Abstract
Chemokine receptors are members of the G protein-coupled receptor (GPCR) family. CCR5 is also the principal co-receptor for macrophage-tropic strains of human immunodeficiency virus, type 1 (HIV-1), and efforts have been made to develop ligands to inhibit HIV-1 infection by promoting CCR5 receptor endocytosis. Given the nature of GPCRs and their propensity to form oligomers, one can consider ligand-based therapies as unselective in terms of the oligomeric composition of complexes. For example, a ligand targeting a CCR5 homomer could likely induce signal transduction on a heteromeric CCR5-CXCR4. Other avenues could therefore be explored. We identified a receptor adaptor interacting specifically with one receptor complex but not others. NHERF1, an adaptor known for its role in desensitization, internalization, and regulation of the ERK signaling cascade for several GPCRs, interacts via its PDZ2 domain with the CCR5 homodimer but not with the CXCR4-CCR5 heterodimer or CXCR4 homodimer. To further characterize this interaction, we also show that NHERF1 increases the CCR5 recruitment of arrestin2 following stimulation. NHERF1 is also involved in CCR5 internalization, as we demonstrate that co-expression of constructs bearing the PDZ2 domain can block CCR5 internalization. We also show that NHERF1 potentiates RANTES (regulated on activation normal T cell expressed and secreted)-induced ERK1/2 phosphorylation via CCR5 activation and that this activation requires NHERF1 but not arrestin2. Taken together, our results suggest that oligomeric receptor complexes can associate specifically with partners and that in this case NHERF1 could represent an interesting new target for the regulation of CCR5 internalization and potentially HIV infection.
Keywords: Adaptor Proteins, Chemokines, ERK, G Protein-coupled Receptors (GPCR), HIV, Receptor Endocytosis, BRET, Arrestin, Bimolecular Fluorescence Complementation, Receptor Dimerization
Introduction
Chemokine receptors are a specialized subset of the superfamily of seven transmembrane proteins, coupled to the heterotrimeric G protein (GPCR).4 Among the chemokine receptors, CXCR4 and CCR5 have been the subject of many studies given their important role as co-receptors for M- and T-tropic HIV infections (1). A major concern with HIV is that it can adapt and become resistant to drugs that target HIV entry at the cell surface (2). GPCRs signal via multiple proteins assembled into a complex, and currently, chemokine receptors are left uncharacterized in terms of their trafficking and association with signaling partners. Although dimerization of GPCRs has been shown for several receptors, including CCR5 and CXCR4 (3–7), very little is known about how receptor dimerization and mainly heterodimerization will affect signal transduction. It was demonstrated that CXCR4 and CCR5 are able to dimerize together. Adenoviral expression of CCR5.32 mutant receptor in primary CD4+ cells was able to down-regulate the cell surface expression of both types of HIV co-receptors and conferred resistance to R5, X4, and R5X4 strains of HIV, type 1 (3). Here we want to explore how expression of a GPCR adaptor will affect the signaling events downstream of CXCR4 and CCR5 homo- or heterodimers.
Adaptors and scaffolding proteins play an important role in G protein-coupled receptor biogenesis, trafficking, and cellular sorting to the plasma membrane (8, 9). Several adaptors assemble into complexes with receptors and downstream effectors to regulate agonist-induced receptor internalization, regulation of kinase activation, regulation of constitutive activity, and coupling to second messengers, as well as spatial organization of synapses (10–13). NHERF1, also known as EBP50, is a phosphoprotein of 50 kDa first identified as a cofactor essential for protein kinase A-mediated inhibition of Na+/H+ exchanger isoform 3 (14). Since then, it has been shown to be a crucial component for recycling and sorting of several receptors, ion channels, and transporters. NHERF1 contains two PDZ (post-synaptic density 95/disc-large/zona occludens-1 domains; PDZ1 and PDZ2) implicated in multiple protein-protein interactions and an ERM (ezrin-radixin-moesin-merlin) domain, which binds to the actin-associated ERM proteins (15, 16). NHERF1 has been found to interact with a variety of proteins such as G proteins (17), receptors (18), effectors (19), as well as other adaptors and scaffolds (20, 21). These interactions are involved in a growing range of functions, including the assembly of signaling complexes, receptor recycling, and transport of proteins to the cell surface (22). Crucial roles of NHERF1 include its involvement in internalization, recycling, and down-regulation or receptors (23–25). A model proposed that the environment of a leucine/isoleucine/valine surrounded by 3–4 hydrophobic residues would be implicated in the specificity of interactions with PDZ domains (26). The chemokine CCR5 receptor C-terminal tail contains a PDZ recognition motif constituted by a serine, valine, glycine, and leucine (SVGL). Interestingly, another chemokine receptor, CXCR4, does not possess any known recognition motif and does not interact with NHERF1 (18). Given that those two chemokine receptors are known to heterodimerize (3), we were then interested to determine whether NHERF1 could interact with chemokine homodimers CCR5 and CXCR4, as well as their heterodimer CXCR4-CCR5. Our results showed that NHERF1 could only bind to the CCR5 homodimer and not to other combinations (CXCR4 homodimer or CXCR4-CCR5 heterodimer), which shows a level of selectivity of receptor signaling complexes to certain partners, therefore increasing the complexity of potential signaling pathways involved with those two receptors. We then pursued the characterization of this interaction. The PDZ2 domain is important for the interaction between NHERF1 and CCR5. We also verified the implication of this domain as well as the WT isoform of NHERF1 in functions of CCR5 such as RANTES-induced ERK phosphorylation and receptor internalization as they can be regulated by NHERF1. Our results demonstrate that NHERF1 plays an important role in the regulation of CCR5 recruitment of arrestin and internalization and that NHERF1 contributes to a more robust activation of ERK.
MATERIALS AND METHODS
Reagents
Reagents were obtained from the following sources: fetal bovine serum, Alexa Fluor 488 IgG and Alexa-Fluor 647 phalloidin, and Lipofectamine 2000 transfection reagents were from Invitrogen; Dulbecco's modified Eagle's medium high glucose and all chemicals were obtained from Sigma, unless noted otherwise. Monoclonal phospho-ERK, polyclonal ERK, and polyclonal GFP and monoclonal NHERF1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Human recombinant RANTES, coelenterazine H, bethyl anti-biotin antibody, and Covance monoclonal anti-HA raw ascites were from Cedarlane Labs (Hornby, Ontario, Canada). Peptides were synthesized by Bio Basic Inc. (Markham, Ontario, Canada).
Constructs
CCR5 and CXCR4 receptors were purchased from Missouri University of Science and Technology cDNA resource center and then transferred into a pcDNA3.1 vector containing fragments of a yellow fluorescent protein, Venus. To construct plasmids coding for human CCR5 and CXCR4 with an N- or C-terminal fragment of the YFP variant Venus, the receptors were amplified by PCR using primers recognizing the receptor sequence along with a restriction site; CXCR4 was cloned into the pcDNA Venus1 or Venus2 vectors using NheI-ClaI, and CCR5 was cloned NheI-BstBI into the same vectors. The receptors were cloned to replace the GCN4 leucine zipper from pcDNA3.1/Zeo(+)-GCN4 leucine zipper-Venus1 and pcDNA3.1/Zeo(+)-GCN4 leucine zipper-Venus2 cDNAs, used as controls in our experiments. All clones were sequenced and verified for exactitude. NHERF1-HA WT and fragment domains (PDZ1-HA, PDZ2-HA, ERM-HA, PDZ1-PDZ2-HA, and PDZ2-ERM-HA) were kind gifts from Dr. Jean-Luc Parent, from the Université de Sherbrooke, Canada. NHERF1-Rluc was constructed by exchanging the HA tag with a full-length Renilla luciferase fragment. All other constructs were obtained from Dr. Terence E. Hébert, McGill University, Canada.
Cell Culture and Transfection
HEK293 cells were grown in Dulbecco's modified Eagle's medium high glucose supplemented with 10% fetal bovine serum and transfected using Lipofectamine 2000 as per the manufacturer's instructions. Cells were plated in 6-well plates. Experiments were carried out 48 h after transfection. When CCR5 was expressed in cells, CD4 was co-expressed as well, because it was shown that CD4 helps CCR5 expression at the plasma membrane (27).
Bioluminescence Resonance Energy Transfer (BRET) and Bimolecular Fluorescence Complementation (BiFC) Experiments
HEK293 cells were co-transfected with vectors expressing the GFP and Rluc fusion proteins (1 μg of each cDNA was transfected into each well of a 6-well plate, and total DNA/dish was kept constant by adding pcDNA vector as required). 48 h after transfection, cells were harvested and washed once with phosphate-buffered saline (PBS). The cells were then suspended in PBS+ (PBS + 0.1% glucose) and distributed into 96-well microplates (white Optiplate; PerkinElmer Life Sciences). Signals were collected on a Packard fusion instrument (PerkinElmer Life Sciences) using coelenterazine H as a substrate. Whether or not BRET occurred was determined by calculating the ratio of the light passed by the 450/58- (luciferase) and 535/25-nm bandpass filters (YFP) for BRET1. This ratio is referred to as the BRET ratio. To avoid possible variations in the BRET signal resulting from fluctuation in the relative expression levels of the energy donor and acceptor, we designed transfection conditions to maintain constant GFP/Rluc expression ratios in each experimental set. BiFC signals were determined by the measurement of the light that passed by the 535/25-nm band pass filters (YFP). BRET background was determined under conditions where resonance energy transfer between Rluc and GFP either could not or did not occur. This was accomplished by expressing Rluc or Rluc-tagged proteins either alone or together with GFP or GFP-tagged proteins, none of which interact physiologically. The background was the same regardless of which of the aforementioned individual proteins or combinations of proteins were expressed, and it has been subtracted to yield net BRET.
Immunoprecipitation and Cell Lysis
48 h after transfection into 100-mm dishes (for these experiments 4 μg of each cDNA was transfected into each dish, and total DNA levels/dish were kept constant by adding pcDNA vector as required) cells were washed with PBS and harvested. Samples were lysed in 0.8 ml of radioimmune precipitation assay buffer (50 mm Tris, pH 7.5, 10 mm MgCl2, 150 mm NaCl, 0.5% sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS, complete protease inhibitors (Roche Applied Science), and DNase I). The lysate was solubilized by incubation at 4 °C for 30 min, precleared with 50 μl of protein A-Sepharose beads at 4 °C for 1 h, and clarified by centrifugation at 14,000 rpm for 10 min. Supernatants were then transferred into another microcentrifuge tube and incubated with an antibody overnight. The immunoprecipitated proteins were eluted from beads with 50 μl of SDS sample buffer and resolved by SDS-PAGE, and Western blots were performed as described previously (28). When immunoprecipitation was not required, cells were lysed in 200 μl of RIPA buffer, precleared with protein A-Sepharose, and then SDS-PAGE loading buffer was added. Immunoblots were probed with either a polyclonal anti-GFP antibody (1:1000), polyclonal ERK (1:5000), monoclonal phospho-ERK (1:5000), or monoclonal anti-HA (Covance, 1:1000 dilution), horseradish peroxidase-conjugated secondary antibodies were also from Santa Cruz Biotechnology (anti-mouse or anti-rabbit, 1:10,000).
Confocal Microscopy
Twenty four hours post-transfection, HEK293 cells were harvested and seeded on laminin-coated coverslips for 4 h at 37 °C. The cells were then fixed for 20 min in PBS, pH 7.4, containing 3% (w/v) paraformaldehyde. The coverslips were washed with PBS, drained, and mounted onto glass slides using a drop of 0.4% 1,4-diazabicyclo[2.2.2]octane/glycerol medium. Coverslips were fixed to the slides with nail polish. Fluorescence microscopy was performed with an Olympus IX81 equipped with a photometrics coolSNAP HQ2 camera and excited series 120Q light source. YFP (Venus) was excited at 488 nm, and image acquisition was done at fluorescence emission 525 nm.
Cell-surface Expression Assay
HEK293 cells were co-transfected with the receptor constructs, NHERF1 WT or fragments of NHERF1 (PDZ domain 1, PDZ2, ezrin-radixin-moesin (ERM), PDZ1-PDZ2, PDZ2-ERM) and/or HA-arrestin2 WT or mutated (V53D). (1 μg of each cDNA was transfected into each well of a 6-well plate, and total DNA/dish was kept constant by adding pcDNA vector as required). 48 h after transfection, cells were stimulated with CCR5 ligand RANTES (regulated upon activation, normal T-cell expressed, and secreted or CCL5) for up to 60 min. Cells were then washed with PBS and fixed with 3.7% formaldehyde in TBS for 5 min. After three washes with TBS, cells were incubated for 45 min in TBS + 1% BSA and then for 1 h in TBS + 1% BSA + relevant primary antibody. Cells were gently washed twice with TBS, incubated, blocked again in TBS + 1% BSA for 15 min, and then with TBS + 1% BSA + the relevant secondary antibody for 60 min. Cells were washed again twice with TBS. o-Phenylenediamine dihydrochloride substrate in a citrate buffer was then prepared and added to the cells to induce the colorimetric reaction. The reaction was stopped with 3 n HCl when color appeared. The colorimetric assay was then read on a plate reader (PerkinElmer Life Sciences) at 492 nm.
RESULTS
Bimolecular Fluorescence Complementation Assay
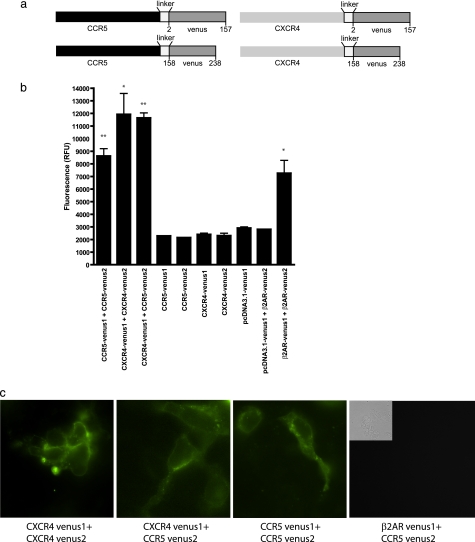
It was described previously that CCR5 and CXCR4 can interact together to form a receptor signaling complex at plasma membrane (3, 29). Up to now, most studies revealing interactions between receptors were done via co-immunoprecipitation or via fluorescence-based two-protein interaction techniques such as FRET or BRET. Here, we decided to use another fluorescence-based technique (BiFC) as a tool to obtain a fluorescence signal when two receptors dimerize. The advantage of using this technique is that it allows another fluorescent- or bioluminescent-tagged protein to be used in FRET or BRET, therefore allowing us to monitor the specific interaction of a given combination of receptor pair with other signaling partners. To do so, we tagged our chemokine receptors with the first 157 amino acids of Venus, a YFP variant, or with the 158–238 remaining amino acids of Venus (Fig. 1a). As described previously for BiFC (30), expression of each construct individually does not produce fluorescence, although expression of both halves, when paired to interacting proteins, will generate a functional fluorescent protein. Fig. 1b shows that 48 h upon expression in HEK293 cells of constructions encoding for CXCR4-Venus1 or CXCR4-Venus2, CCR5-Venus1 or CCR5-Venus2, or the vector pcDNA3.1 encoding only Venus1, very low levels of fluorescence can be detected (2400 ± 105.5, 2302 ± 97.5, 2293 ± 7, 2138 ± 16, and 2909 ± 92 relative fluorescence units, respectively). As a negative control for interactions, we used pcDNA3.1-Venus1 in pair with β2AR-Venus2 (2797 ± 8.5 relative fluorescence units) Another control showing similar plasma membrane expression (β2AR-Venus1 + CCR5-Venus2) was similar to other negative controls (data not shown). As observed for other BiFC pairs, not all receptor pairs were able to generate significant levels of fluorescence compared with controls. Here, we observed that the pair CCR5-Venus1/CXCR4-Venus2 generated very low levels of fluorescence (3734 ± 235; supplemental Fig. 1), which made us select only the CXCR4-Venus1/CCR5-Venus2 pair to perform our studies. The three-dimensional assembly of CCR5-Venus1 and CXCR4-Venus2 probably does not allow the fluorescent protein parts to come in close enough proximity to generate higher levels of fluorescence reconstitution. The β2-adrenergic receptor has been shown to produce dimers and was used as a positive control for our experiments (7257 ± 1019 relative fluorescence units). Finally, we tested our chemokine receptor pairs of interest, CCR5-Venus1/2, CXCR4-Venus1/2, and CXCR4-Venus1/CCR5-Venus2, and our results demonstrate that all these chemokine receptor pairs can interact with each other and allow the reconstitution of the fluorescent signal (8621 ± 587.5, 11925 ± 1657, and 11647 ± 398.5). As indicated in Fig. 1b, these three interactions, along with our positive control, were all significantly different from their negative counterparts, and here the corresponding receptor-Venus parts are expressed alone. Interestingly, all our chemokine receptor pairs were able to generate a higher level of fluorescence than the β2-adrenergic receptors. Fig. 1c shows fluorescence microscopy images of the expression of the different receptor pairs and a control. To perform subsequent experiments for the characterization of interacting partners, we needed to verify the functionality of our antibodies in terms of their capacity to recognize only the fully folded and reconstituted form, and not the nonfluorescent portions of Venus. Having an antibody that recognizes only the reconstituted Venus will allow us to immunoprecipitate only the receptor dimer we are interested in, and therefore eliminate the background noise that we would obtain with the individual constructs. We have tested that when expressed alone, neither CCR5-Venus1 nor CCR5-Venus2 can be immunoblotted with an anti-GFP antibody (supplemental Fig. 2). Given the sequence similarity between GFP and YFP, anti-GFP antibodies can recognize our fully reconstituted Venus, when CCR5Venus1/2 is co-expressed. These results indicate that we have a system that allows us to specifically isolate one receptor pair, among a pool of different receptors expressed in a cell. Similar results were obtained with other receptor pairs, as expected, because all receptors are cloned into the same BiFC vectors (data not shown).
FIGURE 1.
Complementation (BiFC) experiments using various chemokine receptors. HEK293 cells were co-transfected for 48 h with various constructs harboring nonfunctional parts of the YFP variant, Venus. Upon interaction between two receptors, the Venus parts can associate together and regenerate a functional fluorescent signal. a, representation of constructs used in the experiments. b, acquisition of BiFC signals was done in a fusion plate reader (PerkinElmer Life Sciences) bearing filters allowing the measurement of the light that passed by the 535/25-nm bandpass filters (YFP). Results are shown as the relative fluorescence units, and experiments were repeated three times. *, p < 0.05; **, p < 0.01 compared with controls using two-tailed paired Student's t test. c, fluorescence images of bimolecular fluorescence pairs when co-expressed together.
NHERF1 Interaction with CCR5
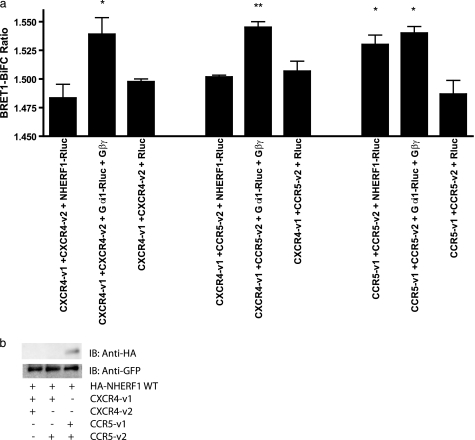
Many GPCRs, including several chemokine receptors, possess a PDZ interaction domain at the very end of their cytoplasmic tail. CCR5 is no exception to this, and earlier reports suggested that CCR5 could interact weakly with NHERF1 (31). Unfortunately, no results were shown, and this interaction was not fully characterized. Fig. 2a shows the interaction, by BRET, of CCR5v1 + CCR5v2 with NHERF1-Rluc (Fig. 2a, 7th column). When compared with its negative control CCR5-v1+CCR5-v2 + Rluc (Fig. 2a, 9th column), our results show that CCR5 can interact with NHERF1 (1.530 ± 0.008 versus 1.487 ± 0.012), as described previously. In fact, this interaction permits levels of BiFC-BRET ratios similar to our positive control, the interaction of CCR5 with its G protein Gαi (Fig. 2a, 8th column; BRET ratio of 1.540 ± 0.057). The interaction between chemokine receptors CXCR4 and CCR5 with Gαi (Fig. 2a, 2nd, 5th, and 8th columns) for signal transduction was chosen as a positive signal due to the receptor coupling to this G protein. Here, we co-expressed Gαi and Gβγ subunits to form a functional G protein. In comparison, neither CXCR4-v1 + CXCR4-v2 (Fig. 2a, 1st column; BRET ratio of 1.483 ± 0.012) nor CXCR4-v1 + CCR5-v2 (4th column; BRET ratio of 1.502 ± 0.001) was able to interact with NHERF1-Rluc as their BRET signals were not significantly different from the negative controls (Fig. 2a, 3rd and 6th columns; BRET ratios of 1.495 ± 0.002 and 1.507 ± 0.008, respectively). We also used a biochemical approach, a co-immunoprecipitation of the various receptor pairs, CXCR4 homodimer, CCR5 homodimer, and CXCR4-CCR5 heterodimer to show the level of interaction by immunoblotting. Fig. 2b shows that no co-immunoprecipitation of NHERF1 was obtained with CXCR4 bearing receptor pairs, although CCR5 can immunoprecipitate NHERF1 weakly, despite consistent levels of receptor expression in HEK293 cells, as revealed by GFP immunoblotting.
FIGURE 2.
NHERF1 interaction with CCR5 homodimers. a, specificity of interaction of NHERF1 with various receptor combinations forming homodimers (CCR5-Venus1/2 or CXCR4 Venus1/2) and heterodimers (CCR5-Venus2 + CXCR4-Venus1) were co-expressed in HEK293 cells for 48 h. BiFC-BRET1 experiments were then performed. BRET values are shown in the text. Results shown are expressed as means ± S.E. of at least three experiments. *, p < 0.05; **, p < 0.01 compared with negative controls using two-tailed paired Student's t test. b, co-immunoprecipitation of NHERF1 with chemokine receptor pairs shows interaction with the CCR5 homodimer only. Results are representative of four independent experiments. IB, immunoblot.
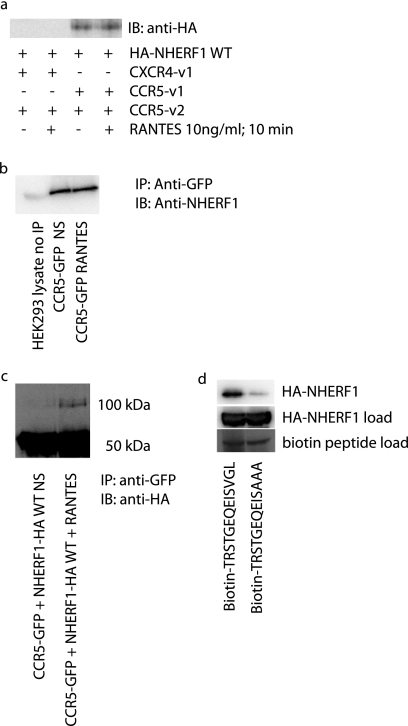
Given that no ligand was present in the previous BiFC-BRET experiments, we decided to test whether the addition of RANTES, a CCR5 ligand, could modulate the interaction between CCR5 containing receptor dimers CCR5-v1 + CCR5-v2 and CXCR4-v1 + CCR5-v2. Potentially, the lack of basal interaction with the heterodimer could be changed upon ligand activation. Fig. 3a shows that it is not the case, as it was impossible to detect an interaction of NHERF1 with the heterodimer, despite a 10-min stimulation with 10 ng/ml RANTES. Also, the basal interaction detected between the CCR5 homodimer and NHERF1 could not be modulated by the addition of RANTES. Therefore, it appears that the interaction between NHERF1 and CCR5 is constitutive. The levels of NHERF1 expressed in the cell might already be sufficient to produce their normal function, and therefore the system might already be saturated when we overexpress NHERF1. That might explain the lack of further recruitment by the receptor. We also verified the effect of receptor stimulation with RANTES on HEK293 endogenous NHERF1 (32) to see if the HA tag added to NHERF1 could modify its activity. As observed previously in Fig. 3a, no change is noted in the levels of NHERF1 interaction with the CCR5 receptor with (3rd lane) or without (2nd lane) stimulation (Fig. 3b). The 1st lane shows a 5% loading of HEK293 cell lysate, showing the 50-kDa band corresponding to NHERF1. NHERF1 has been shown previously to dimerize, although with low affinity (33–35). In previous publications, it was shown that once an NHERF1 protein is immobilized to a receptor, a second NHERF1 could dimerize with the first and then get activated to produce its cellular actions (33, 36). Given the lack of change between basal and stimulated receptor interaction with NHERF1, we wanted to see if NHERF1 dimerization could be changed upon stimulation. Fig. 3c shows that upon stimulation with RANTES, a weak band can be observed in the 100-kDa range, corresponding to an NHERF1 dimer. The NHERF1 dimer is better observed after stimulation, suggesting that there are indeed changes in NHERF1 activity, simply not at the monomer level. In Fig. 3d, we wanted to determine whether disruption of the final few amino acids of the proposed PDZ domain of CCR5 could interfere with the interaction of NHERF1. To do so, we used biotinylated peptides in which the last 13 amino acids of CCR5 were present. One of the peptides had a mutation of the last three amino acids VGL into alanines. Our results show that a reduction in the interaction levels of NHERF1 with the PDZ domain is observed with the mutated domain, suggesting that this is indeed the interaction site of NHERF1 on the CCR5 C-terminal tail.
FIGURE 3.
Effect of RANTES on NHERF1-CCR5 interaction. a, 48 h post-transfection with cDNAs encoding NHERF1 and receptor constructs forming CCR5 homodimers or CXCR4-CCR5 heterodimers, cells were stimulated with 10 ng/ml RANTES, for 10 min. Cells were then lysed and cell lysates migrated on the gel to perform an immunoblot against HA-tagged NHERF1. Results show that RANTES does not modify the extent of interaction between the receptors and NHERF1. Results are representative of three independent experiments. b, effect of RANTES on endogenous NHERF1 recruitment to CCR5. c, effect of RANTES on NHERF1 dimerization. d, 50 μm of each biotin-tagged peptide was incubated in the presence of a cell lysate expressing NHERF1 WT and streptavidin-coupled agarose beads for 1 h, washed with PBS, and loaded on an SDS-polyacrylamide gel for analysis. IB, immunoblot.
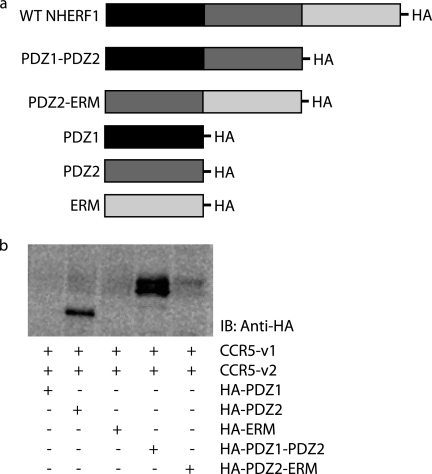
NHERF1 sequences bear three motifs important for protein-protein interactions as follows: PDZ1, PDZ2, and ERM. We used constructs where portions of the NHERF1 domains were deleted and co-expressed those constructs. Deletions mutants correspond to each motif individually (PDZ1, PDZ2, and ERM domains alone), as well as constructs where only one domain was deleted (PDZ1-PDZ2 lacks ERM whereas PDZ2-ERM lacks PDZ1). Those constructs were expressed in HEK293 cells to determine which motif is important for the NHERF1-CCR5 interaction. Fig. 4a is a representation of the different constructs. Fig. 4b shows that with co-expression of CCR5 with PDZ2 alone PDZ1-PDZ2 and PDZ2-ERM allow the interaction to occur, therefore suggesting PDZ2 as the NHERF1 domain interacting with CCR5, as this is the only common motif preserved in all interactions. It has been recently suggested that ERM could inhibit PDZ2 function. The decrease of interaction of the PDZ2 domain in the presence of ERM (PDZ2-ERM) could possibly result from an effect of ERM on the PDZ2 domain capacity to interact with CCR5 in comparison with PDZ2 alone.
FIGURE 4.
PDZ2 mediates NHERF1 interaction with CCR5. a, schematic representation of constructs used in the experiments. b, cells were transfected for 48 h with cDNAs encoding NHERF1 WT or its different domains (PDZ1, PDZ2, ERM, PDZ1-PDZ2, and PDZ2-ERM), CCR5-Venus1 and CCR5-Venus2. Immunoprecipitations using an antibody directed against GFP were performed. An immunoblot (IB) was then performed using an antibody directed against HA-tagged NHERF1 constructs. Results show that PDZ domain 2 is important for the interaction between the receptor and NHERF1. Results are representative of three independent experiments.
NHERF1 Regulation of Arrestin Recruitment to CCR5
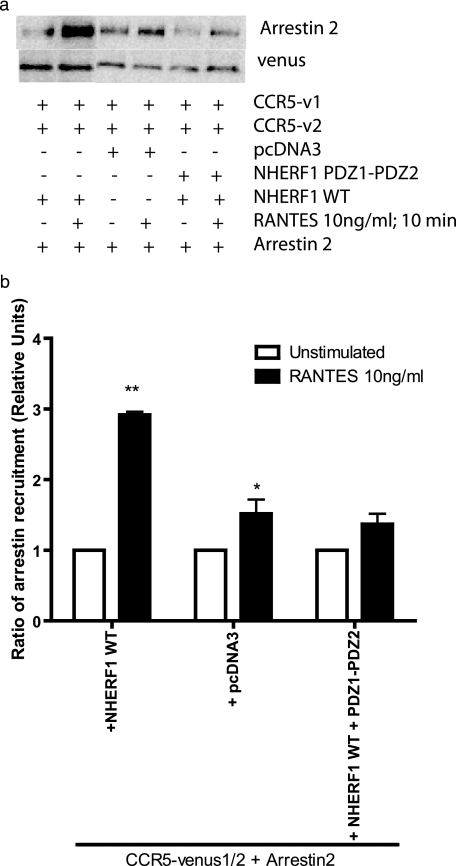
Upon activation, CCR5 normally recruits arrestins, which induce desensitization and internalization of activated receptors. Here, we performed co-immunoprecipitations of the CCR5 homodimer with arrestin, to verify whether NHERF1 affects the levels of arrestin recruited by the receptor. It was shown previously for the parathyroid hormone receptor that NHERF1 could interfere with arrestin recruitment to the receptor during receptor desensitization (37). First, arrestin can interact basally with CCR5. Upon stimulation with RANTES, cells expressing NHERF1 trigger an increase in arrestin recruitment to CCR5 (Fig. 5a, 2nd lane versus control 4th lane). Interestingly, a construction of NHERF1 made of only PDZ1-PDZ2 (Fig. 5a, 5th and 6th lanes), which will compete with WT proteins but not bind to actin-interacting proteins, can diminish the extent of arrestin2 recruitment to the receptor, to levels similar to the control (Fig. 5a). Fig. 5b is a densitometric analysis of several experiments and shows the significance of arrestin recruitment to receptors.
FIGURE 5.
Recruitment of arrestin2 to CCR5 is facilitated by NHERF1. a, HEK293 cells were co-transfected with cDNAs encoding NHERF1 WT or PDZ1-PDZ2, along with CCR5-Venus1 and CCR5-Venus2. Cells were then lysed, and immunoprecipitations using an antibody directed against GFP were performed. An immunoblot was then performed using an antibody directed against arrestin2. b, histogram representation of the results obtained by immunoblotting. *, p < 0.05; **, p < 0.01 compared with negative controls using two-tailed paired Student's t test. Results are representative of three independent experiments.
Regulation of CCR5 Internalization by NHERF1
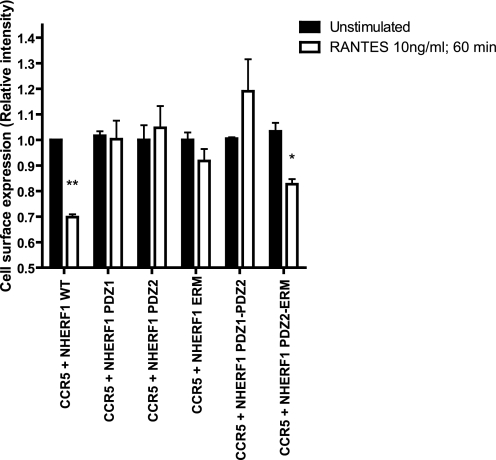
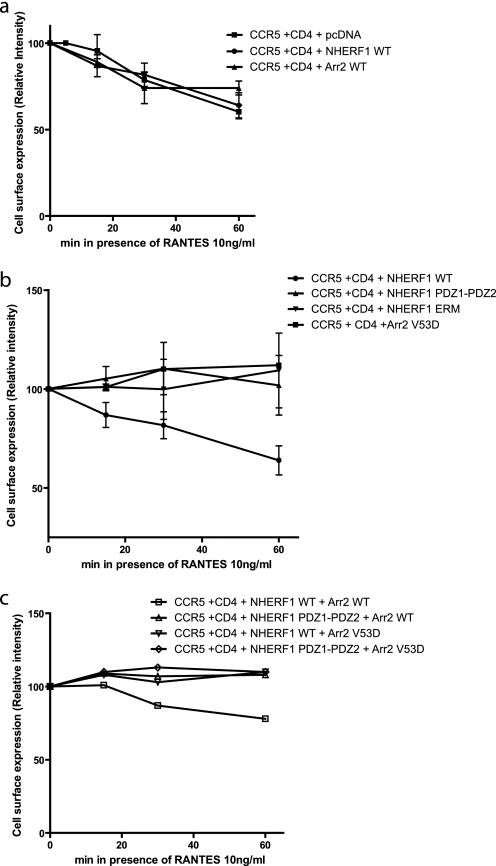
The role of NHERF1 in internalization varies from one interacting protein to another. For some receptors such as the β2-adrenergic receptor and κ-opioid, it will enhance the recycling of receptors (23, 24). Conversely, NHERF1 stabilizes the EGF receptor plasma membrane expression and delays internalization (38). Also, NHERF1 can inhibit PTH1R endocytosis, without affecting its recycling (32). Fig. 6 shows that NHERF1 contributes to the internalization of CCR5 after stimulation with RANTES. When NHERF1 is co-expressed with CCR5, cell surface expression of CCR5 is decreased by ∼30%, as observed with CCR5 internalization without NHERF1 (Fig. 7a). In contrast, co-expression of PDZ1, PDZ2, ERM, or PDZ1-PDZ2 with the CCR5 did not lead to any significant decrease in cell surface expression levels (Fig. 6). The expression of PDZ2-ERM with CCR5 contributed to receptor internalization by ∼20%. Fig. 7 shows the differences in internalization of CCR5 when co-expressed with NHERF1 and/or arrestin2 constructs, WT, or dominant negative. Fig. 7a shows that the extent of internalization does not vary significantly when NHERF1 or arrestin2 WT are co-expressed with the receptor, in comparison with its expression alone. Co-expression of NHERF1 domains PDZ1-PDZ2 or ERM will disrupt the internalization of CCR5. Also, as demonstrated previously (39), an arrestin dominant negative mutant, V53D, can also inhibit CCR5 internalization to a similar extent as the NHERF1 domains (Fig. 7b). Also, we demonstrate here that co-expression of either arrestin2 V53D with WT NHERF1 or PDZ1-PDZ2 with arrestin2 WT will lead to disruption of CCR5 internalization (Fig. 7c), suggesting that both arrestin2 and NHERF1 are important for CCR5 internalization.
FIGURE 6.
NHERF1 effect on CCR5 internalization. 48 h post-transfection, HEK293 cells were incubated with 10 ng/ml RANTES for 60 min and then labeled with an anti-CCR5 antibody targeting an extracellular portion of the receptor to measure cell surface expression. Then an ELISA-type assay was performed to measure cell surface expression assay following RANTES stimulation of CCR5. Results are shown as the relative intensity of expression at the plasma membrane, where unstimulated cells were adjusted empirically to 1, for ease of comparison. Results are representative of at least three independent experiments, and *, p < 0.05; **, p < 0.01 compared with controls using two-tailed paired Student's t test.
FIGURE 7.
Time course of NHERF1 and arrestin2 effects on CCR5 internalization. 48 h post-transfection, HEK293 cells were incubated with 10 ng/ml RANTES for various time frames and then labeled with an anti-CCR5 antibody targeting an extracellular portion of the receptor to measure cell surface expression. Then an ELISA-type assay was performed to measure cell surface assay following RANTES stimulation of CCR5. a, effect on cell surface expression of NHERF1 and arrestin2 WT. b, effect on cell surface expression of NHERF1 PDZ1-PDZ2, NHERF1 ERM, or arrestin2 V53D. c, effect of cell surface expression of combinations of constructs mentioned above. Results are shown as the relative intensity of expression at the plasma membrane, where unstimulated cells were adjusted empirically to 100% plasma membrane expression. Results are representative of three independent experiments.
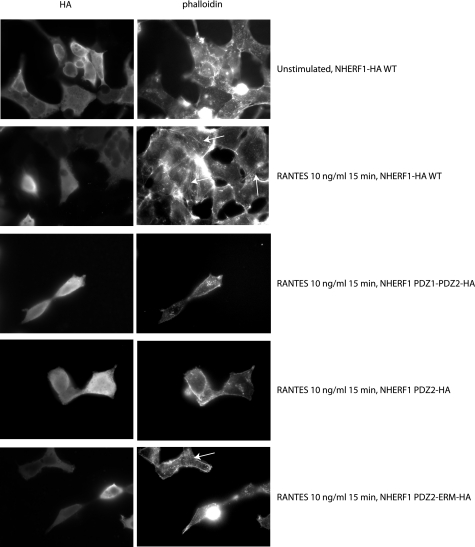
Several studies have shown the importance of actin in CCR5 receptor internalization (40, 41). It was also reported that NHERF1, via its ERM domain, can interact with and promote actin reorganization (42, 43). Here, we wanted to understand whether actin reorganization could be the mechanism by which NHERF1 mediates its action on CCR5 internalization. To do so, we co-expressed CCR5 with various HA-tagged NHERF1 constructs, labeled the cells with phalloidin, and incubated them with an antibody against HA. We then used fluorescence microscopy to analyze the changes in actin remodeling. Fig. 8 shows the expression of HA-tagged NHERF constructs (left panel) and phalloidin labeling (right panel). Upon stimulation with RANTES for 15 min, actin remodeling occurs when WT NHERF1 is co-expressed (Fig. 8, 2nd row, right panel). No actin remodeling is observed with the PDZ1-PDZ2 or PDZ2 domain when co-expressed with the receptor (Fig. 8, 3rd and 4th rows, right panel). Interestingly, when a PDZ2-ERM construct is co-expressed, actin remodeling is observed again (Fig. 8, bottom row, right panel). Those results taken together with our previous results showing PDZ2 as the interaction domain with CCR5 suggest that PDZ2 is required for receptor interaction, whereas ERM is required for actin remodeling. It is only when both are present that modulation of actin remodeling can happen.
FIGURE 8.
Effect of NHERF1 on actin remodeling. Cells were plated on coverslips for 24 h and then transfected with CCR5-GFP and various HA-tagged NHERF1 domain constructs for 48 h. Cells were then stimulated with 10 ng/ml RANTES for 15 min, fixed, and incubated with Alexa Fluor 647 phalloidin and an antibody directed against the epitope HA. Cells were then incubated with a secondary Alexa Fluor 488 antibody. Images were then acquired by fluorescence microscopy.
Effect of NHERF1 on RANTES-induced ERK Activation
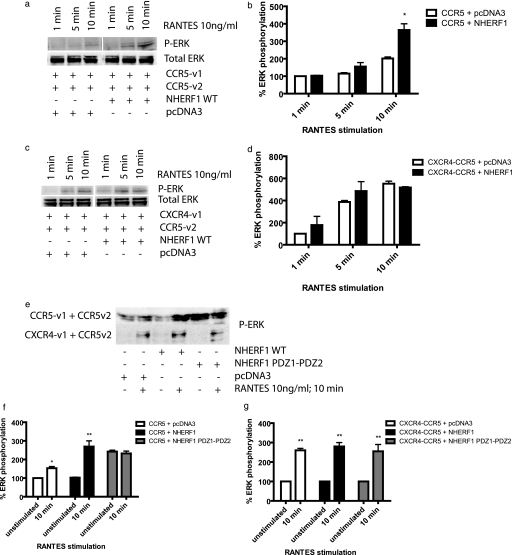
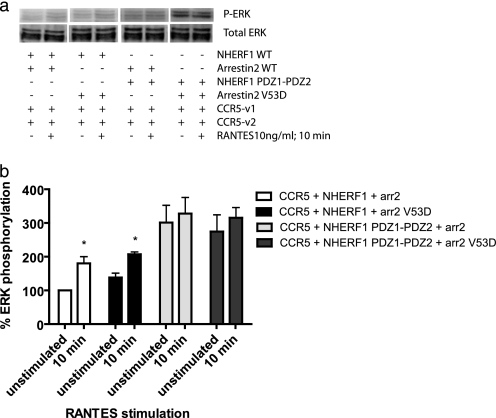
GPCRs activate ERK1/2 through different mechanisms. ERK activation by β-adrenergic receptors occurs in a biphasic manner and involves receptor internalization. An early rapid phase is arrestin-independent, whereas prolonged activation is arrestin-dependent. In the case of PTH1R, parathyroid hormone stimulated ERK1/2 phosphorylation by a PKA-dependent but protein kinase C-independent pathway. Interestingly, NHERF1 blocked parathyroid hormone-induced ERK1/2 phosphorylation downstream of PKA (44). Given the different mechanisms involved in ERK activation, and NHERF1 involvement in the ERK pathway for some receptors, we verified if NHERF1 could be involved in ERK1/2 activation following CCR5 activation by RANTES. Cells were transfected with the receptors dimers CCR5 and CXCR4-CCR5 Venus1/2 and NHERF1 WT cDNAs, and 24 h post-transfection, cells were serum-starved overnight. 48 h post-transfection, cells were stimulated with 10 ng/ml RANTES for 1, 5, or 10 min. Cells were lysed, and then SDS-PAGE and immunoblot were performed. NHERF1 increases the extent of ERK1/2 phosphorylation by the CCR5 homodimer (2-fold after a 10-min stimulation with RANTES), compared with the control co-transfected with pcDNA3 (Fig. 9a, quantified in b). Although NHERF1 did not affect arrestin recruitment and internalization of the heterodimer CXCR4-CCR5, we still verified that it did not have any effect on ERK phosphorylation. We report here that as for arrestin recruitment and heterodimer internalization, no significant effect of NHERF1 was observed for ERK phosphorylation for this receptor pair (Fig. 9, c and d). Fig. 9e (quantification f (top blot) and g (bottom blot)) shows the effect of WT and PDZ1-PDZ2 on ERK phosphorylation following a 10-min RANTES stimulation. WT NHERF1 increases the levels of phosphorylation of ERK induced by CCR5 activation, although no change was observed for the heterodimer. Also of note, PDZ1-PDZ2 increases the basal level of ERK phosphorylation of the CCR5 homodimer but not of the heterodimer. Because this construct was blocking receptor internalization, it is possible that it interferes with desensitization; therefore, once the receptor is stimulated, it does not become rapidly inactivated, desensitized, and internalized. For this reason, we decided to co-express arrestin2 WT or V53D in the presence of NHERF1 WT or PDZ1-PDZ2 to see if they would affect ERK phosphorylation. If ERK gets phosphorylated, despite blockade of arrestin2 in the presence of NHERF1, this would indicate that the pathways used for desensitization and internalization are not linked with ERK activation. Our experiments showed that in the presence of NHERF1 and arrestin2 WT, ERK gets phosphorylated following RANTES stimulation (Fig. 10a, 1st 2 lanes, quantification in b). When NHERF1 WT and arrestin2 V53D are co-expressed, ERK activation via phosphorylation following RANTES stimulation still occurs (Fig. 10a, 3rd and 4th lanes). Those results suggest that as ERK still gets phosphorylated, arrestin2 is not necessary for ERK activation following RANTES stimulation. Upon co-expression of PDZ1-PDZ2 and arrestin2 WT, the basal level of ERK phosphorylation becomes elevated, but there is no change after stimulation (Fig. 10a, 5th and 6th lanes). This result suggests again that arrestin2 cannot promote an increase in ERK phosphorylation following RANTES stimulation. Identical results were obtained when PDZ1-PDZ2 and arrestin V53D were co-expressed with the CCR5 homodimer (Fig. 10a, 7th and 8th lanes). Taken together, those results indicate that ERK phosphorylation following RANTES-induced CCR5 activation is dependent on NHERF1 but is dissociated from the arrestin2 pathway because arrestin2 WT expression was unable to promote an increase in ERK phosphorylation.
FIGURE 9.
ERK1/2 phosphorylation induced by RANTES stimulation of CCR5. Approximately 24 h post-transfection, cell media were changed to DMEM without FBS. 48 h after transfection, the cells were then stimulated with 10 ng/ml RANTES for up to 10 min. After stimulation, cells were harvested in TBS and then lysed with RIPA containing phosphatase inhibitors. Immunoblots were performed using an anti-phospho-ERK antibody or total ERK antibody as a control of loading and of expression. a shows NHERF1 effect on CCR5 dimer. b, quantification of a, where *, p < 0.05 in comparison with CCR5 + pcDNA3. c, NHERF1 effect on the CXCR4-CCR5 heterodimer. d, quantification of c and e, effects of both WT and PDZ1-PDZ2 NHERF1 on ERK phosphorylation in the presence of CCR5 homodimer or the heterodimer. f, quantification of the results in e, top blot. g, quantification of the results from e, bottom blot. Results are representative of three experiments. *, p < 0.05; **, p < 0.01 compared with their unstimulated equivalent, acting as negative controls using two-tailed paired Student's t test.
FIGURE 10.
Effect of NHERF1 and arrestin2 on ERK phosphorylation following RANTES stimulation of CCR5. Approximately 24 h post-transfection with the CCR5 dimer, NHERF1 WT, or PDZ1-PDZ2 and arrestin2 (arr2) WT or V53D, cell media were changed to DMEM without FBS. 48 h after transfection, the cells were then stimulated with 10 ng/ml RANTES for 10 min. After stimulation, cells were harvested in TBS and then lysed with RIPA-containing phosphatase inhibitors. Immunoblots were performed using an anti-phospho-ERK antibody (top panel) or total ERK antibody (bottom panel) as a control of loading and of expression. Results are representative of three experiments. *, p < 0.05 compared with their unstimulated equivalent, acting as negative controls using two-tailed paired Student's t test.
DISCUSSION
Adaptors and scaffolding proteins are highly important for the function of membrane receptors and effectors. Their roles are highly diverse, and among them is the regulation of the spatial and thereby functional association with various co-receptors, G proteins, effectors, and other downstream partners (12). Regulation of GPCRs is frequently governed by adaptors or scaffolding protein interactions with its C-terminal tail, in which several motifs can be identified for interactions. One of those motifs is the sequence XφXφ, where X represents any amino acid, and φ indicates a hydrophobic amino acid, generally leucine, valine, or isoleucine (45, 46). Such a motif is present in the sequence of several GPCRs, and chemokine receptor CCR5 bears the sequence (EISVGL), whereas CXCR4, another chemokine receptor, does not (SFHSS). It was reported previously that CCR5 could interact with a PDZ-containing protein NHERF1 (31). Here, we present the first reported example of an interaction of any NHERF1 PDZ domain via a type two PDZ interacting domain. In comparison with other GPCRs, and mainly to the β2-adrenergic receptor, this interaction is weak (18). One of the reasons for such a weak interaction could come from the PDZ sequence of the CCR5 receptor, as none of the proteins tested up to date was able to interact as strongly as the β2-adrenergic receptor. Yet we were able to show that this interaction is important for CCR5 functions such as internalization and ERK1/2 phosphorylation.
One interesting aspect of GPCRs is their capacity to dimerize or form higher oligomeric complexes. CCR5 and CXCR4 have been shown to form both homo- and heterodimeric complexes. Unfortunately, because both homodimers and heterodimers co-exist in the same cell type when each receptor is co-expressed (each receptor has the capacity to associate with a receptor of the same type or a different one), trying to identify proteins that interact specifically with one signaling complex versus the others was practically unfeasible. Development of technologies such as BiFC (where each receptor bears a complementary portion of a fluorescent protein) and its combination with BRET (another potential interacting partner coupled to Renilla luciferase) has allowed us to identify proteins that can interact specifically with one complex and not with others (Fig. 2a). We present here the results of the interaction of NHERF1 with the CCR5 homodimer but not the CXCR4 homodimer or the CXCR4-CCR5 heterodimer. This has significant value, as this demonstrates that each signaling complex has its own signaling properties, despite similar composition. Because of the incapacity of the CCR5v1 and CXCR4v2 heterodimer to generate significant fluorescence levels, this receptor pair was not studied.
Characterization of the interaction of NHERF1 with CCR5 has shown that the PDZ2 domain is important for the interaction (Fig. 4). This result is surprising, as most other GPCRs interacting with NHERF1 interact with the first PDZ domain, whereas several effectors, cystic fibrosis transmembrane regulator, β-catenin, or other signaling partners, interact by the PDZ2 (47). Interestingly, the PDZ2 domain appears to bind to a protein that has another residue instead of aspartate in the motif, as is the case of the PTH1R and Yes-associated protein YAP-65 (21, 48). CCR5 could potentially be associated with this group of proteins, as its sequence is EISVGL.
Arrestins are cytoplasmic adaptors that bind to phosphorylated GPCRs and uncouple them from their cognate G protein, thereby producing a nonsignaling desensitized receptor. Other roles from arrestins include recruitment of other adaptors required for the formation of clathrin-coated vesicles during internalization as well as mediation of ERK activation (49). It was demonstrated for the PTH1R receptor, the only other GPCR known to interact with the PDZ2 domain, that NHERF1 could regulate PTH1R desensitization via interference with arrestin binding (32). In comparison, CCR5 was not shown to interfere with arrestin2 binding to the receptor. In fact, it appears that co-expression of NHERF1 actually increases the capacity of arrestin2 to be recruited and to interact with CCR5. Competition with a construct bearing PDZ1 and PDZ2 was shown to decrease this increased arrestin recruitment effect (Fig. 5). It appears that the effect of NHERF1 will vary from receptor to receptor, as is the case between PTH1R and CCR5, potentially because they do not belong to the same class of GPCRs (class A for CCR5 and class B for PTH1R).
Binding of arrestins to phosphorylated receptors lessens G protein activation and targets the receptors to clathrin-coated pits for internalization. Class A and class B GPCRs have different affinities for arrestin and therefore have differences in the way they behave once internalized. Here, we wanted to see the effect of NHERF1 on the internalization of CCR5. Contrary to the class B PTH1R, NHERF1 was shown to allow internalization of CCR5. In fact, it appears that a construct composed of the PDZ1-PDZ2 domains, or even only of ERM, will block the internalization of the receptor and promote its accumulation at the plasma membrane (Fig. 7). When the effects of NHERF1 and arrestin2 were combined, no significant changes in internalization kinetics or intensity were observed. Also, we showed that blockade of either arrestin or NHERF1 compromises the internalization process of CCR5. Those results are the opposite of what was observed for the PTH1R receptor. We also demonstrated that actin remodeling is the mechanism by which NHERF1 acts to modulate CCR5 internalization. CCR5 was already shown to induce actin remodeling upon activation.
As mentioned previously, arrestin plays several key roles in GPCR signaling and trafficking regulation. Along with its role in desensitization and internalization, arrestin may act as a scaffold by promoting the activation of the RAS-MEK-ERK signaling cascade. Other signaling pathways, some involving G proteins, can also account for ERK activation. Here, we wanted to determine whether NHERF1 could play a role in the regulation of the activation of the ERK signaling cascade as it was previously shown to be involved in this pathway (50). Our results show that overexpression of NHERF1, in HEK293 cells, leads to an increased phosphorylation of ERK, following RANTES activation of CCR5. When the heterodimer CXCR4-CCR5 was expressed, no increase could be observed. Interestingly, the basal level of ERK phosphorylation was elevated when the CCR5 homodimer was co-expressed with the PDZ1-PDZ2 domains, and no further activation was observed following stimulation. The exact mechanism for such a phenomenon is not clear, but it might be related to the effect of PDZ1-PDZ2 on the internalization and potentially on receptor desensitization, which leads to some activation of signaling by the receptors at the plasma membrane. We were intrigued by the potential mechanism leading to ERK activation. Was the arrestin important for ERK activation or was ERK activation derived from G protein activation? To test this, we co-expressed WT or the mutated isoforms of NHERF1 and arrestin2 and detected ERK phosphorylation following RANTES activation of CCR5. Our results show that ERK phosphorylation is blocked only when a mutated form of NHERF1 is present, and not when arrestin2 is deficient (Fig. 10). Furthermore, we once again detected an elevated level of ERK phosphorylation, even in presence of arrestin2 WT. Those results suggest that the mechanism needed following CCR5 stimulation by RANTES to activate the ERK signaling pathway is independent of arrestin2.
Given the basal interaction of NHERF1 with CCR5, one could suggest that a good strategy would be to attempt to disrupt NHERF1 dimerization, instead of CCR5-NHERF1 interaction. Some attention has been given to the design of small compounds capable of interfering with the capacity of NHERF1 to interact with receptor PDZ domains (47). Because NHERF1 can recruit a second NHERF1 to form a dimer and therefore activate the recruited NHERF1 before it is released in the cytosol where it will perform its actions, disruption of this dimer formation might prove to be a novel therapeutic strategy against HIV infection, which uses both CCR5 internalization and actin polymerization during cell entry. Inhibition of the dimerization of NHERF1 by a small compound or peptide blocking NHERF1 dimerization might be able to disrupt the series of events favoring HIV infection development.
In conclusion, we demonstrated that CCR5, but not CXCR4 or CXCR4-CCR5 dimers, can interact and be regulated by NHERF1. Understanding how CCR5 cell surface expression is regulated is particularly important with regard to HIV-1 entry inhibition. Here, we show that NHERF1 can control the internalization of CCR5 after stimulation by RANTES, as well as its signaling via ERK1/2 phosphorylation. Taken together, our results suggest that identification of specific interacting partners might help develop new therapeutic strategies that might replace or complement current ligand therapies. Identification of unique signaling partners might provide more specific therapeutics and, potentially, fewer side effects.
Supplementary Material
This work was supported in part by Canadian Institutes of Health Research Grant HOP-86863, Natural Sciences and Engineering Research Council of Canada Grant EQPEQ-3591097-2008, and Canada Foundation for Innovation (to D. J. D.).

The on-line version of this article (available at http://www.jbc.org) contains Figs. 1 and 2.
- GPCR
- G protein-coupled receptor
- BiFC
- bimolecular fluorescence complementation
- BRET
- bioluminescence resonance energy transfer
- RANTES
- regulated on activation normal T cell expressed and secreted.
REFERENCES
- 1.Berger E. A., Murphy P. M., Farber J. M. (1999) Annu. Rev. Immunol. 17, 657–700 [DOI] [PubMed] [Google Scholar]
- 2.Esté J. A., Telenti A. (2007) Lancet 370, 81–88 [DOI] [PubMed] [Google Scholar]
- 3.Agrawal L., Lu X., Qingwen J., VanHorn-Ali Z., Nicolescu I. V., McDermott D. H., Murphy P. M., Alkhatib G. (2004) J. Virol. 78, 2277–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margeta-Mitrovic M., Jan Y. N., Jan L. Y. (2000) Neuron 27, 97–106 [DOI] [PubMed] [Google Scholar]
- 5.Deng H., Liu R., Ellmeier W., Choe S., Unutmaz D., Burkhart M., Di Marzio P., Marmon S., Sutton R. E., Hill C. M., Davis C. B., Peiper S. C., Schall T. J., Littman D. R., Landau N. R. (1996) Nature 381, 661–666 [DOI] [PubMed] [Google Scholar]
- 6.Benkirane M., Jin D. Y., Chun R. F., Koup R. A., Jeang K. T. (1997) J. Biol. Chem. 272, 30603–30606 [DOI] [PubMed] [Google Scholar]
- 7.Salahpour A., Angers S., Mercier J. F., Lagacé M., Marullo S., Bouvier M. (2004) J. Biol. Chem. 279, 33390–33397 [DOI] [PubMed] [Google Scholar]
- 8.Brady A. E., Limbird L. E. (2002) Cell. Signal. 14, 297–309 [DOI] [PubMed] [Google Scholar]
- 9.Hall R. A., Lefkowitz R. J. (2002) Circ. Res. 91, 672–680 [DOI] [PubMed] [Google Scholar]
- 10.Hu L. A., Tang Y., Miller W. E., Cong M., Lau A. G., Lefkowitz R. J., Hall R. A. (2000) J. Biol. Chem. 275, 38659–38666 [DOI] [PubMed] [Google Scholar]
- 11.Hu L. A., Chen W., Martin N. P., Whalen E. J., Premont R. T., Lefkowitz R. J. (2003) J. Biol. Chem. 278, 26295–26301 [DOI] [PubMed] [Google Scholar]
- 12.Lezcano N., Mrzljak L., Eubanks S., Levenson R., Goldman-Rakic P., Bergson C. (2000) Science 287, 1660–1664 [DOI] [PubMed] [Google Scholar]
- 13.Sheng M., Kim E. (2000) J. Cell Sci. 113, 1851–1856 [DOI] [PubMed] [Google Scholar]
- 14.Weinman E. J., Steplock D., Wang Y., Shenolikar S. (1995) J. Clin. Invest. 95, 2143–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murthy A., Gonzalez-Agosti C., Cordero E., Pinney D., Candia C., Solomon F., Gusella J., Ramesh V. (1998) J. Biol. Chem. 273, 1273–1276 [DOI] [PubMed] [Google Scholar]
- 16.Reczek D., Berryman M., Bretscher A. (1997) J. Cell Biol. 139, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rochdi M. D., Watier V., La Madeleine C., Nakata H., Kozasa T., Parent J. L. (2002) J. Biol. Chem. 277, 40751–40759 [DOI] [PubMed] [Google Scholar]
- 18.Heydorn A., Søndergaard B. P., Ersbøll B., Holst B., Nielsen F. C., Haft C. R., Whistler J., Schwartz T. W. (2004) J. Biol. Chem. 279, 54291–54303 [DOI] [PubMed] [Google Scholar]
- 19.Moyer B. D., Duhaime M., Shaw C., Denton J., Reynolds D., Karlson K. H., Pfeiffer J., Wang S., Mickle J. E., Milewski M., Cutting G. R., Guggino W. B., Li M., Stanton B. A. (2000) J. Biol. Chem. 275, 27069–27074 [DOI] [PubMed] [Google Scholar]
- 20.Morales F. C., Takahashi Y., Momin S., Adams H., Chen X., Georgescu M. M. (2007) Mol. Cell. Biol. 27, 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohler P. J., Kreda S. M., Boucher R. C., Sudol M., Stutts M. J., Milgram S. L. (1999) J. Cell Biol. 147, 879–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shenolikar S., Weinman E. J. (2001) Am. J. Physiol. Renal Physiol. 280, F389–F395 [DOI] [PubMed] [Google Scholar]
- 23.Cao T. T., Deacon H. W., Reczek D., Bretscher A., von Zastrow M. (1999) Nature 401, 286–290 [DOI] [PubMed] [Google Scholar]
- 24.Li J. G., Chen C., Liu-Chen L. Y. (2002) J. Biol. Chem. 277, 27545–27552 [DOI] [PubMed] [Google Scholar]
- 25.Sneddon W. B., Syme C. A., Bisello A., Magyar C. E., Rochdi M. D., Parent J. L., Weinman E. J., Abou-Samra A. B., Friedman P. A. (2003) J. Biol. Chem. 278, 43787–43796 [DOI] [PubMed] [Google Scholar]
- 26.Sheng M., Sala C. (2001) Annu. Rev. Neurosci. 24, 1–29 [DOI] [PubMed] [Google Scholar]
- 27.Achour L., Scott M. G., Shirvani H., Thuret A., Bismuth G., Labbé-Jullié C., Marullo S. (2009) Blood 113, 1938–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavoie C., Mercier J. F., Salahpour A., Umapathy D., Breit A., Villeneuve L. R., Zhu W. Z., Xiao R. P., Lakatta E. G., Bouvier M., Hébert T. E. (2002) J. Biol. Chem. 277, 35402–35410 [DOI] [PubMed] [Google Scholar]
- 29.Isik N., Hereld D., Jin T. (2008) PLoS One 3, e3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu C. D., Grinberg A. V., Kerppola T. K. (2005) Curr. Protoc. Protein Sci. Unit 19.10.1–19.10.21 [DOI] [PubMed] [Google Scholar]
- 31.Delhaye M., Gravot A., Ayinde D., Niedergang F., Alizon M., Brelot A. (2007) Mol. Pharmacol. 72, 1497–1507 [DOI] [PubMed] [Google Scholar]
- 32.Wang B., Bisello A., Yang Y., Romero G. G., Friedman P. A. (2007) J. Biol. Chem. 282, 36214–36222 [DOI] [PubMed] [Google Scholar]
- 33.Maudsley S., Zamah A. M., Rahman N., Blitzer J. T., Luttrell L. M., Lefkowitz R. J., Hall R. A. (2000) Mol. Cell. Biol. 20, 8352–8363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fouassier L., Yun C. C., Fitz J. G., Doctor R. B. (2000) J. Biol. Chem. 275, 25039–25045 [DOI] [PubMed] [Google Scholar]
- 35.Shenolikar S., Minkoff C. M., Steplock D. A., Evangelista C., Liu M., Weinman E. J. (2001) FEBS Lett. 489, 233–236 [DOI] [PubMed] [Google Scholar]
- 36.Lau A. G., Hall R. A. (2001) Biochemistry 40, 8572–8580 [DOI] [PubMed] [Google Scholar]
- 37.Wang B., Yang Y., Abou-Samra A. B., Friedman P. A. (2009) Mol. Pharmacol. 75, 1189–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazar C. S., Cresson C. M., Lauffenburger D. A., Gill G. N. (2004) Mol. Biol. Cell 15, 5470–5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aramori I., Ferguson S. S., Bieniasz P. D., Zhang J., Cullen B., Cullen M. G. (1997) EMBO J. 16, 4606–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller A., Strange P. G. (2004) Eur. J. Biochem. 271, 243–252 [DOI] [PubMed] [Google Scholar]
- 41.Di Marzio P., Dai W. W., Franchin G., Chan A. Y., Symons M., Sherry B. (2005) Biochem. Biophys. Res. Commun. 331, 909–916 [DOI] [PubMed] [Google Scholar]
- 42.Morales F. C., Takahashi Y., Kreimann E. L., Georgescu M. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17705–17710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmieder S., Nagai M., Orlando R. A., Takeda T., Farquhar M. G. (2004) J. Am. Soc. Nephrol. 15, 2289–2298 [DOI] [PubMed] [Google Scholar]
- 44.Wang B., Yang Y., Friedman P. A. (2008) Mol. Biol. Cell 19, 1637–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Songyang Z., Fanning A. S., Fu C., Xu J., Marfatia S. M., Chishti A. H., Crompton A., Chan A. C., Anderson J. M., Cantley L. C. (1997) Science 275, 73–77 [DOI] [PubMed] [Google Scholar]
- 46.Harris B. Z., Lim W. A. (2001) J. Cell Sci. 114, 3219–3231 [DOI] [PubMed] [Google Scholar]
- 47.Mayasundari A., Ferreira A. M., He L., Mahindroo N., Bashford D., Fujii N. (2008) Bioorg. Med. Chem. Lett. 18, 942–945 [DOI] [PubMed] [Google Scholar]
- 48.Sun C., Mierke D. F. (2005) J. Pept. Res. 65, 411–417 [DOI] [PubMed] [Google Scholar]
- 49.Reiter E., Lefkowitz R. J. (2006) Trends Endocrinol. Metab. 17, 159–165 [DOI] [PubMed] [Google Scholar]
- 50.Sneddon W. B., Friedman P. A. (2007) Endocrinology 148, 4073–4079 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.