Abstract
While it is known that the anti-inflammatory effects of interleukin (IL)-4 require new protein synthesis, the exact mechanisms by which IL-4 suppresses the production of pro-inflammatory cytokines by human monocytes and macrophages is unclear. IL-4 rapidly induced suppressor of cytokine signalling-1 (SOCS1) mRNA and protein, which peaked at 60 min, much earlier than lipopolysaccharide (LPS)-induced SOCS1 mRNA and protein which were consistently maximal 4 hr post-exposure. SOCS1 is a molecule generally considered to be induced for negative feedback of inflammatory processes. We investigated whether the early induction of SOCS1 by IL-4 was responsible for the suppression of LPS-induced tumour necrosis factor (TNF)-α production by IL-4. IL-4 suppressed LPS-induced TNF-α in freshly isolated monocytes at the level of transcription but acted by a different, possibly translational, mechanism in monocytes cultured overnight in macrophage colony-stimulating factor (M-CSF). Despite different modes of regulation by IL-4, the kinetics and magnitude of induction of SOCS1 mRNA and protein by IL-4 in the two cell types were identical. There was no significant difference in the suppression by IL-4 of LPS-induced TNF-α production by bone-marrow derived macrophages from wild-type mice, Ifnγ−/− mice and mice lacking SOCS1 (Socs1−/−Ifnγ−/−). These data suggest that SOCS1 is not involved in the suppression of LPS-induced TNF-α production by IL-4.
Keywords: endotoxin/lipopolysaccharide, inflammation, macrophages/monocytes, signalling/signal transduction
Introduction
Twenty years ago, interleukin (IL)-4 was found to have anti-inflammatory properties, with an ability to suppress the production of tumour necrosis factor (TNF)-α and IL-1β by lipopolysaccharide (LPS)-activated human monocytes.1 Since then, the list of anti-inflammatory properties of IL-4 has grown, with IL-4 also inducing the production of the IL-1 receptor antagonist (IL-1ra)2 and a decoy type II receptor,3 as well as protecting against cartilage and bone erosion.4–6 It is now recognized that IL-4 directs a programme of alternative macrophage activation. In contrast to classically activated (M1) macrophages, which participate in a T helper type 1 (Th1) polarized response and enhance production of pro-inflammatory cytokines, alternatively activated (M2) macrophages counteract inflammation via the release of IL-1ra, IL-10 and transforming growth factor (TGF)-β and promote wound healing and tissue repair.7–9.
IL-4 has been successfully used in the treatment of inflammatory disease in animal models,6,10 and in pilot studies, injections of IL-4 markedly improved the inflammatory skin condition psoriasis.11 Recent studies suggest that therapies that act to increase concentrations of IL-4 in tissues may be useful in the treatment of inflammation.12,13 However, the exact mechanisms by which IL-4 exerts its anti-inflammatory effects are poorly understood. There is a degree of divergence in gene expression profiles of alternatively activated macrophages between species, with many of the murine restricted markers lacking homologues in humans.14 Furthermore, the mechanisms by which IL-4 suppresses pro-inflammatory cytokine production may differ in murine15,16 and human cells.17–19 This highlights the need for further studies in human primary cells. Gaining a better understanding of IL-4 signalling in human cells may allow the design of more effective anti-inflammatory therapies, perhaps by targeting IL-4 signalling intermediates.
Signal transducer and activator of transcription 6 (STAT6) is the primary STAT activated in response to IL-4 stimulation and is critical in the activation or enhanced expression of many IL-4-responsive genes.20 The anti-inflammatory effects of IL-4 may occur directly via STAT6 or via a new protein induced by STAT6. Cytokine-induced signal transduction is regulated via feedback regulatory proteins. One such family of proteins includes the suppressors of cytokine signalling (SOCS) proteins. These are a family of eight proteins [cytokine-inducible Src Homology 2 (SH2)-containing protein (CIS) and SOCS1 to SOCS7]. SOCS1 is an important regulator of STAT6 and IL-4 signalling. There are three functional STAT6 binding motifs of the sequence TTC(N)4GAA located upstream of the transcriptional initiation site in the SOCS1 promoter,21 and overexpression of SOCS1 inhibits STAT6 activation by IL-4 in M1222 and Th2 cells.23 In primary human macrophages, IL-4 induces rapid de novo expression of SOCS1 in a STAT6-dependent manner, and forced expression of SOCS1 inhibits activation of STAT6 by IL-4. In SOCS1-deficient (SOCS1−/−) murine macrophages, STAT6 activation and IL-4-mediated gene expression are much greater and more prolonged than in wild-type macrophages, suggesting that SOCS1 is an important endogenous regulator of IL-4 signalling in macrophages.24
Studies using cycloheximide show that the anti-inflammatory effects of IL-4 in monocytes are dependent on new protein synthesis.17,25 In addition to regulating IL-4 signalling, SOCS1 is also a key regulator of many other cytokines. SOCS1 is a critical inhibitor of the inflammatory cytokine interferon (IFN)-γ,26,27 negatively regulating the phosphorylation and activation of IFN-γ-induced STAT1.28 SOCS1 negatively regulates LPS-induced macrophage activation in mice,29,30 and we have shown that SOCS1 blocks LPS-induced TNF-α production by human monocytes.31 Thus, SOCS1 is a candidate for the mechanism by which IL-4 regulates LPS-induced production of pro-inflammatory cytokines.
To determine whether IL-4-induced SOCS1 is part of the mechanism by which IL-4 suppresses LPS-induced TNF-α production by human monocytes, the kinetics of SOCS1 mRNA induction was examined in primary human monocytes treated with IL-4, LPS and IFN-γ. Responses were compared in freshly isolated human monocytes and monocytes cultured overnight in macrophage colony-stimulating factor (M-CSF). Macrophages from SOCS1-deficient mice were used to demonstrate that SOCS1 is unlikely to be involved in the suppression of LPS-induced TNF-α production by IL-4.
Materials and methods
Isolation of human monocytes
Buffy coats from human blood were kindly provided by the Australian Red Cross Blood Service (Perth, Australia). Mononuclear cells were isolated from buffy coats on density gradients (Lymphopep; Nyegaard, Oslo, Norway). Human monocytes were purified from mononuclear cells to > 80% purity using centrifugal elutriation (JE-6B Rotor; Beckman Coulter, Palo Alto, CA). Purified monocytes were cultured in RPMI-1640 medium (Invitrogen, Mount Waverly, Australia) containing 2 mm glutamine, 50 μm 2-mercaptoethanol, 5 μg/ml gentamycin and 2 mm 3-(N-morpholino)propanesulfonic acid (MOPS), supplemented with 10% fetal calf serum (FCS) (Thermotrace, Melbourne, Australia). Monocytes were either used immediately after isolation (freshly isolated monocytes) or cultured overnight in Teflon Pots (Savillex, Minnetonka, MN) with 25 ng/ml recombinant human M-CSF (PeproTech, Rehovot, Israel). All studies were performed in accordance with National Health and Medical Research Council (Australia) guidelines and approved by the institutional ethics committee.
Monocyte culture and measurement of TNF-α
Human monocytes (5 × 105 in 500 μl) were cultured in flat-bottomed 48-well plates (BD Biosciences, San Diego, CA). TNF-α levels were assayed in culture supernatants harvested 24 hr after stimulation with LPS (500 ng/ml; Sigma, St Louis, MO) with or without recombinant human IL-4 (10 ng/ml; Bender MedSystems, Burlingame, CA) using a human TNF enzyme-linked immunosorbent assay (ELISA) set (BD OPtEIA; BD Biosciences) and DELFIA assay system (Perkin Elmer, Glen Waverley, Australia). IL-4 was added to cultures at the same time as LPS. For each experiment, samples were assayed in triplicate. The assay was sensitive to levels > 10 pg/ml TNF-α.
Real-time polymerase chain reaction (PCR) analysis of TNF-α and SOCS1 mRNA
Human monocytes (106 in 1 ml) were treated with LPS (500 ng/ml), IL-4 (10 ng/ml) or IFN-γ (10 ng/ml) for up to 24 hr. Total RNA was isolated using TRIzol reagent (Invitrogen). Total RNA was reverse-transcribed to cDNA using omniscript II reverse transcriptase (Qiagen, Clifton Hill, Australia) and oligo-dT primers (Promega, Madison, WI) in the presence of RNase inhibitor (Perkin Elmer). Single-stranded cDNA was diluted 1 : 5 and real-time PCR performed using the QuantiTect SYBR green PCR kit (Qiagen) and ABI-PRISM 7900HT (Applied Biosystems, Mulgrave, Australia). SOCS1 mRNA was measured for up to 24 hr after treatment with cytokines. TNF-α was measured for up to 3 hr after treatment with LPS and IL-4 together. To determine the rate of TNF-α mRNA decay, 20 μg/ml actinomycin D (Sigma) was added to cultures to block further mRNA production 60 min after stimulation with LPS with or without IL-4. TNF-α mRNA was measured at 15-min intervals for up 90 min after addition of actinomycin D. PCR conditions involved initial denaturation at 95° for 15 min, followed by 40 cycles of 95° for 15 seconds and 60° for 1 min for TNF-α primers, and 45 cycles of 95° for 15 seconds, 60° for 30 seconds and 72° for 30 seconds for SOCS1 primers. Melting curve analysis was used to assess the specificity of the PCR. Expression levels were determined using a standard curve created from 10-fold serial dilution of a TNF-α plasmid standard or serial dilutions of SOCS1 PCR product, and normalized to the housekeeping gene ubiquitin-conjugating enzyme E2D 2 (UBE2D2).
Western blot analysis
For detection of SOCS1, human monocytes (106 in 1 ml) were treated with LPS (500 ng/ml), IL-4 (10 ng/ml) or IFN-γ (10 ng/ml) for up to 4 hr. IL-4 phosphorylation of STAT6 was examined in the first 2 hr after treatment with LPS plus IL-4. Cells were harvested by centrifugation and monocytes lysed in protein lysis buffer [10 mm Tris, 50 mm NaCl, 5 mm ethylenediaminetetraacetic acid (EDTA) and 1% Triton X-100, pH 7·6] supplemented with 5 mm sodium fluoride, 10 mm sodium molybdate, 1 mm sodium pyrophosphate, 2 mm sodium orthovanadate and 1× protease inhibitors (complete mini, protease inhibitor cocktail tablets; Roche, Penzberg, Germany). Approximately 7·5 μg (STAT6 phosphorylation) or 30 μg (SOCS1) of protein lysate was resolved per lane of a sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to nitrocellulose membrane (Pall Corporation, Pensacola, FL). Membranes were blocked for at least 1 hr in 5% skimmed milk in Tris-buffered saline (TBS)/0·05% Tween-20 (block buffer) followed by overnight incubation at 4° with primary antibodies; phospho-Stat6 (Tyr641) (Cell Signaling Technology, Danvers, MA), anti-SOCS1, clone 4H1 (Millipore, North Ryde, Australia), and anti-α-tubulin clone B-5-1-2 (Sigma) antibodies were either diluted in block buffer or 5% bovine serum albumin (BSA) in TBS/0·05% Tween-20 according to the manufacturer's guidelines. Following four sequential 5-min washes in TBS/0·05% Tween-20, membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit (Rockland Immunochemicals, Gilbertsville, PA) or goat-anti-mouse (Pierce Biotechnology, Rockford, IL) secondary antibodies diluted in block buffer. Bound secondary antibody was detected using chemiluminescence (Roche) and visualized using CL-XPosure film (Pierce).
Nuclear extraction, nuclear factor (NF)-κB and STAT6 electrophoretic mobility shift assays (EMSAs)
Nuclear extracts were prepared from freshly isolated monocytes and monocytes cultured overnight in M-CSF treated with LPS with or without IL-4. Briefly, following stimulation with LPS and IL-4 for 30, 60, 90 and 120 min, 1 ml of ice-cold RPMI was added to each tube and cells were harvested by centrifugation at 335 g for 7 min at 4°. Cells were washed in ice-cold phosphate-buffered saline (PBS) and subsequently lysed in protein lysis buffer (10 mm HEPES.KOH, pH 7·9, 10 mm KCl and 1·5 mm MgCl2) supplemented with 1× protease inhibitor (complete mini, protease inhibitor cocktail tablets; Roche), 0·5 mm dithiothreitol (DTT) and 2 mm sodium orthovanadate prior to use. Lysates were incubated on ice for 20 min before 0·6% NP-40 was added with vortexing to mix. Samples were centrifuged at 14 300 g for 30 seconds at 4°. The cytoplasmic fraction (supernatant) was transferred to a new microfuge tube while nuclear extraction buffer (420 mm NaCl, 20 mm HEPES.KOH, pH 7·9, 1·5 mm MgCl2, 0·2 mm EDTA and 25% glycerol supplemented with 1× protease inhibitor, 0·5 mm DTT and 2 mm sodium orthovanadate) was added to the pellet. Following 20 min of incubation on ice with intermittent agitation, the nuclear fraction was collected by centrifugation (14 300 g for 5 min at 4°) and quantified using the Bradford protein assay method. NF-κB and STAT6 oligos were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Oligos were end-labelled with (γ-32P)-ATP (Perkin-Elmer) and T4 polynucleotide kinase (Promega, Alexandria, Australia). Approximately 1 μg of nuclear extract was incubated with 20 fmol 32P-labelled NF-κB or STAT6 probe and resolved on a non-denaturing 5% polyacrylamide/0·5 × Tris borate-EDTA gel at 180 V for up to 2 hr. The specificity of NF-κB and STAT6 DNA binding was determined by preincubating the nuclear lysates with a 50-fold excess of either unlabelled probe or an unlabelled mutant probe. Gels were dried onto 3 MM Whatman filter paper (Whatman International, Maidstone, UK) and complexes visualized using Super RX X-ray film (Fuji Film Australia, Brookvale, Australia).
Generation and maintenance of Socs1−/−Ifnγ−/− and Ifnγ−/− mice
Mice with a homozygous deletion of both the Socs1 and Ifn-γ genes (Socs1−/−Ifnγ−/−) were generated as described previously27 and were maintained on a C57BL/6 background. C57BL/6 mice with a homozygous deletion of the Ifn-γ gene (Ifnγ−/−) were obtained from The Jackson Laboratory (Bar Harbor, ME). All mice were bred at the animal facilities of The Walter and Eliza Hall Institute of Medical Research (Parkville, Australia). Ethics approval was obtained from the Animal Ethics Committee at the Walter and Eliza Hall Institute of Medical Research.
Isolation, culture of murine bone marrow-derived macrophages (BMMs) and measurement of TNF-α
Murine BMMs were derived from C57BL/6 mice, Ifnγ−/− mice and Socs1−/−Ifnγ−/− mice by culture of whole bone marrow in Dulbecco's modified Eagle's medium (DMEM) supplemented with 100 U/ml penicillin, 0·1 mg/ml streptomycin, 10% FCS and 20% L-cell conditioned medium as a source of M-CSF.32 Independent cultures were generated from three mice of each genotype. Cells (106 in 500 μl) were re-plated in DMEM culture medium supplemented with 10% FCS and 20% L-cell conditioned medium in 24-well plates and incubated overnight before addition of stimuli. TNF-α levels were assayed in culture supernatants harvested 24 hr after stimulation with LPS (10 ng/ml) with or without IL-4 (10 ng/ml) using a murine TNF-α ELISA set (BD OPtEIA; BD Biosciences) and DELFIA assay system (Perkin Elmer). For each experiment, samples were assayed in triplicate. The assay was sensitive to levels > 15 pg/ml TNF-α.
Statistical analysis
Unless otherwise indicated, all values are expressed as mean ± standard error of the mean (SEM) for cells from n donors, using mean values of triplicates for cells from each donor. The significance of results was determined using one-way analysis of variance (ANOVA) and Tukey's post hoc test or a paired Student's t-test. A P value < 0·05 was considered significant.
Results
Suppression of LPS-induced TNF-α production by IL-4
The effect of IL-4 on LPS-induced TNF-α production was compared in monocytes immediately after isolation (freshly isolated monocytes) and monocytes cultured overnight in M-CSF. Monocytes were cultured for 24 hr with 500 ng/ml LPS in the presence or absence of 10 ng/ml IL-4. LPS-induced TNF-α levels in the culture supernatants were significantly greater for the monocytes cultured overnight in M-CSF, compared with the freshly isolated cells (Fig. 1a). For freshly isolated monocytes from 12 donors, IL-4 significantly suppressed LPS-induced TNF-α production by 78 + 2% (mean + SEM). IL-4 also caused significant suppression of LPS-induced TNF-α production by monocytes cultured overnight in M-CSF (n = 9 donors); however, the extent of this suppression (30 + 3%; mean + SEM) was much reduced.
Figure 1.
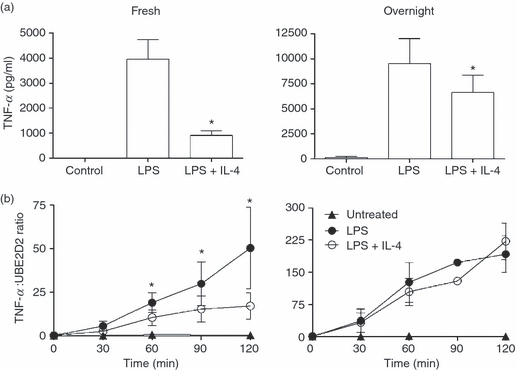
Suppression of lipopolysaccharide (LPS)-induced tumour necrosis factor (TNF)-α production by interleukin (IL)-4 in freshly isolated monocytes (Fresh) and monocytes cultured overnight in macrophage colony-stimulating factor (M-CSF) (Overnight). (a) TNF-α protein in supernatants 24 hr after incubation with LPS (500 ng/ml) with or without IL-4 (10 ng/ml). Values are mean + standard error of the mean (SEM) and have been expressed as a ratio to the mRNA levels of ubiquitin-conjugating enzyme E2D2 (UBE2D2); Fresh, n = 12 donors; Overnight, n = 9 donors. (b) Suppression of LPS-induced TNF-α mRNA production by IL-4. Values are mean ± SEM; Fresh, n = 6 donors; Overnight, n = 3 donors. An asterisk indicates significant suppression by IL-4 (P < 0·05).
To determine whether the effects of IL-4 on LPS-induced TNF-α production were transcriptional, TNF-α mRNA production was quantified using real-time PCR. TNF-α mRNA levels were measured in freshly isolated monocytes (six donors) and monocytes cultured overnight in M-CSF (three donors) after treatment with LPS with or without IL-4 for 2 hr (Fig. 1b). In both cell types, LPS-induced TNF-α mRNA was detectable from 30 min. In monocytes cultured overnight in M-CSF, LPS caused a more rapid accumulation of TNF-α mRNA. By 3 hr, levels of TNF-α mRNA were 60% greater than in freshly isolated monocytes. In freshly isolated monocytes, IL-4 significantly suppressed LPS-induced TNF-α mRNA from 60 min. In contrast, in monocytes cultured overnight in M-CSF, IL-4 had no effect on LPS-induced TNF-α mRNA over the 3 hr time–course (Fig. 1b) and was without effect after 24 hr (results not shown). These results suggest that IL-4 suppresses LPS-induced TNF-α production by different mechanisms in the two cell types.
Effect of IL-4 on TNF-α mRNA stability
IL-4 has previously been shown to suppress LPS-induced TNF-α mRNA production in human monocytes.1,33 However, reports have been conflicting as to whether IL-4 suppression of pro-inflammatory cytokines occurs at the level of transcription18,19,34 or by affecting mRNA stability.17,33 Actinomycin D (20 μg/ml) was added to freshly isolated monocyte cultures 1 hr after stimulation with LPS or LPS plus IL-4 to block further mRNA production. The rate of TNF-α mRNA decay was measured using real-time PCR (Fig. 2). The addition of IL-4 had no effect on the rate of TNF-α mRNA decay in LPS-exposed cells, suggesting that the strong suppression of TNF-α mRNA in freshly isolated monocytes occurs at the level of transcription, not mRNA stability.
Figure 2.
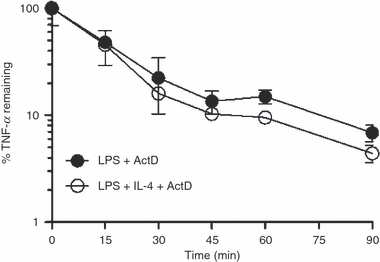
Effect of interleukin (IL)-4 on tumour necrosis factor (TNF)-α mRNA stability in freshly isolated monocytes. Actinomycin D (ActD; 20 μg/ml) was added 1 hr after stimulation with lipopolysaccharide (LPS) with or without IL-4 and the rate of TNF-α decay was measured using real-time polymerase chain reaction (PCR) [mean ± standard deviation (SD); triplicate samples for each time-point from one representative donor].
Effect of IL-4 on STAT6 phosphorylation and STAT6 DNA binding activity
STAT6 is responsible for the activation or enhanced expression of many IL-4-responsive genes, including negative regulatory proteins, such as SOCS1.24 Many of the anti-inflammatory effects of IL-4 are dependent on STAT6.35–37 As the anti-inflammatory effects of IL-4 differed in freshly isolated monocytes and monocytes cultured overnight in M-CSF, we hypothesized that IL-4 activation of STAT6 would be weaker in monocytes cultured overnight in M-CSF. The effect of IL-4 on STAT6 tyrosine phosphorylation and STAT6 DNA binding activity was compared in the two cell types using western blotting on cell lysates and STAT6 EMSAs on nuclear extracts prepared from cells treated with IL-4 over a 2-hr time–course. IL-4 induced phosphorylation of STAT6 and increases in STAT6 DNA binding activity peaked 30 min after treatment. There was no significant difference in STAT6 phosphorylation or nuclear DNA binding activity following treatment with IL-4 between freshly isolated monocytes and monocytes cultured overnight in M-CSF (Fig. 3).
Figure 3.
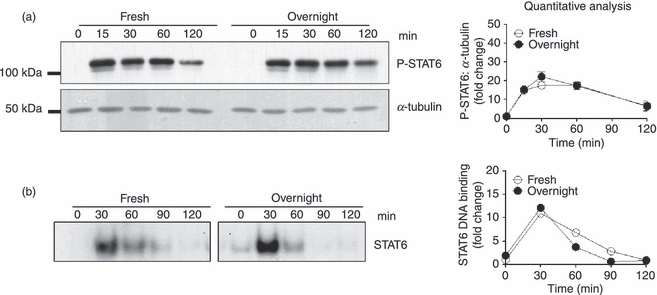
Effect of interleukin (IL)-4 on activation of signal transducer and activator of transcription 6 (STAT6) in freshly isolated monocytes (Fresh) and monocytes cultured overnight in macrophage colony-stimulating factor (M-CSF) (Overnight). (a) Activation of STAT6 by phosphorylation determined in the first 2 hr following IL-4 exposure using western blotting. A representative blot from one donor is shown. A quantitative analysis of phosphorylated STAT6 in protein lysates from three separate donors [mean ± standard error of the mean (SEM)], normalized to the amount of α-tubulin, was performed. Values shown are fold changes relative to phosphorylated STAT6 in untreated samples. (b) STAT6 electrophoretic mobility shift assay (EMSA). The effect of IL-4 on STAT6 DNA binding capacity in nuclear lysates to a radiolabelled STAT6 probe was investigated. A representative EMSA from one donor is shown as well as quantitative analysis of STAT6 DNA binding in nuclear extracts from two separate donors. Values shown are fold changes relative to DNA binding in untreated samples.
Induction of SOCS1 mRNA and protein by IL-4
IL-4 induces SOCS1 in human macrophages, which in turn acts to inhibit IL-4 signalling.24 SOCS1 is also induced by LPS and is a negative regulator of Toll-like receptor (TLR) signalling, suppressing LPS induction of inflammatory cytokines, including TNF-α.29,30 The kinetics of SOCS1 mRNA induction by IL-4 was examined over a 24-hr time–course and compared with the kinetics of SOCS1 induction by LPS or IFN-γ. For cells from four independent donors, IL-4 rapidly induced SOCS1 mRNA production, which peaked after 1 hr. This was much earlier than the induction of SOCS1 mRNA by IFN-γ and LPS, which peaked at 2 and 4 hr, respectively (Fig. 4). Induction of SOCS1 protein by IL-4 closely followed the kinetics of SOCS1 mRNA synthesis. IL-4-induced SOCS1 protein production peaked after 1 hr. This was earlier than the induction of SOCS1 protein by IFN-γ, which had plateaued after 3 hr, and LPS-induced SOCS1 production, which was still increasing 4 hr post-stimulation (Fig. 5).
Figure 4.
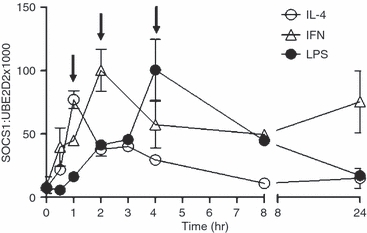
Induction of suppressor of cytokine signalling-1 (SOCS1) mRNA by interleukin (IL)-4, interferon (IFN)-γ and lipopolysaccharide (LPS). Monocytes were cultured overnight in macrophage colony-stimulating factor (M-CSF) and then treated with IL-4 (10 ng/ml), LPS (500 ng/ml) or IFN-γ (10 ng/ml) for up to 24 hr. SOCS1 mRNA levels were quantified using real-time polymerase chain reaction (PCR) and have been expressed as a ratio to the mRNA levels of ubiquitin-conjugating enzyme E2D2 (UBE2D2) (mean ± standard error of the mean; n = 4 donors). Arrows indicate the time-point of maximal induction.
Figure 5.
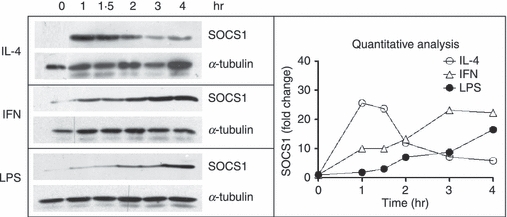
Induction of suppressor of cytokine signalling-1 (SOCS1) protein by interleukin (IL)-4, interferon (IFN)-γ and lipopolysaccharide (LPS). Monocytes were cultured overnight in macrophage colony-stimulating factor (M-CSF) and then treated with IL-4 (10 ng/ml), LPS (500 ng/ml) or IFN-γ (10 ng/ml) for up 4 hr. SOCS1 and α-tubulin protein levels were measured using western blotting. Results of a quantitative analysis are shown upon normalization to the total amount of α-tubulin.
IL-4 had a much weaker effect on LPS-induced TNF-α production in monocytes cultured overnight in M-CSF compared with freshly isolated monocytes. If SOCS1 is part of the mechanism by which IL-4 suppresses LPS-induced TNF-α production, we hypothesized that the induction of SOCS1 mRNA or protein may be stronger, or occur with different kinetics, in freshly isolated monocytes. Induction of SOCS1 mRNA by IL-4, LPS and IFN-γ and induction of SOCS1 protein by IL-4 were compared for 2 hr in freshly isolated monocytes and monocytes cultured overnight in M-CSF. However, there was no significant difference between the two cell types in the kinetics of the induction of SOCS1 mRNA by LPS, IFN-γ and IL-4 and SOCS1 protein by IL-4 (Fig. 6).
Figure 6.
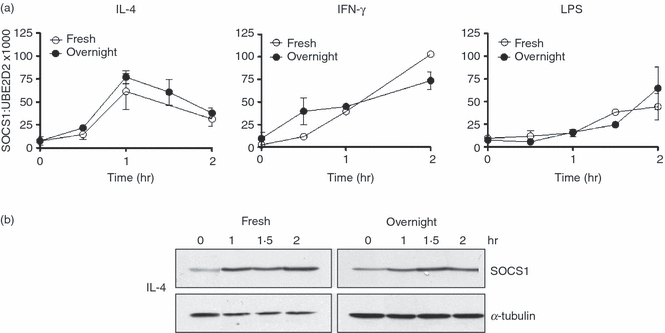
Comparison of the induction of suppressor of cytokine signalling-1 (SOCS1) mRNA and protein in freshly isolated monocytes (Fresh) and monocytes cultured overnight in macrophage colony-stimulating factor (M-CSF) (Overnight). (a) Kinetics of induction of SOCS1 mRNA by interleukin (IL)-4 (10 ng/ml), interferon (IFN)-γ (10 ng/ml) and lipopolysaccharide (LPS) (500 ng/ml) in fresh and overnight cultured monocytes. Values have been expressed as a ratio to the mRNA levels of ubiquitin-conjugating enzyme E2D2 (UBE2D2), (mean ± standard error of the mean; n = 4 donors). (b) Kinetics of induction of SOCS1 protein by IL-4 in fresh and overnight cultured monocytes. Shown is a representative blot from one donor.
Effect of IL-4 on LPS-induced TNF-α production by murine Socs1−/−Ifnγ−/− BMMs
To determine whether IL-4-induced SOCS1 is involved in the suppression of LPS-induced TNF-α production, the effects of IL-4 on BMMs from mice lacking SOCS1 (Socs1−/−Ifnγ−/−) were compared with those on BMMs from Ifnγ−/− and wild-type (C57BL/6) mice (Fig. 7). IL-4 significantly suppressed LPS-induced TNF-α production in wild-type, Socs1−/−Ifnγ−/− and Ifnγ−/− murine BMMs. There was no significant difference in the percentage suppression by IL-4 in Socs1−/−Ifnγ−/− BMMs (34 ± 4%) compared with wild-type (36 ± 6%) and Ifnγ−/− BMMs (31 ± 4%).
Figure 7.
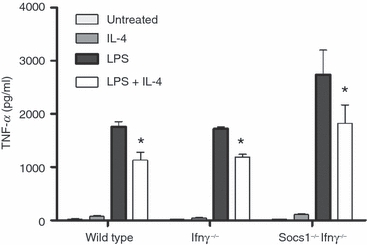
Effect of interleukin (IL)-4 on lipopolysaccharide (LPS)-induced tumour necrosis factor (TNF)-α production by bone marrow-derived macrophages (BMMs) from wild-type, Ifnγ−/− and Socs1−/−Ifnγ−/− mice. BMMs were incubated for 24 hr with IL-4 (10 ng/ml), LPS (100 ng/ml) or LPS plus IL-4 (mean + standard error of the mean; n = 3 individual mice). An asterisk indicates significant suppression relative to LPS-induced levels (P < 0·05).
Effect of IL-4 on LPS activation of NF-κB
LPS signalling through the IκB pathway activates the transcription factor NF-κB, which translocates to the nucleus, activating the transcription of numerous inflammatory cytokines, including TNF-α. We investigated whether the anti-inflammatory effects of IL-4 were mediated by suppressing LPS activation of NF-κB, and whether the effect of IL-4 on NF-κB activation differed between freshly isolated monocytes and monocytes cultured overnight in M-CSF. Monocytes were treated with LPS with or without IL-4 for up to 2 hr and levels of NF-κB were measured in nuclear extracts using an NF-κB EMSA. By 30 min, LPS had strongly activated NF-κB in the nucleus. There was no significant difference in the activation of NF-κB by LPS in freshly isolated monocytes and monocytes cultured overnight in M-CSF. In both freshly isolated monocytes and monocytes cultured overnight in M-CSF, IL-4 had no significant effect on LPS-induced of NF-κB DNA binding activity in the nucleus (Fig. 8).
Figure 8.
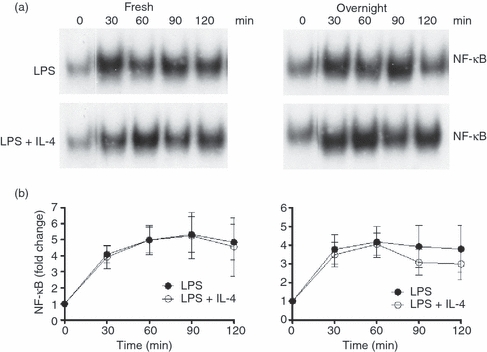
Nuclear factor (NF)-κB electrophoretic mobility shift assay (EMSA). The effect of interleukin (IL)-4 on lipopolysaccharide (LPS) activation of NF-κB was investigated. Nuclear lysates were prepared from freshly isolated monocytes (Fresh) and monocytes cultured overnight in macrophage colony-stimulating factor (M-CSF) (Overnight), cultured for up to 120 min with LPS (500 ng/ml) with or without IL-4 (10 ng/ml). (a) Representative EMSA. (b) Cumulative results from three donors (mean ± standard error of the mean).
Discussion
IL-4 induces SOCS1 mRNA and protein in human monocytes. As the induction of SOCS1 occurred earlier than that induced by LPS and IFN-γ (Figs 4 and 5), SOCS1 was considered potentially responsible for IL-4-mediated suppression of LPS-induced TNF-α production. IL-4 suppressed LPS-induced TNF-α in freshly isolated monocytes at the level of transcription but acted by a different, possibly translational, mechanism in monocytes cultured overnight in M-CSF. However, the kinetics and magnitude of SOCS1 mRNA and protein induction by IL-4, IFN-γ and LPS were very similar in the two cell types. IL-4 was still able to suppress LPS-induced TNF-α production in murine BMMs lacking SOCS1 and, allowing for human/murine differences, this suggests that SOCS1 is not part of the mechanism by which IL-4 suppresses LPS-induced TNF-α production by monocytes.
IL-4 rapidly induced SOCS1 mRNA production, which peaked at 1 hr, much earlier than IFN-γ- and LPS-induced SOCS1 production, which were maximal at 2 and 4 hr post-stimulation, respectively. IL-4-induced SOCS1 protein production peaked at 1 hr, also much earlier than SOCS1 protein production induced by LPS and IFN-γ. The kinetics of induction by IL-4 identified SOCS1 as a potential component of the mechanism by which IL-4 regulated LPS-induced TNF-α production in monocytes. Cross-talk through which SOCS1 is induced by one cytokine and regulates signalling by another has been previously reported. For example, IFNs inhibit IL-4-induced activation of STAT6 and STAT6-dependent gene transcription by inducing expression of SOCS1.38
The anti-inflammatory effects of IL-4 differ between human cells17–19 and murine cells15,16 and change as cells differentiate in vitro.36,39 An advantage of our study is that we examined the effects of IL-4 in human cells in two differentiation states, as well as in murine BMMs. After overnight culture in M-CSF, monocytes have increased expression of integrin receptors.40 They also express higher levels of triggering receptor expressed on myeloid cells-2 (TREM-2) mRNA (results not shown), a marker of newly differentiated macrophages that is up-regulated in macrophages infiltrating tissues from the circulation.41 In the in vivo situation, these cells represent monocytes newly migrated to tissues.
Differences in responses to IL-4 between freshly isolated monocytes and monocytes cultured overnight in M-CSF may be attributable to changes in the IL-4 receptor conformation and signalling. In a previous study, the expression of the γc chain of the IL-4 receptor in monocytes declined after 3 days of culture in M-CSF.36 This decline was associated with reduced IL-4-induced phosphorylation of STAT6 and correlated with an impaired ability of IL-4 to suppress LPS-induced TNF-α production.37 Another possibility is that culture overnight in M-CSF stimulates CD16 expression on monocytes. CD16 expression is induced by, and increases with, in vitro maturation.42 CD14lo CD16hi monocytes have reduced expression of both the IL-4Ra chain and the γc chain of the IL-4 receptor, resulting in diminished sensitivity to IL-4 with reduced activation of STAT6 at low doses of IL-4.43 However, no significant differences in the tyrosine phosphorylation of STAT6, or STAT6 DNA binding activity in the nucleus, were found following IL-4 incubation with monocytes cultured overnight or with freshly isolated monocytes (Fig. 3). Thus, the differences in suppression of TNF-α production by IL-4 may not be caused by differences in signalling through STAT6.
A significant increase in LPS-induced TNF-α production was observed in monocytes cultured overnight in M-CSF, with greater LPS-induced accumulation of TNF-α mRNA. Previous studies have found that increases in TNF-α production during in vitro differentiation of human monocytes are attributable to changes in post-transcriptional processing which increase TNF-α mRNA stability.44 An increase in stability and accumulation of TNF-α mRNA in monocytes cultured overnight in M-CSF may make it more difficult for IL-4 to have its suppressive effects.
Ideally, we would have liked to investigate the effects of IL-4 in human monocytes transfected with a dominant negative form of SOCS1 or a small interfering RNA (siRNA) to SOCS1. However, transfection with adenoviruses encoding potential regulatory molecules enhances type I IFN levels and activation of the IFN-dependent pathway of TLR,31 demonstrating transfection systems to be of little use in human monocytes. Instead we investigated the effects of IL-4 on macrophages from Socs1−/−Ifnγ−/− mice. There was no significant difference in the suppression of LPS-induced TNF-α production by IL-4 between wild-type, Ifnγ−/− and Socs1−/−Ifnγ−/− murine BMMs, suggesting that SOCS1 is not involved in the suppression of LPS-induced TNF-α production by IL-4 in murine macrophages. Despite different modes of regulation by IL-4 in freshly isolated monocytes and monocytes cultured overnight in M-CSF, the kinetics and magnitude of induction by IL-4 of SOCS1 mRNA production in the two cell types were very similar, further suggesting that SOCS1 was not involved.
LPS initiates two distinct signalling pathways downstream of TLR4, the early MyD88-dependent pathway and the later acting MyD88-independent, IFN-dependent pathway. In human monocytes infected with an adenovirus over-expressing SOCS1 (AdV-SOCS1), SOCS1 suppressed LPS-induced TNF-α mRNA and protein production at 24 hr, but had no effect on TNF-α levels in the first 2 hr. SOCS1 did not modulate components of the early Myd88-dependent pathway of LPS signalling. Rather, SOCS1 reduced LPS-induced expression of IFN-β and myxovirus resistance-A (MxA) mRNA and reduced IFN-β production and STAT1 phosphorylation, showing that SOCS1 regulates the later IFN-dependent components of LPS-induced TLR4 signalling.31 In contrast, TNF-α mRNA was reduced by IL-4 by 60 min in freshly isolated monocytes, suggesting regulation of early components of LPS signalling. In monocytes cultured overnight in M-CSF, IL-4 had no effect on TNF-α mRNA over 24 hr. Thus, IL-4, unlike SOCS1, does not act on the IFN-dependent pathways of LPS signalling and is therefore unlikely to be acting via SOCS1.
LPS signalling through the MyD88-dependent pathway activates the transcription factor NF-κB. In both freshly isolated monocytes and monocytes cultured overnight in M-CSF, IL-4 had no effect on LPS activation of the DNA binding capacity of NF-κB (Fig 8). However, it is possible that IL-4 may act downstream of NF-κB binding to inhibit the transcriptional activity of NF-κB once bound to the TNF-α promoter. IL-4 was previously found to regulate NF-κB-dependent transcription without altering activation of NF-κB. For example, IL-4 inhibited NF-κB-dependent transcription in transfected HEK293 cells by sequestering the transcriptional co-activator cAMP response element- binding (CREB)-binding protein.45 Further, IL-4-induced STAT6 competed with NF-κB binding sites in the E-selectin promoter in human vascular endothelial cells.46 IL-4 may also reduce serine 536 phosphorylation of the p65 subunit of NF-κB, as phosphorylation at this site may regulate the transcriptional activity of NF-κB without changing its capacity to bind to promoters.47,48
In a real-time PCR screen for expression of SOCS mRNA, we considered whether IL-4 could be acting via other SOCS proteins. IL-4 also induced CIS mRNA but not SOCS3, SOCS4 and SOCS5 mRNA (results not shown). Another possibility is that IL-4 changes chromatin remodelling of the TNF-α promoter, for example by altering activation of histone deacetylases by LPS which regulate the transcription of inflammatory cytokines.49 Histone acetylation is an important regulatory mechanism for TNF-α production. An 18-hr polarization with IFN-γ, but not IL-4, increases histone acetylation of the TNF-α promoter, enhancing TNF-α production after subsequent exposure to LPS.50 However, it is unclear what effect IL-4 has when added at the same time as LPS. It is also possible that IL-4 reduces transcription of LPS-induced TNF-α mRNA by affecting transcription factors other than NF-κB.
Other IL-4-induced candidate proteins are the peroxisome proliferator-activated receptors (PPARs). PPARs are transcription factors of the nuclear receptor superfamily that modulate inflammatory responses in many cell types, including monocytes.51–53 PPARs are induced by IL-454 and repress transcriptional activity of NF-κB, independently of nuclear translocation and DNA binding.55,56 In macrophages deficient in PPAR-γ, IL-4 failed to suppress the secretion of IL-6.57 In PPAR-δ−/− murine macrophages, IL-4 was unable to suppress LPS-induced secretion of IL-6 and IL-12,58 indicating that a subset of IL-4 dependent anti-inflammatory responses are regulated by PPAR-γ and PPAR-δ. Further studies are needed to clarify the interaction of IL-4 signalling with pathways involved in inflammation in human monocytes and to identify the role of new proteins induced by IL-4.
Acknowledgments
This work was supported by National Health and Medical Research Council (NHMRC) grant #275546 to P.H.H., a State and Territory Affiliate grant and the Adam Gilchrist Trading Challenge Project grant from Arthritis Australia to C.M.P. and NHMRC grant #461232 to S.E.N. E.A.W. was supported by a Murdoch University Research Studentship.
Disclosures
The authors have no conflicts of interest to disclose.
References
- 1.Hart PH, Vitti GF, Burgess DR, Whitty GA, Piccoli DS, Hamilton JA. Potential anti- inflammatory effects of interleukin-4. Suppression of human monocyte TNFα, IL-1 and PGE2 levels. Proc Natl Acad Sci USA. 1989;86:3803–7. doi: 10.1073/pnas.86.10.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fenton MJ, Buras JA, Donnelly RP. IL-4 reciprocally regulates IL-1 and IL-1 receptor antagonist expression in human monocytes. J Immunol. 1992;15:1283–8. [PubMed] [Google Scholar]
- 3.Colotta F, Re F, Muzio M, et al. Interleukin-1 type II receptor: a decoy target for IL-1 that is regulated by IL-4. Science. 1993;23:472–5. doi: 10.1126/science.8332913. [DOI] [PubMed] [Google Scholar]
- 4.Lacraz S, Nicod L, Galve-de Rochemonteix B, Baumberger C, Dayer JM, Welgus HG. Suppression of metalloproteinase biosynthesis in human alveolar macrophages by interleukin-4. J Clin Invest. 1992;90:382–8. doi: 10.1172/JCI115872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cawston TE, Ellis AJ, Bigg H, Curry V, Lean E, Ward D. Interleukin-4 blocks the release of collagen fragments from bovine nasal cartilage treated with cytokines. Biochim Biophys Acta. 1996;1314:226–32. doi: 10.1016/s0167-4889(96)00107-3. [DOI] [PubMed] [Google Scholar]
- 6.Miossec P, Chomarat P, Dechanet J, Moreau JF, Roux JP, Delmas P, Banchereau J. Interleukin-4 inhibits bone resorption through an effect on osteoclasts and proinflammatory cytokines in an ex vivo model of bone resorption in rheumatoid arthritis. Arthritis Rheum. 1994;37:1715–22. doi: 10.1002/art.1780371202. [DOI] [PubMed] [Google Scholar]
- 7.Goerdt S, Politz O, Schledzewski K, et al. Alternative versus classical activation of macrophages. Pathobiology. 1999;67:222–6. doi: 10.1159/000028096. [DOI] [PubMed] [Google Scholar]
- 8.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 9.Mantovani A, Sica A, Locati M. New vistas on macrophage differentiation and activation. Eur J Immunol. 2007;37:14–6. doi: 10.1002/eji.200636910. [DOI] [PubMed] [Google Scholar]
- 10.Lubberts E, Joosten LA, van Den Bersselaar L, et al. Adenoviral vector-mediated overexpression of IL-4 in the knee joint of mice with collagen-induced arthritis prevents cartilage destruction. J Immunol. 1995;163:4546–56. [PubMed] [Google Scholar]
- 11.Ghoreschi K, Thomas P, Breit S, et al. Interleukin-4 therapy of psoriasis induces Th2 responses and improves human autoimmune disease. Nat Med. 2003;9:40–6. doi: 10.1038/nm804. [DOI] [PubMed] [Google Scholar]
- 12.Clarke RM, Lyons A, O’Connell F, Deighan BF, Barry CE, Anyakoha NG, Nicolaou A, Lynch MA. A pivotal role for interleukin-4 in atorvastatin-associated neuroprotection in rat brain. J Biol Chem. 2008;283:1808–17. doi: 10.1074/jbc.M707442200. [DOI] [PubMed] [Google Scholar]
- 13.Aprahamian T, Bonegio R, Rizzo J, Perlman H, Lefer DJ, Rifkin IR, Walsh K. Simvastatin treatment ameliorates autoimmune disease associated with accelerated atherosclerosis in a murine lupus model. J Immunol. 2006;177:3028–34. doi: 10.4049/jimmunol.177.5.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol. 2009;27:451–83. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 15.Clarke CJ, Hales A, Hunt A, Foxwell BM. IL-10-mediated suppression of TNF-alpha production is independent of its ability to inhibit NF kappa B activity. Eur J Immunol. 1998;28:1719–26. doi: 10.1002/(SICI)1521-4141(199805)28:05<1719::AID-IMMU1719>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 16.Mijatovic T, Kruys V, Caput D, Defrance P, Huez G. Interleukin-4 and -13 inhibit tumor necrosis factor-alpha mRNA translational activation in lipopolysaccharide-induced mouse macrophages. J Biol Chem. 1997;272:14394–8. doi: 10.1074/jbc.272.22.14394. [DOI] [PubMed] [Google Scholar]
- 17.Donnelly RP, Fenton MJ, Kaufman JD, Gerrard TL. IL-1 expression in human monocytes is transcriptionally and posttranscriptionally regulated by IL-4. J Immunol. 1991;146:3431–6. [PubMed] [Google Scholar]
- 18.Donnelly RP, Crofford LJ, Freeman SL, Buras J, Remmers E, Wilder RL, Fenton MJ. Tissue-specific regulation of IL-6 production by IL-4. Differential effects of IL-4 on nuclear factor-kappa B activity in monocytes and fibroblasts. J Immunol. 1993;151:5603–12. [PubMed] [Google Scholar]
- 19.Dokter WH, Koopmans SB, Vellenga E. Effects of IL-10 and IL-4 on LPS-induced transcription factors (AP-1, NF-IL6 and NF-kappa B) which are involved in IL-6 regulation. Leukemia. 1996;10:1308–16. [PubMed] [Google Scholar]
- 20.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701–38. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 21.Hebenstreit D, Luft P, Schmiedlechner A, Regl G, Frischauf AM, Aberger F, Duschl A, Horejs-Hoeck J. IL-4 and IL-13 induce SOCS-1 gene expression in A549 cells by three functional STAT6-binding motifs located upstream of the transcription initiation site. J Immunol. 2003;171:5901–7. doi: 10.4049/jimmunol.171.11.5901. [DOI] [PubMed] [Google Scholar]
- 22.Losman JA, Chen XP, Hilton D, Rothman P. Cutting edge: SOCS-1 is a potent inhibitor of IL-4 signal transduction. J Immunol. 1999;162:3770–4. [PMC free article] [PubMed] [Google Scholar]
- 23.Yu CR, Mahdi RM, Ebong S, Vistica BP, Chen J, Guo Y, Gery I, Egwuagu CE. Cell proliferation and STAT6 pathways are negatively regulated in T cells by STAT1 and suppressors of cytokine signaling. J Immunol. 2004;173:737–46. doi: 10.4049/jimmunol.173.2.737. [DOI] [PubMed] [Google Scholar]
- 24.Dickensheets H, Vazquez N, Sheikh F, Gingras S, Murray PJ, Ryan JJ, Donnelly RP. Suppressor of cytokine signaling-1 is an IL-4-inducible gene in macrophages and feedback inhibits IL-4 signaling. Genes Immun. 2007;8:21–7. doi: 10.1038/sj.gene.6364352. [DOI] [PubMed] [Google Scholar]
- 25.Hart PH, Jones CA, Finlay-Jones JJ. Interleukin-4 suppression of monocyte tumour necrosis factor-alpha production. Dependence on protein synthesis but not on cyclic AMP production. Immunology. 1992;76:560–5. [PMC free article] [PubMed] [Google Scholar]
- 26.Starr R, Metcalf D, Elefanty AG, Brysha M, Willson TA, Nicola NA, Hilton DJ, Alexander WS. Liver degeneration and lymphoid deficiencies in mice lacking suppressor of cytokine signaling-1. Proc Natl Acad Sci USA. 1998;95:14395–9. doi: 10.1073/pnas.95.24.14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexander WS, Starr R, Fenner JE, et al. SOCS1 is a critical inhibitor of interferon gamma signaling and prevents the potentially fatal neonatal actions of this cytokine. Cell. 1999;98:597–608. doi: 10.1016/s0092-8674(00)80047-1. [DOI] [PubMed] [Google Scholar]
- 28.Brysha M, Zhang JG, Bertolino P, Corbin JE, Alexander WS, Nicola NA, Hilton DJ, Starr R. Suppressor of cytokine signaling-1 attenuates the duration of interferon gamma signal transduction in vitro and in vivo. J Biol Chem. 2001;276:22086–9. doi: 10.1074/jbc.M102737200. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa R, Naka T, Tsutsui H, et al. SOCS-1 participates in negative regulation of LPS responses. Immunity. 2002;17:677–87. doi: 10.1016/s1074-7613(02)00449-1. [DOI] [PubMed] [Google Scholar]
- 30.Kinjyo I, Hanada T, Inagaki-Ohara K, et al. SOCS-1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity. 2002;17:583–91. doi: 10.1016/s1074-7613(02)00446-6. [DOI] [PubMed] [Google Scholar]
- 31.Prêle CM, Woodward EA, Bisley J, Keith-Magee A, Nicholson SE, Hart PH. SOCS1 regulates the IFN but not NFkappaB pathway in TLR-stimulated human monocytes and macrophages. J Immunol. 2008;181:8018–26. doi: 10.4049/jimmunol.181.11.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wormald S, Zhang JG, Krebs DL, et al. The comparative roles of suppressor of cytokine signaling-1 and -3 in the inhibition and desensitization of cytokine signaling. J Biol Chem. 2006;281:11135–43. doi: 10.1074/jbc.M509595200. [DOI] [PubMed] [Google Scholar]
- 33.Wang P, Wu P, Siegel MI, Egan RW, Billah MM. Interleukin (IL)-10 inhibits nuclear factor kappa B (NF kappa B) activation in human monocytes. IL-10 and IL-4 suppress cytokine synthesis by different mechanisms. J Biol Chem. 1995;270:9558–63. doi: 10.1074/jbc.270.16.9558. [DOI] [PubMed] [Google Scholar]
- 34.Takeshita S, Gage JR, Kishimoto T, Vredevoe DL, Martínez-Maza O. Differential regulation of IL-6 gene transcription and expression by IL-4 and IL-10 in human monocytic cell lines. J Immunol. 1996;156:2591–8. [PubMed] [Google Scholar]
- 35.Ohmori Y, Hamilton TA. STAT6 is required for the anti-inflammatory activity of interleukin-4 in mouse peritoneal macrophages. J Biol Chem. 1998;273:29202–9. doi: 10.1074/jbc.273.44.29202. [DOI] [PubMed] [Google Scholar]
- 36.Bonder CS, Dickensheets HL, Finlay-Jones JJ, Donnelly RP, Hart PH. Involvement of the IL-2 receptor gamma-chain (gammac) in the control by IL-4 of human monocyte and macrophage proinflammatory mediator production. J Immunol. 1998;160:4048–56. [PubMed] [Google Scholar]
- 37.Bonder CS, Hart PH, Davies KV, Burkly LC, Finlay-Jones JJ, Woodcock JM. Characterization of IL-4 receptor components expressed on monocytes and monocyte-derived macrophages: variation associated with differential signaling by IL-4. Growth Factors. 2001;19:207–18. doi: 10.3109/08977190109001087. [DOI] [PubMed] [Google Scholar]
- 38.Dickensheets HL, Venkataraman C, Schindler U, Donnelly RP. Interferons inhibit activation of STAT6 by interleukin 4 in human monocytes by inducing SOCS-1 gene expression. Proc Natl Acad Sci USA. 1999;96:10800–5. doi: 10.1073/pnas.96.19.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hart PH, Jones CA, Finlay-Jones JJ. Monocytes cultured in cytokine-defined environments differ from freshly isolated monocytes in their responses to IL-4 and IL-10. J Leukoc Biol. 1995;57:909–18. doi: 10.1002/jlb.57.6.909. [DOI] [PubMed] [Google Scholar]
- 40.Mayne GC, Borowicz RA, Greeneklee KV, Finlay-Jones JJ, Williams KA, Hart PH. Centrifugation facilitates transduction of green fluorescent protein in human monocytes and macrophages by adenovirus at low multiplicity of infection. J Immunol Methods. 2003;278:45–56. doi: 10.1016/s0022-1759(03)00229-1. [DOI] [PubMed] [Google Scholar]
- 41.Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M, Piccio L, Hernandez M, Colonna M. TREM-2 attenuates macrophage activation. J Immunol. 2006;177:3520–4. doi: 10.4049/jimmunol.177.6.3520. [DOI] [PubMed] [Google Scholar]
- 42.Ziegler-Heitbrock HWL. Heterogeneity of human blood monocytes: the CD14+CD16+ subpopulation. Immunol Today. 1996;17:424–8. doi: 10.1016/0167-5699(96)10029-3. [DOI] [PubMed] [Google Scholar]
- 43.Junttila IS, Mizukami K, Dickensheets H, Meier-Schellersheim M, Yamane H, Donnelly RP, Paul WE. Tuning sensitivity to IL-4 and IL-13: differential expression of IL-4Ralpha, IL-13Ralpha1, and gammac regulates relative cytokine sensitivity. J Exp Med. 2008;205:2595–608. doi: 10.1084/jem.20080452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacKenzie S, Fernàndez-Troy N, Espel E. Post-transcriptional regulation of TNF-alpha during in vitro differentiation of human monocytes/macrophages in primary culture. J Leukoc Biol. 2002;71:1026–32. [PubMed] [Google Scholar]
- 45.Ohmori Y, Hamilton TA. Interleukin-4/STAT6 represses STAT1 and NF-kappa B-dependent transcription through distinct mechanisms. J Biol Chem. 2000;275:38095–103. doi: 10.1074/jbc.M006227200. [DOI] [PubMed] [Google Scholar]
- 46.Bennett BL, Cruz R, Lacson RG, Manning AM. Interleukin-4 suppression of tumor necrosis factor alpha-stimulated E-selectin gene transcription is mediated by STAT6 antagonism of NF-kappaB. J Biol Chem. 1997;272:10212–9. doi: 10.1074/jbc.272.15.10212. [DOI] [PubMed] [Google Scholar]
- 47.Doyle SL, Jefferies CA, O’Neill LA. Bruton's tyrosine kinase is involved in p65-mediated transactivation and phosphorylation of p65 on serine 536 during NFkappaB activation by lipopolysaccharide. J Biol Chem. 2005;280:23496–501. doi: 10.1074/jbc.C500053200. [DOI] [PubMed] [Google Scholar]
- 48.Buss H, Dörrie A, Schmitz ML, Hoffmann E, Resch K, Kracht M. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-κB at serine 536 is mediated by multiple protein kinases including IκB kinase (IKK)-α, IKKβ, IKKε, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J Biol Chem. 2004;279:55633–43. doi: 10.1074/jbc.M409825200. [DOI] [PubMed] [Google Scholar]
- 49.Halili MA, Andrews MR, Sweet MJ, Fairlie DP. Histone deacetylase inhibitors in inflammatory disease. Curr Top Med Chem. 2009;9:309–19. doi: 10.2174/156802609788085250. [DOI] [PubMed] [Google Scholar]
- 50.Garrett S, Dietzmann-Maurer K, Song L, Sullivan KE. Polarization of primary human monocytes by IFN-gamma induces chromatin changes and recruits RNA Pol II to the TNF-alpha promoter. J Immunol. 2008;180:5257–66. doi: 10.4049/jimmunol.180.8.5257. [DOI] [PubMed] [Google Scholar]
- 51.Szanto A, Nagy L. The many faces of PPARgamma: anti-inflammatory by any means? Immunobiology. 2008;213:789–803. doi: 10.1016/j.imbio.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 52.Jiang C, Ting AT, Seed B. PPAR-gamma agonists inhibit production of monocyte inflammatory cytokines. Nature. 1998;391:82–6. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- 53.Bishop-Bailey D, Bystrom J. Emerging roles of peroxisome proliferator-activated receptor-beta/delta in inflammation. Pharmacol Ther. 2009;124:141–50. doi: 10.1016/j.pharmthera.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 54.Ricote M, Welch JS, Glass CK. Regulation of macrophage gene expression by the peroxisome proliferator-activated receptor-gamma. Horm Res. 2000;54:275. doi: 10.1159/000053271. [DOI] [PubMed] [Google Scholar]
- 55.Pascual G, Fong AL, Ogawa S, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–63. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Remels AH, Langen R, Gosker HR, Russell A, Spaapen F, Voncken W, Schrauwen P, Schols AM. PPAR-γ inhibits NF-κB-dependent transcriptional activation in skeletal muscle. Am J Physiol Endocrinol Metab. 2009;297:E174–83. doi: 10.1152/ajpendo.90632.2008. [DOI] [PubMed] [Google Scholar]
- 57.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–20. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Odegaard JI, Ricardo-Gonzalez RR, Red Eagle A, et al. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab. 2008;7:496–507. doi: 10.1016/j.cmet.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]


