Abstract
Epigenetic silencing of RASSF (Ras association domain family) genes RASSF1 and RASSF5 (also called NORE1) by CpG hypermethylation is found frequently in many cancers. Although the physiological roles of RASSF1 have been studied in some detail, the exact functions of RASSF5 are not well understood. Here, we show that RASSF5 plays an important role in mediating apoptosis in response to death receptor ligands, TNF-α and TNF-related apoptosis-inducing ligand. Depletion of RASSF5 by siRNA significantly reduced TNF-α-mediated apoptosis, likely through its interaction with proapoptotic kinase MST1, a mammalian homolog of Hippo. Consistent with this, siRNA knockdown of MST1 also resulted in resistance to TNF-α-induced apoptosis. To further study the role of Rassf5 in vivo, we generated Rassf5-deficient mouse. Inactivation of Rassf5 in mouse embryonic fibroblasts (MEFs) resulted in resistance to TNF-α- and TNF-related apoptosis-inducing ligand-mediated apoptosis. Importantly, Rassf5-null mice were significantly more resistant to TNF-α-induced apoptosis and failed to activate Mst1. Loss of Rassf5 also resulted in spontaneous immortalization of MEFs at earlier passages than the control MEFs, and Rassf5-null immortalized MEFs, but not the immortalized wild type MEFs, were fully transformed by K-RasG12V. Together, our results demonstrate a direct role for RASSF5 in death receptor ligand-mediated apoptosis and provide further evidence for RASSF5 as a tumor suppressor.
Keywords: Apoptosis, Gene Knock-out, Signal Transduction, Tumor Necrosis Factor (TNF), Tumor Suppressor, Hippo, RASSF5/NORE1, TRAIL
Introduction
A small family of genes termed RASSF (Ras association domain family) has been recently described, the members of which are characterized by the presence of a Ras association (RA)4 domain and a novel motif named the SARAH (Salvador, Rassf, Hippo) domain at the C terminus (reviewed in Refs. 1–3). Among the members of the RASSF family, RASSF1 and RASSF5 (also known as NORE1, for novel Ras effector 1) share the closest homology, displaying 49% identity (66% similarity) at the protein level, and are frequently inactivated by CpG hypermethylation in human cancer cell lines and primary tumors (4–6). RASSF1 has been studied extensively (reviewed in Ref. 7) and shown to play important roles in mitosis (8), microtubule and genomic stability (9–11), apoptosis (12, 13), and cell cycle (14), which are consistent with its function as a tumor suppressor. However, not much is known about the physiological roles of RASSF5.
RASSF5 encodes multiple isoforms because of dual promoter usage and alternative splicing (15). The longest isoform, designated RASSF5A (NORE1A), is transcribed from the 5′-most promoter region containing CpG island and encodes a 418-amino acid protein containing the cysteine-rich diacylglycerol/phorbol ester-binding domain (also called protein kinase C conserved region 1, C1), the RA domain, and the C-terminal SARAH domain. Through alternative splicing, another isoform RASSF5B (NORE1Aβ), lacking the SARAH domain is produced. The shortest isoform, RASSF5C (NORE1B, also called RAPL), lacking the N-terminal C1 domain, is produced from a downstream CpG-containing promoter that is less frequently methylated in primary tumors and cancer cell lines than the 5′ upstream promoter (6). Similar to RASSF1A, CpG methylation of RASSF5A has been found in various cancer cell lines and primary tumors of lung, breast, colon, liver, and kidney (6, 16), although less frequently than RASSF1A. These findings suggest that RASSF5A likely functions as a tumor suppressor.
RASSF5A was first identified as the interacting partner of the active GTP-bound form of RAS or other RAS-like GTPases, such as Rap1, M-Ras, and R-Ras/R-Ras3, through the RA domain (17). RASSF5C (NORE1B/RAPL) was also shown to interact with Rap1 following T-cell antigen or chemokine receptor activation, and the RASSF5C-Rap1 complex promotes recruitment of integrin LFA-1 to the T-cell leading edge, enhancing the T-cell affinity for intercellular adhesion molecule (ICAM) (18). Thus, RASSF5C has been identified as a critical molecule for lymphocyte and dendritic cell trafficking. In addition to the RA domain, RASSF5A, through its N-terminal domain, forms a heterodimer with RASSF1A (19), although whether the diacylglycerol/phorbol ester domain of RASSF5A is necessary for this binding is not clear. The C-terminal SARAH domain of RASSF5 also mediates interaction with other SARAH-containing proteins (20), such as the mammalian sterile 20-like kinases MST1/2 (21, 22) and WW45/SAV1 (23), which are the mammalian homologs of Drosophila Hippo and Salvador, respectively. MST1 kinase has been shown to be proapoptotic and is cleaved by Caspase 3 during apoptosis (24, 25). Cleaved MST1 translocates into the nucleus and exhibits significantly higher kinase activity than the unprocessed form (26). Activated MST1 phosphorylates histone H2B on Ser14 (27) and the pro-apoptotic Forkhead box-containing transcription factor, FOXO3a (Forkhead box O3a) (28), in mediating its apoptotic effects. The anti-apoptotic AKT kinase has been shown to phosphorylate MST1 and inhibit MST1-mediated FOXO3a phosphorylation and apoptosis (29). Despite these studies, the exact role of RASSF5 in apoptosis has not been fully demonstrated. In this study, we demonstrate that RASSF5 is required for TNF-α-mediated apoptosis and for full activation of Mst1 in vivo.
EXPERIMENTAL PROCEDURES
Antibodies, Plasmids, and siRNA
The following antibodies were purchased: PARP, Caspase 3, Caspase 8, JNK, phospho-JNK, p38, phospho-p38, p44/42, phospho-p44/42 (Cell Signaling Technology, Danvers, MA); β-actin and TNF-R1 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA); anti-FLAG (M2) and BAX (Sigma-Aldrich); and MST1/2, YAP1, and LATS1 (Bethyl Laboratories, Montgomery, TX). Anti-RASSF5 rabbit polyclonal antibody has been described (17), and anti-WW45 rabbit polyclonal antibody was kindly provided by Dr. Eric W. McIntush (Bethyl Laboratories). Recombinant mouse TNF-α was purchased from PeproTech (Rocky Hill, NJ). pCMV5/FLAG-MST1 (30) were obtained from Addgene (provided by Dr. Joseph Avruch, Harvard Medical School). pCMV6-XL5/TNFRSF1A (TNF-R1) was purchased from OriGene (Rockville, MD), and pCMVSport6/YAP1, pCMVSport6/WW45, and pcDNA3/LATS1 were purchased from Open Biosystems (Huntsville, AL). The following siRNAs were purchased: from Ambion (Austin TX), RASSF5: sense, 5′-CGG GUU UCA UCA AAG UGC Att-3′, and antisense, 5′-UGC ACU UUG AUG AAA CCC Gtg-3′; WW45: sense, 5′-GGU AUU GGG AGA GUU GCU Gtt-3′, and antisense, 5′-CAG CAA CUC UCC CAA UAC Ctg-3′; MST1: sense, 5′-GGG ACU UGA AUA CCU UCA Utt-3′, and antisense, 5′-AUG AAG GUA UUC AAG UCC Ctt-3′; and YAP1: sense, 5′-GGA AUU GAG AAC AAU GAC Gtt-3′, and antisense, 5′-CGU CAU UGU UCU CAA UUC Ctg-3′; and from MWG Operon (Huntsville, AL), LATS1: sense, 5′-AAA CUU UGC CGA GGA CCC GAA tt-3′, and antisense, 5′-UUC GGG UCC UCG GCA AAG UUU tt-3′. Lipofectamine 2000 (Invitrogen) was used for transient transfection of siRNA and plasmid DNA.
Cell Lines
Mouse embryonic fibroblasts (MEFs) were generated from embryonic day 12.5 embryos using a standard protocol and cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Human osteosarcoma U2OS cells were cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Survival Assay
Control or RASSF5 siRNA transfected U2OS cells or wild type or Rassf5−/− MEFs were seeded on a 24-well plate. Mouse TNF-α was treated together with 10 μg/ml cycloheximide for 16 h. Cell survival was measured using the Cell Counting Kit-8 (Dojindo, Rockville, MD) as the manufacturer's protocol. Relative cell survival of control and RASSF5 siRNA transfected cells in the presence of TNF-α treatment was calculated as relative to the respective values obtained in the control and RASSF5 siRNA transfected cells without TNF-α treatment.
Western Blotting and Immunoprecipitation
For Western blotting, the cells were lysed in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) containing protease inhibitor mixture (Roche Applied Science) and phosphatase inhibitors (0.2 mm sodium orthovanadate, 100 mm sodium fluoride). For immunoprecipitation, the cells were lysed in lysis buffer (50 mm Tris-HCl, pH 8.0, 120 mm NaCl, 0.5% Nonidet P-40, 1 mm EDTA) containing protease inhibitor mixture and phosphatase inhibitors, and the cell lysates were precleared with mouse normal IgG and protein G-agarose for 30 min at 4 °C. The cleared lysates were incubated with 2 μg of each antibodies and protein G-agarose at 4 °C overnight. The immunoprecipitates were washed five times with cold lysis buffer and subjected to Western blotting.
Generation of Targeting Vector and Nore1-deficient Mouse
TC-1 mouse ES cell (129SvEv) genomic DNA was used as the template to amplify 4.6 kb of 5′ homology arm using the following primers: Rassf5-LA-S (5′-CCC GCG GCC GCT ACC ACC TAG AGG AGA TGG AGC C-3′) and Rassf5-LA-AS (5′-CCC CTC GAG TAG GGA CAG TTC ATT CAG GCA CC-3′). The 3′ arm was generated by amplifying 3.0 kb of genomic DNA with the primers Rassf5-SA-S (5′-CCC TCT AGA ATT CAG CAA GTT ACA ACC TGC ACA GGG C-3′) and Rassf5-SA-AS (5′-CCC GGA TCC TAG CAC CAT TGT CCA CTT CCT GCC-3′). PCR products were amplified with high fidelity Pfu Turbo DNA polymerase (Stratagene, Cedar Creek, TX), cloned into pCRII-TOPO vector (Invitrogen), and fully sequenced. The 5′ arm was cut with NotI and XhoI and subcloned into the same sites of pLoxP2-neo vector. The 3′ arm was cut with BamHI and XbaI, blunted with T4 DNA polymerase, and subcloned into the blunted ClaI site of pLoxP2-neo vector. Homologous recombination with this targeting vector will result in the deletion of exons 3–5, which encode the RA domain shared by all Rassf5 isoforms. The targeting vector was linearized with NotI and introduced into TC-1 ES cells by electroporation. Ganciclovir- and G418-resistant clones were isolated and screened by PCR (Expand Long Template PCR system; Roche Applied Science) for homologous recombination using the following primers: Rassf5_5′ (5′-GAC ACATCA AGG GCC CAG AGG AC-3′) and RINA (5′-CCA GAC TGC CTT GGG AAA AGC G-3′) for the 5′ region and Rassf5_3′ (5′-ATC AAT CAC CCT GTG CTC TGA GGC-3′) and TWB12 (5′-ATC GCC TTC TAT CGC CTT CTT GAC GAG TTC-3′) for the 3′ region. All of the positive clones were further confirmed by Southern blot analysis using a probe derived from Rassf5 intron2, which was PCR-amplified using the primers Rassf5_probe_S (5′-GGC CAA CAC AAA TTC TGT TCC ATC C-3′) and Rassf5_probe_AS (5′-AGG CAA CAG AGT TGG CTG AAG C-3′). Positive ES clones were injected into C57BL/6J blastocysts, and the resulting chimeras were crossed with C57BL/6J females to generate F1 heterozygotes, which were interbred to generate homozygous Rassf5−/− mice. Genotyping was performed by PCR analysis of tail DNA using two sets of primers: wild type (177-bp PCR product), Rassf5_WT_S1 (5′-CCC AGT GTT CTC ACC TGG AGA ATC-3′), and Rassf5_WT_AS1 (5′-GTC AGC TCA TGT CAC TGG CAA TAA GC-3′); and mutant (228-bp PCR product), Rassf5_MUT_S1 (5′-CCC AGT GTT CTC ACC TGG AGA ATC-3′) and RINA (5′-CCA GAC TGC CTT GGG AAA AGC G-3′). PCR conditions were 35 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s. All of the animal procedures were approved and followed the guidelines provided by the National Institutes of Health Animal Research Advisory Committee.
Immunofluorescence
U2OS cells transfected with either control or RASSF5 siRNA were plated on a four-well chamber slide (Nunc, Rochester, NY). Recombinant mouse TNF-α was added to the culture together with 10 μg/ml cycloheximide for 4 h. The cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with ice cold 0.1% Triton X-100, PBS for 5 min, and blocked with 10% goat serum, and anti-mouse BAX antibody (clone 6A7; Sigma-Aldrich) was applied for 1 h at room temperature. After incubation with anti-mouse Cy3-labeled secondary antibody, the slides were mounted with DAPI containing mounting solution and analyzed by fluorescence microscopy (Leica DMLB; Leica Microsystems).
TNF-α Injection and Western Blot
The mice were pretreated with d-galactosamine (700 mg/kg of body weight) via intraperitoneal injection, and after 20 min, recombinant mouse TNF-α diluted in PBS (20 μg/kg body weight) was injected into 6–8-week-old wild type or Rassf5 mutant mice via tail vein. For Western blotting of liver lysates, the mice were sacrificed at 6 h (before TNF-α-induced death); the livers were dissected and homogenized; and cleared liver lysates were resolved by SDS-PAGE and immunoblotted with indicated antibodies.
Immortalization of MEFs
Standard 3T3 protocol was used for immortalization of MEFs (31). Briefly, 5 × 105 MEFs were seeded initially on a 100-mm dish, and every 3 days, the cells were replated at 5 × 105 cells/plate until immortalization. Immortalization and the passage numbers were recorded at the first sign of cells growing as colonies after crisis.
Transformation Assay
Immortalized wild type and Rassf5−/− MEFs were infected with either control (empty vector) or K-RasG12V retrovirus, and puromycin-resistant cells were selected and pooled. For soft agar assay, the cells were seeded at 1 × 105 cells in 60-mm plates in 0.3% agar in 2× DMEM containing 20% FBS, penicillin/streptomycin, and glutamine. The colonies were counted after 21 days. Experiments were performed in triplicate and repeated three times with two independently derived MEFs for each genotype. For xenograft experiments, one million immortalized wild type or Rassf5 mutant MEFs infected with either control or K-RasG12V retrovirus were injected subcutaneously into nude mice (n = 5 for each MEFs) and analyzed 3–5 weeks after injection.
RESULTS
RASSF5A Mediates TNF-α-induced Apoptosis
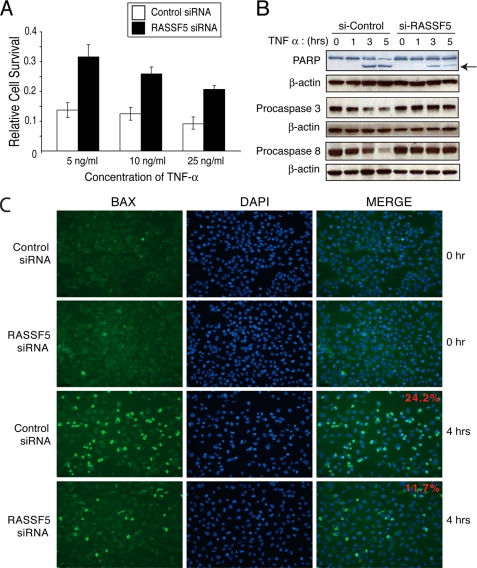
Previous studies have shown that RASSF1A plays an important role in TNF-α- or Fas-mediated apoptosis (12, 32). To investigate whether RASSF5 also plays a role in TNF-α-mediated apoptosis, we depleted RASSF5 using siRNA-mediated knockdown in U2OS cells, which effectively reduced the levels of RASSF5 transcripts (supplemental Fig. S1). Compared with the control siRNA-treated cells, RASSF5-depleted cells showed marked resistance to TNF-α-induced apoptosis, even at the highest dose (25 ng/ml) of TNF-α (Fig. 1A). In control siRNA-treated cells, apoptosis was evident as soon as 3 h after the addition of TNF-α, as demonstrated by the cleavage of PARP (poly-ADP-ribose polymerase), Pro-Caspase 3 (Pro-Casp3), and Pro-Caspase 8 (Pro-CASP8) (Fig. 1B). In contrast, cleavage of PARP, Pro-CASP3, and Pro-CASP8 were markedly reduced in RASSF5-depleted cells in response to TNF-α. TNF-α treatment also led to the oligomerization and activation of BAX, which was diminished by 50% in the RASSF5-depleted cells (Fig. 1C). These results demonstrated that RASSF5 plays an important role in TNF-α-induced apoptosis.
FIGURE 1.
RASSF5 depletion results in reduced TNF-α-mediated apoptosis. A, U2OS cells were transfected with control or RASSF5 siRNA and treated with varying concentrations of TNF-α along with 10 μg/ml cycloheximide. Cell survival was measured with Cell Counting Kit-8 (Dojindo). B, U2OS cells transfected with control or RASSF5 siRNA and treated with 25 ng/ml TNF-α and 10 μg/ml cycloheximide for indicated times were analyzed by Western blotting using PARP, Pro-CASP3, and Pro-CASP8 antibodies. An arrow indicates cleaved PARP. C, U2OS cells transfected with control or RASSF5 siRNA and treated with 25 ng/ml TNF-α and 10 μg/ml cycloheximide for 4 h were fixed, immunostained with anti-BAX antibody, and analyzed by fluorescence microscopy. The percentage of BAX-positive cells/total cells from four or five randomly chosen fields was calculated from three independent experiments, and the representative images are shown.
RASSF5 Interacts with the Proteins in the Hippo Pathway
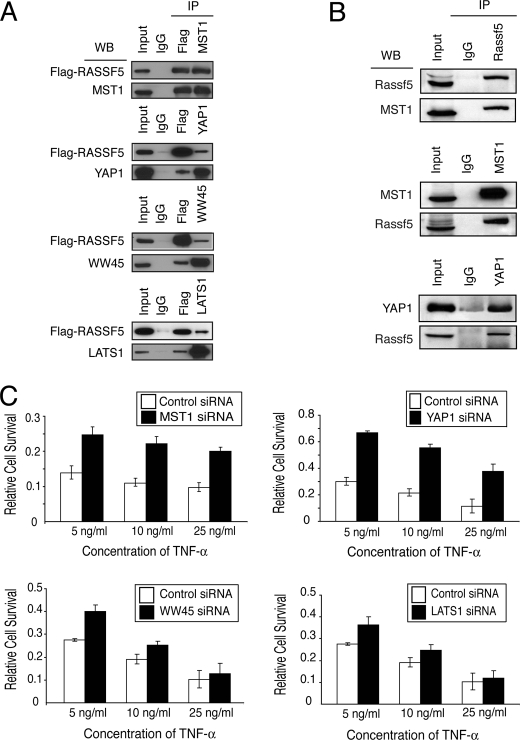
It has been previously shown that RASSF5 interacts with the proapoptotic kinase MST1 (22). Thus, we wanted to confirm the interaction between RASSF5A and MST1. As expected, coexpression of MST1 along with FLAG-RASSF5A in U2OS cells and immunoprecipitation with anti-FLAG antibody followed by immunoblotting with anti-MST1 demonstrated a strong interaction between RASSF5A and MST1 (Fig. 2A). In fact, a reciprocal immunoprecipitation-Western blot analysis demonstrated that nearly all of exogenously expressed MST1 and FLAG-RASSF5A were associated with each other, consistent with the previously reported 1:1 in vitro interaction between RASSF5 and MST1 through their respective SARAH domains (23).
FIGURE 2.
RASSF5 interacts with MST1 and other components of Hippo signaling pathway. A, U2OS cells cotransfected with FLAG-RASSF5 and either MST1, WW45, LATS1, or YAP1 were immunoprecipitated (IP) with either FLAG, MST1, WW45, LATS1, or YAP1 antibodies and analyzed by Western blotting (WB). B, 4-week-old mouse brain lysates were immunoprecipitated with anti-RASSF5, anti-MST1, or anti-YAP1 antibody and analyzed by immunoblotting. C, U2OS cells were transfected with control or MST1, WW45, LATS1, or YAP1 siRNA and treated with varying concentrations of TNF-α along with cycloheximide. Cell survival was measured as described.
Genetic studies in Drosophila have identified other members of the Hippo signaling pathway such as Salvador (WW45 in human), Warts (LATS1/2), and Yorkie (YAP1), which form complexes with Hippo (MST1/2) (reviewed in Ref. 33). We therefore examined RASSF5A interaction with other proteins in the Hippo pathway. U2OS cells were cotransfected with FLAG-RASSF5A and either WW45, LATS1, or YAP1, and cell lysates were immunoprecipitated with anti-FLAG antibody followed by Western blot analysis. The results revealed that WW45, LATS1, and YAP1 were capable of forming complexes with RASSF5A, although at substoichiometric levels (Fig. 2A). The reciprocal immunoprecipitation-Western blotting revealed similar findings. These results demonstrate that only a fraction of WW45, LATS1, or YAP1 is associated with RASSF5A. As a control for specificity, we showed that RASSF5 does not interact with proteins in the NF-κB pathway, IKK, or p65 (supplemental Fig. S2).
To confirm these interactions with endogenous proteins, we used whole mouse brain extracts that contained detectable levels of endogenous Rassf5. Consistent with our previous results, we observed a strong interaction between endogenous Rassf5 and Mst1 (Fig. 2B). However, we could not detect interaction between endogenous Rassf5 and other Hippo proteins, Ww45 and Lats1 (data not shown). This could be due to weak or transient interactions between Rassf5 and other Hippo proteins. The only endogenous interaction we observed was between Rassf5 and Yap1 when we immunoprecipitated mouse brain extracts with anti-Yap1 antibody and immunoblotted with anti-Rassf5 but not in the reciprocal direction (Fig. 2B and data not shown).
It has been previously shown that RASSF1A associates with TNF-R1 in response to TNF-α stimulation (12). To determine whether RASSF5A can also associate with TNF-R1, we cotransfected TNF-R1 along with FLAG-tagged RASSF5A into U2OS cells and examined their interaction. Immunoprecipitation with anti-TNF-R1 antibody followed by immunoblotting with anti-FLAG (FLAG-RASSF5A) revealed that RASSF5A formed a complex with TNF-R1 (supplemental Fig. S3). Interestingly, ectopic expression of MST1, WW45, LATS1, or YAP1 along with TNF-R1 followed by immunoprecipitation with anti-TNF-R1 antibody and immunoblotting with antibodies to various Hippo proteins showed that WW45 and YAP1 associated with TNF-R1, but not LATS1 (supplemental Fig. S3). We could not determine the interaction between TNF-R1 and MST1 because coexpression of TNF-R1 and MST1 resulted in a marked decrease in the TNF-R1 level during immunoprecipitation.
Depletion of MST1 Results in Reduced TNF-α-mediated Apoptosis
We next examined whether the Hippo pathway is involved in TNF-α-mediated apoptosis. We used siRNA to reduce the expression of MST1, WW45, LATS1, and YAP1 and measured TNF-α-mediated apoptosis. Western blotting demonstrated that siRNA treatment effectively reduced the expression of each protein (supplemental Fig. S4). Depletion of MST1 or YAP1 led to a significant reduction in TNF-α-induced apoptosis (Fig. 2C), similar to the level observed with RASSF5A depletion. However, siRNA knockdown of WW45 or LATS1 showed only a moderate inhibition of TNF-α-induced apoptosis. These results demonstrate that MST1 and YAP1, along with RASSF5, are involved in mediating TNF-α-induced apoptosis.
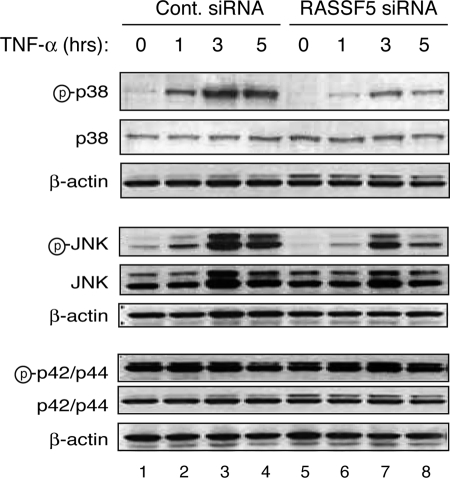
RASSF5A Depletion Results in Reduced JNK and p38 Kinase Activation
Previous reports have shown that MST1 can activate stress-response MAPKs, p38, and JNK during apoptosis (25, 34). Because RASSF5 has also been implicated in the activation of MST1 (21, 35), we next examined the activation of stress-activated kinases following TNF-α treatment and siRNA-mediated depletion of RASSF5. In control siRNA-treated cells, TNF-α treatment led to the activation of both p38 and JNK kinases as early as 1 h after the treatment (Fig. 3). In RASSF5-siRNA-treated cells, however, TNF-α resulted in a substantial reduction in the activation of both p38 and JNK kinases as measured by phosphorylation of p38 and JNK kinases. In contrast, phosphorylation of p42/p44 ERK kinase was not affected by RASSF5 depletion, although we observed a slight decrease in the ERK phosphorylation at 5 h following TNF-α treatment in control siRNA-treated cells (Fig. 3, lane 4) but not in RASSF5-siRNA-treated cells. These results suggest that the stress-response p38 and JNK kinases are activated downstream of RASSF5.
FIGURE 3.
Reduced activation of the stress-activated p38 and JNK kinases in RASSF5 depleted cells treated with TNF-α. U2OS cells transfected with control (Cont.) or RASSF5 siRNA and treated with 25 ng/ml TNF-α and cycloheximide for the indicated times were analyzed by Western blotting with phospho-p38, p38, phospho-JNK, JNK phospho-p42/p44, p42/p44, and β-actin antibodies. Letter p in circle, phospho.
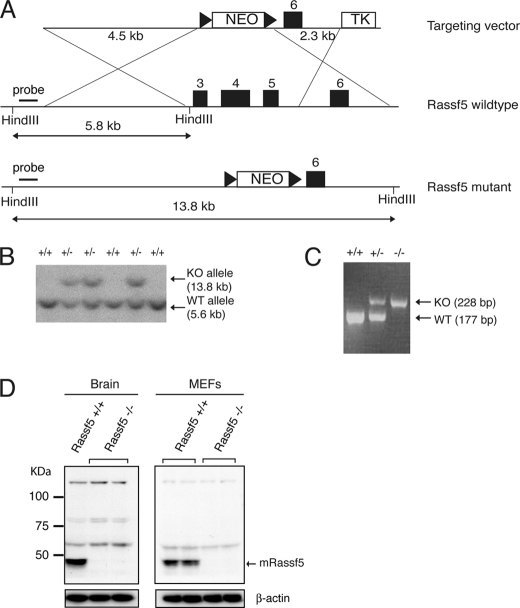
Inactivation of Rassf5 by Gene Targeting
To examine the role of RASSF5 in TNF-α-mediated apoptosis in vivo, we employed gene targeting to inactivate Rassf5 in mouse ES cells. Rassf5 produces at least three isoforms caused by dual promoter usage and alternative splicing (15). To ensure inactivation of all isoforms of Rassf5, we designed our targeting vector to remove the common exons 3–5, which contain the RA domain shared by all Rassf5 isoforms (Fig. 4A). Several mouse ES clones were obtained by homologous recombination as revealed by Southern blot analysis (Fig. 4B). Targeted ES cells were used to generate chimeras that were subsequently crossed with wild type C57BL/6 to obtain offspring with mutant Rassf5 in their germ line. Rassf5 heterozygotes were intercrossed to generate Rassf5 homozygotes (Fig. 4C), which were obtained at a Mendelian ratio (Table 1) and were viable with no gross developmental anomalies. Western blot analysis further confirmed the absence of Rassf5 protein in Rassf5 homozygous mutants (Fig. 4D).
FIGURE 4.
Inactivation of Rassf5 by gene targeting. A, a scheme of Rassf5 targeting strategy. Homologous recombination will delete exons 3–5 and insert the neomycin gene. The triangles indicate LoxP sequences. B, Southern blot analysis of HindIII-digested mouse ES genomic DNA. Wild type (5.6-kb) and targeted (13.8-kb) alleles are indicated. C, PCR analysis of tail DNA. Wild type (177 bp) and targeted (228 bp) bands are indicated. D, mouse brain lysates from 4-week-old Rassf5 wild type and mutant animals or Rassf5+/+ and Rassf5−/− MEFs were analyzed by immunoblotting with anti-RASSF5 antibody. Anti-β-actin was used for loading control. KO, knock-out.
TABLE 1.
Rassf5-null mice are born at Mendelian ratio
| Genotype | Number of mice |
|---|---|
| Rassf5+/+ | 58 (22.6%) |
| Rassf5+/− | 134 (52.1%) |
| Rassf5−/− | 65 (25.3%) |
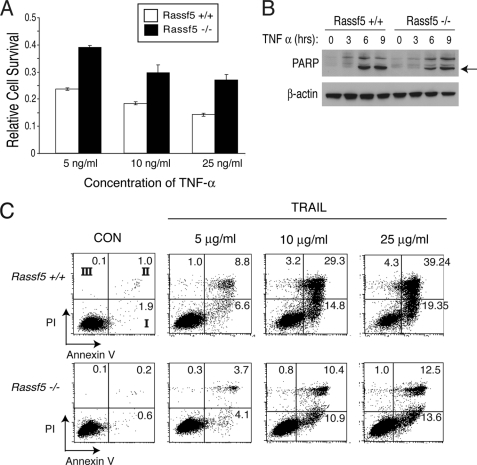
Rassf5-null Mouse Embryonic Fibroblasts Are Resistant to TNF-α- and TRAIL-induced Apoptosis
To determine whether Rassf5-deficient cells derived from Rassf5 mutants are similarly resistant to TNF-α-mediated apoptosis, we generated Rassf5-null MEFs. The addition of TNF-α to the wild type MEFs led to apoptosis in a dose-dependent manner (Fig. 5A). However, Rassf5-deficient MEFs were more resistant to TNF-α-mediated apoptosis. This was further confirmed by annexin V staining on the cell surface, which is a hallmark of apoptosis (supplemental Fig. S5). Western blot analysis of PARP cleavage further demonstrated the resistance to TNF-α-induced apoptosis in Rassf5-null MEFs compared with the control (Fig. 5B). Reintroduction of CMV-driven RASSF5 into the mutant MEFs led to a modest restoration of TNF-α-induced apoptosis (supplemental Table S1). The inability of Rassf5-null cells to undergo apoptosis triggered by TNF-α was not due to reduced expression of TNF-α receptor (TNF-R1) nor its immediate downstream molecules, such as TRADD, TRAF2, and FADD (supplemental Fig. S6). To determine whether Rassf5 can mediate apoptosis induced by another death receptor ligand, Rassf5-deficient cells were treated with increasing doses of TRAIL. Similar to TNF-α, Rassf5-deficient MEFs were also resistant to TRAIL-induced apoptosis compared with wild type cells (Fig. 5C). These results confirm our previous results and further demonstrate that RASSF5 is an important downstream effector of death receptor ligand (TNF-α and TRAIL)-mediated apoptosis.
FIGURE 5.
Rassf5-deficient MEFs are resistant to TNF-α- and TRAIL-induced apoptosis. A, wild type and Rassf5-null MEFs were treated with varying concentrations of TNF-α and cycloheximide, and cell survival was measured as described. B, wild type and Rassf5-null MEFs treated with 25 ng/ml TNF-α and cycloheximide for the indicated times were analyzed by Western blotting with PARP and β-actin antibodies. The arrow indicates cleaved PARP. C, Rassf5+/+ and Rassf5−/− MEFs were treated with varying concentrations of TRAIL (Peprotech) and cycloheximide (10 μg/ml) for 18 h, and cells stained for annexin V and propidium iodide (PI) were analyzed by FACSCalibur (BD Biosciences). The number in each quadrant indicates percentage of cells positive for annexin V, PI or both. Quadrant I, early apoptosis population (with annexin V, without PI); quadrant II, late apoptosis population (with annexin V and PI); quadrant III, necrosis population (without annexin V, with PI). CON, control.
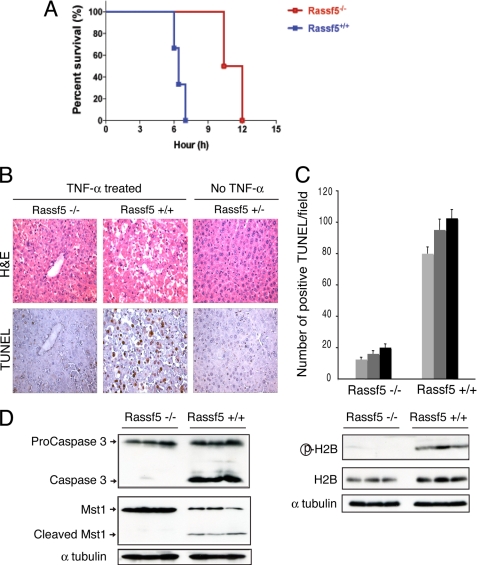
Rassf5-null Mice Are Resistant to TNF-α-induced Apoptosis
Previous studies have shown that administration of TNF-α via tail vein along with the transcriptional inhibitor d-galactosamine causes severe apoptosis in the liver and rapid death in mice (36, 37). We therefore examined TNF-α-induced apoptosis in the livers of wild type and Rassf5 mutant mice. Within 7 h, all of the wild type mice died when injected with d-galactosamine and TNF-α (20 mg/40 g of body weight) (Fig. 6A). Remarkably, Rassf5-null mice remained viable up to 10–11 h after TNF-α injection, and by 12 h, all of the mutants died. For detailed analysis, we administered the same doses of d-galactosamine and TNF-α and isolated the livers at 6 h before the mice succumbed to the effects of TNF-α. A dramatic difference was observed upon gross examination because livers from the wild type mice injected with TNF-α appeared very dark, indicating massive hemorrhage, whereas livers from the Rassf5-null mice treated with the same dose of TNF-α appeared nearly normal, similar to the uninjected control liver (supplemental Fig. S7). Histological analysis demonstrated that the wild type hepatocytes were undergoing massive cell death, which was confirmed by TUNEL staining (Fig. 6B). In contrast, Rassf5-null hepatocytes appeared much more intact, and the Rassf5 mutant hepatocytes showed significantly less apoptosis in response to TNF-α (Fig. 6, B and C).
FIGURE 6.
Rassf5-deficient mice are resistant to TNF-α-induced apoptosis. A, Rassf5 wild type (n = 7) and mutant mice (n = 4) were injected via tail vein with d-galactosamine and TNF-α. The time of death was recorded for each mouse. B, Rassf5 wild type and mutant mice were injected with TNF-α as in A, and all of the animals were sacrificed at 6 h before death (n = 3 for each genotype). Paraffin-embedded sections of liver were analyzed by H & E and TUNEL staining. Control liver (Rassf5+/−) from an uninjected animal is shown. C, positive TUNEL-stained cells (from B) were counted from at least seven randomly chosen fields, and the average number of TUNEL-positive cells/field is shown (n = 3 for each genotype). D, liver extracts from Rassf5 wild type and mutant TNF-α-treated animals (n = 3 for each genotype) were analyzed by Western blotting with Pro-CASP3, MST1, phospho-H2B, H2B, and α-tubulin antibodies. H&E, hematoxylin and eosin.
We further analyzed the livers from TNF-α-injected wild type and Rassf5 mutants by Western blotting. In Rassf5+/+ hepatocytes, TNF-α treatment led to the cleavage and activation of Casp3 (Fig. 6D), whereas Casp3 cleavage was greatly reduced in the TNF-α-injected Rassf5 mutant hepatocytes. Furthermore, in Rassf5+/+ hepatocytes, Mst1 was also cleaved in response to TNF-α, and histone H2B (Ser14) phosphorylation was readily observed, indicating the presence of activated Mst1 in the nucleus. In contrast, TNF-α treatment did not result in the Mst1 cleavage or H2B (Ser14) phosphorylation in Rassf5-null hepatocytes (Fig. 6D), demonstrating that Rassf5 is required for the full activation and cleavage of Mst1. Consistent with this, nuclear translocation of Mst1 following TNF-α treatment was observed in the wild type but not in the Rassf5 mutant hepatocytes (supplemental Fig. S8A). Western blot analysis with phospho-specific Mst1/2 antibody (Ser(P)183/180) (21) further confirmed the cleavage and activation of Mst1/2 in the wild type TNF-α-treated hepatocytes as compared with the mutant (supplemental Fig. S8B).
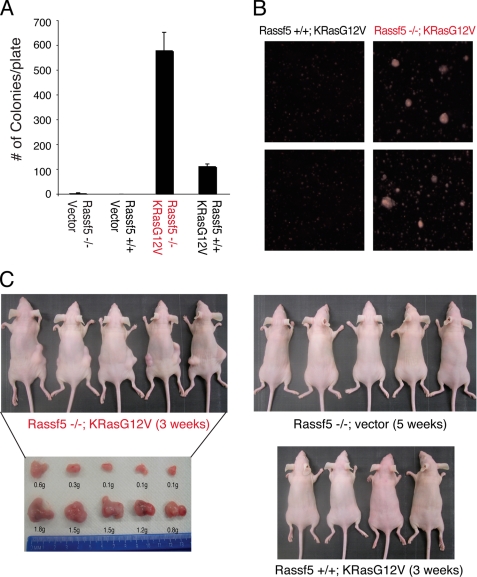
Loss of Rassf5 Cooperates with Activated K-Ras in Cellular Transformation
Epigenetic silencing of RASSF5 in various tumors suggests that RASSF5 might function as a tumor suppressor (6, 16). To examine this possibility, we immortalized Rassf5+/+ and Rassf5−/− MEFs using a 3T3 protocol. During the early passages, Rassf5−/− MEFs displayed slightly faster growth than the wild type littermate MEFs (data not shown), but subsequently, both MEFs reached senescence and crisis at similar passages (data not shown). However, Rassf5−/− MEFs consistently became immortalized earlier (at least 10 passages) than the wild type MEFs (Table 2), demonstrating an increased propensity to spontaneous immortalization in the absence of Rassf5.
TABLE 2.
Immortalization of Rassf5−/− MEFs
a Indicates independent MEFs derived from two littermates.
We next examined the ability of the immortalized MEFs to grow in an anchorage-independent manner. Although immortalized, neither the wild type nor the mutant MEFs were able to grow in soft agar-containing medium (Fig. 7A). However, when the MEFs were infected with the retrovirus expressing constitutively active K-RasG12V, the Rassf5−/− MEFs displayed robust anchorage-independent growth compared with the wild type MEFs, which formed fewer and smaller colonies (Fig. 7, A and B). To further demonstrate the transforming capacity of these MEFs, we injected either wild type MEFs expressing activated K-RasG12V or Rassf5−/− MEFs with or without K-RasG12V subcutaneously into nude mice. Wild type MEFs expressing K-RasG12V or Rassf5−/− MEFs without K-RasG12V failed to form any visible tumors in nude mice up to 3 and 5 weeks, respectively (Fig. 7C). In contrast, Rassf5−/− MEFs expressing K-RasG12V readily formed visible tumors in 3 weeks. Together, these results demonstrate that inactivation of Rassf5 predisposes the MEFs to transformation by oncogenic K-RasG12V and support the role for Rassf5 as a tumor suppressor. However, we note that Rassf5−/− mice did not display a statistically significant increase in spontaneous tumor incidence compared with wild type littermate controls, although we did observe rare cases of liver cancer in Rassf5+/− and Rassf5−/− mice but not in the wild type littermate controls (supplemental Table S2).
FIGURE 7.
Rassf5-deficient MEFs are predisposed to K-RasG12V-induced transformation. A, soft agar assay. Immortalized Rassf5+/+ and Rassf5−/− MEFs stably expressing empty vector or K-RasG12V were grown in soft agar for 21 days, and the colonies were counted. The experiment was performed in triplicate and repeated three times using two independently derived MEFs. B, representative images taken under phase-contrast microscope of immortalized Rassf5+/+ and Rassf5−/− MEFs stably expressing K-RasG12V are shown. C, immortalized Rassf5+/+ and Rassf5−/− MEFs stably expressing empty vector or K-RasG12V were injected subcutaneously into nude mice, and the animals were sacrificed at 3–5 weeks after injection. The tumors were dissected and weighed.
DISCUSSION
In the present study, we demonstrate that RASSF5 is an important downstream effector of TNF-α- and TRAIL-induced apoptosis. Depletion of RASSF5, MST1, and YAP1, but not WW45 and LATS1, resulted in reduced TNF-α-induced apoptosis, indicating that not all components of the Hippo pathway are required for TNF-α-induced apoptosis mediated by RASSF5 and MST1. RASSF5 interacts strongly with MST1 but only weakly with other components of the mammalian Hippo signaling pathway. In Drosophila, dRASSF competes with Salvador (WW45 in human) for Hippo (MST1), and thus, the dRASSF-Hippo complex is thought to be mutually exclusive with the Salvador-Hippo complex (38). However, a recent study has shown that mammalian RASSF6 is capable of forming a tripartite complex with MST1 and WW45 (39). Therefore, further studies will be needed to determine whether RASSF5 is also capable of forming a multi-complex with the members of the Hippo pathway.
The effector role of RASSF5 downstream of TNF-α was further supported by our in vivo studies. Inactivation of Rassf5 in the mouse led to a significant protection from TNF-α-mediated apoptosis of the liver. Our results further showed that Rassf5 is required for the activation of the proapoptotic kinase Mst1 after TNF-α stimulation in vivo, as measured by the absence of Mst1 cleavage and H2B (Ser14) phosphorylation in Rassf5-null cells. A shorter isoform, Rassf5C (Nore1B/Rapl), has also been shown to be necessary for the activation of Mst1 during immune cell trafficking (18, 35). Chemokines and T-cell receptor ligation activate T-cells and result in a rapid phosphorylation and activation of Mst1. It was shown that in the absence of Rassf5C, chemokine- or T-cell receptor-mediated activation of Mst1 in T-cells was undetectable or greatly reduced (18). Therefore, it appears that Rassf5 is necessary for Mst1 activation in response to different stimuli in vivo. Given the previous observations of enhanced MST1 activation when recruited to plasma membrane or in association with activated RAS and RASSF5 (21, 22), it is possible that RASSF5 is responsible for physically recruiting MST1 to the sites where it can be activated via its interaction with activated RAS or with TNF-R1 (in response to TNF-α stimulation). Although the exact mechanisms of how RASSF5 mediates TNF-α-induced apoptosis need further investigation, the downstream activation of the NF-κB pathway following TNF-α stimulation appears unaffected by depletion of RASSF5, either in the siRNA-treated cells or in Rassf5-null MEFs, as evidenced by similar phosphorylation kinetics of IKK and IκB (supplemental Fig. S9). These results suggest that RASSF5 is unlikely to play a role in TNF-α-induced activation of NF-κB pathway.
It was recently shown that following TNF-α stimulation, RASSF1A activated BAX through its interaction with BH3-like protein modulator of apoptosis-1 and subsequently triggered apoptosis (12). RASSF1A also forms a complex with the components of mammalian Hippo pathway (40) and induces apoptosis following stimulation with other death receptor ligands, such as Fas-L or TRAIL (12, 13, 41, 42). Similarly, we also observed reduced apoptosis in response to TRAIL in Rassf5-null MEFs compared with the control. However, deletion of Rassf5 did not have any effects on other apoptotic stimuli such as staurosporine, methyl methanesulfonate, tamoxifen, and nocodazole (supplemental Fig. S10B). Together, these results indicate that death receptor-induced apoptosis, such as TNF-α and TRAIL, is mediated by RASSF5, as well as RASSF1 (12). In this regard, we note that RASSF1 and RASSF5 can form a heterodimer (19), raising the possibility that the two RASSF proteins might share overlapping functions.
The results from this study suggest that inactivation of RASSF5 by epigenetic silencing will likely lead to tumorigenesis in conjunction with other oncogenic events. This is suggested by the precocious spontaneous immortalization of the Rassf5−/− MEFs compared with the wild type MEFs and by enhanced susceptibility of the mutant MEFs to oncogenic transformation by K-RasG12V. These results provide evidence for the role of Rassf5 as a tumor suppressor. However, loss of Rassf5 alone was not sufficient to fully transform MEFs, demonstrating that further genetic and/or epigenetic changes are required for tumorigenesis. This idea is consistent with the low spontaneous tumor incidence observed in Rassf5-null mice (supplemental Table S2). The low tumor incidence in Rassf5-null mice is similar to the Rassf1-null mice (10, 43). Given the high frequency of CpG hypermethylation of RASSF1 and RASSF5 in human cancers, it will be interesting to determine the tumor susceptibility of double Rassf1 and Rassf5 mutant mice. The generation of Rassf5-deficient cells and mice will further help to elucidate the roles of Rassf5 in other cellular processes.
Supplementary Material
Acknowledgments
We thank Drs. Eric McIntush (Bethyl Laboratories) and Daphne Bell (NHGRI, National Institutes of Health) for kindly providing reagents and Richard Proia and the members of the Lee laboratory for helpful comments and suggestions. We also thank Drs. Cuiling Li and Chuxia Deng of the NIDDK Knockout Mouse Core for help in generating the Rassf5 mutant mice and Yun-Ping Wu for the confocal microscopy.
This work was supported, in whole or in part, by a grant the National Institutes of Health NIDDK Intramural Research Program (to S. B. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S10.
- RA
- Ras association
- MEF
- mouse embryonic fibroblast
- PARP
- poly-ADP-ribose polymerase
- CASP
- caspase
- TNF-R1
- TNF receptor 1
- TRAIL
- TNF-related apoptosis-inducing ligand
- ES
- embryonic stem.
REFERENCES
- 1.van der Weyden L., Adams D. J. (2007) Biochim. Biophys. Acta 1776, 58–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richter A. M., Pfeifer G. P., Dammann R. H. (2009) Biochim. Biophys. Acta 1796, 114–128 [DOI] [PubMed] [Google Scholar]
- 3.Avruch J., Xavier R., Bardeesy N., Zhang X. F., Praskova M., Zhou D., Xia F. (2009) J. Biol. Chem. 284, 11001–11005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dammann R., Li C., Yoon J. H., Chin P. L., Bates S., Pfeifer G. P. (2000) Nat. Genet. 25, 315–319 [DOI] [PubMed] [Google Scholar]
- 5.Morris M. R., Hesson L. B., Wagner K. J., Morgan N. V., Astuti D., Lees R. D., Cooper W. N., Lee J., Gentle D., Macdonald F., Kishida T., Grundy R., Yao M., Latif F., Maher E. R. (2003) Oncogene 22, 6794–6801 [DOI] [PubMed] [Google Scholar]
- 6.Hesson L., Dallol A., Minna J. D., Maher E. R., Latif F. (2003) Oncogene 22, 947–954 [DOI] [PubMed] [Google Scholar]
- 7.Donninger H., Vos M. D., Clark G. J. (2007) J. Cell Sci. 120, 3163–3172 [DOI] [PubMed] [Google Scholar]
- 8.Song M. S., Song S. J., Ayad N. G., Chang J. S., Lee J. H., Hong H. K., Lee H., Choi N., Kim J., Kim H., Kim J. W., Choi E. J., Kirschner M. W., Lim D. S. (2004) Nat. Cell Biol. 6, 129–137 [DOI] [PubMed] [Google Scholar]
- 9.Vos M. D., Martinez A., Elam C., Dallol A., Taylor B. J., Latif F., Clark G. J. (2004) Cancer Res. 64, 4244–4250 [DOI] [PubMed] [Google Scholar]
- 10.van der Weyden L., Tachibana K. K., Gonzalez M. A., Adams D. J., Ng B. L., Petty R., Venkitaraman A. R., Arends M. J., Bradley A. (2005) Mol. Cell. Biol. 25, 8356–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L., Tommasi S., Lee D. H., Dammann R., Pfeifer G. P. (2003) Oncogene 22, 8125–8136 [DOI] [PubMed] [Google Scholar]
- 12.Baksh S., Tommasi S., Fenton S., Yu V. C., Martins L. M., Pfeifer G. P., Latif F., Downward J., Neel B. G. (2005) Mol. Cell 18, 637–650 [DOI] [PubMed] [Google Scholar]
- 13.Matallanas D., Romano D., Yee K., Meissl K., Kucerova L., Piazzolla D., Baccarini M., Vass J. K., Kolch W., O'neill E. (2007) Mol. Cell 27, 962–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shivakumar L., Minna J., Sakamaki T., Pestell R., White M. A. (2002) Mol. Cell. Biol. 22, 4309–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tommasi S., Dammann R., Jin S. G., Zhang X. F., Avruch J., Pfeifer G. P. (2002) Oncogene 21, 2713–2720 [DOI] [PubMed] [Google Scholar]
- 16.Calvisi D. F., Ladu S., Gorden A., Farina M., Conner E. A., Lee J. S., Factor V. M., Thorgeirsson S. S. (2006) Gastroenterology 130, 1117–1128 [DOI] [PubMed] [Google Scholar]
- 17.Vavvas D., Li X., Avruch J., Zhang X. F. (1998) J. Biol. Chem. 273, 5439–5442 [DOI] [PubMed] [Google Scholar]
- 18.Katagiri K., Ohnishi N., Kabashima K., Iyoda T., Takeda N., Shinkai Y., Inaba K., Kinashi T. (2004) Nat. Immunol. 5, 1045–1051 [DOI] [PubMed] [Google Scholar]
- 19.Ortiz-Vega S., Khokhlatchev A., Nedwidek M., Zhang X. F., Dammann R., Pfeifer G. P., Avruch J. (2002) Oncogene 21, 1381–1390 [DOI] [PubMed] [Google Scholar]
- 20.Scheel H., Hofmann K. (2003) Curr. Biol. 13, R899–900 [DOI] [PubMed] [Google Scholar]
- 21.Praskova M., Khoklatchev A., Ortiz-Vega S., Avruch J. (2004) Biochem. J. 381, 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khokhlatchev A., Rabizadeh S., Xavier R., Nedwidek M., Chen T., Zhang X. F., Seed B., Avruch J. (2002) Curr. Biol. 12, 253–265 [DOI] [PubMed] [Google Scholar]
- 23.Hwang E., Ryu K. S., Pääkkönen K., Güntert P., Cheong H. K., Lim D. S., Lee J. O., Jeon Y. H., Cheong C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9236–9241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakeya H., Onose R., Osada H. (1998) Cancer Res. 58, 4888–4894 [PubMed] [Google Scholar]
- 25.Graves J. D., Gotoh Y., Draves K. E., Ambrose D., Han D. K., Wright M., Chernoff J., Clark E. A., Krebs E. G. (1998) EMBO J. 17, 2224–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ura S., Masuyama N., Graves J. D., Gotoh Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10148–10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung W. L., Ajiro K., Samejima K., Kloc M., Cheung P., Mizzen C. A., Beeser A., Etkin L. D., Chernoff J., Earnshaw W. C., Allis C. D. (2003) Cell 113, 507–517 [DOI] [PubMed] [Google Scholar]
- 28.Lehtinen M. K., Yuan Z., Boag P. R., Yang Y., Villén J., Becker E. B., DiBacco S., de la Iglesia N., Gygi S., Blackwell T. K., Bonni A. (2006) Cell 125, 987–1001 [DOI] [PubMed] [Google Scholar]
- 29.Jang S. W., Yang S. J., Srinivasan S., Ye K. (2007) J. Biol. Chem. 282, 30836–30844 [DOI] [PubMed] [Google Scholar]
- 30.Lin Y., Khokhlatchev A., Figeys D., Avruch J. (2002) J. Biol. Chem. 277, 47991–48001 [DOI] [PubMed] [Google Scholar]
- 31.Todaro G. J., Green H. (1963) J. Cell Biol. 17, 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foley C. J., Freedman H., Choo S. L., Onyskiw C., Fu N. Y., Yu V. C., Tuszynski J., Pratt J. C., Baksh S. (2008) Mol. Cell. Biol. 28, 4520–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harvey K., Tapon N. (2007) Nat. Rev. Cancer. 7, 182–191 [DOI] [PubMed] [Google Scholar]
- 34.Ura S., Nishina H., Gotoh Y., Katada T. (2007) Mol. Cell. Biol. 27, 5514–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katagiri K., Imamura M., Kinashi T. (2006) Nat. Immunol. 7, 919–928 [DOI] [PubMed] [Google Scholar]
- 36.Leist M., Gantner F., Bohlinger I., Germann P. G., Tiegs G., Wendel A. (1994) J. Immunol. 153, 1778–1788 [PubMed] [Google Scholar]
- 37.Lehmann V., Freudenberg M. A., Galanos C. (1987) J. Exp. Med. 165, 657–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polesello C., Huelsmann S., Brown N. H., Tapon N. (2006) Curr. Biol. 16, 2459–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikeda M., Kawata A., Nishikawa M., Tateishi Y., Yamaguchi M., Nakagawa K., Hirabayashi S., Bao Y., Hidaka S., Hirata Y., Hata Y. (2009) Sci. Signal. 2, ra59. [DOI] [PubMed] [Google Scholar]
- 40.Guo C., Tommasi S., Liu L., Yee J. K., Dammann R., Pfeifer G. P. (2007) Curr. Biol. 17, 700–705 [DOI] [PubMed] [Google Scholar]
- 41.Vichalkovski A., Gresko E., Cornils H., Hergovich A., Schmitz D., Hemmings B. A. (2008) Curr. Biol. 18, 1889–1895 [DOI] [PubMed] [Google Scholar]
- 42.Oh H. J., Lee K. K., Song S. J., Jin M. S., Song M. S., Lee J. H., Im C. R., Lee J. O., Yonehara S., Lim D. S. (2006) Cancer Res. 66, 2562–2569 [DOI] [PubMed] [Google Scholar]
- 43.Tommasi S., Dammann R., Zhang Z., Wang Y., Liu L., Tsark W. M., Wilczynski S. P., Li J., You M., Pfeifer G. P. (2005) Cancer Res. 65, 92–98 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.