Abstract
Trypanosoma brucei, the causative agent of African sleeping sickness, possesses two dithiol glutaredoxins (Grx1 and Grx2). Grx1 occurs in the cytosol and catalyzes protein deglutathionylations with kcat/Km-values of up to 2 × 105 m−1 s−1. It accelerates the reduction of ribonucleotide reductase by trypanothione although less efficiently than the parasite tryparedoxin and has low insulin disulfide reductase activity. Despite its classical CPYC active site, Grx1 forms dimeric iron-sulfur complexes with GSH, glutathionylspermidine, or trypanothione as non-protein ligands. Thus, contrary to the generally accepted assumption, replacement of the Pro is not a prerequisite for cluster formation. T. brucei Grx2 shows an unusual CQFC active site, and orthologues occur exclusively in trypanosomatids. Grx2 is enriched in mitoplasts, and fractionated digitonin lysis resulted in a co-elution with cytochrome c, suggesting localization in the mitochondrial intermembrane space. Grx2 catalyzes the reduction of insulin disulfide but not of ribonucleotide reductase and exerts deglutathionylation activity 10-fold lower than that of Grx1. RNA interference against Grx2 caused a growth retardation of procyclic cells consistent with an essential role. Grx1 and Grx2 are constitutively expressed with cellular concentrations of about 2 μm and 200 nm, respectively, in both the mammalian bloodstream and insect procyclic forms. Trypanothione reduces the disulfide form of both proteins with apparent rate constants that are 3 orders of magnitude higher than those with glutathione. Grx1 and, less efficiently, also Grx2 catalyze the reduction of GSSG by trypanothione. Thus, the Grxs play exclusive roles in the trypanothione-based thiol redox metabolism of African trypanosomes.
Keywords: Glutathion, Iron-Sulfur Protein, Subcellular Fractionation, Sulfhydryl, Thiol, Glutaredoxin, Trypanothione, Tryparedoxin
Introduction
Glutaredoxins (Grxs)2 are ubiquitous small thiol-disulfide oxidoreductases that play crucial roles in the redox homeostasis of the cell. Their considerable variation in specificity allows them to participate in a large number of biological processes. All organisms have an individual set of isoforms that occur in the cytosol, mitochondria, or nucleus of different cells (1–3). Structurally, two main groups are distinguished, dithiol Grxs containing a CXXC active site sequence (mostly CPYC) and monothiol Grxs with a CXXS motif (mostly CGFS) (4). The classical Grx system is composed of a dithiol Grx, GSH, glutathione reductase (GR), and NADPH. Dithiol Grxs act via two distinct mechanisms. Protein disulfides are reduced by a dithiol mechanism requiring both active site cysteines. In contrast, mixed disulfides between GSH and proteins or low molecular mass thiols are mainly reduced by a monothiol mechanism that involves only the more N-terminal redox active cysteine (for reviews, see Refs. 4–6). Important physiological functions are the reversible S-glutathionylation of proteins upon oxidative stress or redox signaling and the delivery of reducing equivalents for the synthesis of DNA precursors by ribonucleotide reductase (RR). Recently, human Grx2 and poplar GrxC1 have been shown to coordinate iron-sulfur clusters (7–11). As monomeric (apo) forms, the proteins are regular Grxs, whereas in the dimeric form with a [2Fe-2S] cluster that bridges the monomers, they are catalytically inactive.
Trypanosomes and Leishmania are the causative agents of several tropical diseases. All trypanosomatid organisms lack GR and thioredoxin reductase but have a trypanothione reductase (TR) instead (for reviews, see Refs. 12 and 13). The parasite thiol redox metabolism is based on trypanothione (T(SH)2), which is formed from spermidine and glutathione by a bifunctional trypanothione synthetase (14). In two consecutive steps, first glutathionylspermidine (Gsp) is formed, which then reacts with a second GSH molecule to yield T(SH)2. A direct linkage of the spermidine and GSH metabolism was first discovered in Escherichia coli, which during stationary phase forms large amounts of Gsp (15). In trypanosomatids, the T(SH)2 system has been shown to deliver the reducing equivalents for the synthesis of DNA precursors by RR (16) as well as the detoxification of hydroperoxides catalyzed by different peroxidases (for a review, see Ref. 17). The reactions are accelerated by tryparedoxin (Tpx), a parasite-specific distant relative of thioredoxins and Grxs (16, 18–20). Despite the absence of thioredoxin reductases, trypanosomes encode a thioredoxin gene, but the protein levels in the parasites are extremely low, and the physiological role of the protein is not known (21, 22). Very recently, it has been shown that the mRNA levels of thioredoxin are elevated upon ER stress (23).
The genome of Trypanosoma brucei (24), the causative agent of African sleeping sickness, revealed the presence of three genes for monothiol Grxs as well as two genes for putative dithiol Grxs. The monothiol Grxs have been partially characterized, and we could show that 1-C-Grx1 is a mitochondrial protein that forms an iron-sulfur cluster (25, 26). Despite the absence of a GR, trypanosomatids contain appreciable concentrations of free GSH (13). Thus, an intriguing question is if in these parasites GSH serves only as a precursor molecule for T(SH)2 or if it has distinct functions, such as protein (de)glutathionylation or as a reducing agent of Grxs.
African trypanosomes occur in two proliferative forms, the bloodstream cells that multiply in the mammalian host and the procyclic insect form in the tsetse fly. Here we report on the in vivo and in vitro characterization of the two dithiol Grxs of these parasites. T. brucei Grx1 is a cytosolic protein that efficiently catalyzes the reduction of protein-GSH mixed disulfides and coordinates iron-sulfur clusters. Grx2 is essential for procyclic parasites and most probably occurs in the intermembrane space of the single mitochondrion. Grx2 catalyzes the reduction of insulin disulfide and has low deglutathionylation activity. The intramolecular disulfide of the Grxs is reduced by T(SH)2 at rate constants that are 3 orders of magnitude higher than those with GSH. Both proteins catalyze the reduction of GSSG by T(SH)2, showing that the dithiol Grxs are closely linked to the unique trypanothione metabolism of these parasites.
EXPERIMENTAL PROCEDURES
Materials
E. coli Grx1 was purchased from IMCO Corp. (Stockholm, Sweden); GSH, hydroxyethyl disulfide (HED), and tetracycline (tet) were from Sigma-Aldrich; and the monoclonal mouse anti-Myc antibodies were from Roche Applied Science. T(SH)2 (27), T. brucei Tpx (19), T. cruzi TR (28), T. brucei glutathione peroxidase type tryparedoxin peroxidase (Px III) (29), T. brucei His6-peroxiredoxin type tryparedoxin peroxidase (TXNPx) (30), the small subunit of T. brucei RR (R2) (31), and His-tagged E. coli cysteine desulfurase (9) were prepared as described. The gene for the large subunit (R1) of RR was cloned in the modified pET vector described below, and the tag-free protein was purified by chromatography on two consecutive nickel-NT-Sepharose columns as outlined below for Grx1 and Grx2.3 Recombinant human GR was a kind gift of Dr. Heiner Schirmer (Heidelberg, Germany). The pETtrx1b vector and the His6-tobacco etch virus (TEV) protease were provided by Gunther Stier (Heidelberg, Germany). Bloodstream and procyclic T. brucei cell lines of strain 449 (strain 427 (MiTat 1) stably transfected with pHD449 encoding the tet repressor) as well as the pHD678 and pHD1700 vectors were provided by Dr. Christine Clayton (Heidelberg, Germany). T. brucei mitoplasts and the cytochrome c antiserum were a kind gift of Dr. André Schneider (Bern, Switzerland). Antibodies against acetate:succinate-CoA-transferase were from Dr. Frédéric Bringaud (Bordeaux, France). Polyclonal guinea pig antisera against recombinant T. brucei Grx1 and Grx2 and rabbit antiserum against T. brucei lipoamide dehydrogenase (LipDH)4 and TXNPx were generated by Eurogentec (Köln, Germany). Genomic DNA was extracted from procyclic T. brucei with the DNeasy® tissue kit (Qiagen, Hilden). Plasmids were purified by the NucleoBond® plasmid purification kit (Macherey und Nagel, Düren).
Cloning and Overexpression of T. brucei Grx1 and Grx2
Total RNA was isolated from procyclic T. brucei using the RNeasy® minikit (Qiagen), and cDNA was prepared by RT-PCR with SuperScriptTM III reverse transcriptase (Invitrogen). The 5′- and 3′-regions of grx1 and grx2 were amplified from the cDNA with gene-specific primers in combination with either the spliced leader primer or a poly(T) primer. The coding region of grx1 was amplified from genomic DNA. The forward primer (5′-G ACC ATG GAA CCC TCT ATC GCT TCG AT-3′) contained an NcoI site (underlined) and GAA encoding an additional glutamate, and the reverse primer (5′-GC GGA TCC TTA GCT CAG CAA ACC ATC-3′) contained a BamHI site directly following the stop codon. The gene was amplified by PCR (94 °C for 2 min; 94 °C for 30 s; 56 °C for 30 s; 72 °C for 2 min; 30 cycles; 72 °C for 10 min; Pfu). The PCR product was cloned into pETtrx1b, a modified pET vector encoding thioredoxin-His6, followed by a TEV-protease cleavage site and the cloning site. E. coli XL1Blue cells were transformed with pETtrx1b/grx1 and grown in 2× YT medium with 50 μg/ml kanamycin. The plasmid was isolated, and the insert was completely sequenced in both directions (GATC Biotech AG, Konstanz, Germany). A 500-ml culture of recombinant BL21(DE3) cells was incubated at 37 °C in 2× YT medium containing 50 μg/ml kanamycin. At an A600 of 0.5, expression was induced by adding 500 μm isopropyl-β-d-thiogalactopyranoside, and the cells were allowed to grow overnight at 18 °C. The coding region of grx2 was amplified from genomic DNA as described for grx1. The forward primer (5′-GC ACC ATG GGA AAT AAC GCA TTG GAT C-3′) contained an NcoI site (underlined) and GGA encoding an additional glycine; the reverse primer (5′-GT GGT ACC TCA GCC CCT TTC AAT TTC G-3′) contained an Acc65I site directly following the stop codon. The PCR product was blunt end-cloned into the pETBlue-1 vector, and E. coli XL1Blue cells were transformed and grown in 2× YT medium. The pETBlue-1/grx2 plasmid was isolated, digested with NcoI and Acc65I, and ligated into the pETtrx1b vector, yielding pETtrx1b/grx2. The plasmid was sequenced and overexpressed exactly as described above for grx1.
Purification of Recombinant T. brucei Grx1 and Grx2
Cells from 500 ml of recombinant bacterial culture were harvested by centrifugation; resuspended in 10 ml of 50 mm sodium phosphate, 300 mm NaCl, pH 7.5 (buffer A) containing 150 nm pepstatin, 4 nm cystatin, and 100 μm PMSF; and disintegrated by sonification in the presence of 1 mg of lysozyme and 0.5 mg of DNase. The cell debris was removed by centrifugation, and the supernatant was applied onto 12 ml of nickel-nitrilotriacetic acid Superflow resin (Qiagen) equilibrated in buffer A and connected to an FPLC system (Amersham Biosciences). After washing with 50 mm imidazole in buffer A, the fusion protein was eluted with 150 mm imidazole. Fractions containing pure fusion protein were concentrated in an Amicon Ultra 5000 molecular weight cut-off concentrator (Millipore). The buffer was exchanged by 50 mm Tris/HCl, pH 7.5, 150 mm NaCl on a PD10 column (GE Healthcare), and the fusion protein was digested overnight at 16 °C with 1 mg of TEV protease in a final volume of 5 ml. The protein solution was applied again onto the nickel-nitrilotriacetic acid column, and the tag-free recombinant Grx1 was obtained in the flow-through. Recombinant T. brucei Grx2 was purified as outlined for Grx1 except that the first nickel column was washed with 20 mm imidazole, and the fusion protein was eluted with 250 mm imidazole. About 30 and 15 mg of pure T. brucei Grx1 and Grx2, respectively, were obtained from 500 ml of bacterial culture.
Determination of Free Thiol Groups
The concentration of free SH groups was determined by reaction with 5,5′-dithiobis-(2-nitrobenzoic acid) (Ellman's reagent; ϵ412 = 13.6 mm−1 cm−1) (32).
DNA Constructs for RNA Interference against Grx1 and Grx2 in T. brucei
Because of the small size of the grx1 (288 bp) and grx2 (324 bp) genes, the complete coding sequences were used to generate the pHD678ms/ri-grx1 (where ms represents “minus stuffer”) and pHD678ms/ri-grx2 constructs. The detailed procedure is provided in the supplemental material.
DNA Constructs for Overexpression of Grx1 and Grx2 in T. brucei
The detailed procedure for cloning the pHD1700/grx1-c-myc2 and pHD1700/grx2-c-myc2 plasmids is provided in the supplemental material.
Cultivation of T. brucei
Bloodstream cells were grown in HMI-9 medium (Invitrogen) at 37 °C in a humidified atmosphere with 5% CO2 and procyclic T. brucei in MEM-Pros medium (Biochrom, Berlin, Germany) containing 14 μm hemin at 27 °C. Both culture media were supplemented with 10% (v/v) heat-inactivated fetal calf serum (Invitrogen), 50 units/ml penicillin, and 50 μg/ml streptomycin as well as 0.2 and 0.5 μg/ml phleomycin for bloodstream and procyclic cells, respectively, as described previously (26). All recombinant cell lines were generated with constructs carrying a hygromycin resistance cassette, and the medium for bloodstream and procyclic cells contained 10 and 50 μg/ml hygromycin, respectively. RNA interference or overexpression of Grx1 or Grx2 was induced by adding 1 μg/ml tet to the medium.
Western Blot Analysis
The parasites were harvested by centrifugation, boiled 5 min in 10–20 μl of 2× Laemmli buffer, and subjected to SDS-PAGE on 15 or 17% gels, followed by Western blot analysis against Grx1 (1:400) and Grx2 (1:200). Horseradish peroxidase-conjugated goat anti-guinea pig IgG served as secondary antibody (1:10,000; Santa Cruz Biotechnology). Bands were visualized by chemiluminescence using the SuperSignal West Pico Kit (Pierce).
Generation and Growth Phenotype Analysis of T. brucei Depleted of Grx1 or Grx2
The transfections were carried out with the human T cell Nucleofactor® kit. 4 × 107 parasites were centrifuged for 10 min at 2000 × g and 4 °C and resuspended in 100 μl of human T cell Nucleofactor® solution. 10 μg of NotI-linearized DNA (pHD678ms/ri-grx1 or pHD678ms/ri-grx2) was added, and electroporation was accomplished in an Amaxa-certified cuvette in the Nucleofactor® II (Amaxa Biosystems, Köln, Germany). Bloodstream transfectants were transferred into 50 ml of prewarmed HMI-9 medium, and procyclic transfectants were seeded in 50 ml of conditioned MEM-Pros medium. 24 h after electroporation, hygromycin was added. Single clones were obtained by serial dilution in prewarmed medium in microtiter plates. Bloodstream (1 × 105 cells/ml) and procylic (5 × 105 cells/ml) T. brucei transfected with the pHD678ms/ri-grx1 or pHD678ms/ri-grx2 plasmid were cultivated in the presence or absence (control) of 1 μg/ml tet. The cells were counted daily in a Neubauer chamber, and the culture was diluted to the initial cell density with fresh medium with or without tet.
Generation and Phenotypic Analysis of T. brucei Cell Lines That Overexpress Grx1-c-Myc2 or Grx2-c-Myc2
4 × 107 Trypanosomes were transfected with 10 μg of the NotI-linearized pHD1700/grx1-c-myc2 or pHD1700/grx2-c-myc2 DNA, and single clones were obtained as outlined above. Expression of the ectopic copy of the genes was induced by 1 μg/ml tet. For continuous growth experiments, the starting density was 1 × 104 and 5 × 105 cells/ml for bloodstream and procyclic parasites, respectively. Every 24 h, the cells were counted, and 1 μg/ml tet was added. For long term cultivation, bloodstream and procyclic parasites were inoculated at a density of 1 × 105 cells/ml and 8 × 105 cells/ml, respectively. The cells were counted every 24 h, and the cultures were diluted to the initial cell density with fresh medium with or without tet. Protein expression was detected by Western blot analyses as described above.
Immunofluorescence Analysis
Trypanosomes transfected with pHD1700/grx1-c-myc2 or pHD1700/grx2-c-myc2 were grown in the presence of tet for 48 h to a density of 1 × 106 cells/ml. The immunofluorescence analysis was carried out essentially as described previously (26) except that the slides were coated 10 min with 0.1 mg/ml polylysine in water (200 μl/well), washed three times for 5 min with 500 μl of water, and dried on air prior to adding the paraformaldehyde-treated cells. An Axiovert 200M microscope equipped with an AxioCam MRm digital camera and the program AxioVision (Zeiss, Jena, Germany) was used for analysis.
Fractionated Digitonin Lysis of T. brucei
The differential membrane permeabilization was done essentially as described by Coustou et al. (33). For each digitonin concentration, 1.4 × 108 procyclic cells were washed once with PBS and twice with 10 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, pH 8.0. A stock solution of 10 mg/ml digitonin (Sigma-Aldrich) was prepared in boiling water, and 300-μl aliquots of various concentrations were made by dilution with buffer. 200 μl of the respective solution was added to each cell pellet, resulting in a fixed protein concentration of 2.8 mg/ml and a ratio of 0–0.55 mg of digitonin/mg of total protein. The samples were incubated at 25 °C for 5 min and centrifuged at 10,000 × g and 4 °C for 5 min, and the supernatants were mixed with 4× sample buffer. The pellets were washed twice with 100 μl of PBS with thorough mixing, dissolved in 200 μl of 1× sample buffer, boiled for 5 min, and stored at −20 °C. Aliquots were subjected to Western blot analysis against Grx1 (1:200), Grx2 (1:100), T. brucei TXNPx (1:3000), LipDH (1:4000), acetate:succinate-CoA-transferase (1:500), and cytochrome c (1:200) as primary antibodies and HRP-conjugated goat anti-guinea pig (1:10,000) and anti-rabbit (1:20,000), respectively, as secondary antibodies.
Determination of the Apparent Rate Constants for the Reduction of Grx1 and Grx2 by T(SH)2 or GSH
Recombinant Grx1 and Grx2 are purified mainly in the dithiol form but upon storage become oxidized to the disulfide form. Incubation of the protein samples with 1.1 eq of hydrogen peroxide for 3 h at room temperature resulted in the fully oxidized form, as shown by reaction with Ellman's reagent. Reduction of oxidized Grx1 and Grx2 by T(SH)2 was monitored in a coupled assay. In a final volume of 200 μl of TR assay buffer (40 mm Hepes, 1 mm EDTA, pH 7.5), the reaction mixture contained 250 μm NADPH, 1.4 units of TR, and 35–130 nm T(SH)2. Control assays confirmed that Grx1 and Grx2 are not directly reduced by TR. Reduction of the Grxs by GSH was measured accordingly in the presence of 250 μm NADPH, 1 unit of GR, and 140–470 μm GSH in GR assay buffer (20.5 mm potassium dihydrophosphate, 26.5 mm dipotassium hydrogen phosphate, 1 mm EDTA, 200 mm KCl, pH 7.0). Oxidized Grx1 or Grx2 was added, and NADPH consumption (ϵ340 = 6.2 mm−1 cm−1) was monitored in a Jasco V650 spectrophotometer recording 1 data point/s. The k1 value for each time point was calculated with the equation, k1 = 2.303/t × log (a/(a − x)) (s−1), where a and (a − x) are the initial and the remaining concentration at time point t of oxidized Grx. The k2 value was obtained by dividing the k1 value by the thiol concentration.
Reduction of GSSG by T(SH)2
The ability of T. brucei Grx1 and Grx2 to catalyze the reduction of GSSG by T(SH)2 was studied in a TR-coupled assay system (19). In a total volume of 1 ml of 100 mm Tris/HCl, 1 mm EDTA, pH 8.0, the reaction mixture contained 200 μm NADPH, 8.4 or 21 μm T(SH)2, 1.6 units of TR, and 20–800 μm GSSG. The reaction was started by adding 10 nm Grx1 or 130 nm Grx2, and consumption of NADPH was monitored at 340 nm.
Reconstitution and Analysis of FeS Grx Clusters
FeS cluster assembly was performed essentially as described (9). 50–230 μm Grx1 or Grx2 in 50 mm sodium phosphate, 300 mm NaCl, pH 8.0, 5 mm DTT was incubated at room temperature under argon atmosphere with Fe(NH4)2(SO4)2, cysteine, Gsp or GSH, cysteine desulfurase, and pyridoxal phosphate in a molar ratio of 1 (Grx):2:2.5:4:0.02:0.04 for at least 1 h. Subsequently, the reaction mixture was centrifuged for 2 min at 13,000 rpm and 4 °C and immediately analyzed by UV-visible spectroscopy and gel filtration chromatography on a Superdex 75 10/300 column (GE Healthcare) in 50 mm sodium phosphate, 300 mm NaCl, pH 8.0. The column was calibrated with the Gel Filtration Calibration Kit LMW (GE Healthcare).
Preparation of Glutathionylated Proteins
In 50 ml of 100 mm potassium phosphate, 1 mm EDTA, pH 7.0, 2 mm BSA was incubated with 20 mm DTT for 1 h at room temperature and dialyzed. Because the protein precipitated during dialysis, only 22 μmol of reduced BSA was recovered. 100 μm pretreated BSA was allowed to react with 17.5 mm GSH and 13 mm diamide for 2 h (34), and the low molecular mass components were removed in an Amicon Ultra 30,000 molecular weight cut-off concentrator. The degree of protein glutathionylation was determined enzymatically. In a total volume of 200 μl of buffer, the cuvette contained 1 mm GSH, 200 μm NADPH, 800 milliunits of GR, and 5 μm E. coli Grx1. The reaction was started by adding 2 μl of the glutathionylated BSA (BSA-SSG) solution, and the absorption decrease at 340 nm was monitored. The concentration of protein-bound GSH groups was calculated from the total NADPH consumption. The BSA concentration was determined at 280 nm (ϵ = 43.6 mm−1 cm−1). Glutathionylated TXNPx was prepared accordingly. A 2 mm protein solution was treated with 20 mm DTT for 30 min at 36 °C, and excess thiol was removed in an Amicon Ultra 10000 concentrator. 1 mm TXNPx was incubated with 114 mm GSH and 86 mm diamide overnight at 20 °C and freed from other reaction components in an Amicon Ultra 10000 concentrator. The procedure resulted in 900 μl of 1.9 mm TXNPx with 3 mm bound GSH corresponding to 1.6 GSH groups/protein monomer.
Reductase Activity of Grx1, Grx2, and Tpx toward Glutathione Mixed Disulfides
The activity of Grx1 and Grx2 in the HED assay was determined essentially as described by Johansson et al. (35). In 1 ml of 100 mm Tris/HCl, 1 mm EDTA, pH 8.0, 0.8 mm HED, 1 mm GSH, 200 μm NADPH, and 300 milliunits of GR were preincubated at 25 °C, resulting in the mixed disulfide between GSH and 2-mercaptoethanol (2-ME-SSG). After 6 min, 5–50 nm Grx1, 20–200 nm Grx2, or 5–25 nm E. coli Grx1 was added, and NADPH consumption was recorded. Grx activity was calculated after subtracting the spontaneous reduction rate.
The kinetic constants for different glutathionylated substrates were determined in a modified HED assay. The reaction mixtures contained, in 200 μl of 100 mm potassium phosphate, 1 mm EDTA, pH 7.0, 200 μm NADPH, 1 mm GSH, 1 unit of GR, and 5–230 μm BSA-SSG, 15–170 μm glutathionylated TXNPx (TXNPx-SSG), or 0.08–3.2 mm HED. Reduction of BSA-SSG and TXNPx-SSG was measured after the addition of 80–550 nm Grx1 and 70 nm to 1.4 μm Grx2, respectively. For the HED assay, all components except Grx were preincubated for 3 min, and the reaction was started by 100 nm Grx. In the Tpx assay, 4.4–14.5 μm Tpx was preincubated with all components except BSA-SSG to ensure complete reduction. After 7 min, the reaction was started by adding 34.5–230 μm BSA-SSG.
Ribonucleotide Reductase Assay
RR activity was determined at 37 °C from the rate of conversion of GDP to dGDP in a photometric assay by coupling the reaction to NADPH consumption catalyzed by TR. The reaction mixtures contained in a total volume of 150 μl of 50 mm Hepes, 100 mm KCl, 7 mm MgCl2, pH 7.6, 200 μm T(SH)2, 200 μm NADPH, 150 milliunits TR, 500 μm dTTP, 1.6 mm ATP, 3 μm R1, 12–18 μm R2, and various concentrations of Grx1 (4–130 μm), Grx2 (3–100 μm), Tpx (4–30 μm), or E. coli Grx1 (2–30 μm). The reaction was started by adding 500 μm GDP, and NADPH oxidation was monitored at 340 nm. The absorption decrease in the absence of GDP was subtracted, and the RR activity was calculated from the ΔΔA/min value.
Peroxidase Assays
A possible peroxidase activity of Grx1 and Grx2 was determined as described (36). The reaction mixtures contained in 150 μl of 100 mm Tris/HCl, 5 mm EDTA, pH 7.6, 240 μm NADPH, 100 μm trypanothione disulfide, 150 milliunits TR, 0.6 μm Px III, and 50 μm Grx1 and Grx2, respectively, or 10 μm Tpx as a positive control. The assay was started by adding 100 μm H2O2, and oxidation of NADPH was monitored. Alternatively, the assays contained 10 μm Tpx plus 10 μm Grx1 or Grx2. In analogy to the peroxidase assays described for yeast Grxs (37), the third type of assays lacked Tpx and Px III but contained 50 μm Grx1 or Grx2.
Insulin Reduction Assay
In a total volume of 200 μl of 100 mm potassium phosphate, 2 mm EDTA, pH 7.0, various concentrations of Grx were incubated with 5 mm thiol (dithioerythritol, T(SH)2, Gsp, and GSH) for 20 min at room temperature. The reaction was started by adding 600 μl of a 1 mg/ml insulin solution in buffer, resulting in a final concentration of 130 μm insulin and 1.25 mm thiol. The increase in turbidity was monitored at 650 nm and 30 °C.
RESULTS
African Trypanosomes Possess Two Only Distantly Related Dithiol Grxs
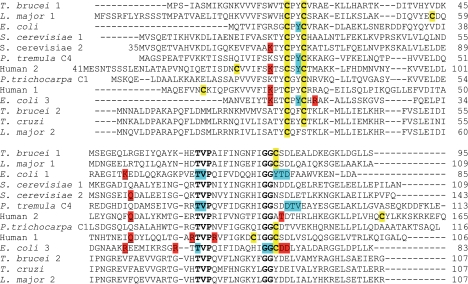
The completely sequenced T. brucei genome revealed two single copy genes for putative dithiol Grxs (24). One gene (Tb 11.47.0012) is located on chromosome 11. Because it encodes a protein with a CPYC active site and its only Cys outside the active site (Cys78) is conserved in human Grx1, we named it T. brucei Grx1. However, although T. brucei Grx1 has the classical active site motif, the overall protein sequence is more similar to human Grx2c (40% identity) than to human Grx1 (31%). The other gene on chromosome 1 (Tb 927.1.1770) encodes a protein with an unusual CQFC motif and only 25% overall sequence identity with Grx1 and was named Grx2. The protein is highly conserved within different trypanosomatids (about 80% identity), but BLAST searches did not reveal any counterpart in organisms outside the order of Kinetoplastida. Remarkably, the Leishmania Grx2 sequences available have a Ser residue instead of the second Cys and thus are monothiol Grxs. The Grx2 proteins do not have any additional Cys residue.
Fig. 1 shows an alignment of the parasite proteins with well characterized Grxs from other organisms (10, 38–42). Besides the redox active motif, a TVP stretch with a cis-Pro residue and a GG motif are characteristic for Grxs (for a review, see Ref. 4). The two glycines together with the Thr of the TVP motif and the Tyr in the active site (corresponding to Gly76, Gly77, Thr64, and Tyr23 in T. brucei Grx1) have been reported to form a binding groove for GSH on the protein surface (10, 38, 39). These residues are conserved in the trypanosomatid sequences. However, the Lys that precedes the active site motif as well as another charged or polar residue found to interact with the glycine carboxylate of GSH in various Grx structures (10, 39, 41, 42) are replaced by hydrophobic residues. Interestingly, in E. coli Grx2, which has the highest activity of the three bacterial dithiol Grxs toward 2-ME-SSG, the Lys is replaced by a Tyr. This supports previous reports that the characteristic γ-glutamate moiety is more important for GSH recognition than the glycine part (43, 44). In T. brucei Grx1 and Grx2, these substitutions probably support the interaction with T(SH)2 (see below), in which the glycine carboxylate does not carry a negative charge but is engaged in an amide bond with the spermidine bridge.
Generation of Recombinant T. brucei Grx1 and Grx2
In the case of the T. brucei monothiol 1-C-Grx1, the starting Met proved not to be encoded by the first ATG following a stop codon (25). Therefore, the 5′-regions of the mRNAs were amplified from cDNA using gene-specific reverse primers in combination with a spliced leader primer. The spliced leader is a 39-bp-long sequence at the 5′-end of all mature kinetoplastid mRNAs that is added by trans-splicing. The PCR yielded a 5′-untranslated region of 197 and 373 bp for Grx1 and Grx2, respectively, and confirmed the start Met depicted in Fig. 1. Amplification of the 3′-untranslated region with an internal primer and a poly(T) primer resulted in fragments with 434 and 409 non-coding bp for Grx1 and Grx2, respectively. The complete mRNA sequences are provided as supplemental Fig. S1. The coding regions of grx1 and grx2 were then amplified from genomic DNA, cloned, and overexpressed in E. coli, and the recombinant proteins were purified as outlined under “Experimental Procedures.” Thiol determinations revealed that the proteins were obtained to ≥90% in the reduced form. Gel chromatography on a Superdex 75 column showed that both recombinant T. brucei Grxs are monomeric proteins (for Grx1, see the inset of Fig. 6B; for Grx2, data not shown).
FIGURE 1.
Comparison of T. brucei Grx1 and Grx2 with dithiol Grxs from other organisms. Strictly conserved residues and cysteines are depicted in boldface type. In addition, all cysteines are highlighted by a yellow background. Residues described as forming a groove for GSH binding on the surface of Grxs (10, 38, 39) or as undergoing, in addition to Cys30, large chemical shift changes in the NMR structure of Populus tremula GrxC4 upon titration with GSH (40) are shown with a blue background. Charged residues engaged in side chain interactions with a bound GSH molecule either in a mixed disulfide with mutants of E. coli Grx1 (38), human Grx1 (41), E. coli Grx3 (39), and yeast Grx2 (42) or in the non-covalent complex of human Grx2 with GSH (10) are highlighted by a red background. Those residues interacting with the glycine carboxylate of GSH are also underlined.
FIGURE 6.
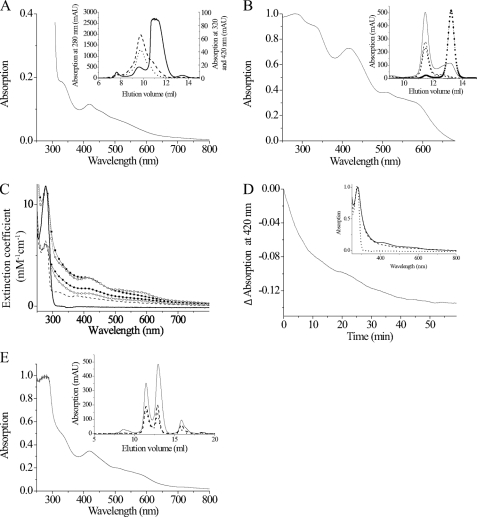
Reconstitution and analysis of the iron-sulfur cluster of Grx1. A, UV-visible spectrum of the Grx1-fusion protein (40 mg/ml) purified from E. coli. Inset, elution profile from the Superdex 75 column recording absorption at 280 nm (solid line), 320 nm (dashed line), and 420 nm (dotted line). The protein eluted in two peaks at 9.74 and 10.96 ml that correspond to 63.5 and 37.6 kDa, respectively. B, UV-visible spectrum of 230 μm tag-free recombinant Grx1 subjected to FeS cluster reconstitution in the presence of Gsp. Inset, gel chromatography profiles of untreated (broken line with squares and dots) and reconstituted protein (solid line) at 280 nm. Grx1 eluted at 13.16 ml and did not show any absorption at 320 or 420 nm. The reconstituted protein eluted in two peaks. The main one at 11.47 ml but not that at 13.16 ml also showed absorption at 320 (dashed line) and 420 nm (dotted line). C, spectra of 50 μm of Grx1 (squares) and Grx2 (circles) after reconstitution in the presence of 100 μm GSH (filled symbols) or Gsp (empty symbols) as well as of free Grx1 (solid line) and Grx2 (broken line). The spectra were recorded 1 h after reconstitution and normalized according to the molar extinction coefficient of the protein. D, 50 μm Grx1 reconstituted in the presence of 100 μm GSH was distributed into two cuvettes, 1 mm Gsp was added to the reference cuvette, and the absorption difference at 420 nm was monitored. Inset, UV-visible spectrum of the sample (dashed line) and the reference (solid line) solution 2 h after exposure to air as well of free Grx1 (dotted line). E, spectrum of 230 mm Grx1 reconstituted in the presence of 1 mm T(SH)2. Inset, the protein eluted from the Superdex 75 column at 11.46 and 12.96 ml. Both protein species absorbed at 320 nm (dashed line) and 420 nm (dotted line) in addition to 280 nm (solid line). The peak at 15.87 ml is a yet to be identified iron-T(SH)2-complex.
RNAi against Grx2 Affects Proliferation of Procyclic Parasites
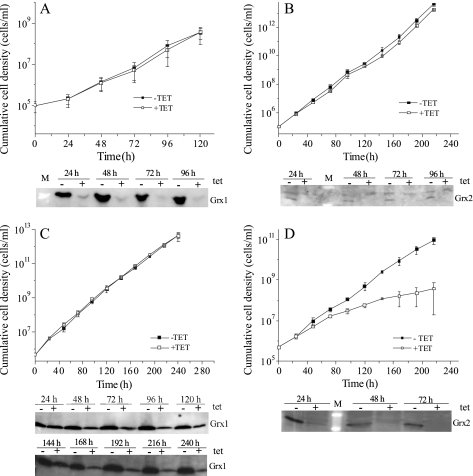
Bloodstream and procyclic T. brucei were transfected with pHD678ms/ri-grx1 or pHD678ms/ri-grx2. 48 h after RNAi induction in bloodstream cells, the protein levels of Grx1 and Grx2 were down-regulated to ≤15% when compared with uninduced cells. This degree of depletion did not affect proliferation (Fig. 2, A and B). In procyclic cells, RNAi against Grx1 resulted in a down-regulation to 20% after 168 h, but again, no growth phenotype was observed (Fig. 2C). In contrast, depletion of Grx2 caused a growth retardation starting 48 h after the onset of RNAi (Fig. 2D); the calculated doubling time was twice that of the non-induced cells, suggesting an essential role.
FIGURE 2.
Depletion of Grx1 and Grx2 in T. brucei by RNA interference. Shown are proliferation profiles of bloodstream (A and B) and procyclic (C and D) T. brucei transfected with pHD678ms/ri-grx1 (A and C) and pHD678ms/ri-grx2 (B and D) and grown in the presence (+tet) or absence (−tet) of 1 μg/ml tet. Every 24 h, the cells were counted, an aliquot was removed for Western blot analysis, and the culture was diluted with fresh medium (±tet) to the initial cell density. Depicted is the mean of the cumulative cell density ± S.D. of three clones and representative Western blots of 1 × 107 cells/lane with the specific antiserum against Grx1 (1:200) (A and C) and Grx2 (1:100) (B and D), respectively.
Overexpression of Grx1 or Grx2 Does Not Affect Proliferation of T. brucei
Parasites transfected with pHD1700/grx1-c-myc2 or pHD1700/grx2-c-myc2 were grown in the presence of tet. Western blot analysis showed that Grx1-Myc2 and Grx2-Myc2 were expressed at levels at least 5–10-fold and 3-fold higher than those of the respective authentic protein (supplemental Fig. S2). This degree of Grx1 and Grx2 overexpression did not affect the proliferation rate of bloodstream and procyclic cells grown continuously for 96 and 168 h, respectively (not shown), or when the cultures were diluted back to the initial cell density every 24 h and were observed for a total of 7 days (supplemental Fig. S2).
In Vivo Concentration and Subcellular Localization of Grx1 and Grx2
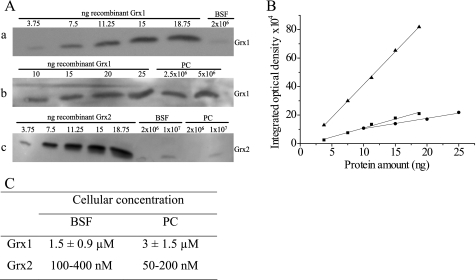
The levels of E. coli Grx1 and Grx2 had been reported to vary during different growth phases (5). Wild type bloodstream and procyclic cells were harvested at early and late logarithmic as well as stationary phase and subjected to Western blot analysis. The intensities of the signals in the different samples were comparable, suggesting that Grx1 and Grx2 are constitutively expressed independent of the growth phase (data not shown). Western blot analysis of total cell lysates in comparison with different amounts of the recombinant protein yielded for Grx1 a cellular concentration of about 2 μm in both parasite forms when assuming an even distribution. For Grx2, a 10-fold lower concentration was obtained (Fig. 3).
FIGURE 3.
Quantification of Grx1 and Grx2 in bloodstream and procyclic T. brucei. A, representative Western blots of total cell lysates from cultured T. brucei in comparison with different amounts of recombinant Grx1 (bloodstream (BSF) (a) and procyclic (PC) (b) cells) and Grx2 (c). B, the integrated optical densities of each gel band were obtained with the BIO-Profil Biolight program and are depicted versus the protein amount (a, ■; b, ●; c, ▴). C, the cellular concentration of both proteins was derived from the standard lines. The values for Grx1 represent the mean ± S.D. of at least three determinations. In the case of Grx2, the values of two independent analyses are shown.
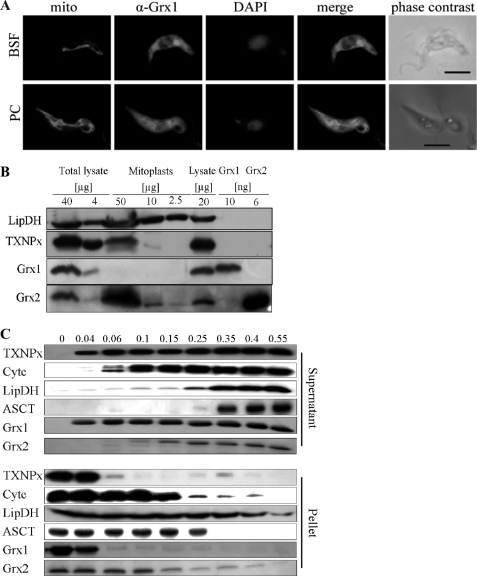
T. brucei that overexpress Grx1-Myc2 or Grx2-Myc2 were subjected to immunofluorescence microscopy as described under “Experimental Procedures.” The staining pattern for Grx1 suggested a mainly cytosolic localization in both developmental states (Fig. 4A) in accordance with the lack of any obvious targeting sequence. The annotated protein sequence of the Leishmania major orthologue (LmjF27.0810) shows an N-terminal extension encoding a putative mitochondrial targeting signal (Fig. 1). However, the start of the coding region should be verified experimentally because an upstream stop codon is missing. The immunofluorescence analysis of Grx2 even in cells overexpressing the protein proved to be difficult. Both the specific Grx2 antiserum and the monoclonal Myc antibody resulted in a weak diffuse staining of the cell (not shown). Because in other organisms Grxs occur in different cellular compartments (2, 3), mitoplasts from procyclic T. brucei were studied by Western blot analysis against Grx1 and Grx2 in comparison with the mitochondrial LipDH and the cytosolic TXNPx. As shown in Fig. 4B, LipDH gave strong signals in both the total lysate and the mitoplast fraction. In contrast, TXNPx showed a strong signal in the cell lysates but only a weak one in the mitoplasts. Grx1 was only detectable in the total lysate. This supports its cytosolic localization and renders an additional mitochondrial occurrence, at least in procyclic cells, unlikely. In contrast, the Grx2 antiserum resulted in a stronger signal in the mitoplast fraction compared with the cell lysate. To confirm the cytosolic and mitochondrial localization of Grx1 and Grx2, respectively, procyclic T. brucei were treated with increasing concentrations of digitonin, which results in the gradual permeabilization of cellular membranes (33). Western blot analysis of the supernatant and pellet fractions of each sample revealed that TXNPx was completely solubilized at a digitonin/protein ratio of 0.06:1 (mg/mg). In contrast, the mitochondrial matrix proteins LipDH and acetate:succinate-CoA-transferase appeared in the supernatant at ratios of ≥0.35:1. Cytochrome c started to be solubilized at a ratio of 0.1:1. Grx1 co-eluted with TXNPx, confirming its cytosolic localization. At low digitonin concentrations, Grx2 remained completely in the pellet fraction, indicating that it does not occur in the cytosol. Interestingly, the solubilization pattern of Grx2 was more similar to that of cytochrome c than of the mitochondrial matrix proteins. Thus, Grx2 occurs probably in the intermembrane space of the single mitochondrion of the parasite, in agreement with the absence of a targeting sequence (45).
FIGURE 4.
Subcellular localization of Grx1 and Grx2. A, immunofluorescence microscopy of bloodstream form (BSF) and procyclic (PC) parasites overexpressing Grx1-c-Myc2. The mitochondrion was stained with MitoTracker® Red (mito). Grx1 was visualized with the polyclonal antiserum (α-Grx1) and Alexa488 anti-guinea pig IgG as secondary antibody. DNA (nucleus and kinetoplast) was stained with DAPI. The images were superimposed with Adobe Photoshop software (merge). The whole parasites are shown on the right (phase contrast). Bar, 5 μm. B, cell lysate (corresponding to 4, 20, and 40 μg of total protein) and isolated mitoplasts (2.5, 10, and 50 μg of protein) of procyclic cells as well as recombinant Grx1 and Grx2 were subjected to Western blot analysis. LipDH and TXNPx served as mitochondrial and cytosolic markers, respectively. C, procyclic T. brucei were treated with increasing digitonin/protein ratios (mg/mg) as indicated at the top. Supernatant and pellet of each sample were subjected to Western blot analysis. In the case of TXNPx, LipDH, and acetate:succinate-CoA-transferase (ASCT), the samples applied onto the gel corresponded to 3.5 × 106 cells. For the Western blots of cytochrome c (Cytc), Grx1, and Grx2, extracts from 1.4 × 107 cells were analyzed.
T(SH)2 Is a Much Better Reductant of Grx1 and Grx2 than GSH
The active site cysteines of Grxs can cycle between the reduced dithiol state and an oxidized form with an intramolecular disulfide bridge. Because reduction of the proteins by T(SH)2 proved to be very fast, nanomolar concentrations of the dithiol were used to allow the spectrophotometric analysis. Assuming that one of the two consecutive steps upon reduction of the protein disulfide by the dithiol is much slower than the other one and determines the overall rate, apparent first order rate constants were calculated for each time point. At least two different concentrations of protein and/or thiol were studied and resulted in very similar values (Table 1). In addition, the log(a/(a − x)) values were plotted versus time, with a and (a − x) being the initial concentration and the remaining concentration at time point t, respectively, of oxidized Grx. A straight line was obtained that within experimental error passed through the origin (46). The slopes that represent the k1 values were in good agreement with the calculated data, supporting the assumption of pseudo-first-order conditions.
TABLE 1.
Apparent rate constants for the reduction of Grx1 and Grx2 by T(SH)2 and GSH, respectively
The values represent the means with S.D. of at least three determinations.
| Protein | Thiol | Concentration |
Apparent rate constants |
||
|---|---|---|---|---|---|
| Protein | Thiol groups | k1 | k2 | ||
| μm | μm | 10−2s−1 | m−1s−1 | ||
| Grx1 | T(SH)2 | 11.9 | 0.125 | 2.2 ± 0.1 | 1.7 ± 0.1 × 105 |
| 11.9 | 0.256 | 3.5 ± 0.5 | 1.4 ± 0.2 × 105 | ||
| GSH | 11.9 | 463 | 1.8 ± 0.2 | 37 ± 3 | |
| 25.5 | 193 | 0.7a | 36a | ||
| Grx2 | T(SH)2 | 13.8 | 0.14 | 1.6 ± 0.1 | 1.1 ± 0.1 × 105 |
| 13.8 | 0.07 | 1.0 ± 0.1 | 1.4 ± 0.2 × 105 | ||
| 27.5 | 0.07 | 0.9 ± 0.1 | 1.3 ± 0.1 × 105 | ||
| GSH | 9.0 | 146 | 1.7 ± 0.1 | 116 ± 9 | |
| 18 | 146 | 1.4 ± 0.2 | 96 ± 14 | ||
a The values are the means of two measurements that differed by less than 7%.
The apparent rate constants for the reduction of Grx1 and Grx2 by GSH were determined as outlined above for T(SH)2. Plotting the data according to a second order reaction resulted in practically identical k2 values (not shown), which supports the assumption that also in the case of GSH, one of the two consecutive reduction steps determines the overall rate. The apparent second order rate constants for the reduction of the proteins by T(SH)2 and GSH were 105 m−1 s−1 and ≤100 m−1 s−1, respectively (Table 1). Thus, T(SH)2 is about 1000-fold more efficient than GSH as reductant of the parasite Grxs. Under the experimental conditions chosen, namely 1.4 μm TR (1.4 units under Vmax conditions), the reaction velocity is supposed to be independent of the trapping reaction. However, if this were not the case, the true second order rate constants are even higher. Taking into account the similar cellular concentrations of GSH and T(SH)2 of about 300 μm in T. brucei (13), the results strongly suggest that the parasite Grxs are kept in the dithiol state by the spontaneous reaction with T(SH)2.
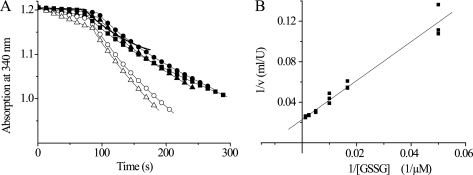
Grx1 Efficiently Catalyzes Reduction of GSSG by T(SH)2
T. brucei lack a specific GR, and GSH is regenerated from GSSG by spontaneous reaction with T(SH)2. Here we studied if at low concentrations of T(SH)2, which might occur under oxidative stress conditions, Grx1 or Grx2 can catalyze this reaction. At GSSG concentrations up to 60 μm, the spontaneous reaction was negligible. However, the presence of 10 nm Grx1 strongly accelerated the reaction (Fig. 5A). Surprisingly, the ΔA/min values remained constant or even decreased when the GSSG concentration was increased, suggesting strong substrate inhibition and a Km value clearly below 20 μm. At 8.4 μm T(SH)2 and 20 μm GSSG, the lowest concentration that allowed a reliably measurement in the coupled assay, Grx1 had 106 ± 0.9 units/mg. In contrast, Grx2 did not reveal any substrate inhibition (Fig. 5B). The kinetic parameters for GSSG in the presence of a fixed concentration of 9.8 μm T(SH)2 were fitted to an exponential curve using the Origin Pro 8.1 program, resulting in a Vmax value of 8.2 ± 0.1 units/mg and a Km value of 71 μm. To allow the direct comparison between both parasite proteins, the activity of Grx2 at 20 μm GSSG was calculated. Here, Grx2 showed 1.8 units/mg and thus a 50-fold lower activity than Grx1.
FIGURE 5.
Reduction of GSSG by T(SH)2 in the absence and presence of Grx1 and Grx2. The reactions were coupled to NADPH consumption catalyzed by TR, as outlined under “Experimental Procedures.” A, the reaction mixtures contained 8.4 μm (filled symbols) or 21 μm (empty symbols) T(SH)2 and 20 μm (−), 60 μm (■), 100 μm (○ and ●), or 200 μm (△ and ▴) GSSG. After about 80 s, Grx1 was added. The activities were calculated from the ΔA/min after subtracting the rate of the spontaneous reaction. B, Lineweaver-Burk plot of the respective analysis of Grx2 at a fixed concentration of 9.8 μm T(SH)2.
Grx1 Forms Dimeric Iron-Sulfur Cluster Complexes
The Trx-His6-TEV-Grx1 fusion protein eluted from the metal affinity column as a light brown solution. The absorption spectrum displayed, in addition to the maximum around 280 nm, peaks at 320 and 420 nm that were reminiscent of Grxs shown to coordinate [2Fe-2S] clusters (Fig. 6A) (7, 9, 11). Size exclusion chromatography revealed two peaks. The main fraction of the protein eluted at a mass of 37.6 kDa, which is higher than the calculated 25.1 kDa, possibly because of a non-ideal shape of the fusion protein. In addition, also tag-free Grx1 with a theoretical mass of 10.831 kDa often runs at higher mass (see below). A small fraction eluted at twice the mass and showed absorption at 320 and 420 nm (Fig. 6A, inset), suggesting formation of a dimeric protein species with a bound FeS cluster. This finding was rather unexpected because T. brucei Grx1 contains the canonical CPYC active site, and the presence of the Pro had been hypothesized to prevent cluster incorporation in other Grxs (9, 11). Unfortunately, the cluster was not stable upon further purification. After TEV cleavage and the second metal affinity chromatography, colorless tag-free Grx1 was obtained. Thus, the FeS cluster was reconstituted in vitro. In the first series of experiments, the reassembly mixtures contained either GSH or Gsp as a non-protein ligand. The absorption spectrum of reconstituted Grx1 (Fig. 6B) was similar to that of the fusion protein isolated from E. coli (Fig. 6A). Furthermore, gel filtration of Grx1 after in vitro reconstitution and of the free protein directly after purification disclosed that the holoprotein eluted as a dimer of apparent 30.2 kDa in contrast to the monomeric apo form with an apparent mass of 14.6 kDa (Fig. 6B, inset). An aliquot of the reconstitution mixture was subjected to the standard HED assay. Under identical assay conditions, free Grx1 showed a ΔA340/min of 0.39, whereas the reconstitution mixture revealed a value of 0.18, which with time increased to 0.27 but did not reach the activity of the free protein. The diminished activity suggests the involvement of one of the redox-active cysteine residues in cluster coordination, as is the case in other Grxs (8, 9).
The fusion protein of T. brucei Grx2 was isolated from E. coli as a colorless monomeric protein. To verify that Grx2 (although it lacks a Pro in the active site) does not form an iron-sulfur complex, both Grxs were subjected to in vitro reconstitution in the presence of GSH and Gsp, respectively. Grx2 showed very low absorption around 400 nm, suggesting that it does not bind an FeS cluster (Fig. 6C). Finally, we studied if Gsp can stabilize the Grx1 cluster upon exposure to air. The reconstitution mixture of Grx1 with GSH was distributed in two cuvettes, Gsp was added to the reference, and the absorption difference was monitored over time (Fig. 6D). The difference in absorption was highest in the first minutes but became less with time. Consequently, after 2 h, the absorption spectra of both solutions showed only a minor, but still characteristic, difference (Fig. 6D, inset). The data revealed that Gsp can slow down cluster disassembly, as has been reported for GSH and other Grxs (7).
In a second type of experiment, the reassembly mixture for Grx1 contained T(SH)2 as a low molecular mass thiol. This resulted in an absorption spectrum very similar to that observed in the presence of GSH or Gsp (Fig. 6, A, B, and E). Surprisingly, T(SH)2 formed an iron-containing complex also in the absence of any Grx5 eluting from the gel chromatography column at 15.87 ml (see inset of Fig. 6E). Interestingly, both protein peaks showed absorption at 320 and 420 nm, suggesting that the chromophore can bind also to the monomeric form of Grx1. This latter protein species probably represents an intermediate form, because incubation of Grx1 with the preformed T(SH)2 complex (generated in a reconstitution mixture lacking Grx and subsequently purified by gel chromatography) resulted nearly exclusively in the dimeric holoprotein species. In contrast, Grx1 that was pretreated with H2O2 to generate the intramolecular disulfide did not bind the preformed complex but eluted as free monomeric protein (data not shown). The reconstitution mixture in the presence of T(SH)2 had the same low HED activity observed for the Gsp-containing complex. In addition, the gel chromatography fraction containing the dimeric holoprotein species had ≤5% HED activity compared with the free protein. This clearly shows that the active site is involved in coordinating this iron-sulfur chromophore. Taken together, T. brucei Grx1 can coordinate iron-sulfur clusters, resulting in the formation of dimeric holoprotein species. Gsp can replace GSH as non-protein ligand and protect the cluster from disassembly under aerobic conditions. In contrast to the monothiols GSH and Gsp, T(SH)2 forms also a free complex that subsequently can react with Grx1 to generate the dimeric holoprotein.
Grx1 and Grx2 as Reductases of Glutathione Mixed Disulfides
The HED assay is the most commonly used Grx-specific enzymatic assay. During an initial preincubation step, HED reacts spontaneously with GSH, yielding 2-mercaptoethanol and 2-ME-SSG, which then acts as the substrate of Grx (47). The Grx-catalyzed reaction generates another molecule of 2-mercaptoethanol and the mixed disulfide between GSH and Grx. This disulfide is subsequently attacked by a second GSH molecule under regeneration of reduced Grx and formation of GSSG. The overall reaction is monitored spectrophotometrically as consumption of NADPH in the GR-catalyzed reduction of GSSG. The reaction scheme is shown in supplemental Fig. S3. To increase sensitivity, the reactions are run at a high pH (8.0) with excess HED (35). Both Grx1 and Grx2 catalyzed the reduction of 2-ME-SSG. The Grx concentrations were chosen so that the ΔA340/min was ≤0.1 to ensure a linear dependence of the reaction from the enzyme concentration (48). Thus, in the standard HED assay at pH 8.0, Grx1 displayed an activity of 26 units/mg, which is nearly 10-fold higher than that of Grx2 (supplemental Table S1). E. coli Grx1 studied as positive control revealed an activity of 100 units/mg, in good agreement with published data (49). In the next step, GSH mixed disulfides of BSA and TXNPx were prepared and studied at pH 7.0. At neutral pH, the background reaction in the absence of Grx is negligible. With both BSA-SSG and TXNPx-SSG, Grx1 showed activities that were 1–2 orders of magnitude higher compared with 2-ME-SSG (Table 2). Toward all three substrates, Grx1 had a 4–18-fold higher catalytic efficiency than Grx2. This mainly reflects differences in the substrate affinity of the proteins because the turnover numbers were identical. The catalytic efficiencies obtained for T. brucei Grx1 are comparable with those reported for human Grx1 (35). In contrast, T. brucei Tpx exerted only very low deglutathionylation activity with BSA-SSG as substrate. Thus, reduction of S-glutathionylated proteins may be a specific physiological function of the parasite Grxs that is not accomplished by Tpx.
TABLE 2.
Reduction of glutathionylated model substrates by different Grxs and Tpx
The activities were measured at pH 7.0 in reaction mixtures containing NADPH, GSH, GR, the respective GSH mixed disulfide, and Grx1, Grx2, or Tpx as described under “Experimental Procedures.” The Km and kcat values were derived from Lineweaver-Burk plots. The values for Grx1 and Grx2 with the protein mixed disulfides are the mean values ± S.D. from at least three independent series of experiments, each measured in duplicate. In the case of 2-ME-SSG, five different concentrations were measured in triplicate. The values for Tpx were obtained from a kinetic analysis with four BSA-SSG concentrations measured in triplicate.
| Protein | BSA-SSG |
TXNPx-SSG |
2-ME-SSG |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| μm | s−1 | m−1s−1 | μm | s−1 | m−1s−1 | mm | s−1 | m−1s−1 | |
| Grx1 | 8.8 ± 1.7 | 1.7 ± 0.1 | 1.9 × 105 | 21 ± 1.3 | 1.5 ± 0.1 | 6.9 × 104 | 0.49 ± 0.25 | 1.5 ± 0.34 | 3.1 × 103 |
| Grx2 | 108 ± 16 | 1.7 ± 0.3 | 1.6 × 104 | 269 ± 128 | 1.0 ± 0.4 | 3.8 × 103 | 1.65 ± 0.19 | 1.22 ± 0.1 | 7.4 × 102 |
| Tpx | 58 ± 16 | 0.02 ± 0.002 | 3.8 × 102 | NDb | ND | ND | ND | ND | ND |
| hGrx1a | 27.7 | 4.66 | 1.68 × 105 | ND | ND | ND | 1.07 | 8.16 | 7.63 × 103 |
| hGrx2a | 4.3 | 1.83 | 4.27 × 105 | ND | ND | ND | 0.11 | 1.30 | 11.68 × 103 |
a hGrx, human Grx, from Johansson et al. (35).
b ND, not determined.
Grx1, but Not Grx2, Can Supply Electrons for Reduction of RR
As shown previously, T. brucei RR obtains its reducing equivalents from the T(SH)2/Tpx system (16). Here we studied the ability of Grx1 and Grx2 to replace Tpx in this reaction. The reaction mixtures contained T(SH)2, NADPH, TR, the R2 subunit of RR in excess over the R1 subunit, and various concentrations of the different oxidoreductases. The assays were started by adding GDP as substrate for RR, and NADPH consumption by TR was monitored. The maximum activity under these conditions was about 25 milliunits/mg R1 obtained in the presence of 7 μm E. coli Grx1. 30 μm Tpx resulted in an RR activity of 15–20 milliunits/mg, in good agreement with previous data from the radioactive assay system (16). In parallel assays with different equimolar concentrations of Grx1 and Tpx, respectively, the RR activity in the presence of Grx1 ranged from 20 to 50% of that with Tpx. With Grx2, RR activity was hardly detectable at all, even when using concentrations of up to 100 μm.
Grx1 and Grx2 Lack Peroxidase Activity
Saccharomyces cerevisiae Grx1 has been reported to be an efficient general glutathione peroxidase (37). In trypanosomes, hydroperoxide reduction is catalyzed by a cascade composed of NADPH, TR, T(SH)2, Tpx, and either TXNPx or Px III, which both act as tryparedoxin peroxidases (17). Thus, the ability of T. brucei Grx1 and Grx2 to reduce H2O2 directly or to replace/support Tpx upon transfer of electrons from T(SH)2 onto Px III was studied. Reaction mixtures that lacked Tpx and Px III but contained 50 μm Grx1 or Grx2 did not show any NADPH consumption exceeding the background reaction between T(SH)2 and hydrogen peroxide. The addition of 10 μm Grx1 or Grx2 to standard peroxidase assays containing 10 μm Tpx (36) did not increase the rate of NADPH consumption. Also, replacement of Tpx by 50 μm Grx1 or Grx2 did not result in hydrogen peroxide reduction. Thus, the parasite Grxs neither function as direct T(SH)2-dependent peroxidases nor replace Tpx as a component of the parasite peroxidase cascade.
Grx1, Grx2, and Tpx as Reductants of Protein Disulfides
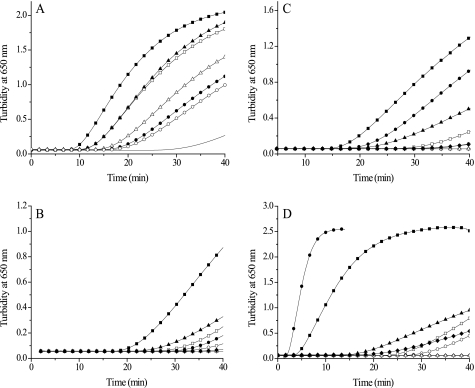
Insulin was used as model substrate to study protein disulfide reduction. Cleavage of the two interchain disulfide bridges of insulin results in formation of the insoluble free B-chain, which is monitored by an increase in turbidity at 650 nm (50). With T(SH)2 as reducing agent, T. brucei Grx1 and Grx2 as well as E. coli Grx1, included as control, catalyzed the reduction of insulin disulfide (Fig. 7A). The highest activity was observed with the E. coli protein, followed by T. brucei Grx2, whereas Grx1 had comparably low activity. Also, in the presence of dithioerythritol, E. coli Grx1 showed the highest activity, followed by T. brucei Grx2, but the Grx1-catalyzed reaction was even slower than that observed with T(SH)2 (Fig. 7, B and C, and supplemental Fig. S4A). On a molar basis, with either T(SH)2 or dithioerythritol as reducing agent, Grx2 was twice as efficient as Grx1 with respect to the time of onset and the rate of precipitation (Fig. 7, B and C, and supplemental Fig. S4A). Next, the reactions of the individual Grx in the presence of dithioerythritol and T(SH)2 as well as the monothiols Gsp and GSH were compared. Grx1 had the highest activity with T(SH)2, followed by Gsp and then dithioerythritol (Fig. 7B). In the case of Grx2, the activity was also highest with T(SH)2, followed by dithioerythritol and Gsp (Fig. 7C). E. coli Grx1 exerted identical activities with both dithioerythritol and T(SH)2 and also showed some activity with Gsp (supplemental Fig. S4B). Under the conditions chosen, none of the proteins catalyzed insulin reduction by GSH.
FIGURE 7.
Comparison of T. brucei Grx1, Grx2, Tpx, and E. coli Grx1 in the insulin reduction assay. The reaction mixtures contained 130 μm insulin, 1.2 mm thiol, and various concentrations of the respective protein. A, T(SH)2-dependent reduction by different Grxs. −, control without dithiol protein; ■, 5 μm and □, 2.5 μm E. coli Grx1; ▴, 22 μm Grx2; ▿, 8 μm Grx2; ●, 43 μm Grx1; ○, 14 μm Grx1. B, Grx1-catalyzed reduction by different thiols. □ and ■, 635 μm T(SH)2; △ and ▴, 1.26 mm Gsp; ○ and ●, 628 μm dithioerythritol; ♢ and ♦, 1.23 mm GSH. Empty and filled symbols display the reaction in the absence and presence of 14 μm Grx1, respectively. C, Grx2-catalyzed reduction by different thiols. □ and ■, 624 μm T(SH)2; △ and ▴, 1.25 mm Gsp; ○ and ●, 628 μm dithioerythritol; ♢ and ♦, 1.3 mm GSH. Empty and filled symbols display the reaction without or with 8 μm Grx2. D, Tpx-catalyzed reduction by different thiols. □ and ■, 635 μm T(SH)2; △ and ▴, 1.26 mm Gsp; ○ and ●, 628 μm dithioerythritol; ♢ and ♦, 1.23 mm GSH. Empty and filled symbols show the reaction in the absence and presence of 3.5 μm Tpx, respectively. The kinetics depicted is representatives of two or three repetitions.
Interestingly, the slow spontaneous reduction of insulin was faster by T(SH)2 than by dithioerythritol (Fig. 7, B–D) which shows that the relative redox potential of the thiol substrates (−242 mV for (TSH)2 (12) and −330 mV for dithioerythritol) cannot be the determining factor for the reaction. In the case of T(SH)2 and Gsp, the thiol pK value probably plays a crucial role. At pH 7.0, T(SH)2 (pK 7.4 (51)) and Gsp are dissociated to about one-third, whereas dithioerythritol (pK values of 9.0 and 9.9) as well as GSH (pK 8.7) are widely present in protonated form. The superior reducing capacity of T(SH)2 is thus due to the fact that it is a dithiol, has a pK value close to neutral pH, and in addition may exert a specific affinity for the proteins. The latter two properties may also explain the unexpected observation that the monothiol Gsp was a more efficient reducing agent of Grx1 than dithioerythritol. T. brucei Tpx catalyzed the reduction of insulin even more efficiently than E. coli Grx1 (Fig. 7D and supplemental Fig. S4B).
DISCUSSION
The two dithiol Grxs of African trypanosomes are constitutively expressed in both the mammalian and insect forms. Neither overexpression nor mRNA depletion of Grx1 affected proliferation of the parasites. A Grx1 null mutant of E. coli as well as the double mutant of Grx1 and Grx2 in yeast cells also did not show any proliferation defect under normal culture conditions (5, 52). In the case of Grx2, RNAi resulted in growth retardation of procyclic T. brucei, which was the first indication that the two Grxs have non-redundant physiological functions. Immunofluorescence microscopy, Western blot analysis of mitoplasts, and fractionated digitonin lysis revealed Grx1 in the cytosol. This is in agreement with the absence of any obvious targeting sequence but contrasts with data from a proteome analysis that identified the protein in the mitochondrial fraction (53). Grx2 also lacks a signal sequence. Upon digitonin lysis of procyclic cells, the elution pattern of Grx2 was very similar to that of cytochrome c. It should be mentioned that the conditions that set free cytochrome c would also release proteins from glycosomes (33). However, Grx2 does not have a peroxisomal targeting signal and, more importantly, was enriched in isolated mitoplasts, which supports a localization in the intermembrane space. Thus, T. brucei contains monothiol Grxs in the mitochondrial matrix (26), but a dithiol Grx has not yet been detected in this compartment. Another Grx that has been found in the mitochondrial intermembrane space is human Grx1, but this protein occurs also in the cytosol (2).
In bloodstream and procyclic T. brucei, the single mitochondrion occupies about 2.3% (54) and 25% (55), respectively, of the total cell volume. If Grx2 is indeed restricted to the intermembrane space, its local concentration would be clearly in the micromolar range. Depletion of the mRNA resulted in a growth phenotype of procyclic cells in accordance with Grx2 playing an essential role in the fully developed mitochondrion of the insect stage. However, the failure to detect a growth phenotype upon RNAi in bloodstream parasites does not necessarily mean that Grx2 is not essential for the rudimentary organelle of these parasites. If also in bloodstream cells Grx2 is restricted to the intermembrane space, its local concentration would be even higher than in procyclic cells, and down-regulation of the mRNA was not efficient enough to affect proliferation under optimal culture conditions. Deletion of the grx2 gene in bloodstream parasites is needed to clarify this point.
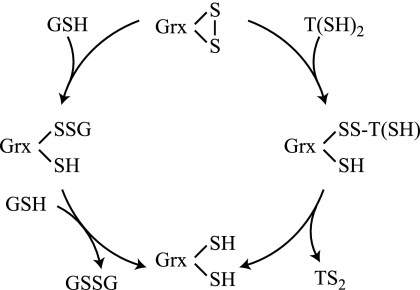
T(SH)2 reduced the disulfide forms of Grx1 and Grx2 with apparent second order rate constants that were 3 orders of magnitude higher compared with GSH. The k1 values for GSH were 10- and 3-fold lower than the observed first order rate constants for E. coli Grx1 obtained by stopped flow kinetics (56). This might be due in part to our less accurate photometric assay, but it could also reflect specific properties of the individual Grxs. Replacement of charged residues (that in other Grxs mediate the interaction with the glycine carboxylate of a bound GSH molecule (Fig. 1) (39, 41, 42)) by hydrophobic ones may weaken GSH binding.
Reduction of Grxs by GSH results in a mixed disulfide that is attacked by a second GSH molecule, leading to the reduced protein and GSSG (Scheme 1, left) (56). For E. coli Grx1, it has been discussed that possibly the Grx1-SSG intermediate is much more reactive toward GSH than the oxidized protein with intramolecular disulfide (56), which would render the latter reaction rate-limiting. The reaction with T(SH)2 should also first generate a mixed disulfide intermediate. In this case, an intramolecular step then generates the reduced protein (Scheme 1, right). With T(SH)2, both reactions, formation and reduction of the mixed disulfide, are expected to be much faster compared with GSH due to its appropriate pK value and because the second step is kinetically favored.
SCHEME 1.
Under normal physiological conditions, the spontaneous reaction with T(SH)2 should be sufficient to recycle GSSG back to GSH. However, this might not be the case under severe oxidative stress conditions when the thiol/disulfide ratios are lowered. As shown here, both parasite Grxs catalyzed the thiol disulfide exchange between T(SH)2 and GSSG. The activity of T. brucei Grx2 was comparable with that reported recently for the corresponding T. cruzi protein despite rather different assay conditions (57). Grx1 exerted a 50-fold higher activity than Grx2 and was also much more effective than Tpx (19). Thus, the cytosolic Grx1 is a strong catalyst of the thiol disulfide exchange between T(SH)2 and GSSG.
To our knowledge, T. brucei Grx1 is the first dithiol Grx with a classical CPYC active site shown to coordinate an iron-sulfur cluster. Other dithiol Grxs reported to bind an iron-sulfur cluster are human Grx2 and poplar GrxC1 (7, 8, 10). Both proteins lack the active site Pro residue but contain a Gly or Ser instead, and it has been hypothesized that this replacement is a prerequisite for iron-sulfur cluster coordination by specific dithiol Grxs (9, 11). In the case of human Grx2, substitution of the Ser by Pro results in a colorless recombinant protein species; however, in vitro, a cluster can be restored (9). T. brucei Grx1 shares 40% of all residues with human Grx2 but has the canonical CPYC active site, suggesting that other structural features play a crucial role for cluster coordination. Comparison of Grx1 with poplar GrxC1 (forming an FeS cluster) and GrxC4 (not forming a cluster) (11, 40) revealed only 31 and 27% identical residues, respectively, and the L. major orthologue showed even an opposite ratio of 28 and 33% overall identity (Fig. 1). These data, therefore, did not unravel the structural basis that allows cluster formation in the parasite Grx1. In human Grx2 and poplar GrxC1, the cluster bridges two monomers and is coordinated by the N-terminal active site Cys of each subunit and two external GSH molecules (8, 10, 11). Most probably, the same type of coordination occurs in T. brucei Grx1. Interestingly, T(SH)2, but not the monothiols Gsp and GSH, yielded a low molecular mass chromophore also in the absence of Grx1. When this preformed ligand was mixed with reduced, but not oxidized, Grx1, the dimeric holoprotein species was obtained. Although the nature of this protein-free iron-containing chromophore is not yet known, the ability of T(SH)2 to act as a strong chelating agent probably plays a major role. As reported recently, T(SH)2 can intercept nitric oxide and labile iron to form a stable free dinitrosyl-trypanothione-iron complex (58). All reconstitution mixtures of Grx1 had diminished activity in the HED assay in accordance with an active site Cys participating in ligand binding. This is further corroborated by the fact that the only additional Cys in T. brucei Grx1 is conserved in human Grx1, which does not bind an iron-sulfur cluster, but does not occur in human Grx2. T. brucei Grx1 is a cytosolic protein and thus cannot be part of the mitochondrial iron-sulfur cluster assembly machinery. Thus, dithiol Grxs, such as T. brucei Grx1, human Grx2c, or poplar GrxC1 may play a yet unknown role as iron-sulfur cluster proteins in the cytosol of different cells.
Protein (de)glutathionylation is a reversible regulatory mechanism in which Grxs play a pivotal role (5, 6). Previously, we demonstrated that several thiol redox proteins of T. brucei are susceptible to specific glutathionylation in vitro (59). As shown here, both parasite Grxs catalyze the reduction of GSH mixed disulfides of BSA, TXNPx, or 2-mercaptoethanol. Toward all three substrates Grx1 exerted a 10-fold higher catalytic efficiency than Grx2 because of lower apparent Km values. In contrast, the turnover numbers were identical, suggesting that the initial interaction between oxidized substrate and enzyme is not rate-determining. Probably, the second step, namely the reaction between GSH and the GSH-Grx mixed disulfide intermediate, is rate-limiting as it has been shown for other Grxs (35, 60). The kcat/Km value of Grx1 with BSA-SSG as substrate was comparable with those reported for human Grx1 and Grx2 (35). This, together with the low micromolar Km values, suggests that protein-GSH mixed disulfides are physiological substrates of Grx1. Although with the substrates tested here, the activity of Grx2 was comparably low, protein deglutathionylation may be an in vivo function also of Grx2, especially when taking into account its distinct subcellular occurrence.
The high rate constants of Grx-catalyzed thiol-disulfide exchange reactions is attributed to the fact that the leaving group in the rate-determining step is Grx-SH (pK values 3.5–4.5), whereas in the non-catalyzed reaction, the leaving group (e.g. BSA-SH) has a pK of 8.5 (61). This explains the minute deglutathionylation activity of thioredoxins with thiol pK values of about 7.0 (6). The same may be the case with Tpx, which has a pK of 7.2 (62) and an extremely low deglutathionylation activity. Protein deglutathionylation seems to be a function specifically exerted by the parasite Grxs, as is the case in mammalian cells (6).
Reduction of insulin disulfide was first shown to be catalyzed by thioredoxins (50), but many Grxs also accelerate the reaction although mostly with lower catalytic efficiencies. T. brucei Grx2 and, to a lesser extent, also Grx1 catalyze insulin disulfide reduction. The localization of Grx2 in the mitochondrial intermembrane space suggests that it is involved in the reduction of highly specific protein disulfides. The Grx2 type protein from T. cruzi has recently been partially characterized but surprisingly was reported not to catalyze the reaction (57). Both T. brucei Grxs exerted higher activities in the presence of T(SH)2 than with dithioerythritol, suggesting that specific interactions between protein and dithiol are of primary importance. In addition, the higher reactivity of T(SH)2 may be explained by its thiol pK of 7.4, which coincides with the physiological pH (51). The observation that Gsp, but not GSH, served as a (albeit weak) reductant of E. coli Grx1 (supplemental Fig. S4B) further supports the notion that the pK value of the thiol plays a major role. In vitro, by far the most efficient parasite protein disulfide reductase proved to be Tpx. It was even more reactive than T. brucei thioredoxin, previously shown to catalyze the reduction of insulin by dithioerythritol or T(SH)2 (63, 64). For T. brucei RR, the T(SH)2/Tpx couple is an efficient reductant (16), and the replacement of Tpx by Grx1 resulted in 50–80% lower RR activities. Because the cellular concentrations of Tpx (50–100 μm in bloodstream and 7–34 μm in procyclic parasites (20)) are significantly higher than those of Grx1, the latter protein does not seem to play a significant role for RR reduction. Grx2 did not support RR activity at all.
Because reduction of insulin disulfide and RR both represent protein disulfide reductions, it was unexpected that Grx1 and Grx2 showed opposite reactivities in the assays. It indicates that the two oxidoreductases have a high specificity toward distinct protein disulfides; the same may be true in the case of glutathionylated proteins as substrates. Future work will aim at identifying physiological substrates of Grx1 and Grx2 and to study their role upon in vivo (de)glutathionylation of proteins.
Supplementary Material
Acknowledgments
Drs. André Schneider (Bern, Switzerland) and Frédéric Bringaud (Bordeaux, France) are acknowledged for kind gifts of purified T. brucei mitoplasts and the antisera against cytochrome c and acetate-succinate-CoA-transferase, respectively. Bruno Manta Porteiro and Dr. Marcelo Comini (Montevideo, Uruguay) are gratefully acknowledged for help in generating the trypanothione-iron complexes. We thank Dr. Michael Boshard (München, Germany) for excellent suggestions regarding construction of the hairpin vectors for RNA interference.
This work was supported by Deutsche Forschungsgemeinschaft SFB 544, “Control of Infectious Tropical Diseases,” Project B3 (to R. L. K.-S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
S. Ceylan and R. L. Krauth-Siegel, unpublished data.
A. Roldan and R. L. Krauth-Siegel, unpublished data.
B. Manta, M. Comini, and R. L. Krauth-Siegel, unpublished data.
- Grx
- glutaredoxin
- GR
- glutathione reductase
- TR
- trypanothione reductase
- T(SH)2
- trypanothione
- RR
- ribonucleotide reductase
- R1
- large subunit of RR
- R2
- small subunit of RR
- tet
- tetracycline
- TEV
- tobacco etch virus
- HED
- hydroxyethyl disulfide
- TXNPx
- 2-Cys-peroxiredoxin type tryparedoxin peroxidase
- Px III
- glutathione peroxidase type tryparedoxin peroxidase
- LipDH
- lipoamide dehydrogenase
- Gsp
- glutathionylspermidine
- Tpx
- tryparedoxin
- 2-ME-SSG
- mixed disulfide between 2-mercaptoethanol and GSH
- BSA-SSG
- glutathionylated BSA
- TXNPx-SSG
- glutathionylated TXNPx.
REFERENCES
- 1.Pedrajas J. R., Porras P., Martínez-Galisteo E., Padilla C. A., Miranda-Vizuete A., Bárcena J. A. (2002) Biochem. J. 364, 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pai H. V., Starke D. W., Lesnefsky E. J., Hoppel C. L., Mieyal J. J. (2007) Antioxid. Redox Signal. 9, 2027–2033 [DOI] [PubMed] [Google Scholar]
- 3.Lönn M. E., Hudemann C., Berndt C., Cherkasov V., Capani F., Holmgren A., Lillig C. H. (2008) Antioxid. Redox Signal. 10, 547–557 [DOI] [PubMed] [Google Scholar]
- 4.Lillig C. H., Berndt C., Holmgren A. (2008) Biochim. Biophys. Acta 1780, 1304–1317 [DOI] [PubMed] [Google Scholar]
- 5.Fernandes A. P., Holmgren A. (2004) Antioxid. Redox Signal. 6, 63–74 [DOI] [PubMed] [Google Scholar]
- 6.Gallogly M. M., Starke D. W., Mieyal J. J. (2009) Antioxid. Redox Signal. 11, 1059–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lillig C. H., Berndt C., Vergnolle O., Lönn M. E., Hudemann C., Bill E., Holmgren A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8168–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Y., Zhong N., Rouhier N., Hase T., Kusunoki M., Jacquot J. P., Jin C., Xia B. (2006) Biochemistry 45, 7998–8008 [DOI] [PubMed] [Google Scholar]
- 9.Berndt C., Hudemann C., Hanschmann E. M., Axelsson R., Holmgren A., Lillig C. H. (2007) Antioxid. Redox Signal. 9, 151–157 [DOI] [PubMed] [Google Scholar]
- 10.Johansson C., Kavanagh K. L., Gileadi O., Oppermann U. (2007) J. Biol. Chem. 282, 3077–3082 [DOI] [PubMed] [Google Scholar]
- 11.Rouhier N., Unno H., Bandyopadhyay S., Masip L., Kim S. K., Hirasawa M., Gualberto J. M., Lattard V., Kusunoki M., Knaff D. B., Georgiou G., Hase T., Johnson M. K., Jacquot J. P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7379–7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fairlamb A. H., Cerami A. (1992) Annu. Rev. Microbiol. 46, 695–729 [DOI] [PubMed] [Google Scholar]
- 13.Krauth-Siegel R. L., Comini M. A. (2008) Biochim. Biophys. Acta 1780, 1236–1248 [DOI] [PubMed] [Google Scholar]
- 14.Comini M., Menge U., Wissing J., Flohé L. (2005) J. Biol. Chem. 280, 6850–6860 [DOI] [PubMed] [Google Scholar]
- 15.Tabor H., Tabor C. W. (1975) J. Biol. Chem. 250, 2648–2654 [PubMed] [Google Scholar]
- 16.Dormeyer M., Reckenfelderbäumer N., Lüdemann H., Krauth-Siegel R. L. (2001) J. Biol. Chem. 276, 10602–10606 [DOI] [PubMed] [Google Scholar]
- 17.Krauth-Siegel L. R., Comini M. A., Schlecker T. (2007) Subcell. Biochem. 44, 231–251 [DOI] [PubMed] [Google Scholar]
- 18.Gommel D. U., Nogoceke E., Morr M., Kiess M., Kalisz H. M., Flohé L. (1997) Eur. J. Biochem. 248, 913–918 [DOI] [PubMed] [Google Scholar]
- 19.Lüdemann H., Dormeyer M., Sticherling C., Stallmann D., Follmann H., Krauth-Siegel R. L. (1998) FEBS Lett. 431, 381–385 [DOI] [PubMed] [Google Scholar]
- 20.Comini M. A., Krauth-Siegel R. L., Flohé L. (2007) Biochem. J. 402, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt A., Clayton C. E., Krauth-Siegel R. L. (2002) Mol. Biochem. Parasitol. 125, 207–210 [DOI] [PubMed] [Google Scholar]
- 22.Piattoni C. V., Blancato V. S., Miglietta H., Iglesias A. A., Guerrero S. A. (2006) Acta Trop. 97, 151–160 [DOI] [PubMed] [Google Scholar]
- 23.Goldshmidt H., Matas D., Kabi A., Carmi S., Hope R., Michaeli S. (2010) PLoS Pathog. 6, e1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D. C., Lennard N. J., Caler E., Hamlin N. E., Haas B., Böhme U., Hannick L., Aslett M. A., Shallom J., Marcello L., Hou L., Wickstead B., Alsmark U. C., Arrowsmith C., Atkin R. J., Barron A. J., Bringaud F., Brooks K., Carrington M., Cherevach I., Chillingworth T. J., Churcher C., Clark L. N., Corton C. H., Cronin A., Davies R. M., Doggett J., Djikeng A., Feldblyum T., Field M. C., Fraser A., Goodhead I., Hance Z., Harper D., Harris B. R., Hauser H., Hostetler J., Ivens A., Jagels K., Johnson D., Johnson J., Jones K., Kerhornou A. X., Koo H., Larke N., Landfear S., Larkin C., Leech V., Line A., Lord A., Macleod A., Mooney P. J., Moule S., Martin D. M., Morgan G. W., Mungall K., Norbertczak H., Ormond D., Pai G., Peacock C. S., Peterson J., Quail M. A., Rabbinowitsch E., Rajandream M. A., Reitter C., Salzberg S. L., Sanders M., Schobel S., Sharp S., Simmonds M., Simpson A. J., Tallon L., Turner C. M., Tait A., Tivey A. R., Van Aken S., Walker D., Wanless D., Wang S., White B., White O., Whitehead S., Woodward J., Wortman J., Adams M. D., Embley T. M., Gull K., Ullu E., Barry J. D., Fairlamb A. H., Opperdoes F., Barrell B. G., Donelson J. E., Hall N., Fraser C. M., Melville S. E., El-Sayed N. M. (2005) Science 309, 416–42216020726 [Google Scholar]
- 25.Filser M., Comini M. A., Molina-Navarro M. M., Dirdjaja N., Herrero E., Krauth-Siegel R. L. (2008) Biol. Chem. 389, 21–32 [DOI] [PubMed] [Google Scholar]
- 26.Comini M. A., Rettig J., Dirdjaja N., Hanschmann E. M., Berndt C., Krauth-Siegel R. L. (2008) J. Biol. Chem. 283, 27785–27798 [DOI] [PubMed] [Google Scholar]
- 27.Comini M. A., Dirdjaja N., Kaschel M., Krauth-Siegel R. L. (2009) Int. J. Parasitol. 39, 1059–1062 [DOI] [PubMed] [Google Scholar]
- 28.Sullivan F. X., Walsh C. T. (1991) Mol. Biochem. Parasitol. 44, 145–147 [DOI] [PubMed] [Google Scholar]
- 29.Melchers J., Krauth-Siegel L., Muhle-Goll C. (2008) Biomol. NMR Assign. 2, 65–68 [DOI] [PubMed] [Google Scholar]
- 30.Budde H., Flohé L., Hecht H. J., Hofmann B., Stehr M., Wissing J., Lünsdorf H. (2003) Biol. Chem. 384, 619–633 [DOI] [PubMed] [Google Scholar]
- 31.Hofer A., Schmidt P. P., Gräslund A., Thelander L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6959–6964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellman G. L. (1959) Arch. Biochem. Biophys. 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 33.Coustou V., Besteiro S., Rivière L., Biran M., Biteau N., Franconi J. M., Boshart M., Baltz T., Bringaud F. (2005) J. Biol. Chem. 280, 16559–16570 [DOI] [PubMed] [Google Scholar]
- 34.Caplan J. F., Filipenko N. R., Fitzpatrick S. L., Waisman D. M. (2004) J. Biol. Chem. 279, 7740–7750 [DOI] [PubMed] [Google Scholar]
- 35.Johansson C., Lillig C. H., Holmgren A. (2004) J. Biol. Chem. 279, 7537–7543 [DOI] [PubMed] [Google Scholar]
- 36.Schlecker T., Comini M. A., Melchers J., Ruppert T., Krauth-Siegel R. L. (2007) Biochem. J. 405, 445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collinson E. J., Grant C. M. (2003) J. Biol. Chem. 278, 22492–22497 [DOI] [PubMed] [Google Scholar]
- 38.Bushweller J. H., Billeter M., Holmgren A., Wüthrich K. (1994) J. Mol. Biol. 235, 1585–1597 [DOI] [PubMed] [Google Scholar]
- 39.Nordstrand K., Aslund F., Holmgren A., Otting G., Berndt K. D. (1999) J. Mol. Biol. 286, 541–552 [DOI] [PubMed] [Google Scholar]
- 40.Noguera V., Walker O., Rouhier N., Jacquot J. P., Krimm I., Lancelin J. M. (2005) J. Mol. Biol. 353, 629–641 [DOI] [PubMed] [Google Scholar]
- 41.Yang Y., Jao S., Nanduri S., Starke D. W., Mieyal J. J., Qin J. (1998) Biochemistry 37, 17145–17156 [DOI] [PubMed] [Google Scholar]
- 42.Discola K. F., de Oliveira M. A., Rosa Cussiol J. R., Monteiro G., Bárcena J. A., Porras P., Padilla C. A., Guimarães B. G., Netto L. E. (2009) J. Mol. Biol. 385, 889–901 [DOI] [PubMed] [Google Scholar]
- 43.Srinivasan U., Mieyal P. A., Mieyal J. J. (1997) Biochemistry 36, 3199–3206 [DOI] [PubMed] [Google Scholar]
- 44.Elgán T. H., Berndt K. D. (2008) J. Biol. Chem. 283, 32839–32847 [DOI] [PubMed] [Google Scholar]
- 45.Herrmann J. M., Hell K. (2005) Trends Biochem. Sci. 30, 205–211 [DOI] [PubMed] [Google Scholar]
- 46.Patton A. R. (ed) (1968) Biochemical Energetics and Kinetics, pp. 56–61, W. B. Saunders Co., Philadelphia [Google Scholar]
- 47.Luthman M., Holmgren A. (1982) J. Biol. Chem. 257, 6686–6690 [PubMed] [Google Scholar]
- 48.Holmgren A. (1979) J. Biol. Chem. 254, 3664–3671 [PubMed] [Google Scholar]
- 49.Porras P., Pedrajas J. R., Martínez-Galisteo E., Padilla C. A., Johansson C., Holmgren A., Bárcena J. A. (2002) Biochem. Biophys. Res. Commun. 295, 1046–1051 [DOI] [PubMed] [Google Scholar]
- 50.Holmgren A. (1979) J. Biol. Chem. 254, 9627–9632 [PubMed] [Google Scholar]
- 51.Moutiez M., Mezaine-Cherif D., Aumercier M., Sergheraert C., Tartar A. (1994) Chem. Pharm. Bull. 42, 2641–2644 [Google Scholar]
- 52.Luikenhuis S., Perrone G., Dawes I. W., Grant C. M. (1998) Mol. Biol. Cell 9, 1081–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panigrahi A. K., Ogata Y., Zíková A., Anupama A., Dalley R. A., Acestor N., Myler P. J., Stuart K. D. (2009) Proteomics 9, 434–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nolan D. P., Voorheis H. P. (1992) Eur. J. Biochem. 209, 207–216 [DOI] [PubMed] [Google Scholar]
- 55.Böhringer S., Hecker H. (1975) J. Protozool. 22, 463–467 [DOI] [PubMed] [Google Scholar]
- 56.Xiao R., Lundström-Ljung J., Holmgren A., Gilbert H. F. (2005) J. Biol. Chem. 280, 21099–21106 [DOI] [PubMed] [Google Scholar]
- 57.Marquez V. E., Arias D. G., Piattoni C. V., Robello C., Iglesias A. A., Guerrero S. A. (2010) Antioxid. Redox Signal. 12, 787–792 [DOI] [PubMed] [Google Scholar]
- 58.Bocedi A., Dawood K. F., Fabrini R., Federici G., Gradoni L., Pedersen J. Z., Ricci G. (2010) FASEB J. 24, 1035–1042 [DOI] [PubMed] [Google Scholar]
- 59.Melchers J., Dirdjaja N., Ruppert T., Krauth-Siegel R. L. (2007) J. Biol. Chem. 282, 8678–8694 [DOI] [PubMed] [Google Scholar]
- 60.Mieyal J. J., Starke D. W., Gravina S. A., Hocevar B. A. (1991) Biochemistry 30, 8883–8891 [DOI] [PubMed] [Google Scholar]
- 61.Gilbert H. F. (1990) Adv. Enzymol. Relat. Areas Mol. Biol. 63, 69–172 [DOI] [PubMed] [Google Scholar]
- 62.Reckenfelderbäumer N., Krauth-Siegel R. L. (2002) J. Biol. Chem. 277, 17548–17555 [DOI] [PubMed] [Google Scholar]
- 63.Reckenfelderbäumer N., Lüdemann H., Schmidt H., Steverding D., Krauth-Siegel R. L. (2000) J. Biol. Chem. 275, 7547–7552 [DOI] [PubMed] [Google Scholar]
- 64.Schmidt H., Krauth-Siegel R. L. (2003) J. Biol. Chem. 278, 46329–46336 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.