Abstract
Stbd1 is a protein of previously unknown function that is most prevalent in liver and muscle, the major sites for storage of the energy reserve glycogen. The protein is predicted to contain a hydrophobic N terminus and a C-terminal CBM20 glycan binding domain. Here, we show that Stbd1 binds to glycogen in vitro and that endogenous Stbd1 locates to perinuclear compartments in cultured mouse FL83B or Rat1 cells. When overexpressed in COSM9 cells, Stbd1 concentrated at enlarged perinuclear structures, co-localized with glycogen, the late endosomal/lysosomal marker LAMP1 and the autophagy protein GABARAPL1. Mutant Stbd1 lacking the N-terminal hydrophobic segment had a diffuse distribution throughout the cell. Point mutations in the CBM20 domain did not change the perinuclear localization of Stbd1, but glycogen was no longer concentrated in this compartment. Stable overexpression of glycogen synthase in Rat1WT4 cells resulted in accumulation of glycogen as massive perinuclear deposits, where a large fraction of the detectable Stbd1 co-localized. Starvation of Rat1WT4 cells for glucose resulted in dissipation of the massive glycogen stores into numerous and much smaller glycogen deposits that retained Stbd1. In vitro, in cells, and in animal models, Stbd1 consistently tracked with glycogen. We conclude that Stbd1 is involved in glycogen metabolism by binding to glycogen and anchoring it to membranes, thereby affecting its cellular localization and its intracellular trafficking to lysosomes.
Keywords: Carbohydrate-binding Protein, Carbohydrate Metabolism, Glycogen, Lysosomes, Trafficking, GABARAP, GABARAPL1, Genethonin 1, Stbd1
Introduction
Glycogen is a branched storage polymer of glucose that serves as an energy reserve in many cell types, with liver and skeletal muscle housing the largest deposits in mammals (1–3). Glycogen metabolism and its regulation have been studied for decades, with most focus on its cytosolic synthesis and degradation in relation to mechanisms of enzyme regulation, intracellular energy metabolism, and whole body glucose homeostasis. Glycogen biosynthesis is initiated by a specialized self-glucosylating protein, called glycogenin, followed by bulk synthesis mediated by glycogen synthase and the branching enzyme. Regulated breakdown of glycogen, to fuel contractile activity in muscle or to generate free glucose in the liver for blood glucose homeostasis, is mediated by glycogen phosphorylase and debranching enzyme. Although glycogen metabolism is usually considered cytosolic, electron microscopy studies generally place glycogen in relative proximity to membranous structures, like the endoplasmic reticulum in liver (4) or the sarcoplasmic reticulum in muscle (5). In several disease states and some genetically modified mouse models, aberrant glycogen metabolism results in the accumulation of abnormal glycogen deposits. Glycogen is also transported to lysosomes where it is directly hydrolyzed to glucose by a lysosomal α-glucosidase (acid maltase) (6). Although probably not the major degradative mechanism under normal circumstances, the significance of this pathway is emphasized by the symptoms of patients with Pompe disease in which the α-glycosidase gene is mutated (7–9). The severity of the phenotype varies with the degree of impairment of glycosidase activity, in the worst cases leading to death within the 1st year after birth. In the disease, undegraded glycogen accumulates in the lysosomes, resulting in potentially fatal tissue damage.
The molecular mechanism by which glycogen is transferred to the lysosome is poorly understood but could involve an autophagy-like pathway. Autophagy describes a set of processes whereby cellular materials are transported to the lysosome for recycling (10–12). In macroautophagy, cytosolic materials are engulfed by a lipid bilayer in structures called autophagosomes that are ultimately delivered to the lysosome. The first genetic link between autophagy and glycogen came from genetic screens in Saccharomyces cerevisiae that linked glycogen accumulation with mutations of Atg1 and Atg13, two proteins implicated in the regulation of autophagy (13). It has also been shown that, in Pompe disease, the inability to dispose of lysosomal glycogen results in a build up of autophagic and late endosomal vesicles (14). In a mouse model of Pompe disease, additional disruption of Atg7, another gene involved in macroautophagy, results in decreased accumulation of lysosomal glycogen (15).
The concept of higher order assemblages of glycogen molecules and associated proteins is not new (5, 16). Fischer and co-workers (17–19) were able to partially purify from muscle what they termed “glycogen particles,” which contained glycogen, several proteins, and elements of sarcoplasmic reticulum. The particles result from the ability of associating proteins to bind to glycogen, sometimes to each other, and sometimes to membranes (Fig. 1B). Known glycogen-associated proteins are glycogenin, the metabolic enzymes glycogen synthase, glycogen phosphorylase, and the debranching enzyme and several regulatory proteins, including phosphorylase kinase and members of the PP1G family of type 1 protein phosphatases (PP1s). PP1G enzymes exist as a PP1 catalytic subunit associated with a glycogen-targeting regulatory subunit (20), of which RGL/GM (PPP1R3A), GL (PPP1R3B), and PTG (PPP1R3C) are the best studied. A more recently identified glycogen-associated protein is laforin, by sequence similarity a member of the atypical dual specificity protein phosphatase subfamily, but which is in fact a glycogen phosphatase involved in maintaining glycogen structural integrity (21, 22). Laforin binds to glycogen via a CBM20 carbohydrate-binding module (23). Virtually all established glycogen-associating proteins have been shown to have a functional role in glycogen metabolism. In this work, we demonstrate that Stbd1, starch binding domain-containing protein 1 (sometimes called genethonin 1),2 can be added to the list of glycogen-binding proteins.
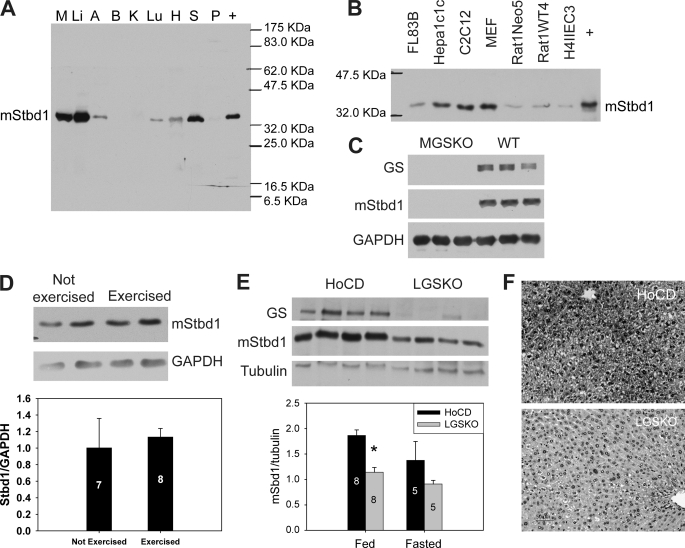
FIGURE 1.
Tissue distribution of Stbd1 protein and correlation with glycogen level in mice. A, Stbd1 protein levels in mouse tissues as judged by Western blotting with anti-mStbd1 antibodies. M, skeletal muscle; Li, liver; A, adipose; B, brain; K, kidney; Lu, lung; H, heart; S, spleen; P, pancreas; +, extract of COS M9 cells overexpressing mStbd1 (running larger because of C-terminal HA tag). Loadings were 10 μg of total protein. B, presence of Stbd1 protein in mouse and rat cell lines, as described in the text. +, COS M9 cells overexpressing mStbd1. C, Stbd1 protein level in skeletal muscle from muscle glycogen synthase knock-out (MGSKO) mice, with the Gys1 gene disrupted, and control WT littermates. GS, glycogen synthase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase, the loading control. D, Stbd1 levels in exercised versus nonexercised wild type mice. A representative Western blot is shown as well as quantitation in the lower bar graph. Numbers within the bars indicate the number of mice analyzed. E, representative Western blots for Stbd1 in livers of fed floxed conditional (HoCD) control mice and liver glycogen synthase knock-out (LGSKO) mice, with Gys2 gene disrupted. Quantitation of Stbd1 levels in control or LGSKO mice under fed or fasted conditions is shown in the bar graph below. *, p < 0.05 with respect to HoCD. F, Stbd1 visualized in liver sections of fed LGSKO and control mice (HoCD) by immunohistochemical staining.
Stbd1 was first identified as being encoded by a novel gene with enriched expression in skeletal muscle and was termed GENX-3414 (24). It was prominently expressed in liver, heart, and placenta. Bouju et al. (24) proposed a membrane association of the protein in muscle tissue, in T-tubules, and sarcoplasmic reticulum. Janecek (25) identified the same protein, which he referred to as genethonin, in a bioinformatics analysis directed at conserved starch binding domains. Stbd1 is one of a small number of proteins in mammalian genomes that contain highly conserved putative CBM20 carbohydrate binding domains (26). Stbd1 has been little studied until recently. During the preparation of this manuscript, Stapleton et al. (27) described a proteomic analysis that identified Stbd1 as a protein associated with glycogen in the liver. Our data also show that Stbd1 binds to glycogen and related polysaccharides in vitro and that in different cell and animal models Stbd1 is linked to glycogen metabolism. Our hypothesis is that Stbd1 functions to tether glycogen to membranes, thereby affecting its localization and possibly its intracellular trafficking to lysosomes.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Mammalian expression vectors containing HA-tagged hStbd1 (hStbd1-HA) and different truncation mutants thereof (ΔN24-HA, ΔN90-HA, and ΔC96-HA) for mammalian expression were made by PCR amplification of a human cDNA with addition of an HA tag at the C terminus. The products were subcloned into BamHI/EcoRI sites of the pcDNA3 vector. Plasmids with point mutations in the carbohydrate binding domain (W293G-HA and W293L-HA) were constructed by site-directed mutation using pcDNA3-hStbd1-HA as template. The primers containing the mutated sequences were designed using the on-line service of PrimerX and were synthesized by Invitrogen. The Pfu Turbo DNA polymerase (Stratagene) was used for the PCR. The PCR product was digested with DpnI (New England Biolabs) at 37 °C for 2–3 h to remove the parental DNA. The PCR product was transformed into competent cells to generate pcDNA3-W293G and pcDNA3-W293L. cDNA encoding GABARAP and GABARAPL1 were respectively subcloned into NotI/XbaI and EcoRI/XbaI sites of pFLAGCMV-2 vectors. His-tagged or GST fusion hStbd1 (human Stbd1) and mStbd1 (mouse Stbd1) vectors for bacterial expression were constructed by subcloning into pET28a or pGEX vectors. Sequences of all constructs were verified by the DNA Sequencing Core Facility, Indiana University School of Medicine.
Expression and Purification of Recombinant Proteins
Escherichia coli BL21 (DE3) competent cells transformed with pET28a-hStbd1, pET28a-mStbd1, and pGEX-mStbd1 vectors were grown at 37 °C until the A600 reached 0.4. Protein expression was induced by 0.4 mm isopropyl β-d-thiogalactoside (IPTG) at 18 °C overnight. The His-tagged hStbd1 and mStbd1 were purified by Ni2+-NTA-agarose chromatography (Qiagen). The proteins were eluted stepwise with 40–200 mm imidazole. hStbd1 and mStbd1 were eluted primarily in fractions with 100 and 200 mm imidazole. The eluted fractions were dialyzed against buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl and 5% 2-mercaptoethanol and stored at −80 °C with 15% glycerol. The His-tagged mStbd1 was dialyzed against phosphate-buffered saline (PBS), pH 7.4, and bound to Affi-Gel 15 for antibody affinity purification. The GST fusion mStbd1 for antibody generation was purified using glutathione-agarose (Sigma). The recombinant protein was eluted with 10 mm glutathione and dialyzed against PBS.
Antibodies
Rabbit polyclonal antibodies were generated against GST-mStbd1 by Cocalico Biologicals. The antibody was affinity-purified by Affi-Gel 15 (Bio-Rad) coupled with His-tagged mStbd1. Rabbit polyclonal anti-HA epitope tag antibodies were from Rockland (Gilbertsville). Mouse monoclonal anti-HA epitope tag antibodies were from Covance (Emeryville). Mouse monoclonal LAMP1 and β-tubulin antibodies were from the Developmental Studies Hybridoma Bank, University of Iowa. Mouse monoclonal HDEL antibodies were from Santa Cruz Biotechnology, Inc. Rabbit monoclonal Syntaxin 6 and glycogen synthase antibodies were from Cell Signaling. Mouse monoclonal glycogen synthase antibodies were from Invitrogen. Rabbit polyclonal anti-hStbd1 antibodies (anti-GENX-3414) used in Fig. 2, A, B, and C were from Protein Tech Group. Rabbit polyclonal GABARAPL1 antibodies were from Protein Tech Group, and mouse monoclonal LC3 antibodies were from NanoTools. Mouse monoclonal anti-FLAG M2 antibodies were from Sigma.
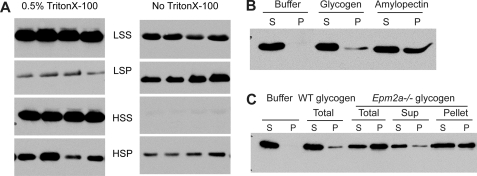
FIGURE 2.
Association of Stbd1 protein with glycogen in vitro. A, fractionation of Stbd1 by centrifugation of wild type mouse muscle extracts in the presence or absence of Triton X-100. Fractions were analyzed by Western blotting using anti-mStbd1 antibodies, as described under “Experimental Procedures.” LSS, supernatant after centrifugation at 8,000 × g centrifugation; LSP, pellet after centrifugation at 8,000 × g; HSS, supernatant after centrifugation at 100,000 × g; HSP, pellet after centrifugation at 100,000 × g. The panels shown were cropped from a single autoradiogram of a single membrane. B, recombinant human Stbd1 protein binding to glycogen and amylopectin. Rabbit liver glycogen and potato amylopectin (Sigma) were incubated with recombinant hStbd1, centrifuged and analyzed by Western blotting using anti-hStbd1 antibodies. S, supernatant after centrifugation at 100,000 × g; P, pellet after centrifugation at 100,000 × g. The negative control contained Stbd1 but without polysaccharide (Buffer). C, recombinant human Stbd1 protein binding to glycogen purified from wild type and laforin knock-out mice. Total glycogen was purified from mouse skeletal muscle of wild type (WT) or laforin knock-out (Epm2a−/−) mice. Muscle glycogen from laforin knock-out mice was additionally fractionated by low speed centrifugation (10,000 × g) to separate supernatant and pellet. Glycogen was purified from this supernatant (Sup) and pellet (Pellet). Purified mouse glycogen (WT total, Epm2a−/− total, Epm2a−/− supernatant, and Epm2a−/− pellet) was incubated with recombinant hStbd1, centrifuged at 100,000 × g, and the supernatant (S) and pellet (P) fractions analyzed by Western blotting. Stbd1 without added glycogen (Buffer) was the negative control.
Cell Culture and Transfections
COS M9 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Mediatech or Sigma) with 25 mm glucose and 10% fetal bovine serum (FBS) (Atalanta Biologicals, Lawrenceville, GA). Transfections were performed with FuGENE 6 (Roche Applied Science) following the manufacturer's instructions. Glucose-starved COS M9 cells were incubated in DMEM without glucose and supplemented with 1 mm sodium pyruvate (Invitrogen) and 10% FBS. The normal “fed” condition had 25 mm d-glucose (Sigma).
Rat1Neo5 and Rat1WT4 cells (28) were grown in DMEM with 25 mm glucose and supplemented with 10% fetal bovine serum and 0.25 mg/ml geneticin. Rat1WT4 is a stably transfected cell line overexpressing wild type rabbit muscle glycogen synthase. Rat1Neo5 cells are a control line transfected with vector. FL83B cells (ATCC) were grown in F-12K medium (ATCC) and supplemented with 10% FBS. C2C12, Hepa1c1c, HepG2, HII4EC3, and mouse embryonic fibroblasts were grown in DMEM with 25 mm glucose with 10% FBS. C2C12 cells were differentiated in DMEM with 25 mm glucose and supplemented with 1% bovine calf serum. All cells were incubated at 37 °C with 5% CO2.
Co-immunoprecipitation of Stbd1, GABARAPL1, and GABARAP
COSM9 cells co-transfected with pcDNA3-hStbd1-HA and pCMVFLAG-GABARAP or with pcDNA3-hStbd1-HA and pCMVFLAG-GABARAPL1 were lysed and centrifuged at 8,000 × g for 15 min at 4 °C. EZview red anti-FLAG gel or EZview red anti-HA affinity gel (Sigma) was equilibrated with lysis buffer before use. The supernatant from cell lysates was mixed with affinity gel and incubated at 4 °C overnight. The samples were centrifuged at 8,000 × g for 30 s to pellet the agarose. The agarose was washed by lysis buffer three times, and loading buffer was added for SDS-PAGE and immunoblotting.
Immunofluorescence Staining and Microscopy
Cells were grown on glass coverslips for 24 or 48 h before fixation. Cells were fixed in PBS with 4% paraformaldehyde. Cells were then quenched and permeabilized in PBS with 100 mm glycine and 0.2% Triton X-100. Nonspecific binding sites were blocked with 5% bovine serum albumin (BSA) (Sigma) in PBS before addition of primary antibodies diluted in PBS with 2% BSA. Antibody dilutions were as follows: anti-mStbd1, 1:200; anti-HA, 1:1000; anti-LAMP1, 1:50; anti-FLAG, 1:1000; anti-GABARAPL1, 1:100; anti-LC3, 1:100; and anti-glycogen synthase, 1:200. Cells were then washed in PBS with 2% BSA and developed with secondary antibodies (1:400, Invitrogen), conjugated with either Texas Red or Alexa488 fluorophores. Nuclei were visualized by staining with 1 μg/ml Hoechst (Invitrogen). The specimens were imaged using a Zeiss Axio Observer Z1 microscope with a Plan Apochromat ×63 oil immersion objective (Zeiss) as structured light acquired via an Apotome (Zeiss). Images were processed with Zeiss Axiovision 4.7.
Periodic Acid-Schiff Reagent Staining and Microscopy
Fixed cells were rinsed with H2O and oxidized with 0.5% periodic acid for 5 min, stained with Schiff reagent for 15 min, and then counterstained in Harris' hematoxylin for 1 min. Negative controls without periodic acid treatment were processed at the same time to ensure staining specificity. The PAS3-stained samples were viewed on a Leica DM3000 microscope (Leica). Images were processed with Leica Application Suite version 3.5.0. When immunofluorescence staining was applied following by PAS staining, the counterstaining step was omitted. The method for the combined PAS and immunofluorescence staining was adapted from Ref. 29. The samples with both PAS and immunofluorescence staining were viewed on a Zeiss Axio Observer Z1 microscope, and images were processed with Zeiss Axiovision 4.7.
Glycogen Purification and Polysaccharide Binding Assay
Glycogen was purified from mouse muscle as described previously (21). Briefly, tissues were boiled in 30% KOH, and lipids were removed by methanol/chloroform extraction. Glycogen precipitated in cold ethanol and redissolved in water was treated with 10% trichloroacetic acid (TCA). After centrifugation, glycogen recovered from the supernatant by ethanol precipitation was subjected to extensive dialysis.
Recombinant hStbd1 was diluted in 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% 2-mercaptoethanol, 0.1% Triton X-100 and subjected to 10,000 × g centrifugation at 4 °C for 20 min. The supernatant was mixed with the supernatant of glycogen/amylopectin centrifuged under the same conditions. The final concentration of protein was 2.5 μg/ml and glycogen/amylopectin was 0.25 mg/ml. hStbd1 without polysaccharide was used as a negative control. The samples were mixed well, incubated at 4 °C for 1 h, and then subjected to ultracentrifugation at 100,000 × g at 4 °C for 90 min. The pellets were resuspended by sonication. The supernatant and pellets were subjected to Western blotting for hStbd1.
Preparation of Tissue and Cell Extracts and Immunoblotting
MGSKO mice (30) and LGSKO mice (31) have been described previously. They carry disruptions of the glycogen synthase genes Gys1 or Gys2, respectively. Animals were maintained in AALAAC approved facilities of the Indiana University School of Medicine Laboratory Animal Research Center, with all experimental protocols approved by the Indiana University School of Medicine IACUC. The exercise protocol followed a previous study (32). Mice were sacrificed by cervical dislocation. Tissues were rapidly frozen in liquid nitrogen and stored at −80 °C. Frozen tissues were powdered and homogenized in buffer containing 50 mm Tris-HCl, pH 7.8, 10 mm EDTA, 2 mm EGTA, 0.1 mm N-p-tosyl-l-lysine chloromethyl ketone (TLCK), 2 mm benzamidine, 0.5 mm phenylmethylsulfonyl fluoride (PMSF), 50 mm 2-mercaptoethanol, and 10 μg/ml leupeptin, with or without 0.5% Triton X-100. Tissue homogenates were used for Western blotting analysis. For analysis of mStbd1 and glycogen fractionation, tissue homogenates were first subjected to low speed centrifugation 10,000 × g at 4 °C for 10 min to generate a low speed pellet (LSP), and the supernatant (LSS) was subjected to further centrifugation. Centrifugation at 100,000 × g at 4 °C for 90 min generated a high speed supernatant (HSS) and glycogen was collected in the high speed pellet fraction (HSP). The HSP was resuspended in the starting volume of homogenization buffer. All fractions were analyzed by Western blotting with anti-mStbd1 antibodies.
Cultured cells were lysed in buffer containing 50 mm Tris-HCl, 100 mm NaCl, pH 7.5, 0.5% Triton X-100, 1 mm PMSF, 0.1 mm TLCK, 1 mm benzamidine, 1 μg/ml aprotinin, pepstatin, and leupeptin. The cell lysates were centrifuged at 10,000 × g for 15 min at 4 °C to pellet insoluble materials.
Protein concentration was determined by the Bradford method using BSA as standard (33). Samples were subjected to 10% SDS-PAGE. Proteins were transferred to nitrocellulose membranes and incubated with antibodies, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies and ECL (Thermo Scientific).
Immunohistochemistry
Fixed mouse liver sections were deparaffinized and then incubated with anti-mStbd1 and HRP-conjugated secondary antibody. The staining was developed with 3,3′-diaminobenzidine reagent in the presence of HRP. The samples were rehydrated before mounting.
Statistical Analysis
The data are presented as means ± S.E. Statistical significance was determined by unpaired Student's t test, and significance was assigned at p < 0.05.
RESULTS
Structure of Stbd1
The architecture of Stbd1 includes an N-terminal hydrophobic segment, a putative leucine zipper domain, and a CBM20 domain at the C terminus (supplemental Fig. S1). Human Stbd1 (hStbd1; 358 residues) is 20 amino acids longer than the mouse protein (mStbd1) with overall 60% identity. Rat Stbd1 is 88 and 63% identical to the mouse and human proteins respectively. Orthologs of Stbd1 are present in mammals but, from bioinformatic analysis, appear to be absent in other vertebrates such as fish and birds, even though the CBM20 domain itself is ancient and can be found in bacteria and archaea. There is a very high degree of conservation in the hydrophobic N terminus and CBM20 domains of Stbd1 (supplemental Fig. S2). A yeast two-hybrid screen using as bait Stbd1 lacking the first 171 residues picked four Stbd1 clones of varying length, with the shortest, identified twice, containing little more than the CBM20 domain (supplemental Fig. S1). It is therefore likely that Stbd1 is a dimer formed by interactions between the CBM20 domains. If the putative leucine zipper has any functional role, it does not appear to be related to oligomerization.
Tissue Distribution of Stbd1
Antibodies raised against recombinant mouse Stbd1 identified a single protein species of ∼36 kDa in extracts of most mouse tissues tested (Fig. 1A), with especially prominent signals in muscle and liver, the main glycogen-accumulating organs, consistent with the earlier report (24). Overexposure of the autoradiogram revealed trace signals in brain, kidney, and pancreas. By Western blotting, we also detected endogenous Stbd1 in several cultured cell lines, including mouse C2C12, Hepa1c1c, embryonic fibroblasts (MEF), and FL83B cells and rat H4IIEC3 and Rat1 cells (Fig. 1B). This anti-mouse Stbd1 antibody, however, did not recognize Stbd1 in human HepG2 or simian COS M9 cells, presumably because of relatively low sequence similarities.
Stbd1 in Mice with Genetically Altered Glycogen Levels
Muscle glycogen synthase knock-out (MGSKO) mice (30) have a disruption of the glycogen synthase gene, Gys1, which is expressed in skeletal muscle and most tissues that produce glycogen other than liver. The MGSKO mice have undetectable glycogen in all tissues analyzed except for liver. Western analysis of muscle extracts from MGSKO mice indicated virtually undetectable Stbd1 protein compared with controls (Fig. 1C). Muscle glycogen can be acutely depleted to ∼10% of the starting level by exercising mice to exhaustion on a treadmill (32). Such an exercise regimen did not affect muscle Stbd1 levels in wild type mice as judged by Western blotting (Fig. 1D).
Liver glycogen synthase knock-out (LGSKO) mice (31) have the second glycogen synthase gene, Gys2, disrupted in the liver so that the liver glycogen content in the fed state is decreased by 95% compared with the wild type. In extracts of livers from fed LGSKO mice, there was a concomitant 40% reduction in Stbd1 protein compared with control mice (Fig. 1E). A reduction in Stbd1 was also observed by immunohistochemical staining of liver sections (Fig. 1F). Liver glycogen can be depleted in wild type mice by overnight fasting, which reduces the level to ∼5% of the fed state, comparable with that of the fed LGSKO liver (31). Fasting did not significantly reduce the liver Stbd1 levels in either wild type or LGSKO mice (Fig. 1E). The data with MGSKO and LGSKO mice indicate a genetic link between glycogen and Stbd1, demonstrating that severely reduced or absent glycogen over the long term correlates with decreased Stbd1 protein. However, short term decreases in either liver or muscle glycogen in wild type mice do not have major effects on Stbd1 protein level.
Interaction of Stbd1 with Glycogen
Glycogen in muscle extracts can be fractionated by centrifugation, typically by first removing gross cellular debris, including contractile proteins and associated structures, with low speed centrifugation followed by ultracentrifugation of the supernatant to sediment the glycogen particles. About half of the Stbd1 was recovered in the low speed pellet (Fig. 2A, LSP). Recovery of Stbd1 in the low speed pellet is most likely due to association with membranous components because inclusion of Triton X-100 in the homogenization buffer greatly increased the fraction of Stbd1 that remained in the supernatant (Fig. 2A, LSS). After high speed centrifugation of the low speed supernatant in the absence of Triton X-100, almost all of the soluble Stbd1 was recovered in the high speed pellet fraction (Fig. 2A, HSP). In the presence of detergent, a substantial amount of Stbd1 was still recovered in the high speed pellet, although some remained in the high speed supernatant. We conclude that a significant fraction of the Stbd1 in a muscle extract co-fractionates with glycogen because of direct binding to the polysaccharide.
Because of the presence of the CBM20 domain, we analyzed the ability of Stbd1 to bind to polysaccharides. Purified recombinant human Stbd1 bound to glycogen and amylopectin in vitro as judged by co-sedimentation during ultracentrifugation (Fig. 2B). Based on this assay, Stbd1 bound more effectively to amylopectin, the predominant polysaccharide of starch, which differs from glycogen in being less branched and more phosphorylated (34). Like amylopectin, glycogen contains small amounts of covalent phosphate, one phosphate per 650–1500 glucose residues, whose function is poorly understood (21, 22). Lafora disease is a progressive myoclonus epilepsy that is characterized by teenage-onset seizures leading to death usually within 10 years (35–37). Lafora disease is characterized by the accumulation of Lafora bodies, deposits that contain poorly branched glycogen. Increased phosphorylation of glycogen in Lafora bodies has been reported (38), and in a mouse model of the disease, glycogen phosphorylation was increased (21, 22). In humans, about 90% of Lafora disease cases can be attributed to mutations in either the EPM2A or the EPM2B gene. EPM2A encodes a glycogen phosphatase, laforin, that we believe functions to limit glycogen phosphorylation to a level permissive of normal glycogen metabolism (21, 22). In the absence of laforin in Epm2a −/− mice, there is a progressive increase in glycogen phosphorylation leading to a derangement of glycogen structure with age, accompanied also by reduced branching and solubility. Stbd1 bound better to glycogen purified from the muscle of 10–12-month-old Epm2a−/− mice than normal mouse muscle glycogen (Fig. 2C). Low speed centrifugation of muscle extracts from Epm2a−/− mice separates the glycogen into supernatant and pellet. The glycogen can be purified from the two fractions to yield relatively normal glycogen in the supernatant and poorly branched, more phosphorylated glycogen in the pellet (22). Stbd1 bound more tightly to glycogen purified from the pellet than the supernatant fraction (Fig. 2C). We conclude that purified Stbd1 can bind to glycogen and interacts preferentially with less branched and/or more phosphorylated polysaccharides.
Subcellular Localization of Stbd1
Detection of endogenous Stbd1 by immunofluorescence in either the FL83B mouse liver cell line (39) or a Rat1 fibroblast cell line, Rat1Neo5 (28), revealed that it predominantly concentrated at perinuclear structures, with diameter up to ∼0.5 μm (Fig. 3A). These structures partly coincided with the presence of LAMP1, a lysosomal marker, and with endoplasmic reticulum detected with antibodies against the residues HDEL that are known to mark the endoplasmic reticulum (supplemental Fig. S3). Stbd1 did not co-localize with microtubules visualized by an antibody directed against β-tubulin but was generally within areas with the densest microtubular network (supplemental Fig. S3).
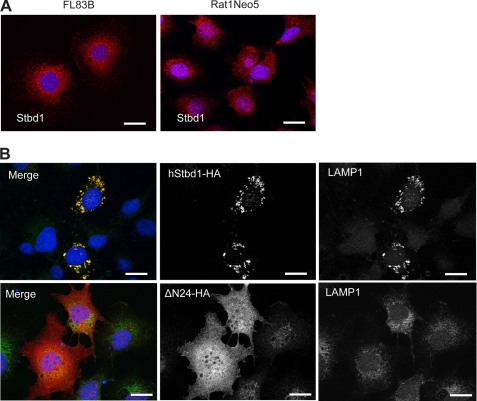
FIGURE 3.
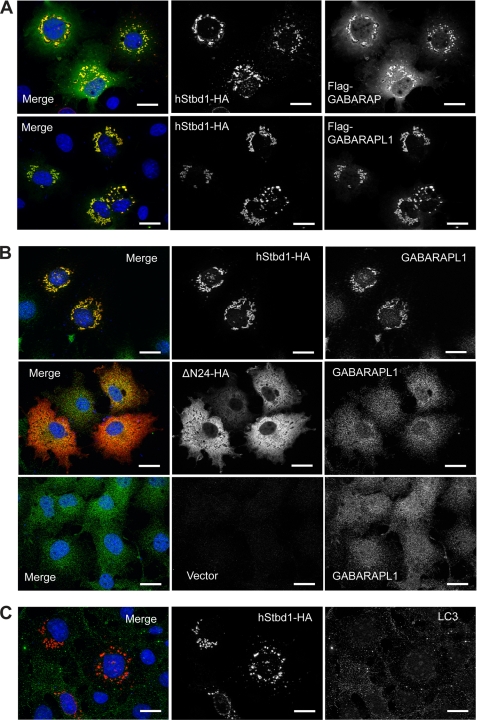
Subcellular localization of Stbd1. A, FL83B (left panel) and RatNeo5 (right panel) cells were immunostained with anti-mStbd1 antibodies (red) to detect endogenous Stbd1. B, full-length Stbd1-HA (upper) or a truncation lacking residues 1–24 (DN24-HA; lower) were overexpressed in COS M9 cells and immunostained with anti-HA antibodies to detect Stbd1 (middle panels) or antibodies directed toward the lysosomal marker LAMP1 (right panels). The images were merged (left panels; Stbd1, red; LAMP1, green). Nuclei were stained with Hoechst (blue). Scale bar, 20 μm.
HA-tagged human Stbd1 transiently expressed in COS M9 cells concentrated at similar perinuclear structures that were enlarged to several micrometers in diameter (Fig. 3B). There was consistent co-localization of LAMP1 in these structures, although often LAMP1 staining was also observed in other regions of the cell. Syntaxin 6, a trans-Golgi marker, as well as anti-HDEL antibodies that mark the endoplasmic reticulum, also co-distributed with these structures but to a lesser degree than LAMP1 (supplemental Fig. S4, B and C). Localization with respect to β-tubulin was similar to what was observed with endogenous Stbd1 (supplemental Fig. S4D). Our interpretation is that the overexpression of Stbd1 impacts the trafficking of various subcellular compartments to promote the accumulation of these marker proteins into the enlarged perinuclear structures containing Stbd1.
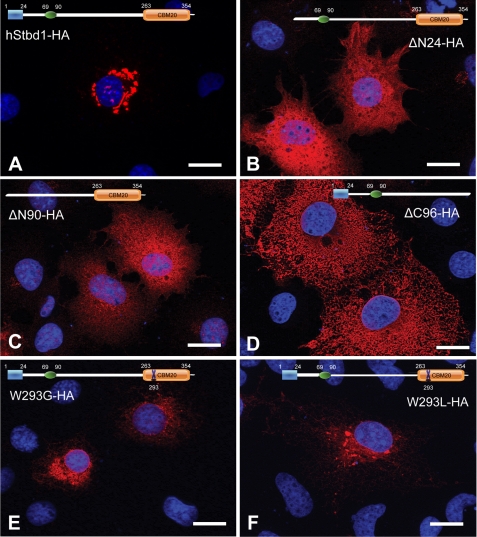
The ectopic expression of Stbd1 allowed us to use mutational analysis to assess the role of different domains of the protein in its subcellular localization. Removal of the conserved hydrophobic N-terminal 24 amino acids resulted in complete loss of the large perinuclear structures and a diffuse cytosolic distribution of Stbd1 (Fig. 4B). Note that expression of this construct did not affect the localization of endogenous LAMP1 and in particular did not provoke LAMP1 appearance in the large perinuclear structures suggesting that membrane association of Stbd1 is necessary for their formation (Fig. 3B). Further N-terminal truncation to delete also the putative leucine zipper domain led to an identical pattern. These results are consistent with the hydrophobic region having an important role in subcellular localization, most likely by directing the protein to membrane compartments. Interestingly, deletion of the carbohydrate binding domain also eliminated the perinuclear localization, although the distribution of the shortened protein is quite different from that of the N-terminal deletion, and had a more reticular appearance. Loss of the CBM20 domain would have disabled oligomerization of Stbd1, which we therefore infer is also required for localization in the perinuclear structures. Subcellular distribution of this truncated Stbd1 may be driven solely by nonspecific insertion of Stbd1 into membranes via the hydrophobic N terminus. We also expressed point mutants of Stbd1 in which a highly conserved Trp residue in the CBM20 domain was changed to either Gly or Leu with the objective of disabling glycogen binding. These mutations were chosen because a mutation in the laforin CBM20 domain, W32G, had been found in a Lafora patient and shown to eliminate glycogen binding (23, 43). The Stbd1 mutants, W293G and W293L, also had a perinuclear localization but did not cause the formation of the large, well defined structures seen with the wild type protein (Fig. 4, E and F).
FIGURE 4.
Mutational analysis of Stbd1 expressed in COS M9 cells. Mutated hStbd1 with a C-terminal HA tag was expressed in COS M9 cells and immunostained with anti-HA antibodies (red). A, full-length human Stbd1. B, deletion of the N-terminal hydrophobic segment (ΔN24-HA). C, deletion of the N-terminal hydrophobic segment and the putative leucine zipper (ΔN90-HA). D, deletion of the C-terminal carbohydrate binding domain (ΔC96-HA). E, mutation of conserved Trp of CBM20 domain to Gly (W293G). F, mutation of conserved Trp of CBM20 domain to Leu (W293L). Nuclei were stained with Hoechst (blue). Scale bar, 20 μm.
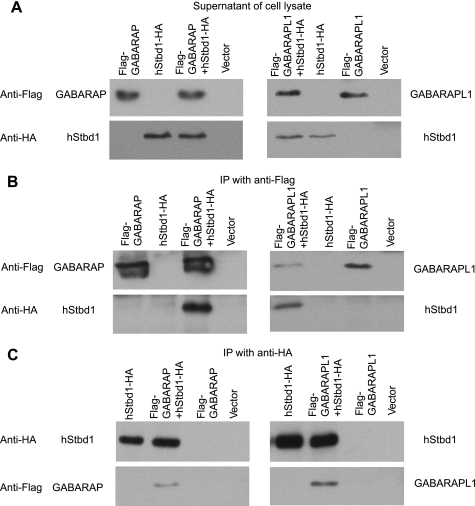
The same yeast two-hybrid screen that suggested that Stbd1 was a dimer also identified as potential interacting proteins GABAA receptor-associated protein (GABARAP) and GABARAP-like 1 (GABARAPL1), two members of the ATG8 family of proteins that are involved in autophagy (40, 41). The interaction of Stbd1 with each protein was confirmed by co-immunoprecipitation of the proteins expressed in COS cells (Fig. 5), whether the pulldown was of the ATG8 protein (Fig. 5B) or of Stbd1 (Fig. 5C). A recent proteomic analysis of human proteins involved in autophagy also reported interaction of Stbd1 with GABARAP and GABARAPL1 (42). In COS cell co-expression, experiments like those used for immunoprecipitation and immunofluorescent staining revealed strong co-localization of GABARAPL1 with Stbd1 in the perinuclear structures described above (Fig. 6A). There was some co-localization of Stbd1 with GABARAP, although not so consistently or strictly as with GABARAPL1 (Fig. 6A). Endogenous microtubule-associated protein-1 light chain 3 (LC3), another member of the ATG8 family, gave a weak signal and did not co-localize with overexpressed Stbd1 (Fig. 6C). To confirm the association of GABARAPL1 with Stbd1, we also analyzed the subcellular distribution of endogenous GABARAPL1. In cells transfected with control vector, GABARAPL1 was present throughout the cytosol, with a punctate appearance (Fig. 6B). When Stbd1 was overexpressed, GABARAPL1 was restricted to the perinuclear structures (Fig. 6B), similar to what was seen with GABARAPL1 overexpression (Fig. 6A). Deletion of the N terminus of Stbd1 eliminates its concentration in the perinuclear structures (Fig. 4), but co-localization with GABARAPL1 was still evident (Fig. 6B); consistent with truncated Stbd1 binding GABARAPL1, the yeast two-hybrid screen that identified GABARAPL1 had used N-terminally truncated Stbd1 as the bait.
FIGURE 5.
Interaction of Stbd1 with GABARAPL1 and GABARAP. hStbd1 with a C-terminal HA-tag, FLAG-tagged GABARAPL1 and FLAG-tagged GABARAP were expressed alone, or in the indicated combination in COS M9 cells. Control cells were transfected with empty pcDNA3 vector (Vector). A, Western blotting of the cell lysates with the indicated antibody. B, immunoprecipitation of GABARAP (left) or GABARAPL1 (right) with Anti-FLAG antibodies covalently bound to agarose followed by Western blotting with the indicated antibody. C, immunoprecipitation of Stbd1 with anti-HA antibodies covalently bound to agarose followed by Western blotting with the indicated antibody.
FIGURE 6.
Subcellular localization of GABARAPL1, GABARAP and LC3 in relation to Stbd1. A, C-terminal HA-tagged hStbd1 was co-expressed in COS M9 cells with N-terminal FLAG-tagged GABARAPL1 or GABARAP, as in Fig. 5, and immunostained with anti-HA antibodies (red) or anti-FLAG antibodies (green). B, HA-tagged Stbd1 or N-terminally truncated Stbd1 was expressed in COS M9 cells and immunostained with anti-HA antibodies to detect Stbd1 (red) and anti-GABARAPL1 antibodies to visualize endogenous GABARAPL1 (green). The bottom row shows cells transfected with empty vector (pcDNA3) to reveal the endogenous GABARAPL1 distribution (green). C, HA-tagged Stbd1 was expressed in COS M9 cells and immunostained with anti-HA antibodies (red) or anti-LC3 antibodies (green) to detect endogenous LC3. Nuclei were stained with Hoechst (blue). Scale bar, 20 μm.
Association of Stbd1 with Glycogen in Cells
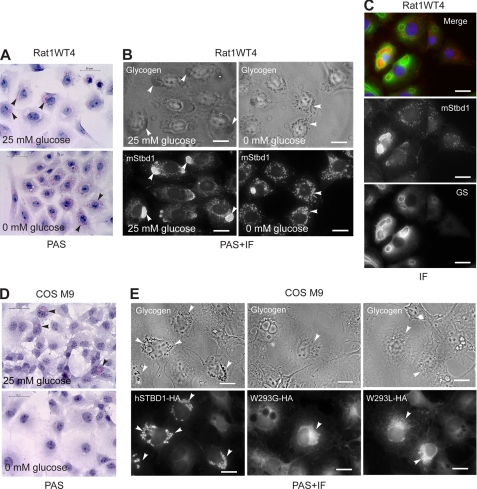
Most cultured cells do not accumulate significant amounts of glycogen, making it difficult to analyze co-localization of glycogen and Stbd1 by light microscopy. Therefore, we utilized a cell line, Rat1WT4, that is stably transfected to overexpress wild type rabbit muscle glycogen synthase (28). The Rat1Neo5 cells described earlier were control cells made in the same study by transformation with empty vector. Glycogen could be detected in the Rat1WT4 cells grown under normal conditions of 25 mm glucose by PAS staining as very large deposits, usually no more than a few per cell and usually proximal to the nucleus (Fig. 7A). These large glycogen deposits were co-localized with Stbd1 (Fig. 7B). If the RatWT4 cells, after initial growth on 25 mm glucose, were transferred to medium lacking glucose for 24 h, the massive glycogen deposits dissipated to generate a larger number of much smaller punctate PAS-positive structures that were strongly co-localized with Stbd1 (Fig. 7, A and B). Immunofluorescent staining for glycogen synthase in Rat1WT4 cells indicated that the enzyme was coincident with the massive glycogen deposits, and especially strong signals were seen surrounding the glycogen giving a clear ring-like appearance in many cells (Fig. 7C). In some cases, the glycogen synthase antibodies defined a ring within which no Stbd1 was detected and which did not have PAS-positive material, as though the glycogen had been evacuated at some stage along with the Stbd1, leaving the glycogen synthase behind.
FIGURE 7.
Co-localization of Stbd1 with glycogen in cells. A, Rat1WT4 cells grown under normal glucose (25 mm; upper panel) or starved (0 glucose; lower panel) conditions for 24h were stained with periodic acid/Schiff reagent (PAS) to visualize glycogen (pink) by light microscopy. Arrowheads indicate examples of glycogen staining. B, co-localization of endogenous Stbd1 protein and glycogen. Rat1WT4 cells were grown as in (A), subjected to PAS staining to visualize glycogen (upper panels) followed by immunofluorescent staining to detect Stbd1 (lower panels). Arrowheads indicate examples of the co-localization of glycogen and Stbd1. C, Rat1WT4 wells were immunostained for Stbd1 (middle panel) and glycogen synthase (GS) (lower panel). The merged image shows Stbd1 (red) and GS (green) with nuclei stained with Hoechst (blue). D, COS M9 cells overexpressing human Stbd1 (hSTBD1-HA) were incubated in medium with 25 mm glucose (upper panel) or no glucose (lower panel) for 24 h and stained with PAS to visualize glycogen by light microscopy. Arrow heads indicate examples of glycogen staining in the fed cells. E, COS M9 cells overexpressed full-length human Stbd1 (hSTBD1-HA; left panels) or the CBM20 point mutants (W293G-HA; middle panels and W293L-HA; right panels). Cells were subjected to PAS staining to visualize glycogen (upper panels) followed by immunofluorescent staining with anti-HA antibodies to visualize Stbd1 (lower panels). Arrowheads indicate Stbd1 localization and, with wild type Stbd1, the co-localized glycogen staining in the upper panels. Scale bars, 20 μm.
PAS staining does not detect the low level of glycogen normally present in COS M9 cells, probably because it is dispersed throughout the cells. However, punctate glycogen deposits are clearly visible by PAS staining of COS cells that overexpress Stbd1 in the perinuclear structures described above (Fig. 7D). Removal of glucose from the medium caused the disappearance of PAS staining consistent with the glycogen being degraded as an energy source (Fig. 7D). Immunofluorescent staining for Stbd1 demonstrated clear co-localization with PAS staining in these perinuclear structures (Fig. 7E). The W293G and W293L mutants were readily detected in a perinuclear location, as in Fig. 4, but there was no associated PAS staining (Fig. 7E), suggesting that the mutant Stbd1 was unable to co-localize with glycogen. The difference in appearance between the Stbd1 and Stbd1-W293G or Stbd1-W293L positive structures is most likely caused by the absence of glycogen. The total glycogen levels in COS cells measured biochemically was unchanged by overexpression of Stbd1 (data not shown). Therefore, Stbd1 causes the concentration of glycogen in these perinuclear structures and modifies their structure and appearance.
DISCUSSION
The most important conclusion from this study is that Stbd1 is involved in glycogen metabolism. Results from several independent experimental approaches support this suggestion. First, Stbd1 physically interacts with glycogen in vitro and in tissue extracts. Second, genetic depletion of glycogen in the muscle or liver of mice correlated with a reduction in Stbd1 protein level, suggesting a genetic link between Stbd1 and glycogen. This is an important point because it proves that the physiological role of Stbd1 is related to glycogen. In other studies, we have observed that the levels of glycogen-metabolizing enzymes, such as glycogen synthase (44) and laforin (45), are reduced when glycogen levels are lowered genetically, probably due to the loss of a stabilizing influence of binding to glycogen. Third, in the cell models where glycogen was visualized, Stbd1 was co-localized with the polysaccharide.
The most obvious structural features of Stbd1 are the N-terminal hydrophobic 24 residues and the C-terminal CBM20 domain, both of which are highly conserved. Two lines of evidence support a membrane localization of Stbd1. First, in muscle extracts, Stbd1 is solubilized from the low speed pellet by detergent during fractionation to isolate glycogen. Second, deletion of the N terminus profoundly alters the subcellular distribution of Stbd1, from the large perinuclear structures, where it is strongly co-localized with the integral membrane protein LAMP1, to a diffuse cytosolic distribution. This result would implicate the N terminus in membrane association. Several experiments suggest that Stbd1 can bind to glycogen. First, in vitro, recombinant Stbd1 co-sedimented with glycogen or the chemically related polysaccharide amylopectin. Second, when muscle extracts were prepared in the presence of detergent to disrupt membranes, a significant proportion of Stbd1 was still recovered in the high speed pellet, indicating that Stbd1 co-sedimented with glycogen independently of association with membranes. Third, in cells where we could visualize glycogen by PAS staining, Rat1WT4 cells or COSM9 cells overexpressing Stbd1, there was co-localization of glycogen and Stbd1, most likely via interaction with the CBM20 domain. Supporting this conclusion, point mutations of a conserved CBM20 residue, Trp-293, implicated in carbohydrate binding, despite retaining a perinuclear Stbd1 staining pattern, totally eliminated perinuclear accumulation of glycogen. The Stbd1-positive perinuclear structures were not seen at all when Stbd1 lacking the CBM20 domain was expressed. These results are consistent with Stbd1 interacting with glycogen in cells. We are able to say less about the other Stbd1 sequence motif, the putative leucine zipper, from our experiments. N-terminal truncation to remove both the hydrophobic tail and the leucine zipper gave the same diffuse subcellular distribution as removal of the hydrophobic segment alone, an essentially negative result. However, from the yeast two-hybrid screen, we can conclude that the leucine zipper region of the molecule is not necessary for oligomerization. Putting all these data together, we propose that Stbd1 serves to tether glycogen, bound to the CBM20 domains, to membranes via interaction of the hydrophobic N terminus (Fig. 8B). Analysis of the Stbd1 sequence for inherent disorder using the PONDR algorithm (46) predicted that the entire region between the N terminus and the CBM20 domain has a high probability to be disordered, potentially providing a flexible connector between the membrane and glycogen interacting regions.
FIGURE 8.
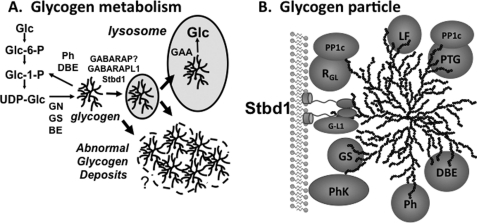
Model for Stbd1 participation in glycogen metabolism and Stbd1 structure. A, classic cytosolic pathway of glycogen synthesis from glucose (Glc) via glucose-6-phosphate (Glc-6-P), glucose-1-phosphate (Glc-1-P) and the glucosyl donor UDP-glucose (UDP-Glc) is mediated by glycogenin (GN), glycogen synthase (GS) and branching enzyme (BE). Degradation to yield Glc-1-P is catalyzed by glycogen phosphorylase (Ph) and debranching enzyme (DBE). A second pathway for glycogen breakdown involves transfer to lysosomes where it is hydrolyzed to glucose by lysosomal α-glycosidase (GAA). From the results of this study, we propose that Stbd1 participates in the vesicular trafficking of glycogen, mediated also by its interaction with GABARABL1 (or maybe GABARAP). In a number of diseases and mouse models, abnormal glycogen deposits can be formed, some possibly with elements of membranes associated. B, a number of the enzymes involved in glycogen metabolism can be partially purified with glycogen. PhK, phosphorylase kinase; PP1, type 1 protein phosphatase; PTG and RGL, glycogen binding PP1 targeting subunits; PP1c, catalytic subunit of PP1; LF, laforin. Shown also is the proposed role for Stbd1, anchoring glycogen to an intracellular membrane and interacting with GABARAPL1 (G-L1).
What is the purpose of localizing glycogen to membranes? As noted in the Introduction, glycogen particles have been found close to endoplasmic reticulum and sarcoplasmic reticulum membranes. Stbd1 could therefore be involved in determining the location of glycogen synthesis. In the case of skeletal muscle, an argument can be advanced that the localization of glycogen close to sites of ATP production may allow rapid retrieval of energy from the glycogen store to fuel muscular activity (5). This hypothesis would in fact be consistent with the localization of Stbd1 in the sarcoplasmic reticulum and T-tubules described in the original paper on Stbd1 (24). Another process that could potentially involve a membrane localization of glycogen is its transport to lysosomes (Fig. 8A), a process that is not really understood but that is well established by the phenotype of Pompe disease. In mouse models of the disease, glycogen is accumulated in lysosomes but also in autophagosome-like vesicles or late endosomes by a process that would involve the intracellular trafficking of glycogen within vesicles (14, 47). The idea is that defective lysosomal disposal of the glycogen causes a back-up of the vesicles delivering glycogen cargo. Such vesicular trafficking of the polysaccharide could well involve the anchoring of glycogen molecules to membranes and require a protein with the properties that we describe here for Stbd1.
Further support for the involvement of Stbd1 in vesicular trafficking comes from its interaction with GABARAPL1 and to a lesser extent GABARAP, which are present in the perinuclear glycogen-enriched structures. GABARAPL1 and GABARAP are two of six mammalian orthologs of Atg8, a protein implicated in autophagosome membrane formation and closure in yeast (40, 41). The better studied members of the mammalian ATG8 family, the microtubule-associated protein-1 light chain 3 (LC3) subfamily, are believed to act similarly to Atg8. However, the functions of GABARAP, GABARAPL1, and GABARAPL2 are not well understood. In yeast, it is clear that autophagy is not completely random and that selectivity can be conferred by interaction of Atg8 with different receptor proteins (48), for example, with Atg32 for disposal of mitochondria (mitophagy) or with Atg30 for disposal of peroxisomes. In mammals, the situation is more complex because of the presence of multiple ATG8 orthologs. Selection of cargo may therefore depend both on the nature of the ATG8 ortholog and its protein targets. For example, in mammals, p62 may mediate trafficking of ubiquitylated proteins via interaction with LC3, whereas GABARAP binding to Nix/Bnip3l has been implicated in mitophagy. Therefore, Stbd1 may act as a glycogen receptor whose interactions with GABARAPL1 and possibly with GABARAP are involved in the vesicular transfer of glycogen to the lysosome (Fig. 8A). Although this process may involve elements of the normal macroautophagic machinery, it is likely distinct from the classic pathway, because LC3 did not co-localize with Stbd1.
In summary, our hypothesis is that Stbd1 localizes glycogen to membranes as a prelude to engulfment in a vesicle that is used to transport glycogen to lysosomes, possibly with participation of GABARAPL1 or GABARAP, acquiring LAMP1 as these vesicles mature prior to lysosomal fusion (Fig. 8). Although LAMP1 is often cited as a lysosomal marker, it is also found in late endosomes, and current thinking is that its intracellular localization is quite dynamic (49). It is of interest that a proteomic analysis of lysosome-related organelles identified Stbd1 in an endosome fraction but not the lysosome itself (50), which would be logical if Stbd1 is degraded in lysosomes. In our experiments, overexpression of Stbd1 in cells drives the formation of glycogen-containing vesicles, resulting in accumulation of the large perinuclear structures positive for LAMP1, GABARAPL1, and Stbd1 that also contain glycogen. Formation of these larger structures may represent an overload of the normal trafficking pathway for lysosomal glycogen disposal with consequent backing up of vesicular glycogen intermediates and the enlargement of these vesicles, somewhat similar to what is seen in Pompe disease.
Stbd1 could target glycogen molecules for lysosomal recycling either as a random housecleaning process or as part of a more directed transport system. With regard to the latter proposition, it is interesting that Stbd1 binds better to plant amylopectin or glycogen from Epm2a−/− mice. Both amylopectin and Epm2a−/− glycogen are less branched than normal glycogen, similar to the polyglucosans associated with other glycogen storage diseases, including Andersen disease, Adult polyglucosan disease, and Tarui disease (51). Evidently, aberrantly branched glycogen in cells is to be avoided, and Stbd1 could selectively target such abnormal glycogen for transport to lysosomes and for subsequent disposal.
Supplementary Material
Acknowledgment
We thank Dr. Keith W. Condon for help with histology.
This work was supported, in whole or in part, by National Institutes of Health Grants DK27221 and NS56454.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
The nomenclature of this protein and gene has been somewhat confusing. We have elected to call the protein Stbd1 rather than genethonin or genethonin 1, to avoid confusion with genethonin 2 and genethonin 3, which are unrelated in sequence but are present in databases. Most genomic databases list the gene as Stbd1.
- PAS
- periodic acid-Schiff
- TLCK
- N-p-tosyl-l-lysine chloromethyl ketone.
REFERENCES
- 1.Roach P. J., Skurat A. V., Harris R. A. (2001) in Handbook of Physiology (Jefferson L. S., Cherrington A. D. eds) Vol. 2, pp. 609–647, Oxford University Press, New York [Google Scholar]
- 2.Roach P. J. (2002) Curr. Mol. Med. 2, 101–120 [DOI] [PubMed] [Google Scholar]
- 3.Greenberg C. C., Jurczak M. J., Danos A. M., Brady M. J. (2006) Am. J. Physiol. Endocrinol. Metab. 291, E1–E8 [DOI] [PubMed] [Google Scholar]
- 4.Cardell R. R., Jr., Michaels J. E., Hung J. T., Cardell E. L. (1985) J. Cell Biol. 101, 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shearer J., Graham T. E. (2004) Exerc. Sport Sci. Rev. 32, 120–126 [DOI] [PubMed] [Google Scholar]
- 6.Rosenfeld E. L. (1975) Pathol. Biol. 23, 71–84 [PubMed] [Google Scholar]
- 7.Raben N., Plotz P., Byrne B. J. (2002) Curr. Mol. Med. 2, 145–166 [DOI] [PubMed] [Google Scholar]
- 8.Reuser A. J., Kroos M. A., Hermans M. M., Bijvoet A. G., Verbeet M. P., Van Diggelen O. P., Kleijer W. J., Van der Ploeg A. T. (1995) Muscle Nerve 3, S61–69 [DOI] [PubMed] [Google Scholar]
- 9.Fukuda T., Roberts A., Ahearn M., Zaal K., Ralston E., Plotz P. H., Raben N. (2006) Autophagy 2, 318–320 [DOI] [PubMed] [Google Scholar]
- 10.Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Nat. Rev. Mol. Cell Biol. 10, 458–467 [DOI] [PubMed] [Google Scholar]
- 11.Levine B., Kroemer G. (2008) Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z., Klionsky D. J. (2009) Curr. Top. Microbiol. Immunol. 335, 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z., Wilson W. A., Fujino M. A., Roach P. J. (2001) Mol. Cell. Biol. 21, 5742–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raben N., Roberts A., Plotz P. H. (2007) Acta Myol. 26, 45–48 [PMC free article] [PubMed] [Google Scholar]
- 15.Raben N., Schreiner C., Baum R., Takikita S., Xu S., Xie T., Myerowitz R., Komatsu M., Van Der Meulen J. H., Nagaruju K., Ralston E., Plotz P. H. (2010) Autophagy, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rybicka K. K. (1996) Tissue Cell 28, 253–265 [DOI] [PubMed] [Google Scholar]
- 17.Meyer F., Heilmeyer L. M., Jr., Haschke R. H., Fischer E. H. (1970) J. Biol. Chem. 245, 6642–6648 [PubMed] [Google Scholar]
- 18.Heilmeyer L. M., Jr., Meyer F., Haschke R. H., Fischer E. H. (1970) J. Biol. Chem. 245, 6649–6656 [PubMed] [Google Scholar]
- 19.Haschke R. H., Heilmeyer L. M., Jr., Meyer F., Fischer E. H. (1970) J. Biol. Chem. 245, 6657–6663 [PubMed] [Google Scholar]
- 20.Ceulemans H., Bollen M. (2004) Physiol. Rev. 84, 1–39 [DOI] [PubMed] [Google Scholar]
- 21.Tagliabracci V. S., Turnbull J., Wang W., Girard J. M., Zhao X., Skurat A. V., Delgado-Escueta A. V., Minassian B. A., Depaoli-Roach A. A., Roach P. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19262–19266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tagliabracci V. S., Girard J. M., Segvich D., Meyer C., Turnbull J., Zhao X., Minassian B. A., Depaoli-Roach A. A., Roach P. J. (2008) J. Biol. Chem. 283, 33816–33825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J., Stuckey J. A., Wishart M. J., Dixon J. E. (2002) J. Biol. Chem. 277, 2377–2380 [DOI] [PubMed] [Google Scholar]
- 24.Bouju S., Lignon M. F., Piétu G., Le Cunff M., Léger J. J., Auffray C., Dechesne C. A. (1998) Biochem. J. 335, 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janecek S. (2002) Bioinformatics 18, 1534–1537 [DOI] [PubMed] [Google Scholar]
- 26.Christiansen C., Abou Hachem M., Janecek S., Viksø-Nielsen A., Blennow A., Svensson B. (2009) FEBS J. 276, 5006–5029 [DOI] [PubMed] [Google Scholar]
- 27.Stapleton D., Nelson C., Parsawar K., McClain D., Gilbert-Wilson R., Barker E., Rudd B., Brown K., Hendrix W., O'Donnell P., Parker G. (2010) Proteomics 10, 2320–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skurat A. V., Dietrich A. D., Roach P. J. (2000) Diabetes 49, 1096–1100 [DOI] [PubMed] [Google Scholar]
- 29.Schaart G., Hesselink R. P., Keizer H. A., van Kranenburg G., Drost M. R., Hesselink M. K. (2004) Histochem. Cell Biol. 122, 161–169 [DOI] [PubMed] [Google Scholar]
- 30.Pederson B. A., Chen H., Schroeder J. M., Shou W., DePaoli-Roach A. A., Roach P. J. (2004) Mol. Cell. Biol. 24, 7179–7187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irimia J. M., Meyer C. M., Peper C. L., Zhai L., Bock C. B., Previs S. F., McGuinness O. P., DePaoli-Roach A., Roach P. J. (2010) J. Biol. Chem. 285, 12851–12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pederson B. A., Cope C. R., Schroeder J. M., Smith M. W., Irimia J. M., Thurberg B. L., DePaoli-Roach A. A., Roach P. J. (2005) J. Biol. Chem. 280, 17260–17265 [DOI] [PubMed] [Google Scholar]
- 33.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 34.Blennow A., Nielsen T. H., Baunsgaard L., Mikkelsen R., Engelsen S. B. (2002) Trends Plant Sci. 7, 445–450 [DOI] [PubMed] [Google Scholar]
- 35.Andrade D. M., Turnbull J., Minassian B. A. (2007) Acta Myol. 26, 83–86 [PMC free article] [PubMed] [Google Scholar]
- 36.Gentry M. S., Dixon J. E., Worby C. A. (2009) Trends Biochem. Sci. 34, 628–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delgado-Escueta A. V. (2007) Curr. Neurol. Neurosci. Rep. 7, 428–433 [DOI] [PubMed] [Google Scholar]
- 38.Sakai M., Austin J., Witmer F., Trueb L. (1970) Neurology 20, 160–176 [DOI] [PubMed] [Google Scholar]
- 39.Breslow J. L., Sloan H. R., Ferrans V. J., Anderson J. L., Levy R. I. (1973) Exp. Cell Res. 78, 441–453 [DOI] [PubMed] [Google Scholar]
- 40.Geng J., Klionsky D. J. (2008) EMBO Rep. 9, 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noda N. N., Ohsumi Y., Inagaki F. (2010) FEBS Lett. 584, 1379–1385 [DOI] [PubMed] [Google Scholar]
- 42.Behrends C., Sowa M. E., Gygi S. P., Harper J. W. (2010) Nature 466, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W., Roach P. J. (2004) Biochem. Biophys. Res. Commun. 325, 726–730 [DOI] [PubMed] [Google Scholar]
- 44.Suzuki Y., Lanner C., Kim J. H., Vilardo P. G., Zhang H., Yang J., Cooper L. D., Steele M., Kennedy A., Bock C. B., Scrimgeour A., Lawrence J. C., Jr., DePaoli-Roach A. A. (2001) Mol. Cell. Biol. 21, 2683–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang W., Parker G. E., Skurat A. V., Raben N., DePaoli-Roach A. A., Roach P. J. (2006) Biochem. Biophys. Res. Commun. 350, 588–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romero P., Obradovic Z., Dunker A. K. (2004) Appl. Bioinformatics 3, 105–113 [DOI] [PubMed] [Google Scholar]
- 47.Fukuda T., Ewan L., Bauer M., Mattaliano R. J., Zaal K., Ralston E., Plotz P. H., Raben N. (2006) Ann. Neurol. 59, 700–708 [DOI] [PubMed] [Google Scholar]
- 48.Komatsu M., Ichimura Y. (2010) Genes Cells 15, 923–933 [DOI] [PubMed] [Google Scholar]
- 49.Saftig P., Klumperman J. (2009) Nat. Rev. Mol. Cell Biol. 10, 623–635 [DOI] [PubMed] [Google Scholar]
- 50.Hu Z. Z., Valencia J. C., Huang H., Chi A., Shabanowitz J., Hearing V. J., Appella E., Wu C. (2007) Int. J. Mass Spectrom. 259, 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DiMauro S., Lamperti C. (2001) Muscle Nerve 24, 984–999 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.