Abstract
3-Deoxy-d-manno-octulosonic acid (Kdo) is an eight-carbon sugar ubiquitous in Gram-negative bacterial lipopolysaccharides (LPS). Although its biosynthesis is well described, no protein has yet been identified as a Kdo hydrolase. However, Kdo hydrolase enzymatic activity has been detected in membranes of Helicobacter pylori and Francisella tularensis and may be responsible for the removal of side-chain Kdo from the LPS core saccharides. We now report the identification of genes encoding a Kdo hydrolase in F. tularensis Schu S4 and live vaccine strain strains, in H. pylori 26695 strain and in Legionella pneumophila Philadelphia 1 strain. We have renamed the genes kdhA for keto-deoxyoctulosonate hydrolase A. Deletion of kdhA abolished Kdo hydrolase activity in membranes of F. tularensis live vaccine strain. The F. tularensis kdhA mutant synthesized a core oligosaccharide containing a Kdo disaccharide with one of the Kdo residues being a terminal side chain. This side-chain Kdo monosaccharide was absent in the wild-type core oligosaccharide. Expression in Escherichia coli of recombinant KdhA from F. tularensis, H. pylori, and L. pneumophila resulted in a reduction of membrane-associated side-chain Kdo. The identification of this previously faceless enzyme will accelerate study of the biosynthetic basis and biologic impact for postbiosynthetic LPS structural modification.
Keywords: Bacteria, Carbohydrate Chemistry, Carbohydrate Metabolism, Hydrolases, Lipopolysaccharide (LPS), 3-Deoxy-D-manno-octulosonic Acid (KDO), Francisella tularensis, Helicobacter pylori, Legionella pneumophila
Introduction
The eight-carbon sugar 3-deoxy-d-manno-octulosonic acid (Kdo)2 is an important component of cell wall polysaccharides of Gram-negative bacteria, of some green algae (1), and of most higher plants (2). In Gram-negative bacteria, Kdo is ubiquitous in lipopolysaccharides (LPS), the major molecule integral to the outer membrane of the organism, and is also present in some capsular polysaccharides (3–5). In LPS, Kdo plays an essential role; in all bacteria, it links the hydrophobic outer membrane anchor, lipid A, with the surface hydrophilic polysaccharide moiety. The latter comprises a core oligosaccharide (linked to lipid A through Kdo) and an outer polysaccharide chain called O-antigen. LPS has been extensively studied due to its medical importance. Lipid A, also called endotoxin, is a bioactive component of LPS and is associated with initiating innate immune signaling through Toll-like receptor (TLR) 4 leading to Gram-negative septic shock (6, 7). The O-antigen contributes to virulence by protecting bacteria from complement-mediated killing (7) unless specific antibodies directed toward the O-polysaccharide are present. Beside its role as a link between carbohydrate and glycolipid (lipid A), Kdo is also important in maintaining outer membrane integrity (8). The medical importance of Kdo prompted investigations into its biosynthesis, now well characterized (9). However, degradation of Kdo is poorly documented. In particular, no Kdo hydrolase enzyme has yet been identified. Kdo hydrolase activity has been described in oysters (10) and in Helicobacter pylori and Francisella tularensis membranes (11, 12), but the gene coding for this enzyme remains unknown.
H. pylori is a major cause of gastritis, gastroduodenal ulcers, and gastric cancer. F. tularensis, one of the most deadly respiratory pathogens in existence, has recently prompted considerable interest as a weaponizable bacterium. Legionella pneumophila is a waterborne organism that can cause a severe pneumonia when spread by aerosol. Interestingly, unlike the LPS of Enterobacteriaceae and most other Gram-negative bacterial pathogens, the LPS of F. tularensis, H. pylori, and L. pneumophila do not trigger any proinflammatory response through TLR4 (13–15). The atypical biological characteristics of these LPS may be attributable to the unusual structure of their lipid A; when compared with Escherichia coli, lipid A from H. pylori and F. tularensis lacks one phosphate group and two acyl chains (7, 16, 17). Lipid A of L. pneumophila possesses long chain fatty acids (28:0(27-oxo) and 27:0-dioic) double the length of enterobacterial acyl groups (18).
The composition of the core oligosaccharide of F. tularensis and H. pylori is unusual as well, with only one Kdo moiety (two in E. coli). H. pylori synthesizes an LPS precursor with two Kdos and then removes the side-chain Kdo with a yet unidentified Kdo hydrolase (11). F. tularensis bacteria, which also express a Kdo hydrolase, are predicted to do the same (12, 19). The gene encoding a Kdo hydrolase in either organism has not yet been identified.
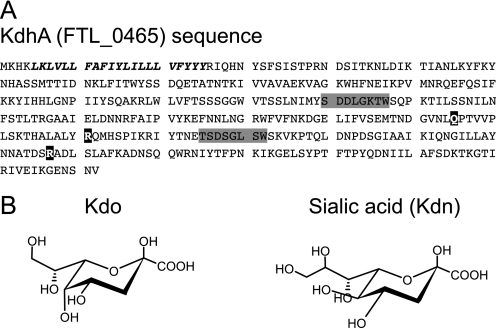
We now describe Kdo hydrolase enzymes from F. tularensis, H. pylori, and L. pneumophila. In F. tularensis live vaccine strain (LVS), the FTL_0465 gene, renamed kdhA, is required for Kdo hydrolase activity in LVS membranes. Expression of KdhA homologs from F. tularensis LVS and Schu S4, H. pylori, and L. pneumophila in E. coli caused reduction of the level of side-chain Kdo in E. coli membrane. Our findings suggest that KdhA is responsible for the removal of side-chain Kdo from Francisella LPS precursors. KdhA proteins were predicted to be sialidases as their sequences contain sialidase motifs (see LVS KdhA, Fig. 1A). The homologies between Kdo hydrolases and sialidases are not surprising given the structural similarities between Kdo and sialic acids. Sialic acids are N- or O-substituted derivatives of neuraminic acid. The most common form, N-acetylneuraminic acid, bears an N-acetyl group at the C-5 position. Kdo is closely related to the sialic acid 2-keto-3-deoxy-d-glycero-d-galacto-nononate (Kdn), which bears a hydroxyl group at C-5 (see Fig. 1B). Kdo differs from Kdn only by the number of carbon atoms (eight in Kdo, nine in Kdn) and their configuration (d-glycero-d-manno in Kdo, d-glycero-d-galacto in Kdn) (see Fig. 1B). A search for other KdhA homologs suggested that numerous sialidase-predicted proteins may actually be Kdo hydrolases.
FIGURE 1.
A sialidase-like protein in F. tularensis LVS. A, amino acid sequence of F. tularensis LVS KdhA showing the transmembrane domain (italics), the BNR/Asp repeats ((S/T)XDXGXT(W/F)) of bacterial sialidase (shaded in gray), and three of the seven residues that compose sialidase catalytic site (in black). Domain predictions were performed using CD-Search (45). B, chemical structure of Kdo and sialic acid Kdn.
EXPERIMENTAL PROCEDURES
Construction of F. tularensis LVS ΔkdhA Deletion Mutant
A ΔkdhA deletion mutant in F. tularensis LVS was constructed by allelic exchange (20). The 1-kb upstream and the 1-kb downstream regions of FTL_0465 were amplified by PCR using primer pairs SIAM_F1/SIAM_R1 and SIAM_F2/SIAM_R2, respectively (supplemental Table 2). Fragments were ligated into the XmaI and KpnI (upstream region) or XmaI and EcoRI (downstream region) sites of pEX18.Kan (21), and the sacB gene from pPV (22) was inserted into the PstI site, yielding pTH49, which was transformed into LVS (23). The transformants were resuspended in 1 ml of tryptic soy broth supplemented with cysteine, incubated at 37 °C without antibiotics on a rotary shaker until an A620 of ∼0.6 was reached, and then plated onto cysteine heart agar containing 2% hemoglobin supplemented with 10% sucrose to induce a second recombination. Individual sucrose-resistant, kanamycin-sensitive colonies were selected as the mutant candidates. PCR was used to screen and select mutants, and the selected mutants were confirmed by genomic sequencing.
Plasmid Construction
Plasmid pET45-KdhA was constructed as follows. The kdhA gene (FTL_0465) from F. tularensis LVS strain was amplified by PCR, using F. tularensis LVS genomic DNA as the template and primers FTL0465_F and FTL0465_R (see supplemental Table 2 for primer sequences). The resulting fragment was digested with KpnI and BamHI and ligated to pET45b.
Plasmid pET45-FTT0399c was constructed as follows. The FTT0399c gene from F. tularensis Schu S4 strain was amplified by PCR, using plasmid pDEST17-FTT0399c, obtained from PlasmID collection (Dana-Farber/Harvard Cancer Center DNA Resource Core) as the template. Forward and reverse primers were the same used for kdhA amplification (see above). The resulting fragment was digested with KpnI and BamHI and ligated to pET45b.
Plasmid pET45-HP0580 was constructed as follows. The HP0580 gene from H. pylori 26695 strain was amplified by PCR, using H. pylori 26695 genomic DNA (a generous gift from M. J. Blaser, New York University) as the template and primers HP0580_F and HP0580_R. The resulting fragment was digested with KpnI and PstI and ligated to pET45b.
Plasmid pET45-lpg2939 was constructed as follows. The lpg2939 gene from L. pneumophila Philadelphia 1 strain was amplified by PCR, using L. pneumophila Philadelphia 1 genomic DNA (a generous gift from R. Isberg, Tufts University) as the template and primers lpg2939_F and lpg2939_R. The resulting fragment was digested with KpnI and PstI and ligated to pET45b.
Preparations of Total Membranes, Outer Membranes, and LPS of F. tularensis
The following procedure was performed to prepare total membranes of F. tularensis strains. Stationary phase cells were prepared as described elsewhere (24). All subsequent steps were carried out at 4 °C or on ice. Cell pellet (5 g) was lysed osmotically by resuspension in 30 ml of ultrapure water and incubation for 30 min. Unbroken cells were removed by centrifugation at 12,000 × g for 20 min. The total membranes were pelleted by centrifugation at 80,000 × g for 1 h, washed once in 50 mm HEPES, pH 7.5, and then resuspended in the same buffer at a concentration of about 2 mg/ml protein, determined by Bradford assay (Bio-Rad).
Outer membranes of F. tularensis strains were prepared by resuspending the total membranes in Dulbecco's PBS (Invitrogen) supplemented with 1% (v/v) sarcosyl (N-lauroylsarcosine sodium salt, Sigma). After incubation at room temperature for 30 min, outer membranes were pelleted by centrifugation at 80,000 × g for 1 h, washed once in Dulbecco's PBS, and then resuspended in the same buffer. LPS was purified from F. tularensis strains as described elsewhere (24), by a modification of the hot phenol-water method (25).
SDS-PAGE, Zinc Stain, and Immunoblotting
To analyze LPS from LVS and ΔkdhA mutant strains, outer membranes were mixed with an equal volume of sample buffer (Bio-Rad) containing 0.1 m Tris-HCl buffer (pH 6.8), 2% (w/v) SDS, 20% (v/v) glycerol, 1% (v/v) 2-mercaptoethanol, and 0.001% (w/v) bromphenol blue. The mixtures were heated at 100 °C for 5 min and electrophoresed in a Tricine SDS-PAGE system using Criterion precast 16.5% acrylamide gel (Bio-Rad). The use of Tricine gels improves the resolution of low molecular LPS bands (26). Gels were stained using the zinc stain procedure described by Hardy et al. (27) or immunoblotted using rabbit polyclonal anti-LVS serum (28) (1:10,000) or mAb 2033 monoclonal antibody (Abcam) (1:5,000).
To determine the subcellular localization of KdhA protein, total membranes from LVS and ΔkdhA mutant strains were resolved in 4–20% Tris-glycine gradient gels (Bio-Rad) and immunoblotted with a rabbit polyclonal anti-KdhASchuS4 serum (1:10,000) or with mAb 2033 monoclonal antibody (1:5,000). To generate a rabbit polyclonal serum against a KdhASchuS4, a pDEST17-kdhA plasmid, ordered from Harvard Institute of Proteomics, was transformed into E. coli BL21(DE3) (Stratagene). N-terminal His-tagged KdhASchuS4 was expressed by induction with 0.5 mm isopropyl-β-d-1-thiogalactopyranoside at A600 = 0.5 for 3 h, solubilized in urea buffer, and purified by affinity chromatography using nickel column. A rabbit polyclonal serum against this protein was generated at Lampire Biological Laboratories (Pipersville, PA) according to a standard protocol established by the vendor.
Lipid A, Core Micropurification, and Identification by MALDI-TOF
The band b and c molecules visualized after zinc stain (see Fig. 2A) were purified from the gel using a procedure described by Pupo and Hardy (29). Pure band b and c molecules were subjected to negative ion matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry analysis. We spotted 1 μl onto the MALDI sample plate followed by 1 μl of 20 mg/ml 5-chloro-2-mercapto-benzothiazole (Sigma) MALDI matrix dissolved in chloroform/methanol (1:1, v/v).
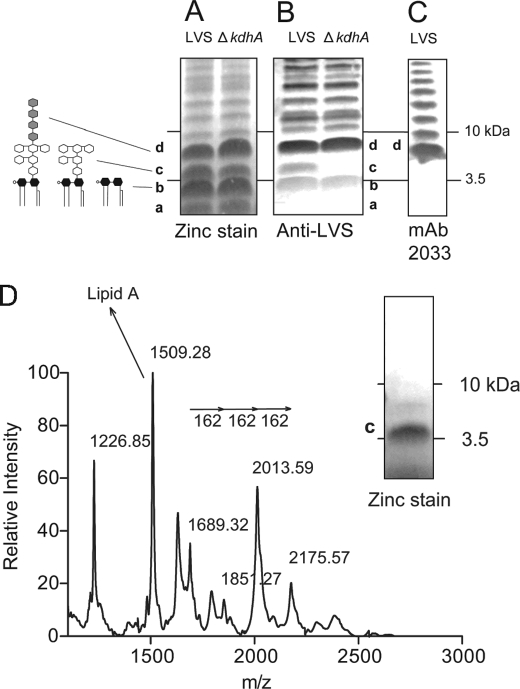
FIGURE 2.
Identification of an altered molecule in ΔkdhA mutant outer membrane. A and B, outer membrane fractions from wild-type LVS and ΔkdhA mutant bacteria were resolved by SDS-PAGE and analyzed by zinc stain (A) and by immunoblot using anti-LVS serum (B). C, identification of band d as lipid A plus core plus the first repeating unit of O-polysaccharide. LVS outer membranes were resolved by SDS-PAGE and immunoblotted with mAb 2033 monoclonal antibody. D, identification of band c as lipid A plus core oligosaccharide by negative-ion MALDI-TOF MS. The band c molecule was micropurified from wild-type outer membranes as described under “Experimental Procedures.” The purity of the band c molecule was monitored by SDS-PAGE followed by zinc stain. Band b was identified as lipid A using the same procedure (supplemental Fig. 2).
Expression of KdhA Orthologs in E. coli BL21
One liter of EPCM1 (30) containing carbenicillin (100 μg/ml) (Sigma) was inoculated using multiple colonies and incubated overnight at 37 °C with 200 rpm of shaking. This was added to 4 liters of warmed EPCM1 containing carbenicillin (100 μg/ml) in a 7-liter fermentor (Applikon Biotechnology, Foster City CA). The starting A600 was ∼1. The culture was grown at 37 °C, pH 7.0, with mixing at 900 rpm and addition of air and oxygen to maintain an excess of oxygen until an A600 between 5 and 7 was reached. Protein expression was induced by lowering the temperature to 25 °C before adding isopropyl-β-d-1-thiogalactopyranoside (0.8 mm). After ∼2 h of induction, the culture was harvested by centrifugation (20 min at 6,620 × g, 4 °C). Culture supernates were discarded, and cell pellets were frozen at −80 °C. Total membranes were prepared using the same procedure as Francisella total membranes (see above), except that cells were broken by sonication.
Analysis of Glycosyl Residues by High-performance Anion-Exchange Chromatography (HPAEC)
Each LPS was dissolved in deionized water (15 mg/ml), acetic acid (Mallinckrodt Baker) was added to 1%, and the solution was heated at 90 °C for 2 h. This procedure hydrolyzes the ketosidic bond between side-chain and inner Kdos and between inner Kdo and lipid A, which precipitates. The lipid A was removed by centrifugation, and the carbohydrate was analyzed by HPAEC on CarboPacTM PA1 column (Dionex Corp.), using 60 mm sodium acetate (Sigma)–100 mm NaOH (Fisher Scientific) for isocratic elution. A Kdo standard (Sigma) was used for peak identification and Kdo amount measures.
LPS delipidated by acetic acid treatment was further hydrolyzed with 0.5 m trifluoroacetic acid (TFA) (Sigma) at 100 °C for 18 h. This procedure hydrolyzes all the glycosidic bonds, releasing LPS monosaccharides. The amount of galactosamine, glucose, and mannose was measured by HPAEC on CarboPacTM PA1 column (Dionex Corp.), with 12 mm NaOH for isocratic elution. Galactosamine, glucose, and mannose standards (Sigma) were used for peak identification and amount measures.
Side-chain Kdo amount was measured in E. coli BL21 strains by treating membranes with 1% acetic acid at 90 °C for 2 h. Kdo released by this mild hydrolysis was analyzed by HPAEC as described above. A further hydrolysis with 0.5 m TFA at 100 °C for 18 h released glucose from E. coli membranes. The amount of released glucose, which most likely comes from LPS molecules, was used to normalize the amount of side-chain Kdo. Other normalization methods (Bradford assay, measures of A280 and A260) yielded similar results.
Sialidase and Kdo Hydrolase Assays
4 × 108 LVS bacteria were osmotically lysed by resuspension in ultrapure water and incubated overnight at 37 °C in 30 μg of 3′- or 6′-sialyllactose (Sigma-Aldrich), 50 mm Tris-HCl (Roche Applied Science), pH 7.2, and 1 mm CaCl2 (Mallinckrodt Baker). Neuraminidase from Clostridium perfringens (1 microunit) (Sigma) was used as positive control. The amount of released sialic acid was measured by the thiobarbituric method (31).
The Kdo hydrolase was assayed as described by Wang et al. (12), with some modifications. Briefly, a 50-μl reaction mixture containing 50 mm potassium phosphate, pH 6, 0.1% Triton X-100 (Sigma), 10 μg of Re LPS from Salmonella enterica serovar Minnesota Re 595 (Sigma), and 0.2 mg/ml Francisella membranes was incubated at 30 °C for 30 min. The amount of released Kdo was measured by HPAEC.
RESULTS
Alteration of the LPS Core Oligosaccharide in an F. tularensis kdhA Deletion Mutant
The KdhA (FTL_0465) protein sequence contains two BNR/Asp repeats ((S/T)XDXGXT(W/F)), a recurring motif in bacterial sialidases (32) (Fig. 1A). Sialidase activity was assessed in F. tularensis LVS. LVS crude extracts were incubated with 3′- or 6′-sialyllactose, and sialic acid release was measured by thiobarbituric assay. No sialidase activity was detected in F. tularensis LVS crude extracts (supplemental Fig. 1). Regarding its homologies with sialidases, we hypothesized that KdhA was a glycosyl hydrolase and first investigated its involvement in LPS synthesis. A ΔkdhA deletion mutant was constructed, LVS and ΔkdhA outer membranes were resolved by Tricine SDS-PAGE, and LPS was visualized by zinc stain. A ladder-like banding pattern characteristic of LPS was observed, with four predominant bands, named a, b, c, and d (Fig. 2A). No difference in these bands was noted between the wild-type and the mutant. However, after probing the gel with an anti-LVS serum, band c was only detected in wild-type outer membranes and not in mutant outer membranes (Fig. 2B). This suggests that the band c molecule is structurally altered in the ΔkdhA mutant and is not recognized by anti-LVS antibodies.
When probed with the monoclonal antibody mAb 2033 specific to the O-antigen, band d is the smallest molecular size band reacting (Fig. 2C) and therefore likely represents lipid A plus core oligosaccharide and the first repeating unit of O-polysaccharide. Therefore, band c is likely to be lipid A or lipid A plus core oligosaccharide. To identify the band c molecule, band c from LVS lane was cut out of the zinc-stained gel, and the molecule was recovered by passive diffusion. The zinc stain procedure does not chemically modify LPS (27), allowing us to perform a MALDI-TOF MS analysis of the wild-type band c molecule. If band c is lipid A linked to the core oligosaccharide, we would expect to observe in the mass spectrum a lipid A fragment and fragments separated by m/z 162, the mass unit of mannose and glucose, the main sugars of F. tularensis core oligosaccharide (Fig. 3D). The species at m/z 1509.3 fits with the monophosphorylated lipid A structure described in LVS by Phillips et al. (17) (Fig. 2D, Table 1). The ion species at m/z 1689.3, 1851.3, 2013.6 and 2175.6 are separated by Δm/z 162 (Fig. 2D, Table 1). Therefore, the band c molecule is lipid A linked to the core oligosaccharide. Following the same procedure, the band b molecule was identified as lipid A (supplemental Fig. 2). Lipid A from wild-type and ΔkdhA bacteria was identical (supplemental Fig. 2), suggesting that the core oligosaccharide and not lipid A is altered in the ΔkdhA mutant strain.
FIGURE 3.
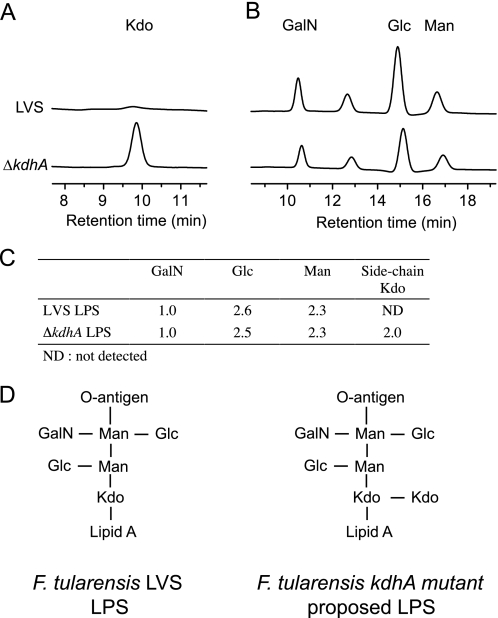
HPAEC analysis of core oligosaccharide from wild-type and ΔkdhA mutant bacteria. A, side-chain Kdo detection after mild hydrolysis of LPS from wild-type and ΔkdhA mutant. Kdo peak was determined by co-chromatography with a Kdo standard. B, galactosamine, glucose, and mannose detection after TFA hydrolysis of LPS from wild-type and ΔkdhA mutant. C, molar ratio of galactosamine, glucose, mannose, and side-chain Kdo in LPS from wild-type and ΔkdhA mutant. D, structure of core oligosaccharide synthesized by F. tularensis LVS strain (16) and proposed structure synthesized by LVS ΔkdhA mutant.
TABLE 1.
Assignment of signals from negative ion MALDI-TOF analysis of band c molecule
| (M-H)−m/z | Proposed compositions |
|---|---|
| 2175.57 | Lipid A, Kdo, Man, Man, Glc |
| 2013.59 | Lipid A, Kdo, Man, Glc or Lipid A, Kdo, Man, Man |
| 1851.27 | Lipid A, Kdo, Man |
| 1689.32 | Lipid A, Kdo |
| 1509.28 | Lipid A |
The LPS Core Region in an F. tularensis kdhA Deletion Mutant Contains a Side-chain Terminal Kdo
It has been proposed that Francisella cells first assemble an LPS precursor that contains a Kdo disaccharide before removing the side-chain Kdo by the action of an unidentified Kdo hydrolase (12, 19). If this Kdo hydrolase is KdhA, LPS from ΔkdhA mutant should contain a side-chain terminal Kdo. To detect it, a common method is to selectively release side-chain Kdo by mild hydrolysis (33). Wild-type and mutant LPS were treated with acetic acid, and the amount of released Kdo was measured by HPAEC. No released Kdo was detected in wild-type LPS hydrolysate, as expected (Fig. 3A). By contrast, Kdo was released in hydrolysates of the mutant LPS, demonstrating that ΔkdhA mutant LPS does contains a terminal side-chain Kdo (Fig. 3A). After TFA hydrolysis, the ratios of other core oligosaccharide sugars (mannose, glucose, and galactosamine) were also measured (Fig. 3B). In wild-type LPS, for one molecule of galactosamine, 2.6 molecules of glucose and 2.3 molecules of mannose were measured (Fig. 3C). This ratio is in agreement with the published core structure (16). A similar ratio of galactosamine, glucose and mannose was measured in the ΔkdhA mutant LPS, which additionally contained 2.0 molecules of side-chain Kdo per molecule of galactosamine (Fig. 3C). This suggests that the structure of the ΔkdhA mutant core oligosaccharide is the one shown in Fig. 3D. Therefore, the alteration of the mutant core oligosaccharide observed by immunoblot (Fig. 2B) may be due to the presence of a terminal side-chain Kdo residue.
KdhA Is a Kdo Hydrolase
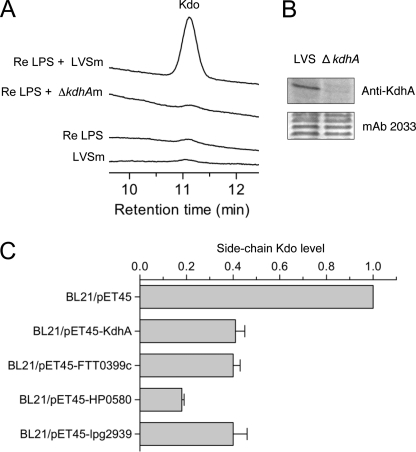
A Kdo hydrolase activity has been detected in total membrane extracts (outer membrane and cytoplasmic membrane) of F. tularensis (12). To test whether KdhA is required for this activity, total membrane extracts of LVS and ΔkdhA mutant were incubated with the rough Re LPS from S. enterica serotype Minnesota Re 595, which is a lipid A molecule linked to one of the Kdo residues in a disaccharide of Kdo. Kdo release was measured by HPAEC. Kdo hydrolase activity was detected in wild-type total membranes by the presence of released Kdo, but not in mutant membranes (Fig. 4A). No Kdo was released from Re LPS alone or LVS total membranes alone (Fig. 4A). These data suggest that KdhA is required for the Kdo hydrolase activity observed in Francisella membranes. Consistent with this hypothesis, KdhA was detected in total membrane extracts, as shown by immunoblot analysis (Fig. 4B).
FIGURE 4.
KdhA is a Kdo hydrolase. A, Kdo hydrolase activity in F. tularensis wild-type and ΔkdhA mutant total membranes. Total membranes of F. tularensis LVS (LVSm) or ΔkdhA (ΔkdhAm) were assayed with Re LPS from S. enterica Re 595 as the substrate. Release of Kdo was measured by HPAEC. B, detection of KdhA protein in LVS total membranes. Total membranes of LVS wild-type and ΔkdhA mutant were analyzed by immunoblotting for the presence of KdhA. Loaded amounts were controlled by probing with mAb 2033 monoclonal antibody, specific to the O-antigen. C, side-chain Kdo trimming in E. coli BL21 expressing the following KdhA orthologs: KdhA from F. tularensis LVS, FTT0399c from F. tularensis Schu S4, HP0580 from H. pylori, and Lpg2939 from L. pneumophila. Total membranes from E. coli BL21 expressing KdhA orthologs were subjected to mild hydrolysis, and released Kdo was measured by HPAEC. Measurements were normalized with the amount of glucose released by TFA treatment and are expressed as -fold induction when compared with BL21/pET45b vector control. The average values ± S.D. of two independent experiments are shown.
To show conclusively that KdhA is a Kdo hydrolase, the kdhA gene from LVS was expressed as an inducible recombinant protein in E. coli. The effect of KdhA expression on the content of side-chain Kdo residues in E. coli total membranes was assessed. Total membranes from the empty vector control and the recombinant KdhA expression strain were isolated and subjected to mild hydrolysis by acetic acid (these conditions release only the terminal side-chain Kdo), and released Kdo was quantified by HPAEC. Terminal side-chain Kdo was detected in membranes of the control strain (Fig. 4C). The release of this sugar was drastically reduced in membranes of the E. coli strain expressing recombinant KdhA; expression of KdhA resulted in removal of 60% of the side-chain Kdo in membranes when compared with the control (Fig. 4C). This result demonstrates that KdhA is a Kdo hydrolase.
Kdo Hydrolases from F. tularensis Schu S4, H. pylori, and L. pneumophila
KdhA is very well conserved among Francisella strains, as shown by a search in the National Center for Biotechnology Information (NCBI) database. In particular, the ortholog from F. tularensis Schu S4, the most virulent Francisella strain, has 369 identical residues over 372 (supplemental Fig. 1). To identify putative Kdo hydrolases in other species, a search was performed using the NCBI server. The 40 most similar proteins in non-Francisella organisms are listed in supplemental Table 1. Putative orthologs were found in the medically important bacteria H. pylori and L. pneumophila (supplemental Table 1). An alignment of KdhA with proteins HP0580 from H. pylori and Lpg2939 from L. pneumophila showed an amino acid similarity of 44 and 48%, respectively (supplemental Table 1, supplemental Fig. 1).
Kdo hydrolase activity was assessed for orthologs from H. pylori and L. pneumophila, as well as from the most virulent strain of F. tularensis, Schu S4 (supplemental Fig. 1). The FTT0399c gene from F. tularensis Schu S4, the HP0580 gene from H. pylori 26695 strain, and the lpg2939 gene from L. pneumophila Philadelphia 1 strain were expressed as recombinant proteins in E. coli. The effect of their expression on side-chain Kdo levels in E. coli total membranes was assessed. Like KdhA from F. tularensis LVS, expression of FTT0399c, HP0580, and lpg2939 resulted in reduction of side-chain Kdo levels in E. coli total membranes when compared with the empty vector control (Fig. 4C). Therefore, Kdo hydrolase enzyme is not restricted to F. tularensis but is also present in H. pylori and L. pneumophila bacteria.
DISCUSSION
This study reports the first identification of genes coding for a Kdo hydrolase enzyme, designated KdhA, in genomes of F. tularensis LVS and Schu S4, H. pylori, and L. pneumophila. Several lines of evidence support this identification. First, deletion of the kdhA gene in F. tularensis LVS strain abolished the Kdo hydrolase activity in membranes. Second, expression of KdhA in E. coli resulted in removal of side-chain Kdo from E. coli membranes.
Several bacterial species synthesize an LPS with only a single Kdo residue by transfer of one Kdo to lipid A (Bordetella, Haemophilus) or of two Kdos to lipid A followed by removal of the side-chain Kdo (Helicobacter (11)). Our data strongly suggest that Francisella is similar to Helicobacter in that regard, as suggested previously (19), and that the enzyme responsible for side-chain Kdo removal is KdhA (FTL_0465 in F. tularensis and HP0580 in H. pylori). Consistent with the previous detection of Kdo hydrolase activity in Francisella membrane (12), KdhA is membrane-bound. Because of its hydrophobicity, we were not able to perform a native purification of KdhA after expression in E. coli. A truncated KdhA that is missing the N-terminal transmembrane helix was purified but showed no Kdo hydrolase activity. This lack of activity may be a result from the truncation or from the need of an unknown co-factor, which may be a protein. Indeed during the reviewing process of this article, new results were published describing the Kdo hydrolase activity in Francisella novicida and H. pylori (34, 35). In these organisms, two subunits were required for Kdo hydrolase activity, the sialidase-like protein (the catalytic subunit) and a small inner membrane protein (a membrane-anchoring subunit) (34–36). It is likely the same in F. tularensis LVS, the second subunit being coded by the FTL_0464 gene, adjacent to kdhA. The reason why the expression of kdhA alone in E. coli was sufficient to remove side-chain Kdo from E. coli membranes (Fig. 4C) remains unknown. It is conceivable that E. coli expresses a protein that can functionally replace FTL_0464.
KdhA was predicted to belong to the sialidase superfamily as its sequence contains two BNR/Asp repeats, common in sialidases (Fig. 1A). Similarities between Kdo hydrolases and sialidases are not surprising given the analogies between Kdo and sialic acids from both a structural (Fig. 1B) and a biosynthetic point of view. The sialic acid biosynthetic pathway may have evolved from the Kdo biosynthetic pathway (37). It is thus tempting to speculate that Kdo hydrolases and sialidases share a common origin and may thus be classified in the same superfamily. Interestingly, Kdo and sialic acids differ considerably in their distribution in the tree of life. Sialic acids are found in the deuterostome lineage of animals and their associated bacteria. Kdo, first discovered in Gram-negative bacteria, is a component of the cell wall of some green algae (1) and of the primary cell wall of most higher plants (2). Both sialic acids and Kdo play essential roles in animals and plants, respectively (38, 39).
Putative Kdo hydrolases were identified in various bacteria and plants by running a BLAST program in the NCBI database. The ones listed in supplemental Table 1 are all predicted to belong to the sialidase superfamily. However, many might be Kdo hydrolases, not sialidases. Indeed, many contain a lower number of BNR/Asp repeats when compared with typical sialidases (2–3, whereas most bacterial sialidases have 4–5 repeats) (supplemental Table 1). They are also smaller than bacterial sialidases, with about 400 amino acids, when compared with 600–1000. Besides, an alignment of 10 of these proteins with LVS KdhA shows that many residues are conserved (supplemental Fig. 1). Finally, these proteins were found almost exclusively in aquatic and plant-associated bacteria, as well as in plants. Plants produce Kdo, but not sialic acid (40). The occurrence of Kdo in aquatic environments has been documented as well; Kdo is a major component of the cell wall of some green algae (1), a group from which higher plants emerged (41). Interestingly, the first Kdo hydrolase activity was discovered in the hepatopancreas of oyster (10), which feeds on microalgae, including green algae. Kdo is an attractive carbon source as the Kdo aldolase-mediated degradation yields a pyruvate (42). Therefore, Kdo hydrolases may function to provide free Kdo for organisms living in contact with plants or microalgae. Putative Kdo hydrolases found in plants may serve the same purpose; under sugar starvation induced by darkness, plants produce glycosyl hydrolases to degrade their own cell wall polysaccharides, which may serve as a carbon source (43).
KdhA from F. tularensis LVS is membrane-bound, and our data suggest that it is targeting Kdo from endogenous LPS. KdhA from H. pylori is likely doing the same. The finding of a Kdo hydrolase in L. pneumophila was surprising as L. pneumophila LPS contains a side-chain Kdo (44). However, the gene coding for the Kdo hydrolase, lpg2939, is adjacent to genes involved in lipid A synthesis, suggesting that L. pneumophila KdhA might indeed be involved in LPS modification. L. pneumophila may express KdhA under certain conditions, keeping or removing the side-chain Kdo depending on the environment. L. pneumophila may also first synthesize an LPS containing a Kdo trisaccharide before removing the terminal Kdo.
Herein, the genes encoding for a Kdo hydrolase enzyme are identified in F. tularensis, H. pylori, and L. pneumophila, and their functional activity is demonstrated. It will be especially interesting to study the role of KdhA in the biology of these pathogens and potentially in the orthologs produced by a number of plants and organisms feeding on plants and algae.
Supplementary Material
Acknowledgments
We are grateful to Laurie Comstock (Brigham and Women's Hospital, Boston, MA) for helpful discussions. We also thank Ralph Isberg (Tufts University School of Medicine, Boston, MA) and Martin Blaser (New York University Langone Medical Center, New York, NY) for providing us with H. pylori 26695 and L. pneumophila Philadelphia 1 genomic DNA, respectively, and Barbara Reinap, Brad Pentelute, Lauren Perry, and Bella Printseva for technical assistance (Harvard Medical School, Boston, MA).
This work was supported, in whole or in part, by National Institutes of Health Grant AI057159 from the NIAID (to D. L. K.). This work was also supported by the New England Center of Excellence in Biodefense and Emerging Infectious Disease.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1 and 2.
- Kdo
- 3-deoxy-d-manno-octulosonic acid
- Kdn
- 2-keto-3-deoxy-d-glycero-d-galacto-nononate
- LVS
- live vaccine strain
- HPAEC
- high-performance anion-exchange chromatography
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- BNR
- bacterial neuraminidase repeat.
REFERENCES
- 1.Becker B., Hård K., Melkonian M., Kamerling J. P., Vliegenthart J. F. (1989) Eur. J. Biochem. 182, 153–160 [DOI] [PubMed] [Google Scholar]
- 2.York W. S., Darvill A. G., McNeil M., Albersheim P. (1985) Carbohydr. Res. 138, 109–126 [Google Scholar]
- 3.Bhattacharjee A. K., Jennings H. J., Kenny C. P. (1978) Biochemistry 17, 645–651 [DOI] [PubMed] [Google Scholar]
- 4.Lenter M., Jann B., Jann K. (1990) Carbohydr. Res. 197, 197–204 [DOI] [PubMed] [Google Scholar]
- 5.Nimtz M., Wray V., Domke T., Brenneke B., Häussler S., Steinmetz I. (1997) Eur. J. Biochem. 250, 608–616 [DOI] [PubMed] [Google Scholar]
- 6.Galanos C., Lüderitz O., Rietschel E. T., Westphal O., Brade H., Brade L., Freudenberg M., Schade U., Imoto M., Yoshimura H., Kusumoto S., Shiba T. (1985) Eur. J. Biochem. 148, 1–5 [DOI] [PubMed] [Google Scholar]
- 7.Raetz C. R., Whitfield C. (2002) Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gronow S., Brade H. (2001) J. Endotoxin Res. 7, 3–23 [PubMed] [Google Scholar]
- 9.Cipolla L., Polissi A., Airoldi C., Galliani P., Sperandeo P., Nicotra F. (2009) Curr. Drug Discov. Technol. 6, 19–33 [DOI] [PubMed] [Google Scholar]
- 10.Li Y. T., Wang L. X., Pavlova N. V., Li S. C., Lee Y. C. (1997) J. Biol. Chem. 272, 26419–26424 [DOI] [PubMed] [Google Scholar]
- 11.Stead C., Tran A., Ferguson D., Jr., McGrath S., Cotter R., Trent S. (2005) J. Bacteriol. 187, 3374–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Karbarz M. J., McGrath S. C., Cotter R. J., Raetz C. R. (2004) J. Biol. Chem. 279, 49470–49478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandström G., Sjöstedt A., Johansson T., Kuoppa K., Williams J. C. (1992) FEMS Microbiol. Immunol. 5, 201–210 [DOI] [PubMed] [Google Scholar]
- 14.Pérez-Pérez G. I., Shepherd V. L., Morrow J. D., Blaser M. J. (1995) Infect. Immun. 63, 1183–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girard R., Pedron T., Uematsu S., Balloy V., Chignard M., Akira S., Chaby R. (2003) J. Cell Sci. 116, 293–302 [DOI] [PubMed] [Google Scholar]
- 16.Vinogradov E., Perry M. B., Conlan J. W. (2002) Eur. J. Biochem. 269, 6112–6118 [DOI] [PubMed] [Google Scholar]
- 17.Phillips N. J., Schilling B., McLendon M. K., Apicella M. A., Gibson B. W. (2004) Infect. Immun. 72, 5340–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zähringer U., Knirel Y. A., Lindner B., Helbig J. H., Sonesson A., Marre R., Rietschel E. T. (1995) Prog. Clin. Biol. Res. 392, 113–139 [PubMed] [Google Scholar]
- 19.Wang X., Ribeiro A. A., Guan Z., McGrath S. C., Cotter R. J., Raetz C. R. (2006) Biochemistry 45, 14427–14440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauriano C. M., Barker J. R., Nano F. E., Arulanandam B. P., Klose K. E. (2003) FEMS Microbiol. Lett. 229, 195–202 [DOI] [PubMed] [Google Scholar]
- 21.Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998) Gene 212, 77–86 [DOI] [PubMed] [Google Scholar]
- 22.Golovliov I., Sjöstedt A., Mokrievich A., Pavlov V. (2003) FEMS Microbiol. Lett. 222, 273–280 [DOI] [PubMed] [Google Scholar]
- 23.Baron G. S., Myltseva S. V., Nano F. E. (1995) Methods Mol. Biol. 47, 149–154 [DOI] [PubMed] [Google Scholar]
- 24.Kim T. H., Sebastian S., Pinkham J. T., Ross R. A., Blalock L. T., Kasper D. L. (2010) J. Biol. Chem. 285, 27839–27849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westphal O., Jann K. (1965) Methods Carbohydr. Chem. 5, 83–91 [Google Scholar]
- 26.Lesse A. J., Campagnari A. A., Bittner W. E., Apicella M. A. (1990) J. Immunol. Methods 126, 109–117 [DOI] [PubMed] [Google Scholar]
- 27.Hardy E., Pupo E., Castellanos-Serra L., Reyes J., Fernández-Patrón C. (1997) Anal. Biochem. 244, 28–32 [DOI] [PubMed] [Google Scholar]
- 28.Sebastian S., Dillon S. T., Lynch J. G., Blalock L. T., Balon E., Lee K. T., Comstock L. E., Conlan J. W., Rubin E. J., Tzianabos A. O., Kasper D. L. (2007) Infect. Immun. 75, 2591–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pupo E., Hardy E. (2007) Electrophoresis 28, 2351–2357 [DOI] [PubMed] [Google Scholar]
- 30.Bernard A., Payton M. (2001) Curr. Protoc. Protein Sci. 5.3.1–5.3.18 [DOI] [PubMed] [Google Scholar]
- 31.Warren L. (1959) J. Biol. Chem. 234, 1971–1975 [PubMed] [Google Scholar]
- 32.Roggentin P., Rothe B., Kaper J. B., Galen J., Lawrisuk L., Vimr E. R., Schauer R. (1989) Glycoconjugate J. 6, 349–353 [DOI] [PubMed] [Google Scholar]
- 33.Kiang J., Szu S. C., Wang L. X., Tang M., Lee Y. C. (1997) Anal. Biochem. 245, 97–101 [DOI] [PubMed] [Google Scholar]
- 34.Zhao J., Raetz C. R. (July20, 2010) Mol. Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stead C. M., Zhao J., Raetz C. R., Trent M. S. (July27, 2010) Mol. Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raetz C. R., Guan Z., Ingram B. O., Six D. A., Song F., Wang X., Zhao J. (2009) J. Lipid Res. 50, S103–S108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angata T., Varki A. (2002) Chem. Rev. 102, 439–469 [DOI] [PubMed] [Google Scholar]
- 38.Varki A. (2008) Trends Mol. Med. 14, 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delmas F., Séveno M., Northey J. G., Hernould M., Lerouge P., McCourt P., Chevalier C. (2008) J. Exp. Bot. 59, 2639–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeleny R., Kolarich D., Strasser R., Altmann F. (2006) Planta 224, 222–227 [DOI] [PubMed] [Google Scholar]
- 41.Palmer J. D., Soltis D. E., Chase M. W. (2004) Am. J. Bot. 91, 1437–1445 [DOI] [PubMed] [Google Scholar]
- 42.Ghalambor M. A., Heath E. C. (1966) J. Biol. Chem. 241, 3222–3227 [PubMed] [Google Scholar]
- 43.Lee E. J., Matsumura Y., Soga K., Hoson T., Koizumi N. (2007) Plant Cell Physiol. 48, 405–413 [DOI] [PubMed] [Google Scholar]
- 44.Moll H., Knirel Y. A., Helbig J. H., Zähringer U. (1997) Carbohydr. Res. 304, 91–95 [DOI] [PubMed] [Google Scholar]
- 45.Marchler-Bauer A., Bryant S. H. (2004) Nucleic Acids Res. 32, W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.