Abstract
Aging affects a myriad of genetic, biochemical, and metabolic processes, and efforts to understand the underlying molecular basis of aging are often thwarted by the complexity of the aging process. By taking a systems biology approach, network analysis is well-suited to study the decline in function with age. Network analysis has already been utilized in describing other complex processes such as development, evolution, and robustness. Networks of gene expression and protein–protein interaction have provided valuable insight into the loss of connectivity and network structure throughout lifespan. Here, we advocate the use of metabolic networks to expand the work from genomics and proteomics. As metabolism is the final fingerprint of functionality and has been implicated in multiple theories of aging, metabolomic methods combined with metabolite network analyses should pave the way to investigate how relationships of metabolites change with age and how these interactions affect phenotype and function of the aging individual. The metabolomic network approaches highlighted in this review are fundamental for an understanding of systematic declines and of failure to function with age.
Introduction
Since the 1950s, much work has been dedicated to understanding the aging process and what affects it (Medawar 1946, 1952; Orgel 1963, 1970, 1973; Medvedev 1990). Although oversimplified, aging can be envisioned as a shift from fully functional to failing-to-function. In youth, organisms operate at their peak performance. Over time, life as a whole begins to lose coordination between the parts, and the systems begin to fail. Reductionist approaches have tested these ideas mechanistically through the failure of genetic replication [e.g., telomere shortening (Oeseburg et al. 2010)], mitochondrial dysfunction (Van Remmen and Jones 2009), accumulation of oxidative stress (Jones 2006), uncontrolled inflammation (Chung et al. 2008), and hormone dysregulation (Panowski and Dillin 2009)) as potential theories of aging. However, the system-wide deterioration that we observe as organisms age likely involves a combination of some or all of these mechanisms working together to create the complex phenomenon that we call aging.
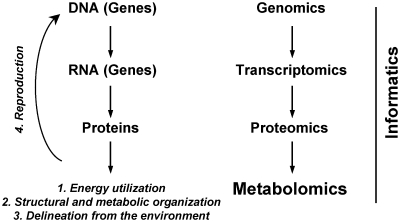
To better understand the complexity of aging, researchers have begun to advocate a systems biology approach, in which we would study both causes and consequences of aging at genome-wide, transcriptome-wide, proteome-wide, and metabolome-wide levels (Barabasi and Oltvai 2004; Hood et al. 2004). Based upon the characteristics of life, there are four general ways ‘systems’ could decline in function with age: (1) decline in the capture and utilization of energy and in the efficiency with which it is extracted; (2) decline in structural or metabolic organization; (3) decline in barrier functions (i.e., delineation from the environment); and (4) decline in the fidelity of storage and transfer of information (i.e., fidelity of DNA during reproduction). Proteomics can be used to investigate the first three characteristics while genomics and gene-expression methods can be used to investigate the fourth characteristic (Fig. 1). In this view, genomics cannot give us a complete picture of aging because it accounts for only reproduction and for DNA encoded in the nucleus and perhaps also for mitochondria. However, the metabolome is the furthest down the line from gene to function and is most characteristic of the entire organismal state and phenotype. Therefore, changes in the metabolome provide the best representation of the characteristic declines in function with age.
Fig. 1.
The central dogma of life studied with “-omics” technologies provide a global perspective on how systems begin to fail with age.
Systems biology teaches us not only that phenotypes are influenced by many important genetic and environmental factors, but also that we need to understand the extremely complex interactions between these components. Interactions between the parts, as well as influences from the environment, give rise to new features, such as network behavior (Alm and Arkin 2003), which are absent in the isolated components. As we become more aware of the roles of the network and its components, we can begin to decode the aging process by connecting the molecular components to discrete or multiple signaling pathways. Here, we argue that network analysis of metabolomic data can help to determine how the failure of networks to maintain stability and homeostasis within living systems can lead to aging.
Network approaches
Systems biologists have made great progress in characterizing the structure and function of molecular networks. For example, gene network structure and function can be used to predict formation of patterns during early development (von Dassow et al. 2000; Salazar-Ciudad and Jernvall 2002; Albert and Othmer 2003), and to explain why networks are resistant to damage (Albert et al. 2000; Wagner 2000; Bergman and Siegal 2003).
To use and understand network analysis in research on aging, we first need a brief explanation of the components and structure of networks. A network is a collection of units potentially interacting as a system, and each network is comprised of a finite set of nodes and edges; nodes are the individual elements (e.g., genes, proteins, and metabolites) within the network, and the edges are the connections (e.g., correlations and enzymatic reactions) that relate the nodes. Edges can be either directed, whereby one node acts upon another (as in gene regulatory networks), or undirected (as in protein–protein interaction [PPI] networks). A node’s connectivity, k, is the number of edges that connect the focal node to other nodes, and nodes with high connectivity are considered hubs. In most biological networks, k is distributed among nodes according to a power-law function P(k)=ζ(−γ)k−γ, where P(k) is the probability that a selected node has exactly k connections (degrees) with other nodes (e.g., proteins), γ is the degree exponent, a characteristic value for a given network which determines many properties of the system, and ζ(−γ), Riemann’s zeta function, is a normalizing constant. The smaller the γ value, the more important is the role of hubs in the network (Barabasi and Oltvai 2004). Mark Newman (2003) provides a thorough review of network structure and function. There are numerous software packages that allow one to draw networks and evaluate network structure, including specific network programs (e.g., Pajek, UCINET, etc.), as well as components of general statistical packages (e.g., BioConductor in R).
The shape of the power law distribution tells us much about networks, but it cannot reveal the underlying hierarchical structure that characterizes many networks. For example, many network analyses have used hierarchical clustering to identify co-regulated groups or modules within the larger network. This concept of modularity assumes that the function of the network can be better understood if we partition the network into a collection of modules. Each module is a discrete entity of several components, which act together to perform a unique function (Hartwell et al. 1999; Lauffenburger 2000; Hasty et al. 2001; Holter et al. 2001; Rao and Arkin 2001; Shen-Orr et al. 2002). This notion of modularity is especially important in biological networks. As Bruce Alberts (1998) noted, we can think of a cell as a collection of machines. Since each machine is made up of interacting molecules, we can imagine that each machine forms a discrete module within the larger network acting within a cell.
In a highly cited article on the topic, Girvan and Newman (2002) refer to this aspect of networks as their “community structure”. The community structure of the network (that is, the modules that underlie the network) can be identified through hierarchical clustering algorithms (Girvan and Newman 2002). More recently, a variety of other approaches have been used, from independent component analysis (e.g., Gong et al. 2007) to iterative machine learning approaches (Segal et al. 2003) to approaches derived from graph theory (Newman’s (2007) eigenvector analysis and Stone & Ayroles’ (2009) Modulated Modularity Clustering). Implementations for many of these methods are available either as packages in R (R Foundation for Statistical Computing, Vienna, Austria), or as standalone software or websites (e.g., Stone and Ayroles 2009). Importantly, to our knowledge, no systematic review has yet been done to determine relative power, speed, biological relevance, and degree of overlap (or lack thereof) in the modules identified by these different statistical methods.
Researchers have used structural analysis of biological networks to better understand a broad range of traits. Studies have found, for example, that the most highly connected nodes are more likely to be essential for organismal function (Jeong et al. 2001), tend to evolve more slowly (Fraser et al. 2002; Hahn et al. 2004; Wuchty 2004), are less likely to be lost over evolutionary time (Krylov et al. 2003), and have a higher probability of being associated with senescence (Promislow 2004; Ferrarini et al. 2005; Budovsky et al. 2007). Results from both theoretical (Albert and Othmer 2003) and empirical (Promislow 2005) studies have shown that network structure is an important predictor of patterns of gene expression. Thus, network studies can provide a unique perspective on the cross-talk between biological modules, identify the most susceptible nodes for loss in communication, and model how changes in these interactions are associated with phenotypes of aging.
Networks and aging
From the above, one can anticipate that highly connected nodes in biological networks would be important in aging. Indeed, proteins associated with aging in yeast have significantly higher connectivity than expected by chance (Promislow 2004), suggesting that a loss in these links would have much greater effects on the entire network than loss of less connected proteins (Albert et al. 2000).
This review focuses on the way in which the structure and function of an organism’s underlying molecular networks can affect its phenotype. In the case of aging, though, we can also reverse the causal arrow, asking instead how aging affects network structure and function. If aging is the gradual failure not simply of individual biological components but rather of complex networks, studies of the structure and function of networks can provide a useful framework to understand both the causes and consequences of aging. Thus, as we turn to a systems biology approach to the study of aging, we shift our focus from studies of network function to studies of network failure.
Ferrarini et al. (2005) developed a theoretical model of network failure, asking in particular what role highly connected nodes, or hubs, play in maintaining network function. Assuming a power-law distribution of network connectivity, their theoretical model showed that restoration of hubs rescued 35% of the lost function, while restoration of weakly connected nodes led to very little improvement in network function. Interestingly, in an analysis of genes that were both highly connected and associated with aging, they found a disproportionate number associated with metabolism—a topic to which we will turn our focus later in this article. In marked contrast with the results of Ferrarini et al. (2005), Csermely and Soti (2006) suggested that the failure of weak links, leading to increased noise and loss of overall network integrity, may account for senescence.
In the previous section, we discussed the community structure of networks. Network approaches have also been used to identify subcomponents of networks associated with aging. As an example of the use of modularity in aging studies, Xue et al. (2007) looked for genes whose expression levels are significantly correlated across age, either positively or negatively. Using expression data from studies of human brains and of whole fruit flies, they were able to identify large modules of co-regulated, age-associated genes. They were further able to show that these modules tended to be associated with particular biological processes, and that expression levels in some genes (such as those associated with cell proliferation) tended to increase with age, while others (such as those associated with differentiation) decreased with age.
Furthermore, Xue et al. (2007) looked at the effect of dietary restriction on network structure. Dietary restriction is the most widely accepted method of extending lifespan in a range of laboratory organisms, from yeast, worms, and flies to mice and rats. Xue et al. (2007) found that dietary restriction altered the structure of the gene co-expression network in Drosophila.
In silico modeling, based on gene arrays or PPIs that have already been established, can also be used to identify components of a network that are associated with lifespan. For example, to construct a “longevity network” via analysis of PPI, Budovsky et al. (2007) used the human orthologs of “longevity assurance genes” that have been established in model organisms (e.g., Saccharomyces cerevisae, Caenorhabditis elegans, and Drosophila melanogaster) in combination with a list of longevity-associated genes to determine whether the encoded proteins could be organized into a network. A core longevity network was constructed that contained 153 of the 211 longevity-associated proteins (LAPs) identified. The core network was characterized by the presence of highly interconnecting nodes (i.e., hubs), in which 15 of the 17 hubs were LAPs. Interestingly, 33 proteins “not” associated with longevity (“non-LAPs”) had connections with several LAPs, such that an extended network including these proteins revealed an additional 40 LAPs that did not appear in the core network (Budovsky et al. 2007).
At this point, we see an urgent need to understand how senescence affects the structural and functional interactions that underlie complex networks. A recent study of age-related changes in gene expression in mice (Southworth et al. 2009) points the way forward. Southworth et al.’s study relied on the ‘AGEMAP’ dataset (Zahn et al. 2007), which measured genome-wide levels of gene expression at four different ages in 16 different tissues in C57BL/6 mice. Using these data, Southworth et al. (2009) constructed gene co-regulatory networks in 16-month-old and 24-month-old mice. They found that the integrity of these networks, measured as the number of connections between genes, declined from 16 to 24 months. In particular, a differential co-expression network analysis from 13 different tissues revealed that 24-month-old mice had 26% fewer total edges than did 16-month-old mice, and large interconnected gene groups were lost with age in a non-uniform fashion. Those genes that tended to lose correlations were associated with specific transcriptional factors and tended to be clustered in specific regions of the chromosome. The clusters of genes that lost correlation with age included genes involved in steroid production, memory, and mitochondrial function while DNA-damaged genes became more correlated with other genes in older mice (Southworth et al. 2009).
Overall, the literature reviewed thus far on genome and proteome network modules demonstrates that aging is associated with (1) failure of hubs; (2) failure of weak links; and (3) redistribution of node structure. The challenge that we now face is to determine what kinds of network components, in terms of both structure and function, are most likely to lose integrity with age.
Metabolic networks and aging
In this section, we will first describe the history of metabolomics and why this particular “-omic” technology has special advantages in the study of aging. Second, we will discuss ways of identifying age-related changes in the structure of any molecular networks (including genomic and proteomic as well as metabolomic networks). Last, we discuss, in particular, the study of senescence in metabolomic networks.
History of metabolomics
Beginning in 1998, the first published study of metabolomic technology emerged as a method to detect hundreds of small molecules, including antioxidants, oxidative stress markers, purines, and indoles, in rat-liver mitochondria (Kristal et al. 1998). Since then, the technology has flourished, increasing the number of publications on the topic from one in 1998 to hundreds by 2003 (Kell 2004), and almost 1000 in 2009 alone. In the early years of metabolomics research, no clear definition of the “metabolome” had yet been claimed. Oliver et al. (1998) made the first attempt by suggesting that the metabolome is a comprehensive measure of low-molecular-weight molecules present in cells in a particular physiological or developmental state. The definition has now evolved into a more generalized term including all small molecules characterizing a biological system (Boccard et al. 2010). In any case, the small molecules detected while measuring the metabolome consist of those generated in vivo, including those affected by diet, drugs, pesticides, and other environmental exposures.
Ideally, a metabolic profile would detect and quantify all small molecules present in a biological system at a given time (i.e., a snapshot of cellular metabolism or of components in circulating blood). However, this is not possible with current technology. As discussed below, the techniques for separation and detection currently in use are unable to identify all in vivo metabolites due to inadequate extraction, chemical diversity, and the dynamic range of the metabolome (Goodacre et al. 2004). These complications have limited our knowledge with respect to known metabolites, but in just 2 years, the number of metabolites identified in humans has more than tripled, from 2180 to almost 7000 by 2009 (Wishart et al. 2009). This number does not include the additional thousands of metabolites from drugs, common toxins or pollutants, and food additives. Thus, it is obvious that we have much to learn about the metabolome.
The progress made thus far in our ability to characterize the metabolome has been made possible by advances in the technology platforms in analytical chemistry. The primary analytical platforms include nuclear magnetic resonance (NMR), mass spectrometry (MS), and a number of separation techniques, such as capillary electrophoresis (CE), gas chromatography (GC), and high performance liquid chromatography (HPLC). Chemical separation by GC or HPLC increases the resolution and sensitivity of MS while enabling selectivity of species based on their chemical properties (Goodacre et al. 2004). In addition, the availability of a variety of columns for HPLC has made the detection of comprehensive metabolic profiles more feasible. NMR spectroscopy has been developed as a valuable resource for metabolomics due its reproducibility and its ability to provide structural information. However, NMR has low sensitivity and low throughput [there are exceptions (Soininen et al. 2009)], which limits our ability to detect the maximum number of metabolites in a short period of time. Mass spectrometry is a more promising technique to generate high-throughput metabolomic data because it overcomes the weaknesses of NMR with higher sensitivity in a relatively shorter time required for analysis. For example, the Fourier transform-MS (FTMS) has a high resolving power and sub-parts-per-million mass accuracy that facilitates identification with less dependence upon chromatography. Further, the measurement of accurate mass by the FTMS has proved useful for characterization of unknown metabolites by the unambiguous assignment of elemental formulas (Aharoni et al. 2002). Thus, the performance characteristics of MS, particularly FTMS, are ideal for the complex mixtures encountered in high-throughput-metabolomics applications.
Metabolomics platforms include both “top–down” versus “bottom–up” approaches. In a bottom–up model, the method is targeted to detect specific molecules or pathways in a hypothesis-driven manner. Instrumentation and bioinformatics are not restrictive for bottom–up models since a finite number of metabolites are targeted and less-sensitive instrumentation is needed. On the other hand, a top–down approach is untargeted and more reliant on sensitivity and robustness of the instrument in order to generate extensive metabolic profiles. Complex bioinformatics are then needed to mine the resultant data and to generate hypotheses. Although both approaches have drawbacks, in systems biology, bottom–up metabolomics is most limiting, as the focus on a few selected metabolites can lead the researcher to ignore critically important interactions with other molecules (Forster et al. 2003). Top–down metabolomic approaches are more useful for drawing general conclusions regarding aging, or any multi-system effect, because they take a global snapshot of the organism’s metabolic state.
The composition of samples selected for metabolomic analysis can also limit and complicate interpretations. Samples of tissues and cells often require rigorous protocols for handling and extraction in order to isolate metabolites; these methods can result in inconsistent and more targeted metabolic profiles. In such a case, one must be careful when drawing conclusions in systems biology studies, since the modeling is not comprehensive and will be biased toward the metabolites that were extracted (Goodacre et al. 2004). On the other hand, biofluids are perhaps the ideal type of sample to be analyzed because they are easily obtained and can be analyzed by MS and NMR with little preparation of the sample (Brindle et al. 2002; Lindon et al. 2003; Nicholson and Wilson 2003). Biofluids, specifically plasma and serum, can provide a clear representation of a global metabolic profile because these fluids have contact with organ systems throughout the body.
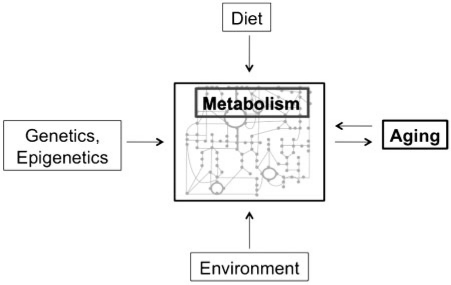
Although the identities of the individual components of the metabolome are less well understood than that of the transcriptome and proteome, metabolomics offers distinct advantages as a means of studying the molecular state of an organism. In particular, theoretical (Fell 1996; Kell and Mendes 2000) and experimental (Raamsdonk et al. 2001) studies of metabolic control reveal that changes in the quantities of individual enzymes have significant effects on the concentrations of numerous individual metabolites. In addition, there are countless chemical alterations and regulatory mechanisms that occur downstream of the genome. As a result, the metabolome is effectively an integration not only of cellular activities at a functional level, but also of extrinsic, environmental factors (Fig. 2). Because the metabolome is essentially the “end of the line” in terms of biological processes, changes in the metabolome may provide a better mirror of the consequences of genotypic and phenotypic changes than do changes of the actual transcriptome or proteome. To summarize, the metabolome is the final fingerprint of genetic regulation and enzymatic activity, and the resulting global metabolic profile can best be measured by metabolomics.
Fig. 2.
Metabolic profiles integrate the effects of diet, environment, and genetics for mechanistic studies of aging.
Recent advances in metabolomics have provided insights into certain aspects of aging, and many trends of metabolites during aging are emerging. Using 1H NMR, urine samples from aging rats, dogs, and humans were analyzed, which showed that resonances associated with creatinine, amino acids, and fatty acids increase with age while glucose/myoinositol (Williams et al. 2005), glycoproteins (Wang et al. 2007), succinate, and other Krebs-cycle intermediates decrease with age (Schnackenberg et al. 2007; Gu et al. 2009). These results demonstrate the utility of NMR to find variability in macromolecules with age, and HPLC–MS studies of aging expand on these results by detecting more than 11,000 ions in a biological sample (Williams et al. 2006). HPLC–MS analysis of urine from aging Sprague Dawley rats not only confirms 1H NMR results of declining Krebs-cycle intermediates but also identifies increased levels of oxidized antioxidants in older rats (Schnackenberg et al. 2007). HPLC–MS mass chromatograms detect significantly more ions in urine from 4-week-old Wistar-derived rats than from 20-week-old rats (Williams et al. 2005). Ions identified as carnitine, ascorbic acid, and urate increase in ion intensity with age, but the large majority of age-associated ions remain unidentified. Another study of aging using HPLC–MS to perform metabolomic characterization in the serum of 4, 10, 18, and 24-month-old rats found that levels of carnosine, cholesterol, and various fatty acids among other unidentified metabolites were associated with age (Yan et al. 2009). These studies demonstrate that HPLC–MS-based metabolomics can provide a wealth of information (e.g., thousands of metabolites detected), although much is to be learned concerning the identity of the ions. One way to attack the problem of unknown molecules is to construct a network connecting metabolites that change with age. The relationships between the known and unknown metabolites may assist in elucidation of pathways and in identification of previously unrecognized ions.
Metabolic networks
While most studies of biological networks and aging have focused on protein–protein or gene regulatory interactions, we suggest here that metabolic networks may prove particularly valuable in furthering our understanding of the biological and phenotypic changes that occur as organisms age. Metabolites are the final fingerprint of genetic regulation and enzymatic reactions, and provide a clear picture of phenotype. The potential value of studying metabolic networks is further supported by the fact that the most widely tested intervention to prolong lifespan, dietary restriction, has obvious effects on energy metabolism (Masoro 2001), as demonstrated by a lifelong metabolomics study of caloric restriction in dogs (Wang et al. 2007).
There are a variety of ways to construct metabolic networks (e.g., Batagelj and Mrvar 1998; Stone and Ayroles 2009). Ravasz et al. (2002) reconstructed a network by using the information about the E. coli enzymes for all metabolic reactions available in the ERGO database. This network includes a few highly connected molecules, such as ATP, that tell us little about the biological functions of the specific nodes and pathways with which they interact. Thus, Ravasz et al. (2002) developed an algorithm that would remove these ‘redundant’ edges, and also simplified the connections between substrates.
Ravasz et al. (2002) suggested that in reconstructing these networks, we need to recognize not only the global patterns of the network (i.e., the number of nodes to which each node is connected), but also the underlying modular substructure. One way that we can measure overall modularity in a network is by estimating a network’s clustering coefficient, C(k). The clustering coefficient measures the degree to which groups of nodes in a network tend to cluster together to form tightly connected neighborhoods. For example, in a network with a high clustering coefficient, if nodes A and B are both connected to node C, there is a high probability that nodes A and B are connected with one another. In a study of metabolic networks in 43 different organisms, Ravasz et al. (2002) found that the clustering coefficient was about an order of magnitude larger than one would expect in a network in which the number of connections is distributed according to a power law, but at random with respect to modularity. This result suggests that metabolic networks are characterized by high intrinsic modularity.
After construction of a network, we can glean important information from analysis of its structure and function. Like gene and protein networks, the most connected modules in metabolic networks play a critical role in overall network size (Jeong et al. 2000; Wagner and Fell 2001) and structure (Wagner and Fell 2001). The diameter of metabolic networks, defined as the average shortest distance between any two nodes in the network, does not change across organisms even with the increasing number of substrates that occur with increasing organismal complexity (Jeong et al. 2000).
In the following section, we describe our current work on the analysis of steady-state metabolic networks. While studies of complex flux-sum (Chung and Lee 2009) or kinetic processes (Coquin et al. 2008) are clearly of interest, a recent study of an in silico metabolic network model by Çakir et al. (2009) found that metabolic variation observed at steady-state, with no perturbation, can reveal connectivity of the underlying metabolic network. Our own work, discussed below, shows how one might use such steady-state comparisons across individuals to assess the effect of aging on metabolomic network structure.
Metabolic network and aging in the common marmoset
We are interested in top–down metabolic networks and how they change with age. Until now, most research on metabolism and aging has been focused on the relationship of dietary restriction to longevity and on mechanisms of age-related metabolic disease. Studies on metabolic mechanisms of aging have focused on a variety of physiological traits, including mitochondrial function, oxidative stress/redox balance and its downstream effects on lipids, proteins, and DNA; signaling in the insulin and insulin-like growth factor (IIS) and TOR pathways; glucocorticoid signaling; and inflammation. However, the overall process of aging likely involves all of these components, so a systems approach using metabolic networking can provide a more comprehensive insight into the interactions between these components.
As we have outlined above, a network approach should help us understand key metabolomic differences in longevity among species, and among individuals within species. In particular, our goal is to identify specific components of the underlying metabolomic network that are most likely to fail with age. Previous progress in metabolic network reconstruction depended critically on the functional annotation of the genes that encode proteins with unknown functions and focused mostly on specific pathways such as fatty acid synthesis or aspartate metabolism (Chen et al. 2009; Curien et al. 2009). However, with the recent advances in metabolomics technologies, we can begin to construct metabolomic networks without relying on data extrapolated from gene-expression assays or on in silico modeling. Steady-state metabolomics can provide the necessary information to elucidate age-related changes in network connectivity and structure.
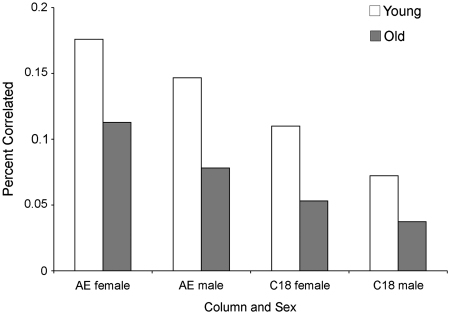
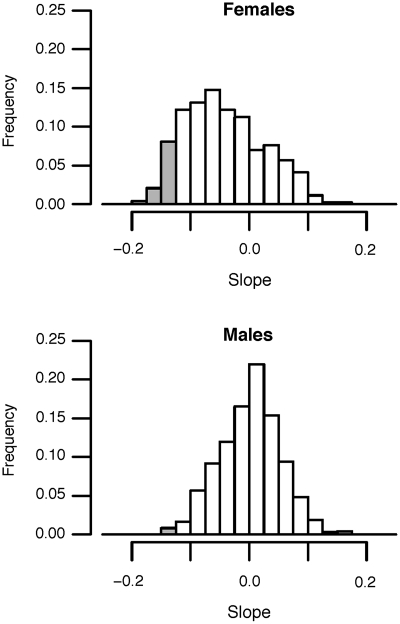
We have used a top–down metabolic profiling technique to compare the steady-state metabolic network of a set (n = 60) of young (<6 years of age) and old (>6 years of age) common marmosets (Callithrix jacchus). The common marmoset has recently emerged as the premiere primate model for studies of aging. Because of their short lifespan (6–15 years) and small size (∼400 g), marmosets are easy to handle, require little space, and make long-term studies of aging more feasible. Using our HPLC–FTMS-based metabolomics platform (Johnson et al. 2008), partnered with a metabolite peak extraction method (Yu et al. 2009), we can identify over 2500 metabolites in a single drop of blood. These metabolites cover the entire profile of the organism, including “dead-end” or inactive ones, as well as metabolites that have yet to be identified. In our preliminary analyses of metabolomic networks in marmosets varying between 2 and 13 years of age, we find that connectivity of metabolites decreases with age (Fig. 3), and that the abundance of numerous metabolites significantly changes with age (Fig. 4). By using these types of analyses (i.e., global metabolic profiling combined with network analysis), well-designed models of aging may reveal biomarkers that correlate with age-related phenotype and disease.
Fig. 3.
Percent of correlated metabolites as a function of extraction column and sex. We calculated correlation coefficients between all possible pairs of metabolites among individuals for both the AE column (n = 765 metabolites) and the C18 column (n = 714 metabolites), for groups of young or old marmosets separately. In each case, there was a total of n(n−1) possible pairwise correlations. We determined the percent of all correlations that was highly significant (P < 10−7). For both columns, and for both sexes, we see more significant correlations among young individuals than among old individuals. In all four comparisons, the difference between old and young groups is highly significant (P < 10−13).
Fig. 4.
Frequency distribution of correlation coefficients between quantity of metabolites and age of the marmoset, using extractions from an anion exchange column (results from a C18 column were comparable). Grey bars indicate correlation coefficients that were significantly different from zero (P < 0.01). Note that for females, a disproportionate number of metabolites decline with age (t-test of average slope across all metabolites, P < 10−7), while on average, males are not significantly different from zero.
To carry out metabolic network analyses of aging, we can use both cross-sectional studies and longitudinal approaches. Cross-sectional approaches can be used to determine how metabolomic networks vary in response to differences in environment, genotype, sex, age, behavior, and so forth. Second, with respect to aging, longitudinal studies, where we track single individuals over time, are particularly valuable, as we can fully control for differences among individuals in genotype or rearing environment. This approach is also a powerful way to monitor effects of experimental manipulations, such as genetic knockdowns or dietary changes, on the metabolome.
Although this review highlights the progress that has been made thus far in using metabolomics data to generate biological networks, there are still key challenges in the field. The field of metabolomics has yet to adopt a set of standardized methods, which involve sampling, sample separation, MS-platform selection, and data extraction. At this point, perhaps the greatest challenge is that curated metabolomic databases are incomplete. Using LC-FTMS, we can obtain estimates of the quantities of thousands of metabolites, each with a unique mass:charge (m/z) and retention time, from a sample of just a few microliters. But without a substantial effort, we would not have detailed understanding of the chemical identities of all of these molecules. Furthermore, methods for quantification of metabolites have yet to be verified.
Relative to work on gene-regulatory and PPI networks, relatively few researchers are working on the construction of metabolomic networks, and on tying together metabolomic network structure and function with physiological traits and environmental perturbations. Fortunately, this gap is likely to be filled in the coming years through research to define the human exposome (Wild 2005). In a recent National Academy of Sciences Workshop sponsored by the National Institute of Environment and Health Sciences, the possibility of creating a “Human Exposome Project” was discussed. This projected could mimic the Human Genome Project to determine how environmental exposures—including diet, physical activity, stress, and drug use—contribute to human phenotype, aging, and disease. The field of metabolomics and network analysis are likely to play a key role in such a project.
We are still in the very early stages of applying metabolomic network analysis to the study of aging. While we know little, the potential for conceptual advances is substantial. We have described a study of aging in primates; we are likely to also reap significant benefits by applying these approaches to short-lived model organisms, such as fruit flies and nematode worms—the stalwarts of research on the biology of aging. Using model systems, we have identified a large number of genes that can extend longevity (reviewed in Kennedy 2008; Shimokawa et al. 2008; Ni and Lee 2010). In humans, however, our current best bet for extending both the quality and quantity of our lives is diet and exercise. As a recent study demonstrates (Lewis et al. 2010), metabolomics may help us to understand just why such environmental interventions work.
The strength of connections between genes, proteins, or metabolites is an important system-wide property of a network that can be used to compare two or more states. Here, we compared young and old marmosets, but this approach can be used to compare many other states such as healthy versus diseased individuals, or wild-type versus mutant strains. Differential network analysis could potentially be used to compare network structure among species. In a study of wing polymorphism in ants, Abouheif and Wray (2002) were able to show that the loss of function in wing development was due to the failure of different network components in different species. As organisms age, do networks fail in similar ways from species to species, and from genotype to genotype, or is each individual unique? With apologies to Tolstoy, is it the case that all young, healthy individuals are alike, but every senescent individual is senescent in its own way? Network analysis should help us to answer this question.
Funding
The National Institutes of Health (P01-ES016731 and T32-ES012870 to D.J., in part).
Acknowledgments
Helpful comments on an earlier version of this manuscript were provided by members of the Promislow laboratory. The authors also acknowledge the generous support of the Division of Comparative Physiology and Biochemistry in the Society for Integrative and Comparative Biology for funding the symposium in which this work was first presented.
References
- Abouheif E, Wray GA. Evolution of the gene network underlying wing polyphenism in ants. Science. 2002;297:249–52. doi: 10.1126/science.1071468. [DOI] [PubMed] [Google Scholar]
- Aharoni A, Ric de Vos CH, Verhoeven HA, Maliepaard CA, Kruppa G, Bino R, Goodenowe DB. Nontargeted metabolome analysis by use of Fourier Transform Ion Cyclotron Mass Spectrometry. OMICS. 2002;6:217–34. doi: 10.1089/15362310260256882. [DOI] [PubMed] [Google Scholar]
- Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–82. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- Albert R, Othmer HG. The topology of the regulatory interactions predicts the expression pattern of the segment polarity genes in Drosophila melanogaster. J Theor Biol. 2003;223:1–18. doi: 10.1016/s0022-5193(03)00035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–4. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Alm E, Arkin AP. Biological networks. Curr Opin Struct Biol. 2003;13:193–202. doi: 10.1016/s0959-440x(03)00031-9. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–13. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Batagelj V, Mrvar A. Pajek—Program for large network analysis. Connections. 1998;21:47–57. [Google Scholar]
- Bergman A, Siegal ML. Evolutionary capacitance as a general feature of complex gene networks. Nature. 2003;424:549–52. doi: 10.1038/nature01765. [DOI] [PubMed] [Google Scholar]
- Boccard J, Veuthey JL, Rudaz S. Knowledge discovery in metabolomics: an overview of MS data handling. J Sep Sci. 2010;33:290–304. doi: 10.1002/jssc.200900609. [DOI] [PubMed] [Google Scholar]
- Brindle JT, et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nat Med. 2002;8:1439–44. doi: 10.1038/nm1202-802. [DOI] [PubMed] [Google Scholar]
- Budovsky A, Abramovich A, Cohen R, Chalifa-Caspi V, Fraifeld V. Longevity network: construction and implications. Mech Ageing Dev. 2007;128:117–24. doi: 10.1016/j.mad.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Çakir T, Hendriks MM, Westerhuis JA, Smilde AK. Metabolic network discovery through reverse engineering of metabolome data. Metabolomics. 2009;5:318–29. doi: 10.1007/s11306-009-0156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, et al. RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: finding potential biomarkers for breast cancer. Analyst. 2009;134:2003–11. doi: 10.1039/b907243h. [DOI] [PubMed] [Google Scholar]
- Chung BK, Lee DY. Flux-sum analysis: a metabolite-centric approach for understanding the metabolic network. BMC Syst Biol. 2009;3:117. doi: 10.1186/1752-0509-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Seo AY, Chung SW, Kim MK, Leeuwenburgh C, Yu BP, Chung HY. Molecular mechanism of PPAR in the regulation of age-related inflammation. Ageing Res Rev. 2008;7:126–36. doi: 10.1016/j.arr.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Coquin L, Feala JD, McCulloch AD, Paternostro G. Metabolomic and flux-balance analysis of age-related decline of hypoxia tolerance in Drosophila muscle tissue. Mol Syst Biol. 2008;4:233. doi: 10.1038/msb.2008.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, Soti C. Cellular networks and the aging process. Arch Physiol Biochem. 2006;112:60–4. doi: 10.1080/13813450600711243. [DOI] [PubMed] [Google Scholar]
- Curien G, Bastien O, Robert-Genthon M, Cornish-Bowden A, Cardenas ML, Dumas R. Understanding the regulation of aspartate metabolism using a model based on measured kinetic parameters. Mol Syst Biol. 2009;5:271. doi: 10.1038/msb.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell DA. London: Portland Press. 1996. Understanding the Control of Metabolism. [Google Scholar]
- Ferrarini L, Bertelli L, Feala J, McCulloch AD, Paternostro G. A more efficient search strategy for aging genes based on connectivity. Bioinformatics. 2005;21:338–48. doi: 10.1093/bioinformatics/bti004. [DOI] [PubMed] [Google Scholar]
- Forster J, Famili I, Palsson BO, Nielsen J. Large-scale evaluation of in silico gene deletions in Saccharomyces cerevisiae. OMICS. 2003;7:193–202. doi: 10.1089/153623103322246584. [DOI] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002;296:750–2. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- Girvan M, Newman ME. Community structure in social and biological networks. Proc Natl Acad Sci USA. 2002;99:7821–6. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong T, Xuan J, Wang C, Li H, Hoffman E, Clarke R, Wang Y. Gene module identification from microarray data using nonnegative independent component analysis. Gene Regul Syst Bio. 2007;1:349–63. [PMC free article] [PubMed] [Google Scholar]
- Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245–52. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Gu H, Pan Z, Xi B, Hainline BE, Shanaiah N, Asiago V, Gowda GA, Raftery D. 1H NMR metabolomics study of age profiling in children. NMR Biomed. 2009;22:826–33. doi: 10.1002/nbm.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Conant GC, Wagner A. Molecular evolution in large genetic networks: does connectivity equal constraint? J Mol Evol. 2004;58:203–11. doi: 10.1007/s00239-003-2544-0. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- Hasty J, McMillen D, Isaacs F, Collins JJ. Computational studies of gene regulatory networks: in numero molecular biology. Nat Rev Genet. 2001;2:268–79. doi: 10.1038/35066056. [DOI] [PubMed] [Google Scholar]
- Holter NS, Maritan A, Cieplak M, Fedoroff NV, Banavar JR. Dynamic modeling of gene expression data. Proc Natl Acad Sci USA. 2001;98:1693–8. doi: 10.1073/pnas.98.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–3. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–4. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Strobel FH, Reed M, Pohl J, Jones DP. A rapid LC-FTMS method for the analysis of cysteine, cystine and cysteine/cystine steady-state redox potential in human plasma. Clin Chim Acta. 2008;396:43–8. doi: 10.1016/j.cca.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DP. Extracellular redox state: refining the definition of oxidative stress in aging. Rejuvenation Res. 2006;9:169–81. doi: 10.1089/rej.2006.9.169. [DOI] [PubMed] [Google Scholar]
- Kell DB. Metabolomics and systems biology: making sense of the soup. Curr Opin Microbiol. 2004;7:296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Kell DB, Mendes P. Snapshots of systems: metabolic control analysis and biotechnology in the post-genomic era. In: Cornish-Bowden A, Cardenas ML, editors. Technological and medical implications of metabolic control analysis. Dordrecht (The Netherlands): Kluwer Academic Publishers; 2000. pp. 3–25. [Google Scholar]
- Kennedy BK. The genetics of ageing: insight from genome-wide approaches in invertebrate model organisms. J Intern Med. 2008;263:142–52. doi: 10.1111/j.1365-2796.2007.01903.x. [DOI] [PubMed] [Google Scholar]
- Kristal BS, Vigneau-Callahan KE, Matson WR. Simultaneous analysis of the majority of low-molecular-weight, redox-active compounds from mitochondria. Anal Biochem. 1998;263:18–25. doi: 10.1006/abio.1998.2831. [DOI] [PubMed] [Google Scholar]
- Krylov DM, Wolf YI, Rogozin IB, Koonin EV. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–35. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger DA. Cell signaling pathways as control modules: complexity for simplicity? Proc Natl Acad Sci USA. 2000;97:5031–3. doi: 10.1073/pnas.97.10.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GD, et al. Metabolic signatures of exercise in human plasma. Sci Transl Med. 2010;2:33–37. doi: 10.1126/scitranslmed.3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindon JC, Holmes E, Nicholson JK. So what's the deal with metabonomics? Anal Chem. 2003;75:384A–391A. doi: 10.1021/ac031386+. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Dietary restriction: current status. Aging (Milano) 2001;13:261–2. doi: 10.1007/BF03353421. [DOI] [PubMed] [Google Scholar]
- Medawar PB. Old age and natural death. Modern Quart. 1946;2:30–49. [Google Scholar]
- Medawar PB. An unsolved problem in biology. London: HK Lewis; 1952. [Google Scholar]
- Medvedev ZA. An attempt at a rational classification of theories of ageing. Biol Rev Camb Philos Soc. 1990;65:375–98. doi: 10.1111/j.1469-185x.1990.tb01428.x. [DOI] [PubMed] [Google Scholar]
- Newman MEJ. The structure and function of complex networks. SIAM Rev. 2003;45:167–256. [Google Scholar]
- Newman MEJ. Finding community structure in networks using the eigenvectors of matrices. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 2007;73:036104. doi: 10.1103/PhysRevE.74.036104. [DOI] [PubMed] [Google Scholar]
- Ni Z, Lee SS. RNAi screens to identify components of gene networks that modulate aging in Caenorhabditis elegans. Brief Funct Genomics. 2010;9:53–64. doi: 10.1093/bfgp/elp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Wilson ID. Opinion: understanding ‘global' systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discov. 2003;2:668–76. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- Oeseburg H, de Boer RA, van Gilst WH, van der Harst P. Telomere biology in healthy aging and disease. Pflugers Arch. 2010;459:259–68. doi: 10.1007/s00424-009-0728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SG, Winson MK, Kell DB, Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16:373–8. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- Orgel LE. The maintenance of the accuracy of protein synthesis and its relevance to ageing. Proc Natl Acad Sci USA. 1963;49:517–21. doi: 10.1073/pnas.49.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE. The maintenance of the accuracy of protein synthesis and its relevance to ageing: a correction. Proc Natl Acad Sci USA. 1970;67:1476. doi: 10.1073/pnas.67.3.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE. Ageing of clones of mammalian cells. Nature. 1973;243:441–5. doi: 10.1038/243441a0. [DOI] [PubMed] [Google Scholar]
- Panowski SH, Dillin A. Signals of youth: endocrine regulation of aging in Caenorhabditis elegans. Trends Endocrinol Metab. 2009;20:259–64. doi: 10.1016/j.tem.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Promislow D. A regulatory network analysis of phenotypic plasticity in yeast. Am Nat. 2005;165:515–23. doi: 10.1086/429161. [DOI] [PubMed] [Google Scholar]
- Promislow DE. Protein networks, pleiotropy and the evolution of senescence. Proc Biol Sci. 2004;271:1225–34. doi: 10.1098/rspb.2004.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raamsdonk LM, et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19:45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- Rao CV, Arkin AP. Control motifs for intracellular regulatory networks. Annu Rev Biomed Eng. 2001;3:391–419. doi: 10.1146/annurev.bioeng.3.1.391. [DOI] [PubMed] [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–5. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- Salazar-Ciudad I, Jernvall J. A gene network model accounting for development and evolution of mammalian teeth. Proc Natl Acad Sci USA. 2002;99:8116–20. doi: 10.1073/pnas.132069499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackenberg LK, Sun J, Espandiari P, Holland RD, Hanig J, Beger RD. Metabonomics evaluations of age-related changes in the urinary compositions of male Sprague Dawley rats and effects of data normalization methods on statistical and quantitative analysis. BMC Bioinformatics. 2007;8:S3. doi: 10.1186/1471-2105-8-S7-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–76. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–8. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Shimokawa I, Chiba T, Yamaza H, Komatsu T. Longevity genes: insights from calorie restriction and genetic longevity models. Mol Cells. 2008;26:427–35. [PubMed] [Google Scholar]
- Soininen P, et al. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst. 2009;134:1781–5. doi: 10.1039/b910205a. [DOI] [PubMed] [Google Scholar]
- Southworth LK, Owen AB, Kim SK. Aging mice show a decreasing correlation of gene expression within genetic modules. PLoS Genet. 2009;5:e1000776. doi: 10.1371/journal.pgen.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EA, Ayroles JF. Modulated modularity clustering as an exploratory tool for functional genomic inference. PLoS Genet. 2009;5:e1000479. doi: 10.1371/journal.pgen.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Remmen H, Jones DP. Current thoughts on the role of mitochondria and free radicals in the biology of aging. J Gerontol A Biol Sci Med Sci. 2009;64:171–4. doi: 10.1093/gerona/gln058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G, Meir E, Munro EM, Odell GM. The segment polarity network is a robust developmental module. Nature. 2000;406:188–92. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- Wagner A. Robustness against mutations in genetic networks of yeast. Nat Genet. 2000;24:355–61. doi: 10.1038/74174. [DOI] [PubMed] [Google Scholar]
- Wagner A, Fell DA. The small world inside large metabolic networks. Proc Biol Sci. 2001;268:1803–10. doi: 10.1098/rspb.2001.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lawler D, Larson B, Ramadan Z, Kochhar S, Holmes E, Nicholson JK. Metabonomic investigations of aging and caloric restriction in a life-long dog study. J Proteome Res. 2007;6:1846–54. doi: 10.1021/pr060685n. [DOI] [PubMed] [Google Scholar]
- Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14:1847–50. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- Williams RE, Lenz EM, Lowden JS, Rantalainen M, Wilson ID. The metabonomics of aging and development in the rat: an investigation into the effect of age on the profile of endogenous metabolites in the urine of male rats using 1H NMR and HPLC-TOF MS. Mol Biosyst. 2005;1:166–75. doi: 10.1039/b500852b. [DOI] [PubMed] [Google Scholar]
- Williams R, Lenz EM, Wilson AJ, Granger J, Wilson ID, Major H, Stumpf C, Plumb R. A multi-analytical platform approach to the metabonomic analysis of plasma from normal and Zucker (fa/fa) obese rats. Mol Biosyst. 2006;2:174–83. doi: 10.1039/b516356k. [DOI] [PubMed] [Google Scholar]
- Wishart DS, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37(Database issue):D603–10. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchty S. Evolution and topology in the yeast protein interaction network. Genome Res. 2004;14:1310–14. doi: 10.1101/gr.2300204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, et al. A modular network model of aging. Mol Syst Biol. 2007;3:147. doi: 10.1038/msb4100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Wu B, Lin Z, Jin H, Huang J, Yang Y, Zhang X, Shen Z, Zhang W. Metabonomic characterization of aging and investigation on the anti-aging effects of total flavones of Epimedium. Mol Biosyst. 2009;5:1204–13. doi: 10.1039/b816407j. [DOI] [PubMed] [Google Scholar]
- Yu T, Park Y, Johnson JM, Jones DP. apLCMS—adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25:1930–36. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, et al. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]