Abstract
Although it is well established that the majority of eukaryotic DNA is sequestered as nucleosomes, the higher-order structure resulting from nucleosome interactions as well as the dynamics of nucleosome stability are not as well understood. To characterize the structural and functional contribution of individual nucleosomal sites, we have developed a chromatin model system containing up to four nucleosomes, where the array composition, saturation, and length can be varied via the ordered ligation of distinct mononucleosomes. Using this system we find that the ligated tetranucleosomal arrays undergo intra-array compaction. However, this compaction is less extensive than for longer arrays and is histone H4 tail-independent, suggesting that well ordered stretches of four or fewer nucleosomes do not fully compact to the 30-nm fiber. Like longer arrays, the tetranucleosomal arrays exhibit cooperative self-association to form species composed of many copies of the array. This propensity for self-association decreases when the fraction of nucleosomes lacking H4 tails is systematically increased. However, even tetranucleosomal arrays with only two octamers possessing H4 tails recapitulate most of the inter-array self-association. Varying array length shows that systems as short as dinucleosomes demonstrate significant self-association, confirming that relatively few determinants are required for inter-array interactions and suggesting that in vivo multiple interactions of short runs of nucleosomes might contribute to complex fiber-fiber interactions. Additionally, we find that the stability of nucleosomes toward octamer loss increases with array length and saturation, suggesting that in vivo stretches of ordered, saturated nucleosomes could serve to protect these regions from histone ejection.
Keywords: Chromatin Regulation, Chromatin Structure, Histones, Nuclear Structure, Nucleosome
Introduction
The nucleosome, composed of a histone octamer wrapped by 147 bp of DNA (1, 2), is the fundamental repeating unit of chromatin. Nucleosomes ubiquitously associate with genomic DNA, occupying the majority of DNA in most cells (3). For example, high-resolution nucleosome mapping of the budding yeast Saccharomyces cerevisiae suggests 69% of the DNA is associated with nucleosomes (4). Nucleosome positioning across the genome is not fully understood but is dictated in part by the underlying genomic sequence by the association and action of non-nucleosomal proteins and potentially by the direct interaction of nucleosomes (5). Positioning of nucleosomes results in free linker DNA between nucleosomes that varies within a genome and between cell types and species and can range from 10 or fewer nucleotides to more than 100 nucleotides (3).
Nucleosomes can interact with one another, generating chromatin structures that affect DNA-based biological processes such as transcription, replication, and repair. Direct nucleosome to nucleosome interactions that have been observed in vitro include short-range nucleosome interactions that form 30-nm fibers and longer range interactions to generate fiber to fiber structures (6–8). The structural details and determinants of these higher-order structures remain an active area of research. However, previous studies have shown that histone tails, N-terminal stretches of amino acids that protrude out from the histone core, play a significant role in these processes, with the H4 tail playing a predominant role in both types of interactions (9–13).
Other nucleosome-associated interactions that affect higher order chromatin structure are mediated by non-histone chromatin associated proteins. Several proteins play a role in the formation and/or maintenance of repressive chromatin, including the polycomb group protein PRC1 (14, 15), MENT (16), MeCP2 (17, 18), and heterochromatin protein 1 (HP1) (19, 20). Although the mechanism by which this repression is achieved is still not well defined in vivo, in vitro these proteins facilitate the creation of short and long range higher order chromatin structures through their interaction with various aspects of histone modifications, histone tails, the nucleosome core, and/or the linker DNA (14–22). Nucleosome to nucleosome interactions may also be present in more transcriptionally active euchromatic regions, as establishment of histone H3 acetylation by the yeast SAGA coactivator complex can be facilitated by what appears to involve long-range bridging of nucleosomes (23).
The presence of nucleosomes can create a physical barrier that limits interaction with underlying DNA by proteins that are involved in transcription, replication, and repair. Because nucleosomes are dynamic structures that can be moved, assembled, and disassembled in a regulated fashion, this access can be controlled. For example, during transcriptional initiation, nucleosomes can be disassembled to generate stretches of DNA that are largely free of nucleosomes (24–26). This disassembly process involves many factors, including nucleosome remodeling complexes, histone modifying enzymes, and histone chaperones (27). Loss of nucleosomes enhances transcription through a number of potential mechanisms, including exposure of stretches of DNA bearing transcriptional activator binding sites (28).
Because the presence and interactions of nucleosomes have important structural and functional consequences but are potentially difficult to dissect into individual nucleosome components with traditional nucleosomal array systems, we sought to develop a new model system in which the composition of individual nucleosomes could be controlled. In this study we ligate mononucleosomes to assemble well ordered di-, tri, and tetranucleosomal arrays to better understand various features of higher-order chromatin structure and nucleosome stability.
EXPERIMENTAL PROCEDURES
601-177-1 Nonpalindromic DNA Template Design and Cloning
Four single repeats of the 601-177-12 positioning sequence, each with a different set of modified ends, were PCR-amplified and cloned into plasmid pRS315. The template used for this amplification was a single copy of the 601–177 sequence, released by ScaI digestion from a plasmid containing 12 contiguous copies of this sequence (10, 29) and purified by size exclusion chromatography. The following primer pairs were used to amplify the 601–177 positioning sequence from the purified 601-177-1 fragment, creating DNA template fragments 1X, 2P, 3X, and 4X: Fragment 1, (5′ XbaI-DraIII, 3′ XbaI-BglI) 5′-GGTATGGTATTCTAGACACTACGTGGGATCTTACATGCACAGGATG-3′ and 5′-GGTATGGTATTCTAGAGCCCTTCTGGCACGGCCGCCCTGGAGAATCC-3′; Fragment 2, (5′ PstI-BglI, 3′ PstI-PflMI) 5′-GGTATGGTATCTGCAGGCCGGAATGGCTCTTACATGCACAGGATG-3′ and 5′-GGTATGGTATCTGCAGCCAATAGTTGGAGGCCGCCCTGGAGAATCC-3′; Fragment 3, (5′ XbaI-PflMI, 3′ XbaI-BspMI) 5′-GGTAGTGTATTCTAGACCAACTAATGGATCTTACATGCACAGGATG-3′ and 5′-GGTATGGTATTCTAGAACCTGCATTCAAAAGGCCGCCCTGGAGAATCC-3′; Fragment 4, (5′ XbaI-BspMI, 3′ XbaI-BstXI) 5′-GGTATGGTATTCTAGATTTTGGAAGCAGGTCTTACATGCACAGGATG-3′ and 5′-GGTAAGGTATTCTAGACCAAAATCGTGGACGCGGCCGCCCTGGAGAATCC-3′. PCR products for fragments 2P and 4X were digested with PstI (fragment 2) or XbaI (fragment 4) and cloned into pRS315. PCR products for fragments 1X and 3X were blunt end-cloned into the SmaI site of pRS315. The resulting plasmids were digested with XbaI (releasing fragments 1X, 3X, and 4X) or PstI (releasing fragment 2P). Fragments were purified by electrophoresis on a 1% agarose gel followed by electroelution and butanol extraction, ethanol precipitation, and quantitation by UV absorbance.
To make isolation of large quantities of template fragment more efficient, multiple copies of each unique fragment were cloned into receiving vectors p601P or p601X, both derivatives of the plasmid containing the dodecameric 601–177 sequence in which only the plasmid backbone remains. Ligations were performed using 0.25–0.5 μg of fragment DNA and 0.1 μg of vector DNA creating a 30:1–60:1 molar excess of fragment to vector DNA. Reactions were in a volume of 20 μl, used 400 units of T4 DNA Ligase (New England Biolabs), and were incubated at 16 °C for 16–20 h. Clones with multiple fragment inserts were linearized by partial digestion with the appropriate enzyme, gel-purified, and ligated as above with additional like fragment creating the following four plasmids: p1X (p601X + 6 copies of fragment 1), p2P (p601P + 12 copies of fragment 2), p3X (p601X + 9 copies of fragment 3), and p4X (p601X + 9 copies of fragment 4). Fragment copy number was determined by digesting plasmids with HindIII and BamHI, which uniquely cut sites framing the set of inserted fragments, and analyzing the size of the products on a 1% agarose gel.
601-177-1 Nonpalindromic Fragment DNA Amplification, Isolation, and Purification
Plasmids p1X, p2P, p3X, and p4X were transformed into DH5α cells, amplified, and purified using the Qiagen Giga Prep plasmid purification system. Purified plasmid DNA was digested in a 1-ml final volume as follows; 1 mg each of p1X and p4X was digested with 140 units each of DraIII and BglI or 35 units each of BspMI and BstXI, respectively. Reactions were incubated at 37 °C for 12 h. 333 μg of p2P was digested with 120 units of PflMI for 12 h at 37 °C at which time an additional 120 units of PflMI was added, and incubation was continued for another 12 h. This was followed by the addition of 60 units of BglI and 12 more hours of incubation at 37 °C. 333 μg of p3X was digested with 12 units of BspMI for 12 h at 37 °C followed by two successive additions of 120 units of PflMI, each incubated 12 h at 37 °C. All enzymes were supplied by New England Biolabs. Digested fragments were purified as above. Purified fragments were tested for self- and ordered ligation using 0.5 μg of each DNA fragment and 400 units of New England Biolabs T4 DNA Ligase.
Histone and Octamer Assembly and Purification
Recombinant Xenopus laevis histones were expressed and purified according to standard methods. Histone octamers were assembled and quantified according to standard methods (30).
Mononucleosome Assembly
Mononucleosomes were prepared by rapid dilution using DNA template and purified histone octamers (31). Briefly, 10 μg of octamer and DNA template at a molar ratio of octamer to DNA ranging from 0.8 to 1.2 were mixed in 1 m NaCl, 10 mm Tris, pH 7.4, 0.1% (v/v) Nonidet P-40, and 25 μg/ml bovine serum albumin (BSA) to a final volume of 100 μl. The mixture was incubated for 30 min at 37 °C, then diluted to 100 mm NaCl in a final volume of 1 ml by 3 successive additions of dilution buffer (20 mm Tris, pH 7.4, 0.1% (v/v) Triton X-100, and 100 μg/ml BSA). Each dilution was followed by 20 min of incubation at room temperature. Assembled mononucleosomes were dialyzed at 4 °C against 2.5 mm NaCl, 10 mm Tris, pH 8.0, 0.25 mm EDTA with three buffer exchanges and concentrated. Saturation was determined by native gel analysis.
Mononucleosome Ligation and Tetranucleosomal Array Purification
Ligations were carried out in a 100-μl final volume, and included ∼2.5 μg of each mononucleosome, New England Biolabs ligation buffer (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 10 mm DTT, 1 mm ATP), and 4000 units of New England Biolabs T4 DNA ligase. Reactions were incubated at 16 °C for 16–20 h. Ligation efficiency was determined by native gel analysis. Under these conditions most nucleosome combinations ligated with 80–90% efficiency. For mononucleosome combinations that ligated with less than 80% efficiency, new ligations were performed using 4MB buffer (50 mm Tris-HCl, pH 7.5, 4 mm MgCl2, 10 mm DTT, 1 mm ATP) instead of New England Biolabs ligation buffer, keeping all other conditions the same, which improved efficiency levels to near 80%. Tetranucleosomal arrays were purified over a 5–25% linear sucrose gradient by centrifugation at 28 K for 23–24 h at 4 °C. Fractions were pooled and dialyzed 2 times against array buffer (2.5 mm NaCl, 10 mm Tris, pH 8.0, 0.25 mm EDTA) + 1 mm DTT followed by one time against array buffer plus 0.1 mm Tris(2-carboxyethyl)phosphine hydrochloride at 4 °C. Dialyzed samples were concentrated using Vivaspin6 30,000 molecular mass cutoff spin columns and quantitated by UV absorbance. Native gel analysis showed >95% of the purified product consisted of the desired tetranucleosomal array species.
Sedimentation Velocity
Sedimentation velocity experiments were performed at 20 °C using a Beckman XL-A ultracentrifuge. Mononucleosomes were sedimented at 35,000 rpm. Tetranucleosomal arrays in the absence of MgCl2 as well as those undergoing intramolecular array compaction were sedimented at 25,000 rpm. Sedimentation of self-associated tetranucleosomal arrays were performed at 3000 rpm. Data were analyzed using the van Holde-Weischet method and Ultrascan data analysis software (Dr. B. Demeler, University of Texas Health Science Center, San Antonio, TX) as previously described (32, 33). The van Holde-Weischet analysis technique is best suited for single species or multiple species with well resolved s values. Because our self-associated tetranucleosome distribution indicated continuous heterogeneity over a range of s values, we have reported the values as apparent s values. All experiments were repeated at least two times with averages, and S.D. were calculated.
Differential Centrifugation
Oligomerization of tetranucleosomal arrays was determined using the intermolecular association assay previously described (7). Briefly, tetranucleosomal arrays (20.0–27.0 ng/μl template DNA) were mixed with an equal volume of 2× MgCl2 in array buffer (2.5 mm NaCl, 10 mm Tris, pH 8.0, 0.25 mm EDTA) plus 0.1 mm Tris(2-carboxyethyl)phosphine hydrochloride to achieve the desired final MgCl2 concentrations. Triton X-100 was added to both sample and buffer to a final concentration of 0.1% before mixing where indicated. After incubation for 15 min at room temperature, samples were centrifuged at 14,000 × g for 10 min. The percentage of tetranucleosomal array remaining in the supernatant was determined by calculating the ratio of the A260 of each sample to the A260 of a sample with no MgCl2 added. All experiments were repeated at least two times. The Mg2+ concentration required for 50% sedimentation was determined for each experiment, and S.D. were calculated.
Nucleosomal Array Sedimentation Coefficient Predictions
Tetranucleosomal array sedimentation coefficients were predicted according to a formula that describes how the sedimentation coefficient of a linear system of multiple repeat units changes with the number of repeats (34): log(sn) = log(smono) +a·log(n). Here, smono is the sedimentation coefficient for a mononucleosome, n is the number of nucleosomes in the array, and sn is the sedimentation coefficient for the array. The equation requires a parameter, a, that characterizes the shape of the repeat unit. This parameter was determined from the measured sedimentation coefficients for the mononucleosome and 12-mer array systems under the relevant assay conditions and then utilized for calculating the predicted sedimentation coefficient for the tetranucleosomal array.
Nucleosome Stability Assays
Nucleosome arrays (18 ng/μl template DNA) were mixed with an equal volume of 2× MgCl2 in array buffer (2.5 mm NaCl, 10 mm Tris, pH 8.0, 0.25 mm EDTA + 0.1 mm Tris(2-carboxyethyl)phosphine hydrochloride) to achieve the desired final MgCl2 concentrations. Triton X-100 was added to both sample and buffer to a final concentration of 0.1%, before mixing where indicated. Reactions were split into two aliquots. Both were incubated for 15 min at room temperature. One aliquot was immediately run on a native 4% gel, whereas the other was incubated for an additional 22–24 h at 4 °C before gel analysis.
RESULTS
Design, Assembly, and Characterization of Tetranucleosomal Arrays
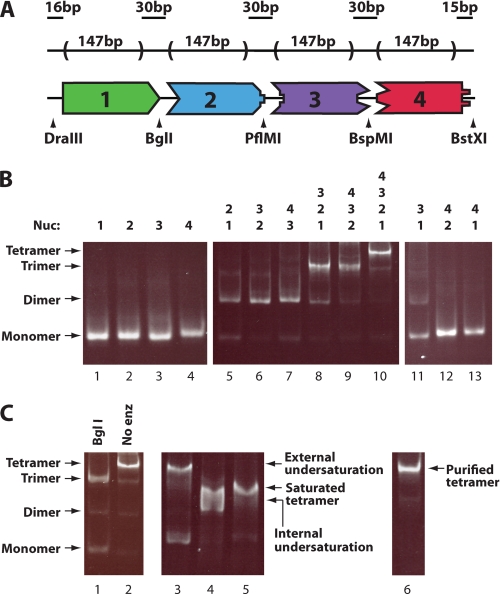
Our aim was to generate four mononucleosomes with different asymmetric sticky ends that can be ligated in only one way. These mononucleosomes would allow us to generate di-, tri-, and tetranucleosomal arrays with specific octamers at defined positions (Fig. 1A). Each of four distinct ∼177-bp fragments were created by PCR amplification of a template containing the 601 positioning sequence (29). This sequence was chosen for the core of each DNA template because its strong octamer positioning character has proven useful in studies from mononucleosomes to extended arrays (10, 35–39). A DNA template length of 177 bp per octamer was selected based on previous structural characterization of compaction properties of 12-mer arrays with this repeat length (10, 37) and because it was found to be the average repeat length between stably positioned nucleosomes in yeast (5).
FIGURE 1.
Generation of tetranucleosomal arrays containing specific histone octamers at defined positions. A, shown is the schematic of the tetranucleosomal array DNA template containing four 601-based nucleosome positioning sequences joined head-to-tail by unique nonpalindromic endonuclease cut sites. Arrowheads indicate the location of designated nonpalindromic cut sites. B, shown is characterization and ligation of the four unique mononucleosomes containing wild type octamer. Fragments utilized are shown above the gel. Unligated mononucleosomes (left panel) and mononucleosomes ligated in combinations expected to result in ligated product (center panel) or those that were not (right panel) are shown. Samples were run on a native 4% polyacrylamide gel and stained with ethidium bromide. C, determination of tetranucleosomal array ligation order, saturation, and purity by native gel analysis is shown. Ligated tetranucleosomal array digested with BglI shows expected monomer and trimer products (lane 1). Ligated tetranucleosomal arrays were made harboring an external position of undersaturation (ligation of mononucleosome one, two, and three, with DNA fragment four), an internal position of undersaturation (ligation of mononucleosomes one, two, and four, with DNA fragment three), or with all positions fully saturated (ligation of mononucleosomes 1–4) and are shown in lanes 3–5, respectively. A purified, fully saturated, tetranucleosomal array is shown in the right panel. Nucleosomal array markers shown on the left apply to all six gel lanes.
To generate large quantities of each fragment, each was cloned in multiple copies. Primer pairs for each fragment introduced unique 5′ and 3′ nonpalindromic restriction endonuclease sites flanked by palindromic sites allowing cloning of multiple like fragments into one vector. Fragments were released from plasmid DNA by cutting at nonpalindromic sequences. After gel purification, each fragment was tested for the ability to ligate to its designed partner fragment(s) and not to itself. Tetranucleosomal arrays were digested with BglI, PflMI, or BspMI. These ligations and digestions confirmed the specificity of ordered ligation (data not shown). Upon ligation of all four fragments, a 30-bp linker region separates each 147-bp 601-positioning sequence, resulting in regularly spaced nucleosomes with the same nucleosome-nucleosome distance present in 601-177-12 arrays.
Mononucleosomes were assembled by rapid dilution using octamer composed of full-length recombinant X. laevis histones. Native gel electrophoresis of these mononucleosomes showed that mononucleosomes with a single octamer position can be generated, making them well suited for ligation (Fig. 1B, lanes 1–4). Mononucleosomes were ligated in all di-, tri-, and tetranucleosomal array combinations and assessed for the specificity and efficiency of their ligation by native gel analysis. For sequential sets of nucleosome ligations, the reaction proceeded nearly to completion (Fig. 1B, lanes 5–10), indicating that each ligation is very efficient. Importantly, when nonsequential nucleosomes were ligated, ligation did not proceed to a significant extent (Fig. 1B, lanes 11–13), suggesting that ligation is occurring with the desired specificity. Further indication of the specificity of the ligation comes from digestion of the ligated array. Tetranucleosomal arrays digested with BglI, the only ligation site that showed a small amount of cross-reactivity with a nonsequential nucleosome (Fig. 1B, lane 11), predominantly showed the expected trinucleosome and mononucleosome products (Fig. 1C, lane 1). Ligated tetranucleosomal arrays were then purified on a sucrose gradient resulting in a uniform product (Fig. 1C, lane 6).
To assess the level of saturation of the tetranucleosomal arrays, subsaturated arrays were created by ligating three mononucleosomes with one DNA positioning fragment lacking histone octamer. Two positions of unsaturation were investigated; one with an internal position and one with an external position of unsaturation. Fully assembled tetranucleosomal arrays were compared with subsaturated arrays by native gel electrophoresis (Fig. 1C, lanes 3–5). On the gel, the migration of the tetranucleosomal array was fully resolved from the externally unsaturated array and partially resolved from the internally unsaturated array, which also exhibited a broadened band. The internally unsaturated array was further tested by sedimentation velocity analysis and gave a profile distinctly different from arrays created with four mononucleosomes (s50% of 14.1 S in low salt, s50% of 15.3 S in 1.75 mm MgCl2; see the next section for comparable data for the analogous saturated array).
Tetranucleosomal Arrays Undergo Compaction and Self-association
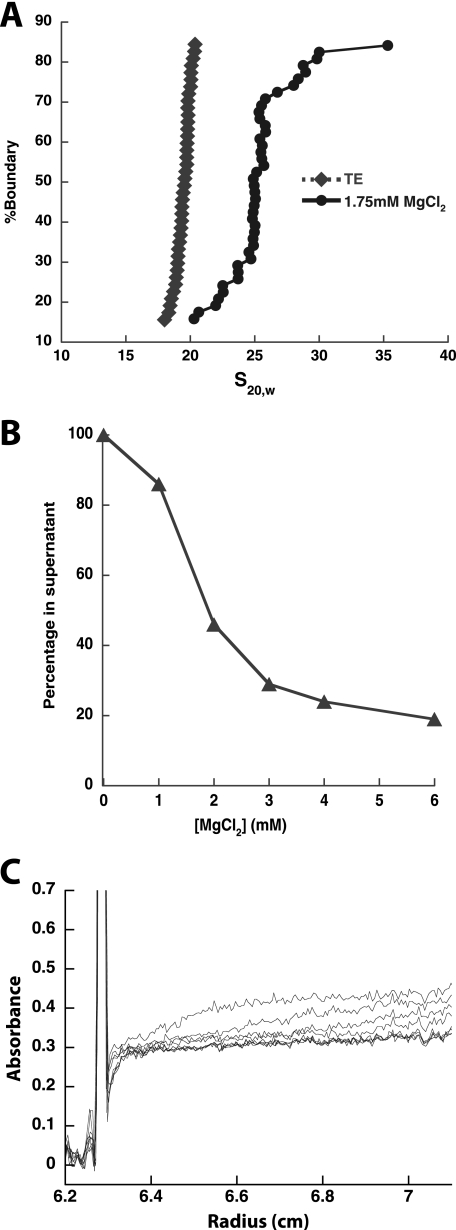
To characterize the structural states adopted by the tetranucleosomal array, sedimentation analysis was employed at varying concentrations of divalent magnesium cation. In the absence of cation, standard nucleosomal arrays (12 or more nucleosomes) adopt an extended conformation (40). To assess the structure of the tetranucleosomal array under these conditions, sedimentation velocity analysis was performed. As seen from the resulting integrated sedimentation coefficient distribution plot (Fig. 2A), the tetranucleosomal array exhibits a tight distribution centered about 19.7 ± 1.0 S. This relative homogeneity in the sedimentation coefficient suggests that the tetranucleosomal array is highly uniform in composition, in agreement with the gel analysis of the array (Fig. 1C). Additionally, the magnitude of the sedimentation coefficient suggests that the tetranucleosomal array adopts an extended conformation. Prior work with polynucleosomes has shown that the sedimentation coefficient of such species can be fit using the sedimentation coefficient of the mononucleosome, a shape factor, and the number of repeated monomer units (34). Using the sedimentation coefficients of a 601-177-1 mononucleosome, 11 S (data not shown), and a saturated 601-177-12 array in its extended form, 34 S (10), we are able to extract the shape factor, and then predict that the extended 601-177-4 tetranucleosomal array would have a sedimentation coefficient of 20.5 S, which is less than a S.D. greater than our measured s (+0.8 S.D.).
FIGURE 2.
Tetranucleosomal array containing wild type octamers show intra- and internucleosomal compaction. A, intra-array compaction of tetranucleosomal arrays is shown. A representative integrated sedimentation coefficient distribution plot of tetranucleosomal arrays with wild type octamers in the presence (1.75 mm MgCl2) and absence (TE) of divalent magnesium ion. s20,w is the sedimentation coefficient corrected for water at 20 °C. B, inter-array association of tetranucleosomal arrays is shown. A representative self-association plot of tetranucleosomal arrays with wild type octamers is shown. The fraction of tetranucleosomal array remaining in the supernatant is plotted as a function of MgCl2 concentration. C, sedimentation velocity analysis of self-associated tetranucleosomal array species generated at 1.75 mm MgCl2 is shown. Shown are 10 boundary plots generated at 3000 rpm from 0.8 to 12.5 min. Of the species that are not already sedimented by the first scan, those with large sedimentation coefficients are observed between 0.3 and 0.4 absorbance units, whereas those with relatively small sedimentation coefficients are observed below 0.3 absorbance units.
To determine whether tetranucleosomal arrays can undergo intra-array compaction, the ability of divalent magnesium to increase the sedimentation coefficient of the array was assessed. For 12-mer arrays, modest concentrations of magnesium ion, 1.0–1.75 mm MgCl2, have been shown to generate intra-array compacted structures similar to the 30 nm fiber (7, 10). Additionally, under these conditions, relatively little inter-array association is observed. To determine magnesium ion conditions where the tetranucleosomal array was not predominantly self-associated, we performed a differential centrifugation experiment (Fig. 2B), where arrays are centrifuged with increasing concentrations of magnesium ion, and the amount of non-sedimented array is assessed (8). We observed that below 1.8 ± 0.1 mm MgCl2, its Mg50%, a majority of the tetranucleosomal array is not sedimented, and sedimentation velocity analysis of the non-differentially sedimented species was performed below this concentration. Initial studies at 1.25 mm MgCl2 suggested that the arrays did not undergo significant intra-array compaction under these conditions (data not shown). To determine whether higher concentrations of MgCl2 were able to induce intra-array compaction, we performed sedimentation velocity analysis of the non-differentially sedimented species at 1.75 mm MgCl2. Under these conditions, for species that were not self-associated, a relatively uniform shift in the sedimentation coefficient distribution was observed, with a median s value of 23.2 ± 1.5 S, indicating some degree of intra-array compaction. To determine how this shift relates to the shift expected for a fully compacted array, we estimated the expected sedimentation constant if array compaction occurs continuously from mononucleosomes to 12-mer arrays, as appears is the case in the absence of divalent magnesium ion. Using the 601-177-1 mononucleosome sedimentation coefficient, 12 S (data not shown), and the sedimentation coefficient of the fully compacted 601-177-12 array, 53 S (10), we calculate that a fully compacted tetranucleosomal array would have a sedimentation coefficient of 27.5 S. This value is almost three S.D. above our measured tetranucleosomal array s (+2.87 S.D.), suggesting that under these divalent magnesium conditions, although arrays do undergo some compaction, they do not appear to exhibit the same extent of compaction as 12-mer arrays.
In addition to this intra-array compaction, our tetranucleosomal arrays also appear to be able to undergo inter-array self-association. Our differential centrifugation magnesium titration results indicated that with increasing MgCl2, part of the array population forms a species with a large sedimentation coefficient (Fig. 2B). To confirm that this species is due to inter-array self-association, similar to longer nucleosomal array systems, we analyzed the differentially sedimented species by sedimentation velocity at a substantially reduced radial acceleration. For a MgCl2 concentration of 1.75 mm we found that the tetranucleosomal arrays sedimented in three classes reflecting the formation of an intra-array compacted species and two types of inter-array conglomerations (Fig. 2C). At these reduced rotational speeds, the majority of the arrays, roughly 63%, did not sediment significantly, consistent with the relatively small sedimentation coefficient of 23 S for the partially intra-array compacted species (Fig. 2A). In contrast, roughly 31% of the arrays exhibited apparent sedimentation coefficients between 20,000 and 160,000 S, which is likely due to inter-array associations (see below). Finally, a small fraction of the arrays (6%) could not be accounted for in the cell and presumably sedimented with an apparent s greater than 160,000 S (together these two groups with large s values constitute the species that were not resolved at higher centrifugation speeds).
The species formed with a sedimentation coefficient of 20,000–160,000 S is consistent with self-association between multiple copies of the tetranucleosomal arrays. Formally, the sedimentation coefficient is the ratio of the buoyant mass divided by the friction coefficient, so an increase in s from the extended, 20 S species could formally occur by either an increase in mass or a decrease in friction coefficient. However, even if the friction coefficient of the array is minimized by assuming it becomes fully spherical, the –fold reduction in the friction coefficient would only be 2.26, suggesting that a significant increase in mass due to inter-array association occurs. Additionally, the sharp demarcation between the unsedimented 23 S species and the 20,000–160,000 apparent s species suggests that like longer nucleosomal arrays (8), the formation of inter-array-associated species is highly cooperative.
Histone H4 Tails Progressively Affect Self-association of Tetranucleosomal Arrays
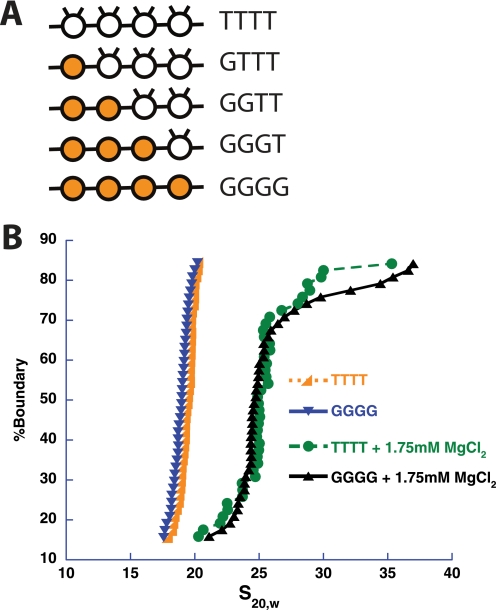
To better characterize the higher-order structures exhibited by our tetranucleosomal array system, we sought to determine how partial and complete loss of the histone H4 tail affected intra-array compaction and inter-array self-association, as this loss negatively impacts both structural transitions in 12-mer arrays (10, 11). Mononucleosomes were assembled, as described above, with wild type octamer (T) or octamer containing globular histone H4 that is missing the first 19 N-terminal amino acids (G). These mononucleosomes were then used to assemble tetranucleosomal arrays containing various degrees of H4 tail loss at specific nucleosomal positions (Fig. 3A).
FIGURE 3.
The histone H4 tails are not essential for intra-array compaction of tetranucleosomal arrays. A, shown is a schematic of tetranucleosomal arrays used in this study with octamers possessing or missing histone H4 tails. The presence of the H4 tails in an octamer (T) is represented in schematic form by a clear circle with two tails. The absence of H4 tails (G) is represented by a shaded circle in the schematic. The octamer letter representations are in ascending order with respect to DNA template fragment number, with the exception of the GTTT array, which is in reverse order, i.e. the tailless nucleosome, G, occurs on DNA template fragment four. B, shown is a representative integrated sedimentation coefficient distribution plot of the tetranucleosomal arrays containing octamers with (TTTT) or without (GGGG) H4 tails in the presence and absence of divalent magnesium ion. s20,w is the sedimentation coefficient corrected for water at 20 °C.
To determine to what extent the H4 tail affects intramolecular tetranucleosomal array compaction, arrays containing four octamers with full-length histones (TTTT) were compared with those with all octamers lacking H4 tails (GGGG) (Fig. 3A). Sedimentation velocity experiments showed no significant difference in compaction between the two array types in the absence or presence of magnesium ion (Fig. 3B). Similarly, several tetranucleosomal arrays lacking fewer H4 tails were tested and showed no difference in sedimentation in the presence of magnesium ion (data not shown). Thus, for the partially compacted state, the H4-tails do not appear to play a significant role in this transition. As the tetranucleosomal arrays did not adopt the full, H4 tail-dependent compaction of longer arrays, intra-array compaction studies with additional tetranucleosomal and shorter arrays were not performed.
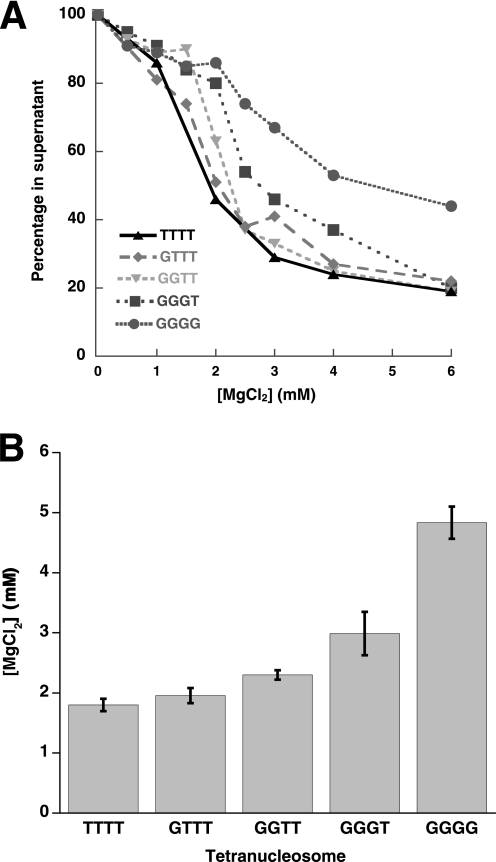
In contrast to intra-array compaction, intermolecular compaction was substantially affected by loss of the H4 tails (Fig. 4A). Compared with a Mg50% of 1.8 ± 0.1 mm seen for the wild type array, the H4 tailless, GGGG, array required 4.8 ± 0.3 mm Mg2+ for the same effect. Additionally, the cooperativity of the self-association curve decreases significantly for the GGGG array.
FIGURE 4.
Inter-array self-association is disrupted by the loss of increasing number of histone H4 tails and is affected by their position within an array. A, the effect of number of octamers without H4 tails in a tetranucleosomal array on inter-array self-association is shown. A representative differential centrifugation plot of tetranucleosomal arrays containing zero, one, two, three, or four tailless octamers is shown. The fraction of tetranucleosomal array remaining in the supernatant is plotted as a function of MgCl2 concentration. B, shown is a histogram representation of the concentration of Mg2+ required for 50% sedimentation for tetranucleosomal arrays depicted in A.
To dissect how the absence of the H4 tail disrupts array self-association, we next examined tetranucleosomal array oligomerization with varying numbers of tails present. Mononucleosomes were ligated to form tetranucleosomal arrays containing one tailless and three wild type (GTTT), two tailless and two wild type (GGTT), or three tailless and one wild type (GGGT) octamer set (Fig. 3A). Differential centrifugation analysis showed an ordered increase in Mg50% as the number of octamers without H4 tails increased (Fig. 4, A and B). This affect was subtle when only one or two octamers present did not have H4 tails (TTTT, Mg50% of 1.8 ± 0.1 mm; GTTT, Mg50% of 2.0 ± 0.1 mm; GGTT, Mg50% of 2.3 ± 0.1 mm) but became more pronounced when 3 and 4 octamers were H4 tailless (GGGT, Mg50% of 3.0 ± 0.4 mm; GGGG, Mg50% of 4.8 ± 0.3 mm).
Nucleosomal Array Length and Saturation Affect Subsequent Octamer Loss
The ability of tetranucleosomal arrays to undergo significant self-association when containing as few as two nucleosomes with H4 tails (Fig. 4, A and B) suggests that this inter-array interaction could require even fewer than four nucleosomes. To explicitly test this idea, we attempted to compare the ability of mono-, di-, tri-, and tetranucleosomal arrays to undergo divalent magnesium ion-induced self-association. Homogeneous nucleosomal systems containing one, two, three, or four nucleosomes were generated via ligation and purification (Fig. 5A, first lane of each panel), and these arrays were subjected to differential centrifugation MgCl2 titration experiments. However, the results of these experiments were initially highly variable, especially for the shorter nucleosomal systems, and therefore inconclusive (data not shown).
FIGURE 5.
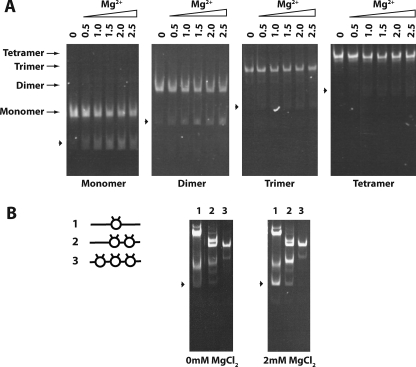
Neighboring nucleosomes enhance nucleosome stability. A, shown is the effect of increasing magnesium concentration on nucleosome stability. Mono-, di-, tri-, and tetranucleosomal arrays were incubated in the presence of increasing concentrations of MgCl2 then run on a native 4% polyacrylamide gel and stained with ethidium bromide. Lanes 0, 0.5, 1.0, 1.5, 2.0,, and 2.5 were incubated in the corresponding mm concentrations of MgCl2. Arrowheads indicate the position of free DNA for each nucleosomal length tested. B, shown is the effect of nucleosome density on nucleosome stability. Trinucleosomal arrays, varying in degree of saturation, were created by ligating combinations of mononucleosomes and free DNA to create trinucleosomal DNA templates with one, two, or three octamers assembled as shown in the schematic, left. Lanes 1–3 show arrays with trinucleosomal DNA template and one, two, or three assembled octamers, respectively. Each array was incubated in the presence or absence of 2.0 mm MgCl2 then run on a native 4% polyacrylamide gel and stained with ethidium bromide. Arrowheads indicate the position of free trinucleosomal DNA.
To better understand the source of this variability, we characterized the integrity of our nucleosomal arrays when treated with divalent magnesium ion. As visualized by native gel-electrophoresis analysis (Fig. 5A), the nucleosomal systems demonstrate increasing stability with increasing length, with the mononucleosomes exhibiting the greatest degree of free DNA in the presence of MgCl2 and the tetranucleosomal arrays showing no significant free DNA. This disassembly of nucleosomes is MgCl2-dependent, as the absence of MgCl2 results in no free DNA for each nucleosomal system, whereas increasing amounts of MgCl2 can generate increasing amounts of free DNA. Additionally, no significant partially saturated array intermediates are generated in producing the fully free DNA state. These trends are observed over multiple trials, although the extent of octamer loss varies slightly. These results suggest that relatively modest concentrations of MgCl2 can induce array disassembly and that disassembly is facilitated by shorter array length or a decrease in array saturation.
To explicitly test the idea that loss of array saturation itself promotes subsequent nucleosome disassembly, we ligated trinucleosomal arrays, where both, one, or neither of the sites flanking the central nucleosome was occupied by octamer (Fig. 5B, left). These three arrays were then purified and analyzed by native gel electrophoresis before addition of MgCl2 (Fig. 5B, center). Despite the fact that during the purification, the desired fractions largely contained one species (data not shown), small amounts of additional species became present when these pooled fractions were analyzed. Nonetheless, it appears that these species are predominantly the parent array in some alternative conformation, as digestion of the arrays at a single site generates only subspecies consistent with digestion of the parent array (supplemental Fig. S1). When equal amounts of these arrays were subjected to MgCl2, like the studies with the arrays of different lengths, disassembly of the arrays to free DNA was observed. Moreover, this disassembly was correlated to the saturation of the arrays, being quite prominent in the array with no octamers flanking the central nucleosome, more moderate with one flanking octamer, and absent with two flanking octamers, reflecting a facilitation of array disassembly by decreasing array saturation.
Dinucleosomes Exhibit Inter-array Self-association
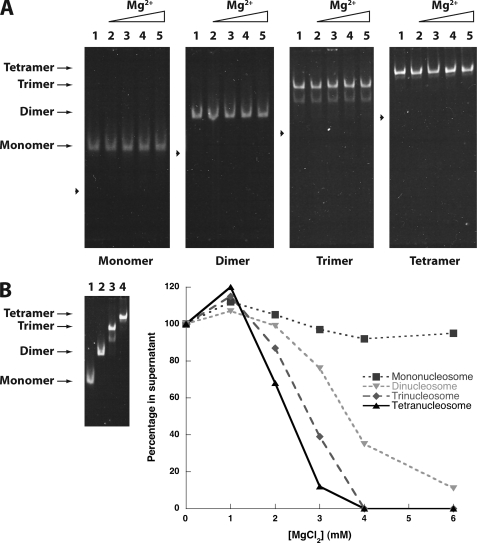
Ion-mediated disassembly of mononucleosomes has been previously observed and can be prevented with the addition of low concentrations of non-ionic detergents (41). Indeed, in the presence of 0.1% Triton X-100 detergent, the mono- di, tri-, and tetranucleosomal systems showed no MgCl2-dependent disassembly (Fig. 6A as compared with Fig. 5A), even at 6 mm MgCl2 (Fig. 6B, left). Thus, these conditions are amenable to determining the minimal nucleosomal array length required to observe array self-association. Performing the differential centrifugation MgCl2 titration experiments, we observed that decreasing nucleosomal array length decreased the propensity of the arrays to generate self-associated species that sediment. Nonetheless, even the dinucleosomal array system showed appreciable differential self-association at modest MgCl2 concentrations, suggesting that chromosomal stretches of nucleosomes as short as two might be sufficient to generate cross-strand interactions.
FIGURE 6.
Neighboring nucleosomes enhance intermolecular compaction. A, shown is the presence of detergent enhances nucleosome stability. Mono-, di-, tri-, and tetranucleosomal arrays were incubated in the presence of 0.1% Triton X-100 and increasing concentrations of MgCl2, then run on a native 4% polyacrylamide gel and stained with ethidium bromide. Lanes 0, 0.5, 1.0, 1.5, and 2.0 were incubated in the corresponding mm concentrations of MgCl2. Arrowheads indicate the position of free DNA for each nucleosomal length tested. B, increasing nucleosomal array length enhances intermolecular compaction. Left, mono-, di-, tri-, and tetranucleosomal arrays were incubated in the presence of 0.1% Triton X-100 and 6.0 mm MgCl2, then run on a native 4% polyacrylamide gel and stained with ethidium bromide. Lanes 1–4 show mono-, di-, tri-, and tetranucleosomal arrays, respectively. Right, a representative self-association plot of mono, di-, tri-, and tetranucleosomal arrays is shown. Nucleosomes were incubated with 0.1% Triton X-100 and increasing concentrations of MgCl2. The fraction of nucleosome remaining in the supernatant is plotted as a function of the array concentration.
DISCUSSION
Intra-array Compaction
Although it was expected that the stability of an intra-array compacted tetranucleosomal array would be less than that for longer nucleosomal arrays, the strong propensity of 601 sequences for compaction as well as the precedent of a fully compacted 601 tetranucleosome in the solid phase (42) indicated that a tetranucleosome might still become predominantly compacted in solution. We found that ligated tetranucleosomal arrays do demonstrate intra-array compaction at 1.75 mm MgCl2 but exhibit less complete compaction than can be achieved for longer nucleosomal arrays and do so in a manner seemingly independent of the histone H4 tail.
A possible interpretation is that chromatin compaction is actually H4 tail-independent, requiring a different model for 30-nm fiber formation. However, in light of the partial compaction of the tetranucleosomal array and the well precedented role of the H4 tail in longer arrays (9, 10, 13, 37), it seems more likely that the observed behavior reflects an inability of the tetranucleosome to readily adopt the 30-nm structure of longer arrays. At least two structural models are consistent with this idea. On one hand, it may be that tetranucleosomal arrays cannot compact to the same degree as longer arrays because they cannot adopt the necessary spatial arrangement of nucleosomes. For example, in a recently proposed version of the solenoid model, these shorter arrays may not be long enough to form the helical repeat unit composed of roughly six nucleosomes (38). In such a case, the compaction observed may simply be due to neutralization of the DNA phosphate backbone, allowing closer approach of the nucleosomes on average without forming the stable, H4 tail-dependent, structure. On the other hand, for models in which fewer than four nucleosomes constitute a repeat unit, such as the two-start helical model (44), it might be that the desired nucleosome arrangement and interactions are present but cannot be sufficiently stabilized unless more of these interactions are summed as is possible with longer arrays. In this case, where the compacted state lies intermediate between the fully extended and compacted state, the nucleosomes may not be in close enough proximity for the H4 tail to exert much of an effect. Both models are possible, and in fact recent structural studies suggest that both states can be adopted (44).
Regardless of which structural intermediates are involved in compaction, this lack of full array compaction even at a MgCl2 concentration significantly above those required for the longer 12-mer arrays (incomplete compaction at 1.75 mm MgCl2 versus complete compaction at 1.0 mm MgCl2) indicates that shorter stretches of nucleosomes have a significantly reduced propensity to undergo intra-array compaction in vitro and potentially reflects a requirement of long stretches of organized nucleosomes to significantly compact in vivo.
Inter-array Self-association
The tetranucleosomal system containing fully intact histones shows self-association properties similar to longer array systems. Like 12-mer arrays, the tetranucleosomal array exhibits magnesium-dependent differential centrifugation, shows decreased self-association with histone H4 tail loss, and generates self-associated species of large size (8, 11, 44). However, this self-associated species displays differences from those of longer arrays. The apparent s values for the self-associated tetranucleosomal arrays are significantly larger than those observed for 12-mer arrays assembled on the 5 S rDNA positioning sequence (8). This is presumably due to augmented inter-array contacts that could arise from a combination of factors, including differences in array features (the tetranucleosomal array contains a stronger positioning sequence, shorter linker DNA lengths, and fewer repeat units with respect to the 5 S rDNA 12-mer array), differences in the level of intra-array compaction attainable before self-association (see below), and differences in assay conditions. Studies on the 12-mer arrays were performed at a divalent magnesium concentration where a lower percentage of arrays was involved in inter-array interactions (∼25% as opposed to ∼50% for the tetranucleosomal array).
The self-association of tetranucleosomal arrays is highly cooperative, with species predominantly existing in either an extensively self-associated state or as individual arrays. This behavior could arise from a number of different sources. Heterogeneity in the array population might cause this behavior, although our gel and centrifugation analyses suggest that the arrays are quite uniform in their structure. Alternatively, formation of the associated arrays may be under kinetic, not thermodynamic, control. Finally, array concentration may play an important role, such that when a sufficient proportion of arrays become self-associated, the concentration of non-associated arrays may drop below a critical concentration required for cooperative association, leaving the remaining population as individual arrays.
From our characterization of tetranucleosomal array self-association, as assessed by differential centrifugation, it appears that relatively few nucleosomes are required for robust self-association. The ligated 601-177-4 array with full-length histones has an Mg50% of 1.8 mm MgCl2 (Fig. 2B). This concentration is only slightly more than that observed for the comparable 601–177 12-mer array (Mg50% of 1.5 mm) (10) and suggests that relatively short stretches of nucleosomes support robust array self-association. Interestingly, this behavior stands in contrast to intra-array compaction, where even larger increases in MgCl2 concentration are not able to recapitulate the behavior of the longer array systems. In fact, this decrease in propensity for tetranucleosomal intra-array compaction may be a contributing cause for the facility with which these shorter arrays self-associate. Our previous work with cross-linked 601-177-12 arrays indicates that intra-array compaction antagonizes inter-array self-association (45), suggesting that the reduced efficacy of tetranucleosomal intra-array compaction may in turn allow inter-array self-association to occur more readily and extensively.
By extending our self-association studies to arrays with varying numbers of H4 tails and varying numbers of nucleosomes, we find that even fewer than four nucleosomes provide significant inter-array associations (Fig. 6B). Although decreasing the number of H4 tails does progressively reduce tetranucleosomal array self-association, tetranucleosomal arrays with as few as two nucleosomes containing intact H4 tails show similar self-association properties to those arrays with all four nucleosomes containing intact H4 tails (Fig. 4). This suggests that only a fraction of the H4 tails might be involved in self-association, in agreement with studies that show that inter-array cross-linking of H4 tails to nucleosomes occurs for only a fraction of the H4 tails (12, 45, 46). Because the H4 tail is not the only factor involved in array self-association, as the tetranucleosomal arrays still exhibit some self-association with the loss of all H4 tails, tailless nucleosomes likely still contribute in some fashion to array self-association. Indeed, the dinucleosome, which contains two-tailed nucleosomes but lacks the contributions of additional H4 tailless nucleosomes, exhibits a decreased propensity for self-association (Figs. 4A and 6B). However, the dinucleosome alone still exhibits a significant degree of self-association.
To what extent this observation extends directly to chromatin in vivo is not currently clear, as the uniformity, composition, and solution context of nuclear chromatin is more complex. Still, it is suggestive that multiple instances of interactions between relatively short stretches of nucleosomes might suffice to generate complicated self-associated structures present in both interphase (2–4 mm MgCl2) and mitotic nuclei (5–17 mm MgCl2) (47). Moreover, this behavior might explain a discrepancy between chromatin structure in vitro and in vivo. In vitro, stretches of nucleosomes have been shown to adopt intra-array compaction to generate 30-nm fibers at moderate ionic strengths and low nucleosome concentrations. Still, the existence of such fibers in vivo has been controversial, with a number of recent in vivo imaging experiments asserting that no such structures exist (48). Because we see that relatively short stretches of nucleosomes can undergo self-association in vitro, inter-array contacts that could form in trans between chromosomes or in cis between distant nucleosomes in the same chromosome may be prevalent under the ionic conditions and high nucleosome concentrations present in the nucleus. Such interactions could either interrupt or mask intra-array compaction, making formation of 30-nm fibers difficult to observe in vivo.
Nucleosome Stability and Disassembly
Changing chromatin structure through nucleosome disassembly constitutes an important mechanism for regulating DNA-dependent processes, such as transcription. In vitro studies of this process have tended to focus on enzyme-mediated disassembly in the context of individual nucleosomes, whereas our studies reveal aspects of the intrinsic stability of nucleosomes within nucleosomal arrays.
Our experiments utilizing arrays that differ in length and initial saturation show that nucleosome disassembly can be facilitated by the addition of MgCl2. It has long been known that the extent of association between histone octamer components and nucleosomal DNA varies with monovalent cation concentrations, with high concentrations of ions being sufficient to completely dissociate histones from DNA (33). The phenomenon is likely due to shielding of the DNA-histone charge interactions that constitute a large component of the free energy of association. Similarly, we believe that the ionic nature of the magnesium divalent cation and monovalent chloride anion can also stabilize the dissociation of the charged nucleosome components. However, we suspect that the divalent magnesium ion interacts with the phosphate backbone of the nucleosomal DNA beyond simple charge shielding in driving nucleosome disassembly, similar to the fact that interactions beyond charge shielding are involved for the increased divalent magnesium cation-driven intra-array compaction and inter-array association over monovalent cations at comparable ionic strength (7, 8).
Our disassembly data also shows that with decreasing array length or saturation, octamer dissociation occurs more readily. This process of octamer loss does not appear to simply be stochastic in nature, as intermediates predicted in such a model are not observed (see supplemental Figs. S2 and S3). A seemingly more appropriate model of octamer loss appears to be one in which the frequency of octamer loss decreases with increasing array length or saturation and where octamer loss is cooperative, meaning that loss of one octamer facilitates subsequent octamer loss. Mechanistically, such behavior might occur by incorporating higher-order chromatin structure into the model. We note that in addition to driving nucleosome dissociation, our other experiments show that MgCl2 can promote intra-array compaction and inter-array associations. Thus, the nucleosome-nucleosome interactions that occur during Mg2+-induced intra-array compaction and inter-array association may increase the free energy required to remove octamers from their nucleosomal DNA. This idea provides an appealing and novel biological role for higher-order chromatin structure. It is known that some remodeling complexes that create a state repressive to transcription act by uniformly spacing nucleosomes (35). Such uniformly spaced nucleosomes presumably have an increased propensity for high-order chromatin structure, which in addition to limiting access may increase the energetic cost of disassembling the nucleosomes and limit their aberrant disassembly. Additionally, during transcription it is known that promoters can experience nucleosomal disassembly (24, 25). Because loss of nucleosome saturation appears to facilitate subsequent nucleosome disassembly, commitment to initial nucleosomal disassembly might cooperatively promote clearance of the neighboring nucleosomes at promoters.
Although MgCl2 can drive nucleosome dissociation, it is important to note that its presence is not always sufficient, as low concentrations of detergent prevent the detection of this disassembly. Nonetheless, we suspect the trends observed in the absence of detergent reflect the relative nucleosome stabilities for the different array species. These trends are likely to become manifest under physiological conditions where molecules such as histone chaperones can lower the energetic cost of histone dissociation from nucleosomal DNA, as chaperones play an essential biological role in disassembly of nucleosomes from transcribed promoters (43).
Overall, the ligatable nucleosomal system developed in this study has allowed us to generate well defined arrays that can be varied with respect to nucleosomal length, composition, and saturation. This has allowed us to probe aspects of higher-order chromatin structure and nucleosome stability that are influenced by the nucleosomes themselves. As the formation of higher-order chromatin structures also often involves nucleosome-interacting proteins, our system should also prove useful for controlling and probing the role of such interactions. Beyond studying higher-order chromatin structure, we expect that this system can also help to clarify mechanistic issues involving proteins and protein complexes that employ chromatin as an enzymatic substrate. Altogether the ligatable nucleosome system should provide a powerful tool to deepening our understanding of a wide range of biological processes involving chromatin.
Supplementary Material
Acknowledgment
We thank Divya Sinha for helpful discussion.
Footnotes
This work was supported, in whole or in part, by National Institutes of Health Grant GM79663 (to M. A. S.-K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
REFERENCES
- 1.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2.Richmond T. J., Davey C. A. (2003) Nature 423, 145–150 [DOI] [PubMed] [Google Scholar]
- 3.van Holde K. E. (1989) in Springer Series in Molecular Biology (Rich A. ed.) pp. 289–354, Springer-Verlag, New York [Google Scholar]
- 4.Yuan G. C., Liu Y. J., Dion M. F., Slack M. D., Wu L. F., Altschuler S. J., Rando O. J. (2005) Science 309, 626–630 [DOI] [PubMed] [Google Scholar]
- 5.Segal E., Fondufe-Mittendorf Y., Chen L., Thåström A., Field Y., Moore I. K., Wang J. P., Widom J. (2006) Nature 442, 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Holde K., Zlatanova J. (2007) Semin. Cell Dev. Biol. 18, 651–658 [DOI] [PubMed] [Google Scholar]
- 7.Schwarz P. M., Hansen J. C. (1994) J. Biol. Chem. 269, 16284–16289 [PubMed] [Google Scholar]
- 8.Schwarz P. M., Felthauser A., Fletcher T. M., Hansen J. C. (1996) Biochemistry 35, 4009–4015 [DOI] [PubMed] [Google Scholar]
- 9.Tse C., Hansen J. C. (1997) Biochemistry 36, 11381–11388 [DOI] [PubMed] [Google Scholar]
- 10.Dorigo B., Schalch T., Bystricky K., Richmond T. J. (2003) J. Mol. Biol. 327, 85–96 [DOI] [PubMed] [Google Scholar]
- 11.Gordon F., Luger K., Hansen J. C. (2005) J. Biol. Chem. 280, 33701–33706 [DOI] [PubMed] [Google Scholar]
- 12.Kan P. Y., Caterino T. L., Hayes J. J. (2009) Mol. Cell. Biol. 29, 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan J. Y., Rangasamy D., Luger K., Tremethick D. J. (2004) Mol. Cell 16, 655–661 [DOI] [PubMed] [Google Scholar]
- 14.Francis N. J., Kingston R. E., Woodcock C. L. (2004) Science 306, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 15.Köhler C., Villar C. B. (2008) Trends Cell Biol. 18, 236–243 [DOI] [PubMed] [Google Scholar]
- 16.Grigoryev S. A., Bednar J., Woodcock C. L. (1999) J. Biol. Chem. 274, 5626–5636 [DOI] [PubMed] [Google Scholar]
- 17.Georgel P. T., Horowitz-Scherer R. A., Adkins N., Woodcock C. L., Wade P. A., Hansen J. C. (2003) J. Biol. Chem. 278, 32181–32188 [DOI] [PubMed] [Google Scholar]
- 18.Nikitina T., Ghosh R. P., Horowitz-Scherer R. A., Hansen J. C., Grigoryev S. A., Woodcock C. L. (2007) J. Biol. Chem. 282, 28237–28245 [DOI] [PubMed] [Google Scholar]
- 19.Hediger F., Gasser S. M. (2006) Curr. Opin. Genet. Dev. 16, 143–150 [DOI] [PubMed] [Google Scholar]
- 20.Kwon S. H., Workman J. L. (2008) Mol. Cells 26, 217–227 [PubMed] [Google Scholar]
- 21.Springhetti E. M., Istomina N. E., Whisstock J. C., Nikitina T., Woodcock C. L., Grigoryev S. A. (2003) J. Biol. Chem. 278, 43384–43393 [DOI] [PubMed] [Google Scholar]
- 22.Dialynas G. K., Vitalini M. W., Wallrath L. L. (2008) Mutat. Res. 647, 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S., Shogren-Knaak M. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18243–18248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinke H., Hörz W. (2003) Mol. Cell 11, 1599–1607 [DOI] [PubMed] [Google Scholar]
- 25.Boeger H., Griesenbeck J., Strattan J. S., Kornberg R. D. (2003) Mol. Cell 11, 1587–1598 [DOI] [PubMed] [Google Scholar]
- 26.Jessen W. J., Hoose S. A., Kilgore J. A., Kladde M. P. (2006) Nat. Struct. Mol. Biol. 13, 256–263 [DOI] [PubMed] [Google Scholar]
- 27.Workman J. L. (2006) Genes Dev. 20, 2009–2017 [DOI] [PubMed] [Google Scholar]
- 28.Lam F. H., Steger D. J., O'Shea E. K. (2008) Nature 453, 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowary P. T., Widom J. (1998) J. Mol. Biol. 276, 19–42 [DOI] [PubMed] [Google Scholar]
- 30.Luger K., Rechsteiner T. J., Richmond T. J. (1999) Methods Enzymol. 304, 3–19 [DOI] [PubMed] [Google Scholar]
- 31.Widlund H. R., Cao H., Simonsson S., Magnusson E., Simonsson T., Nielsen P. E., Kahn J. D., Crothers D. M., Kubista M. (1997) J. Mol. Biol. 267, 807–817 [DOI] [PubMed] [Google Scholar]
- 32.van Holde K. E., Weischet W. O. (1978) Biopolymers 17, 1387–1403 [Google Scholar]
- 33.Carruthers L. M., Tse C., Walker K. P., 3rd, Hansen J. C. (1999) Methods Enzymol. 304, 19–35 [DOI] [PubMed] [Google Scholar]
- 34.Noll H., Noll M. (1989) Methods Enzymol. 170, 55–116 [DOI] [PubMed] [Google Scholar]
- 35.Yang J. G., Madrid T. S., Sevastopoulos E., Narlikar G. J. (2006) Nat. Struct. Mol. Biol. 13, 1078–1083 [DOI] [PubMed] [Google Scholar]
- 36.Zofall M., Persinger J., Kassabov S. R., Bartholomew B. (2006) Nat. Struct. Mol. Biol. 13, 339–346 [DOI] [PubMed] [Google Scholar]
- 37.Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., Peterson C. L. (2006) Science 311, 844–847 [DOI] [PubMed] [Google Scholar]
- 38.Robinson P. J., Fairall L., Huynh V. A., Rhodes D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6506–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Routh A., Sandin S., Rhodes D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8872–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thoma F., Koller T., Klug A. (1979) J. Cell Biol. 83, 403–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thåström A., Gottesfeld J. M., Luger K., Widom J. (2004) Biochemistry 43, 736–741 [DOI] [PubMed] [Google Scholar]
- 42.Schalch T., Duda S., Sargent D. F., Richmond T. J. (2005) Nature 436, 138–141 [DOI] [PubMed] [Google Scholar]
- 43.Adkins M. W., Williams S. K., Linger J., Tyler J. K. (2007) Mol. Cell. Biol. 27, 6372–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorigo B., Schalch T., Kulangara A., Duda S., Schroeder R. R., Richmond T. J. (2004) Science 306, 1571–1573 [DOI] [PubMed] [Google Scholar]
- 45.Sinha D., Shogren-Knaak M. A. (2010) J. Biol. Chem. 285, 16572–16581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng C., Lu X., Hansen J. C., Hayes J. J. (2005) J. Biol. Chem. 280, 33552–33557 [DOI] [PubMed] [Google Scholar]
- 47.Strick R., Strissel P. L., Gavrilov K., Levi-Setti R. (2001) J. Cell Biol. 155, 899–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eltsov M., Maclellan K. M., Maeshima K., Frangakis A. S., Dubochet J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19732–19737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.