Abstract
To understand the role of microRNAs (miRNAs) in pituitary development, a group of pituitary-specific miRNAs were identified, and Dicer1 was then conditionally knocked out using the Pitx2-Cre mouse, resulting in the loss of mature miRNAs in the anterior pituitary. The Pitx2-Cre/Dicer1 mutant mice demonstrate growth retardation, and the pituitaries are hypoplastic with an abnormal branching of the anterior lobe, revealing a role for microRNAs in pituitary development. Growth hormone, prolactin, and thyroid-stimulating hormone β-subunit expression were decreased in the Dicer1 mutant mouse, whereas proopiomelanocortin and luteinizing hormone β-subunit expression were normal in the mutant pituitary. Further analyses revealed decreased Pit-1 and increased Lef-1 expression in the mutant mouse pituitary, consistent with the repression of the Pit-1 promoter by Lef-1. Lef-1 directly targets and represses the Pit-1 promoter. miRNA-26b (miR-26b) was identified as targeting Lef-1 expression, and miR-26b represses Lef-1 in pituitary and non-pituitary cell lines. Furthermore, miR-26b up-regulates Pit-1 and growth hormone expression by attenuating Lef-1 expression in GH3 cells. This study demonstrates that microRNAs are critical for anterior pituitary development and that miR-26b regulates Pit-1 expression by inhibiting Lef-1 expression and may promote Pit-1 lineage differentiation during pituitary development.
Keywords: Antisense RNA, Gene Regulation, MicroRNA, Pituitary Gland, RNA Silencing, Transcription Regulation, Lef-1, Pit-1, Pitx2, Wnt Signaling
Introduction
The anterior pituitary is an essential endocrine organ in the homeostatic regulation of vertebrates. It exerts its versatile functions through five cell types, including corticotropes secreting ACTH, thyrotropes secreting thyroid-stimulating hormone (TSH),2 gondadotropes secreting luteinizing hormone (LH) and follicle-stimulating hormone (FSH), somatotropes secreting growth hormone (GH), and lactotropes secreting prolactin (PRL). The development of the anterior pituitary is dependent on the expression of spatially and temporally regulated factors, most of which are still unknown (1).
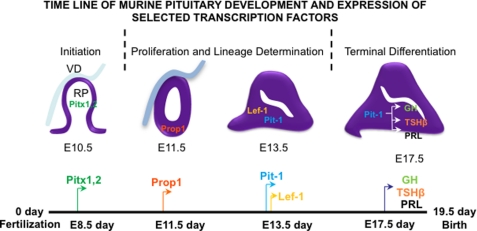
The patterning of the anterior pituitary is directed by a complex mix of morphogenetic signals that regulate development of the endocrine cell type lineages (for a review, see Ref. 1). These signaling molecules include sonic hedgehog (Shh), fibroblast growth factors (FGF), bone morphogenetic proteins, Notch, Wnt factors, and epidermal growth factor (EGF). There are many transcription factors that control early pituitary development and patterning. These include Pitx factors, Lim homeodomain factors (Islet-1, Lhx3, and Lhx4), Hesx1, Prop1, Six homeodomain transcription factor, Pax6, and Lef-1, a component of the Wnt signaling pathway. Pit-1, a POU domain transcription factor, plays a major role in the differentiation of somatotrope, lactotrope, and thyrotrope cell lineages. In concert with these transcription factors, there are other factors that interact to regulate the expression of multiple pituitary hormones (1). A schematic of mouse pituitary development and an expression time line of selected transcription factors are shown in Fig. 1.
FIGURE 1.
Pituitary developmental time line. Shown is a time line of murine pituitary development and expression of selected critical transcription factors and hormones. VD, ventral diencephalons; RP, Rathke's pouch.
The requirement for Lef-1 in pituitary development was shown in the Lef-1−/− null mice with changes in pituitary hormone expression (2, 3). In the Lef-1−/− mutant mouse pituitary, Pit-1 expression was increased at embryonic day 14.5 (E14.5), and the Pit-1 target genes TSHβ and GH were also up-regulated, whereas proopiomelanocortin (POMC) expression (not activated by Pit-1) was unchanged (3). Lef-1 does not activate Pit-1, GH, or TSHβ expression but appears to attenuate Pit-1 expression. β-Catenin is required for Prop1 activation of Pit-1 expression, as noted for Prop1-β-catenin activation of Pit-1 (3).
MicroRNAs (miRNAs) are a class of recently discovered small, endogenous non-coding RNAs that post-transcriptionally regulate expression of protein-coding genes by targeting specific mRNA through complementary sequences in 3′-untranslated regions (3′-UTRs) (4). Animal miRNAs are imperfectly paired to the 3′-UTR of target mRNA and inhibit protein production. These miRNAs can also bind to other regions of the mRNA to degrade the target mRNA or inhibit protein synthesis. Although more than 800 miRNAs have been identified in human and other eukaryotic species, the biological function and the underlying molecular mechanisms of most miRNAs are still unclear. Recent studies revealed expression of many identified miRNAs in distinct areas of the adult mouse central nervous system, which suggests their specific functions within these regions (5). An increasing number of developmental and physiological processes have been found to rely on miRNAs, but the contributions of miRNAs to pituitary development are not known (6–10).
In this report, a global approach was used to study miRNA function in the anterior pituitary by conditionally deleting Dicer1, an endonuclease essential for miRNA maturation. The loss of mature miRNAs leads to dysmorphology and hypoplasia in the anterior pituitary and multiple hormone deficiencies resulting in growth retardation in the Pitx2-Cre/Dicer1 mutant mice. Moreover, we found that Lef-1 was up-regulated in the mutant pituitary, consistent with its role as a repressor of pituitary development (3). We demonstrate that miR-26b directly targets the Lef-1 3′-UTR and represses Lef-1 isoform expression in different cell lines. miR-26b repression of Lef-1 results in an up-regulation of Pit-1 in GH3 pituitary cells. In addition, miR-26b exerts different effects on the Wnt-β-catenin signal response in tissues expressing specific Lef-1 isoforms. These data reveal a new mechanism for the control of Lef-1 expression by miR-26b and a role for multiple miRNAs in regulating hormone expression in the anterior pituitary.
EXPERIMENTAL PROCEDURES
Animals
All animals were housed in the Program of Animal Resources at the Institute of Biosciences and Technology, and were handled in accordance with the principles and procedures of the Guide for the Care and Use of Laboratory Animals (34). All experimental procedures were approved by the Texas A&M Health Science Center Institutional Animal Care and Use Committee. Mice carrying floxed Dicer1 alleles (Dicer1flox/flox) (11) were mated to mice carrying a Pitx2-Cre (12) to generate Pitx2-Cre;Dicer1flox/wt mice. Pitx2-Cre;Dicer1flox/wt mice were subsequently mated to Dicer1flox/flox mice to generate Pitx2-Cre;Dicer1flox/flox mutant mice. Embryos were collected at various time points, considering the day of observation of a vaginal plug to be E0.5. Mice and embryos were genotyped by PCR of DNA extracted from tail biopsies or yolk sacs. Twenty-four mice (mutant, n = 9; wild type, n = 15) from three different litters were subject to the growth curve study. Animals were housed together with littermate controls during the study. The environment of the animal rooms was controlled with a 12-h/12-h light/dark cycle, a relative humidity of 45–55%, and a temperature of 22 °C. The mice had free access to tap water and standard pellet chow. The treatment was blind to the mice genotype. Body weights were taken at noon at the specified time points.
MicroRNA Microarray
Pituitary cell lines were cultured and cells were harvested by scraping and pelleted by centrifugation, and P0 mice tissue was harvested by dissection. Total RNAs, including miRNAs, were prepared using the miRNeasy minikit from Qiagen. LC Sciences performed the miRNA microarray analyses. Microarray assays were performed on a mParaFlo microfluidics chip, and all samples underwent quality control analyses immediately after receipt as well as at each step through the process.
Expression and Reporter Constructs
The expression plasmid containing the cytomegalovirus (CMV) promoter linked to the mmu-miR-26b and mmu-miR-21 precursor was constructed in pSilencer 4.1 (Ambion). Lef-1 ΔN, Lef-1 FL, and β-catenin S37A expression plasmids were constructed in pcDNA 3.1 MycHisC (Invitrogen) as described previously (13). Lef-1 3′-UTR and Lef-1 mutant 3′-UTR generated by mutagenesis were directionally cloned into the pGL3 CXCR4 1P (Addgene, plasmid 11310), (14), replacing the siRNA binding site by XbaI and ApaI. The 7× TopFlash reporter plasmid was constructed into luciferase vector by inserting seven Lef/Tcf binding sites upstream of the minimal thymidine kinase promoter. The Pit-1 promoter and Pit-1 expression vector have been reported previously (15, 16). The shRNA constructs against rat Lef-1 were from Origene (TG712526). All constructs were confirmed by DNA sequencing.
Hematoylin and Eosin (H&E) Staining and Immunohistochemistry
Samples were fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin wax. Sections were cut (7 μm) and stained with H&E. Immunohistochemistry was done on 7-μm paraffin sections stained by an indirect immunoperoxidase method. Peroxidase activity was visualized with AEC stain kit (Sigma). Antibodies were obtained and diluted as follows: Pit-1 (Abcam, ab10623), 1:50; TSHβ (Lifespan, LS-C42182), 1:200; PRL, LHβ (National Hormone and Peptide Program), 1:200; ACTH (Abcam, ab74976), 1:200; GH (Abcam, ab8490), 1:100; Lef-1 (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), sc-8591), 1:200. Secondary antibodies conjugated with biotin were from Vector Laboratories and were used at 1:150 dilution.
Cell Culture, Transient Transfections, Luciferase, and β-Galactosidase Assays
CHO, HEK 293 FT, GH3, αT3, and AtT20 cells were cultured in DMEM supplemented with 5 or 10% FBS and penicillin/streptomycin and transfected by electroporation. The procedures for transient transfections and luciferase and β-galactosidase assays were described previously (17). Transfected cells were incubated for 48 h. LiCl was added to the appropriate cells at a final concentration of 10 mm 23 h before harvest. The pcDNA3.1 empty vector or pSilencer 4.1 negative control vectors were added to equalize the total amount of co-transfected expression vectors.
Chromatin Immunoprecipitation (ChIP) Analysis
The ChIP analysis was performed as described (18) using the ChIP assay kit (Upstate Biotechnology, Inc.) with the following modifications. GH3 cells were fed for 24 h, harvested, and plated in 60-mm dishes. Cells were cross-linked with 1% formaldehyde for 10 min at 37 °C the next day. Samples were incubated with Lef-1 2D12 mouse monoclonal antibody from Millipore overnight at 4 °C. Immune complexes were washed consecutively for 5 min with each of the following solutions: low salt immune complex wash buffer, high salt immune complex wash buffer four times, and LiCl immune complex wash buffer and TE buffer twice. An aliquot of the immunoprecipitated DNA (3 μl) from non-transfected cells was used for PCR (32 cycles). All reactions were done under an annealing temperature of 61 °C. Two primers for amplifying the Lef/Tcf binding site in the Pit-1 promoter are as follows: sense, 5′-TTGCCTTCTCCTCACTTCGT-3′; antisense, 5′-TATCAGCCGAGGTGAAAGGT-3′. All of the PCR products were evaluated on a 1% agarose gel in 1× TBE for appropriate size (322 bp) and confirmed by sequencing. As controls, the primers were used without chromatin, and normal mouse IgG was used replacing the Lef-1 antibody to reveal nonspecific immunoprecipitation of the chromatin.
Real-time PCR Analyses and miRNA Expression
Total RNA, including miRNA, were prepared using the miRNeasy minikit from Qiagen. The amount and integrity of the RNA samples were assessed by measurement at 260 and 280 nm and gel analyses. HEK 293 FT cells transfected with control vector or miR-26b precursor were harvested 48 h post-transfection. Quantitative real-time PCR for miR-26b, miR-24, and miR-375 mature expression was done with TaqMan MicroRNA assay probes (Applied Biosystems), including miR-24 expression as a reference miR. GH3 cells transfected with control vector, miR-26b precursor, Pit-1 expression vector, and miR-21 precursor were harvested 30 h post-transfection. Total RNA was reverse transcribed into cDNA by the iScript Select cDNA Synthesis kit (Bio-Rad). Real-time PCR was carried out in a total reaction of 25 μl containing 12.5 μl of iQ SYBR Green Supermix, 0.1 μm forward primer, 0.1 μm reverse primer, 0.25 μl of cDNA template in the MyiQ Singlecolor real-time detection system and analyzed by the MyiQ optical system software 2.0 (Bio-Rad). β-Actin served as a reference gene for normalization of GH mRNA levels. Rat GH and β-actin primer sequences were from previous reports (19, 20). Lef-1 primer sequences were 5′-GCAGCTATCAACCAGATCC-3′ (forward) and 5′-GATGTAGGCAGCTGTCATTC-3′ (reverse). Dicer1 primer sequences were 5′-GTGGTCTGGCAGGTGTACTA-3′ (forward) and 5′-ACTTCCACGGTGACTCTGAC-3′ (reverse). The thermal cycling profile consisted of 95 °C for 4 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and elongation at 72 °C for 18 s. Samples were run in triplicate. No-template control was run in each experiment. Melting curve analyses were performed to confirm amplification specificity of the PCR products, and all PCR products were sequenced to confirm their identity.
Western Blot Assays
Expression of endogenous expressed Lef-1 FL and Lef-1 ΔN isoforms were identified in CHO, 293, AtT20, and αT3 cells using the Lef-1 antibody (Cell Signaling, C18AT). Lef-1 expression in the rat GH3 cells was detected using the Lef-1 2D12 mouse monoclonal antibody from Millipore. Pit-1 expression was detected in GH3 cells using the Pit-1 mouse monoclonal antibody from Abcam. Approximately 20–30 μg of transfected cell lysates were analyzed in Western blots. Following SDS-gel electrophoresis, the proteins were transferred to PVDF filters (Millipore), immunoblotted, and detected by specific antibodies and ECL Plus reagents from GE Healthcare.
Statistics
Statistics were performed by two-sample t test. p values less than 0.05 were considered to be significant.
RESULTS
Identification of Specific Pituitary miRNAs
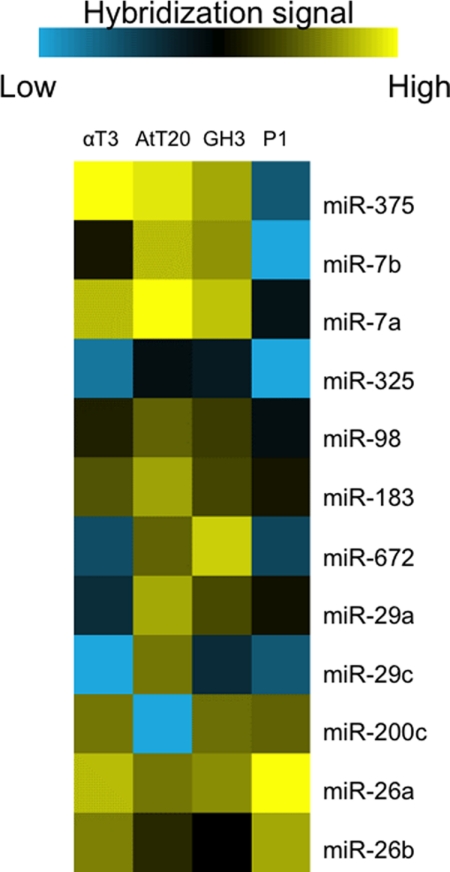
To identify pituitary-specific miRNAs, αT3 (gonadotrope), AtT20 (corticotrope), and GH3 (somatotrope) cell lines were used to harvest miRNAs. Total miRNAs were fluorochrome-labeled and hybridized to spotted miRNA microarrays, comprising probes for all mouse/rat miRNAs in the latest version of the Sanger miRBase database (Release 14.0). We compared the miRNA expression profiles with P1 newborn whole mouse miRNAs and then identified several pituitary-specific miRNAs with high levels of expression in the pituitary cell lines (Fig. 2). We hypothesized that some of the miRNAs directly targeted critical pituitary transcription factors to regulate pituitary development and terminal differentiation. Previous miRNA array analysis on adult mouse pituitary (5) and our miRNA array analysis on the three murine pituitary cell lines identified expression of the miR-26 family in the pituitary.
FIGURE 2.
Microarray analyses of three murine pituitary cell lines. Shown is a heat map of miRNA expression. The miR-26 family is highly expressed in all three pituitary cell lines as well as in the mouse whole body. αT3 (gonadotrope), AtT20 (corticotrope), and GH3 (somatotrope) cell lines were used to harvest miRNAs and compared with P1 newborn whole mouse miRNAs.
Knock-out of Pituitary Mature miRNAs Results in Abnormal Pituitary Development
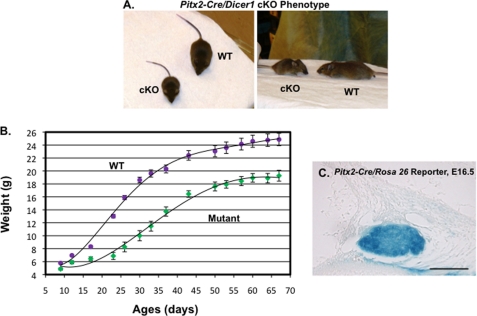
To study the overall function of miRNAs in pituitary development; we mated the mice carrying floxed Dicer1 alleles (Dicer1 flox/flox) (21) to the mice carrying a Pitx2-Cre allele (12) to generate Pitx2-Cre/Dicer1flox/wt mice. Pitx2-Cre/Dicer1flox/wt mice were subsequently mated to Dicer1flox/flox mice to generate Pitx2-Cre/Dicer1flox/flox mutant mice. The mutant mice are born the same size with their wild type littermates, but they begin to show growth retardation immediately after birth with an average body size about half that of the wild type mice at 4 weeks of age (Fig. 3A). The comparison of body weight between wild type and mutant mice at specified time points was recorded on the growth curve (Fig. 3B). Twenty-four mice (mutant, n = 9; wild type, n = 15) from three different litters were subject to the growth curve study. The error bars indicate the S.E. Pitx2-Cre expression occurs specifically in the anterior pituitary, oral epithelium, eyes, and limbs, and the only region affecting growth would be the anterior pituitary gland, which regulates the endocrine system through hormone expression (1, 12). Pitx2-Cre activity was observed in the lineage of all cell types of the anterior pituitary at E16.5, confirmed by analyses of LacZ activation (X-gal stain) in the Pitx2-Cre/Rosa 26 reporter mice (Fig. 3C).
FIGURE 3.
Phenotype of Pitx2-Cre/Dicer1 conditional knock-out (cKO) mouse. A, the mutant mouse (left) at 4 weeks old is significantly smaller compared with the wild type mouse (right). B, growth curve of wild type and Dicer1 mutant mice (mutant, n = 9; wild type, n = 15). Error bars, S.E. The mutant mice are growth-retarded as adults. C, Pitx2-Cre is highly expressed in the anterior pituitary at E16.5 as assayed by analysis of Rosa 26 reporter/LacZ activation (X-gal stain). These data demonstrate that Cre expression driven by Pitx2 is expressed in all pituitary cell lineages.
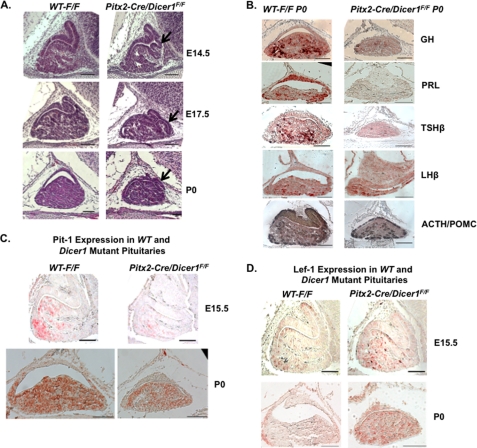
The morphology defects of the anterior pituitary were observed from E14.5. The anterior pituitary in the P0 Dicer1 mutant mouse is hypoplastic. A ventral bulge was observed near the mid-sagittal plate from E14.5 to P0 (Fig. 4A). Immunohistochemistry was performed on P0 wild type (F/F) and Pitx2-Cre/Dicer1 mice pituitaries to determine hormone expression levels using GH, PRL, TSHβ, LHβ, and ACTH antibodies (the ACTH antibody recognizes both ACTH and POMC). GH, PRL, and TSHβ expression in mutant pituitaries was decreased, whereas ACTH/POMC and LHβ expression was not affected compared with wild type pituitaries (Fig. 4B). These results suggest defects in hormone expression in somatotropes, lactotropes, and thyrotropes. The decreased GH and TSHβ correlate with the mice phenotype of growth retardation. The presence of pituitary hormones (albeit at low levels) by specific cell lineages suggests normal cell type differentiation; however, defects in cell proliferation could contribute to the hypoplastic pituitary. The unchanged ACTH/POMC level suggests relatively normal development of the corticotrope cell lineage.
FIGURE 4.
Pituitary dysmorphology, hypoplasia, and hormone deficiencies in the Pitx2-Cre/Dicer1 conditional knock-out (cKO) mouse. A, hematoxylin and eosin-stained pituitaries reveal an abnormal ventral bulge in the anterior lobe and hypoplasia in Pitx2-Cre/Dicer1 mutant pituitaries compared with wild type mice from E14.5 to P0. B, hormone deficiency in P0 Pitx2-Cre/Dicer1 mutant mice. Immunohistochemistry in P0 pituitaries demonstrated decreased expression of GH, PRL, TSHβ, and LHβ in the mutant pituitary, whereas ACTH/POMC expression is similar to wild type. C, immunohistochemistry using a Pit-1 antibody reveals decreased Pit-1 expression at E15.5 and P0 in mutant pituitaries compared with wild type pituitaries. D, immunohistochemistry using a Lef-1 antibody demonstrates increased Lef-1 expression in E15.5 and P0 mutant pituitaries compared with wild type pituitaries. WT, wild type mice; scale bars, 100 μm. All histology panels are sagittal sections of embryonic or P0 pituitary.
The generation of somatotrope, lactotrope, and thyrotrope cell lineages depends on the function of the tissue-specific POU-class homeodomain transcription factor, Pit-1 (officially Pou1f1) (22). The decreased GH, PRL, and TSHβ expression suggested suppressed Pit-1 function, and we found that Pit-1 expression was decreased at E15.5 and P0 (Fig. 4C). Among numerous transcription factors that regulate Pit-1 expression and Pit-1 lineage development, Lef-1 has been reported as a potent repressor (3). Lef-1 is a DNA-binding protein and is expressed in the pituitary gland, teeth, kidney, and skin during mouse embryonic development (2). The Ensemble database records four Lef-1 isoforms transcribed by two different promoters. The P1 promoter generates three long isoforms by alternative splicing (the longest isoform is defined as Lef-1 full-length (FL)), and the P2 promoter generates a short isoform lacking a β-catenin binding domain (Lef-1 ΔN), which is considered a dominant-negative isoform (23, 24).
In the pituitary, Lef-1 expression reappears at E13.5 in the anterior gland following initial transient expression at E9.0 (3). In the Lef-1 knock-out mouse pituitary, not only Pit-1 but also hormones secreted by Pit-1 lineage cells are up-regulated (3). The Prop1 mutant mice phenotype and a previously reported mechanism suggest that Lef-1 represses the activity of the Prop1 and β-catenin complex on the Pit-1 promoter (3, 25). Consistent with this mechanism, Lef-1 expression was up-regulated in the Dicer1 mutant anterior pituitary at E15.5 and P0 (Fig. 4D), which would contribute to decreased Pit-1 expression. Due to the absence of mature miRNAs by pituitary-specific Dicer1 deletion, Lef-1 up-regulation may be attributed to the loss of specific miRNA regulators that repress Lef-1 expression.
Pitx2-Cre Dicer Deletion Knocks Down Mature miRNAs in the Pituitary
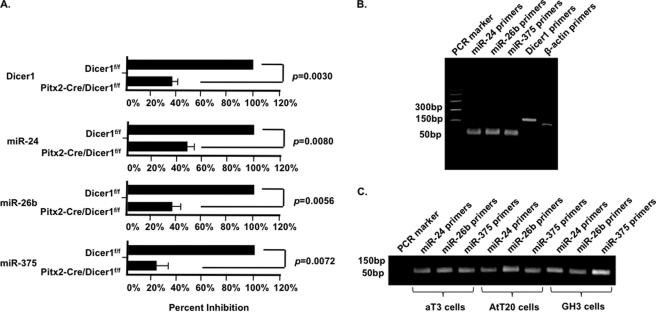
Mature miRNAs are decreased in the Pitx2-Cre/Dicer1f/f mutant mice. The pituitaries of mutant and wild type mice were dissected 4 weeks after birth. Real-time RT-PCR experiments demonstrate reduced Dicer expression in the mutant pituitaries (Fig. 5A). The mature miRNAs of unrelated miRNA families, miR-24, miR-26b, and miR-375, were measured by real-time PCR using Taqman probes (Fig. 5A). β-Actin served as the reference gene for normalization of miRNA and Dicer levels. The PCR products from dissected pituitaries of wild type 4-week mice are shown in Fig. 5B. The PCR products from pituitary cell lines are shown in Fig. 5C. Experiments were repeated three times each from multiple samples, and p values are shown. The residual miRNA and Dicer1 expression is derived from the posterior pituitary where Pitx2-Cre is not expressed, and we are unable to reliably dissect the posterior region from the anterior region.
FIGURE 5.
miRNAs are down-regulated in the Pitx2-Cre Dicer1f/f mutant mice. A, real-time RT-PCR of Dicer1 expression and Dicer1-dependent miRNA expression in normal and mutant pituitaries. Four-week-old Dicerf/f and Pitx2-Cre/Dicerf/f pituitaries were dissected, and total RNA was isolated for real-time RT-PCR using Dicer1 primers and TaqMan probes for miR-24, miR-26b, and miR-375. β-Actin primers were used as a reference. B, real-time PCR products from the analyses in A. C, RT-PCR products using the miRNA probes and RNA extracted from pituitary cell lines. The Dicer1 and Dicer1-dependent miRNAs are significantly down-regulated. The residual miRNA and Dicer1 expression comes from the posterior part of the pituitary, where Pitx2-Cre is not expressed, which was not dissected from the anterior pituitary region. Error bars, S.E.
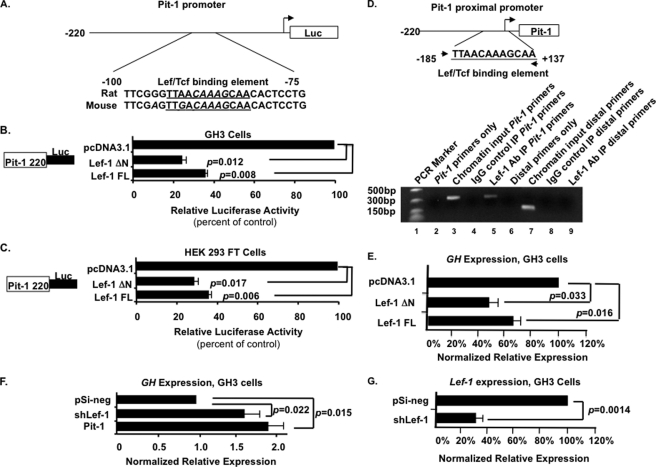
Lef-1 Represses Pit-1 Promoter Activity
The Pit-1 promoter contains a Lef-1/Tcf DNA binding element, which is conserved in rats and mice (Fig. 6A). The Pit-1 220-bp minimal promoter was co-transfected with the Lef-1 FL or Lef-1 ΔN expression plasmids into both GH3 and HEK 293 FT cells. Both isoforms significantly repressed Pit-1 promoter activity ∼3-fold in these cells (Fig. 6, B and C). Because HEK 293 FT cells do not express pituitary factors like Prop1 and Pit-1, the background levels of luciferase activity from the Pit-1 promoter are relatively low. In the GH3 cells, the background levels of luciferase activity from the Pit-1 promoter were increased due to endogenous factors, such as Pit-1. However, because Lef-1 repressed the Pit-1 promoter in both GH3 and HEK 293 FT cells, these data revealed an alternative mechanism of Lef-1 repression independent of Prop1 and other pituitary factors.
FIGURE 6.
Lef-1ΔN and Lef-1 FL isoforms repress Pit-1 promoter activity. A, schematic of the minimal Pit-1 220-bp promoter with the Lef-1 DNA binding element. B, Lef-1 isoforms repress Pit-1 promoter activity. The pA3-Luc reporter plasmid with rat Pit-1 220-bp promoter upstream of the luciferase gene was co-transfected with Lef-1ΔN or Lef-1 FL isoforms in GH3 cells. β-Galactosidase was used as an internal control for transfection efficiency. Error bars, S.E. p values are shown. C, the Pit-1 promoter was co-transfected into HEK 293 FT cells with expression plasmids Lef-1ΔN or Lef-1 FL isoforms. D, a chromatin immunoprecipitation assay reveals endogenous Lef-1 binding to the Pit-1 promoter chromatin in GH3 cells. The Pit-1 promoter with Lef-1 binding site and location of PCR primers are shown at the top. The gel with the specific PCR products from the immunoprecipitated chromatin and controls are shown at the bottom. E, GH expression is decreased in GH3 cells transfected with Lef-1 isoforms. Real-time RT-PCR experiments demonstrate that Lef-1 represses endogenous GH expression. F, GH expression was increased in GH3 cells transfected with shRNA against rat Lef-1. pSilencer 4.1 negative scrambled control vector (pSi-neg) served as a negative control. Pit-1 expression vector served as a positive control. G, shRNA against rat Lef-1 leads to 70% knockdown of endogenous Lef-1 in GH3 cells measured by real-time PCR. Experiments were repeated three times, and all products were confirmed by sequencing. Error bars, S.E. p values are shown.
A ChIP assay demonstrates endogenous Lef-1 binding to the Pit-1 promoter. Non-transfected GH3 cells were used because these cells endogenously express Lef-1 and Pit-1. Mouse Lef-1 monoclonal antibody was used, and Pit-1 promoter chromatin was amplified by PCR using primers specific for the Pit-1 promoter flanking the Lef/Tcf binding site (Fig. 6D). The primers amplified a 322-bp product from the antibody IP and chromatin input (Fig. 6D, lanes 5 and 3, respectively). The primers did not produce the product from normal mouse IgG IP control (Fig. 6D, lane 4). More controls included using primers that amplify a distal downstream region (exon 3) of the Pit-1 gene. The primers recognize the input chromatin (lane 7) but not the Lef-1 antibody IP product (lane 9) or the IgG control (lane 8). These data correlate with the co-transfection data to demonstrate the direct repression of the Pit-1 promoter by Lef-1. Considering this, combined with the previously reported in vivo study and Prop1-related repression mechanism, we conclude that Lef-1 is a potent repressor of Pit-1 expression. The expression of ectopic Lef-1 isoforms has been reported (13). Furthermore, real-time RT-PCR of Lef-1 isoform-transfected GH3 cells reveals a 55% decrease in GH expression by Lef-1 ΔN and a 35% decrease in GH by Lef-1 FL (Fig. 6E). To confirm the specific role of Lef-1 in regulating GH expression, shRNA against rat Lef-1 significantly increased GH expression in GH3 cells (Fig. 6F). As a control we demonstrate that shLef-1 decreases Lef-1 expression by 70% in transfected GH3 cells (Fig. 6G). Thus, Lef-1 represses Pit-1 and subsequently GH expression.
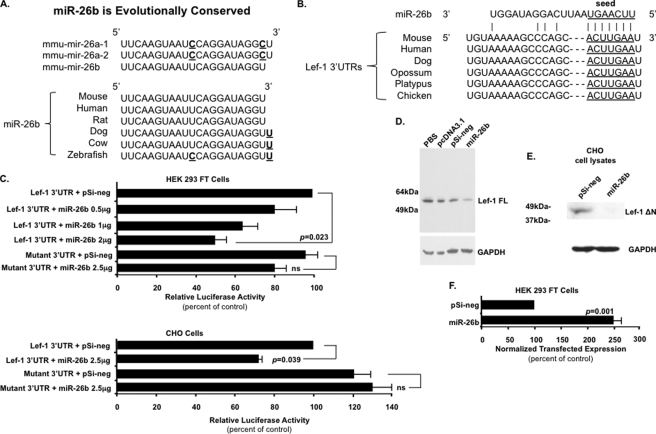
miR-26b Targets Lef-1 Expression
The immunohistochemistry data revealed increased Lef-1 expression in the Dicer1 mutant mice, suggesting a potential role for miRNA regulation of Lef-1. Because we are interested in the role of Lef-1 during developmental processes and Lef-1 plays a major role in pituitary development, we asked if Lef-1 was a target of miRNAs and which miRNAs regulate Lef-1 expression. miRNA target prediction programs like TargetScan and PicTar revealed several miRNAs that have conserved binding sequences in the Lef-1 3′-UTR among vertebrates, including the miR-26 family. The miR-26 family contains miR-26a-1, miR-26a-2, and miR-26b, which have independent transcription mechanisms but highly identical mature sequences, including the same seed sequence (Fig. 7, A and B). The miRNA microarray analyses revealed that the miR-26b mature sequence was among the 20 highest expressed miRNAs in adult mouse pituitary and murine pituitary cell lines. Combined with the significant increase in Lef-1 expression in the mutant mice pituitary, the miR-26 family may be an important miRNA regulator of Lef-1 in mouse pituitary development. To investigate the miR-26 family function and relation with Lef-1, we cloned miR-26b into the pSilencer 4.1 vector and murine Lef-1 3′-UTR (which is shared by all Lef-1 transcripts) into the pGL3 vector between the luciferase ORF and SV40 PA terminator. The luciferase assay revealed that miR-26b repressed the Lef-1 3′-UTR luciferase activity in HEK 293 FT and CHO cells, whereas it did not repress the Lef-1 mutant 3′-UTR, which had six nucleotide mutations in the predicted target site (ACUUGAA to UGAACUU; Fig. 7C). A titration of miR-26b revealed a dose response to the repression of the Lef-1 3′-UTR, as shown in HEK 293 cells (Fig. 7C). In HEK 293 FT cells, only the endogenous Lef-1 FL isoform was detected by Western blot, and transfection of miR-26b knocked down Lef-1 FL expression (Fig. 7D). CHO cells endogenously express only the Lef-1 ΔN isoform (13), and this isoform was knocked down by transfection of miR-26b (Fig. 7E). As a control, the miR-26b expression level in transfected HEK 293 FT cells was assessed by quantitative PCR. Mature miR-26b was significantly increased in cells transfected with the miR-26b precursor vector (Fig. 7F). miR-24 served as the reference gene for normalization of miR-26 expression levels in each experiment.
FIGURE 7.
MicroRNA-26b directly targets the Lef-1 3′-UTR and represses Lef-1 expression. A, microRNA-26 (miR-26) family members are highly identical, and miR-26b is evolutionarily conserved among several vertebrate species. B, the potential miR-26b binding sites in the Lef-1 3′-UTR are highly conserved among different species. C, miR-26b targets the Lef-1 3′-UTR. The 3′-UTR of Lef-1 mRNA, and the Lef-1 mutant 3′-UTR with mutations engineered in the region complementary to the miR-26b seed region (ACUUGAA to UGAACUU), were inserted into the pGL3 vector. Transfections of CHO cells and HEK 293 FT cells were performed using the indicated combinations of plasmids. CHO and HEK 293 FT cells were transfected as previously described (13). To control for transfection efficiency, all transfections included the SV-40 β-galactosidase reporter (0.5 μg). Cells were incubated for 48 h and then assayed for luciferase and β-galactosidase activities. The activities are shown as mean -fold activation compared with the luciferase plasmid without miRNA expression and normalized to β-galactosidase activity ± S.E. from three independent experiments. p values are shown; ns, not significant; pSi-neg, pSilencer 4.1 negative control vector. D, miR-26b represses endogenous Lef-1 FL isoform expression. Western blot of Lef-1 FL protein in control or miR-26b precursor transfected HEK 293 FT cells 48 h post-transfection. Expression of other Lef-1 isoforms were not detected in HEK 293 cells. GAPDH is shown as a loading control. PBS, pcDNA3.1 empty vector, and pSi-neg vector served as controls. E, miR-26b represses endogenous Lef-1 ΔN short isoform (lacks the β-catenin binding domain) expression. Western blot of Lef-1 ΔN protein in control or miR-26b precursor-transfected CHO cells 48 h post-transfection. GAPDH is shown as a loading control. The Lef-1 FL isoform was not detected in CHO cells. F, miR-26b expression level in transfected HEK 293 FT cells assessed by quantitative PCR. Mature miR-26b was significantly up-regulated in cells transfected with miR-26b precursor vector. This experiment was done to demonstrate that transfected miR-26b was expressed at high levels. miR-24 expression level served as the reference gene for normalization of miR-26b expression in each experiment. Error bars, S.E. p values are shown.
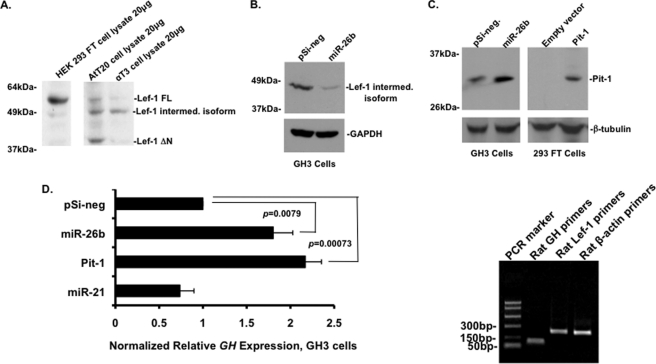
miR-26b Represses Lef-1 and Increases Pit-1 Expression in GH3 Cells
Consistent with the Lef-1 immunohistochemistry data, Lef-1 was widely expressed in pituitary cell lines. The AtT20 mouse pituitary cell line expressed Lef-1 FL, Lef-1 ΔN, and an intermediate isoform, whereas the αT3 pituitary cell line expressed an intermediate isoform (Fig. 8A). The Lef-1 intermediate isoform (containing the β-catenin binding domain) is derived from alternative splicing of the full-length Lef-1 RNA and has been identified in human and mouse tissues and cultured cells (26–28). The increased Lef-1 expression and subsequent decreased Pit-1 expression in the Dicer1 mutant pituitary could be due to several pituitary miRNAs. Thus, we asked if miR-26b effectively repressed Lef-1 expression in a pituitary cell line endogenously expressing Lef-1 and Pit-1. GH3 cells are of rat origin and appear to express only the alternatively spliced Lef-1 intermediate isoform, and transfected miR-26b effectively repressed the endogenous Lef-1 intermediate isoform in GH3 cells (Fig. 8B). miR-26b was transfected in GH3 cells, and the lysates were probed for Pit-1 expression. As expected, Pit-1 expression was increased in the presence of miR-26b in the GH3 cells (Fig. 8C). As a control, we demonstrate that HEK 293 FT cells do not endogenously express Pit-1, but transfected Pit-1 is shown by Western blot. Furthermore, we assessed GH expression levels in miR-26b-transfected cells by real-time PCR (Fig. 8D). GH expression was significantly increased in miR-26b-transfected cells compared with cells transfected with pSilencer negative control or miR-21 (which served as a negative control). GH expression activated by transfected Pit-1 was used as a positive control. β-Actin served as the reference gene for normalization of GH mRNA levels. The PCR products of GH, Lef-1, and β-actin primers were subject to gel analyses to demonstrate primer specificity (Fig. 8D; all PCR products were sequenced to confirm their identity). Taken together, these data reveal a novel pathway and mechanism for the regulation of Lef-1 expression by miR-26b and the subsequent regulation of Pit-1 and GH expression.
FIGURE 8.
miR-26b represses Lef-1 intermediate isoform expression and activates Pit-1 expression in GH3 cells. A, Western blot shows endogenous expression of Lef-1 FL isoform in HEK 293 FT cells. ATt20 cells express both Lef-1ΔN and Lef-1 FL isoforms as well as an intermediate isoform. αT3 cells mainly express the intermediate isoform. 20 μg of cell lysates were loaded in each lane. B, miR-26b repressed Lef-1 intermediate isoform expression in GH3 cells. GH3 cells were transfected with miR-26b, and endogenous Lef-1 expression was detected with the Lef-1 antibody from Millipore. GAPDH expression served as a loading control. C, transfected miR-26b in GH3 cells reveals increased endogenous Pit-1 expression. As a control, Pit-1 was transfected in 293 cells, and Pit-1 was detected using the Pit-1 antibody. β-Tubulin expression acted as a loading control. D, GH mRNA expression was increased in miR-26b-transfected GH3 cells. miR-21 was used as a control and does not affect GH expression. GH expression in Pit-1-transfected cells was used as a positive control. β-Actin served as the reference gene for normalization of GH mRNA levels. Experiments were repeated three times each from multiple samples, and p values are shown. The PCR products from the GH, Lef-1, and β-actin primers are shown, and all products were sequenced to confirm their identity. Error bars, S.E.
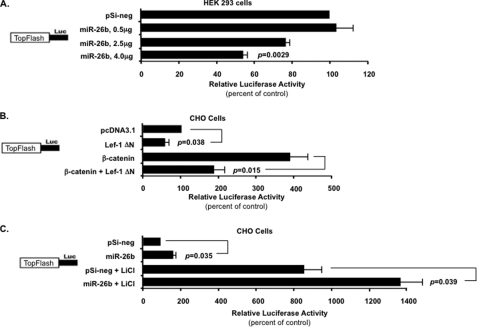
miR-26b Inhibits the TopFlash Reporter in HEK 293 FT Cells but Activates TopFlash in CHO Cells
Lef-1 is an important component involved in the Wnt-β-catenin signal response. Without the Wnt-β-catenin signal, the Lef-1 FL isoform is associated with the TLE-Groucho complex and represses Wnt target gene expression. When the Wnt-β-catenin signal is activated, β-catenin binds to Lef-1 FL and replaces the TLE-Groucho complex to activate Wnt target gene expression (29). The Lef-1 ΔN isoform lacking the N-terminal β-catenin binding domain represses Wnt target gene expression. miR-26b may affect the Wnt-β-catenin signal response by inhibiting Lef-1 expression, whereas other β-catenin targets, such as Tcf factors, may neutralize the effect. To test if the Wnt-β-catenin signal response was affected by miR-26b, we performed the TopFlash assay with the luciferase vector containing seven Lef/Tcf binding sequences upstream of the thymidine kinase minimal promoter. Transfection of the miR-26b expression plasmid in HEK 293 FT cells (which endogenously express the Lef-1 FL isoform) revealed a direct repression of TopFlash reporter activity with increasing concentrations of miR-26b (Fig. 9A). The Lef-1 ΔN isoform was co-transfected in CHO cells with the TopFlash reporter, and as expected, it repressed the TopFlash reporter (Fig. 9B). Furthermore, co-transfection of constitutively active β-catenin activated the TopFlash reporter in CHO cells (Fig. 9B). However, the β-catenin activation is attenuated by Lef-1 ΔN isoform co-expression, which cannot bind β-catenin and competes with other Tcf factors for DNA binding elements in the TopFlash reporter (Fig. 9B). In CHO cells, miR-26b activates the TopFlash reporter by repressing Lef-1 ΔN isoform expression (Fig. 9C). The addition of LiCl to the CHO cell media, which stimulates β-catenin nuclear localization, caused an increase in TopFlash reporter activity. Transfection of miR-26b resulted in a further increase in TopFlash activity due to the repression of Lef-1 ΔN expression (Fig. 9C).
FIGURE 9.
miR-26b represses or activates 7× TopFlash reporter activity dependent on the Lef-1 isoform expressed in a specific cell line. A, the 7× TopFlash reporter plasmid was constructed into a luciferase vector by inserting seven Lef/Tcf binding sites upstream of the minimal thymidine kinase promoter and co-transfected into HEK 293 FT cells (which express only the β-catenin-associated Lef-1 FL isoform) with titrated amounts of miR-26b precursor plasmid. Co-transfection of increased amounts of miR-26b plasmid resulted in a corresponding decrease in TopFlash reporter luciferase activity. The pSilencer 4.1-negative (pSi-neg) control plasmid was transfected to equalize the total amount of co-transfected expression vector. Transfections were performed as described in the legend to Fig. 6. B, the Lef-1 ΔN isoform represses TopFlash reporter activity. The Lef-1 ΔN isoform lacks the β-catenin binding domain and is a dominant negative Lef-1 isoform (only Lef-1 isoform expressed in CHO cells). The TopFlash reporter plasmid was co-transfected in CHO cells with the Lef-1 ΔN isoform expression plasmid and/or the constitutively active β-catenin expression plasmid. The pcDNA3.1 empty vector was added to equalize the total amount of co-transfected expression vectors. C, miR-26b activates 7× TopFlash reporter in CHO cells. The 7× TopFlash reporter plasmid was co-transfected into CHO cells with the pSilencer 4.1-negative control vector or the miR-26b precursor plasmid. LiCl (10 mm) was added to the indicated cell media. CHO cells do not endogenously express the Lef-1 FL isoform but do express the Lef-1 ΔN isoform, which is not responsive to β-catenin (13). Therefore, repression of Lef-1 ΔN expression by miR-26b increases TopFlash reporter activity due to other endogenous Tcf factors. The activities are shown as mean -fold activation compared with the luciferase promoter plasmid without miRNA or β-catenin expression and normalized to β-galactosidase activity ± S.E. (error bars) from 2–3 independent experiments. p values are shown; ns, not significant.
DISCUSSION
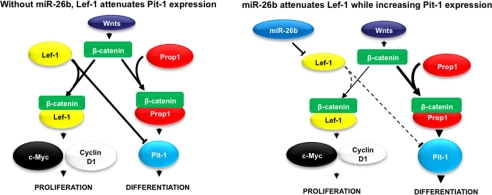
This study reveals an essential role of miRNAs in development of the anterior pituitary. Loss of Dicer1 function in the anterior pituitary progenitor cells leads to anterior pituitary branching; decreased GH, PRL, and TSHβ expression; and growth retardation of the mutant mice. Pit-1 was down-regulated, resulting from increased Lef-1 expression, which can attenuate Pit-1 expression. Loss of mature miRNAs increased Lef-1 expression and revealed that Lef-1 expression in the pituitary was regulated by miRNAs. We demonstrate that miR-26b directly targets the Lef-1 3′-UTR and represses Lef-1 isoform expression in pituitary and non-pituitary cell lines. miR-26b inhibition of Lef-1 increases Pit-1 and GH expression in GH3 cells (Fig. 10).
FIGURE 10.
Model for the mechanism of miR-26b regulation of Lef-1 and Pit-1. Without miR-26b, β-catenin binds Lef-1 or Prop1 in response to Wnt signaling. The Lef-1-β-catenin complex activates c-Myc and cyclin D1 promoters and enhances cell proliferation. The Prop1-β-catenin complex activates the Pit-1 promoter and triggers terminal differentiation of pituitary cell types. However, during normal pituitary development, miR-26b expression reduces Lef-1 attenuation of the Pit-1 promoter by inhibiting Lef-1 expression. This mechanism would promote cell lineage differentiation at later stages of development by Pit-1.
A previous study revealed that Lef-1 interacts with Prop1 and represses the activity of the Prop1-β-catenin complex on the Pit-1 promoter (3). However, an independent Prop1-β-catenin mechanism is revealed, where Lef-1 directly binds the Wnt response element in the Pit-1 promoter and represses Pit-1 activation in concert with other inhibitory factors (Groucho) without β-catenin. The dominant negative Lef-1 ΔN isoform is not responsive to β-catenin and may negatively regulate Pit-1 expression independently of β-catenin. The Lef-1 ΔN isoform may also bind the Wnt response element in the Prop1 promoter and indirectly attenuate Pit-1 expression. Therefore, distinguishing Lef-1 isoform expression patterns during pituitary development would be critical to understanding the overall role of Lef-1 in pituitary development.
Wnt signaling is critical for pituitary cell proliferation and differentiation. β-Catenin is a cofactor for the Lef-1 and Prop1 transcription factors that bind to specific gene targets. The Lef-1-β-catenin complex activates the c-Myc and cyclin D1 promoters to enhance cell proliferation (30, 31), whereas Prop1-β-catenin activates the Pit-1 promoter and triggers terminal differentiation (3). miR-26b may regulate a differentiation pathway by reducing the Lef-1 inhibition of Pit-1 expression (Fig. 10). However, the underlying mechanism may be more complicated. Reducing Lef-1 expression by miR-26b may shift more β-catenin to the Prop1-β-catenin complex and enhance Prop1-specific targets like Pit-1 to promote cell differentiation.
As previously reported, the Lef-1 ΔN isoform competes with the Lef-1 FL isoform and other Tcf factors for DNA binding to their target sites. There is a role for miR-26b in the Wnt-β-catenin signal response due to its repression of specific Lef-1 isoforms, which are differentially expressed in tissues and cells. In HEK 293 FT cells, the Lef-1 FL isoform and β-catenin proteins interact, bind to the seven Lef-1 DNA binding elements, and activate the TopFlash reporter. miR-26b represses Lef-1 FL expression and decreases TopFlash reporter activity. However, in tissues and cells like CHO cells (which only express the Lef-1 ΔN isoform), the Groucho repressor interacts with the Lef-1 ΔN isoform to inhibit the TopFlash reporter, which cannot be derepressed by β-catenin. miR-26b represses Lef-1 ΔN expression and the repressor complex, which allows other Tcf factors to bind (32) and activate the TopFlash reporter.
The branching phenotype of the Pitx2-Cre/Dicer1 mutant pituitary reveals that miRNAs may affect the signal pathways involved in cell proliferation, including the FGF and Wnt signaling pathways. It is reported that Wnt5a is involved in pituitary shaping, and Wnt5a knock-out mice have another type of branching phenotype (33). It is possible that some of the factors in Wnt5a signaling are subject to miRNA regulation, and their expression may be altered in the Dicer1 mutant mice. Experiments are under way to analyze these developmental effects and to determine the molecular mechanisms of other pituitary-specific miRNAs.
This report reveals an essential role for miRNAs in pituitary development and demonstrates a new mechanism for miR-26b regulation of Lef-1 and Pit-1 expression. It is clear that other miRNAs are also required for normal pituitary development, and we are currently identifying other miRNA pituitary target genes. Perturbations in miRNA expression may be causative for clinical pituitary disorders, and screening for specific miRNAs could identify miRNA deficiencies or overexpression in abnormal pituitary tissues. Lef-1 is an important transcription factor involved in many developmental processes, and miR-26b could be a major regulator of Lef-1 in a variety of tissues and cells.
Acknowledgments
We thank Drs. Robert Schwartz, Jianbo Wang, and Shankar Venugopalan and members of the Amendt laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health, NIDCR, Grant DE 13941 (to B. A. A.).
- TSH
- thyroid-stimulating hormone
- LH
- luteinizing hormone
- GH
- growth hormone
- PRL
- prolactin
- miRNA
- microRNA
- En
- embryonic day n
- Pn
- postnatal day n
- FL
- full-length
- IP
- immunoprecipitation
- POMC
- proopiomelanocortin.
REFERENCES
- 1.Zhu X., Gleiberman A. S., Rosenfeld M. G. (2007) Physiol. Rev. 87, 933–963 [DOI] [PubMed] [Google Scholar]
- 2.van Genderen C., Okamura R. M., Fariñas I., Quo R. G., Parslow T. G., Bruhn L., Grosschedl R. (1994) Genes Dev. 8, 2691–2703 [DOI] [PubMed] [Google Scholar]
- 3.Olson L. E., Tollkuhn J., Scafoglio C., Krones A., Zhang J., Ohgi K. A., Wu W., Taketo M. M., Kemler R., Grosschedl R., Rose D., Li X., Rosenfeld M. G. (2006) Cell 125, 593–605 [DOI] [PubMed] [Google Scholar]
- 4.He L., Hannon G. J. (2004) Nat. Rev. Genet. 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 5.Bak M., Silahtaroglu A., Møller M., Christensen M., Rath M. F., Skryabin B., Tommerup N., Kauppinen S. (2008) RNA 14, 432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y., Ransom J. F., Li A., Vedantham V., von Drehle M., Muth A. N., Tsuchihashi T., McManus M. T., Schwartz R. J., Srivastava D. (2007) Cell 129, 303–317 [DOI] [PubMed] [Google Scholar]
- 7.Wang S., Aurora A. B., Johnson B. A., Qi X., McAnally J., Hill J. A., Richardson J. A., Bassel-Duby R., Olson E. N. (2008) Dev. Cell 15, 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C. Z., Li L., Lodish H. F., Bartel D. P. (2004) Science 303, 83–86 [DOI] [PubMed] [Google Scholar]
- 9.Poy M. N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P. E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Stoffel M. (2004) Nature 432, 226–230 [DOI] [PubMed] [Google Scholar]
- 10.Wienholds E., Plasterk R. H. (2005) FEBS Lett. 579, 5911–5922 [DOI] [PubMed] [Google Scholar]
- 11.Harfe B. D., McManus M. T., Mansfield J. H., Hornstein E., Tabin C. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10898–10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W., Selever J., Lu M. F., Martin J. F. (2003) Development 130, 6375–6385 [DOI] [PubMed] [Google Scholar]
- 13.Amen M., Liu X., Vadlamudi U., Elizondo G., Diamond E., Engelhardt J. F., Amendt B. A. (2007) Mol. Cell. Biol. 27, 7560–7573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doench J. G., Petersen C. P., Sharp P. A. (2003) Genes Dev. 17, 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradford A. P., Wasylyk C., Wasylyk B., Gutierrez-Hartmann A. (1997) Mol. Cell. Biol. 17, 1065–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amendt B. A., Sutherland L. B., Semina E. V., Russo A. F. (1998) J. Biol. Chem. 273, 20066–20072 [DOI] [PubMed] [Google Scholar]
- 17.Amendt B. A., Sutherland L. B., Russo A. F. (1999) Mol. Cell. Biol. 19, 7001–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amen M., Espinoza H. M., Cox C., Liang X., Wang J., Link T. M., Brennan R. G., Martin J. F., Amendt B. A. (2008) Nucleic Acids Res. 36, 462–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruszka A., Ren S. G., Dong J., Culler M. D., Melmed S. (2007) Endocrinology 148, 6107–6114 [DOI] [PubMed] [Google Scholar]
- 20.Vadlamudi U., Espinoza H. M., Ganga M., Martin D. M., Liu X., Engelhardt J. F., Amendt B. A. (2005) J. Cell Sci. 118, 1129–1137 [DOI] [PubMed] [Google Scholar]
- 21.Yi R., O'Carroll D., Pasolli H. A., Zhang Z., Dietrich F. S., Tarakhovsky A., Fuchs E. (2006) Nat. Genet. 38, 356–362 [DOI] [PubMed] [Google Scholar]
- 22.Li S., Crenshaw E. B., 3rd, Rawson E. J., Simmons D. M., Swanson L. W., Rosenfeld M. G. (1990) Nature 347, 528–533 [DOI] [PubMed] [Google Scholar]
- 23.Jimenez J., Jang G. M., Semler B. L., Waterman M. L. (2005) RNA 11, 1385–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li T. W., Ting J. H., Yokoyama N. N., Bernstein A., van de Wetering M., Waterman M. L. (2006) Mol. Cell. Biol. 26, 5284–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasonkin I. O., Ward R. D., Raetzman L. T., Seasholtz A. F., Saunders. T. L., Gillespie P. J., Camper S. A. (2004) Hum. Mol. Genet. 15, 2727–2735 [DOI] [PubMed] [Google Scholar]
- 26.Giese K., Amsterdam A., Grosschedl R. (1991) Genes Dev. 5, 2567–2578 [DOI] [PubMed] [Google Scholar]
- 27.Waterman M. L., Fischer W. H., Jones K. A. (1991) Genes Dev. 5, 656–669 [DOI] [PubMed] [Google Scholar]
- 28.Zhou P., Byrne C., Jacobs J., Fuchs E. (1995) Genes Dev. 9, 700–713 [DOI] [PubMed] [Google Scholar]
- 29.Daniels D. L., Weis W. I. (2005) Nat. Struct. Mol. Biol. 12, 364–371 [DOI] [PubMed] [Google Scholar]
- 30.He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. (1998) Science 281, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 31.Tetsu O., McCormick F. (1999) Nature 398, 422–426 [DOI] [PubMed] [Google Scholar]
- 32.Brantjes H., Roose J., van De Wetering M., Clevers H. (2001) Nucleic Acids Res. 29, 1410–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cha K. B., Douglas K. R., Potok M. A., Liang H., Jones S. N., Camper S. A. (2004) Mech. Dev. 121, 183–194 [DOI] [PubMed] [Google Scholar]
- 34.National Research Council (1996) Guide for the Care and Use of Laboratory Animals, National Institutes of Health Publication 86-23, National Institutes of Health, Bethesda, MD [Google Scholar]