit C2 domain). Hence, we named this family the B9-C2 family. B9-C2 domain proteins show a very simple architecture with nearly all members being composed of just the C2 domain (Fig. 2). While some representatives of this family were recognized earlier as displaying relationship to the C2 domains, they were incorrectly described as calcium-binding versions (Avidor-Reiss et al., 2004) (see below).
3.1.3. DOCK-C2 domain in Dock180/Dock1 and Zizimin proteins
The Dock180/Dock1 and Zizimin proteins are atypical GTP/GDP exchange factors for the small GTPases Rac and Cdc42 and are implicated in biological roles related to cell-migration and phagocytosis (Cote and Vuori, 2002; Meller et al., 2005). Two regions were detected as being conserved across all Dock180/Dock1 proteins, one in the middle of the proteins termed CZH1 or DHR1 and another at the C-terminus termed CZH2 or DHR2 (or the Dedicator of cytokinesis). Different Dock proteins have divergent N-termini that might contain SH3 or PH domains (Fig. 2). Of these the region spanning CZH1/DHR1 was shown to bind membrane PtdIns(3,4,5)P3 (Cote et al., 2005). Our sequence analysis revealed that the conserved core of the CZH1/DHR1 region contains a distinctive version of the C2 domain (Fig. 1), which has been confirmed by recent crystallographic evidence (Premkumar et al., 2010), and we accordingly termed this type of C2 domains as the DOCK-C2 family. This family is strictly coupled with the CZH2/DHR2 in the C-terminus and always occurs in the central region of the protein in which it is found.
3.1.4. NT-C2 domain is found in several microfilament/actin and endocytosis related proteins
Another novel version of the C2 domain was identified in the vertebrate Estrogen early-induced gene 1 (EEIG1) (Wang et al., 2004), its Drosophila ortholog required for uptake of dsRNA via the endocytotic machinery to induce RNAi silencing (Saleh et al., 2006) and its C.elegans ortholog Sym-3 (SYnthetic lethal with Mec-3) and the mammalian protein EHBP1 (EH domain Binding Protein-1) that regulates endocytotic recycling. We also detected related C2 domains in two plant proteins, RPG that regulates Rhizobium-directed polar growth (Arrighi et al., 2008) and PMI1 (Plastid Movement Impaired 1) that is essential for intracellular movement of chloroplasts in response to blue light (DeBlasio et al., 2005) and many other uncharacterized proteins across eukaryotes. A subset of these versions has been categorized as a distinct domain, EEIG1, in the PFAM database (PF10358). Our identification of this entire group of domains (Fig. 1) as a distinctive version of the C2 superfamily is supported by both profile-profile comparisons (HHpred p=10-7 corresponding to >97% probability) and meta-fold recognition (Jscores> 60 corresponding to >95% probability). This version of C2 domains shows a diverse range of domain architectures (Fig. 2) but it is nearly always found at the N-termini of proteins that contain it. Hence, we named it the N-terminal C2 (NT-C2) family. It is typically coupled with a coiled-coil domain, that could mediate di/oligo-merization (Parry et al., 2008) and the DIL (Dilute) domain (Ponting, 1995) (Fig. 2). It is also coupled with the Calponin homology (CH) domain (Stradal et al., 1998) in EHBP1 proteins, Filamin/ABP280 repeats (Fucini et al., 1997) and Mg2+ transporter MgtE N-terminal domain (Hattori et al., 2007) in proteins from chlorophyte algae such as Micromonas and Ostreococcus tauri. Thus, a common theme across the NT-C2 domain proteins is the combination to several different domains with microfilament-binding or actin-related roles (i.e. such as CH, DIL, and Filamin). Other conserved groups of the NT-C2 proteins prototyped by EEIG1, PMI1, and SYNC1 have their own distinct C- terminal conserved extensions that are restricted to these groups and might mediate specific interactions (Fig. 2; see supplemental Figs. S5-S7 for sequence alignment).
3.1.5. The apicomplexan specific C2 domain family
This family is typified by the protein PF13_0086 (GI: 124513066; supplementary Fig. S8) from Plasmodium falciparum and contains at least one orthologous representative in all apicomplexan lineages. While it is strongly conserved in apicomplexa, it does not show specific relationships to any of the other more widely distributed C2 families. Hence it appears to have been an apicomplexan innovation, probably emerging via rapid divergence from one of the more prevalent families. It is conceivable that is involved in a role related to one of the unique apicomplexan membrane structures.
3.2. Phyletic patterns and evolutionary relationships within the C2 domain superfamily
Sequence-similarity based clustering, examination of diagnostic sequence conservation patterns and phylogenetic analysis with the approximate likelihood method incorporated in FastTree2 suggested that there are seven major families of the C2 domain (Fig.1, 3, 5). Due to the low sequence similarity between families and the relatively short length of the domain the higher order relationships between the families were not discernable. However, within each family a reasonably resolved phylogenetic picture was observed (Fig. 3). In terms of phyletic patterns six of the seven families were detected in the parabasalid Trichomonas vaginalis, an early-branching eukaryotic lineage (families marked with blue circle in Fig. 3), suggesting that the C2 families had probably differentiated by the time of LECA. However, it should be noted that we could detect only two representatives of the C2 domain (both of the DOCK-C2 family; GIs: 253741934 and 253741923) in another basal eukaryote, the diplomonad Giardia. Under the currently prevalent phylogenetic hypothesis for eukaryotes this would imply massive gene loss in this lineage. The AIDA-C2 family is found only in the metazoan, choanoflagellate, chromist and chlorophyte lineages (Fig. 2, 3) – the most parsimonious explanation for this phyletic pattern is that it was derived via rapid divergence from a pre-existing version prior to the divergence of the plants and animals (perhaps the DOCK- or PTEN- C2 families which it recovers in searches as best hits). Phylogenetic analysis also reveals some clear examples of gene loss among different C2 domain families that could have major consequences for the organization of particular cellular systems. For instance, all the three types of the B9-C2 domain are independently lost in land plants, amoebozoans and dikaryan fungi (Fig. 3, 5). These losses are correlated with the loss of flagella in these lineages (Avidor-Reiss et al., 2004).
Fig. 5.
Divergent evolution of the C2 domain superfamily in eukaryotes. Duplications of the domain and speciation are indicated by red lines whereas domain loss for B9-C2 domains is indicated by blue lines. Dashed red line with question mark (?) indicates unclear point of origination of AIDA-C2. A domain architecture linkage graph for the seven C2 versions is shown in the next column. The arrows indicate the direction of domain connection in a polypeptide with the arrowhead pointing to the C-terminus. Microfilament function-related domains are shown in yellow, phosphatase and phospholipase domains in orange, kinase domains in blue, whereas other domains with enzyme activities in violet. Additional domain abbreviations with PFAM domain ID are as follows: C1, Protein kinase C, C1 domain (PF00130); CalpainIII, Calpain large subunit, domain III (PF01067); DnaJ, DnaJ domain (PF00226); PIP3 phosphatase, including both dual specificity phosphatase, catalytic domain (PF00782) and protein-tyrosine phosphatase (PF00102); Thiored, is a thioredoxin-like domain called DUF547 in PFAM (PF04784); FH2, Formin homology 2 domain (PF02181); HECT, HECT-domain (ubiquitin-transferase) (PF00632); MACPF, MAC/Perforin domain (PF01823); MUNC13, Munc13 (mammalian uncoordinated) homology domain (PF10540); p85, PI3-kinase family, p85-binding domain (PF02192); Papain-like peptidase, calpain family cysteine protease (PF00648); PI34-K, Phosphatidylinositol 3- and 4-kinase (PF00454); PI3Ka, Phosphoinositide 3-kinase family, accessory domain (PF00613); PI-PLC-X, Phosphatidylinositol-specific phospholipase C, X domain (PF00388); PI-PLC-Y, Phosphatidylinositol-specific phospholipase C, Y domain (PF00387); Pkinase, Protein kinase domain (PF00069); PLA2B, Lysophospholipase catalytic domain (PF01735); PLDc, Phospholipase D C terminal (PF12357); PSDcarbxylase, Phosphatidylserine decarboxylase (PF02666); PTB, Phosphotyrosine-binding domain (PF08416); PX, PX domain (PF00787); RasGAP, GTPase-activator protein for Ras-like GTPase (PF00616); Rbd, PI3-kinase family, ras-binding domain (PF00794); SH2, Src homology 2 domain (PF00017); vWA, vWA-like copine domain (PF07002); WW, WW domain (PF00397).
It has been previously noted that classical PKC-C2 domains show expansions in several lineages of eukaryotes, especially in the metazoans. Moreover, multiple copies of the PKC-C2 domains might frequently occur in the same polypeptide (e.g. in the ferlins, Vp115 and tricalbins) (Cho and Stahelin, 2006). While there is a massive lineage-specific expansion of PKC-C2 domains in Trichomonas, at least three distinct types were discerned that appear to have cognates in other eukaryotic lineages – the version fused to the vWA domain (copine-like), a version with multiple C2 domains (a possible precursor of ferlins, tricalbins and Vp115) and another with a standalone C2 domain (Fig. 5). Most of the newly detected families are comprised of relatively few distinct orthologous lineages that are conserved over much of eukaryotic evolution, and almost always present as a single copy in proteins which contain them. For example, the B9-C2 family shows three distinct orthologous lineages, all three of which are always present together in any of the lineages that contain them. Furthermore, trees of each of the three orthologous lineages of B9-C2 domains contain a representative from Trichomonas suggesting that the three copies of the B9-C2 domain had already differentiated from each other in LECA and were largely vertically inherited ever since (Fig. 3, 5). Likewise, within the DOCK-C2 family an ancient orthologous group typified by Zizimin can be discerned in most major eukaryotic groups, whereas the Dock1/Dock180 appears to be restricted to animals and fungi, having probably arisen through a duplication in their common ancestor. In the case of the DOCK-C2 family we observed independent major lineage-specific expansions of Zizimin-like proteins within amoebozoans (Fig. 3) and the heterolobosean Naegleria (supplementary material). This expansion is of interest because a corresponding expansion in the small GTPases of the Rac/CDC42 type has been previously observed in amoebozoans (Anantharaman et al., 2007). Hence, there appears to have been a co-evolutionary diversification of both the GTPases and their GAPs.
In conclusion, the phyletic patterns and phylogenetic analysis suggest that at least 10 distinct copies of the C2 domain were probably present in LECA (three of them B9-C2s), though this number is likely to have been larger because the PKC-C2 was perhaps already present in multiple copies.
3.3. Domain architectures reveal a polarity in the interactions of most C2 domain families
Examination of domain architectures of the PKC-C2 family shows that it is rather promiscuous in its combinations with other domains, being linked both to the N- and C-termini of several domains (Fig. 5). Number of these linked domains are enzymatic domains suggesting that this C2 domain predominantly functions in tethering or localizing different catalytic activities to the membrane. The PTEN-C2 family is similarly linked to several domains both at the N- and the C-terminus, but the enzymatic domains are always found only to N-terminus of the PTEN-C2 domain. Even greater architectural constraints are observed in the remaining families, with the domains of NT-C2, DOCK-C2 and AIDA-C2 families respectively occurring at the N-terminus, middle and C- terminus of the proteins in which they occur. Likewise, the C2 domain of the PI3/4 kinases always occurs in the same position with respect to the kinase catalytic domains (Fig. 5). The B9-C2 domain always occurs as a standalone domain. This, combined with the fact that three orthologous versions of these proteins are always present together in the proteome, and cooperatively localize at base body or centrosome of cilia in both mammal and C. elegans models, (Kyttala et al., 2006; Dawe et al., 2007; Town et al., 2008; Williams et al., 2008; Bialas et al., 2009) implies that they are likely to function as a trimer. The architectures suggest that there have been considerable positional constraints on the location of most families of C2 domains when they are present in multidomain contexts within a polypeptide. As shown in the model of the PH-fold domains in myosins, such a polarity might imply a directional anchoring function with respect to an extended surface like a membrane – i.e. a polypeptide is tethered to the membrane by a specific section (i.e. tail-, head- or centrally- anchored) while its rest is free to hang in the cytosol to mediate other interactions or catalytic activities (Patino-Lopez et al., 2010).
3.4. Diversification of lipid-binding and membrane interaction mode of the C2 domain
One feature that is highly conserved in C2 domains is the pair of hydrophobic residues on the upper part of the β sheet (Fig. 1, 4), which are involved in imparting a curvature of the sheet that allows formation of a concave ligand-binding area. Given that these hydrophobic residues are implicated in particular lipid contacts, and the above observations on the strong architectural polarity in the positions of C2 domains in polypeptides it is likely that the ancestral version of the C2 domain bound membranes. However, beyond the use of the β sheet in forming this concavity, the C2 domains are extremely divergent suggesting that different families might have optimized their interactions with membranes through divergent means.
3.4.1. PKC-C2 structures provide precedence to understand lipid-interactions of the C2 domain
Former structural considerations have primarily concentrated on the PKC-C2s and their Ca2+-binding pocket is believed to make the main contact with the membrane (Rizo and Sudhof, 1998; Cho and Stahelin, 2006). The set of four acidic residues (usually aspartates, labeled D187, D293, D246 and D248 from PDB: 1DSY in Fig. 1 and 4) that are ligands for Ca2+ are highly conserved in this family (Verdaguer et al., 1999), and these residues are found in representatives even from basal eukaryotes. However, all other families of the C2 domain, including the novel ones identified in this study lack this calcium-binding signature or any other set of conserved acidic residues that could form an alternative calcium-binding pocket. These observations suggest that the common ancestor of all C2 domains probably did not bind calcium and this activity was specifically acquired only in last common ancestor of the PKC-C2 family. Hence, the calcium dependent model for membrane interaction cannot be generalized for the remaining representatives of the C2 superfamily. However, certain members of the PKC-C2 family have secondarily lost the Ca2+-binding residues but still bind lipids. While the actual lipid-binding mode of these versions has not been experimentally characterized, based on the structure of such Ca2+-independent PKC-C2 it has been proposed that basic residues in the region that typically contains the Ca2+-binding pocket could make direct membrane contacts (Cho and Stahelin, 2006). Consistent with this, representatives of a distinct Ca2+-independent family, namely the PI3K-C2 family, interact with membranes via a basic loop between β strands 5 and 6 that located in a position comparable to the Ca2+-binding end of PKC-C2s (Fig. 4) (Djordjevic and Driscoll, 2002). In principle, such Ca2+-independent representatives could offer a model for the binding mode of the other families of C2 domains. The concavity in the β sheet on the “upper surface” (Fig. 4) contains a patch of basic and hydrophobic residues (the conserved pair referred to above) that form the PtdIns(4,5)P2-binding surface (Y295, K197, K209, K211, W245 in PDB: 1DSY; Fig. 1 and 4) (Guerrero-Valero et al., 2009). Further, basic residues in this region could mediate membrane penetration (e.g. K205 in PDB: 1DSY). Given the structural conservation of the concavity in the β sheet we reasoned that this surface could indeed contribute to a more general ligand interaction surface, such as in PI3K-C2s. Thus, analysis of the binding modes of the known C2 structures suggested two general regions that could bear determinants for membrane interactions – 1) the loops at one end of the C2 domain that in the classical PKC-C2s contain the Ca2+-binding site and 2) the concave surface of the “upper” β sheet.
3.4.2. Inference of binding modes for other uncharacterized C2 domain families
We used the above determinants identified based on the PKC-C2 family to investigate the binding modes of the other C2 families that have not been completely characterized or were previously unknown. As discussed above, and previously noted by other workers even within the PKC-C2, there is some diversity in terms of lipid interactions (Rizo and Sudhof, 1998; Cho and Stahelin, 2006; Guerrero-Valero et al., 2009). However, our prime objective was to identify the common denominator or the most prevalent mode of interaction for the family. When compared to the PKC-C2 family, the issue of intra-family diversity in the interaction mode is less problematic for the other families because their domain architectures suggest a lower degree of functional diversification. Previous considerations on C2 (Murray and Honig, 2002) and other membrane-binding domains (Blomberg et al., 1999; Stahelin et al., 2004) have suggested that identification of regions of positive electrostatic potential or exposed hydrophobic residues provide useful markers to identify regions that might respectively interact with negatively charged or zwitterionic head-groups and hydrophobic tails of lipids. Hence, we chose representative structures of each C2 family, where available, to compute electrostatic potential surfaces. For families lacking experimental structures we constructed homology models and similar calculated electrostatic potential surfaces. We examined these results in light of the sequence conservation patterns within and between families to predict family-specific interaction surfaces (Fig. 4). In each case we identify the specific residues, which we predict to be involved in interactions with lipids that could be targeted in future experimental studies (Fig. 1, 4)
We specifically asked if the two main interaction regions defined on the basis of the PKC-C2 and PI3K structures might have a role in lipid-interactions in the other families. We observed that the end of the C2 that binds Ca2+ in the PKC-C2 tends to contain a basic patch in each of other families, although it is very poorly developed in the B9-C2 family (Fig. 4). This suggested that end-based binding might be a common mode for most families. However, the residues contributing to this patch are only specific to a given family and not conserved across families (Fig. 1). Moreover, the specific arrangement of the basic patch does differ in each family – for example, in the AIDA and PTEN families the basic patch is bilobed implying that more than one lipid contact could be made via the end-based binding site (Fig. 4). In contrast, in the B9-C2 family the end-based binding site might at best play a minor role in lipid interaction. This would imply that the angle of approach to the membrane would drastically differ between different families. In terms of the concave surface of the “upper” sheet we observed that positively charged regions are well-developed in the case of the PTEN, NT-C2 and B9 families and to a certain degree in the DOCK family. Here again in each of these families the constellation of basic residues contributing to the formation of this basic patch is distinct (Fig. 2, 4). Further, given that the above-noted pair of hydrophobic residues on this surface is also well-conserved, it is likely that they also contribute to an interaction with the lipid as seen in the case of the PKC-C2. Differences in the distribution of basic residues on the concave surface again support differences in the angle of approach to the membrane suggested by differences in the end-based binding mode. For example, NT-C2 domains have a patch of basic residues at the extreme N-terminus of the fold prior to strand-1. These residues are predicted to be in a flexible segment that is likely to be an extension of the concave surface opposite to the end-based binding site (Fig. 1, 4). This, along with the distribution of basic residues on the concave surface in the core NT-C2 domains, suggests that it mainly interacts with the membrane via the concave surface and approaches the membrane in an almost parallel fashion. Similarly, an almost parallel binding is likely for the B9-C2 domains that also have a notable positive charge distribution on the concave surface. B9-C2 domains also conserve an unusual aromatic position in a loop projecting on the concave surface (W33 in Fig. 1, 4). Given that it is predicted to be exposed we suggest that this might represent an additional lipid-interaction element that might penetrate the membrane to interact with the hydrophobic interior.
From these observations we propose that both or at least one of the two major regions implicated in lipid interactions are functional across the C2 domain superfamily. Further, this would also imply that both of these binding regions were probably present in the ancestral C2 domain. One possible consequence of the presence of multiple lipid-interaction regions in the same domain, as also observed in the PH superfamily (Ceccarelli et al., 2007; Jackson et al., 2007), might imply that these modules have a dynamic behavior wherein the different interfaces engage the membrane independently either simultaneously or at different times. This could allow for differences in the approach angle and/or the strength of anchoring of the protein, with important functional consequences that could be experimentally investigated further. However, C2 domains present an interesting situation, where despite the retention of the general lipid-binding interfaces most of the actual residues involved in lipid interactions are different between individual families or even within a family. This is somewhat different from the PH-domain superfamily wherein a set of basic lipid-contacting residues tend to be retained across most of the superfamily despite diversity in rest of the ligand-binding pocket (Lemmon, 2007). At least a subset of these might also be observed in some other lipid-binding superfamilies of the PH-like folds like the GRAM domain, whereas members that have lost these residues have acquired other specificities such as peptide-binding (Geyer et al., 2005). Given the diversity in the predicted lipid interacting residues in the C2 superfamily, it is possible that there are not many constraints for the actual residues involved in the interaction. However, each family tends to conserve its own pattern of basic residues; hence, we argue that the diversity actually represents functional diversification. In addition to the mechanistic considerations such as the angle of approach, it is also possible that the differences represent differences in specificity for particular lipid head-groups (e.g. phosphatidylserine, phosphatidylcholine, phosphatidylglycerol, phosphatidylethanolamine along with inositol and ceramide derivatives; see(Cho and Stahelin, 2006) for summary).
3.5. The newly defined families broaden the functional implications of C2-membrane interactions
The lipid-binding by the previously well-studied PKC, PTEN and PI3K families of C2 domains have been implicated in a number of processes such as vesicular trafficking and diverse lipid-based signaling events. Despite the great diversity their domain architectures suggest that their roles in these processes fall into few basic themes (Fig. 5): 1) Those involved in active membrane dynamics and membrane repair such as the dysferlin group of PKC-C2 domains contain multiple C2 domains; 2) Similarly multiple C2 domains are also a feature of the proteins involved in vesicular dynamics, although these proteins might additionally contain other adaptor domains involved in protein-protein interactions (e.g. the ancient copine lineage of PKC-C2 domains); 3) A large fraction of these C2 domains act as membrane tethers for a slew of enzymatic domains such as protein kinases and phosphatases, peptidases, ubiquitin E3 ligases and phospholipases; 4) Other C2 domains are tethers, which localize domains that regulate small GTPase signaling to membranes (e.g. in Rabphilin). The same theme appears to be the dominant one in the case of the DOCK-C2 family which binds PtdIns(3,4,5)P2 to anchor Rac/CDC42 GTP/GDP exchange factor domains to the membrane (Cote et al., 2005). Rac/Cdc42 GTPases play a role in the dynamics of the actin cytoskeleton (Myers and Casanova, 2008). Our observation that both the DOCK-C2s which are fused to GDP-exchange factor domains and their small GTPase targets are independently expanded in both amoebozoans and Naegleria (an amoeboflagellate) suggests that this might be related to the enhancement of the actin cytoskeleton in these lineages. It is likely that these proteins mediate distinct regulatory effect on these small GTPases at the membrane in relation to amoeboid motility.
While certain previous studies have implicated C2 domains in cytoskeletal functions (Lopez-Lluch et al., 2001; Sklyarova et al., 2002) there is currently not much clarity regarding the roles of C2 domains in the subcellular organization of cytoskeleton. Among the new versions identified in this study tethering of cytoskeletal components to the membrane appears to be an important theme. In the case of the NT-C2 the primary function appears to be linking of actin/microfilament-binding adaptors to the membrane. In support of this prediction, disrupting interactions of the NT-C2 protein EHBP1 with its partner EHD2 results in notable defects in cortical actin rearrangements (Guilherme et al., 2004a). Other data indicate that the NT-C2 domains might also act as a link that tethers endosomal vesicles to the cytoskeleton in course of their intracellular trafficking. This prediction would explain endocytotic defects previously reported upon disruption of NT-C2 domain proteins such as EHBP1 (Guilherme et al., 2004a; Guilherme et al., 2004b; Naslavsky and Caplan, 2005). It is conceivable that in these contexts the NT-C2 might specific bind PtdIns(4)P and PtdIns(4,5)P2 found in the endocytotic recycling vesicle membranes (Blume et al., 2007; Jovic et al., 2009). A similar role for NT-C2 domains in linking membranous structures to the cytoskeleton is also probably required in processes such as the relocation or movement of cellular organelles like chloroplasts, and vesicular movement in Rhizobium-directed root hair growth towards/from plasma membrane (DeBlasio et al., 2005; Arrighi et al., 2008). The Mg2+ transporter MgtE N-terminal domain appears to play a role in the multimerization of these transporters (Hattori et al., 2007). The presence of this domain in certain proteins with NT-C2 suggests that these might have a role in tethering membrane-associated transporters to the cytoskeleton.
A role in linking the cytoskeleton to the membrane appears to be likely even in the case of the AIDA-C2 family, which in animals occurs in the context of the focal adhesion junctions. The presence of a myosin with a C-terminal AIDA-C2 domain in oomycetes is of interest because it points to a second membrane-anchoring mechanism for myosins either in the context of plasma-membrane localization or vesicular transport along microfilaments. Recent studies have indicated that a divergent PH domain play an important role in the membrane localization of several myosins (Patino-Lopez et al., 2010). In the oomycete myosins, in addition to the PH domain, there is a further AIDA-C2 domain that might contribute to a more extensive membrane contact in these proteins. Plant AIDA-C2 proteins, which contain a coiled-coil segment, could also have a cytoskeletal role by interacting with coiled coil segments in other proteins like myosin. The B9-C2 family suggests that not only the actin-myosin based cytoskeleton but also the membrane-anchoring of the microtubule based cytoskeleton might depend on C2 domains. A key step in the maturation of the microtubular cytoskeleton is the directed migration of the centrosome to the apical cell membrane, where it docks and matures to form the basal body of the cilia (Dawe et al., 2007). Given the loss-of-function phenotypes of the B9-C2 proteins (Dawe et al., 2007; Town et al., 2008; Williams et al., 2008; Bialas et al., 2009; Tammachote et al., 2009; Weatherbee et al., 2009) and their interactions with other cytoskeletal proteins like Mechelin/Mks3, Nesprin-2 and γ-tubulin (Town et al., 2008; Dawe et al., 2009) we hypothesize that the trimer comprised of the three paralogous B9-C2 proteins are likely to function as adaptor that mediate anchoring of the centrosome to membrane.
3.6. Evolutionary implications of the C2 superfamily and general conclusions
Classification of the C2 superfamily along with identification of new versions helps in clarifying certain aspects of the emergence of the ancestral eukaryotic cell. As noted before, the C2 domain appears to be part of the proliferation of several families of lipid-binding domain that happened specifically in eukaryotes. While some of these domains like the START, PH and GRAM domains have bacterial antecedents, the C2 domain is apparently a eukaryotic innovation. Although it cannot be ruled out that there are highly divergent bacterial versions that we failed to detect in this study, there are no obvious precursors of the families identified here in bacteria. Furthermore, our classification along with the examination of phyletic patterns (Fig. 3, 5) suggests that the C2 domain were already greatly diversified in LECA itself. The functional predictions presented here show that despite having a common lipid-binding function each family had acquired considerably distinct sub-cellular roles in LECA (Fig. 5). These relate specifically to the anchoring of the both the actin- and tubulin- based cytoskeleton to membranes, organellar movement vesicular trafficking and membrane repair, and membrane-associated phosphoinositide and GTPase signaling. These functions are central to quintessential features of the eukaryotic cell suggesting that the proliferation of the C2 family went hand-in-hand with the emergence of the intracellular complexity of the eukaryotic cell. Thus, the primary diversification of the C2 superfamily can be mapped back to the period between the FECA and LECA, during which features typical of extant eukaryotic cells developed (Mans et al., 2004).
The divergence of the PKC-C2 family, with calcium dependent lipid-binding capability, appears to have been related to the emergence of the vesicular trafficking and membrane repair systems. The corresponding deployment of calcium-dependent signaling mediated by EF-hand domains in the assembly of the centrosome (e.g. the centrin proteins) and the actin-myosin based cytoskeleton (calmodulin) implies that these C2 domains possibly emerged as part of the general adoption of the calcium ion as a messenger to communicate state changes within cells (Clapham, 2007). Emergence of the family of the PKC-C2 domains probably coincided with the emergence of the first endomembrane systems in eukaryotes. The emergence of endomembranes was also accompanied by the divergence of several distinct families of small GTPases that localized to different intracellular compartments and also organized the cytoskeleton as a scaffold for these compartments (Jekely, 2003; Mans et al., 2004; Myers and Casanova, 2008). The origin of the DOCK-C2 family was probably linked to the localization of the Rac/CDC42-like GTPases to cellular membranes. The triplication of B9-C2 domains that also occurred between FECA and LECA appears to have been associated with the emergence of the eukaryotic cilium/flagellum. On the other hand the NT-C2 domains appear to have been associated with the emergence of the microfilament aspect of the cytoskeleton and the need to anchor intra-cellular membrane-bound structures, including organelles to the cytoskeleton. PTEN and PI3K families represent the original recruitment of C2 domains in the context of generating lipid metabolites that could be used as intracellular messengers. The involvement of C2 domains in this aspect appears to have expanded in course of later eukaryotic evolution with other lipid-processing enzymes, namely phospholipases being recruited to membrane via C2 domains. However, this occurred via a distinct family of C2 domains – the PKC-C2 domain. The same domain was also used to recruit other catalytic domain such as peptidases and protein kinases, as well as adaptor domains resulting in a massive expansion of the PKC-C2 domain in signaling contexts in later eukaryotic evolution. In this respect the evolution of the C2 domain closely parallels that of the PH domain superfamily (Lemmon, 2007).
Our analysis and classification of the C2 domain shows that the calcium-dependent membrane binding mode is not the ancestral property of the superfamily, but is limited to just one of the families. Other C2 families, by contrast, possess their own distinctive binding modes that are independent of calcium. We provide specific predictions regarding these binding modes including the actual residues that could be involved in lipid contacts. We hope this analysis might help in further experimental studies testing the hypothesis presented here.
Supplementary Material
Acknowledgments
The first author would like to thank S. Abhiman for his help in data analysis and M. DiPetta for her assistance in analyzing initial sequence alignment of NT-C2 domains. We also thank L.M. Iyer for suggestions on the manuscript. This work is supported by the NIH Postdoctoral Visiting Fellowship and the intramural funds of the National Library of Medicine at the National Institutes of Health, USA.
Footnotes
Appendix A. Supplementary data Supplementary figures are available online: ftp://ftp.ncbi.nih.gov/pub/aravind/temp/C2/C2.html
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alpy F, Latchumanan VK, Kedinger V, Janoshazi A, Thiele C, Wendling C, Rio MC, Tomasetto C. Functional characterization of the MENTAL domain. J Biol Chem. 2005;280:17945–52. doi: 10.1074/jbc.M500723200. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Iyer LM, Aravind L. Comparative genomics of protists: new insights into the evolution of eukaryotic signal transduction and gene regulation. Annu Rev Microbiol. 2007;61:453–75. doi: 10.1146/annurev.micro.61.080706.093309. [DOI] [PubMed] [Google Scholar]
- Aravind L, Neuwald AF, Ponting CP. Sec14p-like domains in NF1 and Dbl-like proteins indicate lipid regulation of Ras and Rho signaling. Curr Biol. 1999;9:R195–7. doi: 10.1016/s0960-9822(99)80127-4. [DOI] [PubMed] [Google Scholar]
- Arrighi JF, Godfroy O, de Billy F, Saurat O, Jauneau A, Gough C. The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc Natl Acad Sci U S A. 2008;105:9817–22. doi: 10.1073/pnas.0710273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–39. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Azzi A, Boscoboinik D, Hensey C. The protein kinase C family. Eur J Biochem. 1992;208:547–57. doi: 10.1111/j.1432-1033.1992.tb17219.x. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner S, Hofmann K, Chiquet-Ehrismann R, Bucher P. The discoidin domain family revisited: new members from prokaryotes and a homology-based fold prediction. Protein Sci. 1998;7:1626–31. doi: 10.1002/pro.5560070717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialas NJ, Inglis PN, Li C, Robinson JF, Parker JD, Healey MP, Davis EE, Inglis CD, Toivonen T, Cottell DC, Blacque OE, Quarmby LM, Katsanis N, Leroux MR. Functional interactions between the ciliopathy-associated Meckel syndrome 1 (MKS1) protein and two novel MKS1-related (MKSR) proteins. J Cell Sci. 2009;122:611–24. doi: 10.1242/jcs.028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg N, Gabdoulline RR, Nilges M, Wade RC. Classification of protein sequences by homology modeling and quantitative analysis of electrostatic similarity. Proteins. 1999;37:379–87. doi: 10.1002/(sici)1097-0134(19991115)37:3<379::aid-prot6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Blume JJ, Halbach A, Behrendt D, Paulsson M, Plomann M. EHD proteins are associated with tubular and vesicular compartments and interact with specific phospholipids. Exp Cell Res. 2007;313:219–31. doi: 10.1016/j.yexcr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Blasutig IM, Goudreault M, Li Z, Ruston J, Pawson T, Sicheri F. Non-canonical interaction of phosphoinositides with pleckstrin homology domains of Tiam1 and ArhGAP9. J Biol Chem. 2007;282:13864–74. doi: 10.1074/jbc.M700505200. [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. Membrane binding and subcellular targeting of C2 domains. Biochim Biophys Acta. 2006;1761:838–49. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Clapham DE. Calcium signaling. Cell. 2007;131:1047–58. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Cote JF, Motoyama AB, Bush JA, Vuori K. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat Cell Biol. 2005;7:797–807. doi: 10.1038/ncb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote JF, Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J Cell Sci. 2002;115:4901–13. doi: 10.1242/jcs.00219. [DOI] [PubMed] [Google Scholar]
- Cozzetto D, Tramontano A. Relationship between multiple sequence alignments and quality of protein comparative models. Proteins. 2005;58:151–7. doi: 10.1002/prot.20284. [DOI] [PubMed] [Google Scholar]
- Cremona O, De Camilli P. Phosphoinositides in membrane traffic at the synapse. J Cell Sci. 2001;114:1041–52. doi: 10.1242/jcs.114.6.1041. [DOI] [PubMed] [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–3. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- Dawe HR, Adams M, Wheway G, Szymanska K, Logan CV, Noegel AA, Gull K, Johnson CA. Nesprin-2 interacts with meckelin and mediates ciliogenesis via remodelling of the actin cytoskeleton. J Cell Sci. 2009;122:2716–26. doi: 10.1242/jcs.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe HR, Smith UM, Cullinane AR, Gerrelli D, Cox P, Badano JL, Blair-Reid S, Sriram N, Katsanis N, Attie-Bitach T, Afford SC, Copp AJ, Kelly DA, Gull K, Johnson CA. The Meckel-Gruber Syndrome proteins MKS1 and meckelin interact and are required for primary cilium formation. Hum Mol Genet. 2007;16:173–86. doi: 10.1093/hmg/ddl459. [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Godi A. PI-loting membrane traffic. Nat Cell Biol. 2004;6:487–92. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- DeBlasio SL, Luesse DL, Hangarter RP. A plant-specific protein essential for blue-light-induced chloroplast movements. Plant Physiol. 2005;139:101–14. doi: 10.1104/pp.105.061887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–7. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Djordjevic S, Driscoll PC. Structural insight into substrate specificity and regulatory mechanisms of phosphoinositide 3-kinases. Trends Biochem Sci. 2002;27:426–32. doi: 10.1016/s0968-0004(02)02136-9. [DOI] [PubMed] [Google Scholar]
- Doerks T, Strauss M, Brendel M, Bork P. GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane-associated proteins. Trends Biochem Sci. 2000;25:483–5. doi: 10.1016/s0968-0004(00)01664-9. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fucini P, Renner C, Herberhold C, Noegel AA, Holak TA. The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold. Nat Struct Biol. 1997;4:223–30. doi: 10.1038/nsb0397-223. [DOI] [PubMed] [Google Scholar]
- Gaullier JM, Simonsen A, D’Arrigo A, Bremnes B, Stenmark H, Aasland R. FYVE fingers bind PtdIns(3)P. Nature. 1998;394:432–3. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- Geyer JP, Doker R, Kremer W, Zhao X, Kuhlmann J, Kalbitzer HR. Solution structure of the Ran-binding domain 2 of RanBP2 and its interaction with the C terminus of Ran. J Mol Biol. 2005;348:711–25. doi: 10.1016/j.jmb.2005.02.033. [DOI] [PubMed] [Google Scholar]
- Gibson TJ, Hyvonen M, Musacchio A, Saraste M, Birney E. PH domain: the first anniversary. Trends Biochem Sci. 1994;19:349–53. doi: 10.1016/0968-0004(94)90108-2. [DOI] [PubMed] [Google Scholar]
- Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003a;19:1015–8. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- Ginalski K, Pas J, Wyrwicz LS, von Grotthuss M, Bujnicki JM, Rychlewski L. ORFeus: Detection of distant homology using sequence profiles and predicted secondary structure. Nucleic Acids Res. 2003b;31:3804–7. doi: 10.1093/nar/gkg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginalski K, Rychlewski L. Detection of reliable and unexpected protein fold predictions using 3D-Jury. Nucleic Acids Res. 2003;31:3291–2. doi: 10.1093/nar/gkg503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginalski K, von Grotthuss M, Grishin NV, Rychlewski L. Detecting distant homology with Meta-BASIC. Nucleic Acids Res. 2004;32:W576–81. doi: 10.1093/nar/gkh370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstadt L, Ponting CP. CHROMA: consensus-based colouring of multiple alignments for publication. Bioinformatics. 2001;17:845–6. doi: 10.1093/bioinformatics/17.9.845. [DOI] [PubMed] [Google Scholar]
- Guerrero-Valero M, Ferrer-Orta C, Querol-Audi J, Marin-Vicente C, Fita I, Gomez-Fernandez JC, Verdaguer N, Corbalan-Garcia S. Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2. Proc Natl Acad Sci U S A. 2009;106:6603–7. doi: 10.1073/pnas.0813099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilherme A, Soriano NA, Bose S, Holik J, Bose A, Pomerleau DP, Furcinitti P, Leszyk J, Corvera S, Czech MP. EHD2 and the novel EH domain binding protein EHBP1 couple endocytosis to the actin cytoskeleton. J Biol Chem. 2004a;279:10593–605. doi: 10.1074/jbc.M307702200. [DOI] [PubMed] [Google Scholar]
- Guilherme A, Soriano NA, Furcinitti PS, Czech MP. Role of EHD1 and EHBP1 in perinuclear sorting and insulin-regulated GLUT4 recycling in 3T3-L1 adipocytes. J Biol Chem. 2004b;279:40062–75. doi: 10.1074/jbc.M401918200. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hattori M, Tanaka Y, Fukai S, Ishitani R, Nureki O. Crystal structure of the MgtE Mg2+ transporter. Nature. 2007;448:1072–5. doi: 10.1038/nature06093. [DOI] [PubMed] [Google Scholar]
- Hayashi I, Vuori K, Liddington RC. The focal adhesion targeting (FAT) region of focal adhesion kinase is a four-helix bundle that binds paxillin. Nat Struct Biol. 2002;9:101–6. doi: 10.1038/nsb755. [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–1. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins. 2001;43:134–44. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Jackson SG, Zhang Y, Haslam RJ, Junop MS. Structural analysis of the carboxy terminal PH domain of pleckstrin bound to D-myo-inositol 1,2,3,5,6-pentakisphosphate. BMC Struct Biol. 2007;7:80. doi: 10.1186/1472-6807-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server for profile--profile sequence alignments. Nucleic Acids Res. 2005;33:W284–8. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jekely G. Small GTPases and the evolution of the eukaryotic cell. Bioessays. 2003;25:1129–38. doi: 10.1002/bies.10353. [DOI] [PubMed] [Google Scholar]
- Jovic M, Kieken F, Naslavsky N, Sorgen PL, Caplan S. Eps15 homology domain 1-associated tubules contain phosphatidylinositol-4-phosphate and phosphatidylinositol-(4,5)-bisphosphate and are required for efficient recycling. Mol Biol Cell. 2009;20:2731–43. doi: 10.1091/mbc.E08-11-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyttala M, Tallila J, Salonen R, Kopra O, Kohlschmidt N, Paavola-Sakki P, Peltonen L, Kestila M. MKS1, encoding a component of the flagellar apparatus basal body proteome, is mutated in Meckel syndrome. Nat Genet. 2006;38:155–7. doi: 10.1038/ng1714. [DOI] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer EL. Kalign--an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–34. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Pleckstrin homology (PH) domains and phosphoinositides. Biochem Soc Symp. 2007:81–93. doi: 10.1042/BSS0740081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Leslie NR, Yang X, Downes CP, Weijer CJ. PtdIns(3,4,5)P(3)-dependent and -independent roles for PTEN in the control of cell migration. Curr Biol. 2007;17:115–25. doi: 10.1016/j.cub.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lluch G, Bird MM, Canas B, Godovac-Zimmerman J, Ridley A, Segal AW, Dekker LV. Protein kinase C-delta C2-like domain is a binding site for actin and enables actin redistribution in neutrophils. Biochem J. 2001;357:39–47. doi: 10.1042/0264-6021:3570039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–37. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–11. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- Meller N, Merlot S, Guda C. CZH proteins: a new family of Rho-GEFs. J Cell Sci. 2005;118:4937–46. doi: 10.1242/jcs.02671. [DOI] [PubMed] [Google Scholar]
- Montaville P, Schlicker C, Leonov A, Zweckstetter M, Sheldrick GM, Becker S. The C2A-C2B linker defines the high affinity Ca(2+)binding mode of rabphilin-3A. J Biol Chem. 2007;282:5015–25. doi: 10.1074/jbc.M606746200. [DOI] [PubMed] [Google Scholar]
- Murray D, Honig B. Electrostatic control of the membrane targeting of C2 domains. Mol Cell. 2002;9:145–54. doi: 10.1016/s1097-2765(01)00426-9. [DOI] [PubMed] [Google Scholar]
- Myers KR, Casanova JE. Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 2008;18:184–92. doi: 10.1016/j.tcb.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalefski EA, Falke JJ. The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci. 1996;5:2375–90. doi: 10.1002/pro.5560051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N, Caplan S. C-terminal EH-domain-containing proteins: consensus for a role in endocytic trafficking, EH? J Cell Sci. 2005;118:4093–101. doi: 10.1242/jcs.02595. [DOI] [PubMed] [Google Scholar]
- Parry DA, Fraser RD, Squire JM. Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure. J Struct Biol. 2008;163:258–69. doi: 10.1016/j.jsb.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Patino-Lopez G, Aravind L, Dong X, Kruhlak MJ, Ostap EM, Shaw S. Myosin 1G is an abundant class I myosin in lymphocytes whose localization at the plasma membrane depends on its ancient divergent PH domain (Myo1PH) J Biol Chem. 2010 doi: 10.1074/jbc.M109.086959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Grishin NV. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics. 2007;23:802–8. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- Perisic O, Fong S, Lynch DE, Bycroft M, Williams RL. Crystal structure of a calcium-phospholipid binding domain from cytosolic phospholipase A2. J Biol Chem. 1998;273:1596–604. doi: 10.1074/jbc.273.3.1596. [DOI] [PubMed] [Google Scholar]
- Ponting CP. AF-6/cno: neither a kinesin nor a myosin, but a bit of both. Trends Biochem Sci. 1995;20:265–6. doi: 10.1016/s0968-0004(00)89040-4. [DOI] [PubMed] [Google Scholar]
- Ponting CP. Novel domains in NADPH oxidase subunits, sorting nexins, and PtdIns 3-kinases: binding partners of SH3 domains? Protein Sci. 1996;5:2353–7. doi: 10.1002/pro.5560051122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Parker PJ. Extending the C2 domain family: C2s in PKCs delta, epsilon, eta, theta, phospholipases, GAPs, and perforin. Protein Sci. 1996;5:162–6. doi: 10.1002/pro.5560050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L, Bobkov AA, Patel M, Jaroszewski L, Bankston LA, Stec B, Vuori K, Cote JF, Liddington RC. Structural basis of membranetargeting by the Dock180 family of Rho-family guanine exchange factors (RhoGEFs) J Biol Chem. 2010 doi: 10.1074/jbc.M110.102517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–52. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Rizo J, Sudhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–82. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- Rui Y, Xu Z, Xiong B, Cao Y, Lin S, Zhang M, Chan SC, Luo W, Han Y, Lu Z, Ye Z, Zhou HM, Han J, Meng A, Lin SC. A beta-cateninindependent dorsalization pathway activated by Axin/JNK signaling and antagonized by aida. Dev Cell. 2007;13:268–82. doi: 10.1016/j.devcel.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O’Farrell PH, Andino R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sklyarova T, De Corte V, Meerschaert K, Devriendt L, Vanloo B, Bailey J, Cook LJ, Goethals M, Van Damme J, Puype M, Vandekerckhove J, Gettemans J. Fragmin60 encodes an actin-binding protein with a C2 domain and controls actin Thr-203 phosphorylation in Physarum plasmodia and sclerotia. J Biol Chem. 2002;277:39840–9. doi: 10.1074/jbc.M207052200. [DOI] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin RV, Ananthanarayanan B, Blatner NR, Singh S, Bruzik KS, Murray D, Cho W. Mechanism of membrane binding of the phospholipase D1 PX domain. J Biol Chem. 2004;279:54918–26. doi: 10.1074/jbc.M407798200. [DOI] [PubMed] [Google Scholar]
- Stradal T, Kranewitter W, Winder SJ, Gimona M. CH domains revisited. FEBS Lett. 1998;431:134–7. doi: 10.1016/s0014-5793(98)00751-0. [DOI] [PubMed] [Google Scholar]
- Tammachote R, Hommerding CJ, Sinders RM, Miller CA, Czarnecki PG, Leightner AC, Salisbury JL, Ward CJ, Torres VE, Gattone VH, 2nd, Harris PC. Ciliary and centrosomal defects associated with mutation and depletion of the Meckel syndrome genes MKS1 and MKS3. Hum Mol Genet. 2009;18:3311–23. doi: 10.1093/hmg/ddp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA)software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Town T, Breunig JJ, Sarkisian MR, Spilianakis C, Ayoub AE, Liu X, Ferrandino AF, Gallagher AR, Li MO, Rakic P, Flavell RA. The stumpy gene is required for mammalian ciliogenesis. Proc Natl Acad Sci U S A. 2008;105:2853–8. doi: 10.1073/pnas.0712385105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Leevers SJ, Panayotou G, Waterfield MD. Phosphoinositide 3-kinases: a conserved family of signal transducers. Trends Biochem Sci. 1997;22:267–72. doi: 10.1016/s0968-0004(97)01061-x. [DOI] [PubMed] [Google Scholar]
- Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC. Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine. Embo J. 1999;18:6329–38. doi: 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DY, Fulthorpe R, Liss SN, Edwards EA. Identification of estrogenresponsive genes by complementary deoxyribonucleic acid microarray and characterization of a novel early estrogen-induced gene: EEIG1. Mol Endocrinol. 2004;18:402–11. doi: 10.1210/me.2003-0202. [DOI] [PubMed] [Google Scholar]
- Weatherbee SD, Niswander LA, Anderson KV. A mouse model for Meckel syndrome reveals Mks1 is required for ciliogenesis and Hedgehog signaling. Hum Mol Genet. 2009;18:4565–75. doi: 10.1093/hmg/ddp422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CL, Winkelbauer ME, Schafer JC, Michaud EJ, Yoder BK. Functional redundancy of the B9 proteins and nephrocystins in Caenorhabditis elegans ciliogenesis. Mol Biol Cell. 2008;19:2154–68. doi: 10.1091/mbc.E07-10-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–71. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- Yabe D, Xia ZP, Adams CM, Rawson RB. Three mutations in sterol-sensing domain of SCAP block interaction with insig and render SREBP cleavage insensitive to sterols. Proc Natl Acad Sci U S A. 2002;99:16672–7. doi: 10.1073/pnas.262669399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.