Abstract
FEN1 cleaves 5′ flaps at their base to create a nicked product for ligation. FEN1 has been reported to enter the flap from the 5′-end and track to the base. Current binding analyses support a very different mechanism of interaction with the flap substrate. Measurements of FEN1 binding to a flap substrate show that the nuclease binds with similar high affinity to the base of a long flap even when the 5′-end is blocked with biotin/streptavidin. However, FEN1 bound to a blocked flap is more sensitive to sequestration by a competing substrate. These results are consistent with a substrate interaction mechanism in which FEN1 first binds the flap base and then threads the flap through an opening in the protein from the 5′-end to the base for cleavage. Significantly, when the unblocked flap length is reduced from five to two nucleotides, FEN1 can be sequestered from the substrate to a similar extent as a blocked, long flap substrate. Apparently, interactions related to threading occur only when the flap is greater than two to four nucleotides long, implying that short flaps are cleaved without a threading requirement.
Keywords: DNA, DNA Enzymes, DNA-Protein Interaction, DNA Repair, DNA Replication, FEN1, Okazaki Fragment Maturation
Introduction
High fidelity DNA replication and repair ensures maintenance of genomic integrity, critical for the viability of eukaryotic cells. Replication on the lagging strand generates short stretches of DNA known as Okazaki fragments that are further processed and finally ligated to form a complete DNA strand. Efficient processing of the Okazaki fragments requires the removal of the RNA/DNA segment that is used to initiate polymerization prior to fragment ligation. Similarly, repair of certain types of DNA damage requires the removal of erroneous or damaged stretches of nucleotides by a process known as long patch-base excision repair (LP-BER).3 Removal of the initiator segment in Okazaki fragment maturation and damaged bases in LP-BER are done by displacing the downstream DNA segment into a 5′ flap structure by replication or repair-associated polymerases (1).
Flap endonuclease 1 (FEN1) is a critical central component of both the replication and repair pathways (1–3). FEN1 is a structure-specific nuclease that recognizes and processes 5′ flap intermediates displaced by replication and repair associated polymerases (1–4). Biochemical analysis shows that FEN1 possesses endonuclease activity and a minor 5′ exonuclease function (5, 6). FEN1 is able to recognize and cleave at the base of the flap, effectively creating a nicked DNA segment. Multiple reports have shown that 5′ flap-bound proteins, annealed DNA segments complementary to the 5′ flap, or large adducts bound to the 5′ flap block FEN1 cleavage in vitro (5, 7–14). Based on results from these 5′ flap blocking experiments, our group proposed a model that FEN1 must first recognize the 5′ end of the flap and track down the unblocked single-stranded 5′ flap before cleaving (7). The steps taken in this tracking model are described in the discussion section. We proposed that the evolutionary development of flap tracking prohibits FEN1 from erroneously binding and cleaving the single-stranded DNA on the lagging strand template.
Early reports showed that FEN1 preferentially binds substrates in which the upstream primer has a one nt single-stranded 3′ overhang (3′ flap) (15). Later reports demonstrated that this 1 nt 3′ overhang and a 5′ flap leads to optimal FEN1 cleavage (6), making the double flap the preferred substrate. Crystal structure data from Archaeoglobus fulgidus FEN1 showed that FEN1 generates a hydrophobic wedge around the 3′ flap which orients FEN1 at the base of the flap (16). The authors proposed that this 3′ flap binding not only stabilizes and orients FEN1 but also acts as a guide such that FEN1 cleaves one nt into the downstream double-stranded region creating a precise nicked structure.
Crystal structure results from the FEN1 homologue bacteriophage T5 5′ exonuclease showed that it contains a helical arch with an inner diameter too small for the passage of double-stranded DNA but wide enough to allow for single-stranded DNA tracking (17). In subsequent reports analyzing flap tracking mechanics, we found that FEN1 is able to tolerate the addition of small adducts and branched DNA modifications within the 5′ flap (8, 9). These data suggest that the FEN tracking capability uses a flexible clamp type mechanism to allow for tracking over base-damaged single-stranded DNA (8, 9). Several reports have shown that when FEN1 binds its DNA substrate, the unstructured arch region of FEN1 becomes structured into an α helical arch (16, 18–20) and moves proximal to a second α-helix-loop-α-helix structure (16). This formation and apparent motion of the helical arch is consistent with the clamp model. Together, these data suggest that FEN1 either uses a tracking or clamp mechanism to guarantee that prior to cleaving, it has bound an unblocked 5′ single-stranded segment of DNA.
Whereas a model in which FEN1 tracks down the 5′ flap, binds at the base, and then cleaves is attractive, it is inconsistent with a report showing FEN1 can bind but not cleave 5′ blocked substrates (21). However, a model consistent with this evidence was proposed based on work using Escherichia coli DNA polymerase I, which has a 5′ nuclease domain with sequence similarity to eukaryotic FEN1. This model envisions that FEN1 initially binds the base of the flap and then threads the 5′ flap prior to cleaving (22).
In the current report, we employed binding and dissociation analyses to clarify the likely sequence of steps employed by FEN1 in its substrate interaction prior to catalysis. Specifically, we attempted to distinguish the interactions that contribute to binding affinity before and after threading and interpret how they relate to the mechanism of FEN1 action.
EXPERIMENTAL PROCEDURES
Materials
Oligonucleotides were synthesized by Integrated DNA Technologies (IDT). Radionucleotides [γ-32P]ATP (6000 Ci/mmol) and [α-32P]dCTP (6000 Ci/mmol) were purchased from PerkinElmer Life Sciences. The Klenow fragment of Escherichia coli DNA polymerase I and polynucleotide kinase used for 3′ and 5′ oligonucleotide substrate labeling, respectively, were purchased from Roche Applied Science. ATP and streptavidin were also purchased from Roche Applied Science. All other reagents were the best commercial available products.
Oligonucleotides
The oligonucleotide sequences used in this study are listed in Table 1. Oligonucleotides were 3′ or 5′ end-radiolabeled, purified, and annealed as previously described (4). Assays that used a streptavidin blocked 5′ flap utilized an oligonucleotide biotinylated on the 5′-end (Table 1 oligonucleotide D1.53B). The location of the radiolabel on the experimental substrate is indicated in each figure with an asterisk. Radiolabeled oligonucleotide substrates were annealed in a ratio of 1:3:6 of labeled primer to template to unlabeled primer, respectively. Unlabeled competitor substrates were annealed in a ratio of 1:1:1 of upstream primer to template to downstream primer.
TABLE 1.
Oligonucleotide sequences
| Oligo | Length (nt) | Sequencea |
|---|---|---|
| Upstream (5′-3′) | ||
| U1 | 26 | CGCCAGGGTTTTCCCAGTCACGACCA |
| U2 | 26 | CGACCGTGCCAGCCTAAATTTCAATA |
| U3 | 26 | CGACCGTGCCAGCCTAAATTTCAATT |
| Downstream (5′-3′) | ||
| D1.2 | 25 | GTCGTTTTACAACGACGTGACTGGG |
| D1.5 | 28 | GCCGTCGTTTTACAACGACGTGACTGGG |
| D1.10 | 33 | CACTGGCCGTCGTTTTACAACGACGTGACTGGG |
| D1.15 | 38 | TAATTCACTGGCCGTCGTTTTACAACGACGTGACTGGG |
| D1.30 | 53 | TTCACGCCTGTTAGTTAATTCACTGGCCGTCGTTTTACAACGACGTGACTGGG |
| D1.53 | 76 | GTACCGAGCTCGAATTCGCCCGTTTCACGCCTGTTAGTTAATTCACTGGCCGTCGTTTTACAACGACGTGACTGGG |
| D1.53B | 76 | βGTACCGAGCTCGAATTCGCCCGTTTCACGCCTGTTAGTTAATTCACTGGCCGTCGTTTTACAACGACGTGACTGGG |
| D2.0 | 28 | CCACCCGTCCACCCGACGCCACCTCCTG |
| D2.27 | 55 | AGGTCTCGACTAACTCTAGTCGTTGTTCCACCCGTCCACCCGACGCCACCTCCTG |
| D3.1 | 27 | CCCCGTCCACCCGACGCCACCTCCTGC |
| D3.2 | 28 | CCCCCGTCCACCCGACGCCACCTCCTGC |
| D3.3 | 29 | ACCCCCGTCCACCCGACGCCACCTCCTGC |
| D3.4 | 30 | AACCCCCGTCCACCCGACGCCACCTCCTGC |
| D3.6 | 32 | ATAACCCCCGTCCACCCGACGCCACCTCCTGC |
| Template (3′-5′) | ||
| T1 | 49 | GCGGTCCCAAAAGGGTCAGTGCTGGGCAAAATGTTGCTGCACTGACCCG |
| T2 | 54 | GCTGGCACGGTCGGATTTAAAGTTAGGTGGGCAGGTGGGCTGCGGTGGAGGACG |
| T3 | 51 | GCTGGCACGGTCGGATTTAAAGTTAGGGCAGGTGGGCTGCGGTGGAGGACG |
| ssDNA (5′-3′) | ||
| D2.F | 30 | AGGTCTCGACTAACTCTAGTCGTTGTTCCA |
a β in the nucleotide sequence represents the location of a biotin-conjugated segment.
Streptavidin Conjugation
To block the 5′ flap end, ×100 molar excess streptavidin, relative to experimental substrate, was incubated with a 5′ biotinylated flap substrate. Streptavidin was preincubated with the substrate for 10 min prior to the addition of FEN1 in the streptavidin “pre-blocked” reactions. Streptavidin was added 10 min after the addition of FEN1 in the streptavidin “post-blocked” reactions.
Enzymes
C-terminal histidine-tagged human FEN1 was cloned into the pET-FCH plasmid, overexpressed in E. coli strain BL21 (DE3)/pLysS, and purified as previously described (9).
FEN1 Substrate Binding Assays
FEN1 substrate binding affinity was measured using electrophoretic mobility shift assay (EMSA). Reactions were performed using a buffer containing 50 mm Tris-HCl (pH 8.0), 2 mm DTT, 0.25 mg/ml bovine serum albumin (BSA), 30 mm NaCl, and 5% glycerol (referred to as reaction buffer). Protein concentrations used are indicated in the figure legends. FEN1 binding assays were carried out in a 20 μl final reaction volume with 0.25 nm experimental substrate. The experimental substrate was pre- or post-blocked as described above. FEN1 was incubated with the experimental substrate for 10 or 15 min at 37 °C as described in the figure legend. Dissociation constant (Kd) experiments were conducted with 0.05 nm labeled experimental substrate per reaction such that [experimental substrate] ≪ Kd. Determination of the Kd values were based on the nonlinear least squares regression fit to the hyperbolic Equation 1,
where, y is the percent of the experimental substrate bound by FEN1; Bmax is the maximum experimental substrate bound by FEN1, [protein] is the FEN1 concentration; and Kd is the dissociation constant.
FEN1 Binding and Cleavage Correlation Assays
The experimental substrate was streptavidin pre- or post-blocked as described above. Reactions were initiated by incubating FEN1 and the 0.05 nm experimental substrate, referred to as the reaction initiation, in reaction buffer at 37 °C for 15 min. Protein concentrations are noted in the figure legend. Fifteen minutes after the reaction initiation, 15 μl from each binding reaction was extracted and added to 0.6 μl of 100 mm MgCl2 to facilitate cleavage, referred to as the cleavage reaction. Twenty minutes after reaction initiation, aliquots from the binding reaction were loaded onto a native gel to measure binding. The cleavage reactions were terminated 25 min after the reaction initiation using 15 μl of 2× termination dye containing 90% formamide (v/v) and 10 mm EDTA with xylene cyanole and bromphenol blue markers and then placed at 95 °C for 5 min. Cleavage reactions were loaded onto a denaturing gel to measure cleavage.
FEN1 Dissociation Assays
The experimental substrate was streptavidin pre- or post-blocked as described above. Dissociation reactions were initiated by incubating 3.75 nm FEN1 with 0.25 nm experimental substrate, referred to as the reaction initiation, in reaction buffer on ice for 10 min in a 20 μl reaction volume. Competitor substrate was then added to the reaction and held at 37 °C. Competitor substrate incubation times as well as protein and competitor substrate concentrations used are shown in the figure legends.
Gel Electrophoresis
FEN1 substrate binding activity was measured using native gels containing 6% polyacrylamide in 1× TBE. Samples were loaded while the gel was being electrophoresed at 200 V and further subject to electrophoresis for 60 min at 200 V. FEN1 nuclease activity was measured using 15% polyacrylamide, 7 m urea denaturing gels containing 1× TBE subjected to electrophoresis for 60 min at 85 watts.
Experimental Result Analysis
Binding, dissociation, and cleavage assays were performed at least in triplicate. Gel drying and phosphor screen exposure were conducted as previously described (4). Binding and cleavage autoradiography measurements were determined using Storm hardware (GE Healthcare) utilizing Molecular Dynamics PhosphorImager technology. Results analysis and quantitation were conducted using Molecular Dynamics ImageQuant software version 5.2. The % DNA substrate bound by FEN1 is defined as [bound/(bound + unbound)] × 100. The % DNA substrate cleaved by FEN1 is defined as [cleaved/(cleaved + uncleaved)] × 100.
RESULTS
FEN1 Binding Affinity on a Flap Substrate Assessed by EMSA Is Similar Irrespective of Accessibility to the 5′ Flap End
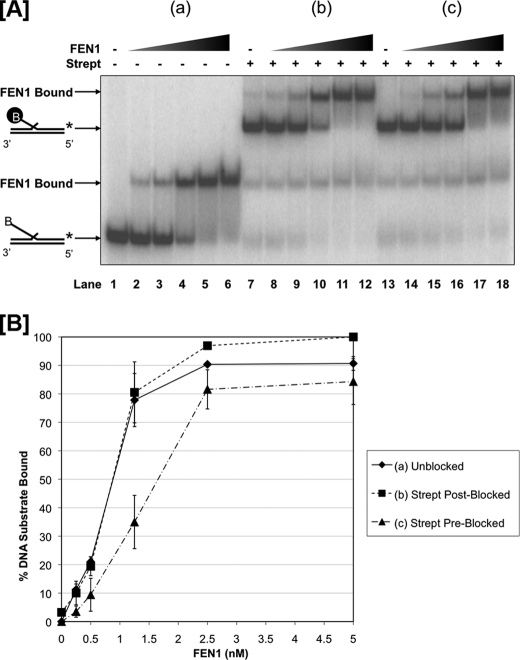
Because it has been previously reported that FEN1 must track on the flap in the 5′-3′ direction to display cleavage activity, we first questioned the relationship between tracking and substrate binding by FEN1. We used EMSA to compare the binding affinity of FEN1 on a 5′ biotinylated 53 nt double flap, that was either unblocked or blocked with streptavidin (Fig. 1A). Based on a tracking model, we anticipated that the amount of FEN1 binding on a blocked substrate would differ from that on an unblocked substrate. To measure binding, we titrated FEN1 with the experimental substrate using the following scenarios: (a) FEN1 was incubated with a 53 nt biotinylated flap as shown in Fig. 1A, lanes 2–6 (referred to as “unblocked”). (b) FEN1 was preincubated with the flap prior to adding streptavidin as shown in Fig. 1A lanes 8–12 (referred to as streptavidin “post-blocked”). (c) Streptavidin was bound to the flap prior to the addition of FEN1 as shown in Fig. 1A lanes 14–18 (referred to as streptavidin “pre-blocked”). Fig. 1A lanes 1, 7, and 13 show the control unblocked, streptavidin post-blocked, and streptavidin pre-blocked flap substrates, respectively, without FEN1. Fig. 1A lanes 2–6 show FEN1 binding to an unblocked flap with respect to protein concentration. When FEN1 was incubated with the unblocked flap substrate for 10 min prior to adding streptavidin, the binding pattern was comparable to the corresponding unblocked reactions (compare Fig. 1A, lanes 2–6 to 8–12, respectively). Interestingly, the binding pattern of FEN1 incubated with the flap substrate pre-blocked with streptavidin was also similar to both the unblocked and streptavidin post-blocked flap scenarios (compare Fig. 1A lanes 14–18 to 2–6 and 8–12, respectively). This similarity in binding affinity irrespective of the accessibility of the 5′ flap end suggests that FEN1 first interacts with the base of the double flap substrate as in the threading model rather than the flap end as in the tracking model.
FIGURE 1.
FEN1 binds similarly to unblocked and blocked 53 nt flap substrates independent of threading. FEN1 binding was measured by EMSA as described under “Experimental Procedures.” Reactions were initiated by incubating increasing concentrations of FEN1 (0.25, 0.5, 1.25, 2.5, and 5 nm) with the experimental substrate for 15 min. A, shows FEN1 bound to a 53 nt 5′ biotinylated flap substrate (U1:T1:D1.53B) when (a) no streptavidin was added to the reaction (lanes 1–6), (b) streptavidin was added 10 min after reaction initiation (lanes 7–12), and (c) streptavidin was added 10 min before reaction initiation (lanes 13–18). Lane 1 shows the substrate alone control. Lanes 7 and 13 show the streptavidin-bound substrate controls. The positions of the substrate alone and FEN1-substrate complex are indicated to the left of the figure. A “B” in the oligonucleotide sequence indicates the location of the 5′ biotin, and the black-circled “B” represents streptavidin-bound biotin. B shows the graphical quantitation of A based on at least three independent EMSA results.
The binding of FEN1 to the substrates in the three different scenarios described above is presented graphically in Fig. 1B. The quantitation shows that FEN1 binding in the streptavidin pre-blocked scenario exhibits only a slight reduction relative to the scenarios in which FEN1 was allowed to thread the flap before blocking the 5′-end.
To confirm this interpretation, we measured the dissociation constants of FEN1 on an unblocked substrate and a streptavidin pre-blocked substrate using a larger range of FEN1 concentrations and a lower concentration of experimental substrate such that [experimental substrate] ≪ Kd. The Kd of FEN1 bound to the unblocked experimental substrate was calculated to be 0.92 + 0.11 nm based on a hyperbolic curve fit (supplemental Fig. S1). Similarly, the Kd of FEN1 bound to the streptavidin pre-blocked experimental substrate was determined to be 0.96 + 0.07 nm (supplemental Fig. S2). The virtually indistinguishable dissociation constants suggest that the binding of FEN1 to a substrate is nearly independent of its ability to thread the 5′ flap.
FEN1 Binding Does Not Correlate with Cleavage
We presumed that FEN1 would thread a flap prior to streptavidin blockage but fail to thread a pre-blocked flap. To confirm this assumption, we compared the binding of FEN1 with the ability to cleave the double flap substrates. Substrates that were either pre- or post-blocked were tested for binding (Fig. 2A) and cleavage (Fig. 2B). As expected, nearly all of the unblocked substrate was cleaved at the lowest FEN1 concentration (Fig. 2B, lane 2) relative to the substrate alone control (Fig. 2B, lane 1). When FEN1 was allowed to incubate with the streptavidin post-blocked substrate, the amount cleaved upon addition of magnesium directly correlated with the amount bound (lanes 3–8 of Fig. 2, B and A, respectively). Conversely, when streptavidin was bound to the flap prior to addition of FEN1, only a small fraction of the substrate was cleaved with addition of magnesium, independent of the FEN1 concentration and the cleaved fraction did not correlate to the amount bound (lanes 9–14 of Fig. 2, B and A, respectively). The small amount of observed cleavage is likely to have occurred on the inevitable minor fraction of substrate that was not biotinylated, ∼9% as determined by gel quantitation. Most importantly, these results confirm that FEN1 was able to thread effectively on a post-blocked substrate but unable to thread a pre-blocked substrate as assessed by the cleavage activity, although FEN1 bound both substrates with similar affinity. Additionally, results show that as FEN1 was titrated onto a substrate that was subsequently blocked, an increasing amount of FEN1 could remain poised for cleavage when the magnesium was added.
FIGURE 2.
Threading of the 5′ flap allows for FEN1 cleavage. The comparison of FEN1 binding and cleavage was made using EMSA and denaturing gel electrophoresis as described under “Experimental Procedures.” Reactions were initiated by incubating increasing concentrations of FEN1 (a) (0.16 nm) and (b), (c) (0.16, 0.31, 0.63, 1.25, 2.5 nm) with the 53 nt 5′ biotinylated experimental flap substrate (U1:T1:D1.53B), referred to as the binding reaction. Similar to Fig. 1A, the experimental flap substrate was (a) not streptavidin blocked (lanes 1–2), (b) streptavidin blocked 10 min after the binding reaction initiation (lanes 3–8), or (c) streptavidin blocked 10 min before the binding reaction initiation (lanes 9–14). Lane 1 shows the substrate alone control. Lanes 3 and 9 show the streptavidin-bound substrate controls. Fifteen minutes after the binding reaction initiation, an aliquot from the reaction was extracted and mixed with Mg2+, referred to as the cleavage reaction. A, shows FEN1 bound to the experimental substrate in the binding reaction and B, shows the corresponding labeled oligonucleotide and FEN1 cleavage products in the cleavage reaction. In A, the positions of the substrate alone and FEN1-substrate complex are indicated to the left of the figure. A “B” in the oligonucleotide sequence indicates the location of the 5′ biotin, and the black-circled “B” represents streptavidin-bound biotin. In B, the position of the labeled oligonucleotide and FEN1 cleavage products are indicated to the left of the figure. Quantitation of the percent experimental substrate bound (A) and cleaved (B) by FEN1 and standard deviation based on at least three independent EMSA results are shown below each figure.
Direct Flap Interactions Improve FEN1 Binding Affinity
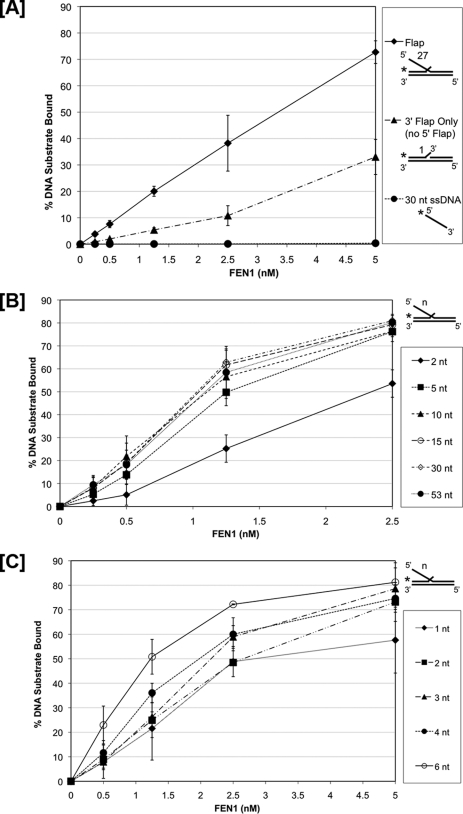
Because 5′ flap threading provided little improvement in binding affinity, we considered the significance of 5′ flap interactions. These were evaluated by comparing FEN1 binding on a 27 nt 5′ flap substrate to a substrate lacking the 5′ flap and a 30 nt ssDNA substrate representing the flap alone (Fig. 3A and supplemental Fig. S3). In this experiment, all substrates but the ssDNA had a one nt 3′ flap. Relative to the 27 nt double flap substrate, FEN1 bound the “3′ flap only” substrate with much lower affinity, and we did not detect any FEN1 binding on the ssDNA. These results show that the presence of the 5′ flap in a double flap substrate is very important for the most stable binding directly from solution.
FIGURE 3.
Long 5′ flaps provide additional FEN1 binding affinity. FEN1 binding was measured by EMSA as described under “Experimental Procedures.” Reactions were initiated by incubating increasing concentrations of FEN1 with the experimental substrates for 10 min. A, shows FEN1 bound to a 27 nt double flap substrate (U2:T2:D2.27), an upstream 1 nt 3′ flap with no downstream 5′ flap substrate (U2:T2:D0), and a 30 nt ssDNA segment (D2.F). B, shows FEN1 bound to substrates with varying 5′ downstream flap lengths of 2 nt (U1:T1:D1.2), 5 nt (U1:T1:D1.5), 10 nt (U1:T1:D1.10), 15 nt (U1:T1:D1.15), 30 nt (U1:T1:D1.30), and 53 nt (U1:T1:D1.53). C, shows FEN1 bound to substrates with varying 5′ downstream flap lengths of 1 nt (U3:T3:D3.1), 2 nt (U3:T3:D3.2), 3 nt (U3:T3:D3.3), 4 nt (U3:T3:D3.4), and 6 nt (U3:T3:D3.6). A–C show the graphical quantitation of the percent experimental substrate bound by FEN1 based on at least three independent EMSA results. In A, the substrate configuration is indicated next to the corresponding legend item to the right of the figure. In B and C, the substrate configurations are indicated to the right of the figure where “n” represents the length of the 5′ flap listed in the figure legend.
The Length of the 5′ Flap Can Add to the Stability of FEN1 Binding
To further explore the contribution of 5′ flap contacts to binding affinity we measured binding to substrates with varying 5′ flap lengths. We used EMSA to visualize FEN1 binding efficiency to double flap substrates (supplemental Fig. S4). A graphical quantitation of FEN1 binding to double flap substrates with 5′ flaps of 2, 5, 10, 15, 30, or 53 nt lengths with increasing FEN1 concentration is shown in Fig. 3B. While FEN1 bound similarly to substrate with flaps of lengths 5, 10, 15, 30, and 53 nt, it bound less when the flap length was 2 nt. These results suggest that FEN1 binds with higher affinity to 5′ flaps of 5 nt or longer. To define the transition more clearly, we narrowed the range of flap lengths (supplemental Fig. S5). The quantitation of FEN1 bound to 1, 2, 3, 4, and 6 nt flaps at varying concentrations of FEN1 is shown in Fig. 3C. FEN1 bound with increasing affinity as the flap length increased. The observed gradual transition to higher affinity binding indicates that there are several relevant binding contacts between the protein and 5′ flap, positioned within the first 6 nt. Because binding to long flaps occurs with similar affinity irrespective of a block at the 5′-end, we presume that the binding contacts that modestly improve affinity to long flap substrates do not require threading.
Substrate Competition Assays Reveal Reduced Dissociation If FEN1 Can Thread
We considered whether small differences in FEN1 binding to blocked and unblocked flap substrates would be enhanced in a substrate competition measurement. This experiment measures the ability of a competing substrate to sequester FEN1 bound to the original experimental substrate. We reasoned that flap threading through the helical arch might not involve traditional charge-charge and hydrophobic contacts measurable by EMSA approaches. Instead the more mechanical threading interaction, likely to uniformly slow association and dissociation rates, leaving the binding equilibrium unaltered, may be detectable by its ability to inhibit FEN1 movement to a competing substrate.
We first assessed the ability of substrates with several structures to sequester FEN1 from the unblocked 53 nt double flap substrate (Fig. 4A). Lanes 3–7 show that the double flap substrate itself is an efficient competitor of the unblocked flap. Neither the “3′ flap only” (lanes 10–14) nor the 30 nt ssDNA alone segment (lanes 17–21) competed as effectively against the double flap substrate. Based on these data, we chose the double flap structure to compete FEN1 from substrates using scenarios similar to those in Fig. 1A.
FIGURE 4.
5′ Flap threading reduces dissociation of FEN1 in the presence of a competitor substrate. FEN1 dissociation was measured by EMSA as described under the “Experimental Procedures.” Reactions were initiated by incubating FEN1 with the experimental substrate for 10 min. Ten minutes after reaction initiation, increasing concentrations of competitor substrate were added. A, shows the FEN1 remaining bound to a 5′ 27 nt flap experimental substrates (U2:T2:D2.27) 10 min after the addition of the competitor substrates (2.5×, 5×, 10×, 25×, and 50× molar excess relative to experimental substrate) having (a) a 27 nt flap (U2:T2:D2.27) (lanes 3–7), (b) an upstream 1 nt 3′ flap with no downstream 5′ flap (U2:T2:U2.0) (lanes 10–14), and (c) a 30 nt ssDNA segment (D2.F) (lanes 17–21). Lanes 1, 8, and 15 show the substrate alone controls. Lanes 2, 9, and 16 show experimental substrate bound by FEN1 in the absence of competitor substrate. B shows the FEN1 remaining bound to a 53 nt 5′ biotinylated flap experimental substrate (U1:T1:D1.53B) 10 min after the addition of increasing concentrations of a 53 nt flap competitor substrate (U1:T1:D1.53) (7.5×, 15×, 30×, 75×, and 150× molar excess relative to experimental substrate) where (a) no streptavidin was added to the reaction (lanes 3–7), (b) streptavidin was added 10 min after reaction initiation (lanes 10–14), or (c) streptavidin was added 10 min before reaction initiation (lanes 17–21). Lane 1 shows the experimental substrate alone control. Lanes 8 and 15 show the streptavidin-bound experimental substrate controls. Lanes 2, 9, and 16 show the experimental substrates bound by FEN1 in the absence of competitor substrate. Quantitation of the percent experimental substrate bound by FEN1 and standard deviation based on at least three independent EMSA results are shown below the figure. The position of the experimental substrate alone and FEN1, experimental substrate complexes are indicated to the left of the figure. The competitor substrate configurations are shown above the figure. In B, a “B” in the oligonucleotide sequence indicates the location of the 5′ biotin, and the black-circled “B” represents streptavidin-bound biotin.
After incubating FEN1 with the 53 nt biotinylated unblocked, streptavidin pre- or post-blocked double flap, an unlabeled 53 nt double flap competitor substrate was added. With unblocked or post-blocked substrates, FEN1 was inefficiently sequestered from the labeled experimental substrate in a competitor concentration-dependent fashion (Fig. 4B lanes 2–7 and 9–14, respectively). Interestingly, FEN1 was efficiently sequestered from the streptavidin pre-blocked experimental substrate that did not allow threading (Fig. 4B, lanes 16–21). These competition results suggest that the ability of FEN1 to thread the 5′ flap reduces dissociation from the experimental substrate. Specifically, the results of this assay suggest that the sequestration of FEN1 from the original substrate is slower if the protein can thread.
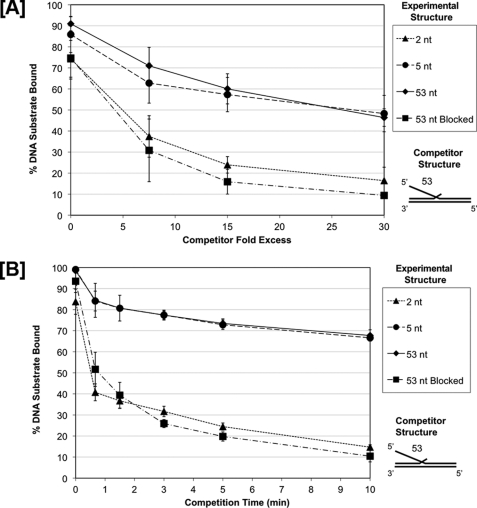
Threading Is Required for Additional Resistance to Binding Competition Observed with Longer Flaps
We hypothesized that allowing threading would generate more protein interactions with the DNA substrate that would resist competition, and that these contacts would occur on longer but not shorter flaps. To test this hypothesis, we incubated FEN1 with unblocked 5′ flap substrates having short flaps of 2 nt or long flaps of 5 nt or 53 nt. Additionally, we incubated FEN1 with a 53 nt biotinylated flap that had been streptavidin pre-blocked. In all four flap-type binding scenarios, we then sequestered FEN1 from these experimental substrates by competing with an unlabeled 53 nt flap competitor substrate. The dissociation of FEN1 from these different experimental substrates is presented graphically in Fig. 5A. As expected, FEN1 was sequestered similarly from both the 5 and 53 nt 5′ long flap experimental substrates (Fig. 5A represented by the circles and diamonds, respectively). These data are consistent with the FEN1 long flap binding curves shown in Fig. 4A. The unblocked 53 nt flap sequestered FEN1 from the 2 nt short flap substrate (Fig. 5A represented by the triangles) more effectively than from substrates with longer flaps, a result also consistent with the binding data in Fig. 4A. The amount of FEN1 that remained bound to the blocked 53 nt experimental substrate (Fig. 5A represented by the squares) was similar to that of the short 2 nt flap substrate in the presence of the 53 nt unblocked competitor.
FIGURE 5.
FEN1 shows slow dissociation from long unblocked 5′ flaps and fast dissociation from short or blocked 5′ flaps in the presence of a competitor substrate. FEN1 dissociation was measured by EMSA as described under the “Experimental Procedures.” Reactions were initiated by incubating FEN1 with various 5′ flap experimental substrate configurations, a 2 nt flap (U1:T1:D1.2), a 5 nt flap (U1:T1:D1.5), a 53 nt flap (U1:T1:D1.53), and a 53 nt streptavidin pre-blocked flap (U1:T1:D1.53B) for 10 min followed by the addition of a 53 nt flap competitor substrate (U1:T1:D1.53). In A, increasing concentrations of competitor substrate were added (7.5×, 15×, and 30× molar excess relative to experimental substrate) to the reaction for 10 min. In B, 30× molar excess competitor substrate was added to the reaction for increasing amounts of time (0.75, 1.5, 3, 5, and 10 min). A and B show the graphical quantitation of the percent experimental substrate remaining bound by FEN1 after competition based on at least three independent EMSA results. The legend indicates the experimental substrate 5′ flap configuration. The competitor substrate configuration is shown to the right of the figure.
Dissociation Rate Is Slow when FEN1 Is Bound to a Long, Unblocked Flap
Because FEN1 binding was less sensitive to binding competition (Fig. 5A) when it could thread flaps 5 nt and longer, we anticipated that contacts made on long flaps slowed the FEN1 dissociation rate. To quantify dissociation rate, we incubated FEN1 with the same four experimental substrates used in the previous competition experiment. The experimental substrates include unblocked 2 nt, 5 nt, or 53 nt 5′ flaps or a streptavidin pre-blocked 53 nt 5′ flap. We then measured the rate of sequestration of FEN1 from the flaps using an unlabeled 53 nt flap competitor. The dissociation rates of FEN1 from the four substrates are graphically represented in Fig. 5B. As predicted, the FEN1 off rate was significantly reduced for the 5 nt and 53 nt flap substrates (Fig. 5B represented by the circles and diamonds, respectively) relative to the rapid off rate of the 2 nt and pre-blocked flap substrates (Fig. 5B represented by triangles and squares, respectively). The slow half-times for dissociation were both greater than 10 min, whereas the rapid half-times were both about 1 min. These results were consistent with the competition results shown in Fig. 5A. Taken together, the binding affinity, competition, and dissociation rate results suggest that the improved resistance to sequestration seen with flaps of 5 nt and longer derives from FEN1 threading. That is, threading must be allowed for additional binding constraints that reduce the sequestration of FEN1 by the competing substrate. The similar rapid off rates with the blocked long, flap and the short, 2 nt flap suggest the striking additional conclusion that FEN1 does not thread the short flap prior to cleaving.
DISCUSSION
Because FEN1 plays a significant role in the maintenance of genomic integrity, it has been the focus of intense research interest for a number of years. However, the exact mechanism by which FEN1 specifically recognizes and interacts with its flap substrates has remained elusive. Current models explaining FEN1-substrate interactions are mainly derived from either biochemical analyses of nuclease function or x-ray crystallographic studies (7, 10, 16, 17, 23, 24). In our current work, using a variety of binding, dissociation, and cleavage assays, we set out to clarify the mechanism of flap substrate recognition by FEN1 and the order of steps in its binding interaction. Using unblocked and blocked flap substrates we showed that FEN1 displays similar binding affinity irrespective of whether the substrate has a free 5′-end (Fig. 1, A and B). To determine whether the 5′ flap nucleotides imparted additional affinity, we compared FEN1 binding affinity to substrates with variable 5′ flap lengths. The results showed that FEN1 makes additional contacts with long 5′ flaps, which contribute to increased binding affinity to the substrate (Fig. 3, A–C).
In an effort to determine whether threading influences binding, we devised substrate competition assays, wherein a competing substrate is added to a complex of FEN1 bound to the experimental substrate. The ability of the competing substrate to sequester FEN1 from the experimental substrate gives an indication of the association and dissociation rates of FEN1. The competition assay showed that the competitor substrate is more effective at sequestering FEN1 from a substrate with a blocked 5′ flap than from an unblocked 5′ flap substrate (Fig. 4B). Our competition results suggest that the threading process influences the dissociation rate. To measure dissociation rate directly, we performed the substrate competition assay in a time-dependent manner. Our results showed that the half-time for dissociation from the long unblocked flap substrate was over 10 min, whereas, the half-time for dissociation for the blocked flap was approximately 1 min. Moreover, FEN1 showed a half-time of dissociation from the short flap of ∼1 min similar to the blocked flap (Fig. 5B).
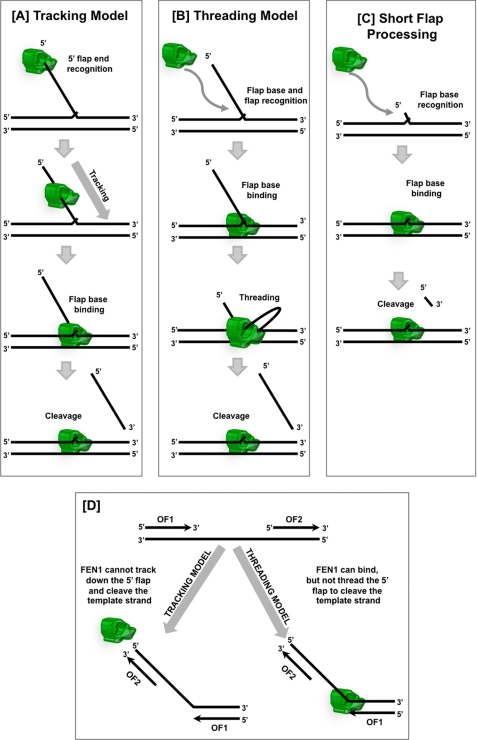
We previously proposed a mechanism of FEN1 action in which the initial contact with the substrate was made at the 5′-end of the flap followed by tracking down to the base of the flap and finally cleavage (Fig. 6A). However, our affinity data are consistent with an alternate mechanism in which the first contact on the substrate by FEN1 is made at the base of the flap (Fig. 6B). Comparative affinity data with variable flap length substrates show that the flap length contributes to the affinity of FEN1 for the substrate. However, the contacts made with the flap leading to the higher affinity do not require threading and may occur prior to threading. Substrate competition experiments with blocked and unblocked flaps indicate that the threading process influences the ability of competitor substrate to sequester FEN1. Moreover, direct measurements of dissociation rate suggest that threading, when allowed, slows the dissociation rate of FEN1. This slowed dissociation of FEN1 from the substrate suggests that the threading process influences binding through a different mechanism than the initial binding process.
FIGURE 6.
Models of FEN1 flap substrate recognition and genome protection. A, FEN1 tracking model. FEN1 identifies the single-stranded 5′ terminus of the downstream flap, tracks down the flap, and binds the base of the flap prior to cleaving the substrate. B, FEN1 threading model. FEN1 initially binds to the flap base and flap nucleotides near the base, threads the single-stranded 5′ flap through the protein, and finally cleaves the substrate. C, FEN1 Short flap processing. In the presence of short flaps, FEN1 binds to the flap base and then directly cleaves the short flap without threading. D, junction between adjacent Okazaki fragments (OF1 and OF2) has most of the elements of a 5′ flap structure. The tracking model provides template protection because FEN1 cannot track onto the template strand because of the annealed upstream Okazaki fragment (OF2). In the threading model, FEN1 binds the template strand between the Okazaki fragments, but cannot thread the template because of OF2. Therefore, the threading model also provides template strand protection.
When considering the threading process for the streptavidin post-blocked substrates, the cleavage assay data (Fig. 2B) suggest that the majority of FEN1 on the streptavidin post-blocked substrate is bound and threaded, poised to cleave with the addition of Mg2+. The competition assay data (Fig. 4B) demonstrate that the unblocked competitor substrate sequesters a similar amount of bound FEN1 from either the unblocked or streptavidin post-blocked substrate. In both cases, the competitor is relatively inefficient at sequestering FEN1. A component of FEN1 may be viewed as trapped on the streptavidin pre-bound substrate. However, trapping may not be absolute. A reasonable explanation is that FEN1 that has bound and threaded on the streptavidin post-blocked substrate has an alternate means of dissociation. For example, although FEN1 behaves like a bead on a string, the protein may periodically open so that it can dissociate directly into solution after threading. Moreover, on an unblocked substrate, although bidirectional threading is likely, the unthreading process coupled with dissociation from the base appears to occur slowly.
Overall, the results are consistent with a mechanism in which FEN1 initially contacts the flap base, and then threads the flap through a chamber in the protein (Fig. 6B). Formation of the chamber is likely to involve the helical arch of FEN1, implicated in threading by structural studies (16–18, 20). The protein-flap structure formed by the threading process disassembles much more slowly than the initial direct binding of FEN1 with the flap base. This dissociation, which we interpret as an unthreading process, results in a very slow dissociation rate. Because the affinity is determined by association and dissociation rates, we hypothesize that the threading process slows association and dissociation equivalently and therefore does not influence the overall binding affinity.
Moreover, similar dissociation characteristics that we measured for blocked long and unblocked short flaps suggest that flaps less than 5 nt are not subject to the threading process (Fig. 6C). Reconstitution reactions in vitro indicate that FEN1 mostly processes flaps less than 5 nt during the Okazaki fragment maturation process (25). Together, these observations suggest that threading is not a requirement for the basic flap removal reaction.
Significantly, data from reconstitution of Okazaki fragment processing show evidence for two pathways of flap removal (12). In the first pathway, FEN1 is the only nuclease that processes the displaced flaps. The vast majority of flaps are cleaved when the they are less than 5 nt long (25). In the second pathway, flaps become sufficiently long to bind replication protein A (RPA), which inhibits FEN1 cleavage. The RPA coated flaps are first processed by a second nuclease, Dna2, which creates flaps that are ∼ 5–6 nt in length. RPA is unable to bind these short flaps, so FEN1 can cleave them to form nicked substrates for ligation (12). In both cases, by the time FEN1 acts, flaps are sufficiently short that nearly all of the flaps do not require threading, and the remaining flaps thread for only 1–2 nucleotides. It could very well be that threading limits the rate of Okazaki fragment processing that would in turn limit the entire replication process. Therefore, the new mechanism is satisfying in that it protects the genome and eliminates a step that could be rate-limiting.
The tracking requirement for FEN1 was hypothesized to evolve as a means to protect the genome. The need for tracking inhibits FEN1 from cleaving the single-stranded template region between Okazaki fragments even though the junction between the single and double strands has most of the elements of the flap substrate structure other than the free 5′-end (Fig. 6D). In our new model, threading is not required for flaps less than 5 nt but is still needed for long flaps. This set of requirements is just as effective in preventing genome cleavage as the original proposed mechanism (Fig. 6D).
Additionally, binding at the base of the flap allows FEN1 to be part of the replication complex in which proteins are constantly or transiently bound to proliferating cell nuclear antigen (PCNA) (26, 27). In this mechanism, PCNA would mediate persistent contacts between FEN1 and DNA polymerase δ reminiscent of the structure of E. coli DNA polymerase I in which a polymerase and a FEN1 homologue are actually part of the same polypeptide chain (11). The new model allows the protein complex to remain intact while the substrate strands move through it, whereas, the old model suggests that FEN1 breaks away to access the 5′ flap end then reforms with the complex prior to cleavage.
Another possible reason to bind the base of the flap is that some flaps are envisioned to be transiently blocked either by hairpin structures in the flap, bubble structures, in which the 5′ flap is bound to a single-stranded region of the template of the replication intermediates, or by recombination intermediates, in which the flap binds to a homologous segment of DNA from an adjacent chromosome. These structured flaps block FEN1 threading and ultimately prohibit cleavage. Thus, binding of FEN1 to these blocked substrates would increase the relative local concentration of FEN1 on the substrate. When these blocked structures are resolved, FEN1 would be able thread and cleave the flap (28–31).
Overall, our results are consistent with a mechanism in which FEN1 binds the base of the flap. FEN1 will cleave without threading while the growing flaps are short. When a flap becomes long, FEN1 will thread the flap through the protein and cleave. The new mechanism fulfills all the biological requirements of FEN1 as did the original mechanism but also has apparent kinetic advantages.
Supplementary Material
Acknowledgments
We thank Dr. Jeffrey J. Hayes and members of the Bambara laboratory for helpful suggestions and discussions. We would like to thank Edward M. Kennedy and Dr. Baek Kim at the University of Rochester for help with active site titration experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant GM024441 (to R. A. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- LP-BER
- long patch-base excision repair
- FEN1
- flap endonuclease 1
- nt
- nucleotide
- EMSA
- electrophoretic mobility shift assay
- ssDNA
- single-stranded DNA
- PCNA
- proliferating cell nuclear antigen.
REFERENCES
- 1.Rossi M. L., Purohit V., Brandt P. D., Bambara R. A. (2006) Chem. Rev. 106, 453–473 [DOI] [PubMed] [Google Scholar]
- 2.Kim K., Biade S., Matsumoto Y. (1998) J. Biol. Chem. 273, 8842–8848 [DOI] [PubMed] [Google Scholar]
- 3.Klungland A., Lindahl T. (1997) EMBO J. 16, 3341–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balakrishnan L., Brandt P. D., Lindsey-Boltz L. A., Sancar A., Bambara R. A. (2009) J. Biol. Chem. 284, 15158–15172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrington J. J., Lieber M. R. (1994) EMBO J. 13, 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kao H. I., Henricksen L. A., Liu Y., Bambara R. A. (2002) J. Biol. Chem. 277, 14379–14389 [DOI] [PubMed] [Google Scholar]
- 7.Murante R. S., Rust L., Bambara R. A. (1995) J. Biol. Chem. 270, 30377–30383 [DOI] [PubMed] [Google Scholar]
- 8.Barnes C. J., Wahl A. F., Shen B., Park M. S., Bambara R. A. (1996) J. Biol. Chem. 271, 29624–29631 [DOI] [PubMed] [Google Scholar]
- 9.Bornarth C. J., Ranalli T. A., Henricksen L. A., Wahl A. F., Bambara R. A. (1999) Biochemistry 38, 13347–13354 [DOI] [PubMed] [Google Scholar]
- 10.Tom S., Henricksen L. A., Bambara R. A. (2000) J. Biol. Chem. 275, 10498–10505 [DOI] [PubMed] [Google Scholar]
- 11.Robins P., Pappin D. J., Wood R. D., Lindahl T. (1994) J. Biol. Chem. 269, 28535–28538 [PubMed] [Google Scholar]
- 12.Bae S. H., Bae K. H., Kim J. A., Seo Y. S. (2001) Nature 412, 456–461 [DOI] [PubMed] [Google Scholar]
- 13.Wu X., Li J., Li X., Hsieh C. L., Burgers P. M., Lieber M. R. (1996) Nucleic Acids Res. 24, 2036–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brosh R. M., Jr., Driscoll H. C., Dianov G. L., Sommers J. A. (2002) Biochemistry 41, 12204–12216 [DOI] [PubMed] [Google Scholar]
- 15.Harrington J. J., Lieber M. R. (1995) J. Biol. Chem. 270, 4503–4508 [DOI] [PubMed] [Google Scholar]
- 16.Chapados B. R., Hosfield D. J., Han S., Qiu J., Yelent B., Shen B., Tainer J. A. (2004) Cell 116, 39–50 [DOI] [PubMed] [Google Scholar]
- 17.Ceska T. A., Sayers J. R., Stier G., Suck D. (1996) Nature 382, 90–93 [DOI] [PubMed] [Google Scholar]
- 18.Hosfield D. J., Mol C. D., Shen B., Tainer J. A. (1998) Cell 95, 135–146 [DOI] [PubMed] [Google Scholar]
- 19.Kim C. Y., Park M. S., Dyer R. B. (2001) Biochemistry 40, 3208–3214 [DOI] [PubMed] [Google Scholar]
- 20.Hwang K. Y., Baek K., Kim H. Y., Cho Y. (1998) Nat. Struct. Biol. 5, 707–713 [DOI] [PubMed] [Google Scholar]
- 21.Hohl M., Dunand-Sauthier I., Staresincic L., Jaquier-Gubler P., Thorel F., Modesti M., Clarkson S. G., Schärer O. D. (2007) Nucleic Acids Res. 35, 3053–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y., Potapova O., Leschziner A. E., Grindley N. D., Joyce C. M. (2001) J. Biol. Chem. 276, 30167–30177 [DOI] [PubMed] [Google Scholar]
- 23.Sakurai S., Kitano K., Yamaguchi H., Hamada K., Okada K., Fukuda K., Uchida M., Ohtsuka E., Morioka H., Hakoshima T. (2005) EMBO J. 24, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finger L. D., Blanchard M. S., Theimer C. A., Sengerová B., Singh P., Chavez V., Liu F., Grasby J. A., Shen B. (2009) J. Biol. Chem. 284, 22184–22194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi M. L., Bambara R. A. (2006) J. Biol. Chem. 281, 26051–26061 [DOI] [PubMed] [Google Scholar]
- 26.Dionne I., Nookala R. K., Jackson S. P., Doherty A. J., Bell S. D. (2003) Mol. Cell 11, 275–282 [DOI] [PubMed] [Google Scholar]
- 27.Li X., Li J., Harrington J., Lieber M. R., Burgers P. M. (1995) J. Biol. Chem. 270, 22109–22112 [DOI] [PubMed] [Google Scholar]
- 28.Wang W., Bambara R. A. (2005) J. Biol. Chem. 280, 5391–5399 [DOI] [PubMed] [Google Scholar]
- 29.Bartos J. D., Wang W., Pike J. E., Bambara R. A. (2006) J. Biol. Chem. 281, 32227–32239 [DOI] [PubMed] [Google Scholar]
- 30.Sharma S., Sommers J. A., Gary R. K., Friedrich-Heineken E., Hübscher U., Brosh R. M., Jr. (2005) Nucleic Acids Res. 33, 6769–6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bae S. H., Kim D. W., Kim J., Kim J. H., Kim D. H., Kim H. D., Kang H. Y., Seo Y. S. (2002) J. Biol. Chem. 277, 26632–26641 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.