Abstract
The genetics of the most common human pathogenic fungus Candida albicans has several unique characteristics. Most notably, C. albicans does not follow the universal genetic code, by translating the CUG codon into serine instead of leucine. Consequently, the use of Saccharomyces cerevisiae as a host for yeast two-hybrid experiments with C. albicans proteins is limited due to erroneous translation caused by the aberrant codon usage of C. albicans. To circumvent the need for heterologous expression and codon optimalization of C. albicans genes we constructed a two-hybrid system with C. albicans itself as the host with components that are compatible for use in this organism. The functionality of this two-hybrid system was shown by successful interaction assays with the protein pairs Kis1–Snf4 and Ino4-Ino2. We further confirmed interactions between components of the filamentation/mating MAP kinase pathway, including the unsuspected interaction between the MAP kinases Cek2 and Cek1. We conclude that this system can be used to enhance our knowledge of protein–protein interactions in C. albicans.
INTRODUCTION
In Candida albicans, protein–protein interactions have not been studied extensively due to the lack of appropriate tools. Recently, a vesicle targeting (1) and several tandem affinity purification methods (2–4) have been developed for application in C. albicans. The transfer of protein–protein interaction technologies to C. albicans is not trivial because of its alternative codon usage, translating 97% of the CUG codons into serine instead of leucine (5,6). For example, the application of the yeast two-hybrid system (7) for detection of interactions between C. albicans proteins can lead to false negative results due to the erroneous translation of C. albicans genes in Saccharomyces cerevisiae. Therefore, it can be necessary to change the CTG sequence by site-directed mutagenesis for application of the yeast two-hybrid system (8,9). Hence, the technique has not been widely used for C. albicans protein–protein interactions despite the existence of a C. albicans genomic DNA library for yeast two-hybrid screening (10).
In a yeast two-hybrid system, a protein of interest is fused to a DNA-binding domain (DBD) of a transcription factor (‘bait’) while the other protein of interest is fused to an activation domain (AD) of a transcription factor (‘prey’). The interaction between the proteins of interest brings the DBD in close proximity to the AD to reconstitute a functional transcription factor, leading to expression of one or more reporter genes. The most commonly used DBDs are the yeast Gal4 DBD [residues 1–147, (7)] and Escherichia coli LexA repressor (11). In the case of the Gal4 DBD, a GAL1, GAL2 or GAL7 promoter is placed upstream of the reporter genes (12), while for the LexA DBD, the reporter genes are preceded by one or more LexA operator sequences (13). As for the AD, the most frequently used are the Gal4 AD [residues 768–881, (7)], the viral protein VP16 (14) and the bacterial B42 AD (15). As shown by Legrain et al. (16), expression levels of the bait and prey proteins are able to affect the outcome of an interaction assay, depending on the nature of the interacting protein pair. Lower expression levels result in lower sensitivity but also reduce possible toxic effects of the protein and increase the specificity of the interaction. Apart from the promoter, the high copy number of the frequently used 2-µm plasmid, with bait or prey, influences the expression. Finally, reporter genes that are applied in a yeast two-hybrid experiment include HIS3, ADE2, LEU2 and E. coli lacZ (12,17,18).
In C. albicans, many signaling pathways are crucial elements for the virulence of this pathogenic fungus by mediating the reversible morphological switch from a unicellular yeast form to a multicellular hyphal form (19,20). These signaling pathways respond to environmental changes in pH, temperature or nutrient sources, and lead to adaptation of the cell in function of the varying conditions in which it has to survive (21). However, the existence of redundancy, parallel pathways and the interconnectivity between these pathways limit the potential to directly connect a protein to a signal or to another protein by traditional genetic analyses. For example, the mating defect in a strain with deletion of the mating MAP kinase Cek2 is only complete when another MAP kinase, Cek1, is also eliminated (22). An alternative strategy in the characterization of signaling pathways consists of establishing the physical interactions between components of the signaling cascade. In this strategy, functional characterization of a signaling protein of interest does not depend on phenotypic analysis but the protein can be linked to a pathway by showing its interaction with another protein of this pathway.
We developed a functional and optimized C. albicans two-hybrid system. All components were chosen for their applicability in C. albicans. The competence of the method was confirmed with two interacting protein pairs, Kis1–Snf4 and Ino4–Ino2. We then applied the system in the context of signaling pathways and identified known and novel interactions between the downstream components of the filamentation/mating MAP kinase pathway. The two-hybrid tool promises to be an integrative part of Candida research and holds great potential in establishing the interactome circuits of this pathogenic fungus.
MATERIALS AND METHODS
Strains
The wild-type strain SC5314 (23) was used for genomic DNA extraction. All genes mentioned originate from C. albicans unless otherwise stated. C. albicans SN152 [arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ URA3/ura3Δ::imm434 IRO1/iro1::imm434; (24)] was used to generate all the strains (Table 1). Saccharomyces cerevisiae strain PJ69-4A (12) was used for the yeast two-hybrid experiment. All strains were cultured on rich agar-medium (2% dextrose, 2% peptone, 1% yeast extract), unless otherwise stated.
Table 1.
Strains
| Strain | Genotype | Source |
|---|---|---|
| Candida albicans | ||
| SN152 | arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ URA3/ura3Δ::imm434 IRO1/iro1::imm434 | (24) |
| SC2H0 | SN152 ADH1b/HIS1 | This study |
| SC2H1 | SN152 LexAOp-ADH1b/HIS1 | This study |
| SC2H2 | SN152 5xLexAOp-ADH1b/HIS1 | This study |
| SC2H3 | SN152 5xLexAOp-ADH1b/HIS1 5xLexAOp-ADH1b/lacZ | This study |
| Saccharomyces cerevisiae | ||
| PJ69-4A | Mata leu2-312 ura3-52 trp1-901 his3-200 gal4Δ gal80Δ GAL2-ADE2 lys2::GAL1-HIS3 met2::GAL7-lacZ | (12) |
Construction of plasmids
Construction of pCOLH0, pCOLH1, pCOLH2 and pC2H-HIS1
Plasmid pCrOpLacZ [(25); Table 2] containing the reporter gene lacZ from Streptococcus thermophilus (26) was modified as followed. CaHIS1 with its terminator was amplified with primers HIS1-F and HIS1-R (all primers are shown in Supplementary Table S1) and cloned into pCrOpLacZ between ClaI and XmaI restriction sites to make pCOLH. The CaNAT1 locus was amplified from plasmid pAU34 (27) with primers NAT1-F and NAT1-R and replaced the ADE2 selection marker in pCOLH, creating pCOLH1 (Supplementary Figure S1A). An oligonucleotide consisting of four LexAOp sequences (4xLexAOp-F with 4xLexAOp-R primers) was integrated in pCOLH1 by ligation at the ApaI site to make pCOLH2 (Supplementary Figure S1B). Plasmid pCOLH0 was created by subcloning the 3343-bp XmaI–AatII fragment from pCrLacZ [(25); Table 2] into pCOLH1, to remove the LexAOp sequence (Supplementary Figure S1C).
Table 2.
Plasmids
| Plasmid | Original plasmid | Source |
|---|---|---|
| pCrLacZ | – | (25) |
| pCrOpLacZ | – | (25) |
| pSFS2 | – | (28) |
| pSN40 | – | (24) |
| pSN69 | – | (24) |
| pSFI1 | – | (29) |
| pAU34 | – | (27) |
| pMET3–VP16 | – | (31) |
| CIp-LexA | – | (25) |
| pGAD424 | – | (12) |
| pGBT9 | – | (12) |
| pCOLH0 | pCrLacZ | This study |
| pCOLH1 | pCrOpLacZ | This study |
| pCOLH2 | pCrOpLacZ | This study |
| pC2H-HIS1 | pSFS2 | This study |
| pC2H-LACZ | pCrOpLacZ | This study |
| pC2HB | pSN40 | This study |
| pC2HP | pSN69 | This study |
The HIS1 reporter construct for the final two-hybrid strain was constructed in two steps. First, the DNA fragment flanked by the restriction sites SacI and HindIII of pCOLH2 (containing the HIS1 reporter gene) was isolated, treated to create blunt ends and subcloned into plasmid pSFS2 (28), which was digested with SacI and SacII before generation of blunt ends. The resulting plasmid was called pSFS2–HIS1. Next, PCR amplification was performed of a 985-bp genomic sequence located on chromosome 4 between the genes PGA59 and PGA62 using primers HIS1IS-F and HIS1IS-R. This sequence, used for integration of the plasmid, was subcloned into the SacI site of pSFS2–HIS1 to create the final plasmid pC2H-HIS1.
Construction of pC2H-LACZ
The ADE2 selection marker in pCrOpLacZ was replaced with the AatII and NarI flanked dominant marker gene MPAR, which was amplified by PCR from plasmid pSFI1 (29) using primers MPA-F and MPA-R, creating pCOLL1. Next, RPS1 was amplified from genomic DNA with primers RPS1-F and RPS1-R and cloned in pCOLL1 with the restriction sites PacI and MluNI, removing the autonomously replicating sequence and generating pCOLL2. Then, the terminator sequence of CaHIS1 was PCR amplified with primers HIS1TLac-F and HISTLac-R, and cloned in pCOLL2 using the restriction site SacI to make pCOLL3. Finally, an oligonucleotide consisting of four LexAOp sequences (4xLexAOp-F with 4xLexAOp-R) was cloned using the restriction site ApaI, to finalize the construction of pC2H–LACZ.
Construction of bait plasmid pC2HB
A double-stranded oligonucleotide containing a multiple cloning site and an HA tag was created by annealing primers MCSB-F and MCSB-R. Following phosphorylation, it was cloned into plasmid pSN40 (24) using restriction sites BspEI and SalI to create pSN40-2. Secondly, the Staphylococcus aureus lexA gene was PCR amplified from plasmid CIp-LexA (25) with primers LexA-F and LexA-R, and cloned with AccIII and BglII restriction enzymes into pSN40-2 to make pSN40-3. Primer LexA-F also contained the nuclear localization sequence of the large T-antigen from simian virus 40 [SV40 NLS; (30)]. Additionally, after amplification with primers MET3P-F and MET3P-R, the 1500-bp MET3 promoter was introduced in pSN40-3 using restriction sites AatII and BspEI to generate pSN40-4. The terminator sequence of ACT1 was amplified with primers ACT1TB-F and ACT1TB-R and cloned into pSN40-4 using the restriction sites SalI and DraIII to create pSN40-5. Then, a 1104-bp fragment of genomic DNA on chromosome 1, between genes XOG1 and HOL1, was PCR amplified with primers BIS-F and BIS-R. This fragment was introduced in pSN40-5 using restriction sites PvuI and SnaBI to make pSN40-6. Subsequently, the unique restriction site NdeI in this last fragment was replaced by NotI, with the double-stranded oligonucleotide NotIB. NotIB was cut at its flanking AseI sites, which bear compatible ends with NdeI, at which position in pSN40-6 NotIB was cloned into to make pSN40-7. Finally, a new multiple cloning site (NheI–StuI–SfuI–AscI–MluI–ScaI) was made by inserting the oligonucleotide NewMCSB-F (with complementary NewMCSB-R sequence) at the AscI site of pSN40-7 after cutting the oligonucleotide with AscI and BsaJI to create the final construct pC2HB.
Construction of prey plasmid pC2HP
A long oligonucleotide (from annealing MCSP-F1 and MCSP-F2 with MCSP-R1 and MCSP-R2) was introduced in plasmid pSN69 (24) digested with AatII and SgrAI, creating pSN69-2. This oligonucleotide consisted of a SV40 NLS, a 3xFLAG tag sequence flanked by two glycine linkers and a multiple cloning site. The MET3 promoter (primers MET3P-F and MET3P-R) was amplified and ligated between the AatII and BspEI sites of pSN69-2 upstream of the SV40 NLS to make pSN69-3. The ACT1 terminator was PCR amplified from genomic DNA with primers ACT1TP-F and ACT1TP-R and cloned into pSN69-3 using SgrAI and BstBI to generate pSN69-4. Next, a sequence located between RXT3 and ORF19.3569 on chromosome 2 was PCR amplified using primers PIS-F and PIS-R and this 850-bp fragment, used for integration purposes, was cloned into pSN69-4 using restriction sites PflMI and ApaI, creating pSN69-5. A NotI restriction site was introduced in the unique SwaI site of pSN69-5, in the middle of this integration sequence, by introducing the phosphorylated oligonucleotide NotIP. The resulting plasmid was called pSN69-6. Additionally, AD VP16 was amplified from pMET3-VP16 (31) and cloned downstream of SV40 NLS between restriction sites SacII and NheI in pSN69-6 to generate pSN69-7. Finally, a multiple cloning site (StuI–XmaI–AscI–MluI–ScaI) was added by inserting an oligonucleotide (NewMCSP-F with its complementary sequence NewMCSP-R) between restriction sites StuI and EcoRI of pSN69-7. The resulting plasmid was called pC2HP.
Yeast two-hybrid plasmids
For two-hybrid analysis in S. cerevisiae PJ69-4A (12), CaHST7 and CaCEK1 were PCR amplified from wild-type SC5314 with the primer pairs HST7-Y2H-F/R and CEK1-Y2H-F/R respectively, and cloned in the yeast two-hybrid plasmids pGAD424 and pGBT9 (12) at the XmaI restriction site. ScTPK1 and ScBCY1 were used as positive controls for the Y2H (32).
Construction of strains
All transformations of C. albicans strains were performed using the lithium acetate method (33). Correct integration of the reporter genes from plasmids pCOLH0, pCOLH1, pCOLH2, pC2H-HIS1 and pC2H-LACZ was verified by PCR diagnosis, and Southern blotting as described earlier (34). Plasmids pCOLH0, pCOLH1 and pCOLH2 were cut with the CelII restriction enzyme for integration in the ACT1 terminator sequence of strain SN152. Transformed cells were grown on YPD containing 200 μg/ml nourseothricin (ClonNat, Werner-Bio). The resulting strains were named respectively SC2H0, SC2H1 and SC2H2.
For the generation of strain SC2H3, HpaI linearized plasmid pC2H-HIS1 was integrated in SN152 between genes PGA59 and PGA62 on chromosome 4 by selective growth on YPD containing nourseothricin and correct integration was verified. The resistance marker cassette was removed as described earlier (28). The nourseothricin sensitive cells were transformed with plasmid pC2H-LACZ linearized at the NcoI site located in the RPS1 gene. The dominant marker gene MPAR (35) was used for selection on synthetic complete (SC) medium with 5 μg/ml mycophenolic acid (Sigma). Correct integration was confirmed by PCR diagnosis and Southern blotting.
Two-hybrid analysis with reporter gene HIS1
Strains SC2H0, SC2H1, SC2H2 or SC2H3 were transformed with the bait plasmid pC2HB and plated on SC medium without leucine. Two independent transformants were subsequently transformed with the prey plasmid pC2HP with selection on SC medium without arginine. PCR analysis was performed to confirm correct integration. Finally, independent transformants were used for a spot assay of fivefold dilution series on SC medium and SC medium without histidine. When indicated, SC medium without methionine was used. Spot assays were performed at 30°C, unless otherwise stated.
β-galactosidase assay
The β-galactosidase assay was accomplished according to the protocol from Rose and Botstein (36) with the only modification that the enzymatic activity was measured at 37°C instead of 28°C. Cells were grown at 30°C in 3 ml SC medium until late-exponential phase (O.D.595 around 5). Statistical analysis was performed using a Student’s t-test (P < 0.01).
Protein extraction, western blotting and co-immunoprecipitation
Cells were grown in 75 ml SC medium lacking methionine at 30°C until late-exponential phase (O.D.595 around 5), collected, and washed in 1× phosphate buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 at pH 7.3). Protein extraction was performed with glass beads in lysis buffer containing 1× PBS, 0.001% Triton X-100, 8.7% glycerol, 25 µM MgCl2, 10 µM EDTA (pH 7), 10 µM dithiotreitol, 100 µM NaF, 4 µM Na3VO4, 1 µM β-glycerophosphate and one tablet of Complete Protease Inhibitor Cocktail (Roche). Protein concentration was measured (37) and 500 µg protein was used for co-immunoprecipitation with horseradish peroxidase-conjugated anti-HA antibodies (Roche) or monoclonal anti-FLAG antibodies (M2, Sigma-Aldrich), by incubation for 2 h at 4°C with Protein G-Agarose beads (Roche). After three wash steps with lysis buffer, SDS sample buffer (5×: 250 mM Tris–HCl, 10% SDS, 0.5% bromophenol blue, 1.4 M β-mercapto-ethanol) was added, samples were heated for 5 min at 95°C and stored at −20°C.
Separation of protein samples was performed by SDS–polyacrylamide gel electrophoresis on NuPAGE Novex Bis–Tris Mini Gels (Invitrogen) followed by transfer to a nitrocellulose membrane (HybondC extra, Amersham). The anti-FLAG antibodies, after addition of horseradish peroxidase-conjugated anti-mouse IgG secondary antibodies (Amersham), and the anti-HA antibodies were detected using Supersignal West Pico Luminol solution (Thermo Scientific). The immunoblots were imaged with Fujifilm LAS-4000 mini, and the accompanying software Image Reader LAS-4000, and Aida Image Analyzer v.4.22 (Life Science Fuji Photofilm Co., Ltd).
RESULTS
Design of a C. albicans two-hybrid strain and compatible bait and prey plasmids
A two-hybrid system contains several components that influence the sensitivity and specificity of the method: a DBD, an AD, the promoter of the bait and prey, and the reporter genes with their promoter regions. The DBD S. aureus LexA (25) and one of the reporter genes, S. thermophilus lacZ (26), were previously shown to be fully functional in C. albicans and were therefore used to construct the Candida two hybrid (C2H) strains and plasmids. A second reporter gene C. albicans HIS1 was selected, in analogy with S. cerevisiae HIS3 in the yeast two-hybrid system. For the choice of the AD, the promoter region of the reporter genes and the promoters of the bait and prey expression constructs, one-hybrid experiments with a DBD (SalexA) as a constant factor were performed. The choice of the integration sites for the four constructs was somewhat more difficult. The RPS1 locus has been extensively used for plasmid integration (38,39) and was used here for integration of the reporter gene lacZ. For the other three plasmids we identified for genomic intergenic regions that were unlikely to be involved in transcriptional regulation of its neighboring genes and containing restriction sites that were compatible with the integrating plasmid. A strain carrying the two reporter constructs and the empty bait and prey plasmids displayed similar growth rates as the SN152 parental strain in rich medium (YPD), SC, and SC–HIS (data not shown).
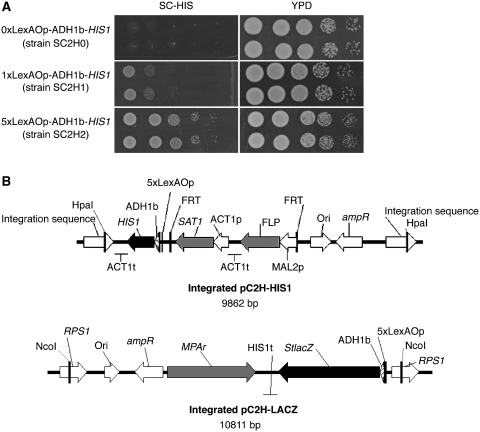
An initial strain, namely SC2H1, was created by integration of plasmid pCOLH1 (Supplementary Figure S1A) in strain SN152. This strain contained one copy of the reporter gene HIS1 cloned downstream of one LexA operator sequence and a basal ADH1 promoter (150 bp). Integration of pCOLH2 (Supplementary Figure S1B) in strain SN152 created strain SC2H2, in which five LexAOp sequences were introduced upstream of HIS1. As a negative control, strain SC2H0 was made by integration of pC2H0 which does not contain any LexAOp sequence (Supplementary Figure S1C). Induction of HIS1 expression by an artificial fusion protein LexA–VP16 was tested in strains SC2H0, SC2H1 and SC2H2. The presence of numerous LexAOp sequences enhanced greatly the growth on selective medium as shown in Figure 1A. This result suggested that binding of LexA-fused sequences was more effective and efficient for growth when multiple LexA operator sequences were present. Similar results were obtained with other ADs than VP16 (data not shown). Additionally, the ADH1 basal promoter by itself did not result in sufficient expression of the reporter gene to sustain the growth of strain SC2H0 on selective medium (Figure 1A). Based upon this result, a third strain, namely SC2H3, was constructed, in which both HIS1 and StlacZ were placed downstream of multiple LexAOp sequences. SC2H3, which originated from SN152, contains the integrated plasmid pC2H–HIS1 (Figure 1B). This plasmid differed from pCOLH2, mainly by the presence of a SAT1 flipper cassette (28) used for selective genomic integration. By the simple removal procedure of the marker in maltose following recombination of the FRT flanking sequences, future genetic manipulations of the two-hybrid strain can be considered. Strain SC2H3 was completed by integration of plasmid pC2H-LACZ (Figure 1B) which includes the second reporter system, StlacZ (26). Two auxotrophic marker genes (LEU2 and ARG4) could still be used in strain SC2H3 for bait and prey integration.
Figure 1.
(A) Influence of LexAOp sequences on reporter gene activation. Strains SC2H0, SC2H1 and SC2H2, carrying respectively no, one and five LexAOp sequences upstream of HIS1 reporter gene were compared in their ability to grow on SC medium lacking histidine. While the presence of at least one LexAOp sequence is necessary for HIS1 expression, the hybrid transcription factor LexA–VP16 clearly takes advantage of the presence of multiple LexAOp sequences. Two independent strains for each of the constructs are shown and grown for 1 day. (B) The two-hybrid strain SC2H3 contains two integrated reporter constructs. Plasmid pC2H–HIS1 includes reporter gene HIS1 in front of a basal ADH1 promoter and five LexAOp sequences for binding of the bait protein. Plasmid pC2H–LACZ contains reporter gene lacZ preceded by a basal ADH1 promoter sequence and five LexAOp sequences for binding of the bait protein. The complete plasmid is flanked by its restriction sites used for integration. Linearized pC2H–HIS1 integrates at the HpaI site located between genes PGA59 and PGA62 on chromosome 4, while pC2H–LACZ integrates at the RPS1 locus on chromosome 1, at the NcoI site. ADH1b: basal promoter of ADH1; ampR: ampicillin resistance gene; Ori: origin of replication; t: terminator; p: promoter.
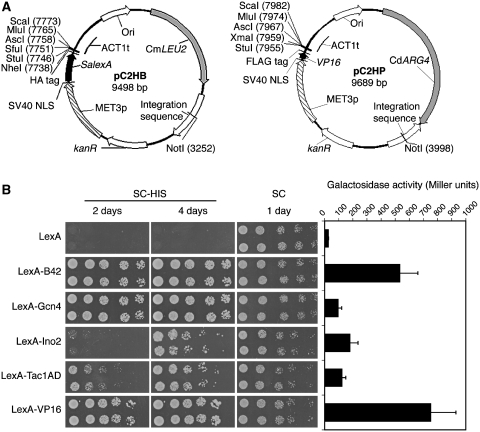
The construction of bait and prey plasmids implied the successive in-frame cloning of the necessary nucleic acid elements. The bait plasmid pC2HB (Figure 2A, left panel) originated from plasmid pSN40 (24), and carried the binding domain SalexA preceded by the nuclear localization sequence of the large T-antigen from simian virus 40 [SV40 NLS; (30)] for specific targeting of the bait to the nucleus. For the purpose of co-immunoprecipitation analyses, an HA tag was added to the bait construct behind the binding site sequence. Glycine linker sequences between the HA tag and SalexA, and between the HA tag and the multiple cloning site were added to enable flexible and independent folding of the different domains of the fusion protein. The auxotrophic marker C. maltosa LEU2 was used to select for transformation of the NotI linearized pC2HB, which is integrated between XOG1 and HOL1 on chromosome 1. The restriction site NotI is present in only four C. albicans genes and therefore is compatible with most of C. albicans genes of interest to be cloned in the bait plasmid. The efficiency of correct integration of the bait plasmid, as determined by PCR diagnosis, was higher than 90% (44 out of 48 transformants that were checked). In order to find a suitable promoter for the bait and prey, three promoters were tested for their reliability in the system. While a minimal promoter sequence (700 bp) of ADH1 did not sustain a reliable expression, the inducible, weak MET3 (40,41) and the strong ACT1 promoter produced positive results (data not shown). The MET3 promoter, turned off by addition of sulfur amino acids to the growth medium, was preferred to minimize side effects and prevent overexpression. Both bait and prey genes were therefore placed under the regulation of the MET3 promoter. Most experiments were performed on SC medium, which contains 0.15 mM methionine and moderately represses the MET3 promoter (40,41). For growth conditions-driven interactions, one would need to consider the usage of media with no or low levels of methionine, which can be seen as a limitation, e.g. for hyphal-specific interactions. For this type of conditional designs, one should consider modifying the background of the two-hybrid strain, rather than altering the growth conditions. By working in a constitutively yeast or filamentous strain background, one should be able to identify cell type specific interactions.
Figure 2.
(A) Bait and prey plasmids, pC2HB and pC2HP. A bait gene of interest (left panel) is cloned into the multiple cloning site downstream of the DNA-binding protein SalexA, and under the control of the MET3 promoter. The pC2HB plasmid is integrated between loci XOG1 and HOL1 on chromosome 1 after linearization at restriction site NotI and selection is obtained with the auxotrophic marker LEU2 from C. maltosa. In the case of prey plasmid pC2HP (right panel), a prey gene of interest is cloned into the multiple cloning site, downstream of AD VP16. The MET3 promoter controls expression of the prey gene. An integration sequence, situated in the intergenic region between genes RXT3 and ORF19.3569 on chromosome 2, makes insertion in the genome possible after linearization of the plasmid at restriction site NotI. C. dubliniensis ARG4 is the auxotrophic marker for transformation. (B) Comparison of activation strength of five different ADs fused to the binding domain LexA in a one-hybrid experiment. Bacterial B42, C. albicans Gcn4, C. albicans Ino2, C. albicans Tac1 hyperactive AD and viral protein VP16 were tested for their ability to stimulate HIS1 and lacZ expression in strain SC2H3. VP16 was chosen to act as the AD in the prey plasmid. kanR: kanamycin resistance gene; Ori: origin of replication; NLS: nuclear localization sequence.
For the construction of the prey plasmid the crucial element was the AD. Five different AD were cloned in fusion with the lexA binding site to compare their activation strength in a one-hybrid experiment (Figure 2B). These five ADs were (i) the codon-optimized bacterial B42 AD, (ii) the transcription factor Gcn4, (iii) the transcription factor Ino2, (iv) the hyperactive AD of transcription factor Tac1 and (v) the codon-optimized viral protein VP16. B42 and VP16 are common ADs for two-hybrid applications in S. cerevisiae (14,15). Gcn4 is a transcription factor of the general amino acid control response (42) and was previously shown to induce gene expression in a one-hybrid assay (25). Ino2 is a transcription factor that presumably regulates ribosomal protein genes (8). Tac1 is a transcriptional activator of drug-responsive genes and a mutation in its AD (N977D) renders it hyperactive (43). All five LexA–AD fusions were transferred to strain SC2H3 and their ability to activate the reporter genes StlacZ and HIS1 was analyzed. Figure 2B shows how the non-Candida ADs VP16 and B42 were the most suitable inducers of the reporter genes in the system. In a spot assay experiment, Gcn4 was comparable with VP16 and B42 in activating HIS1 expression. However, the galactosidase assay favored only VP16 and B42 as very strong activators of StlacZ. The combined results demonstrated that VP16 was the best choice as an AD in the Candida two-hybrid system. VP16 in combination with the weak MET3 promoter was the most likely approach to provide high sensitivity (due to VP16) without losing specificity (due to MET3 promoter). It is noteworthy that the LexA binding sequence by itself caused a basal galactosidase activity of around 20 Miller units. This basal activity was abolished when a non-autoactivating bait gene was introduced in frame with the binding domain (data not shown). The removal of the HA tag also reduced greatly the background activity of the reporter system. This basal activity should therefore be considered as an intrinsic phenomenon of the two-hybrid constructs. Finally, the prey construct pC2HP (Figure 2A, right panel) consisted of, from the N-terminal end, an SV40 NLS for nuclear localization, VP16 as AD, a glycine linker, 3xFLAG tag, a glycine linker and a multiple cloning site. After linearization with restriction enzyme NotI, integration of the plasmid was achieved at an integration site between RXT3 and ORF19.3569 on chromosome 2 using the auxotrophic marker C. dubliniensis ARG4. As with the bait plasmid, the efficiency of correct integration was high (36 out of 40 transformants tested).
Proof-of-principle interactions between Kis1 and Snf4 and between Ino4 and Ino2 show the competence of the system
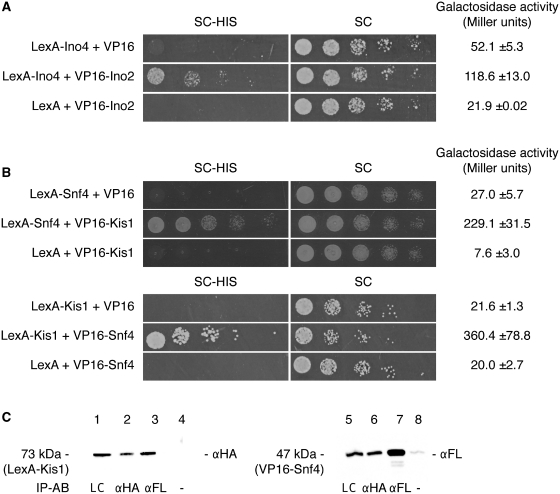
To validate the functionality of the two-hybrid system, interactions between C. albicans proteins that were previously shown to occur were analyzed with the two-hybrid tool. We first examined the heterodimerization of the proteins Ino2 and Ino4. Ino2 is a transcription factor that, together with Ino4, putatively regulates ribosomal protein genes in C. albicans (8). The Ino2–Ino4 dimer binds ICRE (inositol/choline-responsive element)-related motif sequences. After modification of a CTG codon in the basic helix-loop-helix region of Ino4, Ino4 and Ino2 were previously shown to interact in a yeast two-hybrid assay (8). In the Candida background, we have shown that Ino2 could activate HIS1 and StlacZ in a one-hybrid assay (Figure 2A). We cloned full-length INO2 into the prey plasmid pC2HP and full-length INO4 in the bait plasmid pC2HB. The two-hybrid strain SC2H3 was transformed with pC2HB–Ino4 or with the empty control vector pC2HB on medium lacking leucine. A second transformation step on medium lacking arginine was performed with the prey plasmid pC2HP-Ino2 or with the empty control vector pC2HP. As shown in Figure 3A, only the combination pC2HB–Ino4 with pC2HP–Ino2 led to growth on selective medium, indicating interaction between the two proteins. For validation of the binding of Ino4 with Ino2, the second reporter assay, with StlacZ, was performed. As shown in Figure 3A, the values of a galactosidase assay were significantly higher for the interaction compared to the negative controls. For sustainable and strong interactions, growth was clearly visible on selective medium after 1 to 2 days of incubation. For interactions that may be either transient or weaker, one can expect growth on selective medium to occur within 5–7 days. In parallel, LacZ expression levels >100 U can be considered the read-out for a strong and constitutive interaction.
Figure 3.
(A) Two-hybrid interaction of C. albicans Ino4 with Ino2. Ino4 and Ino2 interact strongly, shown by growth of strain SC2H3 on SC–HIS after one day with Ino4 as bait and Ino2 as prey. This interaction is confirmed in the galactosidase assay where the activity measured (in Miller units) was 2.5-fold the background activity. (B) Two-hybrid interaction of C. albicans Kis1 with Snf4. Kis1 and Snf4, two proteins of the Snf1 complex, interact with each other as shown by growth of SC2H3 on SC–HIS and a high galactosidase activity. For the HIS1 reporter assay, cells were incubated for up to 2 days. (C) Co-immunoprecipitation of Kis1 and Snf4. The interaction between bait LexA–HA–Kis1 (73 kDa) and prey VP16–FLAG–Snf4 (47 kDa) is confirmed in a co-IP experiment. Total protein concentrations were equal for each sample. Lane 1: loading control of Kis1; lane 2: IP of Kis1 with anti-HA antibodies; lane 3: co-IP of Kis1 with anti-FLAG antibodies; lane 4: negative control of Kis1 immunoprecipitation without antibodies; lane 5: loading control of Snf4; lane 6: co-IP of Snf4 with anti-HA antibodies; lane 7: IP of Snf4 with anti-FLAG antibodies; lane 8: negative control of Snf4 immunoprecipitation without antibodies. The antibodies used for western blotting are indicated on the right side of each blot and the antibodies for immunoprecipitation are shown below the blot. IP-AB: antibody used for immunoprecipitation; LC: loading control; αFL: anti-FLAG antibody; αHA: anti-HA antibody.
Secondly, we examined the interaction between the proteins Kis1 and Snf4. Kis1 and Snf4 belong to the Snf1 protein complex that is an important regulator of carbohydrate metabolism (3). The α-subunit Snf1, the β-subunit Kis1 and the γ-subunit Snf4 form a complex that directs the cell into adaptation to growth on non-fermentable carbon sources. KIS1 and SNF4 were cloned into the two-hybrid plasmids to check for interaction, and integrated in SC2H3. Despite the presence of glucose, Kis1 and Snf4 strongly interacted in both directions, as seen by the growth on SC–HIS and the high galactosidase activity (Figure 3B). Interaction between Kis1 and Snf4 was also investigated by co-immunoprecipitation experiments in order to confirm the two-hybrid result. SC2H3 containing Kis1–bait and Snf4–prey constructs was grown in non-selective medium to minimize the growth pressure effect on the putative interaction and in the absence of methionine to fully induce the bait and prey expression. Each of the tagged proteins migrated at the appropriate size, as indicated in Figure 3C (lanes 1 and 5). This suggested that the tagged bait and prey proteins were being recognized by their respective antibodies, and that they were being expressed at a level high enough to be detected by immunoprecipitation. When prey FLAG-tagged Snf4 (47 kDa) was precipitated with anti-FLAG antibody coated beads, the bait HA-tagged Kis1 (73 kDa) co-precipitated as shown in Figure 3C (lane 3). The interaction of both proteins was also demonstrated in the other direction with co-precipitated FLAG-tagged Snf4 being identified with immunoprecipitated HA-tagged Kis1 (lane 6). When no antibodies were used to precipitate the proteins, there was often a residual signal of the size of the targeted immunoprecipitated protein (lane 8). One explanation for this could be the unspecific binding of one of more of the artificial sequences added to the proteins to the beads. This co-immunoprecipitation of Kis1 with Snf4 confirmed the interaction observed by the reporter gene assays. By using the reporter systems or the protein tags in the respective constructs, it was therefore demonstrated that the Candida two-hybrid strain and constructs are reliable for the detection of strong positive interactions.
Application of the two-hybrid system on components of the mating/filamentation MAP kinase pathway
The two-hybrid system can be considered a useful tool for the investigation of signaling pathways that are important for virulence of C. albicans. One of these pathways is the MAP kinase pathway responsible for mating and filamentation. This MAPK cascade consists of the kinase Cst20, the MAPKK Hst7 and the MAP kinases Cek2 and Cek1, which are believed to both activate the transcription factor Cph1. Based upon homology with S. cerevisiae Ste11, the uncharacterized C. albicans Ste11 is considered to be the MAPKKK that activates Hst7. All the other components also have homologues in S. cerevisiae and similarly to the pseudohyphal defects observed in S. cerevisiae (44), null mutations of MAPK pathway components in C. albicans give rise to hyphal defects on several hyphae inducing media (45,46). In addition, the MAP kinase pathway plays a crucial role in cell wall integrity (47) and mediation of the mating response triggered by pheromone (22).
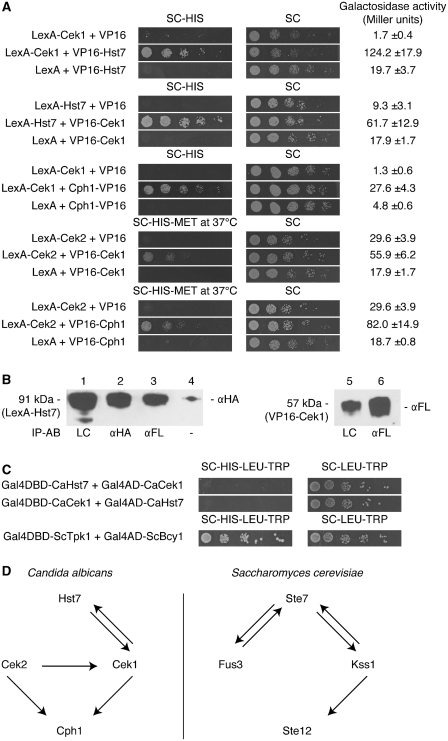
Although the genetic interactions between all components of the Cek1 MAPK pathway strongly suggest they are indeed part of the same phosphorylation-centered signaling cascade no data exist on the physical interactions between the components of the pathway. Therefore, the downstream components of the pathway were used for analysis of protein–protein interactions in the two-hybrid system. The genes HST7, CEK2, CEK1 and CPH1 were cloned in both the bait and prey plasmids. All 16 possible pair-wise combinations of proteins were tested for interaction, together with their respective negative controls. Figure 4A shows the protein pairs that were interacting in the two-hybrid assay. The MAPKK Hst7 and the MAPK Cek1 clearly interact with each other with positive results for both the spot assay and the galactosidase assay. A co-immunoprecipitation experiment with the bait Hst7 and the prey Cek1 further confirmed stable binding (Figure 4B). HA-tagged Hst7 (91 kDa) co-immunoprecipitated with FLAG-tagged Cek1 (57 kDa) using anti-FLAG antibodies at the immunoprecipitation step (lane 3 on Figure 4B). Moreover, a two-hybrid experiment with S. cerevisiae as the host organism failed to detect any interaction between Cek1 and Hst7 (Figure 4C). This result supported the expected positive influence of the endogenous background in identifying protein interactions, as previously demonstrated by Boysen et al. (1) with the VCI system. Further downstream in the cascade, the MAP kinases Cek2 and Cek1 both interacted with Cph1 (Figure 4A). Both interactions were weak as shown by the poor growth on selective medium, but a reproducible significant difference in galactosidase activity compared to the negative controls was observed. In analogy to its yeast homolog Ste12 in a yeast two-hybrid experiment (48), Cph1 was positioned N-terminal from VP16 to improve the interaction efficiency with Cek1. Interestingly, Cek2 and Cek1 also appeared to interact with each other (Figure 4A). This result was unexpected as their yeast homologues Fus3 and Kss1 do not interact with each other in a similar experiment (48). However, the interaction between Cek2 and Cek1 could not be confirmed in a co-IP experiment suggesting that the interaction is only transient. It must be noted that with most interacting protein pairs, not all transformants containing both prey and bait plasmids grew on medium lacking histidine. A protein interaction was considered positive when at least 30% of the transformants grew on selective medium. However, growth of the negative controls was never observed and an overall significant difference was always detected as measured from the galactosidase assay. It is of general interest to mention that some expected interactions, such as between Cek2 and Hst7, did not work in the Candida two-hybrid (data not shown). Although it is relevant to suspect that some interactions will only occur in very specific conditions, e.g. following post translation modifications, one cannot exclude that failure may be a consequence of intrinsic limitations, such as unstable fusion protein, or occlusion of DBD or AD.
Figure 4.
Interactions between downstream targets of the mating/filamentation MAP kinase pathway. (A) All pair-wise combinations of interactions between the MAPKK Hst7, the MAP kinases Cek2 and Cek1, and the transcription factor Cph1 were tested in a two-hybrid assay. Interactions between the proteins pairs Hst7 and Cek1, Cek2 and Cek1, Cek1 and Cph1, and Cek2 and Cph1 were detected. In all cases, a representative transformant is shown for growth for up to 5 days on SC–HIS. At least two independent transformants were used for the galactosidase assay. (B) The interaction between bait LexA–HA–Hst7 (91 kDa) and prey VP16–FLAG–Cek1 (57 kDa) is confirmed in a co-immunoprecipitation (co-IP) experiment. Total protein concentrations were equal for each sample. Lane 1: loading control of Hst7; lane 2: IP of Hst7 with anti-HA antibodies; lane 3: co-IP of Hst7 with anti-FLAG antibodies, visualized with anti-HA antibodies; lane 4: negative control of Hst7 with beads but without antibodies; lane 5: loading control of Cek1; lane 6: IP of Cek1 with anti-FLAG antibodies. IP-AB: antibody used for immunoprecipitation; LC: loading control; αFL: anti-FLAG antibody; αHA: anti-HA antibody. (C) Yeast two-hybrid experiment with Hst7 and Cek1 after 7 days on selective medium. The interacting S. cerevisiae protein pair Bcy1 with Tpk1 was used as positive control. (D) Comparison of pair-wise interactions of C. albicans proteins as shown by a C. albicans two-hybrid experiment (A) and of S. cerevisiae proteins demonstrated in the yeast two-hybrid assay (48,54–56). Arrows are directed from the bait protein to the prey protein.
The Candida two-hybrid system was evaluated by comparison with the yeast two-hybrid system. Interactions between the downstream components of the MAP kinase pathway were compared between C. albicans proteins in the Candida two-hybrid system and yeast proteins in the yeast two-hybrid system (Figure 4D). Both systems were able to detect five interactions from a total of 16 putative interactions. In contrast to its homolog Fus3 in S. cerevisiae, the mating MAP kinase Cek2 interacts with its target transcription factor Cph1 in a two-hybrid experiment while no interaction with its activator Hst7 has been observed.
DISCUSSION
The study on protein–protein interactions can reveal many clues on the function and regulation of a protein. Many protein–protein interaction tools are available but depending on the character of the expected interaction, one method can be more convenient than the other. While tandem affinity purification (49) is the method of preference for detection of stable protein complexes, the yeast two-hybrid system provides a useful tool for the identification of transient binary interactions as seen in signaling pathways. The principle of the traditional two-hybrid system is based on the reconstitution of a transcription factor upon interaction of two proteins of interest. However, many alternative strategies have been described with the most notable being the split ubiquitin two-hybrid system, which allows detection of interactions between membrane proteins (50). Recently, an alternative two-hybrid system was developed for the use in C. albicans (1). In this method, called the vesicle capture interaction assay (VCI), a GFP-fusion protein is targeted to the endocytic vesicle surface upon interaction of two proteins of interest. Applying VCI, the interaction was shown between the MAPK kinase Pbs2 and the MAPK Hog1, both components of the oxidative stress MAP kinase pathway. The quantification procedure for positive interaction relies on the specific localization of the fluorescent signal, which can reliably be applied for two proteins of interest but can hardly be used for screening purposes in high throughput experiments.
Here we propose a traditional two-hybrid system as an additional method for the investigation of protein–protein interactions in C. albicans, which relies on the expression of two reporter systems easily detectable and appropriate to use in large-scale experimental identification of protein interactions. Due to the aberrant codon usage, compatible components that are accurately expressed in C. albicans are required for a C. albicans two-hybrid system. Therefore, we used the CUG codon-free S. aureus lexA as DBD (25) and codon-optimized VP16 as AD. While the use of the reporter gene HIS1 permits the high-throughput screening of DNA libraries against a bait of interest for interaction, the second Candida compatible reporter gene S. thermophilus lacZ can allow the semi-quantitative appreciation of an interaction. It also bears the advantage that false positive interactions caused by selective pressure on prototrophic media can be identified in a galactosidase assay. While the two-hybrid tool brought confirmation to suspected interactions such as Kis1–Snf4 and Ino2–Ino4 interactions, it also highlighted a novel interaction between the MAP kinases Cek2 and Cek1. In yeast, Cek2 homolog Fus3 and Cek1 homolog Kss1 do not interact with each other in a yeast two-hybrid experiment unless their substrate Ste12 is absent (51). The lack of Ste12 might block the pathway, thereby freezing the pathway in a steady-state situation that normally only appears very transiently, for example at the start of the pheromone response. One could suggest that the Cek1–Cek2 and Fus3–Kss1 interactions may play a role in differentially regulating the downstream effectors of the MAP kinase pathway. It is indeed known that the mitogen-activated protein kinase cascade responds to multiple stimuli including pheromones and nutrient starvation signals. In particular, the white and opaque cell responses in C. albicans share the MAP kinase in response to the same pheromone but they do have different transcriptional effectors (52). The fact that Cek2 was only detected in mating competent MTLa or MTLα but not in mating incompetent MTLa/α cell types (22) supports the hypothesis that the Cek2 interaction with other proteins, including Cek1, may be part of the differential mechanism of regulation of the downstream factors of the MAP kinase. Nevertheless, further research on these two MAP kinases in S. cerevisiae and in C. albicans could shed more light on the biological relevance of their interactions.
The most significant advantage of the two-hybrid method lies in the ability to screen for proteins binding a specific protein of interest using a cDNA or genomic DNA library. Recently, the groups of Carol Munro (University of Aberdeen) and Christophe d’Enfert (Pasteur Institute) established a collaborative project for the creation of a complete C. albicans ORFeome library. This ORFeome library could be incorporated in the prey plasmid, thereby greatly reducing the number of transformants that need to be obtained for a full coverage of the interactome using the two-hybrid system as a screening tool. The low transformation efficiency known in C. albicans, may be overcome by the use of this ORFeome library, from which 30 000 transformants would need to be generated for a five time coverage. In addition, screening for protein–protein interactions in C. albicans can serve a more general cause as interactions with S. cerevisiae could be confirmed in C. albicans according to an interspecies application of the paralogous verification method [PVM; (53)]. PVM validates protein–protein binding by comparison of paralogous interactions. Interactions between C. albicans proteins in a C. albicans two-hybrid system could be used as a verification of S. cerevisiae protein–protein interactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Fund for Scientific Research Flanders (FWO) [grant number G.0242.04 to P.V.D.]. Funding for open access charge: The Fund for Scientific Research Flanders (grant number G.0242.04 to P.V.D.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Nico Vangoethem for help with preparation of the figures, Deborah Seys and Cindy Colombo for technical assistance, and Dr Alistair J. Brown (University of Aberdeen), Dr Joachim Morschhäuser (University of Würzburg), Dr Alexander D. Johnson (University of California), Dr Julia R. Köhler (Harvard Medical School) and Dr Dominique Sanglard (University of Lausanne) for their kind gifts of strains and plasmids.
REFERENCES
- 1.Boysen JH, Fanning S, Newberg J, Murphy RF, Mitchell AP. Detection of protein-protein interactions through vesicle targeting. Genetics. 2009;182:33–39. doi: 10.1534/genetics.109.101162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaneko A, Umeyama T, Hanacka N, Monk BC, Uehara Y, Niimi M. Tandem affinity purification of the Candida albicans septin protein complex. Yeast. 2004;21:1025–1033. doi: 10.1002/yea.1147. [DOI] [PubMed] [Google Scholar]
- 3.Corvey C, Koetter P, Beckhaus T, Hack J, Hofmann S, Hampel M, Stein T, Karas M, Entian KD. Carbon source-dependent assembly of the Snf1p kinase complex in Candida albicans. J. Biol. Chem. 2005;280:25323–25330. doi: 10.1074/jbc.M503719200. [DOI] [PubMed] [Google Scholar]
- 4.Blackwell C, Brown JD. The application of tandem-affinity purification to Candida albicans. Methods Mol. Biol. 2009;499:133–148. doi: 10.1007/978-1-60327-151-6_13. [DOI] [PubMed] [Google Scholar]
- 5.Santos MA, Tuite MF. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 1995;23:1481–1486. doi: 10.1093/nar/23.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomes AC, Miranda I, Silva RM, Moura GR, Thomas B, Akoulitchev A, Santos MA. A genetic code alteration generates a proteome of high diversity in the human pathogen Candida albicans. Genome Biol. 2007;8:R206. doi: 10.1186/gb-2007-8-10-r206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 8.Hoppen J, Dietz M, Warsow G, Rohde R, Schüller HJ. Ribosomal protein genes in the yeast Candida albicans may be activated by a heterotrimeric transcription factor related to Ino2 and Ino4 from S. cerevisiae. Mol. Genet. Genomics. 2007;278:317–330. doi: 10.1007/s00438-007-0253-x. [DOI] [PubMed] [Google Scholar]
- 9.Makio T, Nishikawa S, Nakayama T, Nagai H, Endo T. Identification and characterization of a Jem1p ortholog of Candida albicans: dissection of Jem1p functions in karyogamie and protein quality control in Saccharomyces cerevisiae. Genes Cells. 2008;13:1015–1026. doi: 10.1111/j.1365-2443.2008.01223.x. [DOI] [PubMed] [Google Scholar]
- 10.Ni J, Chen X, Yang T, Chen JY. Construction of Candida albicans two-hybrid library and screening for proteins Interacting with Crk1. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao. 2001;33:198–204. [PubMed] [Google Scholar]
- 11.Ruden DM, Ma J, Li Y, Wood K, Ptashne M. Generating yeast transcriptional activators containing no yeast protein sequences. Nature. 1991;350:250–252. doi: 10.1038/350250a0. [DOI] [PubMed] [Google Scholar]
- 12.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golemis EA, Brent R. Fused protein domains inhibit DNA binding by LexA. Mol. Cell. Biol. 1992;12:3006–3014. doi: 10.1128/mcb.12.7.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 15.Ma J, Ptashne M. A new class of yeast transcriptional activators. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 16.Legrain P, Dokhelar MC, Transy C. Detection of protein-protein interactions using different vectors in the two-hybrid system. Nucleic Acids Res. 1994;22:3241–3242. doi: 10.1093/nar/22.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 18.Estojak J, Brent R, Golemis EA. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 20.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentatous forms of Candida albicans during infection. Eukaryotic Cell. 2003;2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants. Microbiol. Mol. Biol. Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Chen J, Lane S, Liu H. A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol. Microbiol. 2002;46:1335–1344. doi: 10.1046/j.1365-2958.2002.03249.x. [DOI] [PubMed] [Google Scholar]
- 23.Gillum AM, Tsay EYH, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 24.Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell CL, Brown AJP. Expression of one-hybrid fusions with Staphylococcus aureus lexA in Candida albicans confirms that Nrg1 is a transcriptional repressor and that Gcn4 is a transcriptional activator. Fungal Gen. Biol. 2005;42:676–683. doi: 10.1016/j.fgb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Uhl MA, Johnson AD. Development of Streptococcus thermophilus lacZ as a reporter gene for Candida albicans. Micriobiology. 2001;147:1189–1195. doi: 10.1099/00221287-147-5-1189. [DOI] [PubMed] [Google Scholar]
- 27.Shen J, Guo W, Köhler JR. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect. Immun. 2005;73:1239–1242. doi: 10.1128/IAI.73.2.1239-1242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reuß O, Vik A, Kolter R, Morschhäuser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 29.Wirsching S, Michel S, Morschhäuser J. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol. Microbiol. 2000;36:856–865. doi: 10.1046/j.1365-2958.2000.01899.x. [DOI] [PubMed] [Google Scholar]
- 30.Schneider J, Schindewolf C, van Zee K, Fanning E. A mutant SV40 large T antigen interferes with nuclear localization of a heterologous protein. Cell. 1988;54:117–125. doi: 10.1016/0092-8674(88)90185-7. [DOI] [PubMed] [Google Scholar]
- 31.Nicholls S, Straffon M, Enjalbert B, Nantel A, Macaskill S, Whiteway M, Brown AJP. Msn2- and Msn4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaryot Cell. 2004;3:1111–1123. doi: 10.1128/EC.3.5.1111-1123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peeters T, Louwet W, Gelade R, Nauwelaers D, Thevelein JM, Versele M. Kelch-repeat proteins interacting with the Galpha protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc. Natl Acad. Sci. USA. 2006;103:13034–13039. doi: 10.1073/pnas.0509644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gietz RD, Schliestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Köhler JR, Fink GR. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc. Natl Acad. Sci. USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose M, Botstein D. construction and use of gene fusions to LacZ (β-galactosidase) which are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- 37.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 38.Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 39.Umeyama T, Nagai Y, Niimi M, Uehara Y. Construction of FLAG tagging vectors for Candida albicans. Yeast. 2002;19:611–618. doi: 10.1002/yea.863. [DOI] [PubMed] [Google Scholar]
- 40.Care RS, Trevethick J, Binley KM, Sudbery PE. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 1999;34:792–798. doi: 10.1046/j.1365-2958.1999.01641.x. [DOI] [PubMed] [Google Scholar]
- 41.Murad AMA, Bakar FDA, Brown AJP. Analysis of Candida albicans MET3 promoter for the expression of a C. albicans gene. Malaysian J. Biochem. Mol. Biol. 2005;12:41–47. [Google Scholar]
- 42.Tripathi G, Wiltshire C, Macaskill S, Tournu H, Budge S, Brown AJP. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 2002;21:5448–5456. doi: 10.1093/emboj/cdf507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coste AT, Turner V, Ischer F, Morschhauser J, Forche A, Selmecki A, Berman J, Bille J, Sanglard D. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2 is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics. 2006;172:2139–2156. doi: 10.1534/genetics.105.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mösch HU, Roberts RL, Fink GR. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Sacccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leberer E, Harcus D, Broadbent ID, Clark KL, Dignard D, Ziegelbauer K, Schmidt A, Gow NA, Brown AJ, Thomas DY. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl Acad. Sci. USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Csank C, Schroppel K, Leberer E, Harcus D, Mohamed O, Meloche S, Thomas DY, Whiteway M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eisman B, Alonso-Monge R, Roman E, Arana D, Nombela C, Pla J. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot. Cell. 2006;5:347–358. doi: 10.1128/EC.5.2.347-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Printen JA, Sprague GFJ. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 50.Johnsson N, Varhavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl Acad. Sci. USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nelson B, Parsons AB, Evangelista M, Schaefer K, Kennedy K, Ritchie S, Petryshen TL, Boone C. Fus1p interacts with components of the Hog1p mitogen-activated protein kinase and Cdc42p morphogenesis signaling pathways to control cell fusion during yeast mating. Genetics. 2004;166:67–77. doi: 10.1534/genetics.166.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yi S, Sahni N, Daniels KJ, Pujol C, Srikantha T, Soll DR. The same receptor, G protein and mitogen-activated protein kinase pathway activate different downstream regulators in the alternative white and opaque pheromone responses of Candida albicans. Mol. Biol. Cell. 2008;19:957–970. doi: 10.1091/mbc.E07-07-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deane CM, Salwiński Ł, Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cel. Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 54.Marcus S, Polverino A, Barr M, Wigler M. Complexes between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc. Natl Acad. Sci. USA. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 56.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.