The structure of maize VP14, a key oxidative enzyme in abscisic acid biosynthesis, provides a framework for understanding the mechanism of this important enzyme. Furthermore, the structure provides a template for the regio- and stereospecificity of VP14 as well as of other plant carotenoid cleavage dioxygenases.
Abstract
The key regulatory step in the biosynthesis of abscisic acid (ABA), a hormone central to the regulation of several important processes in plants, is the oxidative cleavage of the 11,12 double bond of a 9-cis-epoxycarotenoid. The enzyme viviparous14 (VP14) performs this cleavage in maize (Zea mays), making it a target for the rational design of novel chemical agents and genetic modifications that improve plant behavior through the modulation of ABA levels. The structure of VP14, determined to 3.2-Å resolution, provides both insight into the determinants of regio- and stereospecificity of this enzyme and suggests a possible mechanism for oxidative cleavage. Furthermore, mutagenesis of the distantly related CCD1 of maize shows how the VP14 structure represents a template for all plant carotenoid cleavage dioxygenases (CCDs). In addition, the structure suggests how VP14 associates with the membrane as a way of gaining access to its membrane soluble substrate.
INTRODUCTION
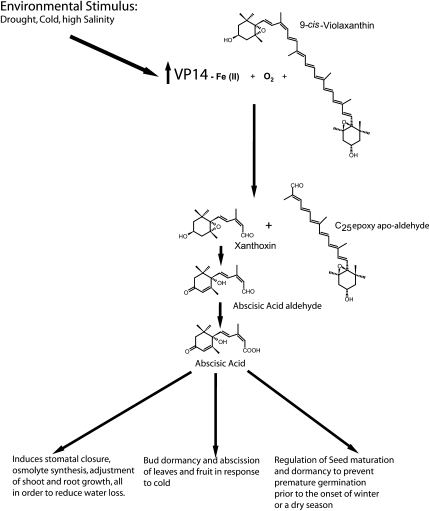
Abscicic acid (ABA) is a key plant hormone involved in a broad spectrum of growth and development processes, including seed maturation and dormancy (Phillips et al., 1997; Finkelstein et al., 2002), shoot and root growth (Sharp, 2002), and responses to drought (Zeevaart and Creelman, 1988; Hasegawa et al., 2000) and nutrient depletion (Gomez-Cadenas et al., 2000) (Figure 1). For example, in conditions of drought, increases in ABA levels lead to restructuring of the cell cytoskeleton and subsequent closure of stomata (Schwartz et al., 1994), biosynthesis of osmolytes (glycine, betaine, etoine, sugar alchohols such as glycerol, and complex sugars) (Hasegawa et al., 2000), and transcription of stress response genes (Bray, 2002).
Figure 1.
Involvement of Abscicic Acid Biosynthetic Pathway in Stress Responses in Plants.
Drought, cold, and/or high salinity induce an increase in VP14 activity. The resulting elevation of ABA levels leads to the appropriate biological responses.
In maize (Zea mays), the world’s most productive cereal crop (United Nations, 2007), vivaparous14 (VP14), a 9-cis-epoxycarotenoid dioxygenase (NCED), catalyzes the rate-limiting step in ABA biosynthesis (Tan et al., 1997; Qin and Zeevaart, 1999)—the oxidative cleavage of the 11,12 carbon-carbon double bond of either 9-cis-violaxanthin or 9-cis-neoxanthin (Schwartz et al., 1997). The C15 aldehyde, xanthoxin, is oxidized and converted through two subsequent reactions to the biologically active ABA (Figure 1) (Taylor et al., 2000; Schwartz et al., 2003a). In conditions of environmental stress, induction of transcription of the NCED gene(s) follows within 15 to 30 min of the onset of the stress (Tan et al., 1997; Qin and Zeevaart, 1999; Thompson et al., 2000). Thus, NCEDs, including VP14, are key regulators that determine ABA levels (Schwartz et al., 2003a), which in turn control ABA-regulated processes. This involvement makes VP14 an attractive target for the development of novel chemical agents and genetic modifications to control ABA levels and improve plant productivity, performance, and architecture (Cutler and Krochko, 1999; Qin and Zeevaart, 1999) (Figure 1). Studies of the mutant vp14-2274 also implicate VP14 in the synthesis of strigolactones (Matusova et al., 2005), signaling molecules involved in shoot branching (Gomez-Roldan et al., 2008; Umehara et al., 2008), presymbiotic growth of arbuscular mycorrhizal fungi (Besserer et al., 2006), and parasitic weed germination (Hauck et al., 1992; Siame et al., 1993).
Recently, progress was made in identifying downstream processes involving the ABA receptors and their partners, including the determination of the structure and the mechanism of activation of the ABA receptor (Miyazono et al., 2009; Nishimura et al., 2009). These results, along with those studies into the physiological effects of ABA, laid a foundation for a more complete understanding of ABA physiology. Yet, although a mechanism has been proposed for the committed step in ABA biosynthesis, the dioxygenase cleavage of the 9-cis-carotenoid double bond (Taylor et al., 2000; Schwartz et al., 2003b), the structure and the nature of the determinants of specificity of the dioxygenase remained unknown.
Using the structure of VP14, determined here to 3.2-Å resolution, we identified amino acid residues that help position the substrate and determine which bond is cleaved. These residues were compared and contrasted with those present in the broadly defined family of plant carotenoid cleavage dioxygenases (CCDs) (von Lintig and Vogt, 2004), which process a wide variety of carotenoid substrates and are involved in the biosynthesis of several plant hormones (Gomez-Roldan et al., 2008; Umehara et al., 2008; Vogel et al., 2008). We tested the importance of these residues by studying the effects of mutations of residues identified by homology with VP14 in a distantly related family member, maize CCD1. Furthermore, the structure led to the identification of a structural motif in the sequence of many plant CCDs that is ideally suited for membrane association, placing these enzymes in close proximity with their lipid-soluble carotenoid substrates.
RESULTS
Overall Structure
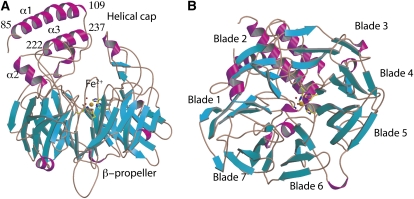
The structure of VP14 (residues 84 to 604 out of 604), determined by multiple wavelength anomalous dispersion (MAD; Hendrickson, 1991), was refined to an Rwork/Rfree of 23.2/28.4% (Table 1; see Methods). VP14 folds as a seven-blade β-propeller with four α-helical inserts that form an α-helical domain on top of the β-propeller (Figure 2). Five of the seven blades of the β-propeller have four locally connected antiparallel strands, while the first and seventh blades have five antiparallel strands. In blades one and seven, the N terminus provides the outermost β-strand (residues 132 to 137 in blade 1; residues 140 to 143 in blade 7), and the C terminus provides the innermost β-strand to blade one. As suggested by Kloer and Schulz (2006), the β-propeller portion of the structure may be a conserved characteristic throughout the CCD family of enzymes, as it is present in both the prokaryotic apocarotenoid 15,15’-oxygenase (ACO) (Kloer et al., 2005) and the eukaryotic VP14.
Table 1.
Data Collection, Phasing, and Refinement Statistics (MAD)
| Data Set | VPSE_inflectiona | VPSE_peak | VPSE_rem | Native |
| Resolution (Å) | 50-3.6 | 50-3.6 | 50-3.6 | 50-3.2 |
| Space group | p6322 | p6322 | p6322 | p6322 |
| Cell dimensions | ||||
| a, b, c (Å) | a=b=161.4 c=149.9 | a=b=161.4 c=149.9 | a=b=161.4 c=149.9 | a=b=161.9 c=149.7 |
| α, β, γ (°) | α=β=90 γ=120 | α=β=90 γ=120 | α=β=90 γ=120 | α=β=90 γ=120 |
| Completeness (%) | 100 (100) | 100 (100) | 100 (100) | 99.9 (99.9) |
| Rsymb | 10.3 (56.4) | 10.5 (46.2) | 11.2 (65.2) | 7.5 (57.3) |
| Redundancy | 21.1 (21.8) | 21.1 (21.8) | 21 (21.6) | 18.5 (18.2) |
| I/sI | 35.6 (6.2) | 41.4 (7.6) | 33.8 (5.2) | 27.1 (3.3) |
| Statistics for refinement | ||||
| Resolution (Å) | 50-3.2 | |||
| No. reflections | 18,636 | |||
| Rwork/Rfreeb | 24.2/28.1% | |||
| No. atoms | 4031 | |||
| Protein | 4018 | |||
| Ligand/ion | 27 | |||
| Water | 11 | |||
| Metals | 1 Fe2+ | |||
| B-factors | 51.9 | |||
| Protein | 51.8 | |||
| Ligand/ion | 77.2 | |||
| Water | 52.1 | |||
| Root mean square deviations | ||||
| Bond lengths (Å) | 0.01 | |||
| Bond angles (°) | 1.3 |
VPSE stands for VP14 protein with all Met residues replaced by selenomethionines.
Rsym = ∑hkl∑j|Ij-<I>|/∑hkl∑J Ij, where <I> is the mean intensity of j observations from a reflection hkl and its symmetry equivalents.
Rwork = ∑hkl ||Fobs| − k|Fcalc| / ∑hkl |Fobs|, where k is a scale factor. Rfree for 5% of all data that were not used in refinement.
Figure 2.
Structure of VP14.
(A) Side view of VP14 with β-strands shown in cyan, α-helices in magenta, and loops in brown.
(B) Top view rotated 90° toward the viewer from (A) showing the seven blades of the propeller with the iron atom at the center.
Catalytic Iron
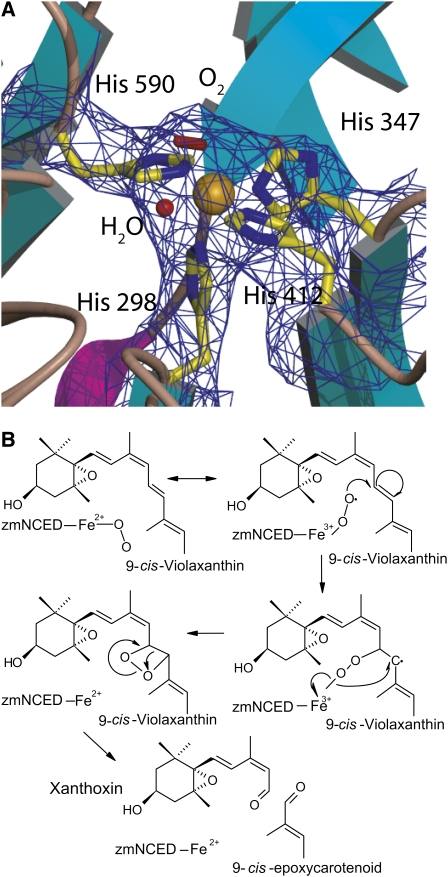
As in other β-propeller structures, a long tunnel surrounded by the blades runs through the center of the structure from one end to the other. The Fe2+ required for dioxygenase activity is located inside the tunnel on the central axis of the β-propeller. It is bound octahedrally by four His residues located on the innermost strands of blades 1, 3, 4, and 5 (His-590 in blade 1, His-298 in blade 3, His-347 in blade 4, and His-412 in blade 5), a water molecule, and an elongated density interpreted as corresponding to a molecule of dioxygen (Figure 3A). His residues 347, 412, and 590 and the water molecule occupy four positions in a plane (distance Fe-Nε = 1.88, 1.86, and 1.87 and Fe-O 2.05 Å, respectively). His-298 (distance Fe-Nε = 1.86 Å) and the dioxygen molecule occupy the positions below and above this plane. His residues 347, 412, and 590 are held in position by hydrogen bonds to Glu residues 264, 477, and 530 (distances O-Nε = 3.24, 2.68, and 3.59, respectively). This type of Fe2+ coordination is found in only in a handful of enzymes, such as superoxide reductase, fumarate reductase, and photosystem II (Gillmor et al., 1997; Lancaster et al., 1999; Katona et al., 2003; Ferreira et al., 2004; Katona et al., 2007). The presence of Fe2+ was confirmed by soaking crystals with orthophenanthroline, which turned the crystals a deep red (Cavallini et al., 1969), indicating the presence of Fe2+.
Figure 3.
Fe2+ Catalytic Binding Center with Oxygen.
(A) 2Fo-Fc electron density map showing the Fe2+ ion (gold) and its octahedral coordination by His residues 298, 347, 412, and 590, a molecule of water, and dioxygen.
(B) Proposed scheme of the reaction.
Mechanism
The putative oxygen molecule is coordinated to the iron in an angular end-on geometry as originally proposed by Pauling (1964), forming an Fe-O-O angle of 131° (proximal oxygen distance Fe-Op = 2.04 Å; distal oxygen Fe-Od = 2.97 Å). The identity of the dioxygen was confirmed by calculating several types of omit maps. Attempts to refine the structure with a water molecule/hydroxide ion instead of dioxygen resulted in large excess positive density in the Fobs-Fcalc maps. Although other interpretations of this density are possible, all our tests suggest that molecular oxygen is the most likely ligand at this position from a crystallographic as well as from a mechanistic point of view.
The observed coordination of the molecular oxygen is compatible with a cleavage mechanism that involves a dioxocyclobutanyl (dioxetane) intermediate (Figure 3B). The need for a highly reactive oxygen species is in agreement with the fact that the nonenzymatic reaction requires singlet oxygen to carry out a 1,2 cycloaddition to carotenoids and other alkenes to form a dioxetane intermediate that collapses into the requisite aldehyde products (Figure 3B) (Que and Ho, 1996).
Also visible in the structure is a shaft lined with charged residues that begins at the bottom of the β-propeller base and ends at the metal coordination center. Kloer et al. (2005) proposed that, in ACO, this tunnel is the path for oxygen access to the active site. In VP14, the tunnel ends short of the iron, blocked by His-298.
Substrate Model
Numerous experiments aimed at obtaining crystalline complexes of VP14 with substrates or inhibitors by soaking and cocrystallization yielded crystals that failed to show density for the ligands. This result is not unusual with enzymes that use large hydrophobic substrates. Nonetheless, the structure of the free enzyme provides strong indications of how the substrate might bind in the active site.
To identify residues that may contribute to substrate specificity and cleavage in VP14, 9-cis-violaxanthin was modeled into the hydrophobic portion of the substrate tunnel, and the energy of the initial model was minimized. The model revealed that the methylenecyclohexane group (carbons 1’ to 6’) (IUPAC-IUB, 1974) of the carotenoid, distal to the 9-cis bond, is within 3 to 5 Å of Leu-218, Leu-227, Val-315, Ile-316, Leu-367, and Met-370. These residues do not make any explicit hydrogen bonds with the substrate molecule but appear to act together to hold the molecule in place through nonspecific van der Waals (hydrophobic) interactions. Over the distance of the isoprene chain from carbon 15’ to 7’, the carotenoid is within 3 to 5 Å of hydrophobic residues Ala-214 and Ile-215, as well as the methylene groups of two hydrophilic amino acids: the β-carbon of Asp-265 and γ-carbon of Glu-264. Leu-218, Met-345, Phe-365, and Val-377 also line the hydrophobic cavity around this portion of the carontenoid, but these residues are 5 to 7 Å from the modeled substrate. Carbon 15 to carbon 9, the key section of the carotenoid that includes the 9-cis bond as well as the 11,12 carbon-carbon double bond cleavage site, are held over the catalytic iron and the oxygen by three Phe residues: Phe-411, Phe-171, and Phe-589. Finally, the 9-cis bond and its proximal methylenecyclohexane group are surrounded by Phe-127, Leu-170, Met-432, Val-478, Trp-501, and Pro-502. These residues not only make nonspecific hydrophobic interactions with the substrate, but also create a key backstop to the molecule such that 11,12 carbon-carbon double bond aligns exactly over the catalytic iron and the oxygen. Val-478 appears to be vital to this arrangement, also providing enough space to accommodate the methyl group protruding from carbon 9. The absence of basic and acidic residues in proximity to the scissile bond further supports the dioxetane-intermediate mechanism.
The Helical Domain and Membrane Penetration
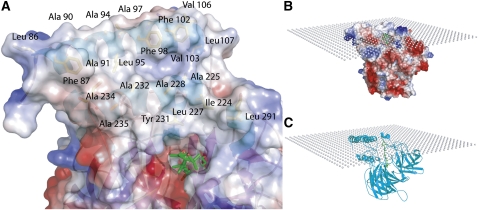
The α-helical domain of VP14 is dominated by two antiparallel α-helices, α1 and α3, which are formed by amino acids 88 to 108 and 222 to 237 (Figure 2A). Analysis of the surface properties of these two helices reveals a hydrophobic patch of 2226 Å2 formed by 25 hydrophobic residues (Leu-86, Phe-87, Ala-90, Ala-91, Ala-93, Ala-94, Leu-95, Ala-97, Phe-98, Gly-101, Phe-102, Val-103, Val-106, Leu-107, Pro-110, Ile-224, Ala-225, Leu-227, Ala-228, Leu-229, Tyr-231, Ala-232, Ala-234, Ala-235, and Gly-237) that may penetrate the membrane beyond the head groups of the membrane phospholipids and interact with fatty acid residues in the membrane interior (Figure 4A). With VP14 in this position, a crown of positive, neutral, and negatively charged residues (Asn-85, Gln-88, Arg-89, Asp-97, Glu-101, Asn-106, Arg-110, Tyr-232, Asp-241, and Arg-373) interact with the charged surface of the membrane directly or through water-mediated contacts. If helix α1 is removed, an area of positive surface charge is exposed, and the hydrophobic patch is disrupted. This observation explains why in genetic and cell biological studies, deleting or disrupting this first helix results in a loss of interaction of VP14 with the thylakoid membrane and loss of ABA biosynthesis (Tan et al., 2001).
Figure 4.
VP14 Helical Domain and Its Interaction with the Thylakoid Membrane.
(A) VP14 is colored according to its electrostatic surface potential and is labeled with key residues depicting the hydrophobic patch used for membrane penetration (red represents negatively charged and blue represents positively charged residues). Key hydrophobic residues from helices α1 and α3 are shown in yellow below the surface (e.g., Phe-87).
(B) Electrostatic surface potential representation of VP14 penetrating the membrane surface, which is represented by a plane.
(C) Same as (B) with the protein shown using a ribbon diagram.
To investigate whether these helices actually could penetrate the membrane, we implemented a modification of the computational/structural approach developed by Mosberg and coworkers (Lomize et al., 2006) to calculate the ΔGtransfer of membrane penetration. Using this procedure, we found for VP14 a maximal membrane penetration of 7 Å with a ΔGtransfer = −8 kcal/mol for VP14 (Figures 4B and 4C). As a positive control, we calculated ΔGtransfer for carnitine palmitoyltransferase 2 (CPT-2) and obtained a value of −9.7 kcal/mol, in good agreement with the reported value of ΔGtransfer = −8.6 (kcal/mol) (Lomize et al., 2006).
The feature proposed for the interaction of CPT-2 with the membrane consists of two antiparallel helices, in a similar arrangement to that found in VP14. It was proposed that in CPT-2, these two helices penetrate the membrane, allowing the enzyme to extract a lipophilic substrate (Rufer et al., 2007). Interestingly, VP14 helix α3 is adjacent to the opening to the substrate tunnel, which is lined by hydrophobic aromatic residues. These observations, coupled with the favorable ΔGtransfer, show how VP14 could use a mechanism of substrate accessibility similar to that proposed for CPT-2. Penetration of the membrane by the hydrophobic patch of VP14 would place the substrate tunnel in close proximity to carotenoids in the thylakoid membrane. Importantly, 9-cis-neoxanthin and 9-cis-violaxanthin, the primary substrates of VP14, are 10 times more abundant in the thylakoid membrane than in other plant membranes (Parry and Horgan, 1991). This mechanism may be consistent throughout the CCD family; a hydrophobic patch, albeit significantly smaller, was observed in the prokaryotic ACO structure (Kloer et al., 2005) (nine residues covering 800 Å2 versus 25 residues and 2226 Å2 in VP14).
VP14 as the Prototype
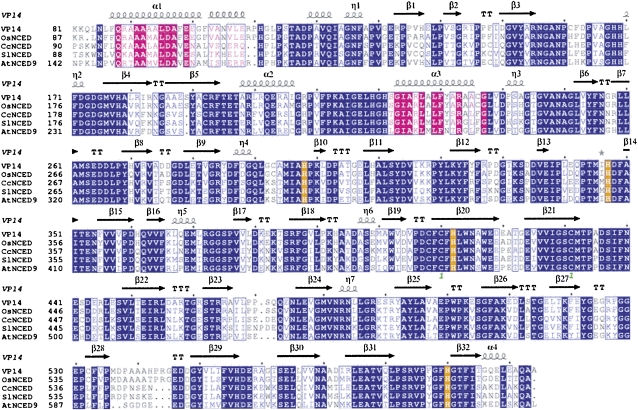
Comparison of the VP14 sequence with those of other plant NCEDs shows 60 to 85% identity across the plant kingdom—from close relatives such as Oryza sativa to distant relatives such as that of Citrus clementina and Arabidopsis thaliana (Figure 5). In addition to similarities in the β-propeller, these sequences show high conservation of the two long antiparallel α-helices that we propose are involved in membrane penetration.
Figure 5.
Sequence Alignment of VP14 and NCED Proteins from Several Plants.
Maize VP14 is aligned with NCED proteins from rice (Os), C. clementina (Cc), tomato (Solanum lycopersicum; Sl), and Arabidopsis (At). Blue background shows sequence identity, and purple background shows identity in the α1 and α3 helices. Blue letters show sequence similarity, and purple letters show similarity in the α1 and α3 helices. Orange background shows identity for the His residues coordinating the catalytic iron. The green number 1 under C410 and C430 indicates a disulfide bond, and T represents turns. The alignment was done with ClustalW and the figure with ESPript (Gouet et al., 1999).
VP14 shows an overall lower, but significant, sequence identity (58 to 16%) with Z. mays CCD1 (Zm CCD1) and the complete family of CCD enzymes of Arabidopsis (see Supplemental Figure 1 online), especially the conservation of key structural elements. This conservation is interesting because members of the CCD family, which encompasses the NCED family, process a more diverse array of carotenoid substrates. For example, maize CCD1 cleaves multiple linear and cyclic carotenoids (Vogel et al., 2008), whereas Arabidopsis CCD7/MAX3 is involved in the synthesis of strigolactones, novel plant signaling molecules that regulate shoot branching (Booker et al., 2004; Gomez-Roldan et al., 2008; Umehara et al., 2008). The significant level of sequence identity between VP14, NCEDs, and the CCDs family in plants makes the VP14 structure an ideal prototype of all these enzymes. VP14 also belongs to a more broadly defined family of CCDs present in bacteria, plants, and animals (von Lintig and Vogt, 2004). These enzymes range from the apocarotenoid-15,15’-oxygenase of Synechocystis spp to the human β-carotene 15,15’-monooxygenase, the enzyme responsible for the production of retinol (vitamin A) (von Lintig and Vogt, 2000; von Lintig et al., 2001).
Modeling of a Distant VP14 Relative: Regions Responsible for Specificity Differences
Using the coordinates of the VP14 substrate model as a template, we constructed a homology model of Zm CCD1. This enzyme was chosen because, in contrast with VP14, its substrate is commercially available, permitting extensive investigation of the effects of selective mutations (see below). A comparison of the VP14 structure and the Zm CCD1 model suggests that the differences in substrate specificity reside in three key regions of the structures.
The first involves the substitution of a Val residue (Val-478) on the innermost strand of blade 6 of VP14 by a Phe (Phe-409) in Zm CCD1. As was observed in the model of VP14 subtrate, this Val creates a small cleft in VP14 that accommodates the protruding methyl group on carbon 9 of the isoprene chain and allows the 11,12 bond to fit over the Fe2+ (Figure 6). The presence of a Phe in Zm CCD1 interferes with this arrangement. This structural difference is highlighted when comparing the sequence of VP14 with those of Zm CCD1 and of the complete family of CCDs from Arabidopsis (At). At NCEDs 2, 3, 5, 6, and 9 have either an Ala or Ile at this position, whereas At CCDs 4, 1, 7, 8, and Zm CCD1 have a Phe or Met (see Supplemental Figure 1 online). This is in agreement with the known activity of these proteins: while At NCEDs 2, 3, 5, 6, and 9 carry out 9-cis-carotenoid cleavage (Iuchi et al., 2001; Tan et al., 2003), At CCDs 4, 1, 7, and 8 and Zm CCD1 are neither involved in ABA biosynthesis nor in the cleavage of other 9-cis-carotenoid (Sorefan et al., 2003; Booker et al., 2004; Vogel et al., 2008).
Figure 6.
Binding Site of VP14 and zmCCD1.
(A) Ribbon diagram of VP14 with a view of 9-cis-violoxanthin (purple) modeled into the substrate binding pocket. Residues equivalent to those mutated in Zm CCD1 are shown in yellow, and His residues binding the catalytic iron (orange) are shown in brown.
(B) Substrate binding pocket of VP14 with electrostatic surface and models of both its 9-cis-violoxanthin substrate (purple) and Zm CCD1 β-apo-8’-carotenal substrate (green).
(C) Electrostatic surface of Zm CCD1 in the same orientation with both its β-apo-8’-carotenal substrate and the VP14 substrate 9-cis-violoxanthin shown as in (B).
The second region involves a loop on the back side of the substrate pocket (residues 499 to 503 in VP14 and 432 to 434 in Zm CCD1) (Figure 6) and a substitution of Leu-170 in VP14 by a Trp (Trp-104) in Zm CCD1. In VP14, this loop and the Leu form a pocket that can accommodate the second ring of the carotenoid in such a way that the 11,12 carbon-carbon double bond of the 9-cis-epoxycarotenoid is close to the Fe2+. This arrangement is not possible in Zm CCD1.
The third region, a feature conserved in the two enzymes, is comprised of three Phe residues (Phe-171, Phe-411, and Phe-589 in VP14 and Phe-105, Phe-343, and Phe-532 in Zm CCD1) important for caging the substrate over the catalytic iron. Comparison of the structures of VP14 modeled with a 9-cis-carotenoid and ACO with substrate reveals that the same three regions account for their differences in substrate specificity.
CCD1 Mutational Studies
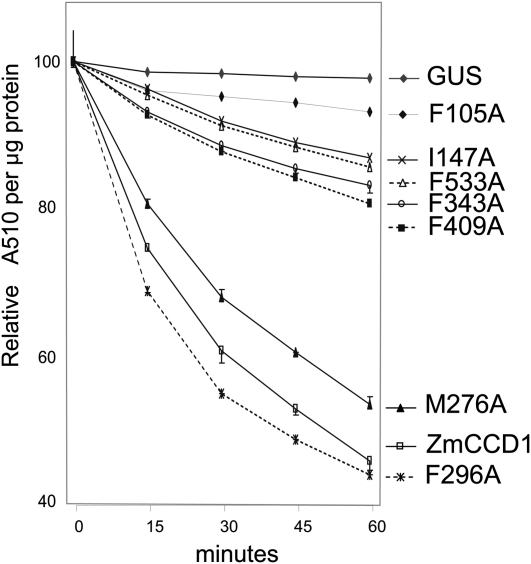
To test the model of Zm CCD1, we mutated to Ala residues indentified to be important for determining specificity and measured the activity of the mutated proteins toward a β-apo-8’-carotenal. Substitution of Phe-105, Phe-343, and Phe-533 of CCD1 by Ala decreased activity to <10, 15, and 30% of the wild type, respectively (Figure 7). As these experiments were carried with high substrate concentrations (72 μM versus a Km of 0.98 μM for the wild type), this result suggests that these effects are a consequence of a reduction in kcat. Detailed kinetic analysis of the F343A mutant showed a 90% decrease in Vmax and threefold increase in Km (Table 2) compared with the wild type that under saturation conditions (72 μM versus a Km mutant of 8.9 μM) predicts an 11% residual activity, which is in good agreement with the 15% of wild-type activity observed in the high substrate concentration experiments. These results are compatible with a 90% reduction in Vmax for P105A and a 67% reduction for P533A. These results underscore the role of these three residues in catalysis, probably by holding the substrate over the active site. Substitution I147A (Ile-215 in VP14) also produced a reduction in activity to 15% of that of the wild type. This corresponds well with the models, as Ile-147 is spatially next to Phe-105 and may be contributing to caging the substrate as well.
Figure 7.
Assays of Activity for Proteins with Various CCD1 Point Mutations.
The consumption of substrate was monitored every 15 min at 510 nm to measure enzyme activity. Assays were performed in triplicate. (Error bars represent the difference between the largest and the smallest value of the triplicate experiments.) The absorbance value at time zero was set to 100, and all other values were scaled accordingly.
Table 2.
Kinetic Parameters of Zm CCD1 Mutants
| Proteina | Vm (pmol s−1 mL−1)b | Km (μM)b |
| Zm CCD1 | 8.45 (±0.83) | 2.95 (±0.09) |
| F343A | 0.97 (±0.08) | 8.90 (±1.7) |
Recombinant protein concentrations in Escherichia coli extracts were normalized by protein immunoblots with anti-glutathione S-transferase antibody.
Kinetic parameters were estimated by nonlinear least square fit to the Michaelis-Menten equation using R software (www.r-project.org). Standard errors for the estimates are in parentheses.
Mutations M276A and F296A in CCD1 (Met-345 and Phe-365 in VP14) did not result in a significant drop in activity. These results are also explained by the models, which show that these residues are 5 to 7 Å from the substrate on the opposite side of the hydrophobic cavity. Mutation of Phe-409 and Phe-533 to Ala (Val-478 and Phe-589 in VP14) resulted in a reduction of activity to 30 and 15% of that of the wild type, reinforcing the observation that the back side of the pocket is important for creating contours to ensure that the C11,C12 carbon-carbon double bond aligns over the catalytic site (Figures 6 and 7). The effect of these mutations on the activity of CCD1 validates the use of VP14 as a template for mapping important residues in the substrate tunnel on distant plant CCDs and for providing a rationale for understanding their substrate specificity.
DISCUSSION
VP14 catalyzes the first committed step in the biosynthesis of ABA in maize: cleavage of the 9-cis-violoxanthin 11,12 double bond by incorporation of the two oxygen atoms of O2 into the two products. The structure of VP14 determined here is fully compatible with the dioxetane mechanism suggested by the uncatalyzed reaction performed with singlet oxygen. The end-on Fe2+-O2 species is in resonance with Fe3+-O2− a radical superoxide ideally suited to attack the double bond resulting in a radical species that can react with the second oxygen to form the dioxetane species that spontaneously produces the two aldehydes. The absence of basic and acidic residues in the proximity of the scissile bond argues against mechanisms that would require proton transfer steps.
VP14 has two domains: a seven-blade β-propeller domain and a second domain comprised largely of α-helices. The β-propeller portion of the structure, present in both the prokaryotic ACO and the eukaryotic VP14 (Kloer et al., 2005), is a conserved characteristic of the CCD family of enzymes, while the helical domain may be a structural feature that differentiates plant carotenoid-cleaving dioxygenases from other members of the family. Our analysis of this domain, in conjunction with previous biochemical studies (Tan et al., 2001), provides a possible mechanism by which the enzyme might penetrate the surface of the thylakoid membrane and extract substrate through a tunnel that extends into the catalytic center.
The broader CCD family plays a vital role in the biosynthesis of other key signaling molecules that regulate growth and development, including strigolactone-related compounds implicated in control of branching (Booker et al., 2004; Gomez-Roldan et al., 2008). Structural comparison of VP14 and a model of CCD1 combined with mutagenesis analysis revealed differences in the active sites, indicating that Phe-171, Phe-411, and Val-478 in VP14 and their equivalent residues in CCD1 are important for discriminating between the 9-cis-carotenoid substrate and other carotenoids. The VP14 structure reported here can be considered a template for all related plant CCDs and provides a blueprint for studies of function in this key family of enzymes. Our studies also show how our VP14 substrate model may be useful not only for the rational design of novel chemical agents but also for engineering of genetically modified organisms for the purpose of controlling ABA levels, an obvious target for crop improvement (Cutler and Krochko, 1999; Qin and Zeevaart, 1999).
METHODS
Expression of Partial VP14
Using the original full-length Zea mays vp14 gene in a pGEX-2T vector (Pharmacia), the DNA sequence representing amino acid residues 75 to 604 (residues 1 to 75 are a signal sequence and were not included in the construct) was subcloned by PCR into pMal-c2x-hv (New England Biolab) using BamHI and EcoRI restriction enzymes (5′ primer, 5′-GCGGCGACTGGATCCAGGAAAGCGGAGGGC-3′; 3′ primer, T7 terminator). M15-(pREP) cells (Qiagen) were transformed with this construct and grown aerobically in 2xTY broth (per liter = 16 g Bacto-tryptone, 10 g yeast extract, and 5 g NaCl) at 15°C to an A600 of ~0.6. Expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside, cells were grown for 16 h, and the cell pellet was frozen at −80°C. All mutants and constructs were expressed in this manner, except for selenomethionine-substituted protein. Selenomethionine-substituted protein in which all Met residues were replaced with selenomethionines was expressed as discussed by Hendrickson (Leahy et al., 1994).
Purification
Cell pellets were resuspended in 20 mM NaH2PO4, pH 7.5, 500 mM NaCl, 15 mM imidazole, 5 mM β-mercaptoethanol buffer (Ni2+ affinity binding buffer), lysed via microfluidization, and centrifuged at 20,000g for 30 min at 4°C. The soluble cytoplasmic fraction was then purified with a Ni2+ affinity column, using an imidazole gradient of 10 to 250 mM as the eluant. The N-terminal tag was cleaved overnight using 100 μL of 1 mg/mL TEV protease to 50 mL Ni2+ affinity column elution at room temperature in a buffer of 50 mM Tris-HCl, 50 mM NaCl, and 1 mM Tris 2-carboxyethyl phosphine, pH 7.5. VP14 in the buffer from the TEV digestion was further purified on an anion exchange column (source 15Q beads from GE Healthcare) using an NaCl gradient of 0 to 1 M as the eluant. Undigested protein was eliminated on a second Ni2+ affinity column using the same. The flow-through was loaded onto a 26/60 Sephacryl S-200 column (GE Healthcare Life Sciences) using a buffer of 50 mM Tris-HCl, 50 mM NaCl, and 1 mM Tris 2-carboxyethyl phosphine, pH 7.5 (crystallization buffer) to elute the protein. The protein was then concentrated to 10 mg/mL using a Vivaspin 20 concentrator (Sartorius Stedim Biotech). Production of the protein with a selenomethionine substitution followed the same protocol.
Crystallization
VP14 was crystallized at 20°C by vapor diffusion in hanging drops of 2 μL of protein at 10 mg/mL in crystallization buffer with an equal volume of a reservoir solution of 0.1 M Na-HEPES, pH 7.5, 1.5 M Li2SO4, 2% hexanediol, and 4% dioxane. Single crystals belonging to P6322 space group grew in 1 to 5 d and contained one monomer in the asymmetric unit
Surface Site Mutagenesis for KE381AA
Because the initial crystallization attempts did not lead to diffracting crystals, the VP14 sequence was analyzed using the Surface-Entropy-Reduction prediction (SERp) server (Goldschmidt et al., 2007). The program predicted Lys-381 and Glu-382 to be in loop regions of the protein and recommended mutation of these residues to Ala to improve the possibility of crystal contacts. These residues were mutated to Ala using the following primer (coding mismatch underlined): 5′-GCCCGTGGTGCTGGACGCGGCGAAGACGTCGCGGTT-3′. The wild-type VP14 gene in the pMal-c2x-hv vector was used as the template for the PCR to generate the double mutant, following the standard QuickChange procedure for site-directed mutagenesis (Stratagene). These mutations lead to diffracting crystals.
Data Collection, Structure Determination, and Refinement
Data to a resolution of 3.6 Å were collected from selenomethionine-substituted crystals at the Brookhaven National Laboratory beamline X6A and processed with the HKL suite (Otwinowski and Minor, 1997). MAD phases (Hendrickson, 1991) were calculated using the program SOLVE (Z score of 36.37, and figure of merit of 0.36 at 3.92 Å) and were improved by density modification with the program RESOLVE (Terwilliger, 2003). The model was built interactively with the program O (Jones et al., 1991; Jones and Kjeldgaard, 1997) and was refined using REFMAC 5.0 (Collaborative Computational Project, Number 4, 1994) with individual restrained B-factors and a solvent correction using the Babinet option. Anisotropic refinement using translation, libration, and screw rotation (TLS) of rigid bodies was performed using each domain as a TLS group (CCP4, 1994). Refinement was monitored using R-free calculated with 5% of the data set aside for cross validation (Brunger, 1997). Drawings were prepared with PYMOL (DeLano, 2002), MOLSCRIPT (Kraulis, 1991), ISIS/DRAW 2.4 (MDL Information Systems, www.mdli.com), and ESPRIPT (Gouet et al., 1999).
Calculation of the ΔG of Transfer into the Membrane
ΔGtransfer, the free energy of insertion of the protein into the membrane from an aqueous solution, was calculated as described by previously (Lomize et al., 2006). Solvation parameters for atoms were obtained from Lomize et al. (2006). The transfer energy for VP14 at different levels of membrane penetration was then calculated by considering the protein as a rigid body and moving it into the membrane in 0.2-Å steps. At each step, the protein was tilted in 0.5° increments in x and y coordinate planes, with a limit of ±5°. The optimal position and orientation was found as the result of the described grid search (Lomize et al., 2006).
Modeling of a VP14 9-cis-Carotene Complex
The VP14 9-cis-carotene model complex was built by manually docking the carotenoid into the active site using the Molecular Operating Environment (MOE; Chemical Computing Group). The coordinates of the resulting model were energy minimized locally (9-cis-carotene and residues within 6 Å were allowed to move subject to tether) with the suite LigEx of MOE. The final model showed no unfavorable interactions.
Model of Maize CCD1
The three-dimensional model of Zm CCD1 was constructed with the MOE program using the structure of VP14 as a template. Following alignment of the two sequences, the homology modeling algorithm was used to build and energy minimize a model of CCD1. The scoring of the model was done using a MOE function that calculates a pseudoenergy of folding as the –kTlog (probability).
Site-Directed Mutagenesis CCD1
Mutations of CCD1 were made using the QuickChange Lighting site-directed mutagenesis kit (Stratagene) following the manufacturer’s instructions. Recombinant proteins were expressed using pGEX2T-ZmCCD1 (Vogel et al., 2008). Mutagenic primers were designed using the QuickChange Primer Design Program (http://www.stratagene.com/qcprimerdesign). The following purified primers were used, and modified codons are underlined and the nucleotide changes are indicated in bold: F296A-sense, 5′-TTATGGACCTCCCTTTATTGGCCCGACCAAAGGAAATGGTG-3′; F296A-antisense, 5′-CACCATTTCCTTTGGTCGGGCCAATAAAGGGAGGTCCATAA-3′; F296L-sense, 5′-TTATGGACCTCCC-TTTATTGTTACGACCAAAGGAAATGG-3′; F296L-antisense, 5′-CCATTTCCTTTGGTCGTAACAATAAAGGGAGGTCCATAA-3′; F343A-sense, 5′-ATCAGATGGTTTCAACTCCCTAATTGTTTCATAGCCCTAATGCTAATGCTT-3′; F343A-antisense, 5′-AAGCATTAGCATTATGGGCTATGAAACAATTAGGGAGTTGAAACCATCTGAT-3′. Mutations were confirmed by complete DNA sequencing of the resulting mutant ccd1 genes.
Recombinant Protein Expression and Extraction of Maize CCD1
pGEX2T-ZmCCD1 and mutant plasmids were cotransformed into BL21(DE3) competent cells (Clontech) with the pGRO7 chaperone (Takara Bio). After 21 h of induction at 20°C, protein was extracted using the B-PER reagent (Pierce) and quantified using the Bio-Rad protein reagent. Lysate was stored on ice at 4°C.
Immunoblot Analysis of CCD1
Protein extracts were resolved by SDS-PAGE using 10% Ready Gel Tris-HCl (Bio-Rad), and immunoblot analysis and estimation of CCD1 concentration by densitometry was performed as previously described (Vogel et al., 2008).
CCD1 Enzyme Assays
Cleavage of β-apo-8’-carotenal (Sigma-Aldrich) by CCD1 was assayed as described (Vogel et al., 2008) with the following modifications. Substrate dilutions were made from a stock solution (50 μM) prepared in buffer containing 1% Tween 20. Triplicate assays were initiated by addition of 100 μg protein extract followed by measurement of absorbance at 510 nm. Production of C17 dialdehyde was confirmed by HPLC as described (Vogel et al., 2008). Absorbance was measured at 30-s intervals for up to 6 min. Reaction velocities were calculated from the slope of absorbance change in the linear range determined for each mutant protein. Kinetic parameters were estimated by nonlinear least-square fit to the Michaelis-Menten equation using R software.
Activity assays in Figure 7 were performed with 72 μM β-apo-8'-carotenal and ~300 μg total protein after normalization of relative recombinant protein based on densitometry of immunoblots. To compare the various mutations, the absorbance value at time zero was set to 100 and all other values were scaled accordingly. All measurements were performed in triplicate with errors bars shown in the figure. Analysis of the completed assays via HPLC revealed that the change in absorbance matched the disappearance of substrate and the production of the C17 product.
Accession Numbers
Sequence data from this article can be found in the GeneBank database under the following accession numbers: Z. mays VP14 (AAB62181.2) and CCD1 (AAZ22349.1), Oryza sativa NCED (NP_001050765.1), Citrus clementina NCED (ABA43901.1), Solanum lycopersicum NCED (CAB10168.1), and Arabidopsis thaliana NCED9 (NP_177960.1). Atomic coordinates and structure factors are deposited in the Protein Data Bank with accession code 3NPE.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Sequence Alignment of VP14 with CCDs from Arabidopsis thaliana and Zm CCD1.
Supplementary Material
Acknowledgments
Use of beamline X6A and X25 of the National Synchrotron Light Source (Brookhaven National Laboratory) and the SGX mail-in program of the Advanced Photon Source (Argonne National Laboratory) are gratefully acknowledged. These studies were supported by Grant MCB-0450465 to L.M.A. from the National Science Foundation and by Grant 0749266 to H.J.K. and D.R.M from the National Science Foundation.
References
- Besserer A., Puech-Pagès V., Kiefer P., Gomez-Roldan V., Jauneau A., Roy S., Portais J.C., Roux C., Bécard G., Séjalon-Delmas N. (2006). Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J., Auldridge M., Wills S., McCarty D., Klee H., Leyser O. (2004). MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 14: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Bray E.A. (2002). Abscisic acid regulation of gene expression during water-deficit stress in the era of the Arabidopsis genome. Plant Cell Environ. 25: 153–161 [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1997). Free R value: Cross-validation in crystallography. Methods Enzymol. 277: 366–396 [DOI] [PubMed] [Google Scholar]
- Cavallini D., Cannella C., Barboni E., Fiori A., Marcucci M. (1969). Interaction of cysteamine oxygenase with o-phenanthroline. Eur. J. Biochem. 11: 360–363 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cutler A.J., Krochko J.E. (1999). Formation and breakdown of ABA. Trends Plant Sci. 4: 472–478 [DOI] [PubMed] [Google Scholar]
- DeLano W.L. (2002). The PyMOL Molecular Graphics System. (San Carlos, CA: DeLano Scientific; ). [Google Scholar]
- Ferreira K.N., Iverson T.M., Maghlaoui K., Barber J., Iwata S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303: 1831–1838 [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R., Gampala S.S., Rock C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.): S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor S.A., Villaseñor A., Fletterick R., Sigal E., Browner M.F. (1997). The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity. Nat. Struct. Biol. 4: 1003–1009 [DOI] [PubMed] [Google Scholar]
- Goldschmidt L., Cooper D.R., Derewenda Z.S., Eisenberg D. (2007). Toward rational protein crystallization: A Web server for the design of crystallizable protein variants. Protein Sci. 16: 1569–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas A., Mehouachi J., Tadeo F.R., Primo-Millo E., Talon M. (2000). Hormonal regulation of fruitlet abscission induced by carbohydrate shortage in citrus. Planta 210: 636–643 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle E., Stuart D.I., Métoz F. (1999). ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308 [DOI] [PubMed] [Google Scholar]
- Hasegawa P.M., Bressan R.A., Zhu J.K., Bohnert H.J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51: 463–499 [DOI] [PubMed] [Google Scholar]
- Hauck C., Muller S., Schildknecht H. (1992). A germination stimulant for parasitic flowing plants from Sorghum bicolor, a genuine host plant. J. Plant Physiol. 139: 474–478 [Google Scholar]
- Hendrickson W.A. (1991). Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science 254: 51–58 [DOI] [PubMed] [Google Scholar]
- Iuchi S., Kobayashi M., Taji T., Naramoto M., Seki M., Kato T., Tabata S., Kakubari Y., Yamaguchi-Shinozaki K., Shinozaki K. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27: 325–333 [DOI] [PubMed] [Google Scholar]
- IUPAC-IUB (1974). Nomenclature of carotenoids. Biochemisty (London: Butterworths; ), pp. 406–431 [Google Scholar]
- Jones T.A., Kjeldgaard M. (1997). Electron-density map interpretation. Methods Enzymol. 277: 173–208 [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. (1991). Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Katona G., Andréasson U., Landau E.M., Andréasson L.E., Neutze R. (2003). Lipidic cubic phase crystal structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.35A resolution. J. Mol. Biol. 331: 681–692 [DOI] [PubMed] [Google Scholar]
- Katona G., Carpentier P., Niviere V., Amara P., Adam V., Ohana J., Tsanov N., Bourgeois D. (2007). Raman-assisted crystallography reveals end-on peroxide intermediates in a nonheme iron enzyme. Science 316: 449–453 [DOI] [PubMed] [Google Scholar]
- Kloer D.P., Schulz G.E. (2006). Structural and biological aspects of carotenoid cleavage. Cell. Mol. Life Sci. 63: 2291–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloer D.P., Ruch S., Al-Babili S., Beyer P., Schulz G.E. (2005). The structure of a retinal-forming carotenoid oxygenase. Science 308: 267–269 [DOI] [PubMed] [Google Scholar]
- Kraulis J. (1991). MOLSCRIPT: A program to produce both detailed and schematic plots of protein structure. J. Appl. Cryst. 24: 946–950 [Google Scholar]
- Lancaster C.R., Kröger A., Auer M., Michel H. (1999). Structure of fumarate reductase from Wolinella succinogenes at 2.2 A resolution. Nature 402: 377–385 [DOI] [PubMed] [Google Scholar]
- Leahy D.J., Erickson H.P., Aukhil I., Joshi P., Hendrickson W.A. (1994). Crystallization of a fragment of human fibronectin: Introduction of methionine by site-directed mutagenesis to allow phasing via selenomethionine. Proteins 19: 48–54 [DOI] [PubMed] [Google Scholar]
- Lomize A.L., Pogozheva I.D., Lomize M.A., Mosberg H.I. (2006). Positioning of proteins in membranes: A computational approach. Protein Sci. 15: 1318–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusova R., Rani K., Verstappen F.W., Franssen M.C., Beale M.H., Bouwmeester H.J. (2005). The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 139: 920–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K., et al. (2009). Structural basis of abscisic acid signalling. Nature 462: 609–614 [DOI] [PubMed] [Google Scholar]
- Nishimura N., Hitomi K., Arvai A.S., Rambo R.P., Hitomi C., Cutler S.R., Schroeder J.I., Getzoff E.D. (2009). Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. (1997). Processing of x-ray diffraction data collected in oscillation mode. Methods in Enzymology, Volume 276: Macromolecular Crystallography, Part A, Carter C.W., Jr, Sweet R.M., eds (New York: Academic Press; ), pp. 307–326 [DOI] [PubMed] [Google Scholar]
- Parry A.D., Horgan R. (1991). Carotenoids and abscisic acid (ABA) biosynthesis in higher plants. Physiol. Plant. 82: 320–326 [Google Scholar]
- Phillips J., Artsaenko O., Fiedler U., Horstmann C., Mock H.P., Müntz K., Conrad U. (1997). Seed-specific immunomodulation of abscisic acid activity induces a developmental switch. EMBO J. 16: 4489–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Zeevaart J.A. (1999). The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. USA 96: 15354–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que L., Jr, Ho R.Y. (1996). Dioxygen activation by enzymes with mononuclear non-Heme iron active sites. Chem. Rev. 96: 2607–2624 [DOI] [PubMed] [Google Scholar]
- Rufer A.C., Lomize A., Benz J., Chomienne O., Thoma R., Hennig M. (2007). Carnitine palmitoyltransferase 2: Analysis of membrane association and complex structure with a substrate analog. FEBS Lett. 581: 3247–3252 [DOI] [PubMed] [Google Scholar]
- Schwartz A., Wu W.H., Tucker E.B., Assmann S.M. (1994). Inhibition of inward K+ channels and stomatal response by abscisic acid: An intracellular locus of phytohormone action. Proc. Natl. Acad. Sci. USA 91: 4019–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S.H., Qin X., Zeevaart J.A. (2003a). Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S.H., Tan B.C., Gage D.A., Zeevaart J.A., McCarty D.R. (1997). Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874 [DOI] [PubMed] [Google Scholar]
- Schwartz S.H., Tan B.C., McCarty D.R., Welch W., Zeevaart J.A. (2003b). Substrate specificity and kinetics for VP14, a carotenoid cleavage dioxygenase in the ABA biosynthetic pathway. Biochim. Biophys. Acta 1619: 9–14 [DOI] [PubMed] [Google Scholar]
- Sharp R.E. (2002). Interaction with ethylene: Changing views on the role of abscisic acid in root and shoot growth responses to water stress. Plant Cell Environ. 25: 211–222 [DOI] [PubMed] [Google Scholar]
- Siame B., Weerasuriya Y., Wood K., Ejeta G., Butler L. (1993). Isolation of strigol, a germination stimulant for Striga asiatica, from host plants. J. Agric. Food Chem. 41: 1486–1491 [Google Scholar]
- Sorefan K., Booker J., Haurogné K., Goussot M., Bainbridge K., Foo E., Chatfield S., Ward S., Beveridge C., Rameau C., Leyser O. (2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17: 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B.C., Cline K., McCarty D.R. (2001). Localization and targeting of the VP14 epoxy-carotenoid dioxygenase to chloroplast membranes. Plant J. 27: 373–382 [DOI] [PubMed] [Google Scholar]
- Tan B.C., Joseph L.M., Deng W.T., Liu L., Li Q.B., Cline K., McCarty D.R. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Tan B.C., Schwartz S.H., Zeevaart J.A., McCarty D.R. (1997). Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 94: 12235–12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I.B., Burbidge A., Thompson A.J. (2000). Control of abscisic acid synthesis. J. Exp. Bot. 51: 1563–1574 [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. (2003). SOLVE and RESOLVE: Automated structure solution and density modification. Methods Enzymol. 374: 22–37 [DOI] [PubMed] [Google Scholar]
- Thompson A.J., Jackson A.C., Parker R.A., Morpeth D.R., Burbidge A., Taylor I.B. (2000). Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol. Biol. 42: 833–845 [DOI] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., Kyozuka J., Yamaguchi S. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- United Nations (2007). Food and Agriculture Organization of the United Nations. http://faostat.fao.org/site/291/default.aspx (February 5, 2008)
- Vogel J.T., Tan B.C., McCarty D.R., Klee H.J. (2008). The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. Journal Biol. Chem. 283: 11364–11373 [DOI] [PubMed] [Google Scholar]
- von Lintig J., Vogt K. (2000). Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving beta-carotene to retinal. J. Biol. Chem. 275: 11915–11920 [DOI] [PubMed] [Google Scholar]
- von Lintig J., Vogt K. (2004). Vitamin A formation in animals: Molecular identification and functional characterization of carotene cleaving enzymes. J. Nutr. 134: 251S–256S [DOI] [PubMed] [Google Scholar]
- von Lintig J., Dreher A., Kiefer C., Wernet M.F., Vogt K. (2001). Analysis of the blind Drosophila mutant ninaB identifies the gene encoding the key enzyme for vitamin A formation invivo. Proc. Natl. Acad. Sci. USA 98: 1130–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J.J. (1964). Nature of the iron–oxygen bond in oxyhæmoglobin. Nature 203: 182–183 [PubMed] [Google Scholar]
- Zeevaart J.A.D., Creelman R.A. (1988). Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39: 439–473 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.