The meiosis II outer plaque (MOP) acts a vesicle tethering complex that is a site for de novo membrane formation. Novel mutants in a MOP protein reveal that interaction of vesicles with the MOP surface is required to recruit a second tethering complex, the exocyst, to the vesicles, suggesting a mechanism by which the MOP promotes vesicle fusion.
Abstract
During meiosis II in Saccharomyces cerevisiae, the cytoplasmic face of the spindle pole body, referred to as the meiosis II outer plaque (MOP), is modified in both composition and structure to become the initiation site for de novo formation of a membrane called the prospore membrane. The MOP serves as a docking complex for precursor vesicles that are targeted to its surface. Using fluorescence resonance energy transfer analysis, the orientation of coiled-coil proteins within the MOP has been determined. The N-termini of two proteins, Mpc54p and Spo21p, were oriented toward the outer surface of the structure. Mutations in the N-terminus of Mpc54p resulted in a unique phenotype: precursor vesicles loosely tethered to the MOP but did not contact its surface. Thus, these mpc54 mutants separate the steps of vesicle association and docking. Using these mpc54 mutants, we determined that recruitment of the Rab GTPase Sec4p, as well as the exocyst components Sec3p and Sec8p, to the precursor vesicles requires vesicle docking to the MOP. This suggests that the MOP promotes membrane formation both by localization of precursor vesicles to a particular site and by recruitment of a second tethering complex, the exocyst, that stimulates downstream events of fusion.
INTRODUCTION
In Saccharomyces cerevisiae, meiosis and sporulation are induced when diploid cells are cultured in the absence of nitrogen and the presence of a nonfermentable carbon source (Esposito and Klapholz, 1981). During sporulation, prospore membrane formation is initiated by the fusion of post-Golgi vesicles into a novel membrane structure through a developmentally controlled rearrangement of the secretory pathway (Neiman, 1998). These vesicles are targeted to accumulate at the four poles of the meiotic spindles where they coalesce into four prospore membranes. As meiosis II progresses, each prospore membrane elongates toward the center of a meiotic spindle, engulfing adjacent nuclear lobes as well as cytoplasm and organelles (Moens and Rapport, 1971; Byers, 1981; Suda et al., 2007). At the completion of meiosis II and nuclear division, each of the four prospore membranes closes, forming four immature, haploid spores within the mother cell cytoplasm (Neiman, 1998).
During meiosis II, each of the four prospore membranes is attached to one of four spindle pole bodies, which are embedded within the nuclear envelope. In vegetative growth, spindle pole bodies serve as microtubule organizing centers, analogous to the centrosome in higher eukaryotes (Robinow and Marak, 1966; Moens and Rapport, 1971; Byers and Goetsch, 1974). Spindle pole bodies are large, cylindrical structures organized into proteinaceous layers called plaques. The central plaque spans the nuclear envelope and forms the stable core of the spindle pole body (Rout and Kilmartin, 1990; Kilmartin et al., 1993; Geissler et al., 1996; Kilmartin and Goh, 1996; Bullitt et al., 1997). The organization of the central plaque has been predicted using fluorescence resonance energy transfer (FRET) analysis (Muller et al., 2005).
The outer plaque forms the cytoplasmic face of the spindle pole body. During mitosis, the outer plaque is composed of Cnm67p, Nud1p, Spc72p, and a microtubule nucleation complex (Bullitt et al., 1997; Adams and Kilmartin, 1999; Schaerer et al., 2001). During meiosis II, the initiation of prospore membrane growth requires a developmentally controlled modification of the outer plaque that confers a new function as a nucleation site for membrane growth (Moens and Rapport, 1971). At this stage, Spc72p is removed from the cytoplasmic surface of the outer plaque and is replaced by the meiotically induced proteins Mpc54p, Spo21p, Spo74p, and Ady4p (Knop and Strasser, 2000; Bajgier et al., 2001; Nickas et al., 2003). Each of the newly modified MOPs serves as the initiation site for the de novo formation of one prospore membrane (Moens and Rapport, 1971).
Yeast sporulation provides an excellent model for the study of vesicle trafficking during cellular differentiation (Neiman, 1998, 2005). The de novo formation of prospore membranes requires both a rerouting of the secretory pathway to target post-Golgi vesicles to the spindle poles and a rewiring of preexisting molecular machineries, such as the fusion machinery. (Neiman, 1998; Jantti et al., 2002; Nakanishi et al., 2006). Post-Golgi vesicles dock at the cytoplasmic surface of the meiosis II outer plaque (MOP), and subsequently fuse to form the prospore membrane (Neiman, 1998; Neiman et al., 2000; Nakanishi et al., 2006). Soluble N-ethylmaleimide–sensitive factor (NSF) attachment protein receptors (SNAREs) are required for vesicle fusion throughout the secretory pathway (Bennett and Scheller, 1993; Weimbs et al., 1998; Chen and Scheller, 2001). Interestingly, fusion at the prospore membrane is mediated by the same SNARE machinery that promotes the fusion of secretory vesicles with the plasma membrane during mitotic growth, with two critical modifications (Neiman, 1998; Nakanishi et al., 2006). The first difference between the mitotic and meiotic SNARE complex is the addition of a meiotically induced t-SNARE, Spo20p, which shares partially redundant function with its mitotic counterpart, Sec9p (Neiman, 1998; Yang et al., 2008) The second difference is the requirement for Sso1p during meiosis II, because its redundant mitotic homologue, Sso2, is not an essential component of the meiotic SNARE complex (Jantti et al., 2002; Nakanishi et al., 2006).
Deletions of genes in the SNARE machinery, such as an sso1Δ, do not disrupt the formation of the MOP, but prevent docked vesicles from fusing together at the MOP's surface (Nakanishi et al., 2006). In contrast, the deletion of the MOP components MPC54, SPO21, or SPO74 leads to the absence of both the MOP structure and docked vesicles (Knop and Strasser, 2000; Bajgier et al., 2001; Nickas et al., 2003). This indicates that the formation of the MOP complex is required for subsequent prospore membrane initiation.
In addition to the SNARE complex, a multiprotein complex called the exocyst is also required for post-Golgi vesicle fusion at the plasma membrane (TerBush et al., 1996). The exocyst docks vesicles to the plasma membrane before fusion (Guo et al., 1999). Two of the exocyst subunits, Sec3p and Exo70p, localize to the plasma membrane by interacting with Rho and Rac family GTPases (Robinson et al., 1999; Guo et al., 2001; Zhang et al., 2001). The remaining exocyst subunits (Sec5p, Sec6p, Sec8p, Sec10p, Sec15p, and Exo84p) are associated with secretory vesicles via interaction of the Sec15 subunit with the Rab GTPase Sec4p (Salminen and Novick, 1989; TerBush et al., 1996; Finger et al., 1998; Guo et al., 1999). Assembly of the complete exocyst by interaction of the plasma membrane– and vesicle-associated subunits tethers the vesicle to the plasma membrane and is necessary for its subsequent fusion (TerBush et al., 1996; Finger et al., 1998; Guo et al., 1999). During prospore membrane initiation, the MOP serves as a vesicle-docking complex (Nakanishi et al., 2006). However, exocyst mutants display defects in prospore membrane formation, suggesting that vesicle docking through the MOP may not be sufficient to promote prospore membrane initiation (Neiman, 1998).
Here we describe the organization of coiled-coil proteins within the MOP complex. On the basis of our model of the MOP, we predicted that the N-termini of Mpc54p and Spo21p are positioned at the cytoplasmic face of the MOP, where vesicles dock during prospore membrane initiation. Phylogenetically conserved residues within the N-terminus of Mpc54p were mutated. Five nonnull mpc54 mutant alleles that formed normal MOP structures were examined for their effect on prospore membrane growth. The characterization of these alleles has revealed that Mpc54p plays a role in docking vesicles to the MOP and that this docking is necessary for vesicle fusion. Additionally, these alleles have allowed us to separate an initial loose tethering of vesicles to the MOP from docking of vesicles to the surface of the MOP. Using an mpc54 mutant, we found that vesicle docking to the MOP is specifically required for recruitment of the exocyst complex to precursor vesicles. Therefore, vesicle docking to the MOP might promote subsequent fusion through the recruitment of a second tethering complex.
MATERIALS AND METHODS
Yeast Strains and Media
Standard S. cerevisiae genetic methods and media were used (Rose and Fink, 1990). The strains used in this study are listed in Table 1. All strains used were in the fast-sporulating SK-1 strain background (Kane and Roth, 1974). Gene insertions and replacements were performed using cassettes amplified by PCR (Longtine et al., 1998) and verified by PCR or phenotype.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| AN117-4B | MATα arg4-NspI his3ΔSK hoΔ::LYS2 leu2 lys2 rme1Δ::LEU2 trp1::hisG ura3 | Neiman et al. (2000) |
| AN117-16D | MATahis3ΔSK hoΔ::LYS2 leu2 lys2 trp1::hisG ura3 | Neiman et al. (2000) |
| NY50 | MATα ura3 his3ΔSK trp1::hisG arg4-NspI lys2 hoΔ::LYS2 rme1Δ::LEU2 leu2 mpc54Δ::his5+ | Nickas et al. (2003) |
| NY51 | MATaura3 leu2 trp1-hisG his3ΔSK lys2 hoΔ::LYS2 mpc54Δ::his5+ | Nickas et al. (2003) |
| NY541 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG mpc54Δ::his5+/mpc54Δ::his5+ | Nickas et al. (2003) |
| HI58 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSKΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG sso1Δ::his5+/sso1Δ::his5+MPC54-RFP::URA3 | Nakanishi et al. (2006) |
| HI66 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG sso1Δ::his5+/sso1Δ::his5+ MPC54-RFP::URA3 SEC8-GFP::LEU2 | This study |
| EMD130 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG mpc54Δ::his5+/mpc54Δ::his5+ SPC42-RFP::HIS3/SPC42-RFP::HIS3 | This study |
| EMD131 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSKh leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG sso1Δ::his5+/sso1Δ::his5+ MPC54-RFP::TRP1 | This study |
| EMD132 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG mpc54Δ::his5+/mpc54Δ::his5+ SEC8-GFP::LEU2 | This study |
| EMD26 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG mpc54Δ::his5+/mpc54Δ::his5+ MPC54-RFP::TRP1/MPC54-RFP::TRP1 | This study |
| EMD27 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG mpc54Δ::his5+/mpc54Δ::his5+ mpc54-RFP-40::TRP1/mpc54-RFP-40::TRP1 | This study |
| EMD28 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG mpc54Δ::his5+/mpc54Δ::his5+ mpc54-RFP-47::TRP1/mpc54-RFP-47::TRP1 | This study |
| EMD29 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG mpc54Δ::his5+/mpc54Δ::his5+ mpc54-RFP-118::TRP1/mpc54-RFP-118::TRP1 | This study |
| EMD30 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG mpc54Δ::his5+/mpc54Δ::his5+ mpc54-RFP-119::TRP1/mpc54-RFP-119::TRP1 | This study |
| EMD31 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG mpc54Δ::his5+/mpc54Δ::his5+ mpc54-RFP-145::TRP1/mpc54-RFP-145::TRP1 | This study |
| AN1107 | MATα arg4-NspI his3ΔSK hoΔ::LYS2 leu2 lys2 rme1Δ::LEU2 trp1::hisG ura3 CNM67-YFP::his5+ | This study |
| AN1108 | MATα arg4-NspI his3ΔSK hoΔ::LYS2 leu2 lys2 rme1Δ::LEU2 trp1::hisG ura3 MPC54-YFP::his5+ | This study |
| AN1117 | MATahis3ΔSK hoΔ::LYS2 leu2 lys2 trp1::hisG ura3 SPO21-CFP::his5+ | This study |
| KBH1 | MATα arg4-NspI his3ΔSK hoΔ::LYS2 leu2 lys2 rme1Δ::LEU2 trp1::hisG ura3 KlURA3-PCNM67-YFP::MPC54 | This study |
| MNH51 | MATahis3ΔSK hoΔ::LYS2 leu2 lys2 trp1::hisG ura3 MPC54::CFP-CgTRP1 | This study |
| MNH52 | MATα arg4-NspI his3ΔSK hoΔ::LYS2 leu2 lys2 rme1Δ::LEU2 trp1::hisG ura3 SPO21::YFP-CgTRP1 | This study |
| MNH53 | MATahis3ΔSK hoΔ::LYS2 leu2 lys2 trp1::hisG ura3 CNM67::CFP-CgTRP1 | This study |
| MNH54 | MATahis3ΔSK hoΔ::LYS2 leu2 lys2 trp1::hisG ura3 SPC42::YFP-KlURA3 | This study |
| MNH55 | MATα arg4-NspI his3ΔSK hoΔ::LYS2 leu2 lys2 rme1Δ::LEU2 trp1::hisG ura3 KlURA3-PCNM67-YFP::CNM67 | This study |
| YS91 | MATα arg4-NspI his3ΔSK hoΔ::LYS2 leu2 lys2 rme1Δ::LEU2 trp1::hisG ura3 KlURA3- PMPC54-YFP::MCP54 | This study |
| YS92 | MATα arg4-NspI his3ΔSK hoΔ::LYS2 leu2 lys2 rme1Δ::LEU2 trp1::hisG ura3 KlURA3-PMPC54-YFP::SPO21 | This study |
| YS236 | MATahis3ΔSK hoΔ::LYS2 leu2 lys2 trp1::hisG ura3 CgTRP1-PMPC54-CFP::SPO21 | This study |
| YS312 | MATahis3ΔSK hoΔ::LYS2 leu2 lys2 trp1::hisG ura3 CgTRP1-PMPC54-CFP::MPC54 | This study |
| YS316 | MATahis3ΔSK hoΔ::LYS2 leu2 lys2 trp1::hisG ura3 CgTRP1-PMPC54-CFP::SPO21 | This study |
| AN310 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG MPC54-YFP::his5+/MPC54-YFP::his5+ SPO21-CFP::his5+/SPO21-CFP::his5+ | This study |
| AN317 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG CNM67-YFP::his5+/CNM67-YFP::his5+ SPO21-CFP::his5+/SPO21-CFP::his5+ | This study |
| EMD52 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG KlURA3-PMPC54-YFP::SPO21/KlURA3-PMPC54-YFP::SPO21 CgTRP1-PMPC54-CFP::MPC54/CgTRP1-PMPC54-CFP::MPC54 | This study |
| EMD100 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG KlURA3-PMPC54-YFP::SPO21/SPO21-CFP::his5+ | This study |
| EMD104 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG MPC54-YFP::his5+/MPC54::CFP-CgTRP1 | This study |
| EMD107 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG MPC54::KlURA3-PMPC54-YFP/CgTRP1-PMPC54-CFP::MPC54 | This study |
| EMD105 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG MPC54::KlURA3-PMPC54-YFP/MPC54::CFP-CgTRP1 | This study |
| KBD3 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG KlURA3-PCNM67-YFP::MPC54/KlURA3- PCNM67-YFP::MPC54 SPO21-CFP::his5+/SPO21-CFP::his5+ | This study |
| MND88 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG CNM67-YFP::his5+/CNM67-YFP::his5+ MPC54::CFP-CgTRP1/MPC54::CFP-CgTRP1 | This study |
| MND100 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG SPC42::YFP-KlURA3/SPC42::YFP-KlURA3 CNM67::CFP-CgTRP1/CNM67::CFP-CgTRP1 | This study |
| MND101 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG KlURA3- PCNM67-YFP::CNM67/KlURA3- PCNM67-YFP::CNM67 MPC54::CFP-CgTRP1/MPC54::CFP-CgTRP1 | This study |
| MND103 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG KlURA3-PCNM67-YFP::CNM67/KlURA3-PCNM67-YFP::CNM67 SPO21-CFP::his5+/SPO21-CFP::his5+ | This study |
| YS118 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG KlURA3-PMPC54-YFP::SPO21/KlURA3-PMPC54-YFP::SPO21 MPC54::CFP-CgTRP1/MPC54::CFP-CgTRP1 | This study |
| YS326 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG CNM67-YFP::his5+/CNM67-YFP::his5+ CgTRP1-PMPC54-CFP::MPC54/CgTRP1-PMPC54-CFP::MPC54 | This study |
| YS329 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG SPO21::YFP-CgTRP/SPO21::YFP-CgTRP1 CgTRP1-PMPC54-CFP::MPC54/CgTRP1-PMPC54-CFP::MPC54 | This study |
| YS335 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG KlURA3- PCNM67-YFP::CNM67/KlURA3- PCNM67-YFP::CNM67 CgTRP1-PMPC54-CFP::MPC54/CgTRP1-PMPC54-CFP::MPC54 | This study |
| YS344 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG KlURA3- PCNM67-YFP::CNM67/KlURA3- PCNM67-YFP::CNM67 CgTRP1-PMPC54-CFP::SPO21/CgTRP1-PMPC54-CFP::SPO21 | This study |
| YS353 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK leu2/leu2 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 ura3/ura3 trp1::hisG/trp1::hisG CNM67-YFP::his5+/CNM67-YFP::his5+ CgTRP1-PMPC54-CFP::SPO21/CgTRP1-PMPC54-CFP::SPO21 | This study |
For the FRET studies, cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) were used as the donor fluorophore and the acceptor fluorophore, respectively. FRET strains were constructed by crossing two haploid strains, one containing a CFP-tagged MOP component (CFP-A) and one containing a YFP-tagged MOP component (YFP-B). These strains were sporulated, tetrads were dissected, and two of the CFP-A/YFP-B segregants were mated. The required haploid strains were constructed as follows. For N-terminal CFP tags: TRP1-PMPC54-CFP was inserted at the 5′ end of the open reading frames (ORFs) of MPC54 and SPO21 in AN117-16D. For C-terminal CFP tags: CFP-TRP1 was inserted at the 3′ end of the ORFs of CNM67 and MPC54 in AN117-16D. CFP-his5+ was inserted at the 3′ end of the ORF of SPO21 in AN117-16D. For N-terminal YFP tags: URA-PCNM67-YFP was inserted at the 5′ end of the ORFs of CNM67 and MPC54 in AN117-4B. URA-PMPC54-YFP was inserted at the 5′ end of the ORF of SPO21 in AN117-4B. For C-terminal YFP tags: YFP-URA3 was inserted at the 3′ end of the ORF of SPC42 in AN117-16D. YFP-his5+ was inserted at the 3′ end of the ORFs of CNM67 and MPC54 in AN117-4B. YFP-TRP1 was inserted at the 3′ end of the ORF of SPO21 in AN117-4B.
To construct EMD26 (MPC54-RFP/MPC54-RFP), EMD27 (mpc54-40-RFP/mpc54-40-RFP), EMD28 (mpc54-47-RFP/mpc54-47-RFP), EMD29 (mpc54-118-RFP/mpc54-118-RFP), EMD30 (mpc54-119-RFP/mpc54-119-RFP), and EMD31 (mpc54-145-RFP/mpc54-145-RFP), the mpc54-RFP allele of interest was inserted into the TRP1 locus of NY51 (MATa, mpc54Δ) and NY50 (MATα, mpc54Δ) using an integrating plasmid containing the mpc54-RFP allele of interest. The resulting haploids were mated.
To construct EMD130 (mpc54Δ/mpc54Δ SPC42-RFP/SPC42-RFP), RFP-HIS3MX6 was inserted at the 3′ end of the ORF of SPC42 in AN117-4B. The haploid was mated with NY51, the resulting diploid was dissected, and haploid segregants with the desired phenotype were mated. To construct EMD131 (sso1Δ/sso1Δ MPC54-RFP), HI3 was transformed with the integrating plasmid, pRS304MPC54RFP.
Plasmids
The plasmids used in this study are listed in Supplementary Table 1. Construction of pFA6a-CFP-CgTRP1, pFA6a-YFP-CgTRP1, and pFA6a-YFP-KlURA3 was as follows: pFA6a-yEGFP-HIS3MX6 was constructed by PCR amplifying yeast-enhanced green fluorescent protein (yEGFP; including the linker region) from pYM12 (Knop et al., 1999) with primers MNO155 and MNO156. The PCR product was digested with PacI and AscI, and the digested fragment was ligated into the similarly digested pFA6a-GFP(S65T)-HIS3MX6 plasmid (Wach et al., 1997). The yEGFP was replaced with CFP or YFP by digesting pDH3 or pDH5 (Muller et al., 2005) with MscI and AscI. These digested fragments were subcloned into the pFA6a-yEGFP-HIS3MX6 backbone, which was similarly digested with MscI and AscI to make pFA6a-CFP-HIS3MX6 and pFA6a-YFP-HIS3MX6. pFA6a-CgTRP1 was constructed by amplifying CgTRP1 from pCgW (gift from L. Huang, University of Massachusetts, Boston) with the primers MNO162 and MNO163, digesting the PCR product with BglII and PmeI, and subcloning the fragment into a similarly digested pFA6a-TRP1. pFA6a-KlURA3 was constructed by amplifying KlURA3 from pKlU (gift from L. Huang) with the primers MNO162 and MNO163, digesting the PCR product with BglII and PmeI, and subcloning the fragment into a similarly digested pFA6a-TRP1 (Longtine et al., 1998). pFA6a-CFP-CgTRP1, pFA6a-YFP-CgTRP1, and pFA6a-YFP-KlURA3 were constructed by subcloning the PacI-BglII fragment from pFA6a-CgTRP1 or pFA6a-KlURA3 into similarly digested pFA6-CFP-HIS3MX6 or pFA6a-YFP-HIS3MX6 to replace HIS3MX6 with CgTRP1 or KlURA3, respectively.
pMN101 (C.g.TRP1-PGAL–GFP(S65T)-TADH) was constructed by PCR amplifying CgTRP1 from pFA6a-CgTRP1 using primers MNO146 and MNO147. The PCR product was cut with PmeI-BglII and replaced the PmeI-BglII fragment of pFA6a-TRP1-PGAL1-GFP(S65T) (Longtine et al., 1998), making pMN101. pMN102 (K.l.URA3-PGAL–GFP(S65T)-TADH) was constructed by PCR amplifying KlURA3 from pFA6a-KlURA3 using primers MNO146 and MNO147. The PCR product was cut with PmeI-BglII and replaced the PmeI-BglII fragment of pFA6a-TRP1-PGAL1-GFP(S65T), making pMN102.
pMN104 (K.l.URA3-PCNM67–GFP(S65T)-TADH), pMN105 (C.g.TRP1-PMPC54–GFP(S65T)-TADH), and pMN106 (K.l.URA3-PMPC54–GFP(S65T)-TADH) were constructed by PCR amplifying the promoters of CNM67 or MPC54 from genomic DNA derived from AN117-4B using the primers MNO226 and MNO227 or MNO228 and MNO229, respectively. PCR products were digested with BglII-PacI and replaced the BglII-PacI fragments of pMN101 and pMN102.
pMN108 (K.l.URA3-PCNM67–YFP-TADH), pMN109 (C.g.TRP1-PMPC54–CFP-TADH), and pMN110 (K.l.URA3-PMPC54 –YFP-TADH) were constructed by PCR amplifying CFP or YFP from pFA6a-CFP-CgTRP1 or pFA6a-YFP-KlURA3, respectively, using the primers MNO224 and MNO225. PCR products were digested with PacI-AscI and replaced the PacI-AscI fragment of pMN104, pMN105, or pMN106.
pRS314MPC54-RFP was constructed by cloning a SpeI-XhoI fragment carrying MPC54-RFP from pRS423-MPC54-RFP (gift from H. Nakanishi, Stony Brook University, Stony Brook, NY) into similarly digested pRS314. pRS316MPC54-RFP was constructed by cloning a SpeI-XhoI fragment carrying MPC54-RFP from pRS314MPC54-RFP into similarly digested pRS316. QuickChange mutagenesis was targeted against the pRS316-MPC54-RFP plasmid to construct the mpc54 mutant plasmids: pRS316-MPC54-40-RFP, pRS316-MPC54-47-RFP, pRS316-MPC54-118-RFP, pRS316-MPC54-119-RFP, and pRS316-MPC54-145-RFP (primers used for each mutation are listed in Supplementary Table 2). pRS426-MPC54-RFP, pRS426-MPC54-40-RFP, pRS426-MPC54-47-RFP, pRS426-MPC54-118-RFP, pRS426-MPC54-119-RFP, and pRS426-MPC54-145-RFP were constructed by cloning a NotI-XhoI fragment from pRS316- MPC54-RFP carrying the appropriate mpc54 allele into a similarly digested pRS426. pRS304-MPC54-RFP, pRS304-MPC54-40-RFP, pRS304-MPC54-47-RFP, pRS304-MPC54-118-RFP, pRS304-MPC54-119-RFP, and pRS304-MPC54-145-RFP were constructed by cloning a NotI-XhoI fragment from pRS316- MPC54-RFP carrying the appropriate mpc54 allele into a similarly digested pRS304.
pRS424-SEC3-GFP was constructed by cloning a KpnI/BamHI fragment carrying SEC3-GFP from pRS316-Sec3-GFP (Finger et al., 1998) into similarly digested pRS424. pRS426-GFP-EXO70 was constructed by cloning a KpnI/BamHI fragment carrying GFP-EXO70 from pRS314-GFP-EXO70 (a gift from D. TerBush USUHS, Bethesda, MD) into similarly digested pRS426.
Site-Directed Mutagenesis
Mutagenic primers were designed based on the guidelines of the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Oligos are listed in Supplementary Table 2. PCR using PfuTurbo DNA polymerase, and the mutagenic oligos altered codons of phylogenetically conserved residues in plasmids containing MPC54-RFP. Conserved residues were changed to alanines. PCR products were digested with DpnI. The digestion product was analyzed by electrophoresis. Samples for which a band was visible after DpnI digestion were transformed into BSJ72 E. coli cells. Samples for which no band was visible after DpnI digestion were transformed into XL2-Blue MRF Escherichia coli. Plasmids were recovered, and mutations were confirmed by sequencing and restriction analysis.
Sporulation Assays
Cells were induced to sporulate in liquid medium essentially as described previously (Neiman, 1998). Briefly, cells were cultured to saturation in either rich medium or synthetic medium selective for plasmids, then cultured overnight to midlog phase in yeast extract-peptone-acetate medium, and then transferred to 2% potassium acetate at a concentration of 3 × 107 cells/ml.
Fluorescence Microscopy
For direct detection of fluorescent proteins in fixed cells, cells were fixed with 3.7% formaldehyde for 5 min and mounted with mounting medium containing 4,6-diamidino-2-phenylindole (DAPI; Vectashield; Vector Laboratories, Peterborough, United Kingdom). Images were acquired using two microscope setups. The first was a Zeiss Axioplan2 microscope (Carl Zeiss, Thornwood, NY) with a Zeiss mRM Axiocam. The second was a Zeiss Observer.Z1 microscope (Carl Zeiss, Thornwood, NY) with an Orca II ERG camera (Hamamatsu, Bridgewater, NJ). Images were deconvolved using Axiovision 4.8 software.
For the FRET studies, yeast strains containing spindle pole body proteins tagged with CFP and YFP were analyzed at ∼4 h after transfer to sporulation medium. Progression through meiosis was assessed by the number of the spindle pole bodies seen with CFP and YFP fluorescence. Cells that were in meiosis II were analyzed.
Image acquisition for the FRET studies was conducted on microscopes at two sites: University of Washington, Seattle, and Stony Brook University, Stony Brook. For the Seattle acquisitions, 200 μl of sporulating culture was spun down at 2300 × g. Cells were resuspended in 30 μl of fresh 2% potassium acetate and sonicated to reduce flocculence. A 3-μl aliquot was mounted on a pad of 0.9% SeaKem LE agarose (FMC BioProducts, Rockland, ME) in SDC medium as described (Sundin et al., 2004). Microscopy was performed on a DeltaVision system manufactured by Applied Precision (Issaquah, WA). The microscope was equipped with an Uplan Apo 100× oil objective (1.35 NA), a CoolSnap HQ digital camera from Roper Scientific (Tucson, AZ), and optical filter sets from Omega Optical (Brattleboro, VT). For each strain 20–225 images were captured. Exposure times were 0.4 s with 2 × 2 binning and a final image size of 512 × 512 pixels. Using the differential interference contrast (DIC) channel, fields were focused manually before an automated capture of a single focal plane in the order YFP, FRET, CFP, and DIC images. Because YFP photobleaches rapidly when exposed to CFP excitation light, the order of capture is critical (Hailey et al., 2002).
For the Stony Brook acquisitions, 700 μl of sporulating culture was spun down at 2300 × g. Cells were resuspended in 70 μl of fresh 2% potassium acetate and then sonicated. A 2-μl aliquot was mounted on a pad of 1% agarose Low EEO (US Biologicals, Swampscott, MA) in S Media (0.17% yeast nitrogen base without amino acids [US Biologicals] and 0.5% ammonium sulfate) as described (Sundin et al., 2004). Microscopy was performed on a Zeiss Axioplan2 microscope (Carl Zeiss, Thornwood, NY) equipped with a 100× oil objective (Plan-Neofluar, NA 1.46) and a Zeiss mRM Axiocam. For each strain 20–225 images were captured. Exposure times were 0.8 s with 2 × 2 binning and a final image size of 650 × 514 pixels. Image acquisition was as described above.
FRET Analysis
For the Seattle acquisitions, the 12-bit images were converted into 16-bit TIFF format by the Java program R3D Converter (Riffle, unpublished data). For the Stony Brook acquisitions, the Zeiss-formatted images were converted to TIFFs using ImageJ (http://rsb.info.nih.gov/ij/). In both cases, these TIFF images were then analyzed by a custom MatLab program (MathWorks, Natick, MA), FretSCal, that evaluates certain regions of the images based on user-defined criteria (Muller and Ess, unpublished data). FretSCal was used to identify candidate spindle pole bodies in each image, defined as bright fluorescence of a diameter of <5 pixels. The image around each candidate spindle pole body was then displayed in the DIC, FRET, CFP, and YFP channels and examined manually for three criteria: 1) whether the putative spindle pole body was in the cytoplasm of a cell; 2) whether it was round and in focus in all channels; and 3) that there were no more than four bright candidate spindle pole bodies per cell. Images meeting all three criteria were collected and the average FRET value for the entire accepted dataset was calculated. FRET values were expressed as a FretR index, which measures the fold increase of fluorescence intensity in the FRET channel over a baseline determined from the fluorescence in the CFP and YFP channels (Muller et al., 2005). Based on this index, the predicted value of FretR is 1 if no energy is transferred. FretR values above 1.2 were considered true interactions.
Transmission Electron Microscopy
Sporulating cells were fixed for 1 h in 3% glutaraldehyde in cacodylate buffer (100 mM sodium cacodylate, 5 mM CaCl2, pH 6.8), washed once in cacodylate buffer, left overnight at 23°C, resuspended in 4% KMnO4 in distilled water, and incubated for 30 min at 23°C. The cells were then washed with distilled water until the supernatant was clear, resuspended in saturated uranyl acetate for 2 h, and dehydrated by 10-min incubations in a graded acetone series: two incubations each in 30, 50, 70, and 95% acetone and four incubations in 100% acetone. For embedding, the cells were first incubated twice in acetonitrile for 10 min and then transferred to a 1:1 solution of acetonitrile:Epon mix (50% Epon 812, 15% dodecenyl succinic anhydride, 35% nadic methyl anhydride) for 4 h. The cells were incubated in Epon mix for 12 h with changes every 4 h and, finally, the catalyst 2,4,6-tris(dimethylaminomethyl)phenol (DMP-30) was added, and samples were placed into a vacuum oven at 60°C for 2 d to harden. After the samples were sectioned, images were collected on an FEI BioTwin G2 microscope (FEI, Hillsboro, OR) at 80 kV using an AMT XR-60 camera (AMT, Danvers, MA).
RESULTS
The Organization of Subunits within the MOP Structure
MOP proteins form the interface between the spindle pole body and the prospore membrane and are essential for prospore membrane assembly (Knop and Strasser, 2000; Bajgier et al., 2001; Nickas et al., 2003). Using FRET analysis, the organization of subunits within the MOP structure was examined. A similar approach had been used to determine the relative positioning of the central core proteins of the mitotic spindle pole body (Muller et al., 2005). However, the organization of proteins within the MOP remained unknown.
To determine whether the constitutive outer plaque component Cnm67p maintains its position within the spindle pole body during meiosis II, the FRET between Cnm67-CFP and Spc42-YFP was measured in mitotic and meiotic outer plaques. The FRET ratio (FretR) is a measurement of the transfer of energy from an excited donor fluorophore (e.g., CFP) to an adjacent acceptor fluorophore (e.g., YFP; Muller et al., 2005). The FretR between the C-terminus of Cnm67p and the C-terminus of Spc42p was similar in mitotic and meiotic cells, suggesting that in both conditions the C-terminus of Cnm67p is proximal to the outer surface of the central plaque (i.e., distal to the nucleus; Figure 1, Table 2). The FretR between these termini was in agreement with previously published FretR data (Muller et al., 2005; Table 2). Cnm67p contains a central coiled-coil motif that forms the intermediate layer between the central and the outer plaques (Schaerer et al., 2001). Because the C-terminus of Cnm67p is located near the central plaque and its coiled-coil domain spans the gap between the central and outer plaques, this indicates that the N-terminus of Cnm67p is positioned at the lower surface of the MOP structure (Figure 1). Therefore, by determining the FRET interactions between the N-terminus of Cnm67p and other MOP proteins, the orientation of these proteins within the MOP structure could be predicted.
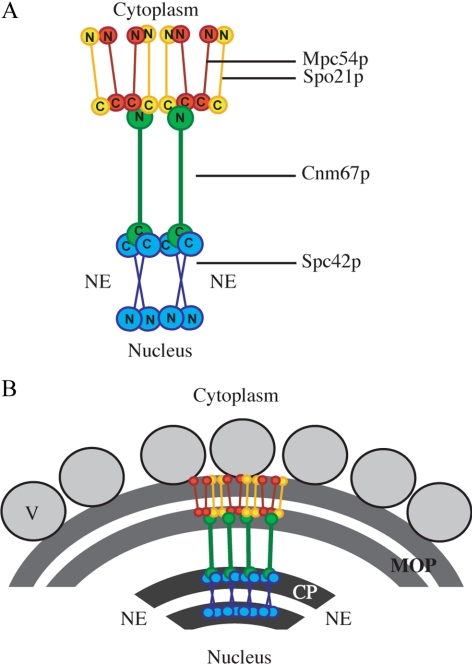
Figure 1.
Model of MOP organization based on FRET. (A) Model of the relative positions of the termini of Mpc54p (red), Spo21p (yellow), Cnm67p (green), and Spc42 (blue) based on FRET data. The location of the nuclear envelope is indicated (NE). (B) Model of the organization of the MOP proteins within the MOP structure. Vesicles are shown docking to the outer surface of the MOP (V).
Table 2.
FRET interactions between the MOP components
| Strain | YFP acceptor | CFP donor | FretR | Interactiona |
|---|---|---|---|---|
| MND100b | Spc42-YFP | Cnm67-CFP | 2.42 ± 0.15 | High |
| MND100 | Spc42-YFP | Cnm67-CFP | 2.33 ± 0.18 | High |
| MND88 | Cnm67-YFP | Mpc54-CFP | 0.98 ± 0.08 | None |
| YS326 | Cnm67-YFP | CFP-Mpc54 | 1.10 ± 0.22 | None |
| AN317 | Cnm67-YFP | Spo21-CFP | 1.01 ± 0.10 | None |
| YS353 | Cnm67-YFP | CFP-Spo21 | 1.08 ± 0.24 | None |
| MND101 | YFP-Cnm67 | Mpc54-CFP | 1.35 ± 0.11 | Low |
| MND103 | YFP-Cnm67 | Spo21-CFP | 1.47 ± 0.14 | Low |
| YS335 | YFP-Cnm67 | CFP-Mpc54 | 1.03 ± 0.08 | None |
| YS344 | YFP-Cnm67 | CFP-Spo21 | 1.02 ± 0.27 | None |
| AN310 | Mpc54-YFP | Spo21-CFP | 1.24 ± 0.12 | Low |
| YS118 | YFP-Spo21 | Mpc54-CFP | 1.11 ± 0.14 | None |
| EMD104 | Mpc54-YFP | Mpc54-CFP | 1.26 ± 0.14 | Low |
| EMD105 | YFP-Mpc54 | Mpc54-CFP | 0.91 ± 0.11 | None |
| EMD100 | Spo21-YFP | Spo21-CFP | 1.34 ± 0.13 | Low |
| KBD3 | YFP-Mpc54 | Spo21-CFP | 1.05 ± 0.09 | None |
| EMD52 | YFP-Spo21 | CFP-Mpc54 | 1.23 ± 0.11 | Low |
| EMD107 | YFP-Mpc54 | CFP-Mpc54 | 0.96 ± 0.13 | None |
a FretR values are grouped based on their intensity. FretR above 2.0 is considered high. FretR between 1.2 and 1.5 is considered low. FretR lower than 1.15 indicates no interaction.
b This experiment was performed in vegetative cells. All other FretR values were derived from cells in meiosis II.
Like Cnm67p, Mpc54p and Spo21p are both coiled-coil MOP proteins (Knop and Strasser, 2000). Interestingly, in transmission electron microscopy (TEM) images an electron lucent layer in the MOP structure resembles the region between the central plaque and the outer plaque, a region composed of the coiled-coil domain of Cnm67p (Schaerer et al., 2001). The electron lucent region of the MOP may similarly correspond to the coiled-coil domains of Mpc54p and Spo21p, suggesting that these proteins may also have a vertical orientation typical of coiled-coil proteins within the spindle pole body structure.
To analyze the organization of these coiled-coil proteins by FRET, Cnm67p, Mpc54p, and Spo21p were tagged with CFP or YFP at either the N- or C-terminus. Different combinations of CFP- and YFP-tagged proteins were integrated into cells to produce strains containing one CFP donor and one YFP acceptor. The FretR value of each CFP–YFP combination was then determined (Table 2), and a model of the relative positions of the subunits was constructed (Figure 1A). In general, the FretR values were lower for proteins in the MOP compared with proteins in the central core of the spindle pole body (Table 2). This disparity has two possible sources. First, the proteins in the MOP may not be as tightly packed as the proteins in the central plaque. Second, the central plaque is primarily composed of an array of Spc42p, whereas the MOP is composed of at least six different proteins. Therefore, the fluorophore tags of two specific MOP proteins may be further apart. However, even though the FretR values were lower, significant interactions allowed us to predict the relative positions of Mpc54p and Spo21p.
Mpc54p and Spo21p were analyzed individually for their ability to FRET with Cnm67p. The C-termini of both Mpc54p and Spo21 showed FRET interaction with the N-terminus of Cnm67p (Table 2). In contrast, neither the N-terminus of Mpc54p nor the N-terminus of Spo21p displayed interactions with the N-terminus of Cnm67p (Table 2). As predicted, neither end of Mpc54p or Spo21p FRET with the distant C-terminus of Cnm67p which localizes near the central plaque (Table 2). Therefore, FRET data indicates that both Mpc54p and Spo21p are oriented with their N-termini toward the outer face of the MOP and their C-termini toward Cnm67 and the central plaque of the spindle pole body (Figure 1).
Mpc54p and Spo21p were also analyzed for their ability to FRET with each other. The C-termini of Spo21p and Mpc54p both self-FRET and FRET with each other (Table 2). As expected, the N-terminus of each protein did not FRET with its own C-terminus, because these termini are separated by a central coiled-coil motif (Figure 1A, Table 2). At the outer surface of the MOP, the N-terminus of Mpc54p FRETs with the N-terminus of Spo21p but not with itself (Table 2). This suggests that at the outer surface of the MOP the N-termini of the proteins may be more widely splayed, which may account, in part, for the curving of the MOP structure during meiosis II (Figure 1B). These data suggest that Mpc54p and Spo21 form hetero-oligomers in which the two proteins have a head-to-head parallel arrangement (Figure 1A). Additionally, these data indicate that the N-termini of Mpc54p and Spo21p form the outer surface of the MOP where vesicles dock and fuse, and suggesting that the N-termini of these proteins might be directly involved in vesicle docking during prospore membrane initiation (Figure 1B).
Isolating Nonnull Alleles of mpc54
Position-specific iterated BLAST (basic local alignment search tool; http://blast.ncbi.nlm.nih.gov/Blast.cgi; Altschul et al., 1990) identified several regions that were conserved between the S. cerevisiae Mpc54p and Spo21p and their orthologues in other yeasts. The conserved residues in Spo21p were enriched at the C-terminus, which is buried within the MOP structure and therefore is unlikely to play a role in events at the surface of the MOP (data not shown). Significantly, the regions of conservation within Mpc54p were located in the N-terminus of the protein, which is oriented toward the outer surface of the MOP (Figure 2A). As this is the surface where the prospore membrane assembles, conserved residues in Mpc54p may be important to promote vesicle docking and fusion during prospore membrane initiation.
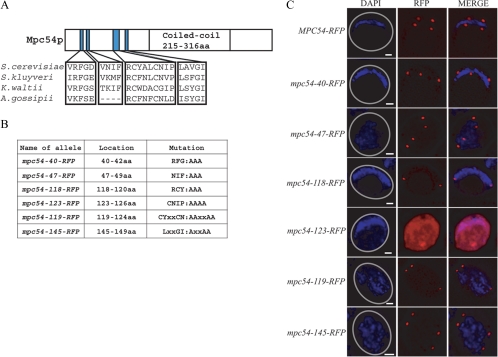
Figure 2.
Construction of mutant alleles based on conservation of residues. (A) Conserved residues of Mpc54. The Mpc54p schematic is oriented with the N-terminus to the left. (B) Conserved residues in an RFP-tagged version of MPC54 were mutated to alanines to construct mpc54-RFP mutant alleles. (C) Analyzing the localization of RFP-tagged alleles to the spindle pole bodies in meiosis II cells. Five of the six mpc54-RFP mutant alleles were stable and localized appropriately. The number of visible spindle pole bodies differs in the images shown due to the random distribution of the four spindle pole bodies between the chosen focal planes. Gray circles in the DAPI images indicate the outline of the imaged cell.
To test the importance of these conserved patches, mutations were introduced into Mpc54p in which clusters of conserved residues were changed to alanines (Figure 2B). To determine if the mutations created null alleles, these mutations were directed against a functional RFP-tagged version of Mpc54p. Therefore, the presence of RFP at the spindle pole bodies during meiosis II indicates that the Mpc54p alleles are stable and localize appropriately. Plasmids containing either MPC54-RFP, mpc54-40-RFP, mpc54-47-RFP, mpc54-118-RFP, mpc54-123-RFP, mpc54-119-RFP, or mpc54-145-RFP were transformed into an mpc54Δ strain and analyzed by fluorescence microscopy. Five of the six alleles (the exception being mpc54-123-RFP, which did not form RFP foci and was designated null) encoded versions of Mpc54p that were stable and localized to the spindle pole body (Figure 2C).
mpc54 Mutant Alleles Disrupt Spore Formation
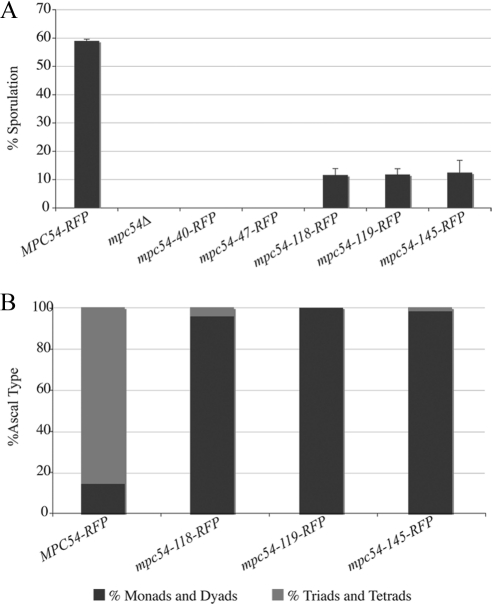
If the mpc54 mutants disrupt the function of Mpc54p during prospore membrane formation, then transforming these alleles into an mpc54Δ strain would not rescue sporulation. All strains expressing the Mpc54p mutant proteins on a plasmid displayed a dramatic reduction or a complete loss of sporulation compared with wild-type Mpc54p-RFP (Figure 3A). Asci that did form in the mutants were monads or dyads, also indicating a defect in sporulation (Figure 3B). Assembly of the MOP structure is particularly susceptible to expression levels of the MOP components (Bajgier et al., 2001; Nickas et al., 2003). To ensure that the mutant phenotypes observed were not an indirect effect of reduced expression levels, the mutant alleles were placed on high-copy plasmids or were integrated into chromosomes. Under these different conditions the sporulation efficiency was similar for each mpc54 mutant (Supplementary Table 3), indicating that these phenotypes are due to changes in the functionality of the mutant forms of Mpc54p and not simply due to reduced expression of the protein. This suggests that the mutations in the different alleles directly disrupt spore formation.
Figure 3.
The mpc54-RFP mutant alleles disrupt spore formation. (A) Sporulation efficiency of an mpc54Δ strain transformed with Mpc54p mutants under the MPC54 promoter was monitored using light microscopy. Values shown are the average of three experiments in which 200 cells were analyzed for sporulation in each experiment. Error bars, 1 SD. (B) The distribution of ascal types in an mpc54Δ strain transformed with Mpc54p mutants under the MPC54 promoter was monitored using light microscopy.
mpc54 Mutant Alleles Promote Wild-Type MOP Composition
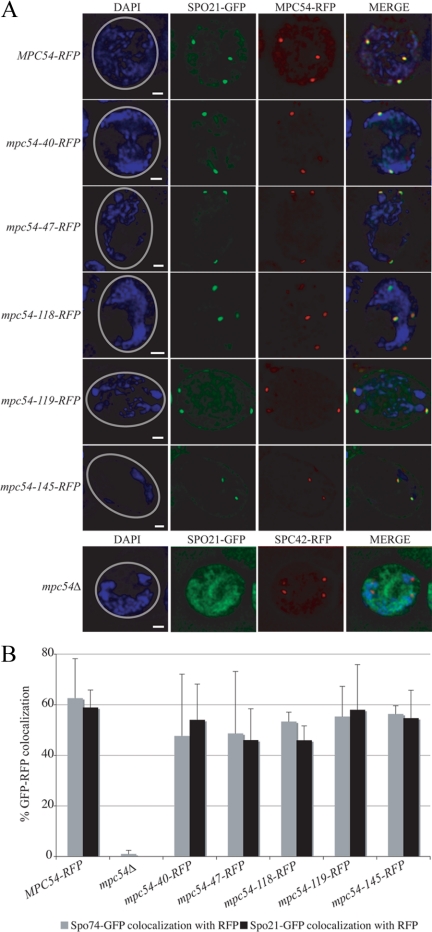
The recruitment of Mpc54p, Spo21p, and Spo74p to the MOP is required both for the formation of the MOP structure and for prospore membrane initiation (Knop and Strasser, 2000; Bajgier et al., 2001; Nickas et al., 2003). To determine whether the sporulation defect of the mpc54 mutant alleles was due to a defect in forming the MOP complex, the recruitment of Spo21p and Spo74p to the MOP was analyzed. Either Spo21-GFP or Spo74-GFP was coexpressed with RFP-tagged versions of the various Mpc54p mutants, and colocalization of RFP and GFP dots was analyzed (Figure 4). Colocalization in the mutants was compared both to wild-type Mpc54-RFP in which GFP-RFP colocalization was observed and to an mpc54Δ in which GFP did not frequently colocalize with the RFP-tagged spindle pole body (in the mpc54Δ, spindle pole bodies were marked by Spc42-RFP; Figure 4A). The recruitment of Spo21-GFP and Spo74-GFP in cells expressing mpc54-40-RFP, mpc54-47-RFP, mpc54-118-RFP, mpc54-119-RFP, and mpc54-145-RFP was comparable to that of wild-type MPC54-RFP (Figure 4B). These data indicate that the mutant proteins not only localized to the MOPs, but also recruited other MOP components.
Figure 4.
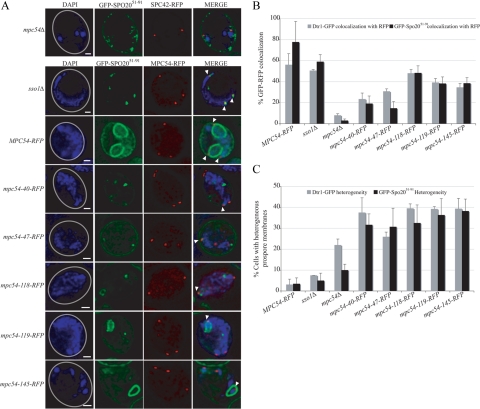
mpc54-RFP mutant alleles maintain the composition of the MOP. (A) Representative examples of the colocalization between spindle pole bodies (identified by Mpc54-RFP or Spc42-RFP dots) and Spo21-GFP in meiosis II cells. The number of visible spindle pole bodies differs in the images shown due to the random distribution of the four spindle pole bodies between the chosen focal planes. Gray circles in the DAPI images indicate the outline of the imaged cell. Scale bars, 1 μm. (B) Colocalization between RFP-tagged spindle pole bodies and Spo74-GFP or Spo21-GFP during meiosis II in various strains. At least 100 RFP dots were analyzed for GFP colocalization for each experiment. Error bars, 1 SD.
mpc54 Mutant Alleles Disrupt Early Stages of Prospore Membrane Formation
Sporulation defects can result from disruptions at any stage of prospore membrane development or spore maturation. To determine at which stage of prospore membrane growth the Mpc54p mutant proteins impaired sporulation, the progress of prospore membrane growth was analyzed by fluorescence microscopy. The prospore membrane markers Dtr1-GFP and GFP-Spo2051-91 were transformed into cells expressing an mpc54 mutant allele (Figure 5A). For comparison, the GFP-tagged prospore membrane markers were also transformed into an MPC54-RFP strain, an mpc54Δ strain, and an sso1Δ strain, in which vesicles dock at the surface of the MOP but do not fuse into a membrane (Nakanishi et al., 2006; Figure 5A).
Figure 5.
mpc54-RFP mutant alleles have defects in prospore membrane formation. (A) Representative examples of the colocalization between RFP-tagged spindle pole bodies and GFP-Spo2051-91 in MPC54-RFP, sso1Δ, mpc54Δ, and mpc54 mutant strains. GFP-Spo2051-91 signals appear as curved lines that indicate prospore membrane growth (MPC54-RFP) or as dots that indicate a stalling of prospore membrane initiation (sso1Δ). The number of visible spindle pole bodies differs in the images shown due to the random distribution of the four spindle pole bodies between the chosen focal planes. Gray circles in the DAPI images indicate the outline of the imaged cell. Arrowheads highlight the position of spindle pole bodies in the merged images. Scale bars, 1 μm. (B) Colocalization between RFP-tagged spindle pole bodies and GFP-Spo2051-91 or Dtr1-GFP during meiosis II in various strains. In the GFP-Spo2051-91 colocalization experiments, mpc54-40-RFP and mpc54-47-RFP were determined by a Student's t test to be statistically different from wild-type at a p < 0.003, but mpc54-118-RFP, mpc54-119-RFP, and mpc54-145-RFP were not. In the Dtr1-GFP colocalization experiments, mpc54-40-RFP and mpc54-47-RFP were statistically different from wild-type at a p < 0.03, but mpc54-118-RFP, mpc54-119-RFP, and mpc54-145-RFP were not. At least 160 RFP dots were analyzed for GFP colocalization for each experiment. Error bars, 1 SD (C) The heterogeneity of prospore membrane progression. For a single cell, the GFP-Spo2051-91 or Dtr1-GFP signals that localized at the four RFP dots were categorized as linear, dots, or absent. Cells in which RFP dots colocalized with GFP signals of different categories were considered heterogeneous. Cells in which all RFP colocalized with GFP or all RFP dots lacked a GFP signal were considered homogeneous. For all strains, n > 50. Error bars, 1 SD.
For cells expressing mpc54-40-RFP and mpc54-47-RFP, the association of GFP-tagged prospore membranes with RFP-tagged spindle pole bodies was reduced compared with wild type, but the reduction was not as severe as in the mpc54Δ (Figure 5B). Additionally, the GFP signal was limited to dots in these strains, similar to sso1Δ (data not shown). Taken together, this suggests that both the recruitment of precursor vesicles to the cytoplasm around the spindle pole body and the subsequent initiation of prospore membrane formation are disrupted in mpc54-40-RFP and mpc54-47-RFP alleles. In contrast, the association between GFP-tagged prospore membranes and RFP-tagged spindle pole bodies in cells expressing mpc54-118-RFP, mpc54-119-RFP, and mpc54-145-RFP was similar to wild type (Figure 5B). Additionally, prospore membranes were able to form in these strains, though at a reduced rate (data not shown).
In wild-type cells, Dtr1-GFP and GFP-Spo2051-91 recruitment to the MOP was typically homogeneous, meaning that the progress of prospore membrane growth was synchronized at all of the MOPs within a single cell. Therefore, all the prospore membranes in a given cell appeared to be at the same stage of development (Figure 5, A and C). In contrast, each of the mpc54 mutants displayed some degree of heterogeneity in the size and morphology of prospore membranes within a single cell (Figure 5, A and C). This suggests that although the mutant MOPs are able to occasionally recruit precursor vesicles, recruitment was asynchronous and the coordination of prospore membrane growth at all four MOPs was disrupted by the mpc54 mutations.
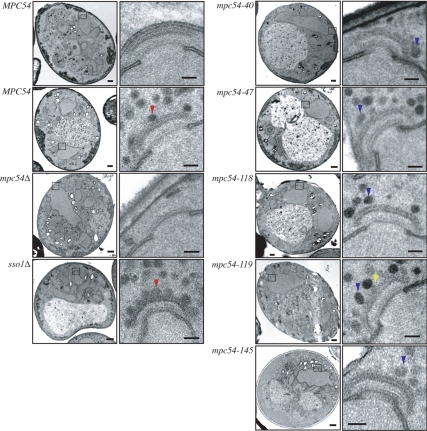
mpc54 Mutant Alleles Maintain Proper MOP Ultrastructure
The fluorescence data suggested that the composition of the MOP was not disrupted by the mpc54 point mutations (Figure 4B). To examine this possibility more closely, thin-section TEM analysis was used to analyze the structure of the MOP as well as the stages of prospore membrane formation in fine detail. Analysis of cells expressing the mpc54 mutants revealed that the MOPs displayed the distinct, layered morphology of wild-type MOPs (Figure 6). This clearly defined MOP structure was absent in the mpc54Δ mutant (Figure 6). These results support the finding that mpc54 mutant alleles maintain the composition of the MOP by recruiting Spo21p and Spo74p (Figure 4B). Taken together, these data suggest that the Mpc54p mutations maintain the composition and the ultrastructure of the MOP complex.
Figure 6.
Mpc54p plays a role in vesicle docking. Representative examples of TEM images of the MOP in single sections of meiosis II spindle pole bodies. Vesicles flush against the surface of the MOP are indicated with a red arrow. Loosely tethered vesicles are indicated with a blue arrow. Electron-dense projections between the MOP and tethered vesicles are indicated with a yellow arrow. Scale bars for whole cell images, 500 nm. Scale bars for higher magnification images, 100 nm.
Mpc54p Plays a Role in Vesicle Docking
Despite the presence of an intact MOP structure, vesicles frequently were found close to, but not in contact with, the surface of the MOP in the mpc54 mutants at an average distance of ∼12 nm between the surface of the vesicle and the surface of the MOP (Figure 6, Table 3). This consistent distance between MOP and the vesicle suggests that vesicles are loosely tethered to the MOP, even though they cannot dock to its surface. The gap observed in the mpc54 mutants is in contrast to the phenotype of the SNARE mutant, sso1Δ, where vesicles are in close contact with the surface of the MOP (Figure 6, Table 3). These data suggest that the Mpc54p N-terminus plays a role in vesicle docking at the surface of the MOP. Moreover, engagement with the MOP surface is critical to prospore membrane initiation.
Table 3.
The effect of the Mpc54p mutant proteins on vesicle docking at the MOP
| Genotype | Vesicles |
PsMs at MOPs | |||
|---|---|---|---|---|---|
| Docked, 0–35 nm | Tethered, 36–60 nm | Accumulated, 61–300 nm | Total, 0–300 nm | ||
| sso1Δ | 6.4 ± 1.8 | 1.1 ± 1.2 | 13.8 ± 5.8 | 21.3 ± 6.7 | N |
| MPC54 | 3.3 ± 0.6 | 2.3 ± 1.5 | 8.0 ± 2.7 | 13.7 ± 1.2 | Y |
| mpc54Δ | 0.9 ± 1.2 | 0.8 ± 0.6 | 5.2 ± 1.4 | 6.9 ± 2.0 | N |
| mpc54-40-RFP | 0.8 ± 1.1 | 1.6 ± 1.6 | 8.9 ± 2.9 | 11.2 ± 2.8 | N |
| mpc54-47-RFP | 0.1 ± 0.4 | 1.9 ± 1.4 | 5.6 ± 3.1 | 7.6 ± 3.8 | N |
| mpc54-118-RFP | 0.2 ± 0.4 | 2.8 ± 1.7 | 7.5 ± 5.2 | 10.5 ± 5.6 | Y |
| mpc54-119-RFP | 0.3 ± 0.5 | 3.8 ± 1.8 | 7.4 ± 4.3 | 11.4 ± 4.9 | Y |
| mpc54-145-RFP | 0.5 ± 1.0 | 3.1 ± 1.4 | 7.4 ± 4.0 | 10.9 ± 4.8 | Y |
For all samples, except mpc54Δ, the distance between the surface of the MOP and the center of the vesicles was measured. For mpc54Δ, the distance between the surface of the central plaque and the center of the vesicles was measured and then corrected based on the average distance between the surface of the central plaque and the surface of the MOP in wild-type cells. Each vesicle was categorized according to its distance from the MOP surface (Supplementary Figure 1). The numbers displayed were derived from averaging the number of vesicles in each category for each MOP analyzed. Only MOPs that displayed proper electron-dense layering but lacked prospore membranes were analyzed. The number of spindle pole bodies examined for each strain was (from top to bottom) as follows: 16, 11, 3, 9, 21, 15, 12, and 11. The number of docked vesicles in the mpc54 mutants was significantly different from both the sso1Δ strain (p < 0.0001 for all mutants) and the MPC54-RFP strain (p < 0.0001 for all mutants except mpc54-47-RFP where p < 0.003).
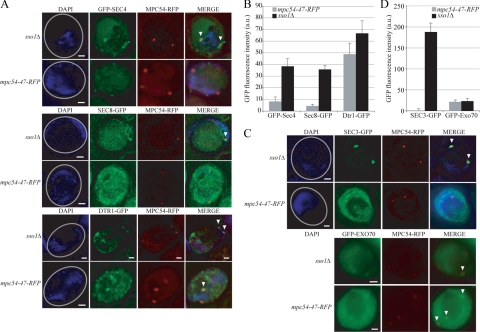
Vesicle Docking to the MOP Promotes the Recruitment of Exocyst Effectors and Components
These data show that the mpc54 alleles separate the steps of vesicle tethering to the MOP from docking to the MOP. Therefore, these alleles can be used to examine what changes occur on the vesicles after they dock to the MOP. The localization of additional proteins required for prospore membrane growth was examined in the mpc54-47-RFP mutant, which retains the structure and composition of the MOP, while exhibiting a severe defect in vesicle docking. For comparison with the mpc54-47-RFP strain, where vesicles loosely tether to the MOP, an sso1Δ strain, where vesicles dock to the MOP, was also examined.
In Schizosaccharomyces pombe, one MOP protein, Spo13p, serves as a guanine nucleotide exchange factor for Ypt2p, the ortholog of S. cerevisiae Sec4p (Yang and Neiman, 2010). Conversion of Sec4p to its GTP-bound form is necessary for its association with membranes (Goud et al., 1988). Although there is no SPO13 ortholog in S. cerevisiae, if the nucleotide exchange function of the MOP is conserved one might expect activation of Sec4p and its recruitment to the vesicles to be dependent on contact with the MOP. The fluorescence signal of GFP-Sec4 adjacent to the spindle pole bodies was readily visible in sso1Δ cells, indicating that Sec4 has been recruited to the membrane in these cells (Figure 7A). However, in mpc54-47-RFP cells, clear GFP-Sec4 fluorescence was not seen near the spindle pole bodies (Figure 7A). This observation was corroborated by quantitation of the GFP fluorescence intensity adjacent to the spindle pole bodies (Figure 7B). Our TEM data indicate that, on average, there are slightly fewer vesicles located near the MOP in mpc54-47-RFP cells than in sso1Δ cells (Table 3). However, the difference in GFP-Sec4 fluorescence is not due simply to differences in the number of vesicles as a marker for the precursor vesicles, Dtr1-GFP is comparably localized in both cell types (Figure 7, A and B). Thus, the difference in GFP-Sec4 localization suggests that docking to the MOP is necessary for recruitment of Sec4p to precursor vesicles.
Figure 7.
Vesicle docking recruits Sec3p. (A) Representative examples of the association of GFP-Sec4, Sec8-GFP, and Dtr1-GFP with an RFP-tagged MOP. The number of visible spindle pole bodies differs in the images shown due to the random distribution of the four spindle pole bodies between the chosen focal planes. Gray circles in the DAPI images indicate the outline of the imaged cell. Arrowheads highlight colocalization of the GFP marker with the spindle pole body in the merged images. Scale bars, 1 μm. (B) The intensity of the GFP-Sec4, Sec8-GFP, and Dtr1-GFP signals at RFP-tagged MOPs. For each MOP examined: The GFP intensity within a 0.2-μm area of interest that included the RFP-tagged MOP was measured in arbitrary units. The background intensity was determined by averaging the GFP intensities within three 0.2-μm areas in the cytoplasm adjacent to the spindle pole body examined. This background GFP intensity value was then subtracted from the GFP intensity value at the MOP to acquire a final value for the GFP intensity at each MOP examined. n = 100. Error bars, 1 SE of the mean. (C) Representative examples of the association of Sec3-GFP and GFP-Exo70 with an RFP-tagged MOP. Image specifications are the same as in A. (D) The intensity of the Sec3-GFP and GFP-Exo70 signals at RFP-tagged MOPs. GFP intensities were acquired as in B. n = 100. Error bars, 1 SE of the mean.
In vesicle trafficking to the plasma membrane, Sec4p on vesicles recruits a subset of the exocyst complex to the vesicles by interaction with the exocyst subunit Sec15p (Salminen and Novick, 1989). We were unable to see clear localization of Sec15-GFP in either sso1Δ or mpc54-47-RFP cells (data not shown). However, a second exocyst subunit, Sec8p, displayed a similar result to GFP-Sec4 in that it was clearly present near the MOP in sso1Δ cells but not in mpc54-47-RFP cells (Figure 7, A and B). This result provides further evidence that the vesicles in mpc54-47-RFP cells lack activated Sec4p.
Sec3p and Exo70p are distinct components of the exocyst in that they are not part of the subcomplex that associates with vesicles, but instead localize independently to the plasma membrane (Boyd et al., 2004). To determine whether the localization of Sec3p or Exo70p is affected by vesicle docking at the MOP, Sec3-GFP and GFP-Exo70 signals were examined in sso1Δ and mpc54-47-RFP cells. GFP-Exo70p was faintly visible adjacent to the MOPs in both conditions, indicating that docking to the MOP is not required for Exo70 association with precursor vesicles (Figure 7, C and D). In contrast, Sec3-GFP colocalized with the spindle pole bodies only in sso1Δ cells, similar to GFP-Sec4 (Figure 7, C and D). Because Sec3p localization to membranes is thought to be independent of Sec4p, these observations suggest that vesicle docking to the MOP is required both for Sec4p and Sec3p recruitment to precursor vesicles.
DISCUSSION
The Organization of the MOP
The formation of a normal MOP structure is required for prospore membrane initiation (Knop and Strasser, 2000; Bajgier et al., 2001; Nickas et al., 2003). Here we present a model for the organization of two MOP proteins based on FRET interactions between MOP components. In this model, the N-termini of Spo21p and Mpc54p are proximal to the outer surface of the MOP. In contrast, Cnm67p forms a bridge between the central plaque and the lower surface of the MOP (Schaerer et al., 2001; Muller et al., 2005). The vegetative spindle pole body has been proposed to be organized around sequential layers of vertically organized coil-coil proteins that interact at their globular head and tail domains. Our results indicate that the MOP extends this stacking by adding an additional Mpc54p/Spo21p layer. The S. pombe MOP has also been suggested, based on two-hybrid data, to consist of similarly stacked coiled-coil proteins (Nakase et al., 2008). Thus, vertically organized layers of such proteins may be a general organizational strategy for the spindle pole body of ascomycetes.
How the other components of the MOP are organized on the Cnm67p/Mpc54p/Spo21p scaffold remains to be determined. Ady4p, we have recently shown, is not a stable component of the complex (Mathieson et al., 2010). Preliminary FRET data examining the orientation of Nud1p suggests that it also localizes to the lower surface of the MOP, near the N-terminus of Cnm67p and the C-termini of Mpc54p and Spo21p, consistent with its proposed location in vegetative cells (data not shown; Muller et al., 2005). We were unable to position Spo74p within the MOP using FRET. Two-hybrid data suggest that Spo74p binds near the C-terminal end of Spo21p, suggesting that it too may be located more toward the interior of the structure. Therefore, our data indicating that the N-termini of Spo21p and Mpc54p are oriented outward from the center of the spindle pole body strongly implicate these ends of the proteins as the likely site of interaction between the MOP and the prospore membrane.
Mpc54p Is Involved in Vesicle Docking
Previously, only null alleles of the MOP components had been constructed and analyzed. These null alleles grossly affect the structure of the MOP and are therefore too blunt an instrument for defining the discrete functions for the MOP proteins. Consistent with the idea that the N-terminus of Mpc54p is a site of membrane interaction, we have isolated several alleles mutant in this domain that display membrane assembly defects. Though the alleles vary in their severity, they share the general features of 1) assembling MOPs that appear normal in composition and ultrastructure and 2) impairing vesicle docking onto the surface of the MOP. In TEM images, prospore membrane precursor vesicles appear to be loosely tethered to the MOP, but rather than binding to it, they remain separated from its surface. One simple interpretation of these results is that the mutations have impaired a site on Mpc54p that is directly required for docking of vesicles to the MOP. However, in this case some other factors must be responsible for the looser association of vesicles with the MOP seen in the TEM images.
How do vesicles tether to the MOP? Based on earlier work, one likely candidate is Ssp1p (Maier et al., 2007). Ssp1p and Ady3p are members of the leading edge protein complex that coats the growing lip of the prospore membrane (Moreno-Borchart et al., 2001). Ady3p associates with the MOP before membrane formation, at which point it forms a complex with Ssp1p and other leading edge proteins (Moreno-Borchart et al., 2001). It has been suggested that Ssp1p is associated with precursor vesicles and is important for their aggregation near the spindle pole body before membrane formation (Maier et al., 2007). However, we found that in an ady3Δ ssp1Δ mpc54-47-RFP triple mutant, vesicles remained associated with the MOP as in the mpc54-47-RFP single mutant (Mathieson, unpublished data), indicating that Ssp1p is not required for the connection between vesicles and the MOP that is seen in the mpc54 mutants.
An alternative possibility is that one of the MOP proteins may mediate this tethering. For example, before docking, Mpc54p or Spo21p might “reach out” and bind to vesicles at a distance from the MOP surface. In such a model, the mpc54 mutations described here would interfere not with vesicle binding per se, but rather with some structural rearrangement necessary to bring the vesicle down to the MOP surface. Interestingly, TEM images of MOPs in the mpc54 mutants occasionally revealed an electron density connecting the MOP and the associated vesicles (Figure 6). These projections were the same density as the MOP, suggesting that they were continuous with the surface of the MOP complex.
Vesicle Docking to the MOP Recruits Components of the Exocyst
Earlier work has shown that prospore membrane precursor vesicles dock onto the MOP surface before fusion (Nakanishi et al., 2006). This result suggested that docking to the MOP might be a prerequisite to fusion. The mpc54 alleles described here demonstrate that docking is indeed necessary for fusion to occur. Moreover, they allowed us to examine what other factors might be altered on the vesicles as a consequence of docking to the MOP. Based on studies of the MOP in the fission yeast S. pombe, one possible role for docking would be to trigger activation and recruitment of the Rab protein Sec4p (Yang and Neiman, 2010). Interestingly, we found that GFP-Sec4 appeared to be present at the spindle pole bodies with docked (sso1Δ) but not undocked (mpc54-47-RFP) vesicles, suggesting that activation and recruitment of Sec4p does require docking to the MOP. Thus, activation and recruitment of Rab proteins to precursor vesicles as a consequence of docking may be an evolutionarily conserved function of the spindle pole body.
One effect of Sec4p activation during vesicle trafficking to the plasma membrane is to recruit subunits of the exocyst complex to the vesicle. Like GFP-Sec4, the exocyst subunit Sec8p localized strongly adjacent to the MOP in sso1Δ cells, presumably to the vesicles accumulated there, but did not do so in the mpc54-47-RFP cells. Thus activation of Sec4p is likely required to recruit the exocyst to prospore membrane precursor vesicles as it is to secretory vesicles in vegetative cells. This recruitment of the exocyst may explain why docking to the MOP is required for subsequent fusion of the vesicles.
Complete assembly of the exocyst requires the six subunits associated with the vesicles as well as Sec3p and Exo70p, which localize independently to the plasma membrane (Boyd et al., 2004). We have shown that GFP-Exo70 is localized adjacent to the spindle pole bodies regardless of whether vesicles have docked. In contrast, Sec3p localization appears to require vesicle docking. How might docking of the vesicles lead to Sec3p recruitment? Sec3p is targeted to the plasma membrane through interactions with both the lipid phosphatidylinositol 4,5-bisphosphate and the GTPases Rho1p or Cdc42p (Guo et al., 2001; Zhang et al., 2001, 2008). Docking might therefore affect one or both of these factors. Given that the S. pombe MOP has guanine nucleotide exchange factor activity toward a Rab family GTPase, one interesting possibility is that the S. cerevisiae MOP might have an analogous exchange activity toward the Rho/Rac GTPases Rho1p or Cdc42p. In this instance, vesicle docking would lead to activation of one of these GTPases on the docked vesicles, which would in turn recruit Sec3p.
Two Tethering Complexes?
Different vesicle tethering complexes function at different steps in the secretory pathway (TerBush et al., 1996; Conboy and Cyert, 2000; Sacher et al., 2000). These complexes act upstream of the SNAREs to physically connect the two membranes that will fuse. The S. cerevisiae MOP links precursor vesicles before fusion and so fulfills the first criterion. However, our results suggest that one role of the MOP is to recruit a second tethering complex to the vesicles. Why would two different vesicle tethers be necessary for prospore membrane assembly? In addition to linking membranes, different tethering complexes also have physical interactions with Rab GTPases, the SNAREs, or both. Although our data imply that the S. cerevisiae MOP can promote Rab (Sec4p) activation, it may be that the S. cerevisiae MOP lacks the capacity to promote SNARE assembly and so must recruit the exocyst for this purpose. In this model, docking to the MOP does not directly serve to stimulate downstream events of fusion but rather to control where in the cell membrane formation will occur—only at vesicles physically linked to the MOP.
A Parallel in Higher Cells
The recruitment of the exocyst by the MOP is similar to the events of abscission during cytokinesis in higher cells. In mammalian cells, centriolin, a Nud1 ortholog, is found at the centrosome and also at the midbody during cytokinesis (Gromley et al., 2003). Centriolin at the midbody helps recruit the exocyst, which in turn participates in the membrane fusion events leading to cell separation (Gromley et al., 2005). Although Nud1 is not directly required for prospore membrane assembly, it interacts with the MOP proteins (Knop and Strasser, 2000; Nickas et al., 2003). It is possible therefore, that functional analogs of the MOP are involved in centriolin-mediated exocyst recruitment in mammalian cells and the subsequent fusion of recruited vesicles to complete cell division.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Huang (University of Massachusetts, Boston), H. Tachikawa (University of Tokyo, Japan), D. TerBush, and H. Nakanishi (Fukushima Medical University, Japan) for plasmids. This work was supported by NIH grant GM62184 to A.M.N. and grant P41-RR011823 to T.N.D.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-07-0563) on September 8, 2010.
REFERENCES
- Adams I. R., Kilmartin J. V. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bajgier B.K., Malzone M., Nickas M., Neiman A.M. SPO21 is required for meiosis-specific modification of the spindle pole body in yeast. Mol. Biol. Cell. 2001;12:1611–1621. doi: 10.1091/mbc.12.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M. K., Scheller R. H. The molecular machinery for secretion is conserved from yeast to neurons. Proc. Natl. Acad. Sci. USA. 1993;90:2559–2563. doi: 10.1073/pnas.90.7.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd C., Hughes T., Pypaert M., Novick P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell Biol. 2004;167:889–901. doi: 10.1083/jcb.200408124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullitt E., Rout M. P., Kilmartin J. V., Akey C. W. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 1997;89:1077–1086. doi: 10.1016/s0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- Byers B. Cytology of the yeast life cycle. In: Strathern J. N., Jones E. W., Broach J. R., editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1981. pp. 59–96. [Google Scholar]
- Byers B., Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb. Symp. Quant. Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Chen Y. A., Scheller R. H. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- Conboy M. J., Cyert M. S. Luv1p/Rki1p/Tcs3p/Vps54p, a yeast protein that localizes to the late Golgi and early endosome, is required for normal vacuolar morphology. Mol. Biol. Cell. 2000;11:2429–2443. doi: 10.1091/mbc.11.7.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito R. E., Klapholz S. Meiosis and ascospore development. In: Strathern J. N., Jones E. W., Broach J. R., editors. The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1981. pp. 211–287. [Google Scholar]
- Finger F. P., Hughes T. E., Novick P. Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell. 1998;92:559–571. doi: 10.1016/s0092-8674(00)80948-4. [DOI] [PubMed] [Google Scholar]
- Geissler S., Pereira G., Spang A., Knop M., Soues S., Kilmartin J., Schiebel E. The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Goud B., Salminen A., Walworth N. C., Novick P. J. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- Gromley A., Jurczyk A., Sillibourne J., Halilovic E., Mogensen M., Groisman I., Blomberg M., Doxsey S. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 2003;161:535–545. doi: 10.1083/jcb.200301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A., Yeaman C., Rosa J., Redick S., Chen C. T., Mirabelle S., Guha M., Sillibourne J., Doxsey S. J. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Guo W., Roth D., Walch-Solimena C., Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Tamanoi F., Novick P. Spatial regulation of the exocyst complex by Rho1 GTPase. Nat. Cell Biol. 2001;3:353–360. doi: 10.1038/35070029. [DOI] [PubMed] [Google Scholar]
- Hailey D. W., Davis T. N., Muller E. G. Fluorescence resonance energy transfer using color variants of green fluorescent protein. Methods Enzymol. 2002;351:34–49. doi: 10.1016/s0076-6879(02)51840-1. [DOI] [PubMed] [Google Scholar]
- Jantti J., Aalto M. K., Oyen M., Sundqvist L., Keranen S., Ronne H. Characterization of temperature-sensitive mutations in the yeast syntaxin 1 homologues Sso1p and Sso2p, and evidence of a distinct function for Sso1p in sporulation. J. Cell Sci. 2002;115:409–420. doi: 10.1242/jcs.115.2.409. [DOI] [PubMed] [Google Scholar]
- Kane S. M., Roth R. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J. V., Dyos S. L., Kershaw D., Finch J. T. A spacer protein in the Saccharomyces cerevisiae spindle poly body whose transcript is cell cycle-regulated. J. Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J. V., Goh P. Y. Spc110p: assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO J. 1996;15:4592–4602. [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Knop M., Strasser K. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 2000;19:3657–3667. doi: 10.1093/emboj/19.14.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maier P., Rathfelder N., Finkbeiner M. G., Taxis C., Mazza M., Le Panse S., Haguenauer-Tsapis R., Knop M. Cytokinesis in yeast meiosis depends on the regulated removal of Ssp1p from the prospore membrane. EMBO J. 2007;26:1843–1852. doi: 10.1038/sj.emboj.7601621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson E. M., Schwartz C., Neiman A. M. Membrane assembly modulates the stability of the meiotic spindle pole body. J. Cell Sci. 2010;123:2418–2490. doi: 10.1242/jcs.062794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens P. B., Rapport E. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen) J. Cell Biol. 1971;50:344–361. doi: 10.1083/jcb.50.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Borchart A. C., Strasser K., Finkbeiner M. G., Shevchenko A., Knop M. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 2001;20:6946–6957. doi: 10.1093/emboj/20.24.6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller E. G., Snydsman B. E., Novik I., Hailey D. W., Gestaut D. R., Niemann C. A., O'Toole E. T., Giddings T. H., Jr, Sundin B. A., Davis T. N. The organization of the core proteins of the yeast spindle pole body. Mol. Biol. Cell. 2005;16:3341–3352. doi: 10.1091/mbc.E05-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., Morishita M., Schwartz C. L., Coluccio A., Engebrecht J., Neiman A. M. Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J. Cell Sci. 2006;119:1406–1415. doi: 10.1242/jcs.02841. [DOI] [PubMed] [Google Scholar]
- Nakase Y., Nakamura-Kubo M., Ye Y., Hirata A., Shimoda C., Nakamura T. Meiotic spindle pole bodies acquire the ability to assemble the spore plasma membrane by sequential recruitment of sporulation-specific components in fission yeast. Mol. Biol. Cell. 2008;19:2476–2487. doi: 10.1091/mbc.E08-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 1998;140:29–37. doi: 10.1083/jcb.140.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M., Katz L., Brennwald P. J. Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics. 2000;155:1643–1655. doi: 10.1093/genetics/155.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas M. E., Neiman A. M. Ady3p links spindle pole body function to spore wall synthesis in Saccharomyces cerevisiae. Genetics. 2002;160:1439–1450. doi: 10.1093/genetics/160.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas M. E., Schwartz C., Neiman A. M. Ady4p and Spo74p are components of the meiotic spindle pole body that promote growth of the prospore membrane in Saccharomyces cerevisiae. Eukaryot. Cell. 2003;2:431–445. doi: 10.1128/EC.2.3.431-445.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow C. F., Marak J. A fiber apparatus in the nucleus of the yeast cell. J. Cell Biol. 1966;29:129–151. doi: 10.1083/jcb.29.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N. G., Guo L., Imai J., Toh E. A., Matsui Y., Tamanoi F. Rho3 of Saccharomyces cerevisiae, which regulates the actin cytoskeleton and exocytosis, is a GTPase which interacts with Myo2 and Exo70. Mol. Cell. Biol. 1999;19:3580–3587. doi: 10.1128/mcb.19.5.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Fink G. R. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rout M. P., Kilmartin J. V. Components of the yeast spindle and spindle pole body. J. Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge S. A., Morris A. J., Engebrecht J. Relocalization of phospholipase D activity mediates membrane formation during meiosis. J. Cell Biol. 1998;140:81–90. doi: 10.1083/jcb.140.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M., Barrowman J., Schieltz D., Yates J.R., 3rd, Ferro-Novick S. Identification and characterization of five new subunits of TRAPP. Eur J. Cell Biol. 2000;79:71–80. doi: 10.1078/S0171-9335(04)70009-6. [DOI] [PubMed] [Google Scholar]
- Salminen A., Novick P. J. The Sec15 protein responds to the function of the GTP binding protein, Sec4, to control vesicular traffic in yeast. J. Cell Biol. 1989;109:1023–1036. doi: 10.1083/jcb.109.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerer F., Morgan G., Winey M., Philippsen P. Cnm67p is a spacer protein of the Saccharomyces cerevisiae spindle pole body outer plaque. Mol. Biol. Cell. 2001;12:2519–2533. doi: 10.1091/mbc.12.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda Y., Nakanishi H., Mathieson E. M., Neiman A. M. Alternative modes of organellar segregation during sporulation in Saccharomyces cerevisiae. Eukaryot. Cell. 2007;6:2009–2017. doi: 10.1128/EC.00238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin B. A., Chiu C. H., Riffle M., Davis T. N., Muller E. G. Localization of proteins that are coordinately expressed with Cln2 during the cell cycle. Yeast. 2004;21:793–800. doi: 10.1002/yea.1133. [DOI] [PubMed] [Google Scholar]
- TerBush D. R., Maurice T., Roth D., Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Alberti-Segui C., Rebischung C., Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Weimbs T., Mostov K., Low S. H., Hofmann K. A model for structural similarity between different SNARE complexes based on sequence relationships. Trends Cell Biol. 1998;8:260–262. doi: 10.1016/s0962-8924(98)01285-9. [DOI] [PubMed] [Google Scholar]
- Yang H. J., Nakanishi H., Liu S., McNew J. A., Neiman A. M. Binding interactions control SNARE specificity in vivo. J. Cell Biol. 2008;183:1089–1100. doi: 10.1083/jcb.200809178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. J., Neiman A. M. A guanidine nucleotide exchange factor is a component of the meiotic spindle pole body in Schizosaccharomyces pombe. Mol. Biol. Cell. 2010;21:1272–1281. doi: 10.1091/mbc.E09-10-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bi E., Novick P., Du L., Kozminski K. G., Lipschutz J. H., Guo W. Cdc42 interacts with the exocyst and regulates polarized secretion. J. Biol. Chem. 2001;276:46745–46750. doi: 10.1074/jbc.M107464200. [DOI] [PubMed] [Google Scholar]
- Zhang X., Orlando K., He B., Xi F., Zhang J., Zajac A., Guo W. Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J. Cell Biol. 2008;180:145–158. doi: 10.1083/jcb.200704128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.