Abstract
The endoplasmic reticulum (ER) has a sophisticated quality control (QC) system to eliminate improperly folded proteins from the secretory pathway. Given that protein folding is such a fastidious process and subject to adverse environmental conditions, the ER QC system appears to have been usurped to serve as an environmental sensor and responder in plants. Under stressful conditions, the ER protein folding machinery reaches a limit as the demands for protein folding exceed the capacity of the system. Under these conditions, misfolded or unfolded proteins accumulate in the ER, triggering an unfolded protein response (UPR). UPR mitigates ER stress by upregulating the expression of genes encoding components of the protein folding machinery or the ER-associated degradation system. In Arabidopsis thaliana, ER stress is sensed and stress signals are transduced by membrane-bound transcription factors, which are activated and mobilized under environmental stress conditions. Under acute or chronic stress conditions, UPR can also lead to apoptosis or programmed cell death. Despite recent progress in our understanding of plant protein QC, discovering how different environmental conditions are perceived is one of the major challenges in understanding this system. Since the ER QC system is one among many stress response systems in plants, another major challenge is determining the extent to which the ER QC system contributes to various stress responses in plants.
INTRODUCTION
The concept of quality control (QC) was first applied over two decades ago by Hurtley and Helenius (1989) to the mechanisms that safeguard the correct folding and assembly of secreted and membrane proteins in the endoplasmic reticulum (ER). In Arabidopsis thaliana, somewhat over 17% of all proteins have predicted signal peptides and 33% have at least one transmembrane domain, many of which are likely to be associated with ER membranes or other organelles on the secretory pathway (Arabidopsis Genome Initiative, 2000). All of the cell wall protein components and many of the proteins that populate the plasma membrane or that are found in vacuoles/lysosomes, protein bodies, etc., are proteins derived from the secretory pathway (Sanderfoot and Raikhel, 2003). In the synthesis of secreted proteins, N-terminal signal peptides are cleaved cotranslationally by signal peptidases in the ER lumen, a step that can be critical to the protein folding process (Coleman et al., 1995; Vitale and Denecke, 1999). Upon entering the ER lumen, chaperones and cochaperones bind to protect growing polypeptide chains from aggregation, giving them time to fold and to prevent them from collapsing into non-native structures (Figure 1). Oligosaccharide units are added onto proteins bearing glycosylation recognition sites, and the presence and modification of these units serve as signals for the entry to and exit from cycles in the calnexin/calreticulin protein folding machine. The monitoring of protein folding and the modification of oligosaccharide side chains is a QC process that dictates whether a protein is exported to the Golgi or targeted for degradation (Hubbard and Ivatt, 1981). A few protein species are retained in the ER through retention signals that interact with integral membrane or soluble receptors (Pagny et al., 1999). A partial list of genes known to be involved in Arabidopsis protein QC, with accession numbers and references, is provided in Supplemental Table 1 online.
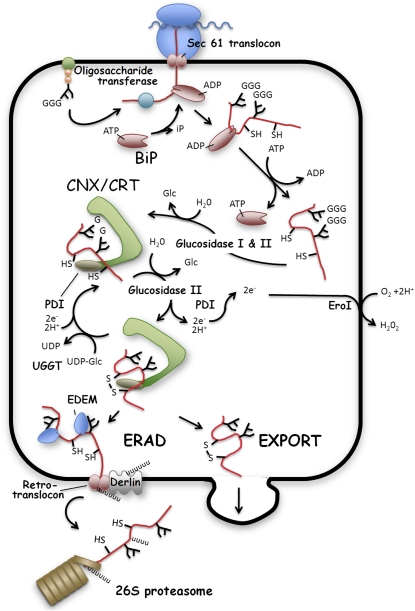
Figure 1.
Protein Folding and Modification in the ER.
The signal peptide sequence directs the translocation of polypeptides through the Sec61 translocon into the ER lumen. The ADP-bound state of the chaperone BiP binds to nascent polypeptides to prevent protein aggregation. If an N-glycosylation site is detected, a preassembled oligosaccharide core (Glc3Man9GlcNAc2) is transferred to the nascent protein catalyzed by the OST. Subsequently, rapid deglucosylation of the two outermost glucose residues (G) on the oligosaccharide core structures permits the entry of the nascent protein into the CNX/CRT protein folding machinery. PDI family proteins accelerate the folding by catalyzing formation of intra- and intermolecular disulfide bonds on the nascent proteins. The oxidizing environment in the ER lumen is maintained by Ero1. Removal of the innermost glucose residues by glucosidase II releases correctly folded proteins for export from the ER. Incorrectly folded proteins are reglucosylated by UGGT and are cycled again through the CNX/CRT apparatus. Terminal improperly folded proteins are recognized by EDEM and targeted to the retrotranslocon for ERAD. The functions of the illustrated proteins in Arabidopsis have not, in large part, been demonstrated experimentally but have been inferred from their homology to proteins in yeast and mammalian cells.
While the ER QC system plays a vital role in assuring the quality of proteins that enter the secretory pathway, the QC system also appears to endow plants with the capacity to adapt to stressful environmental conditions. It is not clear how or why that happens, but it is speculated that protein folding is so finicky that environmental conditions such as pathogen attack, drought, heat stress, and salinity can easily upset the process, triggering ER stress responses. Another possibility is that many genes involved in stress responses are secreted proteins and, as a result, ER stress responses and responses to environmental conditions have evolved as coregulated systems. One of the most highly conserved ER stress response mechanisms in eukaryotic organisms is the unfolded protein response (UPR). UPR is part of a QC mechanism optimizing the operation of the protein folding machinery in the ER under differing conditions. The protein folding and ER-associated degradation (ERAD) machinery has limited capacity, and UPR is induced when the folding demands in the ER exceed the capacity (Malhotra and Kaufman, 2007). This can happen when large amounts of secreted proteins are produced or the folding environment in the ER is disturbed under environmental stress conditions, leading to the accumulation of unfolded and/or misfolded proteins.
The mechanisms of QC and UPR in embryophytes and their importance in seed development have been recently reviewed (Urade, 2007; Tajima et al., 2008a; Vitale and Boston, 2008). The involvement of QC and UPR in environment stresses, including biotic and abiotic stresses, has received much attention and has shed new light on understanding and improving environmental stress tolerance in land plants. This review describes the complexity of the protein folding and degradation process and recent progress in identifying in plants the components of the QC system, which have been previously described in yeast and mammalian cells. In this review, we emphasize the importance of QC components in environmental stress tolerance and discuss how plants cope with the ER stress induced by adverse environmental conditions.
PROTEIN QC IN EMBRYOPHYTES
Protein Folding and Misfolding in the ER
Understanding protein folding has been one of the grand challenges of biology. Misfolding of proteins occurs even though the native state of a protein generally represents a low free energy form. In the process of folding, large proteins (in particular) pass through folding intermediates in which elementary folding units, so called foldons (Englander et al., 2007; Lindberg and Oliveberg, 2007), collapse into non-native forms. The energy landscape for protein folding can have many hills and valleys, and proteins en route to a native state may pass through or get stuck in one of many kinetically stable misfolded forms (Hartl and Hayer-Hartl, 2009). The smoothness or roughness of the energy landscape may dictate the stability of intermediate, misfolded, and native forms (Bryngelson et al., 1995).
Protein folding in the ER, in particular, is aided by chaperones and cochaperones. Chaperones are proteins that interact transiently with other non-native proteins to help them acquire a native state. Chaperones do not direct the outcome of the folding process; instead, they enhance the efficiency of the process (Hartl and Hayer-Hartl, 2009). A major role of chaperones is to prevent protein aggregation. Protein folding in the ER occurs in a crowded environment in which the concentration of proteins may be 100 g L−1 (Schroder and Kaufman, 2005). In such an environment, protein folding intermediates can interact through nonspecific hydrophobic interactions leading to the formation of protein aggregates. Chaperones are also thought to delay the folding of proteins during synthesis. Protein synthesis is relatively slow, proceeding at a rate of ~4 amino acids s−1, while the hydrophobic collapse of a protein can occur in nanoseconds (Stevens and Argon, 1999). Rapid collapse could prevent the intervention of other slower processes required for successful protein modification folding such as Pro isomerization. Furthermore, during the synthesis and translocation into the ER lumen of a large protein, if the N-terminal portion were to collapse, it might prevent the C-terminal portion of the protein from contributing to the global pattern of folding.
Binding protein (BiP), a member of the HSP70 family, is the most abundant chaperone protein in the ER. It is associated with the Sec61 translocon complex and interacts cotranslationally with nascent proteins (Figure 1; Kleizen and Braakman, 2004). BiP has the ability to bind folding intermediates of a large number of the proteins because of its capacity to bind hydrophobic peptides that in the folded state form β-strands buried in the interior of a protein in its native state (Flynn et al., 1991).
BiP has a nucleotide binding site in its N-terminal domain and has ATPase activity. In the ADP-bound state, BiP has high affinity for binding proteins in its C-terminal substrate binding domain (Flynn et al., 1989). The binding of BiP to proteins is released by ATP binding through nucleotide exchange (Wei et al., 1995); therefore, cycles of nucleotide hydrolysis and exchange drive the binding and release BiP from unfolded or misfolded protein substrates, a process that terminates when the hydrophobic sequences in the protein substrate are buried (Gething, 1999). One of the best examples of the role of BiP in the biogenesis of a secreted protein is the binding of BiP to sites in monomers of the trimeric bean protein Phaseolin that are buried in the mature protein (Vitale et al., 1995; Foresti et al., 2003). The binding and release cycles are regulated by cofactors, such as DNAJ proteins that promote ATP hydrolysis or ATP:ADP exchange (Cheetham and Caplan, 1998).
Arabidopsis has three BiP genes: BiP1, BiP2, and BiP3. At-BiP1 and At-BiP2 are nearly identical in protein sequence. Both are expressed at fairly high levels throughout the plant and are induced by ER stress agents, such as tunicamycin and DTT (Liu et al., 2007a; Iwata et al., 2008), and by heat (Gao et al., 2008). At-BiP3 is normally expressed at much lower levels and because of that is highly induced (in terms of fold change in expression) by ER stress agents in seedlings (Martínez and Chrispeels, 2003; Liu et al., 2007a; Iwata et al., 2008). Plant BiP expression has also been shown to be upregulated by other environmental stresses, such as drought (Figueiredo et al., 1997), cold (Anderson et al., 1994), and insect and pathogen attack (Jelitto-Van Dooren et al., 1999). Also, overexpression of plant BiP has been reported to confer drought tolerance in soybean (Glycine max; Valente et al., 2009) and tobacco (Nicotiana tabacum) plants (Alvim et al., 2001). Mutation of At-BiP2 makes the plant more sensitive to pathogen attack resulting from the impaired induction of PR1, linking the secretory pathway to the systemic acquired resistance pathway (Wang et al., 2005). In addition, BiP gene expression has been shown to be induced in specific cell types at developmental stages associated with high secretory activity (Boston et al., 1996) or in plants that have expressed assembly-defective proteins (Boston et al., 1991), further implicating the role of BiP in protein folding. Arabidopsis also has homologs of the yeast DNAJ protein Scj1p (Schlenstedt et al., 1995), named At-J1, At-J2, and At-J3. However, the roles of these DNAJ proteins in plants are less well studied. Yang et al. (2009) reported that the protein THERMOSENSTITIVE MALE STERILE1, which contains DNAJ domain and ERdj5C domain, functions in thermotolerance of pollen tubes in Arabidopsis.
Similar to BiP, endoplasmin/GRP94 is also an abundant ER molecular chaperone and one of the glucose-regulated proteins. SHEPHERD (SHD) appears to encode the only ortholog of GRP94 in Arabidopsis, which is highly induced by tunicamycin and DTT (Martínez and Chrispeels, 2003; Liu and Howell, 2010). Biochemical and cell biology studies indicated that At-SHD has in vivo chaperone activity (Klein et al., 2006). Interestingly, the substrates/targets of At-SHD seem to be very specific. Null mutants of At-SHD have defects in meristem size control, resembling the phenotypes of clavata mutants. Genetic analysis showed that At-SHD is essential for the synthesis of CLV proteins, especially at high temperature (Ishiguro et al., 2002).
Protein N-Glycosylation
Most proteins targeted to the secretory pathway are glycoproteins modified by the addition of N-linked oligosaccharides. Glycosylation is an important posttranslational modification because it stabilizes some proteins against denaturation and proteolysis, enhances their solubility, and serves as a recognition signal for QC in the ER (Hammond et al., 1994; Ceriotti et al., 1998; Helenius and Aebi, 2004). It is clear from the action of tunicamycin, an inhibitor of N-glycosylation and a potent inducer of UPR, that failure to N-glycosylate rapidly generates ER stress.
Nascent plant proteins entering the ER lumen via the Sec61 translocon complex are scanned by oligosaccharyltransferase (OST) for the presence of sequons or glycosylation site recognition motifs (Asn-X-Ser/Thr; Figure 1). If sequons are detected, then presynthesized oligosaccharides linked to lipid carriers (dolichol-pyrophosphate) are transferred en bloc by OST to the γ-amido group of Asn residues in the sequon (Kornfeld and Kornfeld, 1985). OST is a heteromeric, multisubunit protein associated with the translocon complex and consists of eight subunits in yeast, five of which are essential.
Arabidopsis has homologs to the five essential OST subunits (homologous yeast proteins are in parentheses): DAD1 and DAD2 (yeast Ost1 and Ost2, respectively), STT3A and STT3B (yeast Stt3), DGL1 (yeast Wbp1), and HAP6 (yeast Swp1). Mammalian cells have two OST isoforms with different catalytic subunits (STT3A and STT3B) (Ruiz-Canada et al., 2009). This also appears to be the case for Arabidopsis, since it has homologs of both forms (Koiwa et al., 2003). The mammalian OST isoform STT3A is primarily responsible for cotranslational glycosylation of nascent polypeptides as they enter the ER lumen, while the STT3B isoform is capable of posttranslational glycosylation of skipped sequons (Ruiz-Canada et al., 2009). In Arabidopsis, a T-DNA insertion in STT3A results in an osmotically sensitive root phenotype and induction of UPR due to protein hypoglycosylation. The single mutants stt3a-1 and stt3b-1 are both viable, but the double mutant is a gametophytic lethal, suggesting that the plant OST isoforms have partially overlapping roles (Koiwa et al., 2003). At-DAD1 and At-DAD2 are thought to be involved in OST anchoring (Gallois et al., 1997), and overexpression of these genes suppressed the onset of DNA fragmentation under UV-C exposure in Arabidopsis (Danon et al., 2004). DGL1-1 (for Defective Glycosylation 1-1) is an essential OST subunit in that mutations in this gene impair glycosylation and affect cell differentiation and growth in Arabidopsis (Lerouxel et al., 2005).
In most eukaryotes, the oligosaccharide units attached to N-glycosylated proteins are branched structures (Figure 2) composed of three glucoses, nine mannoses, and two N-acetyl-glucosamines (Glc3Man9GlcNAc2) (Helenius and Aebi, 2004). The oligosaccharide units are formed on the cytoplasmic side of the ER membrane by the attachment of two N-acetyl-glucosamines and five mannoses to lipid dolichol (Dol), which is a polyisoprenoid lipid comprised of ~20 isoprene units (Swiezewska and Danikiewicz, 2005). This sugar precursor is flipped over to the ER lumen where four mannose and three glucose residues are added subsequently to produce the core oligosaccharide (Helenius and Aebi, 2004). Leaf Wilting 1 protein is responsible for the Dol biosynthesis in Arabidopsis, and mutations in this gene result in leaf wilting, lower leaf turgor, and drought resistance (Zhang et al., 2008).
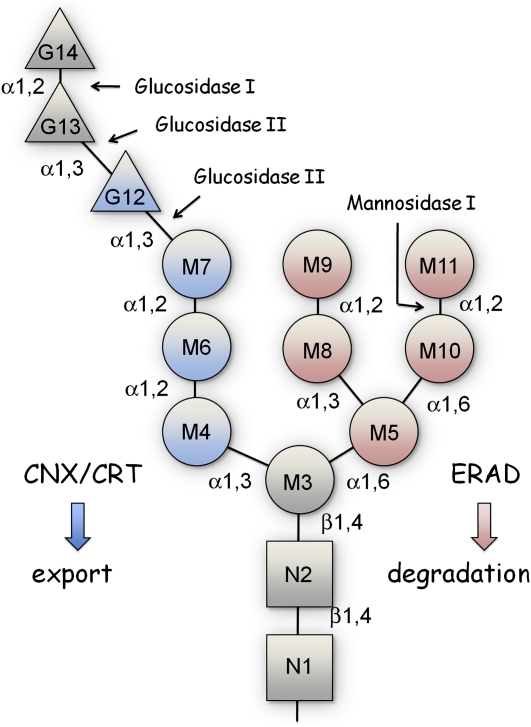
Figure 2.
The Structure of Lipid-Linked Oligosaccharide Unit Glc3Man9GlcNAc2.
Two N-acetylglucosamine (N1 and N2, gray squares) and five mannose residues (M3 to M7, gray, pink, and blue circles) are added subsequently to the polyisoprenoid lipid dolichol (Dol) on the cytoplasmic side of the ER. The above lipid-linked oligosaccharide (LLO) unit Man5GlcNAc2 is then flipped over to face the ER lumen, where four additional mannose (M8 to M11, pink circles) and three glucose residues (G12 to G14, blue and gray triangles) are added to form the LLO unit Glc3Man9GlcNAc2. The moieties containing 14 monosaccharide units are then transferred from the lipid Dol to proteins and then subjected to further glucose and/or mannose trimming at bond sites indicated by arrows. The enzymes that catalyze the respective hydrolysis reactions are indicated. The folding status of the protein is recognized either by the CNX/CRT through residues indicated in blue or EDEM through residues indicated in pink. The properly folded proteins are secreted out or the unfolded/misfolded proteins are retrieved for degradation through ERAD.
The core oligosaccharide structure is subject to glucose trimming and further modification as the glycoprotein passes through the secretory pathway (Figures 1 and 2). The modifications that occur on the core structure in the ER are important because they serve as recognition tags for ER QC and reflect the folding status of proteins (Helenius and Aebi, 2004). In particular, protein glycans are recognition signals for the entry to and exit from the ER protein folding apparatus involving calnexin (CNX) and calreticulin (CRT). Many reviews have been written on the subject, including Helenius and Aebi (2004), Moremen and Molinari (2006), Caramelo and Parodi (2008), and a recent article about CRT in plants (Jia et al., 2009).
CNX-CRT Protein Folding Cycle
The principal protein folding machine in the ER is the CNX/CRT protein folding apparatus (Figure 1). CNX and CRT are lectins, and the oligosaccharide side chains on glycoproteins are the tickets for the entry and exit from the protein folding machinery. Under stress conditions, in particular, glycoproteins repeatedly fold and refold constituting a CNX/CRT folding cycle (Hammond and Helenius, 1994). Entry of a glycoprotein into the cycle begins with the successive removal of the outermost and the next glucose residue (G14 and G13) from the core glycan by glucosidase I and II (Figure 2). Glucosidase I is a type II membrane glycoprotein closely associated with OST and the translocon complex (Caramelo and Parodi, 2008). Glucosidase I is capable of hydrolyzing α-1,2 glycosidic bonds, cleaving off glucose 14 (G14) from the core oligosaccharide structure. Removal of the terminal glucose residues from N-linked oligosaccharide chains can be critical to protein folding. When the removal was blocked in bean (Phaseolus vulgaris) by glucosidase inhibitors castanospermine and N-methyldeoxynojirimycin, phaseolin polypeptides with partially trimmed glycans were unable to form mature trimeric proteins (Lupattelli et al., 1997).
Arabidopsis has two glucosidase I genes that are close homologs of the enzymes in yeast (Matsushima et al., 2003). Glucosidase II is a soluble luminal enzyme that hydrolyzes α-1,3 linkages and cleaves off glucose 13 (G13) from the core oligosaccharide. Glucosidase II is composed of two glycoprotein subunits, a catalytic subunit α, and a β-subunit. The β-subunit appears to have a subcellular localization and/or regulatory role (Caramelo and Parodi, 2008). Arabidopsis has a close homolog of the yeast α-subunit, but the two annotated β-subunit genes in Arabidopsis (Burn et al., 2002) are more distantly related to their yeast and mammalian counterparts.
The monoglucosylated core glycan (containing G12) so generated binds to CNX and/or CRT, which are folding cages protecting nascent proteins from forming homo- or hetero-oligomeric aggregates (Figure 1; Ruddock and Molinari, 2006). CRT is a soluble protein in the ER lumen, and CNX is a type I membrane protein. In Arabidopsis, there are three isoforms of CRT and two of CNX. Both CNXs ectodomain and CRT have similar structures with globular N-domains, an extended Pro-rich hairpin structure called the P domain, and a C domain. The principal difference between CRT and CNX is that CNX has a transmembrane domain interposed between the P and C domains.
The N-terminal regions of the two lectins are globular β-sandwich domains that interact with glucose moieties on the monoglucosylated oligosaccharides of nascent glycoproteins. The N domain is highly conserved in plants and has two signature motifs, KHEQKLDCGGGYVLL and IMFGPDICG. A unique feature of CRTs in various plants is that they are glycosylated with high mannose-containing oligosaccharides (Nardi et al., 1998; Pagny et al., 2000; Navazio et al., 2002). This has not yet been demonstrated for Arabidopsis CRTs, although the N domain sequences have potential N-glycosylation sites (Jia et al., 2009). The P domain contains two triplet repeats that confer an extended hairpin structure maintained by the binding of Ca2+. A wheat (Triticum aestivum) CRT (Ta-CRT) is induced by drought, and overexpression of Ta-CRT in tobacco plants enhances drought resistance (Jia et al., 2008).
Glycoproteins are released from CNX or CRT by the further action of glucosidase II, which cleaves off G12 by hydrolyzing the α-1,3 glycosidic bond leaving a nonglucosylated oligosaccharide (Figures 1 and 2). Released proteins that are not yet properly folded are reglucosylated by UDP-glucose:glycoprotein glucosyltransferase (UGGT) so that they can reenter the CNX/CRT cycle. Thus, UGGT plays a key role in sensing protein misfolding and sending proteins back for additional rounds of folding. UGGT is a large soluble protein in the ER lumen with a C-terminal catalytic segment and large N-terminal domain that is thought to recognize the folding state of proteins.
A UGGT homolog is found in Arabidopsis, and mutations in the gene have been picked up in genetic screens. Li and colleagues (Jin et al., 2007, 2009) reported that BRI1-9, a structurally imperfect but functionally competent brassinosteroid receptor, is retained and then disposed of by the ER QC system in wild-type plants. However, in an ebs1-1 suppressor with a defect in UGGT, BRI1-9 apparently is not reglucosylated and fails to be retained in the CNX/CRT cycle, allowing it to escape QC and emerge to the cell surface as a functional receptor (Jin et al., 2007). Another suppressor, ebs2, with a mutation in At-CRT3, also restores considerable BR sensitivity of the bri1-9 mutant. The authors demonstrated in a pull-down assay that only At-CRT3 among the CRTs coimmunoprecipitated with the mutant BRI1-9 receptor. As for the interaction with CNXs, they found that neither single or double CNX mutations suppressed the bri1-9 mutations and suggested that the CNX interaction might be involved in BRI1 receptor folding but was not responsible for retaining the mutant BRI1-9 receptor in the ER (Jin et al., 2009).
A different genetic screen conducted by Li et al. (2009) picked up mutations in these and other ER QC genes. This screen was set up to detect Arabidopsis innate immunity mutants with defects in the perception of pathogen-associated molecular patterns (PAMPs). PAMPs are recognized by pattern recognition receptors (PRRs), leucine-rich receptor kinases that activate PAMP-triggered immunity (Zipfel, 2008). Three PRRs have been identified in Arabidopsis: FLS2, which recognizes bacterial flagellin; EFR, which perceives the bacterial elongation factor-Tu; and CERK1, which recognizes fungal chitin. One of the mutations showing a defect in PAMPs mapped to a member of the CRT family, At-CRT3, the same gene affected by the ebs2 suppressor in the studies described above by Jin et al. (2007). Nine crt alleles were found and most blocked the accumulation of the CRT3 protein. Unexpectedly, the crt mutants were still sensitive to the surrogates for bacterial flagellin or fungal chitin, indicating that At-CRT3 is required for EFR, but not for FLS2 or CERK1. Recently, Christensen et al. (2010). reported that At-CRT3 has plant-specific properties. While other Arabidopsis CRTs (At-CRT1a and b) fully complemented defective functions in CRT-deficient mouse fibroblasts, At-CRT3 did not restore all mutant defects, such as cell adhesiveness. Thus, At-CRT3 appears to be an unusual, plant-specific CRT that plays a role in the QC of cell surface hormone receptors and in the biogenesis of PAMPs.
Other EFR mutations mapped to Arabidopsis UGGT and to the H/KDEL ER retention receptor ERD2 (Li et al., 2009). Like the crt mutants, all five erd2 mutations were insensitive to the surrogate peptide elf18 but were sensitive to surrogates for bacterial flagellin or fungal chitin. The erd2 mutations were unexpected; why would a function that retains proteins in the ER be required for the operation of a PRR? It was found that the At-CRT3 protein failed to accumulate in the erd2 mutations and, as described above, CRT3 is required for EFR accumulation. Still other EFR mutations mapped to stromal-derived factor-2 (SDF2) (Nekrasov et al., 2009). In mammalian cells, SDF2 is in a complex with other chaperones and cochaperones, including ERdj3B and BiP (Meunier et al., 2002). In Arabidopsis, SDF2 and ERdj3B are required for the function of the membrane glycoprotein EFR. Loss of At-SDF2 resulted in ER retention and degradation of EFR (Nekrasov et al., 2009). At-SDF2 transcription is activated upon UPR induction by DTT and tunicamycin treatment, and consistently At-SDF2 protein increases. Mutations in At-SDF2 did not affect ER stress sensing and perception but resulted in pronounced plant growth impairment upon DTT treatment, indicating that SDF2 not only interacts with EFR but also with other proteins. The three-dimensional structure of At-SDF2 suggests that it may act as the adaptor for the interaction with different partners or ligands (Schott et al., 2010).
Protein Oxidation
In the CNX/CRT cages, proteins in the act of folding are thought to form and reform disulfide bonds through repeated oxidation and reduction catalyzed by protein disulfide isomerase (PDI) and other thiodisulfide oxidoreductases (Figure 1). In yeast, ERp57, a thioredoxin like protein, is bound to CNX-CRT folding cages and specifically interacts with glycoproteins (Holst et al., 1997). It is not known in Arabidopsis whether one or more PDIs function as ERp57. ERp57 homologs in Arabidopsis are members of the PDI family (Lu and Christopher, 2008).
The formation of protein disulfide bonds in the ER requires oxidizing equivalents, which are supplied in yeast by ER oxidoreductase 1 (Ero1p). Arabidopsis has two ERo1p homologs, AERO1 and AERO2 (Dixon et al., 2003). Ero1p in yeast is a flavin-containing ER membrane-associated protein that transfers electrons directly to molecular oxygen in the cytoplasm during disulfide bond formation in the ER. It is thought that oxidizing equivalents flow from O2 to the flavin cofactor in Ero1, then through an intercysteine relay to dithiol/disulphide sites on the surface of Ero1p (Sevier and Kaiser, 2006). ER oxidoreductin 1 (Ero1), in turn, oxidizes members of the family of ER oxidoreductases (Frand and Kaiser, 1999; Gross et al., 2006). The oxidizing character of the ER compartment is buffered by a high ratio of oxidized to reduced glutathione (GSSG/GSH), much of which is found in mixed disulfides with proteins (Hwang et al., 1992; Bass et al., 2004).
ERAD
The protein folding machinery attempts to fold proteins properly. However, proteins that do not achieve native forms can accumulate in the ER as aggregates or are eliminated by the ERAD system, which recognizes, targets, retrotranslocates, ubiquitinates, and degrades misfolded proteins (Figure 1). Aggregates of the seed storage proteins irreversibly associated with BiP accumulate in the ER of lima beans (Phaseolus lunatus) when glycosylation is blocked by the action of tunicamycin (Sparvoli et al., 2000). The ability of the cell to discriminate between native and misfolded proteins is of great importance in order to send properly folded proteins along their way through the secretory pathway, but to eliminate potentially toxic, misfolded forms.
No single feature, including time spent in folding and refolding proteins, determines whether proteins will be eliminated or exported from the ER down the secretory pathway (Wiseman et al., 2007). This is because the cell needs to protect inherently slow-folding proteins but eliminate misfolded forms of other proteins (Quan et al., 2008). The efficiency of the ERAD system is relevant to ER stress because ER stress occurs if and when the system is overloaded with misfolded proteins.
The recognition of folded and misfolded forms is challenging given the different types of protein substrates with which the ER QC system must contend; nonetheless, it has been possible in yeast to classify substrates into categories based on the subcellular location of their misfolded domains (Brodsky and Wojcikiewicz, 2009). Proteins with misfolded domains in the different subcellular locations are inspected by different components of the QC surveillance system (Huyer et al., 2004). Misfolded soluble proteins in the ER lumen and membrane proteins with misfolded domains projecting into the ER lumen are ERAD-L substrates and membrane proteins with misfolded domains within the membrane are ERAD-M substrates, while membrane proteins with misfolded domains projecting into the cytoplasm are ERAD-C substrates (Vashist and Ng, 2004).
In yeast, the ERAD-L pathway uses a bipartite recognition mechanism that monitors both the folding and the glycosylation states of protein substrates (Knop et al., 1996; Spear and Ng, 2005; Denic et al., 2006; Gauss et al., 2006a). The system includes a large membrane complex involved in the retrotranslocation of the proteins and Hrd1/Hrd3/Yos9 components of an ubiquitin ligase complex that recognizes both misfolded proteins and glycans (Denic et al., 2006; Gauss et al., 2006b). Hrd3 is an E3 ubiquitin ligase that recognizes misfolded proteins, and Yos9 is a lectin that recognizes a terminal α-1,6-linked mannose (M10) (Quan et al., 2008). The terminal α-1,6-linked mannose (M10) on N-linked oligosaccharide chains are exposed in yeast by the action of Htm1, an α-1,2-specific exomannosidase that removes M11 (Clerc et al., 2009). It is thought that Htm1 then marks the oligosaccharide component of misfolded glycoproteins for destruction by the ERAD-L system (Quan et al., 2008). Arabidopsis lacks homologs of the critical recognition components of this system, Hrd3 and Yos9.
ER degradation–enhancing α-mannosidase-like lectins (EDEMs) in mammalian cells play a similar role to Htm1 in yeast. EDEM1 is thought to interrupt futile folding cycles, extract the unfolded or misfolded proteins, shield them from reglucosylation by UGGT, and divert them to the ERAD pathway (Kanehara et al., 2007). EDEM1 is also thought to be an α-1,2-mannosidase, but that assertion is somewhat controversial because the protein lacks a critical Cys at the active site. Nonetheless, overexpression of EDEM1 accelerates demannosylation of misfolded substrates by means that may be either direct or indirect (Olivari et al., 2006). In addition, EDEM1 is a lectin that binds oligosaccarharides with terminal α-1,6-linked mannose, and binding of EDEM1 to demannoyslated glycoproteins prevents their aggregation with other proteins prior to retrotranslocaion. Mammalian cells have two EDEMs, EDEM1 and 2. EDEM1 is thought to escort proteins destined for degradation to the retrotranslocon apparatus because EDEM1 coprecipitates with Derlin-2 and -3 (Oda et al., 2006), proteins considered to be part of the retrotranslocon channel. Arabidopsis has homologs to EDEM1 and EDEM2 and likely uses these proteins in extracting escorting proteins to a retrotranslocon (Figure 1). In addition, maize (Zea mays) has four Derlin-like genes that encode homologs of yeast Der1p protein implicated in ERAD (Kirst et al., 2005), and Arabidopsis has three yeast Der1p homologs.
Proteins misfolded in the ER are disposed of by the 26S proteosome in the cytoplasm (Figure 1). That means that ERAD-L substrates need to be transported across the ER membrane for ubiquitination and disposal in the cytoplasm. The structure of retrotranslocon is unsettled, particularly the role of sec61, the major component of the ER translocon. It is thought that sec61 also serves as the retrotranslocon channel or is a component of the channel (Nakatsukasa and Brodsky, 2008). Evidence supporting the role of sec61 as a retrotranslocon includes the fact that sec61 coimmunoprecipitates with certain protein substrates destined for ERAD elimination (Wiertz et al., 1996; Pilon et al., 1997). On the other hand, complexes containing the major E3 ubiquitin ligases associated with ERAD, Hrd1 and Doa10, do not include sec61 (Carvalho et al., 2006; Denic et al., 2006; Gauss et al., 2006a). Instead, in yeast, Hrd1p and Doa10p appear to act both as E3 ubiquitin ligases and membrane channels. The catalytic RING domains of both E3 ligases are located on the cytoplasmic side of the ER membrane and both have multiple membrane-spanning domains (Nakatsukasa and Brodsky, 2008). Hrd1p also interacts with Hrd3p, Der1p, and Usa1p, which are all thought to make up the retrotranslocon complex for ERAD-L substrates in yeast. However, Arabidopsis does not have convincing homologs for these factors. Arabidopsis has several sec61 homologs that may function as translocons or retrotranlocon channels. The evidence for reverse translocation in plants comes from studies on the toxin ricin, a ribosome-inactivating protein from castor beans (Frigerio and Roberts, 1998; Frigerio et al., 1998). The mature ricin comprises a catalytic A chain and lectin B chain linked by a disulfide bridge. Both chains are derived from a single precursor polypeptide and matured in seed storage vacuoles. When engineered ricin, dispossessed of its B chain sequences, is overexpressed, the orphan A chain is recognized as the defective protein and transported to the cytosol for ERAD (Di Cola et al., 2001). Ubiquitination does not appear to be required for retrotranslocation of ricin A chains in tobacco protoplasts because depletion of Lys residues (polyubiquitin sites) in the A chains does not affect their recognition in the ER or retrotranslocation but does affect their degradation following retrotranslocation (Di Cola et al., 2005).
The ERAD-C pathway degrades proteins with misfolded cytoplasmic domains. In yeast, a pathway much less complex than ERAD-L mediates ERAD-C degradation of substrates such as Ste6-166p (Huyer et al., 2004). The pathway involves Doa10p, a membrane multispanning ubiquitin ligase, interacting proteins, Ubc7p, an ubiquitin conjugating enzyme, and its membrane anchor, Cue1p (Carvalho et al., 2006). Both the ERAD-C and -L pathways converge on Cdc48p, a protein extracting AAA-ATPase, and its cofactor Npl4 and Ubx2p, a membrane-recruiting factor (Carvalho et al., 2006). Cdc48 associates with the 26S proteasome cap (Verma et al., 2000) and in doing so delivers the polyubiquitinated protein to the proteosome. In the ricin expression system described above, retrotranslocation of the ricin A chain also requires CDC48 as demonstrated by the action of a dominant negative CDC48 (Marshall et al., 2008).
There is no evidence yet for an ERAD-C pathway in plants; however, Arabidopsis has three very close homologs with Cdc48 (Rancour et al., 2002), but not for other components, which either may be more specialized or may not exist in plants. Much less is known about the ERAD-M pathway, but its substrates also seem to be ubiquitinated by the Hrd1 complex. Barley (Hordeum vulgare) powdery mildew resistance o (MLO) is the founder of a sequence-diversified protein family with seven-transmembrane helices. Using a series of single amino acid substitution mutants of the MLO protein, Muller et al. (2005) demonstrated that ERAD-M-like QC mechanisms are conserved across yeast, plant, and mammals. Degradation of the mutant MLOs in Arabidopsis was dependent on the function of At-CDC48 and the 26S proteasome.
UPR
UPR, which is conserved from yeast to mammalian cells, brings protein folding and degradation capacity in line with demands either by enhancing the protein folding or degradation machinery or by slowing protein production. UPR signaling in mammalian cells has three pathway branches, each involving different classes of ER stress transducers. One branch is mediated by membrane-associated transcription factors, such as activating transcription factor 6 (ATF6), which targets stress response genes. Another branch involves an RNA splicing factor, inositol-requiring protein 1 (IRE1), which splices an mRNA encoding another transcription factor that also targets stress response genes. The third branch involves a protein kinase-like ER kinase (PERK) that regulates translation. In each pathway, a membrane-associated protein, either on its own or through its association with other factors, senses misfolded protein accumulation and transmits the signal from the ER lumen to the cytosol (Ron and Walter, 2007).
The molecular signature for UPR in plants is the transcriptional upregulation of genes related to protein folding and degradation (Martínez and Chrispeels, 2003; Kamauchi et al., 2005). Although UPR in plants was reported nearly two decades ago (Boston et al., 1991; Denecke et al., 1991; D'Amico et al., 1992), the membrane-associated transcription factor branch of the UPR signaling pathway in plants was discovered only recently (Figure 3; Liu et al., 2007a). A key component of UPR is bZIP28, an ER-localized membrane-associated bZIP transcription factor in Arabidopsis, which transduces stress signals from the ER to the nucleus during UPR (Liu et al., 2007a; Tajima et al., 2008b). Although At-bZIP28 has very little sequence similarity to ATF6, both proteins are structurally conserved. Like ATF6, At-bZIP28 is a type II membrane protein, with its N-terminal bZIP domain facing the cytosol and its C-terminal regulatory domain facing the ER lumen. Under normal conditions, At-bZIP28 is localized in the ER, but when misfolded proteins accumulate following treatment by ER stress agents, such as tunicamycin, it is translocated to the Golgi where At-S1P (a Ser protease) is localized. At-bZIP28 has a predicted S1P cleavage site and likely is processed by At-S1P and apparently also by the metalloprotease At-S2P (our unpublished data; Che et al., 2010).
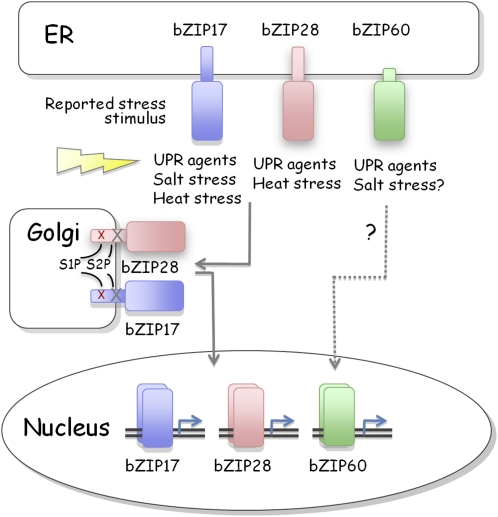
Figure 3.
ER Stress Response Pathways in Plants.
Three bZIP membrane-bound transcription factors (MTFs) are localized on the ER membrane under normal conditions. Under environmental stress conditions as indicated, the MTFs are transported to the Golgi apparatus and proteolytically processed by Golgi-localized proteases, site-1 protease (S1P; cleavage site indicated by red x) and site-2 protease (S2P; cleavage site indicated by black x). The processed forms of MTFs are imported into the nucleus to activate stress response genes. The activation mechanism for At-bZIP60 is not known.
The released N-terminal component of At-bZIP28 (At-bZIP28n), bearing the transcriptional activation domain and DNA binding domain, relocates to the nucleus (Figure 3) where it interacts with CCAAT box proteins, which are trimeric proteins composed of nuclear factors At-NF-YA4/At-NF-YB3/At-NF-YC2 (Liu and Howell, 2010). Interestingly, the At-NF-Y factors are also regulated by stress. The At-NF-YC2 gene is upregulated by ER stress, and the At-NF-YB3 protein relocates from the cytoplasm to the nucleus in response to stress (Liu and Howell, 2010). It was shown in a yeast two-hybrid system that At-bZIP28 homodimerizes and forms heterodimers with At-bZIP60, another UPR signaling component in plants (Liu and Howell, 2010). At-bZIP60 is also predicted to be an ER membrane protein and reported to be proteolytically activated (Iwata and Koizumi, 2005; Iwata et al., 2008). The C-terminal region of At-bZIP60 is much shorter than that of At-bZIP28, which is assumed to be important for sensing the stress in the ER lumen. Also unlike At-bZIP28, the C-terminal region of At-bZIP60 does not contain a canonical At-S1P cleavage site. Although the activation mechanism of At-bZIP60 is unresolved, the N-terminal component of At-bZIP60 has transcriptional activity in protoplast reporter assays (Iwata and Koizumi, 2005).
At-bZIP28 was originally identified as a factor activated by UPR stress agents, such as tunicamycin and DTT, but it was later reported to respond also to heat stress (Gao et al., 2008). Since UPR stress agents are thought to promote the accumulation of unfolded proteins and since these stress agents are surrogates for heat stress, it could be argued that heat triggers UPR by also promoting the accumulation of unfolded proteins. That idea is speculative and needs to be tested because it is also possible that heat and ER stress agents activate UPR by different mechanisms. Koizumi et al. (1999) demonstrated that tunicamycin does, indeed, elicit UPR by blocking protein glycosylation, not by some undescribed cytotoxic mechanism. They found that overexpression of UDP-N-acetylglucosamine:dolichol phosphate N-acetylglucosamine-1-P transferase, the enzyme that transfers the core oligosaccharide onto glycoproteins, counteracts the activation of UPR by tunicamycin.
Salt stress was found to activate bZIP17 in Arabidopsis. At-bZIP17 is similar in structure to At-bZIP28 (i.e., both are type II membrane proteins and have the same domain distribution and canonical At-S1P site). At-bZIP17 relocates from the ER to the nucleus under salt stress through At-S1P-dependent proteolysis (Liu et al., 2007b). The null mutants of At-S1P and At-bZIP17 are both salt sensitive. ER stress–related salt stress genes differ from those reported for UPR (Martínez and Chrispeels, 2003; Kamauchi et al., 2005). Upregulation of BiP and ER-associated calcium-dependent folding proteins are characteristic of UPR, but these genes are not upregulated soon after the imposition of salt stress (Liu et al., 2007b). The genes upregulated by At-bZIP17 appear to be capable of mitigating the effects of salt stress because overexpression of the N-terminal component of At-bZIP17 driven by a stress-inducible promoter modestly enhances tolerance to salt stress in Arabidopsis (Liu et al., 2008). Fujita et al. (2007) found that overexpression of the full length of At-bZIP60 also enhanced salt tolerance in Arabidopsis. The expression of At-bZIP60 is also upregulated by salt stress, which further suggests that At-bZIP60 is activated during salt stress response. Recently, At-bZIP17 was shown also to be activated by heat stress and UPR stress agent tunicamycin (Che et al., 2010). However, At-bZIP17 does play the same role as At-bZIP28 in upregulating target genes in response to heat and UPR agents in that unlike At-bZIP28, overexpression of the N-terminal component of At-bZIP17 in protoplasts does not activate the BiP3 promoter (Tajima et al., 2008b).
As yet, there is no clear understanding as to how environmental stresses induce UPR in plants and how the membrane-associated transcription factor system discriminates different environmental stresses. ER stress agents that affect N-glycosylation, cellular redox changes, or Ca2+ homeostatic balance are surrogates for environmental stresses, and all of these agents induce UPR. However, the agents have somewhat different effects. For example, Martínez and Chrispeels (2003) showed that the set of genes induced by tunicamycin and DTT in Arabidopsis are different (i.e., the sets only partially overlap). Therefore, it is possible that closer analysis of the effects of the different stress agents on UPR signaling in plants might provide greater insight into the mechanisms by which different environmental stresses activate UPR.
Less attention has been paid to other branches of the UPR signaling pathway in plants, although IRE1 homologs have been reported in plants (Koizumi et al., 2001). IRE1 is a transmembrane protein kinase/ribonuclease that plays a critical role as an ER stress sensor/transducer in yeast and mammalian cells. IRE1 acts by splicing an mRNA encoding a transcription factor (Hac1 and XBP1 in yeast and mammals, respectively), which then in turn, activates the expression of stress response genes (for recent reviews, see Schroder and Kaufman, 2005; Bernales et al., 2006; Ron and Walter, 2007). IRE1-mediated mRNA splicing is unconventional because it occurs in the cytoplasm, not in the nucleus where other mRNAs are normally spliced. Arabidopsis has at least two genes with IRE1-related sequences: IRE1a (formerly At-IRE1-2) and IRE1b (formerly At-IRE1-1), which were first described by Koizumi et al. (2001). They found that IRE1a and b green fluorescent protein fusions were located in the perinuclear ER. They also demonstrated functional complementation of the Arabidopsis IRE1a luminal domain (sensor domain) in yeast and that the plant IRE1a has autophosphorylation activity in vitro, but they were unable to demonstrate endonuclease activity. Noh et al. (2002) confirmed several observations made by Koizumi et al. (2001), but they also attempted to show unsuccessfully whether IRE1a could splice yeast Hac1 RNA in tunicamycin-treated Arabidopsis protoplasts. Similar results were also reported in rice (Oryza sativa; Okushima et al., 2002). However, the target of plant IRE remains elusive to date. It is of considerable interest to know whether IRE1a and/or b play a role in ER stress responses and what that role might be.
PERK is another ER membrane–bound sensor/transducer mediating the third branch of the ER stress response in mammalian cells. Unlike IRE1 and ATF6, which act at the transcriptional level, PERK alleviates ER stress by attenuating bulk protein synthesis through the phosphorylation of eukaryotic translation initiation factor 2 α-subunit (Rutkowski and Kaufman, 2004). This type of UPR control has not been found in Arabidopsis. First, a PERK ortholog has not been identified in Arabidopsis. Second, the promoter element AARE (amino acid response element), which in mammals is characteristic of genes regulated through the PERK-ATF4 pathway, is not found in Arabidopsis genes upregulated by UPR (Martínez and Chrispeels, 2003).
UPR and Programmed Cell Death
Plants have signaling pathways leading from UPR to programmed cell death (PCD; Figure 4). The Arabidopsis heterotrimeric G protein plays a role in stress signaling leading to PCD as well as in plant development (Wang et al., 2007). Gβ forms a stable heterodimer with the Gα subunit, involved in the signaling events that trigger UPR-associated cell death. A null mutation in the Gβ subunit, but not Gα subunit, is more resistant to tunicamycin and has an alleviated UPR response, with attenuated induction of BiP and apoptosis (Wang et al., 2007). In mammals, apoptosis is prominently controlled through functionally conserved proteins, such as CED9/BCL-2 (anti-apoptotic protein) and BAX (pro-apoptotic protein). Although core regulators of PCD are conserved in mammals (e.g., the Bcl-2 family and caspases), they have not been identified in plants.
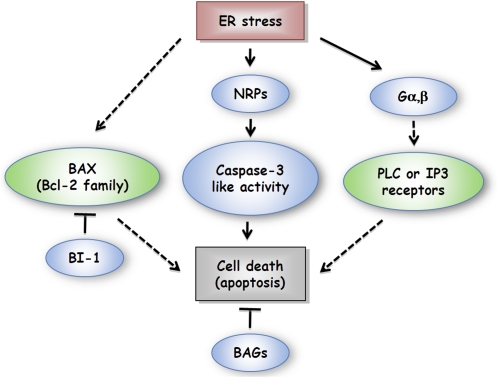
Figure 4.
ER Stress–Induced Cell Death (Apoptosis) in Plants.
ER stress activates the expression of NRPs, which induces the caspase-3–like activity to trigger cell death. ER stress promotes the formation of Gαβ heterodimer to induce PCD, presumably by activating PLC or IP3 receptor pathways. Negative regulators, such as BAGs and BI-1, repress PCD in plants. Note that the counterparts of mammalian BAX or BCL-2 have not been identified in plants so far.
The Bcl-2–associated athanogene (BAG) family is a multifunctional group of cytoprotective proteins that behave as molecular cochaperones (Briknarová et al., 2001) during UPR. The ER-localized BAG7 was shown to specifically interact with chaperone BiP2 in Arabidopsis and considered as an essential component of the UPR during heat and cold tolerance to delay the ER stress–induced cell death (Williams et al., 2010). Overexpression of At-BAG4 in tobacco plants confers tolerance to a wide range of abiotic stresses, such as UV light, cold, oxidants, and salt treatments (Doukhanina et al., 2006). BAX inhibitor-1 (BI-1) was first identified as a suppressor of cell death activated by BAX in yeast or mammalian cells (Xu and Reed, 1998). In mammalian cells, BI-1 was shown to interact with BCL-2 but not BAX or BAK. The ER-localized Arabidopsis BI-1 (At-BI-1) appears to play a similar role as a survival factor under multiple stress conditions that could trigger PCD, including cell death induced by fungal toxins, Pseudomonas infection, heat shock, or tunicamycin (Watanabe and Lam, 2008; Kawai-Yamada et al., 2009).
It is now clear that the regulation of cell death pathways is relevant to both abiotic and biotic stress responses in plants (Lam et al., 2001). Recently, N-rich proteins (NRPs) were identified as targets of a novel adaptive pathway that integrates ER and osmotic stress signals in soybean based on coordinate regulation and synergistic upregulation by tunicamycin (UPR inducer) and polyethylene glycol (osmotic stress inducer) treatments. The cell death domains containing NRPs were demonstrated to induce caspase activity and to be the critical mediators of osmotic- and ER stress–induced cell death in plants (Costa et al., 2008). Metacaspases are believed to be the functional homologs of animal caspases in plants, although metacaspases do not have caspase-like activities. Arabidopsis has nine metacaspases, and one of them, MC8, is strongly upregulated by oxidative stress induced by UVC and H2O2 treatment and mediates the activation of subsequent cell death (He et al., 2008).
What conditions induce cytoprotective activities and what conditions trigger PCD? This question was addressed in human cells in a study by Lin et al. (2007) wherein they examined the time course for the onset of PCD with continued application of stress. They found that signaling on the three branches of the UPR pathway showed differing degrees of persistence in the face of continued stress. All three branches were activated upon introduction of stress, but IRE1 signaling was attenuated first followed by ATF6 and lastly by PERK. The onset of apoptosis largely corresponded to the fall in IRE1 signaling. The authors attributed this to cytoprotective effects of IRE1 signaling and found when IRE1 was put under the control of a drug-inducible promoter, allowing for its signals to be sustained, cell survival was enhanced.
Similar time course studies need to be conducted in plants. What stress responses are short-term responses and what responses derive from chronic stress? Are some of the short-term responses to stress cytoprotective but lead to PCD when the stress persists? What might be the adaptive function in plants if short-term cytoprotection fails and long-term stress elicits PCD? Perhaps the collapse of cells overburdened with misfolded and potentially noxious proteins might spare others from the same fate.
Plant stress responses may confer tolerance and spare plants growing in the wild, but such responses may be counterproductive for crop plants. Plant stress responses generally slow plant growth and delay development. Constitutive expression of an activated bZIP17 construct in Arabidopsis results in dwarfed plants delayed in development (Liu et al., 2008). Only when the transgene is regulated by a stress-activated promoter does it confer any added measure of salt tolerance. In any case, it needs to be tested whether plants with disabled or altered stress responses are more productive under ideal growth conditions and when faced with environmental stress.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Table 1. A Partial List of Genes Involved in Arabidopsis Protein QC.
Supplementary Material
Acknowledgments
We acknowledge support from Fudan University (to J.-X.L.), the National Natural Science Foundation of China (31070233 to J.-X.L.), the U.S. National Science Foundation (IOS0919707 to S.H.H.), and the Iowa State University Plant Sciences Institute.
References
- Anderson J.V., Li Q.B., Haskell D.W., Guy C.L. (1994). Structural organization of the spinach endoplasmic reticulum-luminal 70-kilodalton heat-shock cognate gene and expression of 70-kilodalton heat-shock genes during cold acclimation. Plant Physiol. 104: 1359–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvim F.C., Carolino S.M.B., Cascardo J.C.M., Nunes C.C., Martinez C.A., Otoni W.C., Fontes E.P.B. (2001). Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiol. 126: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bass R., Ruddock L.W., Klappa P., Freedman R.B. (2004). A major fraction of endoplasmic reticulum-located glutathione is present as mixed disulfides with protein. J. Biol. Chem. 279: 5257–5262 [DOI] [PubMed] [Google Scholar]
- Bernales S., Papa F.R., Walter P. (2006). Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22: 487–508 [DOI] [PubMed] [Google Scholar]
- Boston R.S., Fontes E.B., Shank B.B., Wrobel R.L. (1991). Increased expression of the maize immunoglobulin binding protein homolog b-70 in three zein regulatory mutants. Plant Cell 3: 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boston R.S., Viitanen P.V., Vierling E. (1996). Molecular chaperones and protein folding in plants. Plant Mol. Biol. 32: 191–222 [DOI] [PubMed] [Google Scholar]
- Briknarová K., et al. (2001). Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat. Struct. Biol. 8: 349–352 [DOI] [PubMed] [Google Scholar]
- Brodsky J.L., Wojcikiewicz R.J. (2009). Substrate-specific mediators of ER associated degradation (ERAD). Curr. Opin. Cell Biol. 21: 516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryngelson J.D., Onuchic J.N., Socci N.D., Wolynes P.G. (1995). Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins 21: 167–195 [DOI] [PubMed] [Google Scholar]
- Burn J.E., Hurley U.A., Birch R.J., Arioli T., Cork A., Williamson R.E. (2002). The cellulose-deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. Plant J. 32: 949–960 [DOI] [PubMed] [Google Scholar]
- Caramelo J.J., Parodi A.J. (2008). Getting in and out from calnexin/calreticulin cycles. J. Biol. Chem. 283: 10221–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T.A. (2006). Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126: 361–373 [DOI] [PubMed] [Google Scholar]
- Ceriotti A., Duranti M., Bollini R. (1998). Effects of N-glycosylation on the folding and structure of plant proteins. J. Exp. Bot. 49: 1091–1103 [Google Scholar]
- Che P., Bussell J.D., Wenxu Zhou W., Estavillo G., Pogson B.J., Smith S.M. (2010). Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Sci. Signal., in press [DOI] [PubMed] [Google Scholar]
- Cheetham M.E., Caplan A.J. (1998). Structure, function and evolution of DnaJ: Conservation and adaptation of chaperone function. Cell Stress Chaperones 3: 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen A., et al. (2010). Higher plant calreticulins have acquired specialized functions in Arabidopsis. PLoS ONE 5: e11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc S., Hirsch C., Oggier D.M., Deprez P., Jakob C., Sommer T., Aebi M. (2009). Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 184: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.E., Lopes M.A., Gillikin J.W., Boston R.S., Larkins B.A. (1995). A defective signal peptide in the maize high-lysine mutant floury 2. Proc. Natl. Acad. Sci. USA 92: 6828–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M.D., Reis P.A., Valente M.A., Irsigler A.S., Carvalho C.M., Loureiro M.E., Aragão F.J., Boston R.S., Fietto L.G., Fontes E.P. (2008). A new branch of endoplasmic reticulum stress signaling and the osmotic signal converge on plant-specific asparagine-rich proteins to promote cell death. J. Biol. Chem. 283: 20209–20219 [DOI] [PubMed] [Google Scholar]
- D’Amico L., Valsasina B., Daminati M.G., Fabbrini M.S., Nitti G., Bollini R., Ceriotti A., Vitale A. (1992). Bean homologs of the mammalian glucose-regulated proteins: Induction by tunicamycin and interaction with newly synthesized seed storage proteins in the endoplasmic reticulum. Plant J. 2: 443–455 [DOI] [PubMed] [Google Scholar]
- Danon A., Rotari V.I., Gordon A., Mailhac N., Gallois P. (2004). Ultraviolet-C overexposure induces programmed cell death in Arabidopsis, which is mediated by caspase-like activities and which can be suppressed by caspase inhibitors, p35 and Defender against Apoptotic Death. J. Biol. Chem. 279: 779–787 [DOI] [PubMed] [Google Scholar]
- Denecke J., Goldman M.H., Demolder J., Seurinck J., Botterman J. (1991). The tobacco luminal binding protein is encoded by a multigene family. Plant Cell 3: 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V., Quan E.M., Weissman J.S. (2006). A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 126: 349–359 [DOI] [PubMed] [Google Scholar]
- Di Cola A., Frigerio L., Lord J.M., Ceriotti A., Roberts L.M. (2001). Ricin A chain without its partner B chain is degraded after retrotranslocation from the endoplasmic reticulum to the cytosol in plant cells. Proc. Natl. Acad. Sci. USA 98: 14726–14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cola A., Frigerio L., Lord J.M., Roberts L.M., Ceriotti A. (2005). Endoplasmic reticulum-associated degradation of ricin A chain has unique and plant-specific features. Plant Physiol. 137: 287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D.P., Van Lith M., Edwards R., Benham A. (2003). Cloning and initial characterization of the Arabidopsis thaliana endoplasmic reticulum oxidoreductins. Antioxid. Redox Signal. 5: 389–396 [DOI] [PubMed] [Google Scholar]
- Doukhanina E.V., Chen S., van der Zalm E., Godzik A., Reed J., Dickman M.B. (2006). Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J. Biol. Chem. 281: 18793–18801 [DOI] [PubMed] [Google Scholar]
- Englander S.W., Mayne L., Krishna M.M. (2007). Protein folding and misfolding: Mechanism and principles. Q. Rev. Biophys. 40: 287–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo J.E.F., Cascardo J.C.M., Carolino S.M.B., Alvim F.C., Fontes E.P.B. (1997). Water-stress regulation and molecular analysis of the soybean BiP gene family. Braz. J. Plant Physiol. 9: 103–110 [Google Scholar]
- Flynn G.C., Chappell T.G., Rothman J.E. (1989). Peptide binding and release by proteins implicated as catalysts of protein assembly. Science 245: 385–390 [DOI] [PubMed] [Google Scholar]
- Flynn G.C., Pohl J., Flocco M.T., Rothman J.E. (1991). Peptide-binding specificity of the molecular chaperone BiP. Nature 353: 726–730 [DOI] [PubMed] [Google Scholar]
- Foresti O., Frigerio L., Holkeri H., de Virgilio M., Vavassori S., Vitale A. (2003). A phaseolin domain involved directly in trimer assembly is a determinant for binding by the chaperone BiP. Plant Cell 15: 2464–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Mizukado S., Fujita Y., Ichikawa T., Nakazawa M., Seki M., Matsui M., Yamaguchi-Shinozaki K., Shinozaki K. (2007). Identification of stress-tolerance-related transcription-factor genes via mini-scale Full-length cDNA Over-eXpressor (FOX) gene hunting system. Biochem. Biophys. Res. Commun. 364: 250–257 [DOI] [PubMed] [Google Scholar]
- Frand A.R., Kaiser C.A. (1999). Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol. Cell 4: 469–477 [DOI] [PubMed] [Google Scholar]
- Frigerio L., Roberts L.M. (1998). The enemy within: Ricin and plant cells. J. Exp. Bot. 49: 1473–1480 [Google Scholar]
- Frigerio L., Vitale A., Lord J.M., Ceriotti A., Roberts L.M. (1998). Free ricin A chain, proricin, and native toxin have different cellular fates when expressed in tobacco protoplasts. J. Biol. Chem. 273: 14194–14199 [DOI] [PubMed] [Google Scholar]
- Gallois P., Makishima T., Hecht V., Despres B., Laudié M., Nishimoto T., Cooke R. (1997). An Arabidopsis thaliana cDNA complementing a hamster apoptosis suppressor mutant. Plant J. 11: 1325–1331 [DOI] [PubMed] [Google Scholar]
- Gao H., Brandizzi F., Benning C., Larkin R.M. (2008). A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 16398–16403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R., Jarosch E., Sommer T., Hirsch C. (2006b). A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat. Cell Biol. 8: 849–854 [DOI] [PubMed] [Google Scholar]
- Gauss R., Sommer T., Jarosch E. (2006a). The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 25: 1827–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M.J. (1999). Role and regulation of the ER chaperone BiP. Semin. Cell Dev. Biol. 10: 465–472 [DOI] [PubMed] [Google Scholar]
- Gross E., Sevier C.S., Heldman N., Vitu E., Bentzur M., Kaiser C.A., Thorpe C., Fass D. (2006). Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. USA 103: 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C., Braakman I., Helenius A. (1994). Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. USA 91: 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C., Helenius A. (1994). Folding of VSV G protein: Sequential interaction with BiP and calnexin. Science 266: 456–458 [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Hayer-Hartl M. (2009). Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16: 574–581 [DOI] [PubMed] [Google Scholar]
- He R., Drury G.E., Rotari V.I., Gordon A., Willer M., Farzaneh T., Woltering E.J., Gallois P. (2008). Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J. Biol. Chem. 283: 774–783 [DOI] [PubMed] [Google Scholar]
- Helenius A., Aebi M. (2004). Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73: 1019–1049 [DOI] [PubMed] [Google Scholar]
- Holst B., Tachibana C., Winther J.R. (1997). Active site mutations in yeast protein disulfide isomerase cause dithiothreitol sensitivity and a reduced rate of protein folding in the endoplasmic reticulum. J. Cell Biol. 138: 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard S.C., Ivatt R.J. (1981). Synthesis and processing of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 50: 555–583 [DOI] [PubMed] [Google Scholar]
- Hurtley S.M., Helenius A. (1989). Protein oligomerization in the endoplasmic reticulum. Annu. Rev. Cell Biol. 5: 277–307 [DOI] [PubMed] [Google Scholar]
- Huyer G., Piluek W.F., Fansler Z., Kreft S.G., Hochstrasser M., Brodsky J.L., Michaelis S. (2004). Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 279: 38369–38378 [DOI] [PubMed] [Google Scholar]
- Hwang C., Sinskey A.J., Lodish H.F. (1992). Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257: 1496–1502 [DOI] [PubMed] [Google Scholar]
- Ishiguro S., Watanabe Y., Ito N., Nonaka H., Takeda N., Sakai T., Kanaya H., Okada K. (2002). SHEPHERD is the Arabidopsis GRP94 responsible for the formation of functional CLAVATA proteins. EMBO J. 21: 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y., Fedoroff N.V., Koizumi N. (2008). Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y., Koizumi N. (2005). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. USA 102: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelitto-Van Dooren E.P., Vidal S., Denecke J. (1999). Anticipating endoplasmic reticulum stress. A novel early response before pathogenesis-related gene induction. Plant Cell 11: 1935–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X.Y., He L.H., Jing R.L., Li R.Z. (2009). Calreticulin: Conserved protein and diverse functions in plants. Physiol. Plant. 136: 127–138 [DOI] [PubMed] [Google Scholar]
- Jia X.Y., Xu C.Y., Jing R.L., Li R.Z., Mao X.G., Wang J.P., Chang X.P. (2008). Molecular cloning and characterization of wheat calreticulin (CRT) gene involved in drought-stressed responses. J. Exp. Bot. 59: 739–751 [DOI] [PubMed] [Google Scholar]
- Jin H., Hong Z., Su W., Li J. (2009). A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 106: 13612–13617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Yan Z., Nam K.H., Li J. (2007). Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell 26: 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamauchi S., Nakatani H., Nakano C., Urade R. (2005). Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J. 272: 3461–3476 [DOI] [PubMed] [Google Scholar]
- Kanehara K., Kawaguchi S., Ng D.T. (2007). The EDEM and Yos9p families of lectin-like ERAD factors. Semin. Cell Dev. Biol. 18: 743–750 [DOI] [PubMed] [Google Scholar]
- Kawai-Yamada M., Hori Z., Ogawa T., Ihara-Ohori Y., Tamura K., Nagano M., Ishikawa T., Uchimiya H. (2009). Loss of calmodulin binding to Bax inhibitor-1 affects Pseudomonas-mediated hypersensitive response-associated cell death in Arabidopsis thaliana. J. Biol. Chem. 284: 27998–28003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst M.E., Meyer D.J., Gibbon B.C., Jung R., Boston R.S. (2005). Identification and characterization of endoplasmic reticulum-associated degradation proteins differentially affected by endoplasmic reticulum stress. Plant Physiol. 138: 218–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein E.M., Mascheroni L., Pompa A., Ragni L., Weimar T., Lilley K.S., Dupree P., Vitale A. (2006). Plant endoplasmin supports the protein secretory pathway and has a role in proliferating tissues. Plant J. 48: 657–673 [DOI] [PubMed] [Google Scholar]
- Kleizen B., Braakman I. (2004). Protein folding and quality control in the endoplasmic reticulum. Curr. Opin. Cell Biol. 16: 343–349 [DOI] [PubMed] [Google Scholar]
- Knop M., Finger A., Braun T., Hellmuth K., Wolf D.H. (1996). Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 15: 753–763 [PMC free article] [PubMed] [Google Scholar]
- Koiwa H., Li F., McCully M.G., Mendoza I., Koizumi N., Manabe Y., Nakagawa Y., Zhu J., Rus A., Pardo J.M., Bressan R.A., Hasegawa P.M. (2003). The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15: 2273–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N., Martinez I.M., Kimata Y., Kohno K., Sano H., Chrispeels M.J. (2001). Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 127: 949–962 [PMC free article] [PubMed] [Google Scholar]
- Koizumi N., Ujino T., Sano H., Chrispeels M.J. (1999). Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiol. 121: 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R., Kornfeld S. (1985). Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54: 631–664 [DOI] [PubMed] [Google Scholar]
- Lam E., Kato N., Lawton M. (2001). Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411: 848–853 [DOI] [PubMed] [Google Scholar]
- Lerouxel O., Mouille G., Andème-Onzighi C., Bruyant M.P., Séveno M., Loutelier-Bourhis C., Driouich A., Höfte H., Lerouge P. (2005). Mutants in DEFECTIVE GLYCOSYLATION, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein underglycosylation and defects in cell differentiation and growth. Plant J. 42: 455–468 [DOI] [PubMed] [Google Scholar]
- Li J., Zhao-Hui C., Batoux M., Nekrasov V., Roux M., Chinchilla D., Zipfel C., Jones J.D. (2009). Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. USA 106: 15973–15978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B., Shokat K.M., Lavail M.M., Walter P. (2007). IRE1 signaling affects cell fate during the unfolded protein response. Science 318: 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg M.O., Oliveberg M. (2007). Malleability of protein folding pathways: A simple reason for complex behaviour. Curr. Opin. Struct. Biol. 17: 21–29 [DOI] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Che P., Howell S.H. (2007a). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19: 4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Che P., Howell S.H. (2007b). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 51: 897–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Howell S.H. (2008). Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ. 31: 1735–1743 [DOI] [PubMed] [Google Scholar]
- Lu D.P., Christopher D.A. (2008). Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol. Genet. Genomics 280: 199–210 [DOI] [PubMed] [Google Scholar]
- Lupattelli F., Pedrazzini E., Bollini R., Vitale A., Ceriotti A. (1997). The rate of phaseolin assembly is controlled by the glucosylation state of its N-linked oligosaccharide chains. Plant Cell 9: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra J.D., Kaufman R.J. (2007). The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 18: 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R.S., Jolliffe N.A., Ceriotti A., Snowden C.J., Lord J.M., Frigerio L., Roberts L.M. (2008). The role of CDC48 in the retro-translocation of non-ubiquitinated toxin substrates in plant cells. J. Biol. Chem. 283: 15869–15877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I.M., Chrispeels M.J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R., Kondo M., Nishimura M., Hara-Nishimura I. (2003). A novel ER-derived compartment, the ER body, selectively accumulates a beta-glucosidase with an ER-retention signal in Arabidopsis. Plant J. 33: 493–502 [DOI] [PubMed] [Google Scholar]
- Meunier L., Usherwood Y.K., Chung K.T., Hendershot L.M. (2002). A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell 13: 4456–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen K.W., Molinari M. (2006). N-linked glycan recognition and processing: the molecular basis of endoplasmic reticulum quality control. Curr. Opin. Struct. Biol. 16: 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Piffanelli P., Devoto A., Miklis M., Elliott C., Ortmann B., Schulze-Lefert P., Panstruga R. (2005). Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell 17: 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Brodsky J.L. (2008). The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9: 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi M.C., Giacomelli E., Dainese P., Fitchette-Laine A.C., Faye L., Baldan B., Navazio L., Mariani P. (1998). Ginkgo biloba express calreticulin, the major calcium-binding reticuloplasmin in eucaryotic cells. Bot. Acta 111: 66–70 [Google Scholar]
- Navazio L., Miuzzo M., Royle L., Baldan B., Varotto S., Merry A.H., Harvey D.J., Dwek R.A., Rudd P.M., Mariani P. (2002). Monitoring endoplasmic reticulum-to-Golgi traffic of a plant calreticulin by protein glycosylation analysis. Biochemistry 41: 14141–14149 [DOI] [PubMed] [Google Scholar]
- Nekrasov V., et al. (2009). Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh S.J., Kwon C.S., Chung W.I. (2002). Characterization of two homologs of Ire1p, a kinase/endoribonuclease in yeast, in Arabidopsis thaliana. Biochim. Biophys. Acta 1575: 130–134 [DOI] [PubMed] [Google Scholar]
- Oda Y., Okada T., Yoshida H., Kaufman R.J., Nagata K., Mori K. (2006). Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 172: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Koizumi N., Yamaguchi Y., Kimata Y., Kohno K., Sano H. (2002). Isolation and characterization of a putative transducer of endoplasmic reticulum stress in Oryza sativa. Plant Cell Physiol. 43: 532–539 [DOI] [PubMed] [Google Scholar]
- Olivari S., Cali T., Salo K.E., Paganetti P., Ruddock L.W., Molinari M. (2006). EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem. Biophys. Res. Commun. 349: 1278–1284 [DOI] [PubMed] [Google Scholar]
- Pagny S., Cabanes-Macheteau M., Gillikin J.W., Leborgne-Castel N., Lerouge P., Boston R.S., Faye L., Gomord V. (2000). Protein recycling from the Golgi apparatus to the endoplasmic reticulum in plants and its minor contribution to calreticulin retention. Plant Cell 12: 739–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagny S., Lerouge P., Faye L., Gomord V. (1999). Signals and mechanisms for protein retention in the endoplasmic reticulum. J. Exp. Bot. 50: 157–164 [Google Scholar]
- Pilon M., Schekman R., Römisch K. (1997). Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 16: 4540–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan E.M., Kamiya Y., Kamiya D., Denic V., Weibezahn J., Kato K., Weissman J.S. (2008). Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol. Cell 32: 870–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancour D.M., Dickey C.E., Park S., Bednarek S.Y. (2002). Characterization of AtCDC48. Evidence for multiple membrane fusion mechanisms at the plane of cell division in plants. Plant Physiol. 130: 1241–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Walter P. (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Ruddock L.W., Molinari M. (2006). N-glycan processing in ER quality control. J. Cell Sci. 119: 4373–4380 [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada C., Kelleher D.J., Gilmore R. (2009). Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136: 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski D.T., Kaufman R.J. (2004). A trip to the ER: Coping with stress. Trends Cell Biol. 14: 20–28 [DOI] [PubMed] [Google Scholar]
- Sanderfoot A.A., Raikhel R.V. (2003). The secretory system of Arabidopsis. The Arabidopsis Book, Somerville C.R., Meyerowitz E.M., eds (Rockville, MD: American Society of Plant Biologists; ), doi/http://www.aspb.org/publications/arabidopsis/ [Google Scholar]
- Schlenstedt G., Harris S., Risse B., Lill R., Silver P.A. (1995). A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/Kar2p via a conserved domain that specifies interactions with Hsp70s. J. Cell Biol. 129: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott A., Ravaud S., Keller S., Radzimanowski J., Viotti C., Hillmer S., Sinning I., Strahl S. (2010). Arabidopsis stromal-derived Factor2 (SDF2) is a crucial target of the unfolded protein response in the endoplasmic reticulum. J. Biol. Chem. 285: 18113–18121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder M., Kaufman R.J. (2005). ER stress and the unfolded protein response. Mutat. Res. 569: 29–63 [DOI] [PubMed] [Google Scholar]
- Sevier C.S., Kaiser C.A. (2006). Disulfide transfer between two conserved cysteine pairs imparts selectivity to protein oxidation by Ero1. Mol. Biol. Cell 17: 2256–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparvoli F., Faoro F., Daminati M.G., Ceriotti A., Bollini R. (2000). Misfolding and aggregation of vacuolar glycoproteins in plant cells. Plant J. 24: 825–836 [DOI] [PubMed] [Google Scholar]
- Spear E.D., Ng D.T. (2005). Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J. Cell Biol. 169: 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens F.J., Argon Y. (1999). Protein folding in the ER. Semin. Cell Dev. Biol. 10: 443–454 [DOI] [PubMed] [Google Scholar]
- Swiezewska E., Danikiewicz W. (2005). Polyisoprenoids: Structure, biosynthesis and function. Prog. Lipid Res. 44: 235–258 [DOI] [PubMed] [Google Scholar]
- Tajima H., Iwata Y., Iwano M., Takayama S., Koizumi N. (2008b). Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochem. Biophys. Res. Commun. 374: 242–247 [DOI] [PubMed] [Google Scholar]
- Tajima H., Iwata Y., Koizumi N. (2008a). Endoplasmic reticulum stress response and regulated intramembrane proteolysis in plants. Plant Biotechnol. 25: 271–278 [Google Scholar]
- Urade R. (2007). Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J. 274: 1152–1171 [DOI] [PubMed] [Google Scholar]
- Valente M.A., et al. (2009). The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. J. Exp. Bot. 60: 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Ng D.T. (2004). Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 165: 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., Deshaies R.J. (2000). Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11: 3425–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A., Bielli A., Ceriotti A. (1995). The binding protein associates with monomeric phaseolin. Plant Physiol. 107: 1411–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A., Boston R.S. (2008). Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic 9: 1581–1588 [DOI] [PubMed] [Google Scholar]
- Vitale A., Denecke J. (1999). The endoplasmic reticulum-gateway of the secretory pathway. Plant Cell 11: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Narendra S., Fedoroff N. (2007). Heterotrimeric G protein signaling in the Arabidopsis unfolded protein response. Proc. Natl. Acad. Sci. USA 104: 3817–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Weaver N.D., Kesarwani M., Dong X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. (2008). BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J. Biol. Chem. 283: 3200–3210 [DOI] [PubMed] [Google Scholar]
- Wei J., Gaut J.R., Hendershot L.M. (1995). In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J. Biol. Chem. 270: 26677–26682 [DOI] [PubMed] [Google Scholar]
- Wiertz E.J., Tortorella D., Bogyo M., Yu J., Mothes W., Jones T.R., Rapoport T.A., Ploegh H.L. (1996). Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384: 432–438 [DOI] [PubMed] [Google Scholar]
- Williams B., Kabbage M., Britt R., Dickman M.B. (2010). AtBAG7, an Arabidopsis Bcl-2-associated athanogene, resides in the endoplasmic reticulum and is involved in the unfolded protein response. Proc. Natl. Acad. Sci. USA 107: 6088–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman R.L., Powers E.T., Buxbaum J.N., Kelly J.W., Balch W.E. (2007). An adaptable standard for protein export from the endoplasmic reticulum. Cell 131: 809–821 [DOI] [PubMed] [Google Scholar]
- Xu Q., Reed J.C. (1998). Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol. Cell 1: 337–346 [DOI] [PubMed] [Google Scholar]
- Yang K.Z., Xia C., Liu X.L., Dou X.Y., Wang W., Chen L.Q., Zhang X.Q., Xie L.F., He L., Ma X., Ye D. (2009). A mutation in Thermosensitive Male Sterile 1, encoding a heat shock protein with DnaJ and PDI domains, leads to thermosensitive gametophytic male sterility in Arabidopsis. Plant J. 57: 870–882 [DOI] [PubMed] [Google Scholar]
- Zhang H., Ohyama K., Boudet J., Chen Z., Yang J., Zhang M., Muranaka T., Maurel C., Zhu J.K., Gong Z. (2008). Dolichol biosynthesis and its effects on the unfolded protein response and abiotic stress resistance in Arabidopsis. Plant Cell 20: 1879–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2008). Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20: 10–16 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.