Abstract
Background
Within the current worldwide epidemic of community-acquired Staphylococcus aureus infections, attention has focused on the role of methicillin-resistant strains. We characterized methicillin-susceptible strains that also contribute.
Methods
We tracked cultures from abscesses submitted to the microbiology laboratory at St. Louis Children’s Hospital. We also sought Panton-Valentine leukocidin (PVL) genes in methicillin-susceptible Staphylococcus aureus (MSSA) isolates, and we further characterized some isolates by multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), antibiotic susceptibility, accessory gene regulator (agr) allele, and presence of the arcA gene of the arginine catabolic mobile element (ACME).
Results
From 1999 to 2007, we detected a 250-fold increase in cultures of abscesses yielding methicillin-resistant Staphylococcus aureus (MRSA) and a 5-fold increase in abscess cultures yielding MSSA. MSSA isolates from abscesses and wounds were more likely to encode PVL than isolates from other sources. In contrast to PVL-negative isolates of MSSA which were genetically diverse, PVL-positive isolates were predominantly MLST 8, Agr type 1. More than half of PVL-positive MSSA isolates were resistant to erythromycin and susceptible to clindamycin with absence of inducible resistance, a pattern uncommon in PVL-negative MSSA but frequent in the USA300 clone of MRSA. In addition, PFGE of PVL-positive MSSA strains revealed the USA300 pattern.
Conclusions
In addition to methicillin-resistant strains, the current epidemic of Staphylococcus aureus infections includes infections caused by methicillin-susceptible strains that are closely related genetically and share phenotypic characteristics other than susceptibility to methicillin. These findings suggest that factors other than methicillin resistance are driving the epidemic.
Keywords: Staphylococcus aureus, Panton-Valentine leukocidin, methicillin resistance
INTRODUCTION
Prior to the late 1990s, methicillin-resistant Staphylococcus aureus (MRSA) infections occurred almost exclusively in association with hospitals, nursing homes, and other health care institutions. In the last decade, community-acquired (CA) MRSA infections have become prevalent in many locations around the world [1–3]. Circulating CA strains of MRSA are genetically distinct from those that were traditionally detected in health care-associated (HA) infections. In the US, traditional HA-MRSA strains typically possess the type II staphylococcal chromosomal cassette mec (SCC), a mobile element carrying the mecA gene that encodes penicillin-binding protein 2a, a cell-wall transpeptidase with low affinity for methicillin and other β-lactamase-resistant semisynthetic penicillins [4]. In contrast, strains typically detected in the current epidemic of CA-MRSA infections carry smaller SCCs, designated type IV or V [5, 6]. In addition, circulating CA-MRSA strains often carry genes (lukFS-PV) encoding Panton-Valentine leukocidin (PVL), a bi-component, pore-forming staphylococcal exotoxin [7, 8]. These strains are now also being recognized as causes of health care-associated MRSA infections [9].
PVL was first described in 1932 [10], years before the first appearance in 1961 of methicillin resistance in staphylococci [11]. Intradermal injection of PVL in rabbits produces severe necrotizing skin lesions [12, 13], consistent with the association between PVL-positive staphylococcal strains and the formation of furuncles, cutaneous abscesses, and severe necrotic skin lesions [12, 14–17]. Interestingly, the strains of S. aureus responsible for widespread epidemics in newborn nurseries during the 1950s and 1960s were recently shown to produce PVL [18]. The importance of PVL as a virulence factor has recently been demonstrated in a mouse model of pneumonia [19]; however, it was dispensable for virulence in mouse models of sepsis and skin infection [20]. Recent clinical experience strongly suggests that PVL-positive strains of S. aureus exhibit enhanced virulence [16, 21–24], although it is unclear whether this phenotype is attributable to PVL itself, to linked and yet unidentified virulence determinants, or to altered regulation of toxin expression.
Previous studies of both methicillin-susceptible Staphylococcus aureus (MSSA) and MRSA isolated sequentially in hospital laboratories from all specimen types indicated that 2 to 17% of MSSA strains were positive for PVL [16, 25, 26]. However, the prevalence of PVL genes among MSSA strains isolated from abscesses or furuncles has been as high as 93% [16]. As in many other US cities, cutaneous infections caused by CA-MRSA strains at our center have risen dramatically in the last decade; however, cutaneous infections with MSSA at our center also have increased. In this study, we sought to determine the prevalence of PVL genes among isolates of MSSA recovered in our laboratory, as well as to define the clinical and molecular features of PVL-positive MSSA in our community.
METHODS
Patient and isolate identification
All study procedures were approved by the institutional Office of Human Research Protection. In order to examine the magnitude of increase in abscesses, we queried the result database of the clinical microbiology laboratory at St. Louis Children’s Hospital to ascertain all routine and anaerobic cultures submitted from January 1999 through December 2007 that yielded MSSA or MRSA. From these, we counted S. aureus isolates for which the specimen source was listed as “abscess” or “wound” when the cultures were ordered. Isolates recovered within 30 days of a prior isolate in the same patient were excluded.
Frequency of PVL genes
In order to examine the frequency of PVL in MSSA, we evaluated 214 sequential isolates of MSSA from clinical specimens from all sources submitted to the SLCH clinical microbiology laboratory between October 2005 and March 2006. Patient data collected for each specimen included date of service, source and body site of the culture. Duplicate isolates (within 30 days in the same patient) were excluded. The presence of the lukF-PV gene was detected by multiplex PCR as previously described [27].
Characterization of MSSA strains
From within the 214 sequential isolates of MSSA, those with specimen source listed as “abscess” or “wound” (n = 81) were further characterized (see Supplementary Figure 1). Antimicrobial susceptibility testing, including the D-test for inducible clindamycin resistance, was performed by disk diffusion in accordance with procedures of the Clinical Laboratory Standards Institute (CLSI) [28]. Isolates were tested by PCR for the presence of the accessory gene regulator (agr) allele group as previously described [29] and for the presence of the arcA gene of the arginine catabolic mobile element (ACME) originally identified in a USA300 isolate of MRSA [30]. All PVL-positive MSSA isolates from abscesses and wounds (n= 29) and a subset of PVL-negative isolates from abscesses and wounds (n= 31; selected to represent different body sites) were analyzed by multilocus sequence typing (MLST) performed at the Washington University Genome Sequencing Center [31]. A subset of the PVL-positive isolates which were MLST 8 was further analyzed by pulsed-field gel electrophoresis (PFGE) after SmaI digestion, performed at the Centers for Disease Control and Prevention (Atlanta, GA). To minimize potential bias toward genetic relatedness, we selected for PFGE six of the PVL-positive MSSA strains that represented maximally diverse patterns of antimicrobial susceptibility. The PFGE patterns were analyzed using Bionumerics (version 5.10, Applied Maths, Sint-Marten-Latem, Belgium) and grouped into pulsed-field types using Dice coefficients and 80% similarity as previously described [32].
Medical record review
To permit assessment of epidemiologic associations with PVL-positive MSSA infection, we reviewed clinical and epidemiologic characteristics of patients with MSSA isolated from abscesses or wounds. Medical records were available for 69 of 81 patients, including 28 whose isolates were PVL positive and 41 whose isolates were PVL negative. Chart abstraction was performed using a standardized form that captured demographic data, health history information, and illness characteristics, including demonstrated and putative risk factors for CA-MRSA colonization and infection ([33] and S. Fritz, unpublished data).
Statistical analysis
Differences between proportions were tested for statistical significance using Fisher’s exact test, and differences in means were tested with the Student’s t-test. A p-value of <0.05 was considered statistically significant.
RESULTS
Number of MRSA and MSSA cutaneous infections
We counted isolates of MRSA and MSSA from abscesses and wounds recovered annually in the clinical microbiology laboratory at SLCH. In keeping with the current and well-recognized epidemic of CA-MRSA, the number of MRSA isolates from abscesses increased 250-fold between 1999 and 2007, following an approximately exponential curve (Figure 1). During the same period, the number of MSSA isolates from abscesses increased fivefold. A review of medical records data indicated a five-fold increase in incision and drainage procedures performed between 1999 to 2003 (data not shown), suggesting that the increase in S. aureus isolation was not attributable only to a change in practice (increased culturing of abscesses), but also reflected an actual increase in the incidence of abscesses.
Figure 1.
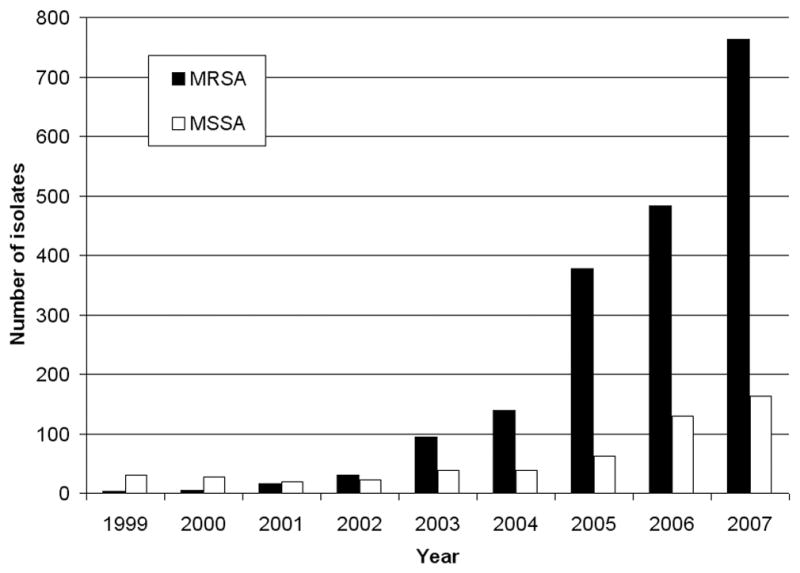
Number of S. aureus isolates from abscesses at SLCH by year. While the number of MRSA isolates increased nearly exponentially over the time period studied, there was also a 5-fold increase in MSSA isolates from abscesses.
Frequency of PVL genes in MSSA
Of the 214 consecutive MSSA isolates recovered in the SLCH clinical microbiology laboratory in the period October 2005 – March 2006, 31 (14.5%) were positive for the lukF-PV gene. Twenty-nine of the 31 PVL-positive MSSA isolates were from specimens with “abscess” or “wound” listed as the source. Specimens from these sources were significantly more likely to carry PVL (29 of 81, 36%) than isolates from other sources (2 of 133, 1.5%; p <0.001) (Table 1).
Table 1.
Presence of PVL Genes among 214 MSSA Isolates by Specimen Source
| Specimen Source | Number Tested | PVL-Positive n (%) | Combined PVL-Positive n (%) |
|---|---|---|---|
| Abscess | 31 | 16 (52 %) | 29 / 81 (36%) |
| Wound | 50 | 13 (26%) | |
| Drainage/discharge | 28 | 2 (7.1%) | 2 / 133 (1.5%) |
| Tissues/aspirates | 14 | 0 | |
| Blood | 7 | 0 | |
| Urine | 4 | 0 | |
| Respiratory | 66 | 0 | |
| Other | 14 | 0 | |
| Total | 214 | 31 (14.5%) |
Antibiotic susceptibility of MSSA isolates from abscesses and wounds
The antibiotic susceptibility patterns of the 81 MSSA isolates (29 PVL-positive and 52 PVL-negative) grown from abscess or wound cultures were reviewed. Of the PVL-positive isolates, 55% displayed a susceptibility profile defined by resistance to erythromycin, susceptibility to clindamycin, and a negative D-test for inducible clindamycin resistance (Table 2). This susceptibility profile is consistent with the presence of the msrA gene that accounts for isolated erythromycin resistance, and is frequently present in CA-MRSA, specifically USA300 strains, in the United States [24, 34, 35]. In contrast, only 2% of the PVL-negative MSSA isolates displayed this same antibiotic susceptibility pattern.
Table 2.
Antibiotic Susceptibility Profiles of 83 MSSA Isolates from Abscesses and Wounds
| Antibiotic Susceptibility Profile | PVL-Negative (n = 52) | PVL-Positive (n = 29) | ||
|---|---|---|---|---|
| Disk Diffusion Interpretation | D-Test | (n/% having the indicated antibiotic susceptibility profile) | ||
| Erythromycin | Clindamycin | |||
| S | S | ND | 36 (69 %) | 12 (41 %) |
| R | S | Negative | 1 (2%)* | 16 (55%)* |
| R | S | Positive | 12 (23%) | 0 |
| R | S | ND | 1 (2%) | 0 |
| R | R | ND | 2 (4%) | 1 (3%)** |
p < 0.001 by Fisher’s exact test
Represents an isolate whose disk diffusion test result was intermediate to erythromycin and clindamycin
S, susceptible; R, resistant; ND, not done
Molecular typing of MSSA isolates
The 81 isolates from abscesses and wounds were characterized further by Agr typing and arcA PCR. Of the 29 PVL-positive isolates, 25 (86%) were of Agr type 1, compared with 20 (38%) of the 52 PVL-negative isolates (p < 0.001). The arcA gene of the ACME was present in 3 (10%) of the PVL-positive isolates and in 2 (4%) of the PVL-negative isolates.
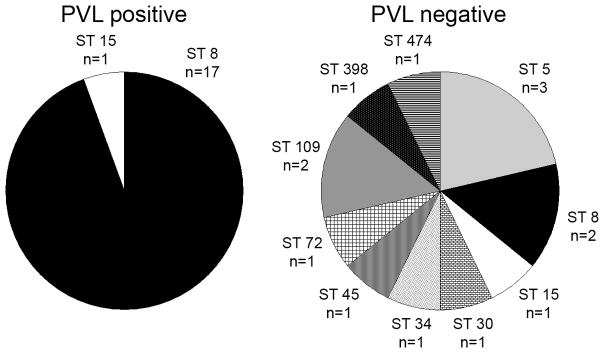
MLST results were available for 32 MSSA isolates from abscesses and wounds (18 of 29 PVL-positive isolates and 14 of 31 PVL-negative isolates) (Figure 2). An MLST assignment was unable to be made in other strains due to technical issues or lack of an allelic match in the existing type database. Of note, the antimicrobial susceptibilities of the MLST-assignable isolates paralleled those of the entire respective PVL-positive or PVL-negative group (data not shown). Of the 18 PVL-positive isolates for which results were available, 17 (94%) were MLST type 8, compared to 2 (14%) of the 14 PVL-negative abscess isolates for which MLST results were available (p < 0.001). There was substantial diversity among the PVL-negative isolates, as nine different MLST sequence types were represented in this group of strains.
Figure 2.
Multilocus sequence types for PVL-positive (left panel) and PVL-negative (right panel) MSSA strains obtained from abscesses and wounds. ST, sequence type.
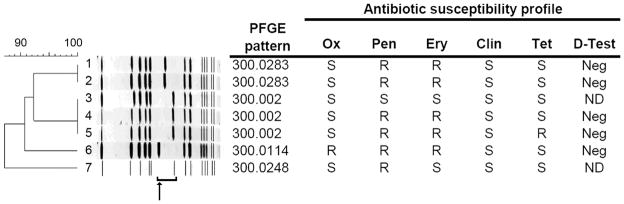
Six of the 18 PVL-positive MSSA isolates of MLST type 8 were further analyzed by pulsed-field gel electrophoresis (PFGE). For this analysis we chose strains with distinct antibiotic susceptibility patterns in an effort to represent phenotypic diversity. All six of these PVL-positive MSSA isolates showed PFGE patterns consistent with that of the USA300 clone of MRSA (Figure 3). Interestingly, the six isolates varied in the migration of the DNA fragment thought to contain the SCCmec cassette.
Figure 3.
Pulsed-field gel electrophoresis banding patterns of PVL-positive Staphylococcus aureus isolates. Antibiotic susceptibility profiles and D-test results, as measured by standard disk-diffusion techniques, are shown to the right of each gel lane. Lanes 1–5 and 7 represent PVL-positive MSSA isolates with representative antibiotic susceptibility patterns. Lane 6, shown for comparison, is a PVL-positive CA-MRSA isolate. The bracket indicates a DNA fragment thought to include the SCCmec cassette, and the arrow its location in CA-MRSA USA300. The dendrogram to the left of the gel shows the genetic relatedness among the strains; as a group, the seven isolates are > 87% similar. Ox, oxacillin; Pen, penicillin; Ery, erythromycin; Clin, clindamycin; Tet, tetracycline; Neg, negative; R, resistant; S, susceptible; ND, not done.
Epidemiologic associations with PVL-positive MSSA infection
Of the 81 MSSA isolates from abscesses or wounds, 80 (99%) were collected in outpatient settings. When comparing the abscess patients according to the presence or absence of PVL genes in their MSSA isolates, there were more patients in the PVL-positive group who were identified as of African-American race (p = 0.011). Differences in the clinical presentation or course between patients with PVL-positive and PVL-negative isolates were not statistically significant, with the exception that patients with PVL-negative isolates were more likely to be admitted to the hospital (p = 0.002; Table 3).
Table 3.
Demographic Characteristics, Medical History, and Clinical Findings in Patients with PVL-positive and PVL-negative MSSA Abscesses
| Characteristic/Finding | PVL-Positive | PVL-Negative | P-value |
|---|---|---|---|
| n/#* (%) | n/#* (%) | ||
| Age, years (mean ± SD) | 7.7 (6.1) | 9.0 (6.4) | 0.390 |
| Male sex | 17/28 (61) | 15/38 (40) | 0.135 |
| African-American | 21/27 (78) | 17/38 (45) | 0.011 |
| Prior abscess | 7/27 (26) | 6/40 (15) | 0.349 |
| Chronic health problem | 8/28 (29) | 20/39 (51) | 0.081 |
| Hospitalizations in last year | 2/28 (7) | 6/40 (15) | 0.455 |
| Fever at presentation | 0/18 (0) | 1/23 (4) | 1.00 |
| Hospital admission | 4/24 (17) | 18/31 (58) | 0.002 |
| Leukocytosis (WBC >15,000) | 2/9 (22) | 3/21 (14) | 0.622 |
| Positive blood culture | 1/4 (25) | 2/15 (13) | 0.530 |
| Multiple synchronous abscesses | 5/27 (19) | 12/39 (31) | 0.391 |
| Abscess located on lower body site | 13/28 (46) | 12/41 (29) | 0.203 |
| Abscess diameter, cm (mean ± SD) | 2.88 (1.34) | 2.77 (1.34) | 0.834 |
| Developed subsequent abscess (within 12 months) | 4/22 (18) | 7/39 (18) | 1.00 |
| Either a prior OR subsequent abscess | 10/23 (44) | 10/39 (26) | 0.169 |
| Both a prior AND subsequent abscess | 1/21 (5) | 3/39 (8) | 1.00 |
The denominator represents the number of patients for whom the information was available.
DISCUSSION
During the past decade, the emergence and spread of CA-MRSA has been observed in countries throughout the world. This development is a dramatic departure from the previous pattern of very close linkage of MRSA to health care institutions, particularly hospitals and nursing homes. The strains of MRSA associated with community-acquired infection differ from most healthcare-associated strains in a number of respects, including having the type IV or V SCCmec and possessing genes encoding PVL. In some studies, a close genetic relationship between some strains of CA-MRSA and circulating community strains of MSSA has been demonstrated [36, 37]. Here we report an increase in detection of disease-causing isolates of MSSA that also possess the genes that encode PVL. Among the 214 MSSA isolates studied, prevalence of PVL was 14.5%, while in the subset of 81 isolates recovered from abscesses and wounds, PVL prevalence was 36%. In comparison, PVL prevalence among MSSA isolates colonizing the nares of healthy children in our community was 1.5% ([33] and S. Fritz, unpublished data).
Our data also suggest that the present group of PVL-positive MSSA isolates are genetically restricted and closely related to epidemic strains of community-acquired MRSA. The majority of our PVL-positive MSSA isolates were of multilocus sequence type 8, and the subset analyzed by PFGE all were highly related to USA300. In addition, most carried the agr type 1 allele that has been associated with CA-MRSA [35]. PVL-positive MSSA often exhibited an antibiotic resistance pattern typical of CA-MRSA, characterized by constitutive resistance to erythromycin and susceptibility to clindamycin. This pattern suggests the presence of the msrA gene and is uncommon, in our laboratory, among healthcare-associated MRSA or in PVL-negative strains of MSSA [34, 35].
In addition, epidemiologic parallels between PVL-positive MSSA and PVL-positive MRSA are apparent. Abscesses caused by PVL-positive MSSA were more frequent in African-American patients, similar to our finding in a recent study of increased MRSA colonization in African-American compared to Caucasian children in the St. Louis area [33]. This is also consistent with the finding in other studies that CA-MRSA infection is observed more frequently in African-American patients [38, 39] Likewise, McCaskill and coworkers found that invasive infections due to MSSA of USA300 clonal origin were more common in African-Americans [24]. Though most of the studied PVL-positive MSSA isolates had the same MLS type and were very closely related by PFGE, they do not appear to be strictly clonal, based on modest variation among the isolates in antibiotic susceptibility profiles, presence of the arcA gene of the ACME, and their agr allele groups. This is consistent with a recent examination of the genetic diversity among MRSA USA300 clones [40].
The results of our study are provocative because they have implications for theories regarding the impetus behind the current worldwide outbreak of community-acquired MRSA. If methicillin-susceptible as well as methicillin-resistant strains are spreading, the driving force is likely unrelated to methicillin resistance and might instead be related to other undefined fitness characteristics of the MLST type 8 USA300 clone. The observation that the emergence of PVL-positive MSSA is occurring several years after the proliferation of PVL-positive MRSA suggests that the mec gene or perhaps a larger portion of the SCCmec element on which it is carried may be unstable, leading to its loss from some strains of MRSA. Indeed, a recent investigation suggested that the SCCmec element does not contribute to staphylococcal pathogenesis in the absence of beta-lactam antibiotics [41].
Our study has several limitations. It is based on data from a single center, and thus must be replicated at other sites before the findings are considered generalizable. Since PCR for PVL genes was performed only on isolates recovered in 2005 and 2006, it was not possible to quantify the contribution of PVL-positive MSSA to the 5-fold increase in MSSA isolates we observed in the larger time period from 1999–2007. The conclusion that PVL-positive MSSA isolates are related to the common clone associated with community-acquired MRSA is based on characterization by MLST and PFGE; inclusion of more strains or detailed genomic analysis would further illuminate the genetic relatedness of these strains. Our finding that the PVL-positive MSSA isolates were homogeneous suggests that there may be specific epidemiologic characteristics that would be better characterized by a larger study. We suspect that there are also clinical differences between PVL-positive and PVL-negative MSSA infections, as has been demonstrated in other studies [16, 21–24]. It is likely that studies of larger numbers of strains will allow these differences to be discerned, as they have been for MRSA strains.
The history of staphylococcal infections has been marked by rapid emergence of new strains with distinctive epidemiologic and clinical characteristics. We are currently in such a period at the present time. What appeared to be an epidemic of community-acquired PVL-positive MRSA may now evolve as an epidemic of community-acquired PVL-positive S. aureus infections. Continued epidemiologic surveillance will be important to monitor the course of this development as well as to anticipate additional changes that are likely to occur.
Supplementary Material
Characterization of 214 consecutive MSSA isolates submitted to the SLCH Bacteriology Laboratory. PVL detection was performed on all isolates, while the 81 isolates with source listed as “abscess” or “wound” were subjected to further molecular typing and analysis.
Acknowledgments
This work was supported by National Institutes of Health grants K08-DK067894 and UL1-RR024992 and by the Washington University Department of Pediatrics. We thank David Warren, MD, MPH, for thoughtful review of the manuscript; and Gregory Fosheim, MPH, and Roberta B. Carey, PhD, at the Centers for Disease Control and Prevention for their assistance with the performance and interpretation of the pulsed-field gel electrophoresis.
References
- 1.Fridkin SK, Hageman JC, Morrison M, et al. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med. 2005;352:1436–44. doi: 10.1056/NEJMoa043252. [DOI] [PubMed] [Google Scholar]
- 2.Tenover FC, McDougal LK, Goering RV, et al. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol. 2006;44:108–18. doi: 10.1128/JCM.44.1.108-118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tristan A, Bes M, Meugnier H, et al. Global distribution of Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus, 2006. Emerg Infect Dis. 2007;13:594–600. doi: 10.3201/eid1304.061316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito T, Katayama Y, Asada K, et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1323–36. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma XX, Ito T, Tiensasitorn C, et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2002;46:1147–52. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother. 2004;48:2637–51. doi: 10.1128/AAC.48.7.2637-2651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baba T, Takeuchi F, Kuroda M, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–27. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 8.Vandenesch F, Naimi T, Enright MC, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003;9:978–84. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis. 2008;46:787–94. doi: 10.1086/528716. [DOI] [PubMed] [Google Scholar]
- 10.Panton PN, Valentine FCO. Staphylococcal toxin. Lancet. 1932;1:506–8. [Google Scholar]
- 11.Jevons MP. “Celbenin”-resistant staphylococci. Brit Med J. 1961;i:124–25. [Google Scholar]
- 12.Cribier B, Prevost G, Couppie P, Finck-Barbancon V, Grosshans E, Piemont Y. Staphylococcus aureus leukocidin: a new virulence factor in cutaneous infections? An epidemiological and experimental study. Dermatology. 1992;185:175–80. doi: 10.1159/000247443. [DOI] [PubMed] [Google Scholar]
- 13.Ward PD, Turner WH. Identification of staphylococcal Panton-Valentine leukocidin as a potent dermonecrotic toxin. Infect Immun. 1980;28:393–7. doi: 10.1128/iai.28.2.393-397.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baggett HC, Hennessy TW, Leman R, et al. An outbreak of community-onset methicillin-resistant Staphylococcus aureus skin infections in southwestern Alaska. Infect Control Hosp Epidemiol. 2003;24:397–402. doi: 10.1086/502221. [DOI] [PubMed] [Google Scholar]
- 15.Boubaker K, Diebold P, Blanc DS, et al. Panton-Valentine leukocidin and staphyloccoccal skin infections in schoolchildren. Emerg Infect Dis. 2004;10:121–4. doi: 10.3201/eid1001.030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lina G, Piemont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–32. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 17.Prevost G, Bouakham T, Piemont Y, Monteil H. Characterisation of a synergohymenotropic toxin produced by Staphylococcus intermedius. FEBS Lett. 1995;376:135–40. doi: 10.1016/0014-5793(95)01260-9. [DOI] [PubMed] [Google Scholar]
- 18.Robinson DA, Kearns AM, Holmes A, et al. Reemergence of early pandemic Staphylococcus aureus as a community-acquired meticillin-resistant clone. Lancet. 2005;365:1256–8. doi: 10.1016/S0140-6736(05)74814-5. [DOI] [PubMed] [Google Scholar]
- 19.Labandeira-Rey M, Couzon F, Boisset S, et al. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science. 2007;315:1130–3. doi: 10.1126/science.1137165. [DOI] [PubMed] [Google Scholar]
- 20.Voyich JM, Otto M, Mathema B, et al. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J Infect Dis. 2006;194:1761–70. doi: 10.1086/509506. [DOI] [PubMed] [Google Scholar]
- 21.Dohin B, Gillet Y, Kohler R, et al. Pediatric bone and joint infections caused by Panton-Valentine leukocidin-positive Staphylococcus aureus. Pediatr Infect Dis J. 2007;26:1042–8. doi: 10.1097/INF.0b013e318133a85e. [DOI] [PubMed] [Google Scholar]
- 22.Bocchini CE, Hulten KG, Mason EO, Jr, Gonzalez BE, Hammerman WA, Kaplan SL. Panton-Valentine leukocidin genes are associated with enhanced inflammatory response and local disease in acute hematogenous Staphylococcus aureus osteomyelitis in children. Pediatrics. 2006;117:433–40. doi: 10.1542/peds.2005-0566. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez BE, Hulten KG, Dishop MK, et al. Pulmonary manifestations in children with invasive community-acquired Staphylococcus aureus infection. Clin Infect Dis. 2005;41:583–90. doi: 10.1086/432475. [DOI] [PubMed] [Google Scholar]
- 24.McCaskill ML, Mason EO, Jr, Kaplan SL, Hammerman W, Lamberth LB, Hulten KG. Increase of the USA300 clone among community-acquired methicillin-susceptible Staphylococcus aureus causing invasive infections. Pediatr Infect Dis J. 2007;26:1122–7. doi: 10.1097/INF.0b013e31814536e0. [DOI] [PubMed] [Google Scholar]
- 25.Hsu LY, Koh TH, Kurup A, Low J, Chlebicki MP, Tan BH. High incidence of Panton-Valentine leukocidin-producing Staphylococcus aureus in a tertiary care public hospital in Singapore. Clin Infect Dis. 2005;40:486–9. doi: 10.1086/427033. [DOI] [PubMed] [Google Scholar]
- 26.Prevost G, Couppie P, Prevost P, et al. Epidemiological data on Staphylococcus aureus strains producing synergohymenotropic toxins. J Med Microbiol. 1995;42:237–45. doi: 10.1099/00222615-42-4-237. [DOI] [PubMed] [Google Scholar]
- 27.Elizur A, Orscheln RC, Ferkol TW, et al. Panton-Valentine Leukocidin-positive methicillin-resistant Staphylococcus aureus lung infection in patients with cystic fibrosis. Chest. 2007;131:1718–25. doi: 10.1378/chest.06-2756. [DOI] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Seventeenth International Supplement. 2007;M100–S17:27. [Google Scholar]
- 29.Lina G, Boutite F, Tristan A, Bes M, Etienne J, Vandenesch F. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl Environ Microbiol. 2003;69:18–23. doi: 10.1128/AEM.69.1.18-23.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diep BA, Gill SR, Chang RF, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–9. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 31.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–15. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–20. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fritz SA, Garbutt J, Elward A, Shannon W, Storch GA. Prevalence of and risk factors for community-acquired methicillin-resistant and methicillin-sensitive Staphylococcus aureus colonization in children seen in a practice-based research network. Pediatrics. 2008;121:1090–8. doi: 10.1542/peds.2007-2104. [DOI] [PubMed] [Google Scholar]
- 34.Liao RS, Storch GA, Buller RS, et al. Blinded comparison of repetitive-sequence PCR and multilocus sequence typing for genotyping methicillin-resistant Staphylococcus aureus isolates from a children's hospital in St. Louis, Missouri J Clin Microbiol. 2006;44:2254–7. doi: 10.1128/JCM.00690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bogdanovich T, Aydin N, Chavez-Bueno S, McCracken G, Bozdogan B, Appelbaum PC. Genetic characterization of erythromycin- and methicillin-resistant community-acquired Staphylococcus aureus isolated from children in Texas. Diagn Microbiol Infect Dis. 2007;59:231–3. doi: 10.1016/j.diagmicrobio.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 36.Fey PD, Said-Salim B, Rupp ME, et al. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:196–203. doi: 10.1128/AAC.47.1.196-203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mongkolrattanothai K, Boyle S, Kahana MD, Daum RS. Severe Staphylococcus aureus infections caused by clonally related community-acquired methicillin-susceptible and methicillin-resistant isolates. Clin Infect Dis. 2003;37:1050–8. doi: 10.1086/378277. [DOI] [PubMed] [Google Scholar]
- 38.Sattler CA, Mason EO, Jr, Kaplan SL. Prospective comparison of risk factors and demographic and clinical characteristics of community-acquired, methicillin-resistant versus methicillin-susceptible Staphylococcus aureus infection in children. Pediatr Infect Dis J. 2002;21:910–7. doi: 10.1097/00006454-200210000-00005. [DOI] [PubMed] [Google Scholar]
- 39.Moran GJ, Krishnadasan A, Gorwitz RJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–74. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 40.Kennedy AD, Otto M, Braughton KR, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A. 2008;105:1327–32. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diep BA, Stone GG, Basuino L, et al. The arginine catabolic mobile element and staphylococcal chromosomal cassettemec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197:1523–30. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of 214 consecutive MSSA isolates submitted to the SLCH Bacteriology Laboratory. PVL detection was performed on all isolates, while the 81 isolates with source listed as “abscess” or “wound” were subjected to further molecular typing and analysis.