Abstract
Most human B-cell non-Hodgkin’s lymphomas arise from germinal centers. Within these sites, the mismatch repair factor MSH6 participates in antibody diversification. Reminiscent of the neoplasms arising in patients with Lynch syndrome III, mice deficient in MSH6 die prematurely of lymphoma. In this study, we characterized the B-cell tumors in MSH6-deficient mice and describe their histological, immunohistochemical, and molecular features, which include moderate microsatellite instability. Based on histological markers and gene expression, the tumor cells seem to be at or beyond the germinal center stage. The simultaneous loss of MSH6 and of activation-induced cytidine deaminase did not appreciably affect the survival of these animals, suggesting that these germinal center-like tumors arose by an activation-induced cytidine deaminase-independent pathway. We conclude that MSH6 protects B cells from neoplastic transformation by preserving genomic stability.
DNA mismatch repair (MMR) is a highly conserved process that provides protection from errors made during normal DNA replication and from environmental genotoxic agents.1,2,3,4 Heterozygous germline mutations in the human MMR genes MSH2, MSH6, MLH1, or PMS2 lead to hereditary nonpolyposis colorectal cancer, also called Lynch syndrome.5,6 When the normal allele is lost, the resulting neoplastic defect in MMR is often accompanied by deleterious mutations of proto-oncogenes and tumor suppressor genes, as well as instability in microsatellites. Recent studies have defined an additional familial cancer susceptibility syndrome tentatively named Lynch syndrome III7 that is caused by inheritance of homozygous or compound heterozygous mutations of MMR genes. This syndrome, also referred to as CoLoN (colon tumors or/and leukemia/lymphoma or/and neurofibromatosis),8 childhood cancer syndrome,9 or constitutional MMR-deficiency syndrome,10 is associated with hematological malignancies in almost half of the known families11 with T-cell lymphomas occurring more commonly than B-cell non-Hodgkin’s lymphomas. Neurological cancers also occur, most often in children but can also appear during the second and third decades of life.
Efforts to understand the normal biology of MMR in mammalian cells, including B lymphocytes in which MMR plays unique roles, have centered on mice with null or mutant alleles of MMR genes.12,13 MSH2 and MSH6 form a heterodimer that recognizes base-base mismatches arising during DNA replication, as well as mismatches caused by alkylated DNA adducts formed by chemotherapeutic agents or oxidative stress.1,2,3,4 Msh2−/− mice succumb rapidly to T-cell lymphomas,14,15,16 whereas Msh6-deficient mice die at a somewhat older age, primarily of B-cell lymphomas.17 That animals lacking both alleles of an MMR factor develop hematological malignancies is reminiscent of patients with Lynch syndrome III, and these animals provide a potential model system for examining this emerging disease.
B-cell lymphomas in Msh6−/− mice have not yet been characterized in detail, but the observation that MSH6 deficiency is associated primarily with B-cell lymphomas suggests that MSH6 might play a special role in protecting B cells from transformation. This finding is of interest because a high proportion of human B-cell non-Hodgkin’s lymphomas are believed to arise from germinal center (GC) B cells,18,19,20 in which MSH6 directly contributes to the genetic instability required for class switch recombination (CSR) and somatic hypermutation (SHM).21,22,23
SHM and CSR are mechanisms of antibody diversification that occur in GC B cells.13,24 Both are initiated by activation-induced cytidine deaminase (AID), which deaminates cytidines in single-stranded DNA at a very high rate to create uridines that are processed in different ways. When a uridine is converted to a thymidine during replication, this is referred to as a phase 1 mutation. On the other hand, these mutations can be processed by base excision repair or recognized as G:U mismatches and processed by MMR.13,24 Paradoxically, the same base excision repair and MMR factors that typically promote genomic stability are recruited to these AID-induced lesions in Ig genes in which they mediate error-prone repair processes that contribute more than half of the mutations that arise in Ig loci. These additional mutations are referred to as phase 2 mutations.13 Unlike its heterodimeric partner MSH2, MSH6 physically interacts directly with mismatched bases25 and bears within its N-terminal domain a binding motif that probably mediates the interaction of the MMR complex with proliferating cell nuclear antigen (PCNA), which itself recruits error-prone polymerases to Ig variable (V) and switch region sequences.26,27 Besides recruiting the subsequent enzymes in MMR, MSH6 can also signal to other cellular programs, such as the apoptosis machinery, although the details of this are not well understood.28 These MSH6-specific structural and functional features raise the possibility that it may serve functions that are independent of MSH2 and that are distinct from its paralog MSH3. In MMR, MSH2 can form a heterodimer with either MSH6, which recognizes single-base mismatches, or with MSH3, which recognizes larger mismatches associated with insertions and deletions. Nevertheless, the stable expression of MSH6 requires heterodimerization with MSH2, so most, if not all, of the functions of MSH6 must depend on the presence of MSH2.
Derangements of SHM and CSR are believed to cause many lymphomas. AID-induced mutations in Ig loci and proto-oncogenes can lead to double-stranded DNA breaks and chromosomal rearrangements that deleteriously place certain genes under the control of Ig regulatory sequences.29 SHM can be mistargeted to cancer-related genes, such as Bcl6 and Myc, contributing to malignant transformation.30 In normal GC B cells, MMR contributes to the error-free repair of AID-induced mutations at many different genomic loci, including some oncogenes, but also carries out error-prone repair of the V and switch regions within the Ig locus as well as to oncogenes such as Bcl6 and a few other genes.31 The role of MMR in CSR-mediated pathogenic translocations or aberrant SHM, however, is not known.
Here we describe our characterization of the stage of development from which B-cell lymphomas arise in Msh6−/− mice and our search for the mechanism of neoplastic transformation. We report that the lymphomas arising in Msh6−/− mice have properties of cells at or near the GC stage of B-cell development. In addition, the tumors are quite heterogeneous in their morphology and expression of GC markers. Genomic analysis of expression profiles, karyotype, and copy number alterations are described. We also tested a hypothesis that MSH6 might perform its protective role by repairing AID-induced mutations in oncogenes or tumor suppressors. A genetic cross revealed instead that the events causing malignant transformation of B cells in these mice are not dependent on AID. We conclude that Msh6 protects GC B cells from transformation through its general role in preserving genomic stability rather than through its specific role in contributing to the genomic instability of Ig genes that is associated with SHM and CSR.
Materials and Methods
Mice and Survival Curve
Msh6−/− mice were described previously17 and housed in a barrier facility at the Albert Einstein College of Medicine. Aicda−/− mice were the kind gift from Dr. T. Honjo. Because studies in C57BL/6 mice and humans have reported no differences between Aicda+/+ and Aicda+/− individuals,32,33,34 Msh6−/−Aicda+/− were included in the cohort of Msh6−/− mice. Although a recent study35 reported that loss of a single copy of AID attenuated plasma cell tumors in BALB/c-Bcl-xl transgenic mice, the dosage effects may reflect stain differences.36 Survival was analyzed with GraphPad Prism software (GraphPad Software Inc., San Diego, CA). All experiments were approved by the Albert Einstein College of Medicine Institutional Animal Committee.
Histopathology and Immunohistochemistry
Formalin-fixed paraffin-embedded sections were stained with H&E or antibodies as follows. For AID immunohistochemistry (IHC), antigen retrieval was performed in 1 mmol/L EDTA (pH 7.5) in a microwave oven. After standard blocking steps, slides were incubated overnight at 4°C with undiluted supernatant from rat anti-AID hybridoma 328.15 (M. Scharff, manuscript in preparation) and visualized by standard methods. Antigen retrieval before B220 (BD Biosciences, San Jose, CA), PNA (Vector Laboratories, Burlingame, CA), CD3 (Dako, Carpintera, CA), Pax5 (BD Biosciences), MAC387 (Dako), Igκ (Dako), and IgM (Southern Biotechnology, Birmingham, AL) staining was performed in a vegetable steamer in Antigen Unmasking Solution (Vector Laboratories), and Pax5 IHC was performed using a Mouse-On-Mouse kit (Vector Laboratories). Bcl6 IHC (Santa Cruz Biotechnology, Santa Cruz, CA) was performed as described.37
Microsatellite Instability, V(D)J Recombination, and Somatic Hypermutation
Microsatellite instability (MSI) assays were performed as described in Kuruguchi et al38 and in some cases were confirmed by direct sequencing. For somatic hypermutation (SHM), a 0.5-kb portion of the Ig JH2-JH4 intron was sequenced as described27 and analyzed using SHMTool.39
Spectral Karyotype Analysis, Array Comparative Genomic Hybridization, Expression Profiling
For spectral karyotype analysis, lymphomas were dissociated, filtered, and resuspended in RPMI 1640 completed with fetal calf serum to 10% and with 4 mmol/L l-glutamine, 1 mmol/L sodium pyruvate, 100 IU/ml penicillin, 100 μg/ml streptomycin, 0.1 mg/ml gentamicin, and 55 μmol/L β-mercaptoethanol and supplemented with 100 ng/ml Colcemid (Gibco, Carlsbad, CA), 50 ng/ml interleukin-4 (R&D Systems, Minneapolis, MN), and 50 μg/ml lipopolysaccharide (Sigma-Aldrich, St. Louis, MO). After 6 hours of Colcemid treatment, lymphoma cells were swelled in 75 mmol/L KCl and gradually transferred to a fixative (3:1 ratio of methanol and glacial acetic acid). Spectral karyotyping was performed as described40 at the Genome Imagine Facility of Albert Einstein College of Medicine. Between five and eight metaphases were analyzed for each tumor, and abnormalities were then defined as described.41
For array comparative genomic hybridization, genomic DNA from lymphomas and sex-matched C57BL/6J kidney was submitted to Roche NimbleGen (Madison, WI) for array comparative genomic hybridization on a mouse whole genome tiling array (385,000 probes). Analysis was performed as per Roche NimbleGen and with Pathway Analysis software (Ingenuity Systems, Redwood City, CA).
For expression profiling, RNA was isolated from frozen tumor samples as described previously,42 and amplified RNA from tumors or normal spleen and from reference RNA (Universal Mouse Reference RNA, Stratagene, La Jolla, CA) were labeled to Cy3 and Cy5, respectively, according to the manufacturer’s protocols (Quick-Amp Labeling Kit, Agilent Technologies, Santa Clara, CA). Hybridizations were performed onto a customized mouse Agilent array (Mmeb 4x44K) covering approximately 36,000 genes produced specifically for the National Institute of Allergy and Infectious Diseases Microarray Research Facility. Data from the scanned chips are stored at the microarray database (mAdb) maintained by the Center for Information Technology, NIH. Image analysis and normalization were done with Feature Extraction Software (Agilent Technologies). Clustering and principal component analysis were performed with Genesis43 and significance analysis of microarrays was applied as described.44
Results
Msh6-Deficient Mice Develop B-Cell Lymphomas
Although previous studies have reported that Msh6−/− mice develop B-cell lymphomas, the stage of B-cell differentiation reflected by those tumors has not been examined.17,45,46 We therefore studied new tumors generated from fully backcrossed Msh6−/− mice. The mice were regularly monitored and necropsied when moribund. Findings at necropsy included marked splenomegaly, frequent enlargement of mesenteric and cervical lymph nodes, and, less frequently, enlarged mediastinal nodes. Some mice exhibited hepatic involvement with multiple small white foci.
By histological criteria,47,48 10 of the 12 cases studied in detail were diagnosed as B-cell lineage neoplasms with another two diagnosed as T-cell lymphoblastic lymphomas. The B-cell neoplasms included plasmacytomas, B-cell lymphoblastic lymphomas, immunoblastic lymphoma, and follicular lymphoma (Table 1). The degree to which neoplastic populations ablated normal histological architecture was variable both between mice and across different anatomical sites within a given mouse. In some instances, an entire lymph node was replaced by sheets of neoplastic cells, whereas in others the tumor cells comprised a minor population. In addition, some tumors were associated with large populations of histiocytes.
Table 1.
Histological, Immunohistochemical, and Clonal Analysis of Msh6−/− Lymphomas
| Case | Classification* | Clonality† | B220‡ | CD3 | BCL6 | PAX5 | PNA | AID | IgM | Igκ |
|---|---|---|---|---|---|---|---|---|---|---|
| 1125 | DLBCL | Oligo | + | + | + | + | + | + | − | + |
| 1301 | HS + APCT | Mono | + | − | + | + | + | − | + | + |
| 1281 | FBL | ND | + | − | + | + | + | + | − | − |
| 850 | LL | Oligo | + | +/− | − | + | − | − | − | − |
| 480 | HS + LL | Mono | + | +/− | − | + | − | − | + | − |
| 624 | HS + PB PCT | Oligo | + | − | + | + | + | − | +/− | + |
| 599 | LL | Oligo | + | + | − | + | − | − | − | − |
| 720 | LL | Mono | + | + | − | + | − | − | + | − |
| 981 | LL | Oligo | + | + | + | − | +/− | +/− | − | − |
| 164 | IB-APCT | Mono | + | + | + | + | +/− | + | +/− | − |
Classification based on H&E analysis: DLBCL, diffuse large B cell lymphoma; HS, histiocytic sarcoma; FBL, follicular B cell lymphoma; LL, lymphoblastic lymphoma; PB PCT, plasmablastic plasmacytoma; APCT, anaplastic plasmacytoma; IB-APCT, mixed immunoblastic lymphoma and APCT. Mice 599, 720, 981, and 164 were generated from an Msh6+/− by Aicda+/− cross and were heterozygotic at the Aicda locus.
PCR products from tumors were compared with those from wild-type spleen, in which all possible rearranged alleles were detected. Oligo, oligoclonal; mono, monoclonal; ND, not determined.
When both B220 (B cells) and CD3 (T cells) staining was observed, the dominant stain among neoplastic cells was used to assign lineage; two cases scored as T-cell lymphomas are not listed.
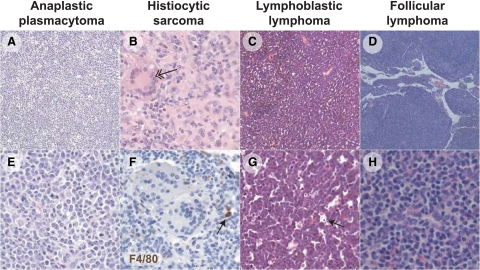
The most common findings were mixed populations of neoplastic cells with features of immunoblasts of GC or early post-GC origin, as well as anaplastic and plasmablastic plasma cells, but only rare mature plasma cells. Depending on the dominance of a particular cell subset, these presentations were consistent with diagnoses of immunoblastic lymphoma and anaplastic and plasmablastic plasmacytoma (APCT)49 (Figure 1, A and E). Histiocytic infiltrates, including some suggestive of true histiocytic sarcoma including multinucleate giant cells (Figure 1B, arrow) were also common. In cases classified as lymphoblastic lymphoma (LL), spleen and peripheral lymph nodes and sometime submandibular and perirenal lymph nodes were densely populated by lymphoblasts associated with frequent mitotic figures and a typical starry sky appearance (Figure 1, C and G) because of the presence of tingible body macrophages containing apoptotic bodies (Figure 1G, arrow). The histiocytic-like neoplastic cells did not stain with the F4/80 antibody (Figure 1F). Finally, one case was diagnosed as follicular B-cell lymphoma with a dense mixture of centrocytes and centroblasts in greatly enlarged splenic follicles (Figure 1, D and H).
Figure 1.
Loss of Msh6 leads largely to B-cell lymphomas of diverse morphology. All panels were stained with H&E, except for F, which was stained by F4/80 (brown) and hematoxylin (blue). A: Tumor 1301 is an example of an anaplastic plasmacytoma that effaced the splenic follicular architecture with a uniform population of round cells (E) with ample pale basophilic cytoplasm with round nuclei (plasmacytoid) admixed with histiocytes. B: Tumor 624 is an example of a histiocytic sarcoma in which nuclei are large and oval, and large multinucleated forms are apparent (double arrow). F: The histiocytic-like neoplastic cells do not stain with the F4/80 antibody; a reactive macrophage in the micrograph (arrow) demonstrates effective immunohistochemistry with this antibody. C: Tumor 981 is an example of lymphoblastic lymphoma, with splenic architecture effaced by small dark round cells with uniform round stippled nuclei with one to two generally centrally placed nucleoli and scant cytoplasm (G). Throughout the lesions there were many tingible body macrophages carrying apoptotic bodies (arrow), endowing these tumors with a “starry sky” appearance. D: Tumor 1281 is an example of follicular lymphoma, in which neoplastic B cells have formed nodules composed of generally small cells with round-to-ovoid stippled nuclei, with numerous cells which are histiocytic in appearance (H). Original magnification: ×5 (D); ×10 (A and C); ×10 (B, E–H).
Although the B-cell lymphomas identified in the original report of MSH6-deficient mice were from stock on a mixed genetic background,17 a reexamination of these cases identified four APCTs, three LLs, one compound APCT and histiocytic sarcoma, and three diffuse large B-cell lymphomas (DLBCLs) with a variable background of histiocytes (not shown). This finding is consistent with previous reports17,45,46 in which most tumors of Msh6−/− mice were histologically of B-cell origin but were frequently associated with large accumulations of normal-appearing or possibly transformed histiocytes.
Many Msh6−/− Lymphomas Share Features of GC and Post-GC B Cells
To further investigate the stage of differentiation of MSH6-deficient B-lineage lymphomas, we analyzed the tumors by immunohistochemistry. As suggested by the histological analysis, 10 tumors classified as B-cell lineage lymphomas were confirmed to be of B-cell origin on the basis of B220 staining, and 9 of the 10 were PAX5+ (Table 1). Many cases had prominent populations of CD3+ T cells that were rarely dominant over the B220+ cells and probably represented infiltrating reactive T cells. Variable staining for IgM was observed in four cases and staining for κ light chains was observed in three of the cases diagnosed with populations of plasmablastic and anaplastic plasma cells, as expected from previous studies.49 Three cases (1125, 1281, and 164) were strongly positive for both BCL6 and PAX5, which are highly expressed in normal GC B cells, and they were also positive for expression of two other GC B-cell markers, PNA and AID. One tumor (1301) was positive for BCL6, PAX5, and PNA, and another (981) was strongly positive for BCL6 and weakly positive for PNA and AID. The expression of multiple GC markers suggests that at least six (1125, 1301, 1281, 624, 981, and 164) (Table 1) of the ten tumors were GC-experienced B cells, even though they varied somewhat in their histological appearance.
Microarray expression profiling was performed on three LLs (850, 981, and 480) and one mixed immunoblastic tumor with anaplastic plasmacytoma (IB-APCT) (164). The expression profiles for the LLs were very similar to each other and distinct from those of normal spleens, and the profiles for the LLs differed from that for the IB-APCT by hierarchical clustering (Supplemental Figure S1A, heat map, see http://ajp.amjpathol.org) and principal component analysis (Supplemental Figure S1B, see http://ajp.amjpathol.org) in keeping with their distinct histopathological presentations. To determine whether the pattern of gene expression for the LLs could be associated with a particular normal cell of origin, we compared the pattern for LLs with profiles established by microarray analyses of sorted purified splenic GC B cells, class-switched memory B cells, and plasma cells.50 As detailed in Supplemental Table 1 (see http://ajp.amjpathol.org), expression of a number of genes was significantly increased or decreased in the LLs. These genes were uniformly over- or underexpressed in all three LLs compared with normal spleen (Supplemental Figure S1, B and C, see http://ajp.amjpathol.org). The results did not reveal a clear relationship of the LLs to a specific normal cell type but did suggest a state of differentiation between GC and plasma cells. This was most easily seen with the “splitting” of canonical GC genes (eg, Aicda increased and Bcl6 decreased) and plasma cell genes (eg, Atf6 increased and Prdm1 decreased).
Msh6−/− Lymphomas Bear Fully Recombined V(D)J Heavy and Light Chain Alleles and Are Monoclonal or Oligoclonal
Although most tumors displayed histological, phenotypic, and molecular features consistent with GC or post-GC B-cell lineage lymphomas, these characteristics are not predictive of clonality. Therefore, we assessed the status of V(D)J recombination of the IgH and κ chain locus by PCR with a panel of degenerate primers that recognize the vast majority of VH and D families and κ light chains.51,52 Fully rearranged alleles were amplified from DNA of all but one (1281) of the lymphomas tested (Table 1). In four tumors (720, 164, 1301, and 480), the presence of a single amplicon was indicative of monoclonality. In the remaining five tumors, several VHDJH amplicons or VHDJH and DJH amplicons were identified, compatible with the presence of a single clone with two rearranged heavy chain alleles or of a nonproductive DJ and a productive VDJ rearrangement. In some of the tumors that appeared oligoclonal for IgH (850, 624, and 599), a single κ chain rearrangement was detected, effectively ruling out a polyclonal tumor pattern (data not shown). Although we were unable to clearly distinguish monoclonality from oligoclonality in some tumors, polyclonal patterns similar to those seen in wild-type spleens were not observed in any of the cases. Thus, the Msh6−/− B-lineage neoplasms appeared to be monoclonal or oligoclonal.
Microsatellite Instability in Msh6−/− B-Cell Lymphomas
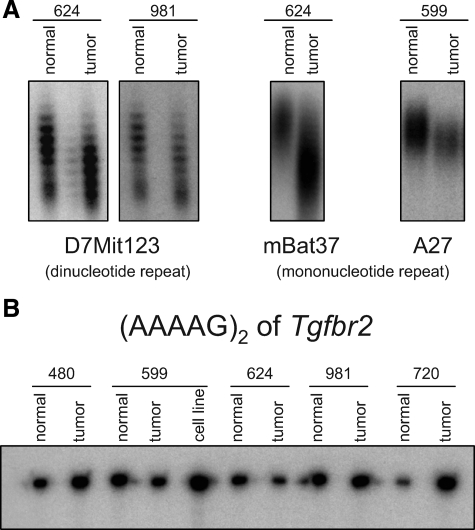
Loss of MSH6 enzymatic activity is associated with MSI in mouse and human tumors.46 To determine whether MSI is also a feature of the tumors studied here, paired DNAs prepared from tumors and tails of individual mice were assayed for MSI using a panel of five mononucleotide and dinucleotide repeats. These markers are among those commonly used to evaluate MSI in mice, and two were selected for their high sensitivity (Table 2).53,54,55,56 All tumors showed clear instability for at least one locus; instability was observed in 4 of 12 assays at mononucleotide tracts and in 4 of 7 assays at dinucleotide tracts (Figure 2A and data not shown). These results suggest that MSI could contribute to lymphoma initiation or progression in Msh6−/− mice.
Table 2.
PCR Primers
| Assay | Primer names | Sequence or reference | Notes |
|---|---|---|---|
| Microsatellite instability | |||
| mBat-37 | Bacher et al.53 | ||
| A27 | Kabbarah et al.54 | ||
| (GT)3 tract of Tgfbr2 | Lowsky et al.55 | ||
| (AAAAG)2 tract of Tgfbr2 | TGFBR2-Fwd | 5′-GACCCCAAGCTCACCTACCAC-3′ | 35 cycles of 95°C for 1 minute, 62°C for 30 seconds, 72°C for 30 seconds |
| TGFBR2-Rev2 | 5′-TACCCGGGATAACTACTCACCTTC-3′ | ||
| D7Mit123 | UniSTS Accession No. 116680 | 50 cycles of 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds | |
| D7Mit91 | UniSTS Accession No. 131298 | 30 cycles of 94°C for 60 seconds, 56°C for 60 seconds, and 72°C for 30 seconds | |
| U12235 | A hemi-nested PCR was performed using primers P1 and P256 for 25 rounds of 94°C for 30 seconds, 60°C for 1 minute, and 72°C for 2 minutes, followed by 35 rounds with primers P1 and P3 of 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds | ||
| Quantitative RT-PCR | |||
| Mouse AID | QRT-PCR-mAID.up2 | 5′-CTGGGAAGGGCTACATGAAA-3′ | 95°C for 15 minutes followed by 40 cycles of 94°C for 15 seconds, 60°C for 30 seconds, 72°C for 30 seconds |
| QRT-PCR-mAID.down2 | 5′-GAAATGCATCTCGCAAGTCA-3′ | ||
| Mouse GAPDH | QRT-PCR-mGAPDH.fwd | 5′-GGCATTGCTCTCAATGACAA-3′ | |
| QRT-PCR-mGAPDH.rev | 5′-CCCTGTTGCTGTAGCCGTAT-3′ | ||
| Somatic hypermutation | |||
| JH2-JH4 intron | JH2-JH4.fwd | 5′-GGCACCACTCTCACAGTCTCCTCAGG-3′ | 95°C for 5 minutes followed by 35 cycles of 95°C for 30 seconds, 60°C for 1 minute, 72°C for 1 minute |
| JH2-JH4.Adown | 5′-CCTGACCCAGACCCATGTCTCAACTTT-3′ | ||
| V(D)J recombination | |||
| Heavy chain locus | As per Ehlich et al.51 | Hot-start PCR | |
| Light chain V-Jκ | As per Yamagami et al.52 | ||
Figure 2.
Msh6−/− B-cell lymphomas display microsatellite instability. Differences in the size of the amplicon between normal tail DNA and tumor DNA from the same mouse, resolved on sequencing gels, indicate instability not encoded in the germline. A: Examples of instability at dinucleotide repeat D7Mit123 in tumors 624 and 981 and of mononucleotide repeat mBat37 in tumor 624 and of mononucleotide repeat A27 in tumor 599. B: Five Msh6−/− tumors, as well as a cell line derived from tumor 599, were stable at the (AAAAG)2 tract within the coding region of Tgfbr2. Stability was also observed at the (GT)3 repeat tract of the same gene (not shown).
MSI occurs within the coding region of the TGFBR2 gene in human colon cancers57 and in T-cell lymphomas of MSH2-deficient mice.58 Although the (AAAAG)2 sequence in the murine coding region would be expected to provide less opportunity for slippage events than its human counterpart, which is a true mononucleotide repeat, this site has nevertheless been observed to be unstable in murine Msh2−/− lymphomas. Assays of Msh6−/− tumors for MSI in the repeat tracts within the coding sequence of Tgfbr2 were negative (Figure 2B), and this result was confirmed by direct sequencing (data not shown).
Chromosomal Changes in Msh6−/− Tumors
Many types of human B-cell non-Hodgkin’s lymphomas exhibit recurrent translocations that place proto-oncogenes under the control of regulatory sequences in the IgH or IgL loci.19 In mouse B-cell lineage neoplasms, T(12;15) translocations juxtaposing Myc and the IgH locus are a feature of more than 90% of pristane-induced plasmablastic plasmacytomas,59 whereas IgH/Bcl6 translocations have been identified in a minority of mouse DLBCLs.60 Here, we used spectral karyotyping to determine whether tumors of Msh6-deficient mice exhibited translocations or other gross chromosomal anomalies. Spectral karyotyping of four primary tumors and a cell line derived from one of those tumors revealed that all cases were diploid or near diploid with chromosome numbers ranging from 38 to 42 (Supplemental Figure S2A, see http://ajp.amjpathol.org). No chromosomal translocations were observed. Losses and/or gains of whole chromosomes were observed in most spreads from each tumor and the cell line, but there were no obvious recurrent patterns of gain or loss.
Segmental aneuploidies have been implicated in colon cancers in MMR-deficient mice and microsatellite-unstable human colon cancers.61,62 To test for tumor-specific losses or gains in chromosomal regions beyond the resolution of spectral karyotype, we used array comparative genomic hybridization (Table 3). Excluding physiological V(D)J recombination events within the IgH locus, seven copy number alterations (CNA) involving segmental losses and one involving a gain were detected in the four tumors tested. Among these, one alteration was observed in three of four tumors, four were observed in two of four tumors, and three were observed in only one tumor. The coordinates in Table 3 are based on the shortest instance of each CNA. The deletion found in three of four tumors (CNA03) on chromosome 11 (Supplemental Figure S3, see http://ajp.amjpathol.org) includes one known gene, SfiI, which has a predicted function in centrosome assembly based on its yeast homolog63 and has been reported to be expressed in T cells by expression profiling studies.64,65
Table 3.
Copy Number Alterations in Msh6−/− Lymphomas
| CNA | Chr | Tumor no. | Band | Start | Stop | Length (bp) | Type |
|---|---|---|---|---|---|---|---|
| CNA01 | Chr10 | 164, 480 | qA4 | 27,519,847 | 27,577,357 | 57,510 | Loss |
| CNA02 | Chr7 | 164, 981 | qA3 | 26,605,452 | 26,657,288 | 51,836 | Loss |
| CNA03 | Chr11 | 164, 480, 981 | qA1 | 3001,791 | 3099,730 | 97,939 | Loss |
| CNA04 | Chr14 | 981 | qC2 | 52,652,778 | 53,130,162 | 477,384 | Loss |
| CNA05* | Chr12 | 164, 480 | qF1, qF2 | 114,068,720 | 114,971,457 | 902,737 | Loss |
| CNA06 | Chr4 | 480 | qC4, qC5 | 88,688,443 | 91,879,594 | 3,191,151 | Loss |
| CNA07 | Chr6 | 164, 480 | qF3 | 129,938,558 | 130,203,359 | 264,801 | Loss |
| CNA08 | Chr11 | 164 | qA1, qA2 | 11,201,260 | 16,186,605 | 4,985,345 | Gain |
CNA, arbitrary identification assigned to the alterations in this study; Chr, chromosome; Tumor no., the tumors that exhibited the alteration; Start, Stop, and Length coordinates are based on mouse genome assembly February 2006 (mm8); chromosomal bands are from the UCSC Genome Browser.
This deletion occurred in the IgH locus.
Survival of Msh6−/− Mice Is Not Affected by AID Deficiency
Because the highly mutagenic enzyme AID is strongly expressed in normal GC B cells and we observed AID protein in a few of the samples, we asked whether any of the lymphomas exhibited AID-induced SHM. The JH2-JH4 intron of the Igh locus is highly targeted by AID in normal B cells. We amplified and sequenced genomic DNA from this region66 from four tumors. Mutations at frequencies that were higher than the PCR error rate were detected in RGWY hotspot sequences preferentially targeted by AID13 in tumor 624 that had BCL6+ and PAX5+ cells and in tumor 850 that was PAX5+ but BCL6− (Table 1). In contrast, the frequency of mutations was not above the PCR error rate in tumor 1125, which was BCL6+, PAX5+, PNA+, and AID+, nor in tumor 1301, which was BCL6+, PAX5+, and PNA+. The finding of mutations even in non-AID-expressing tumors suggests that AID was functional at some point during the lifespan of at least some of the MSH6-deficient tumors.
AID has been implicated in B-cell lymphomagenesis by virtue of its expression in tumors67,68 and its ability to introduce double-stranded breaks and point mutations in or near proto-oncogenes.29 The lack of chromosomal translocations in the Msh6−/− lymphomas suggested that inappropriately resolved CSR was not a mechanism of neoplastic transformation in these animals. A well recognized alternative mechanism is that perturbations in the targeting of SHM contribute to B-cell lymphomagenesis. Aberrant SHM has been reported in human DLBCLs in several oncogenes,30 and AID induces phase 1 mutations in scores of highly expressed genes in normal GC B cells31 including CD95,69 Igα and Igβ,70 and BCL6.71 When MMR encounters AID-induced mutations within the Ig locus of GC B cells, it recruits error-prone repair, leading to the further introduction of phase 2 mutations. Outside of the Ig locus, MMR recruits error-prone repair to only a few genes such as BCL6 but mostly mediates error-free repair of non-Ig genes including oncogenes targeted by AID within GC B cells.31 The mechanisms that regulate this targeting are poorly understood but may involve ubiquitylation of proliferating cell nuclear antigen (PCNA).26,27
We hypothesized that MSH6 participates in this error-free repair of AID mutations at oncogenes or tumor suppressors and that this is the way in which it might protect B cells from neoplastic transformation. Rather than proceeding to phase 2 mutagenesis, as it does in the Ig locus, MSH6 may recruit error-free repair to these AID-induced lesions. If this were true, it would provide an explanation for why B cells are the lineage that becomes malignant in the absence of MSH6, since AID expression is largely limited to B cells. In addition, it might shed light on why Msh2−/− mice do not develop B-cell lymphomas, because it is a domain within the MSH6 protein that mediates physical recognition and binding of the MSH2/MSH6 complex to G:U mismatches.25,72 Finally, it might also help explain why Msh3−/− mice do not develop B-cell lymphomas either, because MSH3 does not recognize single base pair mismatches.
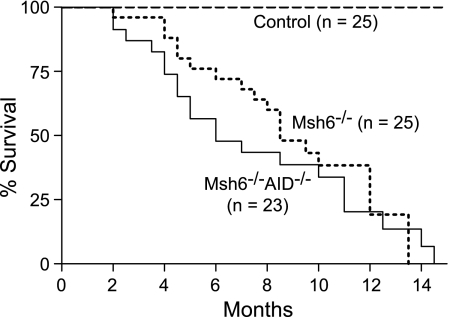
To test this hypothesis, we generated mice doubly deficient in MSH6 and AID (Msh6−/−Aicda−/−) and compared their survival with that of mice deficient in MSH6 alone for development of disease (Figure 3). If the hypothesis were false, then withdrawal of AID from the Msh6−/− mice should have no effect on survival. If the hypothesis were true, then removing AID would eliminate the initial hit, the repair function of MMR would be rendered unnecessary, and the mice should exhibit normal survival. Mutant mice of both genotypes began to die at approximately 2 months of age and all were dead at approximately 14 months. Although the median survival for the double knockout mice was 6.0 months versus 8.5 months for Msh6−/− animals, suggesting a trend toward shorter latency in the absence of AID, the survival curves were not significantly different (n = 48, log-rank test P = 0.456). The Msh6−/−Aicda−/− mice died of lymphomas that histologically and by IHC were largely B-cell LLs (data not shown). Thus, the hypothesis that MSH6 protects B cells by repairing AID-induced lesions is incorrect and B-cell lymphomas arise in Msh6−/− mice through an AID-independent pathway.
Figure 3.
Survival curves for control, Msh6−/−, and Msh6−/−Aicda−/− mice. Animals were fully backcrossed to C57BL/6, housed in the same barrier facility and monitored daily for morbidity and mortality. Log-rank test P = 0.456.
Discussion
Chromosomally unstable mouse tumors can serve as informative models for human malignancies and facilitate the identification of new loci that contribute to malignant transformation.73 In humans, defects in MMR predispose to colon and hematopoietic malignancies. In mice, MSH2-deficiency is associated mostly with T-cell lymphomas, whereas defects in MSH6 result primarily in B-cell lymphomas. In some strains of mice that develop both T- and B-cell lymphomas, the latency for T-cell tumors is shorter.74,75 The scarcity of B-cell lymphomas in MSH2-deficient mice might simply be due to the longer latency required for the appearance of B-cell lymphomas. This view is supported by the observation that MSH2-deficient nude mice that lack T cells develop B-cell lymphomas.45
MSH6 contains an N-terminal domain that is absent from MSH2 and differs in primary sequence from that of MSH3.76 This domain of MSH6 contains intrinsic DNA-binding activity, mediates the interaction with proliferating cell nuclear antigen (PCNA), and is likely to mediate other protein-protein interactions as well.76,77,78 Whereas single mismatched base pairs are recognized by the MSH2/MSH6 heterodimer, both smaller and larger stretches of mismatches or loops resulting from deletions or insertions are recognized by a heterodimer formed by MSH2 and MSH3. Whereas MSH3 and MSH6 compete for interaction with MSH2 to recruit downstream MMR pathway members to their respective types of DNA lesions, there is in fact some functional redundancy between them.21,79 That AID-generated lesions in the Ig locus are single-base G:U mismatches provides an explanation for why MSH2/MSH6, and not MSH2/MSH3, is important for SHM and CSR in normal GC B cells. Msh3−/− mice are not especially prone to developing neoplasms. Doubly deficient Msh3−/− Msh6−/− mice have shorter survival and increased intestinal tumor development compared with Msh6−/− mice, but interestingly, loss of MSH3 does not affect lymphomagenesis.80,81 This observation suggests that the role MSH6 plays in protecting B cells from neoplastic transformation or progression is distinct from MSH3 and depends on its ability to recognize single-base mismatches.
In addition to the correction of mispaired bases, MMR is involved in the processing of DNA damage and the subsequent induction of an apoptotic response.28 An inability to mount an apoptotic response to DNA damage is unlikely to be a factor in the genesis of the B-cell lymphomas described here, since mice with a point mutation in MSH6 that markedly interferes with repair function, but not with apoptotic signaling, also die of B-cell lymphomas with similar latencies.46 Likewise the recently reported ability of MSH2 to suppress Eμ-myc-driven B-cell lymphomas seems to be unrelated to MMR-dependent apoptosis.82 On the other hand, the unique genotoxic stress to which GC B cells are subjected18 and the intrinsic MSI exhibited by these cells83 raise the possibility that MSH6 protects B cells through its traditional enzymatic role in mismatch repair.
A protective role for MSH2 and MSH6 in B cells is somewhat paradoxical because the MSH2:MSH6 heterodimer contributes ∼50% of the AID-dependent mutations in SHM (phase 2) and plays an important role in the recombination events that are involved in CSR,13,24 activities that might be expected to contribute to B-cell transformation. Most of the tumors in the Msh6−/− mice were diagnosed histologically as B-cell lineage lymphomas belonging to different categories including LLs, DLBLCs with centroblastic or immunoblastic features, PCT with anaplastic, plasmablastic, and plasmacytic features, and histiocyte-associated DLBCLs. Histiocyte-rich B-cell lymphomas represents another clinical entity that would benefit from an animal model,84 and it would be interesting in future studies to examine closely the histiocytic component in these tumors. Recent studies suggest that APCT with anaplastic and plasmablastic cytologies are more closely related to memory B cells than to mature plasma cells or GC B cells, even though they exhibit expression of genes that might traditionally be associated with GC or plasma-cell differentiation programs (H. Morse, unpublished observations).
In addition, we studied a cell line (designated “599”) cloned from one of the oligoclonal B-cell lymphoblastic lymphomas (tumor 599) whose karyotype is listed in Supplemental Figure S2B (see http://ajp.amjpathol.org). We presume it arose from a minor population of the tumor, as its surface molecule phenotype by flow cytometry (B220+, IgM+, and PNA+) was closer to that of normal GC B cells than most of the tumor cells visualized by IHC in the sectional stains of its parent tumor (not shown). This cell line readily formed DLBCL-like tumors in recipient immunosuppressed mice, and AID expression could be induced in culture (not shown). Despite the fact that we were unable to detect ongoing SHM or CSR in this line, these cells may be a potentially useful tool for future studies. Moreover, these observations also suggest that some tumor cells may have expressed GC markers at levels below the sensitivity of IHC or that only minor populations of cells within the tumors have characteristics of GC cells. This notion is corroborated by the finding that tumor 850 exhibited signature AID-like somatic mutations even though GC markers were not detected by IHC (Table 1 and Supplemental Table 2, see http://ajp.amjpathol.org).
AID-dependent balanced translocations that juxtapose proto-oncogenes to Ig gene regulatory elements are the initiating events in mature B-cell neoplasms of humans and mice as well as mouse plasmacytomas.59,85,86 By spectral karyotype, the B-cell lineage tumors of MSH6-deficient mice did not bear chromosomal translocations, although chromosomal aneuploidies were common. Similar abnormalities have been observed in embryonic fibroblasts from mice deficient in MSH282,87 and in colon cancers in MMR-deficient mice and humans with microsatellite instability.61,62
Array comparative genomic hybridization, with its greater resolution, did reveal chromosomal regions that were amplified or deleted, and some of these were common to two or three of the four tumors examined (Table 3). Excluding the IgH locus, the seven CNAs identified in this study included the coding regions of 29 genes, of which Ingenuity Pathway Analysis identified 5 (Cdkn2a, Cdkn2b, Klra4, Ikzf1, and Grb10) with known roles in cancer. The tumor suppressor genes Cdkn2a (p16Ink4a) and Cdkn2b (p15Ink4b) are frequently lost in human leukemias and lymphomas, including DLBCLs,88,89 and they were deleted in one tumor (480, CNA06) in this study. Ikzf1 (Ikaros), a regulator of lymphocyte differentiation that participates in translocations with BCL6 in DLBCLs,90 was amplified in tumor 164 (CNA08) but its expression was decreased in most of the tumors (Supplemental Table 1, see http://ajp.amjpathol.org). The CNA08 region also includes Grb10, a gene shown to limit in vitro transformation of fibroblasts.91 The fifth gene with an annotated cancer function was Klra4 (Ly49D) on chromosome 6 that was deleted in two tumors (164 and 480, CNA07). In vitro studies have suggested that Ly49D inhibits chemotaxis of natural killer cells,92 a finding of possible relevance to immune surveillance of tumors. The copy number alteration that recurred in three of four tumors (CNA03) in band 11qA1 is in a region syntenic with human chromosome 22q12.2 that includes the human gene neurofibromatosis 2 (NF2). This finding is interesting in light of the clinical overlap between Lynch syndrome III and neurofibromatosis type 1,10 although it is unclear whether there is a mechanistic link between these two types of neurofibromatosis.93
Like the MSI observed in these tumors (Figure 2), the presence of recurrent losses and gains of small chromosomal segments suggests that absence of Msh6 is associated with increased chromosomal instability in B cells. This instability may contribute to or even cause the tumors, as has been reported for MMR-deficient murine colon cancer.61 However, it is also possible that the CNAs simply accumulated during the course of the neoplastic proliferation. Although MSH6-deficient patients do not exhibit the severe MSI phenotype seen in other MMR-defective hereditary nonpolyposis colorectal cancer,94 approximately half of MSH6-mutated hereditary nonpolyposis colorectal cancer tumors exhibit low levels of MSI,95,96 and a few MSH6-deficient human tumors have been reported to have high levels of MSI.97,98 We have previously reported low-to-moderate levels of MSI in murine tumors deficient in MSH617 or carrying inactivating point mutations of MSH6.46 The partially redundant activities of MSH6 and MSH3 are likely to explain why MSH6-deficient tissues only accumulate low amounts of MSI. These observations are interesting in light of the report that normal GC B cells themselves accumulate instability at microsatellites83 (see below).
AID is responsible not only for initiating point mutations during SHM as a canonical function but is also involved in noncanonical “off target” SHM activity affecting non-Ig genes that may introduce activating mutations in BCL6, MYC, and other proto-oncogenes. The GC and post-GC stage of differentiation of the tumors from MSH6-deficient mice and the heightened expression of Aicda transcripts seen in the LL prompted us to ask whether AID might contribute to the pathogenesis of these neoplasms, perhaps by introducing phase 1 mutations in oncogenes or tumor suppressors around the genome that are normally repaired in a high-fidelity fashion by MSH6. To address this issue genetically, we generated mice that were both AID-deficient and MSH6-deficient and compared their survival with that of mice deficient for MSH6 alone. The survival of Msh6−/−Aicda−/− mice was somewhat but not significantly shorter than that of Msh6−/− mice (Figure 3), suggesting that, although AID was not playing a critical role in B-cell neoplasia in Msh6−/− mice, its presence might affect some events late in the transformation process, perhaps even exerting a subtle protective effect. Similar studies of AID in Eμ-myc and λMYC transgenic mice also showed no significant differences in survival, although there was not a consensus as to whether tumors in AID-deficient mice were less mature.86,99,100 In Bcl6-Myc double transgenic mice, the presence or absence of AID also had no effect on overall mortality, but the tumors of AID-deficient mice were of pre-GC origin compared with the GC origins of the tumors in mice that were AID-sufficient.86 These findings raise the possibility that in B-cell precursors having already experienced a tumor-initiating event early in development,82 there may be conditions or factors besides AID that are unique to this lineage and are normally repaired or modulated by MSH6 to protect them from undergoing additional transforming events. Non-neoplastic MMR-deficient GC B cells display high levels of MSI,83 and GC B cells generate abnormally high numbers of point mutations and double-stranded breaks even in the absence of AID.101,102,103,104 GC B cells are subjected to genotoxic stress due to their rapid division times105 and to the suppression of DNA damage-response pathways.106,107,108,109 In addition, in the absence of AID, GC B cells seem less likely to undergo apoptosis.110 We speculate that these or other special mechanisms that B cells have evolved have allowed them to tolerate mutagenic and oncogenic events triggered by AID and explain why the AID-sufficient mice survived slightly longer than the doubly deficient mice. Although the molecular nature of these mechanisms is not known, our characterization of Msh6−/− mice supports the notion that B cells are particularly dependent on MMR to maintain genomic stability and prevent lymphomagenesis and that this role for MMR is either not dependent on AID or perhaps an alternative scenario in which the absence of MSH6 allows so much instability that the contributions of AID cannot be detected. This study did reveal that AID is not playing a critical role in MSH6-deficient B-cell lymphoma; however, we were unable to clearly explain why loss of MSH6 predisposes to B-cell lymphoma and loss of MSH2 causes T-cell lymphoma. Further examination of the role of MSH6 would provide important insights into the origins of B-cell lymphomas.
Acknowledgments
We authors thank Tasuku Honjo for his generous gift of the Aicda−/− mice, Fei Li Kuang and Elena Tosti for experimental assistance and reagents, Lily Chen for assistance with expression profiling, Kan Yang and Steven Brunnert for pathological assistance, John Manis, Elena Avdievich, Sonja Schaetzlein, Radma Mahmoud, and Jacob Glass for technical advice, and Barbara Birshtein and Moshe Sadofsky for helpful discussions. Spectral karyotyping was performed in the Genome Imaging Facility of Albert Einstein College of Medicine.
Footnotes
Address reprint requests to Matthew D. Scharff, M.D., Cell Biology Department, Chanin 403, Albert Einstein College of Medicine, 1300 Morris Park Ave., Bronx, NY 10461; or Herbert C. Morse III, M.D., Chief, Laboratory of Immunopathology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, 5640 Fishers Lane, Room 1421, Rockville, MD 20852. E-mail: matthew.scharff@einstein.yu.edu and hmorse@niaid.nih.gov.
Supported in part by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases (D.-M.S. and H.C.M.) and in part by the National Cancer Institute (award P30CA013330 to R.S.S. and C.M.). J.U.P. is supported by the Medical Scientist Training Program at Albert Einstein College of Medicine (grant T32-GM007288). M.D.S. is supported by grants R01-CA72649 and R01-CA102705 and by the Harry Eagle Chair provided by the National Women’s Division of the Albert Einstein College of Medicine. W.E. is supported by grants R01-CA76329 and R01-CA93484.
None of the authors disclosed any relevant financial relationships.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the NIH.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
A guest editor acted as editor-in-chief for this manuscript. No person at Thomas Jefferson University or Albert Einstein College of Medicine was involved in the peer review process or final disposition for this article.
Current address of M.D.I.-U.: AIDS Research Alliance, Los Angeles, California of Z.L.: Cell Biology, Novo Nordisk, Beijing, China; of J.U.P.: Internal Medicine, Massachusetts General Hospital, Boston, MA.
References
- Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- Watson P, Lynch HT. The tumor spectrum in HNPCC. Anticancer Res. 1994;14:1635–1639. [PubMed] [Google Scholar]
- Felton KEA, Gilchrist DM, Andrew SE. Constitutive deficiency in DNA mismatch repair: is it time for Lynch III? Clin Genet. 2007;71:499–500. doi: 10.1111/j.1399-0004.2007.00801.x. [DOI] [PubMed] [Google Scholar]
- Bandipalliam P. Syndrome of early onset colon cancers, hematologic malignancies and features of neurofibromatosis in HNPCC families with homozygous mismatch repair gene mutations. Fam Cancer. 2005;4:323–333. doi: 10.1007/s10689-005-8351-6. [DOI] [PubMed] [Google Scholar]
- Kruger S, Kinzel M, Walldorf C, Gottschling S, Bier A, Tinschert S, von Stackelberg A, Henn W, Gorgens H, Boue S, Kolble K, Buttner R, Schackert HK. Homozygous PMS2 germline mutations in two families with early-onset haematological malignancy, brain tumours. HNPCC-associated tumours, and signs of neurofibromatosis type 1. Eur J Hum Genet. 2008;16:62–72. doi: 10.1038/sj.ejhg.5201923. [DOI] [PubMed] [Google Scholar]
- Wimmer K, Etzler J. Constitutional mismatch repair-deficiency syndrome: have we so far seen only the tip of an iceberg? Hum Genet. 2008;124:105–122. doi: 10.1007/s00439-008-0542-4. [DOI] [PubMed] [Google Scholar]
- Ripperger T, Beger C, Rahner N, Sykora KW, Bockmeyer CL, Lehmann U, Kreipe HH, Schlegelberger B. Constitutional mismatch repair deficiency and childhood leukemia/lymphoma—report on a novel biallelic MSH6 mutation, Haematologica. 2010;95:841–844. doi: 10.3324/haematol.2009.015503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Edelmann W. Loss of DNA mismatch repair function and cancer predisposition in the mouse: animal models for human hereditary nonpolyposis colorectal cancer. Am J Med Genet C Semin Med Genet. 2004;129C:91–99. doi: 10.1002/ajmg.c.30021. [DOI] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- Reitmair AH, Redston M, Cai JC, Chuang TC, Bjerknes M, Cheng H, Hay K, Gallinger S, Bapat B, Mak TW. Spontaneous intestinal carcinomas and skin neoplasms in Msh2-deficient mice. Cancer Res. 1996;56:3842–3849. [PubMed] [Google Scholar]
- Reitmair AH, Schmits R, Ewel A, Bapat B, Redston M, Mitri A, Waterhouse P, Mittrucker HW, Wakeham A, Liu B, Thomason A, Griesser H, Gallinger S, Ballhausen WG, Fishel R, Mak TW. MSH2 deficient mice are viable and susceptible to lymphoid tumours. Nat Genet. 1995;11:64–70. doi: 10.1038/ng0995-64. [DOI] [PubMed] [Google Scholar]
- de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- Edelmann W, Yang K, Umar A, Heyer J, Lau K, Fan K, Liedtke W, Cohen PE, Kane MF, Lipford JR, Yu N, Crouse GF, Pollard JW, Kunkel T, Lipkin M, Kolodner R, Kucherlapati R. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91:467–477. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- Küppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Rosenwald A, Staudt LM. Lymphoid malignancies: the dark side of B-cell differentiation. Nat Rev Immunol. 2002;2:920–933. doi: 10.1038/nri953. [DOI] [PubMed] [Google Scholar]
- Li Z, Scherer SJ, Ronai D, Iglesias-Ussel MD, Peled JU, Bardwell PD, Zhuang M, Lee K, Martin A, Edelmann W, Scharff MD. Examination of Msh6- and Msh3-deficient mice in class switching reveals overlapping and distinct roles of MutS homologues in antibody diversification. J Exp Med. 2004;200:47–59. doi: 10.1084/jem.20040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesendanger M, Kneitz B, Edelmann W, Scharff MD. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J Exp Med. 2000;191:579–584. doi: 10.1084/jem.191.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo SA, Yang WW, Gearhart PJ. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J Exp Med. 2004;200:61–68. doi: 10.1084/jem.20040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutS[α] DNA lesion recognition complex. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Langerak P, Nygren AO, Krijger PH, van den Berk PC, Jacobs H. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J Exp Med. 2007;204:1989–1998. doi: 10.1084/jem.20070902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa S, Avdievich E, Peled JU, MacCarthy T, Werling U, Kuang FL, Kan R, Zhao C, Bergman A, Cohen PE, Edelmann W, Scharff MD. Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proc Natl Acad Sci USA. 2008;105:16248–16253. doi: 10.1073/pnas.0808182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien V, Brown R. Signalling cell cycle arrest and cell death through the MMR System. Carcinogenesis. 2006;27:682–692. doi: 10.1093/carcin/bgi298. [DOI] [PubMed] [Google Scholar]
- Pérez-Durán P, de Yebenes VG, Ramiro AR. Oncogenic events triggered by AID, the adverse effect of antibody diversification. Carcinogenesis. 2007;28:2427–2433. doi: 10.1093/carcin/bgm201. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Lagelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Unniraman S, Zhou S, Schatz DG. Identification of an AID-independent pathway for chromosomal translocations between the Igh switch region and Myc. Nat Immunol. 2004;5:1117–1123. doi: 10.1038/ni1127. [DOI] [PubMed] [Google Scholar]
- Takizawa M, Tolarova H, Li Z, Dubois W, Lim S, Callen E, Franco S, Mosaico M, Feigenbaum L, Alt FW, Nussenzweig A, Potter M, Casellas R. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J Exp Med. 2008;205:1949–1957. doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski DA, Stavnezer J. Antibody class switching differs among SJL. C57BL/6 and 129 mice. Int Immunol. 2007;19:545–556. doi: 10.1093/intimm/dxm020. [DOI] [PubMed] [Google Scholar]
- Cattoretti G, Angelin-Duclos C, Shaknovich R, Zhou H, Wang D, Alobeid B. PRDM1/Blimp-1 is expressed in human B-lymphocytes committed to the plasma cell lineage. J Pathol. 2005;206:76–86. doi: 10.1002/path.1752. [DOI] [PubMed] [Google Scholar]
- Kuraguchi M, Yang K, Wong E, Avdievich E, Fan K, Kolodner RD, Lipkin M, Brown AM, Kucherlapati R, Edelmann W. The distinct spectra of tumor-associated Apc mutations in mismatch repair-deficient Apc1638N mice define the roles of MSH3 and MSH6 in DNA repair and intestinal tumorigenesis. Cancer Res. 2001;61:7934–7942. [PubMed] [Google Scholar]
- MacCarthy T, Roa S, Scharff MD, Bergman A. SHMTool: a webserver for comparative analysis of somatic hypermutation datasets. DNA Repair. 2009;8:137–141. doi: 10.1016/j.dnarep.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage M, Coleman A, Manoir SD, Veldman T, McCormack S, Dickson RB, Barlow C, Wynshaw-Boris A, Janz S, Wienberg J, Ferguson-Smith MA, Schrock E, Ried T. Multicolour spectral karyotyping of mouse chromosomes. Nat Genet. 1996;14:312–315. doi: 10.1038/ng1196-312. [DOI] [PubMed] [Google Scholar]
- Davisson MT. Rules and guidelines for nomenclature of mouse genes. International Committee on Standardized Genetic Nomenclature for Mice. Gene. 1994;147:157–160. doi: 10.1016/0378-1119(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Lee CH, Melchers M, Wang H, Torrey TA, Slota R, Qi C-F, Kim JY, Lugar P, Kong HJ, Farrington L, van der Zouwen B, Zhou JX, Lougaris V, Lipsky PE, Grammer AC, Morse HC., III Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J Exp Med. 2006;203:63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MR, Nation PN, Andrew SE. A lack of DNA mismatch repair on an athymic murine background predisposes to hematologic malignancy. Cancer Res. 2005;65:2626–2635. doi: 10.1158/0008-5472.CAN-04-3158. [DOI] [PubMed] [Google Scholar]
- Yang G, Scherer SJ, Shell SS, Yang K, Kim M, Lipkin M, Kucherlapati R, Kolodner RD, Edelmann W. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell. 2004;6:139–150. doi: 10.1016/j.ccr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Fredrickson TN, Harris AW. Atlas of Mouse Hematopathology. London: Informa Healthcare,; 2000 [Google Scholar]
- Morse HC, 3rd, Anver MR, Fredrickson TN, Haines DC, Harris AW, Harris NL, Jaffe ES, Kogan SC, MacLennan IC, Pattengale PK, Ward JM. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100:246–258. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- Qi CF, Zhou JX, Lee CH, Naghashfar Z, Xiang S, Kovalchuk AL, Fredrickson TN, Hartley JW, Roopenian DC, Davidson WF, Janz S, Morse HC., 3rd Anaplastic, plasmablastic, and plasmacytic plasmacytomas of mice: relationships to human plasma cell neoplasms and late-stage differentiation of normal B cells. Cancer Res. 2007;67:2439–2447. doi: 10.1158/0008-5472.CAN-06-1561. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Cheah MT, Franco CB, Hosen N, Pin CL, Sha WC, Weissman IL. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J Immunol. 2007;179:6808–6819. doi: 10.4049/jimmunol.179.10.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlich A, Martin V, Muller W, Rajewsky K. Analysis of the B-cell progenitor compartment at the level of single cells. Curr Biol. 1994;4:573–583. doi: 10.1016/s0960-9822(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Yamagami T, ten Boekel E, Schaniel C, Andersson J, Rolink A, Melchers F. Four of Five RAG-Expressing JC[κ]−/− small pre-BII cells have no L chain gene rearrangements: detection by high-efficiency single cell PCR. Immunity. 1999;11:309–316. doi: 10.1016/s1074-7613(00)80106-5. [DOI] [PubMed] [Google Scholar]
- Bacher JW, Abdel Megid WM, Kent-First MG, Halberg RB. Use of mononucleotide repeat markers for detection of microsatellite instability in mouse tumors. Mol Carcinog. 2005;44:285–292. doi: 10.1002/mc.20146. [DOI] [PubMed] [Google Scholar]
- Kabbarah O, Mallon MA, Pfeifer JD, Edelmann W, Kucherlapati R, Goodfellow PJ. A panel of repeat markers for detection of microsatellite instability in murine tumors. Mol Carcinog. 2003;38:155–159. doi: 10.1002/mc.10157. [DOI] [PubMed] [Google Scholar]
- Lowsky R, DeCoteau JF, Reitmair AH, Ichinohasama R, Dong WF, Xu Y, Mak TW, Kadin ME, Minden MD. Defects of the mismatch repair gene MSH2 are implicated in the development of murine and human lymphoblastic lymphomas and are associated with the aberrant expression of rhombotin-2 (Lmo-2) and Tal-1 (SCL). Blood. 1997;89:2276–2282. [PubMed] [Google Scholar]
- Yao X, Buermeyer AB, Narayanan L, Tran D, Baker SM, Prolla TA, Glazer PM, Liskay RM, Arnheim N. Different mutator phenotypes in Mlh1- versus Pms2-deficient mice. Proc Natl Acad Sci USA. 1999;96:6850–6855. doi: 10.1073/pnas.96.12.6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- Lowsky R, Magliocco A, Ichinohasama R, Reitmair A, Scott S, Henry M, Kadin ME, DeCoteau JF. MSH2-deficient murine lymphomas harbor insertion/deletion mutations in the transforming growth factor β receptor type 2 gene and display low not high frequency microsatellite instability. Blood. 2000;95:1767–1772. [PubMed] [Google Scholar]
- Potter M. Neoplastic development in plasma cells. Immunol Rev. 2003;194:177–195. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- Qi CF, Hori M, Coleman AE, Torrey TA, Taddesse-Heath L, Ye BH, Chattopadhyay SK, Hartley JW, Morse HC., 3rd Genomic organisation and expression of BCL6 in murine B-cell lymphomas. Leuk Res. 2000;24:719–732. doi: 10.1016/s0145-2126(00)00028-x. [DOI] [PubMed] [Google Scholar]
- Chen P-C, Kuraguchi M, Velasquez J, Wang Y, Yang K, Edwards R, Gillen D, Edelmann W, Kucherlapati R, Lipkin SM. Novel roles for MLH3 deficiency and TLE6-like amplification in DNA mismatch repair-deficient gastrointestinal tumorigenesis and progression. PLoS Genet. 2008;4:e1000092. doi: 10.1371/journal.pgen.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann K, Terdiman JP, French AJ, Roydasgupta R, Sein N, Kakar S, Fridlyand J, Snijders AM, Albertson DG, Thibodeau SN, Waldman FM. Chromosomal instability in microsatellite-unstable and stable colon cancer. Clin Cancer Res. 2006;12:6379–6385. doi: 10.1158/1078-0432.CCR-06-1248. [DOI] [PubMed] [Google Scholar]
- Jones MH, Winey M. Centrosome duplication: is asymmetry the clue? Curr Biol. 2006;16:R808–R810. doi: 10.1016/j.cub.2006.08.041. [DOI] [PubMed] [Google Scholar]
- Chtanova T, Newton R, Liu SM, Weininger L, Young TR, Silva DG, Bertoni F, Rinaldi A, Chappaz S, Sallusto F, Rolph MS, Mackay CR. Identification of T cell-restricted genes, and signatures for different T cell responses. Using a comprehensive collection of microarray datasets. J Immunol. 2005;175:7837–7847. doi: 10.4049/jimmunol.175.12.7837. [DOI] [PubMed] [Google Scholar]
- Martinez-Sanz J, Yang A, Blouquit Y, Duchambon P, Assairi L, Craescu CT. Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J. 2006;273:4504–4515. doi: 10.1111/j.1742-4658.2006.05456.x. [DOI] [PubMed] [Google Scholar]
- Bardwell PD, Woo CJ, Wei K, Li Z, Martin A, Sack SZ, Parris T, Edelmann W, Scharff MD. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat Immunol. 2004;5:224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- Smit LA, Bende RJ, Aten J, Guikema JEJ, Aarts WM, van Noesel CJM. Expression of activation-induced cytidine deaminase is confined to B-cell non-Hodgkin’s lymphomas of germinal-center phenotype. Cancer Res. 2003;63:3894–3898. [PubMed] [Google Scholar]
- Greeve J, Philipsen A, Krause K, Klapper W, Heidorn K, Castle BE, Janda J, Marcu KB, Parwaresch R. Expression of activation-induced cytidine deaminase in human B-cell non-Hodgkin lymphomas. Blood. 2003;101:3574–3580. doi: 10.1182/blood-2002-08-2424. [DOI] [PubMed] [Google Scholar]
- Müschen M, Re D, Jungnickel B, Diehl V, Rajewsky K, Kuppers R. Somatic mutation of the CD95 gene in human B cells as a side-effect of the germinal center reaction. J Exp Med. 2000;192:1833–1840. doi: 10.1084/jem.192.12.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MS, Kanegai CM, Doerr JR, Wall R. Somatic hypermutation of the B cell receptor genes B29 (Igβ, CD79b) and mb1 (Igα, CD79a). Proc Natl Acad Sci USA. 2003;100:4126–4131. doi: 10.1073/pnas.0735266100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- Shell SS, Putnam CD, Kolodner RD. Chimeric Saccharomyces cerevisiae Msh6 protein with an Msh3 mispair-binding domain combines properties of both proteins. Proc Natl Acad Sci USA. 2007;104:10956–10961. doi: 10.1073/pnas.0704148104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, O'Neil J, Gutierrez A, Ivanova E, Perna I, Lin E, Mani V, Jiang S, McNamara K, Zaghlul S, Edkins S, Stevens C, Brennan C, Martin ES, Wiedemeyer R, Kabbarah O, Nogueira C, Histen G, Aster J, Mansour M, Duke V, Foroni L, Fielding AK, Goldstone AH, Rowe JM, Wang YA, Look AT, Stratton MR, Chin L, Futreal PA, DePinho RA. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DP, Bath ML, Harris AW, Cory S. T-cell lymphomas mask slower developing B-lymphoid and myeloid tumours in transgenic mice with broad haemopoietic expression of MYC. Oncogene. 2005;24:3544–3553. doi: 10.1038/sj.onc.1208399. [DOI] [PubMed] [Google Scholar]
- Hartley JW, Chattopadhyay SK, Lander MR, Taddesse-Heath L, Naghashfar Z, Morse HC, 3rd, Fredrickson TN. Accelerated appearance of multiple B cell lymphoma types in NFS/N mice congenic for ecotropic murine leukemia viruses. Lab Invest. 2000;80:159–169. doi: 10.1038/labinvest.3780020. [DOI] [PubMed] [Google Scholar]
- Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol Cell. 2007;26:565–578. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AB, Deterding L, Tomer KB, Kunkel TA. Multiple functions for the N-terminal region of Msh6. Nucleic Acids Res. 2007;35:4114–4123. doi: 10.1093/nar/gkm409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguri C, Duband-Goulet I, Friedrich N, Axt M, Belin P, Callebaut I, Gilquin B, Zinn-Justin S, Couprie J. Human mismatch repair protein MSH6 contains a PWWP domain that targets double stranded DNA. Biochemistry. 2008;47:6199–6207. doi: 10.1021/bi7024639. [DOI] [PubMed] [Google Scholar]
- Umar A, Risinger JI, Glaab WE, Tindall KR, Barrett JC, Kunkel TA. Functional overlap in mismatch repair by human MSH3 and MSH6. Genetics. 1998;148:1637–1646. doi: 10.1093/genetics/148.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann W, Umar A, Yang K, Heyer J, Kucherlapati M, Lia M, Kneitz B, Avdievich E, Fan K, Wong E, Crouse G, Kunkel T, Lipkin M, Kolodner RD, Kucherlapati R. The DNA mismatch repair genes Msh3 and Msh6 cooperate in intestinal tumor suppression. Cancer Res. 2000;60:803–807. [PubMed] [Google Scholar]
- de Wind N, Dekker M, Claij N, Jansen L, Klink YV, Radman M, Riggins G, Valk MVD, van 't Wout K, Riele HT. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat Genet. 1999;23:359–362. doi: 10.1038/15544. [DOI] [PubMed] [Google Scholar]
- Nepal RM, Tong L, Kolaj B, Edelmann W, Martin A. Msh2-dependent DNA repair mitigates a unique susceptibility of B cell progenitors to c-Myc-induced lymphomas. Proc Natl Acad Sci USA. 2009;106:18698–18703. doi: 10.1073/pnas.0905965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Bertocci B, Delbos F, Quint L, Weill J-C, Reynaud C-A. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity. 1998;9:127–134. doi: 10.1016/s1074-7613(00)80594-4. [DOI] [PubMed] [Google Scholar]
- Abramson JS. T-cell/histiocyte-rich B-cell lymphoma: biology, diagnosis, and management. Oncologist. 2006;11:384–392. doi: 10.1634/theoncologist.11-4-384. [DOI] [PubMed] [Google Scholar]
- Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, Mo T, Murty VV, Dalla-Favera R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC, 3rd, Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- Campbell MR, Wang Y, Andrew SE, Liu Y. Msh2 deficiency leads to chromosomal abnormalities, centrosome amplification, and telomere capping defect. Oncogene. 2006;25:2531–2536. doi: 10.1038/sj.onc.1209277. [DOI] [PubMed] [Google Scholar]
- Drexler HG. Review of alterations of the cyclin-dependent kinase inhibitor INK4 family genes p15, p16, p18 and p19 in human leukemia-lymphoma cells. Leukemia. 1998;12:845–859. doi: 10.1038/sj.leu.2401043. [DOI] [PubMed] [Google Scholar]
- Elenitoba-Johnson KSJ, Gascoyne RD, Lim MS, Chhanabai M, Jaffe ES, Raffeld M. Homozygous deletions at chromosome 9p21 involving p16 and p15 are associated with histologic progression in follicle center lymphoma. Blood. 1998;91:4677–4685. [PubMed] [Google Scholar]
- Hosokawa Y, Maeda Y, Ichinohasama R, Miura I, Taniwaki M, Seto M. The Ikaros gene, a central regulator of lymphoid differentiation, fuses to the BCL6 gene as a result of t(3;7)(q27;p12) translocation in a patient with diffuse large B-cell lymphoma. Blood. 2000;95:2719–2721. [PubMed] [Google Scholar]
- Morrione A, Valentinis B, Resnicoff M, Xu S, Baserga R. The role of mgrb10α in insulin-like growth factor I-mediated growth. J Biol Chem. 1997;272:26382–26387. doi: 10.1074/jbc.272.42.26382. [DOI] [PubMed] [Google Scholar]
- Inngjerdingen M, Rolstad B, Ryan JC. Activating and inhibitory Ly49 receptors modulate NK cell chemotaxis to CXC chemokine ligand (CXCL) 10 and CXCL12. J Immunol. 2003;171:2889–2895. doi: 10.4049/jimmunol.171.6.2889. [DOI] [PubMed] [Google Scholar]
- Ferner RE. Neurofibromatosis 1 and neurofibromatosis 2: a twenty first century perspective. Lancet Neurol. 2007;6:340–351. doi: 10.1016/S1474-4422(07)70075-3. [DOI] [PubMed] [Google Scholar]
- Hewish M, Lord CJ, Martin SA, Cunningham D, Ashworth A. Mismatch repair deficient colorectal cancer in the era of personalized treatment, Nat Rev Clin Oncol. 7:197–208. doi: 10.1038/nrclinonc.2010.18. [DOI] [PubMed] [Google Scholar]
- Wu Y, Berends MJW, Mensink RGJ, Kempinga C, Sijmons RH, van der Zee AGJ, Hollema H, Kleibeuker JH, Buys CHCM, Hofstra RMW. Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations. Am J Hum Genet. 1999;65:1291–1298. doi: 10.1086/302612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berends MJW, Wu Y, Sijmons RH, Mensink RGJ, van der Sluis T, Hordijk-Hos JM, de Vries EGE, Hollema H, Karrenbeld A, Buys CHCM, van der Zee AGJ, Hofstra RMW, Kleibeuker JH. Molecular and clinical characteristics of MSH6 variants: an analysis of 25 index carriers of a germline variant. Am J Hum Genet. 2002;70:26–37. doi: 10.1086/337944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks YMC, Wagner A, Morreau H, Menko F, Stormorken A, Quehenberger F, Sandkuijl L, Møller P, Genuardi M, van Houwelingen H, Tops C, van Puijenbroek M, Verkuijlen P, Kenter G, van Mil A, Meijers-Heijboer H, Tan GB, Breuning MH, Fodde R, Winjen JT, Bröcker-Vriends AHJT, Vasen H. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: impact on counseling and surveillance. Gastroenterology. 2004;127:17–25. doi: 10.1053/j.gastro.2004.03.068. [DOI] [PubMed] [Google Scholar]
- Palomaki GE, McClain MR, Melillo S, Hampel HL, Thibodeau SN. EGAPP supplementary evidence review: DNA testing strategies aimed at reducing morbidity and mortality from Lynch syndrome. Genet Med. 2009;11:42–65. doi: 10.1097/GIM.0b013e31818fa2db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani A, Kakazu N, Tsuruyama T, Okazaki IM, Muramatsu M, Kinoshita K, Nagaoka H, Yabe D, Honjo T. Activation-induced cytidine deaminase (AID) promotes B cell lymphomagenesis in Emu-cmyc transgenic mice. Proc Natl Acad Sci USA. 2007;104:1616–1620. doi: 10.1073/pnas.0610732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepal RM, Zaheen A, Basit W, Li L, Berger SA, Martin A. AID and RAG1 do not contribute to lymphomagenesis in Emu c-myc transgenic mice. Oncogene. 2008;27:4752–4756. doi: 10.1038/onc.2008.111. [DOI] [PubMed] [Google Scholar]
- Ouchida R, Ukai A, Mori H, Kawamura K, Dollé MET, Tagawa M, Sakamoto A, Tokuhisa T, Yokosuka T, Saito T, Yokoi M, Hanaoka F, Vijg J, Wang J-Y. Genetic analysis reveals an intrinsic property of the germinal center B cells to generate A:T mutations. DNA Repair. 2008;7:1392–1398. doi: 10.1016/j.dnarep.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Longo NS, Satorius CL, Plebani A, Durandy A, Lipsky PE. Characterization of Ig gene somatic hypermutation in the absence of activation-induced cytidine deaminase. J Immunol. 2008;181:1299–1306. doi: 10.4049/jimmunol.181.2.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J Exp Med. 2002;195:1193–1198. doi: 10.1084/jem.20011858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bross L, Muramatsu M, Kinoshita K, Honjo T, Jacobs H. DNA double-strand breaks: prior to but not sufficient in targeting hypermutation. J Exp Med. 2002;195:1187–1192. doi: 10.1084/jem.20011749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, Green R, Carroll M, Melnick A. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- Phan RT, Saito M, Kitagawa Y, Means AR, Dalla-Favera R. Genotoxic stress regulates expression of the proto-oncogene Bcl6 in germinal center B cells. Nat Immunol. 2007;8:1132–1139. doi: 10.1038/ni1508. [DOI] [PubMed] [Google Scholar]
- Zhou JX, Lee CH, Qi CF, Wang H, Naghashfar Z, Abbasi S, Morse HC., 3rd IFN regulatory factor 8 regulates MDM2 in germinal center B cells. J Immunol. 2009;183:3188–3194. doi: 10.4049/jimmunol.0803693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaheen A, Boulianne B, Parsa JY, Ramachandran S, Gommerman JL, Martin A. AID constrains germinal center size by rendering B cells susceptible to apoptosis. Blood. 2009;114:547–554. doi: 10.1182/blood-2009-03-211763. [DOI] [PubMed] [Google Scholar]