Abstract
Autophagy plays an important role in maintaining intracellular homeostasis by promoting the transit of cytoplasmic material, such as proteins, organelles and pathogens, for degradation within acidic organelles. Yet, in immune cells, autophagy pathways serve an additional role in facilitating intracellular surveillance for pathogens and changes in self. Autophagy pathways can modulate key steps in the development of innate and adaptive immunity. In terms of adaptive immunity, autophagy regulates the development and survival of lymphocytes as well as the modulation of antigen processing and presentation. Specialized forms of autophagy may be induced by some viral pathogens, providing a novel route for major histocompatibility complex (MHC) class I antigen presentation and enhanced CD8+ T-cell responses. Autophagy induction in target cells also increases their potential to serve as immunogens for dendritic cell cross-presentation to CD8+ T cells. The requirement for autophagy in MHC class II presentation of cytoplasmic and nuclear antigens is well established, yet recent studies also point to a critical role for autophagy in modulating CD4+ T-cell responses to phagocytosed pathogens. Autophagy pathways can also modulate the selection and survival of some CD4+ T cells in the thymus. However, much still remains to be learned mechanistically with respect to how autophagy and autophagy-linked genes regulate pathogen recognition and antigen presentation, as well as the development and survival of immune cells.
Keywords: antigen presentation, autophagy, histocompatibility antigens, T cells
Introduction
Autophagy has been defined as an ‘auto-digestive’ process that promotes the delivery of intracellular components from the cytoplasm to lysosomal or vacuolar compartments for terminal degradation and recycling. Thus, in many cells, autophagy functions in the removal of aggregated and misfolded proteins, in the turnover of senescent or damaged organelles, in cellular remodelling and in regenerating molecular building blocks during conditions of stress. Autophagy is also now widely recognized as playing an important role in modulating several pathways involved in immune recognition and responsiveness. Cells of the immune system exploit autophagy as a means to detect invading pathogens or changes in the status of self, and to initiate innate and/or adaptive immune responses. This review will focus on the importance of autophagic processes within antigen-presenting cells (APC) and how these pathways facilitate immune surveillance to influence T-cell responses. Autophagy-related genes may also function in processes that are only indirectly linked to degradation, and the potential importance of these findings in terms of host immunity is briefly discussed.
Pathways for autophagy
Bulk and selective transport via autophagy
In eukaryotic cells, multiple constitutive and inducible pathways for autophagy promote cargo delivery from the cytoplasm to vacuolar organelles for degradation. Two of these routes – microautophagy and macroautophagy – were initially described as bulk processes that indiscriminately capture cytoplasmic components, including proteins, ribosomes and organelles, for digestion in acidic vacuolar compartments such as lysosomes.1 Microautophagy has been extensively studied in yeast but few reports have been published addressing the significance of this pathway in mammalian cells. During microautophagy, lysosomal membrane invaginations directly engulf cytoplasmic components, including organelles such as nuclei, peroxisomes and mitochondria, for digestion. Notably, while some pharmacological agents have been used to distinguish microautophagy from macroautophagy, both pathways appear to utilize a conserved family of autophagy-related (Atg) proteins.2,3
Macroautophagy is constitutively active at low levels in many mammalian cells, promoting the degradation of long-lived proteins and organelles.1 During cellular-nutrient or growth-factor deprivation, bulk macroautophagy is rapidly induced to salvage amino acids. While macroautophagy was classically defined as indiscriminate, there may be some selectivity in directing molecules for digestion via this pathway. Degradation of polyubiquitinated protein aggregates by macroautophagy is dependent on their ability to bind a conserved adaptor protein, p62/SQSTM1.3 This may facilitate the clearance of misfolded protein aggregates, which are unable to access the proteasome for conventional cytoplasmic degradation. p62/SQSTM1 also binds light chain-3 (LC3), a homologue of yeast Atg8 that is important in macroautophagy.4 Antigenic epitopes linked to LC3 are digested by macroautophagy and transported to major histocompatibility complex (MHC) class II+ compartments within APC for presentation to CD4+ T cells.5 Specialized forms of macroautophagy have also evolved to promote the selective encapsulation of cytoplasmic microorganisms (xenophagy), the turnover of mitochondria (mitophagy), the clearance of peroxisomes (pexophagy) and nuclear degradation (nucleophagy). The steps imparting specificity to each of these pathways still remain poorly defined. p62/SQSTM1 has been implicated in conferring specificity during xenophagy, as autophagic capture of some cytoplasmic bacteria was linked to this adaptor protein and polyubiquitinated bacterial proteins.6 In xenophagy, activation of autophagy pathways in response to innate signalling may be important in selectively promoting pathogen sequestration or destruction. Interferon-γ (IFN-γ) treatment of macrophages stimulates autophagy and the destruction of phagocytosed pathogens, which is dependent on the expression of Irgm1, an immunity-related guanosine triphosphatase that regulates autophagy.7 Surprisingly, Irmg1 also functions in promoting T-cell survival following stimulation with IFN-γ, probably by preventing autophagy-induced T-cell death.8 Polymorphisms at the immunity-related GTPase family, M loci in humans are linked to susceptibility to Crohn's disease.9 The selective autophagy of mitochondria (mitophagy) is mediated by Ulk1 and multiple conserved Atg proteins, including Atg5 and Atg7.3,10 In mice, deletion of the autophagy genes Atg7 or Atg5 results in severe developmental consequences and impairs the generation and survival of B lymphocytes and mature T lymphocytes.11–13 Analysis of T lymphocytes from Atg7-deficient mice revealed an increased amount of cellular reactive oxygen species, which was attributed to impaired mitophagy.14 It is therefore important to recognize that autophagy pathways and Atg proteins play multiple, and sometimes counterintuitive, roles in immune cells.
The ins and outs of macroautophagy
In immune cells, macroautophagy can be up-regulated by the activation of innate immune receptors, including toll-like receptor (TLR) ligation, nucleotide-binding oligomerization domain protein (NOD)1 and NOD2 engagement, and receptors for cytokines such as IFN-γ.7,8,11,15–18 Yet, mechanistically how these signals promote autophagy is not well defined. By contrast, the initiation of macroautophagy in response to nutrient/growth factor depletion has been studied in detail. Depletion of cellular growth factors disrupts signalling through growth factor receptors and prevents the activation of a class I phosphatidylinositol-3 phosphatase (PI3P) kinase, which blocks the activation of mammalian target of rapamycin (mTOR).19 This failure to activate mTOR induces macroautophagy, a condition that is mimicked by treating cells with rapamycin. Macroautophagy is initiated in the cytoplasm by the formation of a crescent-shaped isolation membrane (IM) (Fig. 1). The class III PI3P kinase, Vps34, binds to Beclin 1, Vps15 and other cofactors to promote formation of the IM. This kinase complex is sensitive to 3-methyladenine and wortmannin, agents used to inhibit macroautophagy in cells. Yet, Vsp34 is not a perfect target for selectively disrupting autophagy because this kinase also associates with endosomes and phagosomes, regulating the fusion and transit of molecules between these organelles.19 Confirming the inhibition of macroautophagy using gene targeting, as well as pharmacological agents, is therefore important.11 The IM enlarges to capture and enclose its cytoplasmic cargo in a double membrane vesicle known as an autophagosome. This requires two protein-conjugation systems: the first consisting of Atg12-Atg5-Atg16L; and the second a complex to promote LC3 lipidation. Atg4, Atg7 and Atg3 promote cytoplasmic LC3-I processing and phosphatidylethanolamine conjugation, giving rise to LC3-II, which associates with the membranes of autophagosomes.
Figure 1.
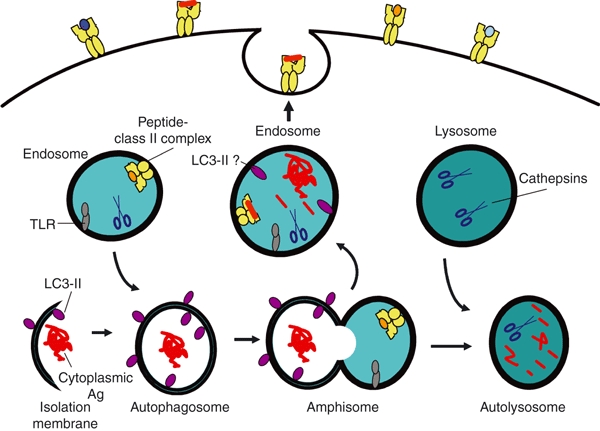
Macroautophagy modulates the degradation of long-lived proteins and organelles. Macroautophagy is initiated in the cytoplasm by the formation of an isolation membrane that engulfs macromolecules and organelles to yield a double-membrane vesicle known as an autophagosome. Treatment of cells with 3-methyadenine or RNA interference (RNAi), to perturb the expression of several different Atg proteins, blocks the formation of these vesicles. Autophagosomes readily fuse with endosomes, giving rise to amphisomes. In many cases the fusion of these vesicles is transient, with an exchange of contents and dissociation of each organelle. This temporary vesicle docking may promote the delivery of cytoplasmic or nuclear antigens (Ags), as well as toll-like receptor (TLR) ligands, into endosomes. In endosomes, some TLR ligands intersect their receptors to initiate pathways for innate signalling. Upon transport into acidic endosomes, cytoplasmic and nuclear Ags are proteolytically processed by cathepsins. The resulting antigenic peptides bind major histocompatibility complex (MHC) class II molecules, followed by the transit of these complexes to the cell surface for presentation to CD4+ T cells. Amphisomes can also fuse with lysosomes to form a terminal digestive compartment, known as an autolysosome. Upon induction of autophagy, cytoplasmic light chain-3 (LC3)-I is modified to yield LC3-II, which preferentially associates with membrane vesicles such as autophagosomes. LC3-II may also be exchanged during vesicle fusion. During the fusion of autophagosomes with lysosomes, proteases are transferred to the resulting autolysosomes to facilitate the digestion of captured macromolecules such as LC3-II.
Autophagosomes can fuse with early endosomes, multivesicular bodies, or late endosomes, giving rise to organelles known as amphisomes that subsequently fuse with lysosomes to form the terminal digestive compartment known as an autolysosome.19 During these fusion processes, proteases and lipases from endosomes and lysosomes are delivered into these autophagic vesicles to promote digestion of the contents, along with the transfer of proteins mediating endosome/lysosome fusion. However, in several cases, molecules captured from the cytoplasm into autophagosomes appear to reach TLR or MHC class II molecules within endosomes and lysosomes to promote innate or adaptive immunity.5,11,15 This would imply the transit of molecules from autophagosomes or amphisomes back into the endocytic network. Indeed, such transit for membrane-associated proteins was detected by live cell imaging.20 Stable autophagosome and lysosome fusion was observed approximately one-third of the time in cells, while, more frequently, these organelles docked, transferred their contents and then separated in a process dubbed ‘kiss and run fusion’. This may also explain reports of the co-localization of LC3 with class II molecules, the MHC encoded peptide editor (DM) and proteins resident in mature endosomes and phagosomes.5,18 The fusion of autophagosomes/amphisomes with phagosomes has yet to be documented, and much remains to be established in terms of defining how autophagy and Atg proteins promote phagosome maturation.16,21–23
To ascertain a requirement for macroautophagy in immunological processes, investigators have frequently disrupted the expression of proteins linked to this pathway, including Beclin1, Atg12, Atg5, Atg7 or Atg16L1.11 However, caution is necessary as molecules such as Atg5 function in protein transport, separately from conventional macroautophagy and macromolecular degradation.11,22 Thus, the use of multiple complementary approaches to demonstrate a role for autophagy in immunology pathways is highly recommended. These include testing the effects of targeted disruption of several autophagy-linked or Atg proteins, biochemical assays to track the autophagy-linked conversion of LC3-I to LC3-II, morphological detection of double-membrane autophagosomes, or pharmacological disruption of macroautophagy.24,25 The detection of LC3 or fluorescent-tagged LC3 proteins bound to intracellular vesicles has been used as a measure of autophagosome formation; however, such results must be interpreted with considerable care as LC3 molecules function in membrane expansion and associate with other cellular structures, including RNA virus-replication complexes, the endoplasmic reticulum (ER), phagosomes, protein aggregates and lipid vacuoles.11,16–21,26–28 A recent report also documented the formation of autophagic vesicles in cells lacking both Atg5 and Atg7.29
Chaperone-mediated autophagy
Chaperone-mediated autophagy (CMA) is a highly selective pathway promoting protein delivery from the cytoplasm to endosomes and lysosomes.30 While constitutively active in many mammalian cells, this pathway is significantly up-regulated following prolonged cell duress as a result of nutrient depletion, upon oxidative or chemically induced stress and in response to protein misfolding. Macroautophagy and CMA are linked via their sequential induction in response to starvation. Macroautophagy is rapidly induced upon serum starvation but decreases after 24 hr, while cellular levels of CMA increase as macroautophagy subsides. Disruption of cellular Atg proteins that block macroautophagy results in increased protein transport and degradation via CMA.30 CMA is dependent upon a molecular chaperone complex composed of several heat shock proteins, including constitutively expressed heat shock 70-kDa protein (hsc70) and lysosome-associated membrane protein 2A (LAMP2A).31 Cytoplasmic proteins and peptides bearing a pentapeptide motif loosely homologous to the sequence KFERQ, associate with hsc70 and are delivered to a membrane translocation complex containing LAMP2A (Fig. 2). It is unclear whether additional molecular signals target proteins for CMA. CMA is ATP-dependent, and cytoplasmic hsc70 probably serves to select and promote the unfolding of substrates for translocation. hsc70 resident in endosomes and lysosomes is also required for translocation, yet whether this chaperone influences the processing of the transported proteins and peptides within these organelles is unknown. hsp90 in the cytoplasm also modulates CMA, although the precise role of this heat shock protein in substrate selection versus translocation is unclear. CMA diminishes with aging, and prolonged life span in rodents was observed with inducible expression of LAMP2A in the liver.32 In terms of innate immunity, mice lacking both LAMP2A and the highly homologous protein, LAMP2B, display reduced neutrophil phagosome maturation, probably as a result of defects in late endosome or lysosome fusion with phagosomes.33 As discussed next, both CMA and macroautophagy play important roles in modulating antigen (Ag) presentation.
Figure 2.
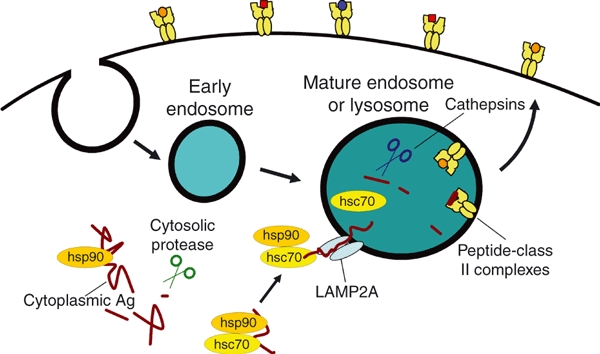
Chaperone-mediated autophagy (CMA) is a regulated pathway that promotes protein delivery from the cytoplasm to endosomal compartments. In the cytoplasm, heat shock proteins, such as hsc70 and hsp90, selectively bind proteins or antigens (Ags) via the recognition of loosely conserved peptide motifs or structures. Complex formation with these heat shock proteins probably promotes Ag unfolding and may direct these Ags for processing by cytoplasmic proteases such as the proteasome and calpain, although whether these chaperones act in concert or sequentially remains untested. Cytoplasmic hsc70 also binds to a transmembrane protein, lysosome-associated membrane protein 2A (LAMP2A), which is found in mature endosomes and lysosomes. Thus, hsc70 may play a critical role in guiding Ags or antigenic peptides to LAMP2A for membrane translocation and entry into endosomes or lysosomes. Within these acidic compartments, the translocated Ags and peptides are further processed by cathepsin proteases before binding major histocompatibility complex (MHC) class II molecules. The resulting peptide–MHC class II complexes are subject to editing by the MHC encoded peptide editor (DM) within late or mature endosomal vesicles before transport to the cell surface for presentation to CD4+ T cells.
Autophagy and Ag presentation
MHC class I-restricted Ag presentation and autophagy
CD8+ T cells recognize antigenic peptides in the context of MHC class I molecules expressed on the surface of nucleated cells. Epitopes from cytoplasmic and nuclear Ags, including viral proteins, endogenous tumour Ags and self-Ags, access class I molecules for presentation via a conventional pathway that is dependent upon the proteasome and a transporter for antigenic peptides (TAP).34,35 In the cytoplasm of cells, Ags are processed by the proteasome, with the resulting peptides translocating into the ER via TAP. In the ER these peptides may be further processed or directly bind newly synthesized MHC class I complexes with the aid of chaperone proteins. Peptide-loaded MHC class I complexes transit to the Golgi and gain entry into secretory vesicles for delivery to the cell surface. Using this pathway, MHC class I molecules access peptides from the cytoplasm and nucleus as a means to detect infection or cell transformation.
In dendritic cells and macrophages, an alternate path, termed cross-presentation, facilitates MHC class I presentation of epitopes from exogenous Ags and phagocytosed material. Cross-presentation is critical in promoting CD8+ T-cell responses to lipid-encapsulated Ags, tumour cells and bacteria.36,37 There are multiple pathways that promote Ag cross-presentation via MHC class I molecules, with the majority of these routes involving Ag capture by phagocytosis. Both TAP-dependent and TAP-independent routes for cross-presentation have been reported, as well as peptide loading by MHC class I molecules in the ER or phagosome before the egress of these resulting MHC class I–peptide complexes to the cell surface.
Evidence that autophagy directly influences the conventional pathway for MHC class I presentation is very limited, yet studies do suggest that the induction of autophagy may play a role in alternate pathways for MHC class I presentation. Inhibition of macroautophagy in APC by disrupting Atg12 or Atg5 expression, or by using pharmacological agents, failed to perturb MHC class I presentation of multiple endogenous and exogenous protein Ags.23,38,39,40 Conjugation of LC3 to a viral epitope similarly did not alter MHC class I presentation of this epitope.5 Yet, inhibition of macroautophagy in IFN-γ-treated B16 murine melanoma cells diminished MHC class I protein surface expression and tumour cell cytolysis by CD8+ T lymphocytes.41 MHC class I presentation by these IFN-γ-treated tumours was proteasome-dependent, suggesting a possible connection between autophagy and the conventional pathway for MHC class I presentation. Herpes simplex virus (HSV) infection of macrophages induces a novel autophagy pathway modulating MHC class I presentation. Initially, viral capsid Ag presentation in infected macrophages proceeds via the conventional MHC class I pathway.42 During the late stages of infection, viral capsid presentation is dependent upon Ag processing in acidic organelles as well as Atg5 expression. These HSV-infected cells contain LC3+ double-membrane vesicles that are closely associated with the nuclear envelope, consistent with virus induction of a novel form of xenophagy. HSV-1 encodes the ICP34.5 protein, which is known to inhibit classical macroautophagy, and in macrophages infected with an ICP34.5-deficient virus the accumulation of autophagosomes containing LC3 and the capsid Ag is detected. These studies suggest a potential role for some forms of autophagy in modulating MHC class I presentation by some tumours or virus-infected cells.
A small number of studies have examined the in vivo requirements for autophagy in MHC class I cross-presentation, and intriguing results were obtained. Selective deletion of Atg5 in dendritic cells did not alter the cross-presentation of phagocytosed Ags by MHC class I, as demonstrated using Ag-coated splenocytes.23 Yet, induction of autophagy in target cells did promote their cross-presentation to Ag-specific CD8+ T cells. CD8+ T-cell responses were enhanced using dendritic cells that had phagocytosed tumour cells treated with rapamycin or starvation to induce autophagy.43 Immunization of mice with purified autophagosomes from these tumour cells similarly promoted the activation of tumour-specific T cells. Using a related approach, in vivo immunization of mice with virus-infected ultraviolet (UV)-irradiated target cells, which were undergoing autophagy before cell death, also proved superior for inducing viral Ag cross-presentation to CD8+ T cells.44 This effect was dependent upon target-cell Atg5 expression, suggesting a requirement for target-cell autophagy induction. Dendritic cell production of type I IFNs in response to these virus-infected target cells was proposed as one possible explanation for this enhancement of CD8+ T-cell responses in vivo. Further mechanistic studies are needed to define how autophagic cells and autophagosomes serve as targets for MHC class I cross-presentation and to identify potential applications of these approaches for vaccine development.
MHC class II-restricted Ag presentation and autophagy
CD4+ T cells recognize antigenic peptides bound to MHC class II molecules expressed on APC and thymic epithelial cells (TEC). MHC class II protein expression can also be induced on non-professional APC and tumour cells following exposure to cytokines, TLR ligands and other activators of innate signalling pathways. Traditionally, MHC class II molecules were viewed as promoting CD4+ T-cell responses to extracellular pathogens and protein Ags, as well as inactivated viral vaccines. Yet, studies of human T-cell responses also suggested a role for MHC class II molecules in promoting immunity to infectious viruses and tumour Ags.45–47 Sequencing of MHC class II ligands from a variety of APC revealed predominantly self-Ags found either on the cell surface or within the endosomal network.48,49 Yet, 10–30% of the ligands identified after release from MHC class II molecules are derived from cytoplasmic or nuclear Ags, suggesting that alternate pathways, such as autophagy, may deliver ligands to MHC class II molecules.48,50,51
MHC class II α and β subunits assemble in the ER with a conserved chaperone, the invariant chain (Ii), and transit to endosomal compartments. Within acidic endosomes, Ii is proteolytically cleaved, releasing peptide-receptive MHC class II αβ complexes. The binding of antigenic peptides to these MHC class II proteins is modulated by the editing molecules DM and peptide editor. MHC class II complexes travel through early endosomes, multivesicular bodies or mature endosomes (termed MIIC), lysosomes and phagosomes to acquire peptides before transit to the cell surface for display to CD4+ T cells.37,52 Ligand binding to MHC class II complexes at the cell surface is also observed, probably as a result of peptide exchange. Ags, including self- and foreign proteins, continuously traffick into the endosomal network for processing by cellular cathepsins and reductases. Remarkably, exposure of APC to cytokines such as IFN-γ and TLR ligands regulates autophagy pathways as well as cellular endocytosis.11,15,52,53–56 The physical intersection of autophagy pathways with endosomes and lysosomes is critical in promoting cytoplasmic and nuclear Ag processing and presentation by MHC class II molecules (Figs 1 and 2).
A wide variety of short- and long-lived cytoplasmic and nuclear Ags, including viral, tumour, self- and ectopically expressed bacterial proteins, are presented by MHC class II molecules on APC and tumours, probably via several distinct autophagy pathways.57,58 Presentation of these Ags is typically sensitive to choloroquine or lysomotrophic amines, as these agents block the acidification of endosomal compartments through which MHC class II molecules transit. Remarkably, MHC class II presentation of some cytoplasmic Ags is dependent upon proteolysis by the proteasome or calpains,59–61 while the processing of other nuclear and cytoplasmic Ags is mediated by cathepsin proteases in acidic compartments.38,39,59–62 In contrast to MHC class I proteins, TAP is not required for MHC class II presentation of cytoplasmic or nuclear Ags.60,63,64 While cytoplasmic and nuclear Ags display some heterogeneity in their processing, these endogenous Ags also appear to access distinct pathways for autophagy. Exposure of APC to 3-methyladenine failed to perturb MHC class II presentation of a number of some cytoplasmic Ags, suggesting a role for pathways other than macroautophagy.60,61 Yet, for several nuclear and cytoplasmic Ags, 3-methyladenine was a potent inhibitor of MHC class II presentation (Fig. 1).5,38,39,61 Disrupting Atg12 or Atg7 expression via RNA interference (RNAi) also confirmed a requirement for macroautophagy in the presentation of two distinct nuclear Ags.5,38,40,65 MHC class II presentation of influenza matrix protein is enhanced by conjugation to LC3 before expression in APC.5 Whether nuclear localization favours Ag processing via macroautophagy, remains controversial. MHC class II presentation of a bacterial Ag via macroautophagy was found to be equally efficient regardless of whether this protein was ectopically expressed in the cytoplasm or in the nucleus of renal tumour cells.65 By contrast, MHC class II presentation of the viral Epstein Barr virus nuclear Ag 1 (EBNA1) by macroautophagy was enhanced upon deletion of its nuclear targeting sequence.40 Notably, macroautophagy is not involved in the presentation of epitopes from two other viral nuclear Ags – EBNA2 and EBNA3C – which rely on cross-presentation and processing via the conventional MHC class II pathway.66 Several cytoplasmic Ags access the selective autophagy pathway of CMA for MHC class II presentation (Fig. 2).63 Peptides from the cytoplasmic Ags glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and aspartate aminotransferase are frequently detected bound to MHC class II molecules, with the transit of both these Ags from the cytoplasm to lysosomes occurring via CMA.30,50,58 We showed previously that MHC class II presentation of cytoplasmic GAD65, a key diabetes autoantigen, is dependent upon CMA by manipulating the expression of LAMP2A and hsc70 in human B lymphoblasts.63 A recent study also demonstrated that both α and β isoforms of hsp90 are required for MHC class II presentation of cytoplasmic GAD Ag.67 Misfolded proteins in the ER are frequently translocated to the cytoplasm for proteolysis, and MHC class II presentation of one of these Ags was also dependent on CMA.63 Together these studies suggest that several autophagy pathways, which intersect the endosomal network, facilitate MHC class II presentation of cytoplasmic and nuclear Ags.
MHC class II presentation, autophagy and innate signalling
Several studies point to a role for autophagy or Atg proteins in promoting MHC class II presentation following innate signalling and phagocytosis (Fig. 3). Much like TLR ligands, muramyldipeptides can activate an innate signalling cascade mediated by NOD2, which induces autophagy and mobilizes the transit of intracellular MHC class II to the surface of dendritic cells.18 NOD2 activation also results in co-localization of the MHC class II editor, DM, with LC3 in intracellular vesicles. Disrupting dendritic cell expression of Atg5, Atg7 or Atg16L1 impairs the transit of MHC class II to the cell surface, DM co-localization with LC3 and MHC class II presentation of a bacterially delivered Ag in response to muramyldipeptides. Notably, genetic studies in humans have linked variants of NOD2 and ATG16L1 with susceptibility to Crohn's disease. Using dendritic cells from individuals expressing these genetic variants of NOD2 or ATG16L, muramyldipeptide treatment failed to induce cellular LC3+ vesicle accumulation, MHC class II transit to the cell surface and MHC class II presentation of bacterial Ags. Together these studies suggest roles for autophagy and Atg proteins in modulating innate immune responses to NOD2 ligands, as well as promoting the presentation of a phagocytosed bacterial Ag.
Figure 3.
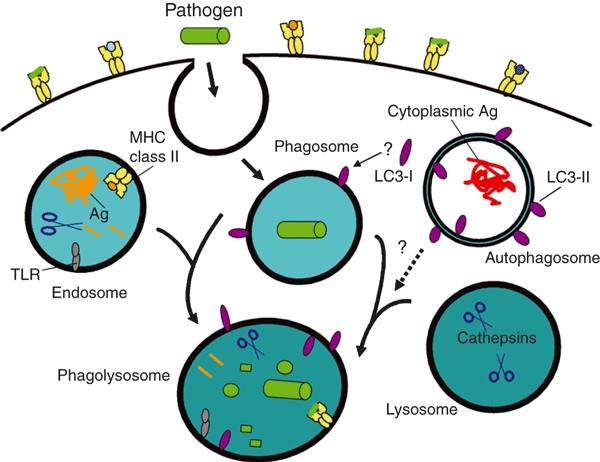
Autophagy, Atg proteins and pathogen phagocytosis. During phagocytosis, extracellular pathogens, such as bacteria, are internalized into phagosomes which fuse with endosomes and lysosomes to form phagolysosomes to promote pathogen degradation. Endosomes enriched for major histocompatibility complex (MHC) class II molecules, and select proteases, fuse with phagosomes, facilitating the association of antigenic peptides from the pathogen with class II molecules for presentation to CD4+ T cells. Disrupting the expression of Atg5, Atg7 or Atg16L1 proteins can perturb this pathway for MHC class II antigen (Ag) presentation. Lysosomes contribute digestive enzymes to the phagosome to promote its maturation into a phagolysosome. Signalling through toll-like receptor (TLR) or other innate receptors during phagocytosis can induce autophagy along with enhanced surface expression of MHC class II protein. Light chain-3 (LC3)-II is detected on phagolysosomes, yet there is currently no firm evidence to suggest that this is mediated by autophagosome fusion with phagosomes or phagolysosomes. Rather, LC3-I in the cytoplasm may be converted to LC3-II and sequestered onto phagocytic vesicles, along with other cytoplasmic cofactors that regulate organelle fusion and membrane remodelling. Alternatively, LC3-II may be delivered to endosomes (Fig. 1), which fuse with phagocytic organelles. Defining mechanistically how Atg proteins and autophagy pathways regulate phagocytosis and Ag cross-presentation remain important priorities.
Studies of HSV also suggest a connection among innate signalling, Atg proteins and Ag cross-presentation. HSV infection triggers innate signalling via TLRs, as well as the induction of adaptive CD4+ and CD8+ T-cell responses. In vivo activation of virus-specific CD4+ T cells was significantly impaired upon HSV infection of mice with Atg5-deficient dendritic cells.23 HSV-infected dendritic cells are unable to directly prime CD4+ T cells, leading this team of investigators to speculate that Atg5 expression in dendritic cells may modulate Ag cross-presentation. Indeed, MHC class II cross-presentation of phagocytosed Ags is significantly reduced in Atg5-deficient dendritic cells, but this is only observed in the presence of lipopolysaccharide (LPS). In the absence of TLR ligands, MHC class II cross-presentation of phagocytosed Ags was independent of Atg5 expression. Dendritic cells lacking Atg5 expression also displayed defects in MHC class II presentation of soluble protein Ags. Surprisingly in this model system, the induction of classical macroautophagy by dendritic cell starvation or treatment with rapamycin reduced MHC class II presentation of phagocytosed Ags. These studies reveal a role for Atg5 in MHC class II Ag presentation which is dependent on TLR activation but probably distinct from classical macroautophagy.
By contrast, the induction of autophagy using rapamycin or starvation enhanced MHC class II presentation of a secreted mycobacterial Ag following the phagocytosis of bacille Calmette–Guérin (BCG) by macrophages and dendritic cells.68 The induction of macroautophagy in these APC results in the enhanced delivery of lysosomal cathepsins and LC3 to mycobacteria-positive phagosomes. A secreted mycobacterial Ag (Ag85B) appears to localize in LC3+ autophagosomes, suggesting perhaps Ag release from phagosomes and recapture in autophagosomes. In vivo the immunization of mice with rapamycin-treated dendritic cells infected with mycobacteria increases the priming of IFN-γ-positive mycobacteria-specific CD4+ T cells. Whether these observations are specific for mycobacteria or are more broadly applicable, remains to be tested. These studies also suggest a role for classical macroautophagy in capturing intracellular Ags released from pathogens for MHC class II presentation. Yet, the recruitment or activation of Atg proteins may also play a critical role here in promoting phagosome maturation and pathogen killing. Much remains to be established in terms of understanding this latter process, as well as the multiple functions of Atg proteins in immune cells.
Autophagy and thymic selection
Developing T lymphocytes in the thymus interact with TEC, including cortical cells (cTEC), which promote positive selection, and medullary cells (mTEC), which function in negative selection to delete autoreactive T cells. In contrast to professional APC, TEC are inefficient at capturing and internalizing Ags by endocytosis.69 This has given rise to suggestions that TEC may rely heavily on intracellular pathways, such as autophagy, to acquire epitopes from endogenous or intracellular Ags for presentation in the context of MHC proteins. Analysis of thymus tissue from mice expressing a green fluorescent protein-conjugated LC3 (GFP–LC3) transgene expressed under the control of the actin promoter, revealed constitutive activation of macroautophagy in TEC, in contrast to tissues such as skeletal muscle.70 Nearly 70% of cTEC in murine thymic tissue sections contain GFP–LC3+ autophagosomes, compared with < 1% of dendritic cells and mTEC.71 A recent study also confirmed the detection of LC3-II in both cTEC and mTEC, along with the co-localization of MHC class II in vesicles with LC3 in immortalized TEC lines treated with IFN-γ.72 To test if macroautophagy in TEC influences T-cell selection, thymi from embryonic Atg5-deficient or wild-type mice were engrafted into adult wild-type mice or T-cell-receptor (TCR) transgenic mice.71 The cellular organization of thymi from Atg5-deficient mice is similar to that in wild-type animals, while TEC from these mice display normal levels of MHC class I expression and slightly reduced levels of MHC class II expression. Remarkably, positive selection of some, but not all, CD4+ TCR transgenic cell lines is impacted by the loss of thymic Atg5. Depending on the TCR transgenic line tested, no change, an increase, or a decrease, in the generation of single positive transgenic T cells is observed in animals lacking thymic Atg5. By contrast, no change in the selection of CD8+ T cells is observed with the deletion of Atg5 in the thymus. These studies are provocative in suggesting that Atg5, and potentially macroautophagy, can regulate the positive selection of T cells. Yet, neonatal lethality is observed in Atg5-deficient animals, as well as defects in T-cell development and survival.11 Further studies are needed to ensure that the loss of Atg5 in TEC selectively perturbs only macroautophagy to influence T-cell selection, especially in light of studies suggesting Atg5 regulation in multiple intracellular processes.11–13,22,23
Concluding remarks
Immune cells, and in particular APC, exploit multiple intracellular pathways, including secretion, endocytosis, phagocytosis and autophagy, to survey the host and to modulate the activation of innate and adaptive responses. Evidence now suggests that in many cases CD4+ T-cell responses to pathogens are dependent on cellular autophagy pathways. Yet, much still remains to be ascertained to determine whether these observations are linked to a requirement for classical degradative autophagy, or reflect novel roles for autophagy-related proteins in innate signalling or in trafficking regulatory proteins to organelles such as phagosomes. Further studies are also required to define the role of autophagy in modulating CD8+ T-cell-mediated immunity, and in particular why phagosome maturation linked to autophagy enhances MHC class II, but not MHC class I, cross-presentation. The potential that autophagy induction in target cells may be useful in the development of vaccines that activate both CD4+ and CD8+ T cells is intriguing, particularly in light of the need for novel protocols for inducing protective immunity to tumours as well as emerging infectious diseases.
Acknowledgments
Grants from the National Institutes of Health and NIAID provide support to J.S.B. and V.L.C. for their work on autophagy and viral pathogens.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunz JB, Schwarz H, Mayer A. Determination of four sequential stages during microautophagy in vitro. J Biol Chem. 2004;279:9987–96. doi: 10.1074/jbc.M307905200. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 5.Schmid D, Pypaert M, Münz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–16. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 7.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–41. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 8.Feng CG, Zheng L, Jankovic D, et al. The immunity-related GTPase Irgm1 promotes the expansion of activated CD4+ T cell populations by preventing interferon-gamma-induced cell death. Nat Immunol. 2008;9:1279–87. doi: 10.1038/ni.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kundu M, Lindsten T, Yang CY, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–70. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller BC, Zhao Z, Stephenson LM, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–14. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 14.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046–55. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 15.Tal MC, Iwasaki A. Autophagy and innate recognition systems. Curr Top Microbiol Immunol. 2009;335:107–21. doi: 10.1007/978-3-642-00302-8_5. [DOI] [PubMed] [Google Scholar]
- 16.Sanjuan MA, Dillon CP, Tait SW, et al. Toll-like receptor signaling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–7. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 17.Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 18.Cooney R, Baker J, Brain O, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 19.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–82. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008;9:574–87. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Z, Fux B, Goodwin M, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–69. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HK, Mattei LM, Steinberg BE, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–39. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korkhov VM. GFP-LC3 labels organized smooth endoplasmic reticulum membranes independently of autophagy. J Cell Biochem. 2009;107:86–95. doi: 10.1002/jcb.22103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–8. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 28.Shibata M, Yoshimura K, Tamura H, et al. LC3, a microtubule-associated protein1A/B light chain3, is involved in cytoplasmic lipid droplet formation. Biochem Biophys Res Commun. 2010;393:274–9. doi: 10.1016/j.bbrc.2010.01.121. [DOI] [PubMed] [Google Scholar]
- 29.Nishida Y, Arakawa S, Fujitani K, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–8. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 30.Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–35. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- 31.Kaushik S, Massey AC, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19:2179–92. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–65. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beertsen W, Willenborg M, Everts V, Zirogianni A, Podschun R, Schröder B, Eskelinen EL, Saftig P. Impaired phagosomal maturation in neutrophils leads to periodontitis in lysosomal-associated membrane protein-2 knockout mice. J Immunol. 2008;180:475–82. doi: 10.4049/jimmunol.180.1.475. [DOI] [PubMed] [Google Scholar]
- 34.Donaldson JG, Williams DB. Intracellular assembly and trafficking of MHC class I molecules. Traffic. 2009;10:1745–52. doi: 10.1111/j.1600-0854.2009.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raghavan M, Del Cid N, Rizvi SM, Peters LR. MHC class I assembly: out and about. Trends Immunol. 2008;29:436–43. doi: 10.1016/j.it.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amigorena S, Savina A. Intracellular mechanisms of antigen cross presentation in dendritic cells. Curr Opin Immunol. 2010;22:109–17. doi: 10.1016/j.coi.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 37.Ramachandra L, Simmons D, Harding CV. MHC molecules and microbial antigen processing in phagosomes. Curr Opin Immunol. 2009;21:98–104. doi: 10.1016/j.coi.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, Mautner J. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol. 2003;33:1250–9. doi: 10.1002/eji.200323730. [DOI] [PubMed] [Google Scholar]
- 39.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Munz C. Endogenous MHC class II processing of a viral nuclear antigen afer autophagy. Science. 2005;307:593–6. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 40.Leung CS, Haigh TA, Mackay LK, Rickinson AB, Taylor GS. Nuclear location of an endogenously expressed antigen, EBNA1, restricts access to macroautophagy and the range of CD4 epitope display. Proc Natl Acad Sci USA. 2010;107:2165–70. doi: 10.1073/pnas.0909448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B, Lei Z, Lichty BD, Li D, Zhang GM, Feng ZH, Wan Y, Huang B. Autophagy facilitates major histocompatibility complex class I expression induced by IFN-gamma in B16 melanoma cells. Cancer Immunol Immunother. 2009;59:313–21. doi: 10.1007/s00262-009-0752-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.English L, Chemali M, Duron J, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10:480–7. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Wang LX, Yang G, Hao F, Urba WJ, Hu HM. Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res. 2008;68:6889–95. doi: 10.1158/0008-5472.CAN-08-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uhl M, Kepp O, Jusforgues-Saklani H, Vicencio JM, Kroemer G, Albert ML. Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell Death Differ. 2009;16:991–1005. doi: 10.1038/cdd.2009.8. [DOI] [PubMed] [Google Scholar]
- 45.Jacobson S, Sekaly RP, Jacobson CL, McFarland HF, Long EO. HLA class II-restricted presentation of cytoplasmic measles virus antigens to cytotoxic T cells. J Virol. 1989;63:1756–62. doi: 10.1128/jvi.63.4.1756-1762.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malnati MS, Ceman S, Weston M, DeMars R, Long EO. Presentation of cytosolic antigen by HLA-DR requires a function encoded in the class II region of the MHC. J Immunol. 1993;151:6751–6. [PubMed] [Google Scholar]
- 47.Wang RF, Wang X, Atwood AC, Topalian SL, Rosenberg SA. Cloning genes encoding MHC class II-restricted antigens: mutated CDC27 as a tumor antigen. Science. 1999;284:1351–4. doi: 10.1126/science.284.5418.1351. [DOI] [PubMed] [Google Scholar]
- 48.Chicz RM, Urban RG, Gorga JC, Vignali DA, Lane WS, Strominger JL. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J Exp Med. 1993;178:27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hunt DF, Michel H, Dickinson TA, et al. Peptides presented to the immune system by the murine class II major histocompatibility complex molecule I-Ad. Science. 1992;256:1817–20. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 50.Dongre AR, Kovats S, deRoos P, et al. In vivo MHC class II presentation of cytosolic proteins revealed by rapid automated tandem mass spectrometry and functional analyses. Eur J Immunol. 2001;31:1485–94. doi: 10.1002/1521-4141(200105)31:5<1485::AID-IMMU1485>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 51.Dengjel J, Schoor O, Fischer R, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci USA. 2005;102:7922–7. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rocha N, Neefjes J. MHC class II molecules on the move for successful antigen presentation. EMBO J. 2008;27:1–5. doi: 10.1038/sj.emboj.7601945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krawczyk M, Leimgruber E, Seguín-Estévez Q, Dunand-Sauthier I, Barras E, Reith W. Expression of RAB4B, a protein governing endocytic recycling, is co-regulated with MHC class II genes. Nucleic Acids Res. 2007;35:595–605. doi: 10.1093/nar/gkl980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsang AW, Oestergaard K, Myers JT, Swanson JA. Altered membrane trafficking in activated bone marrow-derived macrophages. J Leukoc Biol. 2000;68:487–94. [PubMed] [Google Scholar]
- 55.Watts C, West MA, Zaru R. TLR signalling regulated antigen presentation in dendritic cells. Curr Opin Immunol. 2010;22:124–30. doi: 10.1016/j.coi.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 56.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–89. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 57.Zhou D, Blum JS. Presentation of cytosolic antigens via MHC class II molecules. Immunol Res. 2004;30:279–90. doi: 10.1385/IR:30:3:279. [DOI] [PubMed] [Google Scholar]
- 58.Münz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423–49. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- 59.Lich JD, Elliott JF, Blum JS. Cytoplasmic processing is a prerequisite for presentation of an endogenous antigen by major histocompatibility complex class II proteins. J Exp Med. 2000;191:1513–24. doi: 10.1084/jem.191.9.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mukherjee P, Dani A, Bhatia S, et al. Efficient presentation of both cytosolic and endogenous transmembrane protein antigens on MHC class II is dependent on cytoplasmic proteolysis. J Immunol. 2001;167:2632–41. doi: 10.4049/jimmunol.167.5.2632. [DOI] [PubMed] [Google Scholar]
- 61.Dörfel D, Appel S, Grünebach F, Weck MM, Müller MR, Heine A, Brossart P. Processing and presentation of HLA class I and II epitopes by dendritic cells after transfection with in vitro-transcribed MUC1 RNA. Blood. 2005;105:3199–205. doi: 10.1182/blood-2004-09-3556. [DOI] [PubMed] [Google Scholar]
- 62.Jaraquemada D, Marti M, Long EO. An endogenous processing pathway in vaccinia virus-infected cells for presentation of cytoplasmic antigens to class II-restricted T cells. J Exp Med. 1990;172:947–54. doi: 10.1084/jem.172.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou D, Li P, Lin Y, Lott JM, Hislop AD, Canaday DH, Brutkiewicz RR, Blum JS. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 2005;22:571–81. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 64.Dissanayake SK, Tuera N, Ostrand-Rosenberg S. Presentation of endogenously synthesized MHC class II-restricted epitopes by MHC class II cancer vaccines is independent of transporter associated with Ag processing and the proteasome. J Immunol. 2005;174:1811–9. doi: 10.4049/jimmunol.174.4.1811. [DOI] [PubMed] [Google Scholar]
- 65.Riedel A, Nimmerjahn F, Burdach S, Behrends U, Bornkamm GW, Mautner J. Endogenous presentation of a nuclear antigen on MHC class II by autophagy in the absence of CRM1-mediated nuclear export. Eur J Immunol. 2008;2038:2090–5. doi: 10.1002/eji.200737900. [DOI] [PubMed] [Google Scholar]
- 66.Taylor GS, Long HM, Haigh TA, Larsen M, Brooks J, Rickinson AB. A role for intercellular antigen transfer in the recognition of EBV-transformed B cell lines by EBV nuclear antigen-specific CD4+ T cells. J Immunol. 2006;177:3746–56. doi: 10.4049/jimmunol.177.6.3746. [DOI] [PubMed] [Google Scholar]
- 67.Houlihan JL, Metzler JJ, Blum JS. HSP90alpha and HSP90beta isoforms selectively modulate MHC class II antigen presentation in B cells. J Immunol. 2009;182:7451–8. doi: 10.4049/jimmunol.0804296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009;15:267–76. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- 69.Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9:833–44. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 70.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 72.Kasai M, Tanida I, Ueno T, Kominami E, Seki S, Ikeda T, Mizuochi T. Autophagic compartments gain access to the MHC class II compartments in thymic epithelium. J Immunol. 2009;183:7278–85. doi: 10.4049/jimmunol.0804087. [DOI] [PubMed] [Google Scholar]


