Abstract
Eosinophils are granular leukocytes that have significant roles in many inflammatory and immunoregulatory responses, especially asthma and allergic diseases. We have undertaken a fairly comprehensive proteomic analysis of purified peripheral blood eosinophils from normal human donors primarily employing 2-dimensional gel electrophoresis with protein spot identification by matrix-assisted laser desorption/ionization mass spectrometry. Protein subfractionation methods employed included isoelectric focusing (Zoom® Fractionator) and subcellular fractionation using differential protein solubilization. We have identified 3,141 proteins which had Mascot expectation scores of 10−3 or less. Of these 426 were unique and non-redundant of which 231 were novel proteins not previously reported to occur in eosinophils. Ingenuity Pathway Analysis showed that some 70% of the non-redundant proteins could be subdivided into categories that are clearly related to currently known eosinophil biological activities. Cytoskeletal and associated proteins predominated among the proteins identified. Extensive protein posttranslational modifications were evident, many of which have not been previously reported that reflected the dynamic character of the eosinophil. This dataset of eosinophilic proteins will prove valuable in comparative studies of disease versus normal states and for studies of gender differences and polymorphic variation among individuals.
Keywords: Asthma, 2-DE, Eosinophils, Phosphoproteins, Protein expression
1 Introduction
Eosinophils are pleiotropic multifunctional granulocytic leukocytes that function in diverse inflammatory and immunoregulatory responses [see reviews 1, 2]. Important pathologies associated with eosinophils include asthma, allergy, and parasitic helminth infections [1, 3]. In normal adults eosinophils are produced, differentiated, and matured in the bone marrow, migrate into the peripheral blood, and subsequently target to different tissues including mammary gland, thymus, uterus, lung, and especially the gastrointestinal tract [1]. However, in many inflammatory pathologies eosinophils can also be associated with numerous other organs and tissues. Eosinophil trafficking into inflammatory sites involves a number of cytokines, chemokines, growth factors, lipids, as well as four major cationic proteins (EPO, MBP, ECP, EDN) packaged within four types of cytoplasmic granules each with unique morphologies [1, 4].
Most current established methods of preparing eosinophils from peripheral blood utilizing anti-CD16 immunomagnetic beads to remove neutrophils yield a population of cells which is relatively homogenous typically greater than 98% [5]. However, density gradient centrifugation can further distinguish two populations of eosinophils termed “normodense” (specific gravity > 1.085 g/l) and “hypodense” (specific gravity < 1.085 g/l) [6]. Hypodense eosinophils typically account for 5–10% of the total peripheral blood eosinophil population in normal adults and represent activated and degranulated cells [7]. Normodense eosinophils can be converted to a hypodense form in vitro by a number of different stimuli; e.g., GM-CSF, IL-3, eotaxin and IL-5 [8]. Moreover, in many inflammatory pathologies the hypodense population of cells is increased; however, the composition of these hypodense eosinophils can differ when compared with in vitro stimulated cells [4, 8, 9]. Therefore, within the hypodense population of eosinophils there is indication of further microheterogeneity likely due to differential degranulation depending on the nature of the stimulus employed or the pathology involved. Thus, the molecular basis for all of the observed eosinophil heterogeneity is not fully established and clearly requires further investigation. A comparative proteome map of eosinophils from normal adults would be considerably important toward defining eosinophil heterogeneity, especially in inflammatory diseases.
It is clear that eosinophils play an important role in the etiology of bronchial asthma which is one of the fastest growing diseases in the Western world. The prevalence of asthma has increased steadily over the last two decades and in 2006 an estimated 16.1 million adults (7.3 % > 18 y) and some 9.4% or 6.8 million children were affected by the disease in the United States [10]. The rapid increase in the incidence of asthma and its resulting consequence on the healthcare system underscores the need for the identification of new therapeutic targets for the treatment of allergic inflammation. None of the currently available treatments for asthma and allergic diseases are curative. Recently, a number of factors have been proposed which appear to control eosinophil trafficking, survival and activation in animal models; however, to date, no therapeutic target has been developed which successfully eliminates tissue-activated eosinophils in human trials. Furthermore, the dual role of eosinophils in pro-inflammation and anti-inflammation has yet to be elucidated fully. Together these findings clearly emphasize the current status of an incomplete understanding of eosinophil activation and function, and argue for more comprehensive studies of the eosinophil phenotype.
We hypothesize that an unbiased characterization of a comprehensive set of proteins expressed by eosinophils will provide novel insights into the molecular circuitry, signaling pathways, proinflammatory mediators and cytokines that may play a role in the pathogenesis of eosinophil inflammation. Furthermore, the map will be important in comparative eosinophil studies of disease or therapeutic treatments. In the present work we initiated the proteomic characterization of peripheral blood eosinophils obtained from healthy, non-allergic donors. Furthermore, in order to decrease the complexity of the cell lysate and maximize the total number of proteins identified in the study, we fractionated the cell lysate into cytoplasmic, membrane, organelle, granule, and nuclear fractions, and resolved the proteins by 2-DE. We report a number of proteins that are unique to eosinophils and are associated with eosinophil effector functions. We posit that the present data will therefore serve as an important reference database for the discovery of markers of activated eosinophils and for future studies investigating therapeutic targets for eosinophilia-related inflammatory diseases.
2 Materials and Methods
2.1 Materials
Histopaque®-1077, α-cyano-4-hydroxycinnamic acid, benzamidine, leupeptin, aprotinin, microcystin, and dextran (Fluka) were obtained from Sigma-Aldrich (St. Louis, MO). Sodium orthovanadate and PMSF were products of Fisher Scientific (Fair Lawn, NJ). Thiourea, CHAPS, iodoacetamide, IPG strips (pH 5–8), Precision Plus molecular weight standards, Protean II XL Tris-HCL precast gels (8–16%), Criterion Tris-HCl precast gels (8–16%), RC DC protein assay kit (Lowry method with reduction compatibility (RC) and detergent compatibility (DC), Criterion Dodeca electrophoretic 13.3 cm × 8.7 cm multi-gel cell (12 gels, 11 cm strips) and Protean II XL electrophoretic 19.3 cm × 18.3 cm cell (2 gels, 18 cm strips) were products from BioRad (Hercules, CA.). IPG strips (11 and 18 cm, pH 3–10, 4–7, and 6–11), DeStreak rehydration buffer, IPG buffer/ampholytes, and Ettan DALT IPGphor II isoelectric focusing cell were obtained from GE Healthcare. TCEP and Perfect-FOCUS (protein precipitation reagent for 2-DE samples) were purchased from G-Biosciences (St. Louis, MD). Sypro Ruby fluorescent protein gel stain, Pro-Q Diamond fluorescent phosphoprotein gel stain, Peppermint Stick phosphoprotein molecular weight markers, and a Zoom® IEF fractionator were obtained from Invitrogen (Carlsbad, CA). HBSS without Mg 2+ or Ca 2+ was from Gibco. The ProteoExtract® Subcellular Proteome Extraction kit was from Calbiochem (San Diego, CA). VarioMACS separation columns, MACS Separator (magnetic), and CD16 MicroBeads for eosinophil isolation were purchased from Miltenyi Biotec (Auburn, CA).
2.2 Eosinophil isolation from peripheral human blood
Blood donors for eosinophil isolation included three female and five male non-smoking donors (ages 18–50 y) who showed neither asthmatic nor allergic symptoms [5]. Briefly, 1.5 ml of 15% Dextran and 1.5 ml of 0.25 M EDTA was immediately added to 60 ml of collected blood in two 50 ml polypropylene conical centrifuge tubes and allowed to sediment for 30 min at RT. After sedimentation, the leukocyte-containing layer was overlayed onto Histopaque®-1077 (15 ml leukocyte/7.5 ml Histopaque® and centrifuged (720 × g) at RT for 40 min. Following washing at 4°C with 20 mM HEPES-1X HBSS, granulocytes were recovered by centrifugation (300 × g for 7 min) and any remaining erythrocytes were lysed with consecutive additions of 5 ml of ice-cold 0.2% NaCl for 30 s followed by 5 ml of 1.8% NaCl. Following the further addition of 20 ml of HBSS, cells were centrifuged at 300 × g for 7 min at 4°C. Eosinophils were subsequently isolated by negative selection using CD16+ MicroBeads as instructed by the manufacturer (Miltenyi Biotec). Briefly, CD16+ cells (essentially neutrophils) were labeled with CD16+ MicroBeads and the cell suspension was loaded onto a VarioMACS column. The column was placed in the magnetic field of a MACS Separator and the labeled CD16+ cells were retained on the column. Unlabeled purified eosinophils were not retained and were washed out and collected. After removal of the column from the magnetic field, the neutrophil fraction was eluted from the column. Eosinophil purity was consistently monitored by Hansel staining and typically ranged above 98%. The levels of activated eosinophils in our preparations ranged between 1% to 3% as estimated by measurement of the activation marker CD69 [11].
2.3 2-DE sample preparation
Following CD16+ MicroBead eosinophil isolation, cells were pelleted at 300 × g and washed with 30 ml of 1X HBSS. After additional centrifugation at 300 × g for 7 min cells were solubilized for 2-DE in DeStreak rehydration buffer to a protein concentration of 1 µg/µl. If not used immediately for IEF, samples were stored at −80°C. Eosinophils were also subjected to subcellular fractionations using Calbiochem’s ProteoExtract® Subcellular Proteome Extraction kit according to the manufacturer’s provided protocol. Some samples were also subjected to IEF fractionation using a ZOOM® IEF Fractionator (Invitrogen, Carlsbad, CA) following the manufacturer’s provided protocol. Ranges of pH collected were 3.0–5.4, 5.4–7.0, and 7.0–10.0. Protein concentrations were established using the RC DC protein assay kit (BioRad). Typically, 10 × 106 cells yielded ~500 µg of cell lysate protein.
2.4 2-D gel electrophoresis
Eosinophil protein samples (200 µg for 11 cm and 350 µg for 18 cm IPG strips) were adjusted to 200 µl (11 cm strips) and 350 µl (18 cm strips) with DeStreak rehydration buffer and buffer/ampholyte was added to give a final concentration of 0.5%. The mixtures were microcentrifuged at 2000 rpm for 2 min. IEF was performed with a multi-sample IPGphor (GE Healthcare). Different pH gradient IPG strips were investigated. Paper wicks were placed over each electrode of the ceramic strip holders and 8 µl of Milli-Q H2O was added to the wicks just prior to the addition of sample/DeStreak buffer mixtures. Dry IPG strips were added to each sample mixture with the gel side of the strip facing down and the strips were covered with mineral oil. The strip holders were placed in an IPGphor IEF cell and focused at 20°C with the following protocols: for 11 cm IPG strips: 50 V for 11 h (active rehydration), 250 V gradient for 1 h, 500 V gradient for 1 h, 1000 V gradient for 1 h, 8000 V gradient for 2 h, and held at 8000 V for 6 h; for 18 cm IPG strips: 50 V for 11 h, 250 V for 1 h, 500 V for 1 h, 1000 V for 1 h, 8000 V for 2 h, and held at 8000 V for 8 h. IPG strips were then removed and carefully blotted with damp filter paper to remove excess mineral oil. After IEF the strips were equilibrated for 15 min in 4 ml of an equilibration buffer (50 mM Tris-HCL, pH 8.8, containing 6 M urea, 2% SDS, 20% glycerol and 10 µl/ml TCEP), followed by 15 min of equilibration with 4 ml of the above buffer containing 25 mg iodoacetamide/ml buffer. Strips were then rinsed with 1X Tris-glycine-SDS second dimension electrophoresis running buffer, pH 8.3, and placed in IPG wells of gels with the positive end of the strip toward the left side of the gels. Strips were subsequently overlayed with 0.5% molten agarose. Criterion gels were then placed in a second dimension electrophoresis cell and electrophoresis was conducted using pre-chilled 1X electrophoresis running buffer and 150 V for about 2¼ h or until the bromophenol blue dye reached the gel bottom. The electrophoresis (10°C) protocol for the Protean II gels was as follows: 35 V for 30 min, 50 V for 1 h, 70 V for 1 h, 100 V for 2 h, and 120 V for 12 h or until the dye front reached the bottom of the gel. After the second dimension of electrophoresis the gels were removed from their plates and rinsed with Milli-Q H2O prior to staining.
2.5 Fluorescent staining of 2-D gels
Gels were fixed, stained, and destained essentially according to the manufacture’s (Invitrogen) recommendations. Briefly, gels intended for Pro-Q Diamond staining were fixed in a solution of 50% methanol and 10% acetic acid in double distilled H2O overnight with shaking at RT then washed 3X in double distilled H2O, stained in Pro-Q Diamond stain at RT for 90 min, and destained in a solution of 20% acetonitrile, 50 mM sodium acetate (pH 4.0) in double distilled H2O. Gels intended for Sypro Ruby staining were fixed in 10% methanol, 7% acetic acid, in double distilled H2O for 2 h at RT. Subsequently, the Sypro Ruby stain was applied overnight at RT followed by destaining (10% ethanol) for 1 h. Some gels underwent both staining processes first with Pro-Q Diamond followed by Sypro Ruby.
2.6 Imaging of 2-D gels
Sypro stained gels were imaged at 100 µm resolution with a ProExpress 2-D Proteomic Imaging System (PerkinElmer Life and Analytical Sciences, Waltham, MA) at 460/80 nm excitation and 650/150 nm emission wavelengths. Pro-Q Diamond stained gels were imaged with a Fuji FLA-5100 (Fujifilm USA, Inc., Valhalla, NY) at 532 nm excitation (laser) and 575 nm longpass emission.
2.7 2-D gel image analysis
2-DE gel images were analyzed using Progenesis SameSpots software v2.0 (Nonlinear USA, Inc., Durham, NC). This software automatically detects individual protein spots within each image and matches identical protein spots across all images. It also removes noise from measurements of spot volumes using a proprietary algorithm for noise determination and correction. After automatic matching, manual review and adjustments were done to confirm proper spot detection and matching. The intensity of each protein spot was normalized based on the total spot volume of each gel, that is, the spot volume of each spot area was divided by the sum of all spot volumes in the gel. Spots present on less than two gels or with normalized volumes less than 150 were filtered out. Selected spots were robotically picked (Genomic Solutions, Ann Arbor, MI), trypsin digested, and robotically processed (Genomic Solutions) according to the manufacture’s recommendations prior to protein identification by MALDI-MS. Tryptic peptide samples were robotically transferred to MALDI-MS target plates. About 1 µl of MALDI matrix solution (α-cyano-4-hydroxycinnamic acid in 50:50 acetonitrile/H2O, 5 mg/ml) was also added robotically to the tryptic samples.
2.8 Manual gel sample preparation for MS
When many protein spots were to be picked, we employed the Genomic Solutions robotics instrumentation as described above. However, for those gels with few spots to be picked we used the following manual procedure. Gel samples were cut into 1 mm size pieces and placed into separate 0.5 ml polypropylene tubes. Ammonium bicarbonate buffer (100 µl of 50 mM, pH 8.0) was added to each tube and the samples were then incubated at 37°C for 30 min. After incubation, the buffer was removed and 100 µl of water was added to each tube. The samples were then incubated again at 37°C for 30 min. After incubation, the water was removed and 100 µl of acetonitrile was added to each tube to dehydrate the gel pieces. The samples were vortexed and after 5 min the acetonitrile was removed. Acetonitrile (100 µl) was again added to each of the sample tubes, vortexed, and acetonitrile removed after 5 min. The samples were then placed in a speedvac for 45 min to remove any excess solvent. To a 20 µg vial of lyophilized trypsin (Promega Corp., Madison, WI) was added 2 ml of 25 mM ammonium bicarbonate (pH 8.0). The trypsin solution was then vortexed and added to each sample tube in an amount required to just cover the dried gel (about 10 µl) and the samples were subsequently incubated at 37°C for 6 h. After digestion, the samples were removed from the oven and 1 µl of sample solution was spotted directly onto a MALDI target plate and allowed to dry. Subsequently, 1 µl of α-cyano-4-hydroxycinnamic acid matrix solution (50:50 acetonitrile/water at 5 mg/ml) was applied on the sample spot and allowed to air dry.
2.9 Mass spectrometry
MALDI TOF/MS was used to analyze tryptic peptide samples and identify proteins. Data were acquired with an Applied Biosystems (Foster City, CA) 4800 MALDI-TOF/TOF Proteomics Analyzer. Applied Biosystems software package included the 4000 Series Explorer (v3.6 RC1) with Oracle Database Schema (v3.19.0), and Data v3.80.0 to acquire both MS and MS/MS spectral data. The instrument was operated in positive ion reflectron mode with a mass range of 850–3000 Da and with the focus mass set at 1700 Da. For MS data, 1000–2000 laser shots were acquired and averaged from each sample spot. Automatic external calibration was performed using a peptide mixture with reference masses 904.468, 1296.685, 1570.677, and 2465.199. Following MALDI-MS analysis, MALDI- MS/MS was performed on several (5–10) of the most abundant ions from each sample spot. A 1 kV positive ion MS/MS method was used to acquire data under post-source decay conditions. The instrument precursor selection window was +/−3 Da. For MS/MS data, 2000 laser shots were acquired and averaged from each sample spot. Automatic external calibration was performed using reference fragment masses of 175.120, 480.257, 684.347, 1057.475, and 1441.635 (from precursor mass 1570.700). Applied Biosystems GPS Explorer ™ (v3.6) software was used in conjunction with MASCOT (Matrix Science, London, UK) to search the respective protein databases using both MS and MS/MS spectral data for protein identification. Protein match probabilities were determined using expectation values and/or MASCOT protein scores. The expectation value is the number of matches with equal or better scores that are expected to occur by chance alone. The default significance threshold was typically p<0.05; however, for protein identifications herein we used a more stringent threshold of 10−3. The lower the expectation value, the more significant the score. Expectation values were derived from Mascot scores (see www.matrixscience.com). MS peak filtering included the following parameters: mass range 800 Da to 4000 Da, minimum S/N filter = 10, mass exclusion list tolerance = 0.5 Da, and mass exclusion list (for some trypsin and keratin-containing compounds) included masses 842.51, 870.45, 1045.56, 1179.60, 1277.71, 1475.79, and 2211.1. For MS/MS peak filtering, the minimum S/N filter was set to 10. For protein identification, the human taxonomy was searched in either the NCBI or SwissProt databases. Other parameters included the following: selecting trypsin; maximum missed cleavages = 1; fixed modifications included carbamidomethyl (C) for 2-D gel analyses only; variable modifications included oxidation (M); precursor tolerance was set at 0.2 Da; MS/MS fragment tolerance was set at 0.3 Da; mass = monoisotopic; and peptide charges were only considered as +1.
3 Results
3.1 General characterization of the expression dataset
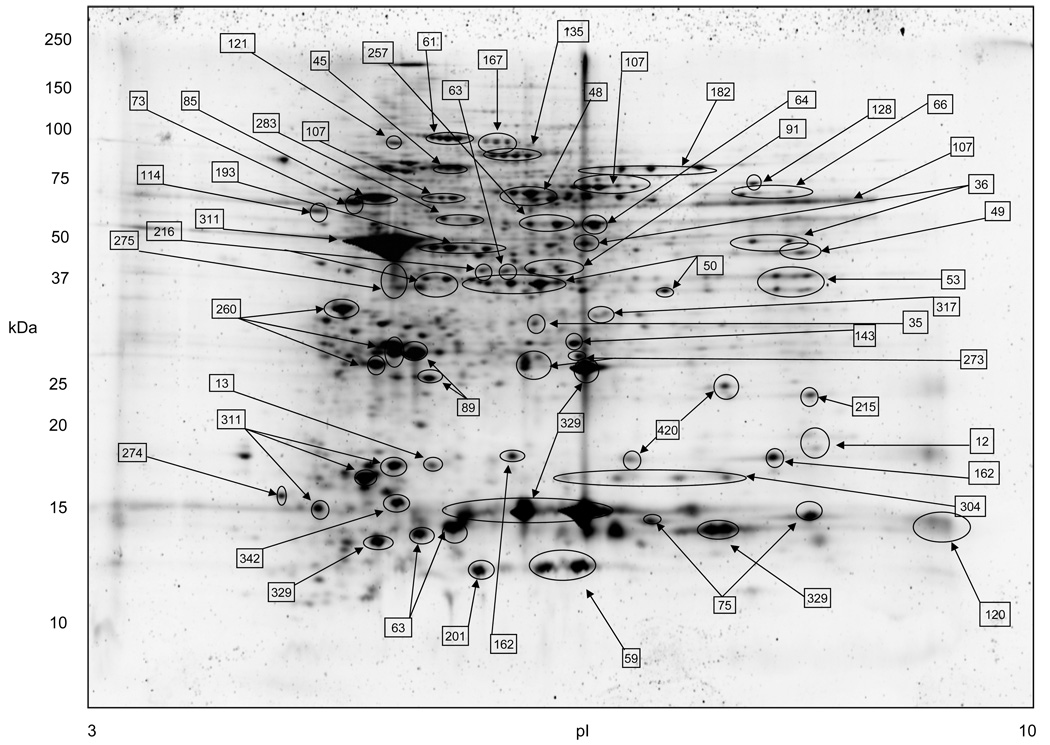
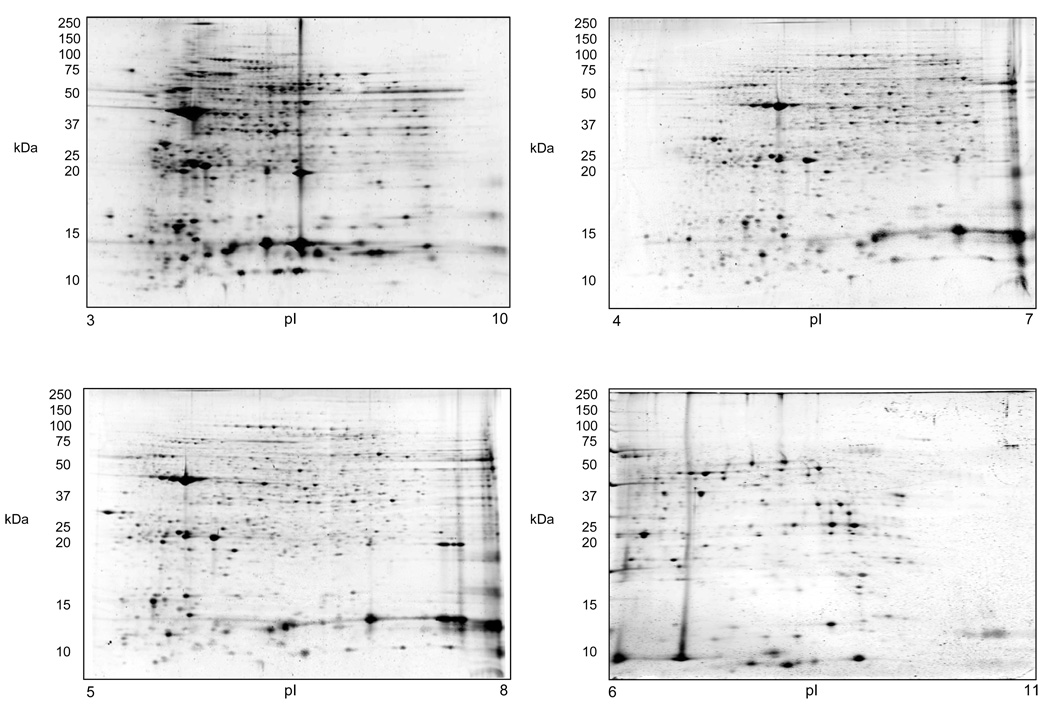
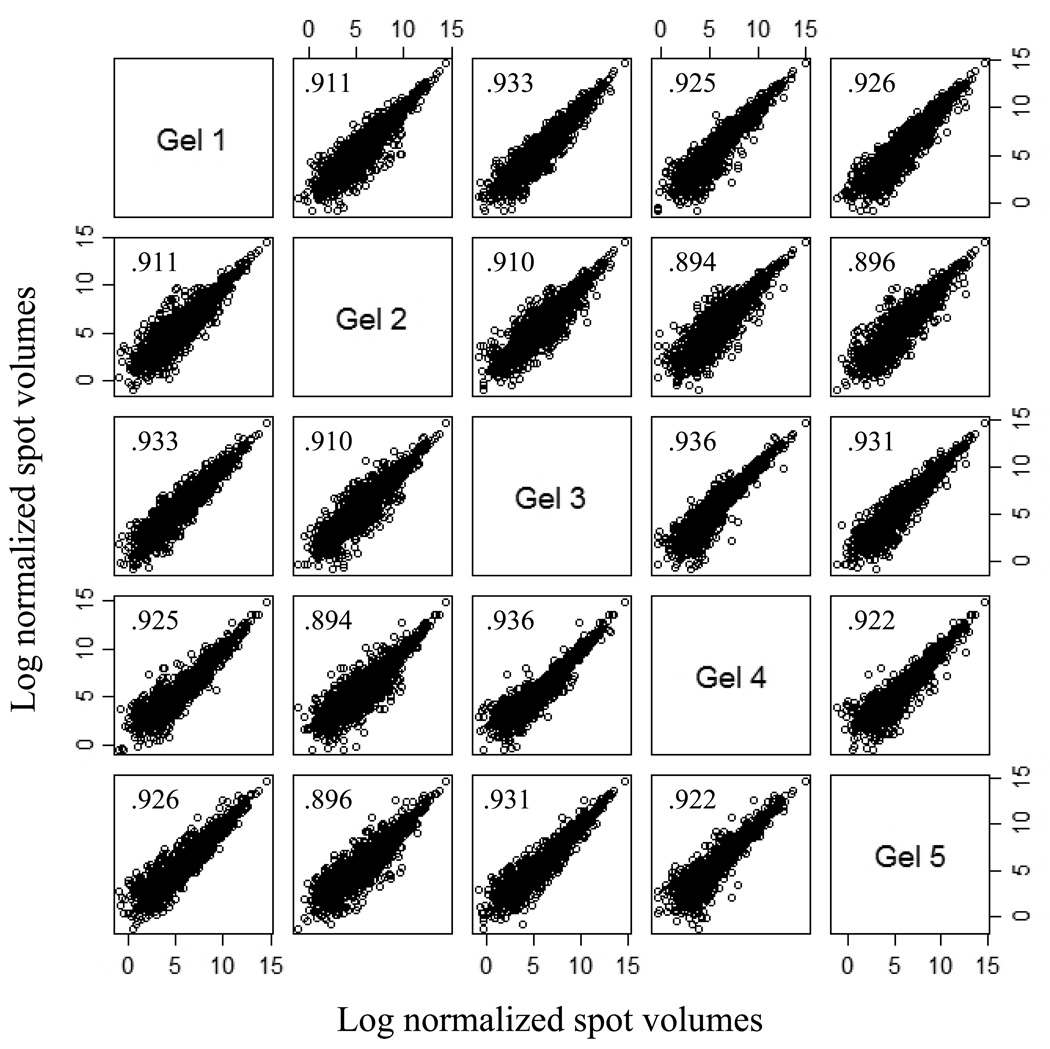
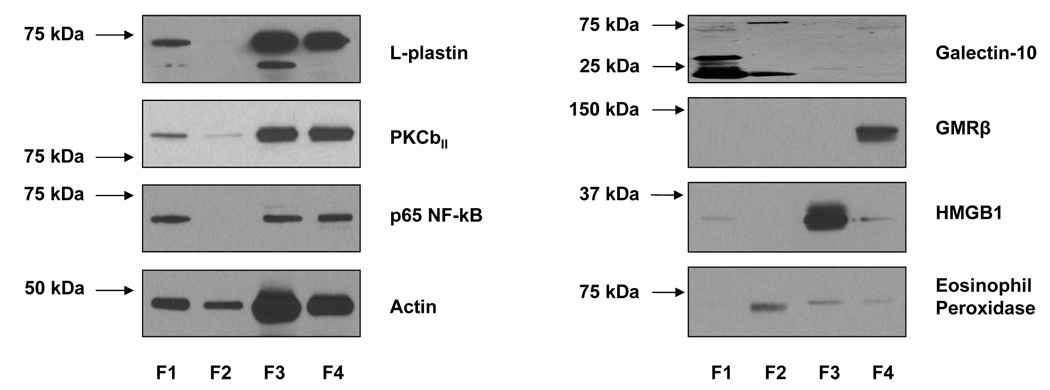
A representative 2-DE separation of an eosinophil whole cell lysate sample is shown in Fig. 1. The identities of selected non-redundant prominent protein spots are indicated in Table 1. Example of 2-D gels focused over the pH ranges 3–10, 4–7, 5–8, and 6–11 are shown in Fig. 2. In general, eosinophil lysates focused reasonably well in the pH range 3–10. To demonstrate gel to gel reproducibility five gels were selected and the log of normalized spot volumes from gel 1 was plotted pairwise versus gels 2 to 5 as shown in Fig. 3 and the Pearson’s correlation co-efficient (r2) was calculated. As evident in Fig. 3, the r2 values indicated that the 2-DE analyses were reasonably reproducible as conducted (mean ± SD = 0.91844 ± 0.01490). The distribution of proteins identified in the various fractions analyzed (subcellular, IEF, and total lysate) is given in Table 2. Overall, some 3,141 proteins from the 2-DE gels were identified by MALDI-TOF/MS from fractions summarized in Table 2. All of these 3,141 protein spots gave protein IDs with an expectation score of < 10−3. Of these, 426 proteins identified had unique non-redundant SwissProt identifiers and 231 proteins of the 426 were classified as novel proteins not previously reported to be expressed in eosinophils (Table 1). In general, the fractionation of proteins into the four commercially designated subcellular fractions shown using the ProteoExtract Subcellular Proteome Extraction procedure was quite useful even though some proteins did not distribute authentically; i.e., some proteins distributed correctly according to their known literature localization whereas other proteins did not. For example, many granular proteins (e.g., EPO, ECP, EDN, and MBP) distributed into the cytoskeletal fraction (F4) and most of the actin was in the nuclear fraction (F3). However, in general, the subcellular fractionation method principally proved valuable in reducing protein complexity and increasing low abundance proteins. Fig. 4 is a Western blot analysis that shows the distribution of eight randomly selected proteins into the four subcellular fractions (F1 to F4) demonstrating in part the effectiveness of the differential fractionation method.
Figure 1.
The proteome map of human peripheral blood eosinophils. Isoelective focusing was conducted in the pH range 3 to 10 in the first dimension. Major protein spots indicated by arrows are identified in Table 1. Protein vertical streak was due to galectin-10 insolubility (see Discussion). 200 µg of total eosinophil cell lysate was loaded to the gel.
Table 1.
List of identified proteins with significant SwissProt IDs
| ID No. |
Protein Name | Swiss Prot Access. no. a |
Theoret./ observ. pIb |
Theoret./ Observ. Mrc (kDa) |
Pept. Matc h (n) |
Seq. cov.d (%) |
Mascot expect. scoree |
Sub. cell. Fract. No. f |
|---|---|---|---|---|---|---|---|---|
| 1 | Actin capping protein | A4D0V4 | NDg | ND | ND | ND | 8.70E-19 | 6 |
| 2 | AH receptor-interacting protein | O00170 | 6.09/6.44 | 38.10/64.84 | 11 | 39 | 1.50E-06 | 2 |
| 3 | Hunc18b2 | O00184 | 4.40E-08 | 6 | ||||
| 4 | 26S proteasome non-ATPase regulatory subunit 11 |
O00231 | 6.09/6.39 | 47.72/85.60 | 4 | 10 | 4.90E-03 | 3 |
| 5 | Chloride intracellular channel | O00299 | 5.09/5.68 | 27.25/53.71 | 5 | 19 | 6.90E-04 | 5 |
| 6 | 26S proteasome non-ATPase regulatory subunit 14 |
O00487 | 6.06/6.34 | 34.73/68.86 | 8 | 35 | 2.70E-04 | 3 |
| 7 | Lysosomal alpha-mannosidase precursor | O00754 | 6.84/5.95 | 114.36/97.37 | 13 | 15 | 2.70E-28 | 4 |
| 8 | Proteasome subunit alpha type 7 | O14818 | 8.60/8.50 | 28.04/58.91 | 11 | 50 | 2.90E-16 | 5 |
| 9 | Ras-related protein Rab-7L1 | O14966 | 6.73/6.66 | 23.43/40.18 | 9 | 52 | 2.20E-03 | 3 |
| 10 | Actin-related protein 2/3 complex subunit 1B | O15143 | 8.69 | 41.72 | 6 | 22 | 9.98E-05 | 6 |
| 11 | Actin-related protein 2/3 complex subunit 2 | O15144 | 6.84/6.90 | 34.43/57.04 | 22 | 63 | 5.80E-48 | 3 |
| 12 | Actin-related protein 2/3 complex subunit 3 | O15145 | 8.82/8.49 | 20.76/28.61 | 7 | 35 | 1.90E-06 | 1 |
| 13 | Actin-related protein 2/3 complex subunit 5 | O15511 | 5.47/5.87 | 16.37/27.15 | 7 | 56 | 1.30E-17 | 3 |
| 14 | Thioredoxin-like protein 1 | O43396 | 4.84/5.50 | 32.63/68.45 | 11 | 50 | 1.30E-11 | 3 |
| 15 | Mitotic checkpoint protein | O43684 | 6.36/6.68 | 37.59/70.09 | 15 | 46 | 2.90E-19 | 2 |
| 16 | Alpha-actinin-4 | O43707 | 5.27/5.72 | 105.24/184.80 | 35 | 46 | 1.20E-29 | 3 |
| 17 | Keratin, type II cuticular Hb6 | O43790 | 5.56/6.24 | 55.12/215.17 | 22 | 48 | 2.70E-16 | 1 |
| 18 | Glia maturation factor gamma | O60234 | 5.18/5.48 | 16.96/28.19 | 11 | 58 | 1.50E-23 | 1 |
| 19 | Sorting nexin-3 | O60493 | 8.73/8.41 | 18.81/24.55 | 9 | 51 | 1.10E-04 | 5 |
| 20 | Docking protein 2 | O60496 | 6.58/5.93 | 45.75/84.31 | 16 | 32 | 2.10E-18 | 2 |
| 21 | Protein diaphanous homolog 1 | O60610 | ND | ND | ND | ND | 4.80E-03 | 6 |
| 22 | Mannose-6-phosphate receptor-binding protein 1 |
O60664 | ND | ND | ND | ND | 6.10E-11 | 6 |
| 23 | Histone H2B type 1-K | O60814 | 10.32/9.09 | 13.75/27.17 | 7 | 53 | 2.70E-15 | 3 |
| 24 | WD repeat protein 1 | O75083 | 6.18/6.60 | 66.84/126.16 | 16 | 40 | 4.30E-18 | 3 |
| 25 | Copine-3 | O75131 | 5.60/5.98 | 60.95/112.67 | 14 | 26 | 1.20E-13 | 1 |
| 26 | SH3 domain-binding glutamic acid-rich-like protein |
O75368 | 5.22/5.79 | 12.76/20.10 | 3 | 35 | 3.20E-04 | 1 |
| 27 | Citrate synthase, mitochondrial precursor | O75390 | 8.45/8.20 | 51.91/71.91 | 12 | 20 | 2.30E-34 | 4 |
| 28 | Protein CREG1 precursor | O75629 | 7.05/6.61 | 24.17/39.66 | 3 | 15 | 1.90E-03 | 4 |
| 29 | Protein XRP2 | O75695 | 5.00/5.41 | 40.47/73.37 | 12 | 30 | 3.60E-06 | 3 |
| 30 | 6-Phosphogluconolactonase | O95336 | 5.70/6.09 | 27.81/52.24 | 14 | 37 | 1.80E-35 | 1 |
| 31 | Ras-related protein Rab-3D | O95716 | 4.76/5.01 | 24.48/46.39 | 14 | 49 | 5.80E-13 | 3 |
| 32 | L-Lactate dehydrogenase A chain | P00338 | 8.44/6.11 | 36.95/106.74 | 14 | 33 | 2.10E-17 | 5 |
| 33 | Glutamate dehydrogenase 1, mitochondrial precursor |
P00367 | 7.66/7.01 | 61.70/95.99 | 14 | 29 | 2.90E-11 | 3 |
| 34 | Glutathione reductase, mitochondrial precursor |
P00390 | 8.74 | 56.79 | 14 | 34 | 1.26E-29 | 6 |
| 35 | Purine nucleoside phosphorylase | P00491 | 6.45/6.57 | 32.33/58.66 | 14 | 75 | 5.80E-45 | 5 |
| 37 | Carbonic anhydrase 1 | P00915 | 6.59/8.34 | 28.91/71.39 | 14 | 58 | 3.40E-28 | 4 |
| 38 | Carbonic anhydrase 2 | P00918 | 6.87/7.13 | 29.28/58.26 | 14 | 40 | 2.70E-12 | 1 |
| 39 | Alpha-1-antitrypsin precursor | P01009 | 5.37/5.40 | 46.88/89.30 | 14 | 21 | 1.20E-07 | 1 |
| 40 | Alpha-1-antichymotrypsin precursor | P01011 | 5.33/4.77 | 47.49/111.08 | 14 | 30 | 3.60E-09 | 3 |
| 41 | Hemoglobin subunit delta | P02042 | 7.85/8.67 | 16.16/16.71 | 14 | 95 | 8.50E-23 | 4 |
| 42 | Spectrin alpha chain, erythrocyte | P02549 | 4.96/5.37 | 280.88/219.26 | 14 | 15 | 1.30E-04 | 1 |
| 43 | Fibrinogen beta chain precursor | P02675 | 8.54 | 56.58 | 14 | 43 | 2.51E-14 | 6 |
| 44 | Transthyretin precursor | P02766 | 5.52/6.13 | 15.99/25.35 | 14 | 63 | 5.80E-13 | 3 |
| 45 | Serum albumin precursor | P02768 | 5.92/6.30 | 71.32/34.20 | 14 | 34 | 9.20E-28 | 2 |
| 46 | Lactotransferrin precursor | P02788 | 8.50/7.16 | 80.01/167.43 | 14 | 61 | 1.50E-89 | 3 |
| 47 | Ferritin light chain | P02792 | 5.51/5.97 | 20.06/93.00 | 14 | 46 | 1.30E-30 | 3 |
| 48 | Catalase | P04040 | 6.95/6.73 | 59.82/118.28 | 14 | 29 | 2.10E-64 | 6 |
| 49 | Fructose-bisphosphate aldolase A | P04075 | 8.39/8.45 | 39.85/73.95 | 14 | 36 | 1.10E-12 | 5 |
| 50 | Annexin A1 | P04083 | 6.64/6.39 | 38.92/66.84 | 14 | 70 | 2.10E-92 | 1 |
| 51 | Superoxide dismutase [Mn], mitochondrial precursor |
P04179 | 8.35/6.15 | 24.88/42.73 | 14 | 16 | 1.90E-03 | 4 |
| 52 | Keratin, type II cytoskeletal 1 | P04264 | 8.16/6.09 | 66.02/53.75 | 14 | 32 | 5.40E-49 | 5 |
| 53 | Glyceraldehyde-3-phosphate dehydrogenase |
P04406 | 8.58/8.52 | 36.21/57.25 | 14 | 57 | 8.50E-14 | 5 |
| 54 | Calpain small subunit 1 | P04632 | 5.05/5.29 | 28.47/51.56 | 14 | 30 | 7.30E-09 | 3 |
| 55 | Cytochrome b-245 heavy chain | P04839 | 8.90/7.20 | 66.21/26.85 | 14 | 17 | 1.30E-05 | 4 |
| 56 | Guanine nucleotide-binding protein G(i), alpha-2 subunit |
P04899 | ND | ND | ND | ND | 2.20E-45 | 6 |
| 57 | Aldehyde dehydrogenase, mitochondrial precursor |
P05091 | 6.63/7.06 | 56.86/62.30 | 15 | 30 | 2.30E-35 | 4 |
| 58 | Integrin beta-2 precursor | P05107 | 6.69/6.22 | 87.98/88.07 | 29 | 39 | 2.70E-23 | 4 |
| 59 | Protein S100-A8 | P05109 | 6.51/6.39 | 10.85/45.31 | 10 | 82 | 1.80E-20 | 5 |
| 60 | Myeloperoxidase precursor | P05164 | 9.19/9.75 | 84.78/90.76 | 23 | 31 | 3.60E-24 | 5 |
| 61 | Gelsolin precursor | P06396 | 5.90/6.32 | 86.04/131.40 | 21 | 29 | 1.50E-52 | 5 |
| 62 | ATP synthase subunit beta, mitochondrial precursor |
P06576 | 5.26/5.40 | 56.52/95.54 | 20 | 52 | 1.20E-59 | 3 |
| 63 | Protein S100-A9 | P06702 | 5.71/8.70 | 13.29/282.65 | 6 | 50 | 5.80E-22 | 5 |
| 64 | Alpha-enolase | P06733 | 6.99/5.82 | 47.48/145.56 | 18 | 44 | 8.50E-15 | 5 |
| 65 | Glycogen phosphorylase, liver form | P06737 | 6.71/6.84 | 97.49/184.17 | 32 | 41 | 6.80E-50 | 5 |
| 66 | Glucose-6-phosphate isomerase | P06744 | 8.43/8.23 | 63.34/101.91 | 25 | 51 | 2.10E-32 | 5 |
| 67 | Tropomyosin alpha-3 chain | P06753 | ND | ND | ND | ND | 3.00E-33 | 6 |
| 68 | L-Lactate dehydrogenase B chain | P07195 | 5.72/6.02 | 36.90/62.40 | 14 | 47 | 6.80E-24 | 1 |
| 69 | Glutathione peroxidase 1 | P07203 | 6.15/5.97 | 22.27/36.59 | 10 | 69 | 7.30E-11 | 2 |
| 70 | Protein disulfide-isomerase precursor | P07237 | 4.76/5.17 | 57.48/107.61 | 25 | 57 | 5.80E-79 | 3 |
| 71 | Cathepsin D precursor | P07339 | 6.10/5.98 | 45.04/52.24 | 14 | 40 | 2.90E-15 | 1 |
| 72 | Annexin A2 | P07355 | 7.57/7.62 | 38.81/72.03 | 19 | 49 | 2.70E-26 | 1 |
| 73 | Tubulin beta-2 chain | P07437 | 4.78/5.29 | 50.10/97.65 | 22 | 55 | 8.50E-28 | 3 |
| 74 | Beta-hexosaminidase beta chain precursor | P07686 | 6.29 | 63.53 | 11 | 19 | 9.98E-05 | 6 |
| 75 | Profilin-1 | P07737 | 8.48/8.53 | 15.22/25.17 | 9 | 62 | 4.30E-52 | 3 |
| 76 | Adenine phosphoribosyltransferase | P07741 | 5.78/5.66 | 19.77/34.90 | 6 | 32 | 5.40E-24 | 3 |
| 77 | Heat shock protein HSP 90-alpha | P07900 | ND | ND | ND | ND | 3.50E-42 | 6 |
| 78 | Heterogeneous nuclear ribonucleoproteins C1/C2 |
P07910 | 4.95/8.19 | 33.71/65.31 | 7 | 25 | 5.50E-03 | 2 |
| 79 | Heat shock 70 kDa protein 1 | P08107 | 5.48/6.18 | 70.29/78.53 | 17 | 26 | 2.30E-47 | 5 |
| 80 | Annexin A6 | P08133 | 5.42/6.02 | 76.17/132.77 | 41 | 65 | 6.80E-63 | 3 |
| 81 | Beta-glucuronidase precursor | P08236 | 6.54/6.94 | 75.01/132.90 | 9 | 24 | 8.00E-03 | 1 |
| 82 | Heat shock protein HSP 90-beta | P08238 | 4.97/5.38 | 83.55/154.23 | 24 | 35 | 1.10E-20 | 1 |
| 83 | Leukocyte elastase precursor | P08246 | 9.71/5.65 | 29.13/70.56 | 7 | 34 | 2.30E-07 | 3 |
| 84 | Glutathione S-transferase A1 | P08263 | ND | ND | ND | ND | 6.10E-18 | 6 |
| 85 | Vimentin | P08670 | 5.06/5.24 | 5.38/86.36 | 25 | 57 | 6.80E-29 | 5 |
| 86 | Guanine nucleotide-binding protein G(k) subunit alpha |
P08754 | ND | ND | ND | ND | 2.60E-03 | 6 |
| 87 | Annexin A5 | P08758 | 4.94/5.40 | 35.97/63.04 | 18 | 57 | 6.80E-79 | 3 |
| 88 | 40S Ribosomal protein SA | P08865 | 4.79/5.11 | 32.95/75.93 | 10 | 45 | 1.30E-14 | 3 |
| 89 | Glutathione S-transferase P | P09211 | 5.43/6.01 | 23.44/37.00 | 11 | 53 | 5.40E-53 | 1 |
| 90 | High mobility group protein B1 | P09429 | 5.62/6.56 | 25.05/57.42 | 11 | 46 | 1.10E-36 | 3 |
| 91 | Fructose-1,6-bisphosphatase 1 | P09467 | 6.54/6.75 | 37.19/74.75 | 20 | 54 | 1.30E-65 | 3 |
| 92 | Annexin A4 | P09525 | 5.84/5.97 | 36.09/61.14 | 22 | 63 | 6.80E-57 | 1 |
| 93 | Heterogeneous nuclear ribonucleoprotein A1 | P09651 | 9.26/9.08 | 38.81/51.48 | 15 | 46 | 3.40E-11 | 5 |
| 94 | U2 small nuclear ribonucleoprotein A | P09661 | 8.72/8.52 | 28.51/44.45 | 8 | 28 | 7.80E-04 | 6 |
| 95 | Leukotriene A-4 hydrolase | P09960 | 5.80/6.23 | 69.87/125.95 | 27 | 51 | 1.70E-34 | 1 |
| 96 | Histone H2A.Z | P0C0S5 | 10.58/5.63 | 35.67/25.67 | 2 | 12 | 4.40E-06 | 3 |
| 97 | Eosinophil-derived neurotoxin | P10153 | 9.10/8.89 | 18.86/28.78 | 5 | 22 | 2.30E-24 | 1 |
| 98 | Lysosomal alpha-glucosidase precursor | P10253 | 5.62/5.89 | 106.13/41.83 | 13 | 15 | 1.00E-03 | 4 |
| 99 | Thioredoxin | P10599 | 4.82/5.31 | 12.01/17.70 | 8 | 55 | 5.40E-11 | 1 |
| 100 | Lysosomal protective protein precursor | P10619 | 6.16/5.20 | 54.94/55.37 | 8 | 14 | 9.20E-13 | 5 |
| 101 | Esterase D | P10768 | 6.54/6.57 | 31.96/56.58 | 11 | 49 | 7.30E-14 | 1 |
| 102 | 78 kDa glucose-regulated protein precursor | P11021 | ND | ND | ND | ND | 3.00E-38 | 6 |
| 103 | Heat shock cognate 71 kDa protein | P11142 | 5.37/5.85 | 71.08/142.38 | 22 | 35 | 9.20E-62 | 3 |
| 104 | Integrin alpha-M precursor | P11215 | 6.88/6.85 | 128.41/234.21 | 30 | 56 | 1.83E-21 | 4 |
| 105 | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor |
P11310 | 8.61/6.84 | 47.01/80.06 | 13 | 35 | 7.30E-11 | 3 |
| 106 | Glucose-6-phosphate 1-dehydrogenase | P11413 | 6.39/6.88 | 59.68/112.14 | 32 | 70 | 2.90E-28 | 3 |
| 107 | Eosinophil peroxidase precursor | P11678 | 10.31/7.62 | 81.96/113.08 | 13 | 18 | 4.60E-07 | 6 |
| 108 | Proliferating cell nuclear antigen | P12004 | 4.57/5.12 | 29.09/64.84 | 7 | 31 | 3.80E-05 | 3 |
| 109 | Annexin A3 | P12429 | 5.63/8.74 | 36.52/68.97 | 10 | 36 | 1.10E-37 | 4 |
| 110 | Eosinophil cationic protein precursor | P12724 | 10.31/9.53 | 18.94/35.5 | 7 | 42 | 1.20E-20 | 3 |
| 111 | Alpha-actinin-1 | P12814 | 5.25 | 103.56 | 25 | 35 | 1.50E-14 | 2 |
| 112 | Myosin heavy chain, cardiac muscle beta isoform |
P12883 | 5.63/3.60 | 223.76/32.64 | 17 | 13 | 3.00E-03 | 6 |
| 113 | ATP-dependent DNA helicase 2 subunit 1 | P12956 | 6.23/6.59 | 70.08/135.42 | 22 | 49 | 8.50E-42 | 3 |
| 114 | Ribonuclease inhibitor | P13489 | 4.71/5.11 | 51.77/85.58 | 21 | 62 | 5.40E-56 | 1 |
| 115 | Elongation factor 2 | P13639 | ND | ND | ND | ND | 1.40E-16 | 6 |
| 116 | Keratin, type I cytoskeletal 10 | P13645 | 5.13/7.93 | 59.711/50.00 | 19 | 29 | 7.30E-33 | 3 |
| 117 | Protein disulfide-isomerase A4 precursor | P13667 | 4.96/5.51 | 73.23/143.51 | 34 | 48 | 5.80E-46 | 3 |
| 118 | Translationally-controlled tumor protein | P13693 | 4.84/5.25 | 19.70/39.23 | 10 | 47 | 1.50E-19 | 1 |
| 119 | Delta-aminolevulinic acid dehydratase | P13716 | 6.32/6.57 | 36.73/70.58 | 11 | 34 | 5.50E-09 | 3 |
| 120 | Bone marrow proteoglycan precursor | P13727 | 6.23/9.39 | 25.90/20.82 | 6 | 26 | 1.10E-14 | 1 |
| 121 | Plastin-2 (L-plastin) | P13796 | 5.20/5.63 | 70.82/121.32 | 30 | 57 | 5.40E-56 | 1 |
| 122 | Acylamino-acid-releasing enzyme | P13798 | 5.29/5.71 | 82.14/152.88 | 19 | 31 | 4.60E-18 | 3 |
| 123 | Macrophage migration inhibitory factor | P14174 | 8.24/7.90 | 12.64/18.94 | 5 | 24 | 2.70E-11 | 1 |
| 124 | Hematopoietic lineage cell-specific protein | P14317 | 4.74/7.11 | 50.08/73.33 | 8 | 17 | 2.20E-04 | 6 |
| 125 | Farnesyl pyrophosphate synthetase | P14324 | ND | ND | ND | ND | 1.70E-08 | 6 |
| 126 | Alcohol dehydrogenase [NADP+] | P14550 | 6.32/6.66 | 36.89/71.94 | 18 | 55 | 3.40E-40 | 1 |
| 127 | Neutrophil cytosol factor 1 | P14598 | 9.12/7.87 | 44.88/101.38 | 11 | 31 | 7.30E-21 | 6 |
| 128 | Pyruvate kinase isozymes M1/M2 | P14618 | 7.96/7.90 | 58.47/104.26 | 19 | 42 | 5.40E-13 | 1 |
| 129 | Endoplasmin precursor | P14625 | 4.76/5.21 | 92.67/180.61 | 28 | 38 | 3.60E-56 | 3 |
| 130 | Heterogeneous nuclear ribonucleoprotein L | P14866 | 6.65/6.86 | 60.72/122.75 | 12 | 27 | 4.60E-08 | 3 |
| 131 | Aspartyl-tRNA synthetase, cytoplasmic | P14868 | 6.11 | 57.5 | 22 | 44 | 9.98E-24 | 6 |
| 132 | Ras-related C3 botulinum toxin substrate 2 precursor |
P15153 | 7.52/7.87 | 21.81/37.71 | 8 | 39 | 3.60E-44 | 1 |
| 133 | Ezrin | P15311 | 5.94 | 69.48 | 20 | 30 | 3.15E-19 | 6 |
| 134 | Nucleoside diphosphate kinase A | P15531 | 5.83/6.16 | 17.31/29.25 | 6 | 42 | 1.80E-16 | 1 |
| 135 | Arachidonate 15-lipoxygenase | P16050 | 6.14/6.58 | 75.50/114.56 | 21 | 40 | 3.40E-19 | 1 |
| 136 | Histone H2A.x | P16104 | 10.74/5.55 | 15.14/27.56 | 4 | 48 | 5.80E-04 | 3 |
| 137 | Carbonyl reductase [NADPH] 1 | P16152 | 8.55/9.67 | 30.64/49.27 | 8 | 41 | 2.00E-04 | 6 |
| 138 | Beta-galactosidase-related protein precursor | P16279 | 6.5/6.23 | 60.86/123.48 | 11 | 24 | 4.60E-11 | 3 |
| 139 | Stathmin | P16949 | 5.76/6.01 | 17.29/28.28 | 9 | 42 | 2.10E-26 | 3 |
| 140 | Galectin-3 | P17931 | 8.61/8.20 | 26.23/57.44 | 5 | 22 | 4.30E-03 | 1 |
| 141 | T-complex protein 1 subunit alpha | P17987 | ND | ND | ND | ND | 3.90E-04 | 6 |
| 142 | Vinculin | P18206 | 5.50/6.47 | 124.29/176.11 | 38 | 36 | 1.30E-32 | 1 |
| 143 | Phosphoglycerate mutase 1 | P18669 | 6.67/6.54 | 28.90/53.87 | 14 | 56 | 4.30E-31 | 1 |
| 144 | Myosin regulatory light chain 2, nonsarcomeric |
P19105 | 4.67/5.04 | 19.84/30.71 | 9 | 63 | 1.70E-23 | 2 |
| 145 | Neutrophil cytosol factor 2 | P19878 | 5.88/6.22 | 60.24/119.95 | 21 | 42 | 9.20E-23 | 1 |
| 146 | Annexin A7 | P20073 | 5.52/6.26 | 52.99/86.00 | 10 | 23 | 2.30E-09 | 1 |
| 147 | Azurocidin precursor | P20160 | 9.75/9.47 | 27.32/57.23 | 5 | 30 | 1.70E-13 | 3 |
| 148 | Proteasome subunit beta type 1 | P20618 | 8.27/7.84 | 26.70/43.24 | 10 | 52 | 2.90E-17 | 3 |
| 149 | Lamin-B1 | P20700 | ND | ND | ND | ND | 1.70E-21 | 6 |
| 150 | Vacuolar ATP synthase subunit B, brain isoform |
P21281 | 5.57/6.03 | 56.81/101.80 | 10 | 25 | 1.80E-08 | 3 |
| 151 | Iron-responsive element-binding protein 1 | P21399 | ND | ND | ND | ND | 1.40E-06 | 6 |
| 152 | Voltage-dependent anion-selective channel protein 1 |
P21796 | 8.62/5.30 | 30.87/51.41 | 9 | 41 | 2.10E-19 | 4 |
| 153 | Ubiquitin-like modifier-activating enzyme 1 | P22314 | 5.49/5.74 | 118.86/171.81 | 29 | 40 | 7.30E-30 | 1 |
| 154 | Nucleoside diphosphate kinase B | P22392 | 8.52/8.39 | 17.40/26.76 | 13 | 87 | 2.30E-28 | 1 |
| 155 | Heterogeneous nuclear ribonucleoproteins A2/B1 |
P22626 | 8.97/7.79 | 37.46/45.38 | 16 | 42 | 3.60E-30 | 3 |
| 156 | Cytochrome b-c1 complex subunit 2, mitochondrial precursor |
P22695 | 8.74/7.67 | 48.58/81.54 | 9 | 30 | 4.60E-06 | 3 |
| 157 | Liver carboxylesterase 1 precursor | P23141 | 6.15/6.11 | 62.77/95.84 | 13 | 26 | 2.80E-05 | 4 |
| 158 | Splicing factor, proline- and glutamine-rich | P23246 | 9.45/7.89 | 76.21/228.51 | 9 | 17 | 2.90E-20 | 6 |
| 159 | Peptidyl-prolyl cis-trans isomerase B precursor |
P23284 | 9.33/9.23 | 22.79/25.86 | 7 | 41 | 1.80E-09 | 6 |
| 160 | Tryptophanyl-tRNA synthetase | P23381 | 5.83/6.25 | 53.47/98.77 | 18 | 50 | 4.30E-24 | 1 |
| 161 | Adenosylhomocysteinase | P23526 | 5.92/6.28 | 48.26/80.14 | 20 | 43 | 2.10E-36 | 1 |
| 162 | Cofilin-1 | P23528 | 8.22/8.13 | 18.72/28.60 | 10 | 65 | 3.40E-26 | 1 |
| 163 | Myeloblastin precursor | P24158 | 8.72/9.04 | 28.25/25.26 | 3 | 11 | 4.60E-14 | 4 |
| 164 | Proteasome subunit alpha type 1 | P25786 | 6.15/6.45 | 29.82/61.79 | 10 | 47 | 2.90E-16 | 3 |
| 165 | Proteasome subunit alpha type 2 | P25787 | 6.92/6.82 | 26.00/45.64 | 13 | 61 | 1.70E-34 | 3 |
| 166 | Proteasome subunit alpha type 4 | P25789 | 7.57/7.57 | 29.75/59.21 | 13 | 59 | 4.60E-33 | 3 |
| 167 | Moesin | P26038 | 6.08/6.51 | 67.89/150.87 | 30 | 50 | 4.30E-51 | 3 |
| 168 | Protein S100-A4 | P26447 | 5.85/5.87 | 11.95/13.94 | 6 | 36 | 2.90E-17 | 1 |
| 169 | Elongation factor 1-gamma | P26641 | 6.25 | 50.43 | 15 | 33 | 3.97E-36 | 6 |
| 170 | Annexin A13 | P27216 | 5.47/5.71 | 35.54/65.47 | 8 | 30 | 3.00E-04 | 3 |
| 171 | 14-3-3 protein theta | P27348 | ND | ND | ND | ND | 1.50E-08 | 6 |
| 172 | Replication protein A 70 kDa | P27694 | 6.92/6.93 | 68.72/144.08 | 14 | 30 | 5.80E-07 | 3 |
| 173 | Calreticulin precursor | P27797 | 4.29/4.77 | 48.28/134.58 | 14 | 44 | 4.60E-40 | 3 |
| 174 | Histone H2A type 1-E | P28001 | 11.05/6.25 | 14.10/26.03 | 7 | 43 | 3.40E-20 | 3 |
| 175 | Proteasome subunit beta type 8 precursor | P28062 | 7.63/8.11 | 30.68/56.66 | 10 | 29 | 2.30E-30 | 3 |
| 176 | Proteasome subunit beta type 9 precursor | P28065 | 4.93/5.25 | 23.36/38.82 | 7 | 40 | 3.80E-05 | 3 |
| 177 | Proteasome subunit alpha type 5 | P28066 | 4.74/5.18 | 26.57/55.00 | 8 | 43 | 3.50E-06 | 3 |
| 178 | Proteasome subunit beta type 4 precursor | P28070 | 5.72/5.93 | 29.23/51.19 | 7 | 37 | 1.50E-16 | 3 |
| 179 | Mitogen-activated protein kinase 1 | P28482 | 6.53/6.73 | 41.76/76.16 | 5 | 15 | 6.80E-08 | 3 |
| 180 | Grancalcin | P28676 | 5.02/5.23 | 24.22/47.52 | 10 | 52 | 2.90E-18 | 3 |
| 181 | Tyrosine-protein phosphatase non-receptor type 6 |
P29350 | 7.65/6.32 | 67.92/101.19 | 15 | 33 | 7.30E-34 | 3 |
| 182 | Transketolase | P29401 | 7.58/7.41 | 68.52/144.42 | 14 | 25 | 2.30E-58 | 6 |
| 183 | Endoplasmic reticulum protein ERp29 precursor |
P30040 | 6.77/6.53 | 29.03/61.97 | 11 | 36 | 7.30E-41 | 3 |
| 184 | Peroxiredoxin-6 | P30041 | 6.02/6.49 | 25.13/40.62 | 5 | 21 | 3.00E-04 | 1 |
| 185 | Flavin reductase | P30043 | 7.13/7.48 | 22.22/42.00 | 10 | 67 | 1.30E-30 | 1 |
| 186 | Peroxiredoxin-5, mitochondrial precursor | P30044 | 8.85/7.04 | 23.00/27.77 | 11 | 49 | 4.60E-26 | 1 |
| 187 | Thioredoxin-dependent peroxide reductase, mitochondrial precursor |
P30048 | 7.67/6.06 | 28.02/35.25 | 5 | 21 | 5.80E-21 | 2 |
| 188 | UMP-CMP kinase | P30085 | 5.44/5.82 | 22.44/37.00 | 9 | 58 | 2.90E-08 | 1 |
| 189 | Phosphatidylethanolamine-binding protein 1 | P30086 | 7.42/7.63 | 21.16/35.70 | 13 | 67 | 6.80E-34 | 1 |
| 190 | Protein disulfide-isomerase A3 precursor | P30101 | 5.98/5.95 | 57.46/108.29 | 26 | 49 | 1.80E-60 | 3 |
| 191 | Ser/Thr-protein phosphatase 2A 65 kDa | P30153 | 4.96/5.51 | 66.07/121.85 | 18 | 35 | 1.10E-16 | 3 |
| 192 | Ser/Thr Sorcin | P30626 | 5.32/5.49 | 21.95/33.15 | 7 | 40 | 3.60E-11 | 2 |
| 193 | Ser/Thr Leukocyte elastase inhibitor | P30740 | 5.90/6.20 | 42.83/74.25 | 13 | 38 | 1.30E-05 | 2 |
| 194 | Succinate dehydrogenase mitochondrial precursor |
P31040 | 7.06/5.83 | 73.67/33.63 | 12 | 19 | 1.10E-05 | 4 |
| 195 | Coronin-1A | P31146 | 6.25/6.76 | 51.68/98.51 | 14 | 32 | 5.40E-08 | 1 |
| 196 | Rab GDP dissociation inhibitor alpha | P31150 | 5.00/5.37 | 51.12/113.09 | 22 | 55 | 4.60E-22 | 3 |
| 197 | Heterogeneous nuclear ribonucleoprotein H3 | P31942 | 6.37/5.73 | 36.96/69.00 | 6 | 26 | 1.80E-14 | 6 |
| 198 | Heterogeneous nuclear ribonucleoprotein H | P31943 | 5.89 | 49.48 | 10 | 33 | 3.15E-09 | 6 |
| 199 | 14-3-3 protein beta/alpha | P31946 | 4.76/4.09 | 28.05/59.13 | 8 | 32 | 5.60E-03 | 6 |
| 200 | Stress-induced-phosphoprotein 1 | P31948 | ND | ND | ND | ND | 1.40E-04 | 6 |
| 201 | Protein S100-A11 | P31949 | 6.51/7.33 | 10.88/15.94 | 9 | 60 | 5.40E-31 | 6 |
| 202 | Peroxiredoxin-2 | P32119 | 5.68/5.46 | 22.65/36.44 | 7 | 27 | 3.40E-22 | 2 |
| 203 | Cytidine deaminase | P32320 | 6.55/6.16 | 16.69/24.75 | 5 | 48 | 6.80E-06 | 2 |
| 204 | N-acetylgalactosamine-6-sulfatase precursor | P34059 | 6.25/6.42 | 58.45/135.28 | 13 | 24 | 5.40E-26 | 4 |
| 205 | Heat shock 70 kDa protein 4 | P34932 | 5.18/5.22 | 95.10/175.14 | 17 | 20 | 3.60E-07 | 3 |
| 206 | Prohibitin | P35232 | 5.57/7.38 | 29.84/61.79 | 5 | 24 | 4.20E-03 | 4 |
| 207 | Keratin, type I cytoskeletal 9 | P35527 | 5.19/5.72 | 62.32/122.35 | 15 | 35 | 1.20E-14 | 3 |
| 208 | Phosphoenolpyruvate carboxykinase, cytosolic [GTP] |
P35558 | ND | ND | ND | ND | 8.70E-45 | 6 |
| 209 | Myosin-9 | P35579 | 5.50/5.51 | 227.65/214.11 | 33 | 23 | 2.10E-12 | 2 |
| 210 | Myosin-11 | P35749 | 5.42/5.77 | 228.05/131.40 | 25 | 16 | 3.10E-04 | 2 |
| 211 | Keratin, type II cytoskeletal 2 epidermal | P35908 | 8.07/6.14 | 66.11/43.76 | 12 | 22 | 2.90E-12 | 1 |
| 212 | Phosphoglucomutase-1 | P36871 | 6.32/6.48 | 61.56/105.99 | 21 | 43 | 3.60E-20 | 1 |
| 213 | Phospholipid hydroperoxide glutathione peroxidase, mitochondrial precursor |
P36969 | 8.64/7.50 | 22.68/29.12 | 11 | 60 | 9.20E-20 | 1 |
| 214 | Hippocalcin-like protein 1 | P37235 | 5.21/5.36 | 22.41/31.71 | 6 | 32 | 2.10E-05 | 1 |
| 215 | Transgelin-2 | P37802 | 8.41/8.07 | 22.55/35.68 | 17 | 75 | 5.40E-35 | 1 |
| 216 | Transaldolase | P37837 | 6.36/5.89 | 37.69/63.54 | 18 | 45 | 2.30E-38 | 4 |
| 217 | Vacuolar ATP synthase catalytic subunit A, ubiquitous isoform |
P38606 | 5.35/5.72 | 68.66/137.78 | 15 | 32 | 6.80E-09 | 3 |
| 218 | Stress-70 protein, mitochondrial precursor | P38646 | 5.87/5.80 | 73.92/131.24 | 18 | 32 | 2.90E-37 | 3 |
| 219 | Eukaryotic initiation factor 4A–III | P38919 | 6.30/6.43 | 47.13/88.36 | 7 | 14 | 3.40E-12 | 3 |
| 220 | Acidic leucine-rich nuclear phosphoprotein 32 | P39687 | 3.99/4.48 | 28.68/56.85 | 9 | 31 | 1.80E-18 | 3 |
| 221 | Macrophage capping protein | P40121 | 5.88/6.17 | 38.78/75.25 | 11 | 43 | 2.30E-07 | 1 |
| 222 | Malate dehydrogenase, cytoplasmic | P40925 | ND | ND | ND | ND | 6.90E-15 | 6 |
| 223 | Malate dehydrogenase, mitochondrial precursor |
P40926 | 8.92/9.11 | 35.96/61.23 | 13 | 50 | 1.50E-14 | 6 |
| 224 | Myeloid cell nuclear differentiation antigen | P41218 | 9.77/7.92 | 46.09/111.49 | 8 | 23 | 1.80E-19 | 6 |
| 225 | Tyrosine-protein kinase CSK | P41240 | 6.62/6.92 | 51.24/99.43 | 15 | 44 | 2.90E-10 | 3 |
| 226 | Caspase-3 precursor | P42574 | 6.09/6.50 | 32.04/56.82 | 19 | 63 | 5.80E-21 | 2 |
| 227 | Lysosomal Pro-X carboxypeptidase precursor | P42785 | 6.75/6.47 | 56.28/109.73 | 14 | 26 | 2.30E-21 | 4 |
| 228 | Platelet-activating factor acetylhydrolase IB subunit alpha |
P43034 | 6.97 | 47.18 | 13 | 34 | 9.98E-25 | 6 |
| 229 | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor |
P43304 | 7.23 | 81.3 | 20 | 26 | 3.97E-14 | 6 |
| 230 | 26S protease regulatory subunit 6B | P43686 | ND | ND | ND | ND | 4.00E-04 | 6 |
| 231 | Ubiquitin carboxyl-terminal hydrolase 5 | P45974 | 4.91/5.42 | 96.64/168.41 | 19 | 25 | 3.60E-07 | 1 |
| 232 | Crk-like protein | P46109 | 6.26/6.47 | 33.87/67.18 | 14 | 57 | 7.30E-11 | 2 |
| 233 | Vesicle-fusing ATPase | P46459 | 6.52 | 83.02 | 17 | 24 | 1.26E-07 | 6 |
| 234 | F-actin capping protein subunit beta | P47756 | 5.36/5.88 | 31.62/57.61 | 19 | 65 | 2.10E-27 | 2 |
| 235 | 26S proteasome non-ATPase regulatory subunit 8 |
P48556 | 6.85/6.51 | 30.16/52.13 | 11 | 33 | 4.30E-20 | 2 |
| 236 | Serpin B10 | P48595 | 5.80/5.97 | 45.49/241.04 | 21 | 59 | 3.60E-42 | 1 |
| 237 | Glutathione synthetase | P48637 | ND | ND | ND | ND | 5.50E-15 | 6 |
| 238 | T-complex protein 1 subunit epsilon | P48643 | ND | ND | ND | ND | 3.00E-04 | 6 |
| 239 | Keratin, type II cytoskeletal 6E | P48668 | 8.14/6.99 | 60.27/44.52 | 16 | 29 | 2.10E-09 | 2 |
| 240 | Isocitrate dehydrogenase [NADP], mitochondrial precursor |
P48735 | 8.88/6.53 | 51.33/116.03 | 20 | 47 | 1.50E-19 | 4 |
| 241 | CD97 antigen precursor | P48960 | 6.50/5.39 | 94.60/133.76 | 7 | 9 | 9.20E-07 | 2 |
| 242 | Calcium signal-modulating cyclophilin ligand | P49069 | ND | ND | ND | ND | 9.30E-06 | 6 |
| 243 | Ribose-5-phosphate isomerase | P49247 | 8.78/7.18 | 33.53/54.80 | 8 | 27 | 4.30E-11 | 1 |
| 244 | T-complex protein 1 subunit gamma | P49368 | 6.10/6.42 | 61.07/109.15 | 16 | 33 | 4.60E-12 | 2 |
| 245 | Elongation factor Tu, mitochondrial precursor |
P49411 | 7.26 | 49.85 | 8 | 23 | 9.98E-06 | 6 |
| 246 | Proteasome subunit beta type 3 | P49720 | 6.14/6.31 | 23.22/48.25 | 11 | 50 | 1.50E-23 | 3 |
| 247 | Proteasome subunit beta type 2 | P49721 | 6.51 | 22.99 | 10 | 46 | 2.51E-19 | 6 |
| 248 | Rab GDP dissociation inhibitor beta | P50395 | 6.11/6.37 | 51.09/82.02 | 30 | 73 | 9.20E-59 | 1 |
| 249 | Vasodilator-stimulated phosphoprotein | P50552 | 9.05/6.34 | 39.98/56.07 | 8 | 23 | 8.50E-11 | 4 |
| 250 | Dynamin-2 | P50570 | 7.04/6.38 | 98.45/131.31 | 24 | 29 | 1.80E-18 | 3 |
| 251 | Annexin A11 | P50995 | 7.53/8.48 | 54.68 | 24 | 41 | 1.50E-40 | 4 |
| 252 | Ras-related protein Rab-5C | P51148 | 8.64/5.23 | 23.70/56.07 | 9 | 44 | 4.40E-06 | 4 |
| 253 | Ras-related protein Rab-7 | P51149 | 6.4 | 23.76 | 12 | 66 | 6.80E-37 | 3 |
| 254 | Ras-related protein Rab-27A | P51159 | 5.19/5.50 | 25.13/52.12 | 9 | 35 | 4.60E-10 | 2 |
| 255 | Galactokinase | P51570 | 6.04/5.90 | 42.70/77.82 | 11 | 28 | 1.50E-38 | 6 |
| 256 | Heterogeneous nuclear ribonucleoprotein A3 |
P51991 | 9.10/9.66 | 39.80/63.97 | 11 | 31 | 2.80E-04 | 6 |
| 257 | 6-phosphogluconate dehydrogenase | P52209 | 6.80/7.07 | 53.62/83.71 | 21 | 46 | 6.80E-51 | 1 |
| 258 | Heterogeneous nuclear ribonucleoprotein M | P52272 | 8.85/8.12 | 7762/79.83 | 16 | 26 | 9.30E-04 | 6 |
| 259 | Rho GDP-dissociation inhibitor 1 | P52565 | 5.03/5.43 | 23.25/51.19 | 10 | 43 | 1.10E-32 | 3 |
| 260 | Rho GDP-dissociation inhibitor 2 | P52566 | 5.10/5.61 | 23.03/49.74 | 12 | 80 | 6.80E-19 | 3 |
| 261 | Hexokinase-3 | P52790 | 5.27/5.57 | 100.51/166.43 | 20 | 26 | 2.30E-21 | 1 |
| 262 | F-actin capping protein alpha-1 subunit | P52907 | 5.45/5.74 | 33.07/64.26 | 16 | 69 | 1.70E-15 | 2 |
| 263 | Biliverdin reductase A precursor | P53004 | 6.06/6.35 | 33.69/69.46 | 13 | 42 | 5.80E-34 | 1 |
| 264 | ATP-citrate synthase | P53396 | 6.95/7.17 | 121.66/214.05 | 34 | 39 | 9.20E-30 | 3 |
| 265 | Dipeptidyl-peptidase 1 precursor | P53634 | 6.54/5.37 | 52.61/21.11 | 5 | 10 | 8.70E-05 | 3 |
| 266 | Tyrosyl-tRNA synthetase, cytoplasmic | P54577 | 6.64/6.84 | 56.45/116.20 | 13 | 27 | 5.40E-10 | 3 |
| 267 | Adenylate kinase isoenzyme 2, mitochondrial |
P54819 | 7.85/8.25 | 26.69/55.06 | 9 | 41 | 8.50E-11 | 4 |
| 268 | Alpha-soluble NSF attachment protein | P54920 | 5.23/5.57 | 33.68/63.46 | 12 | 44 | 4.60E-12 | 3 |
| 269 | Transitional endoplasmic reticulum ATPase | P55072 | ND | ND | ND | ND | 6.60E-05 | 6 |
| 270 | Histone H2B type 1-D | P58876 | 10.32/9.56 | 13.98/27.17 | 7 | 52 | 1.10E-11 | 3 |
| 271 | Neutrophil defensin 1 precursor | P59665 | 6.54/8.33 | 10.54/55.89 | 5 | 26 | 4.60E-17 | 4 |
| 272 | Actin-related protein 2/3 complex subunit 4 | P59998 | 8.53/8.48 | 19.67/31.69 | 10 | 44 | 3.60E-16 | 3 |
| 273 | Triosephosphate isomerase | P60174 | 6.45/6.82 | 26.94/47.85 | 16 | 70 | 2.30E-42 | 1 |
| 274 | Myosin light polypeptide 6 | P60660 | 4.56/3.51 | 16.96/18.16 | 10 | 62 | 4.30E-24 | 6 |
| 275 | Actin, cytoplasmic 1 | P60709 | 5.29/5.8 | 42.02/63.77 | 6 | 23 | 7.30E-21 | 2 |
| 276 | Eukaryotic initiation factor 4A–I | P60842 | 5.32/5.8 | 46.35/92.56 | 18 | 50 | 1.20E-09 | 3 |
| 277 | Ribose-phosphate pyrophosphokinase I | P60891 | 6.56/6.94 | 35.33/68.86 | 8 | 28 | 1.70E-07 | 3 |
| 278 | Proteasome subunit alpha type 6 | P60900 | 6.34/6.47 | 27.84/50.00 | 7 | 31 | 2.90E-09 | 3 |
| 279 | Cell division control protein 42 homolog precursor |
P60953 | 5.76/6.85 | 21.70/34.13 | 6 | 29 | 9.20E-15 | 3 |
| 280 | Destrin | P60981 | 8.06/7.95 | 18.95/28.60 | 1 | 6 | 2.30E-03 | 3 |
| 281 | Ras-related protein Rab-2A | P61019 | 6.08/5.83 | 23.70/47.74 | 9 | 50 | 5.80E-11 | 3 |
| 282 | Ubiquitin-conjugating enzyme E2 N | P61088 | 6.13/6.22 | 17.18/25.71 | 11 | 57 | 9.20E-15 | 1 |
| 283 | Actin-related protein 3 | P61158 | 5.61/6.12 | 47.67/84.00 | 17 | 48 | 6.80E-40 | 1 |
| 284 | Actin-like protein 2 | P61160 | 6.30/6.58 | 45.02/79.74 | 17 | 50 | 3.60E-17 | 3 |
| 285 | Alpha-centractin | P61163 | 6.19 | 42.7 | 12 | 37 | 1.80E-16 | 2 |
| 286 | ADP-ribosylation factor 3 | P61204 | 6.84/7.07 | 20.46/56.97 | 11 | 69 | 5.40E-16 | 4 |
| 287 | Ras-related protein Rap-1b precursor | P61224 | 5.65/5.82 | 21.04/57.68 | 8 | 43 | 1.50E-10 | 4 |
| 288 | Transforming protein RhoA precursor | P61586 | 5.83/6.34 | 22.10/42.90 | 8 | 39 | 2.30E-11 | 1 |
| 289 | 10 kDa heat shock protein, mitochondrial | P61604 | 8.91/6.94 | 10.92/169.95 | 9 | 66 | 6.80E-26 | 4 |
| 290 | Lysozyme C precursor | P61626 | 9.38/8.95 | 16.98/23.91 | 6 | 38 | 7.30E-12 | 1 |
| 291 | Beta-2-microglobulin precursor | P61769 | 6.06/6.43 | 13.82/28.62 | 3 | 37 | 3.40E-04 | 4 |
| 292 | Heterogeneous nuclear ribonucleoprotein K | P61978 | 5.39/5.88 | 51.30/107.85 | 14 | 34 | 1.80E-12 | 2 |
| 293 | 14-3-3 protein gamma | P61981 | 4.8 | 28.46 | 9 | 36 | 3.40E-10 | 2 |
| 294 | Ser/Thr protein phosphatase alpha catalytic subunit |
P62136 | 5.94/6.15 | 38.23/71.15 | 14 | 32 | 7.30E-18 | 3 |
| 295 | Ser/Thr protein phosphatase beta catalytic subunit |
P62140 | 5.84/6.18 | 37.69/72.20 | 17 | 58 | 1.30E-12 | 3 |
| 296 | Calmodulin | P62158 | 4.09/3.38 | 16.70/23.48 | 5 | 45 | 1.10E-19 | 6 |
| 297 | 14-3-3 protein epsilon | P62258 | 4.63 | 29.33 | 13 | 53 | 2.30E-18 | 3 |
| 298 | 26S protease regulatory subunit S10B | P62333 | 7.1 | 44.43 | 12 | 35 | 5.00E-09 | 6 |
| 299 | Ras-related protein Rab-11A | P62491 | 3.12/8.88 | 24.49/28.69 | 9 | 33 | 4.90E-06 | 4 |
| 300 | Histone H4 | P62805 | 11.36/9.50 | 11.36/24.82 | 7 | 56 | 3.40E-33 | 3 |
| 301 | Histone H2B type 1-C/E/F/G/I | P62807 | 10.31/8.47 | 13.98/26.16 | 9 | 61 | 4.30E-21 | 3 |
| 302 | GTP-binding nuclear protein Ran | P62826 | 7.01/7.14 | 24.57/43.15 | 15 | 56 | 1.70E-28 | 1 |
| 303 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta 2 |
P62879 | 5.60/5.87 | 38.05/67.26 | 8 | 32 | 2.00E-03 | 3 |
| 304 | Peptidyl-prolyl cis-trans isomerase A | P62937 | 7.68/8.09 | 18.23/27.67 | 11 | 61 | 4.30E-46 | 1 |
| 305 | FK506-binding protein 1A | P62942 | 7.88/8.05 | 12.00/20.77 | 6 | 50 | 6.80E-12 | 1 |
| 306 | Ubiquitin | P62988 | 6.56/6.97 | 8.56/23.81 | 6 | 61 | 2.30E-24 | 3 |
| 307 | Growth factor receptor-bound protein 2 | P62993 | 5.89/6.2 | 25.30/47.79 | 15 | 61 | 9.20E-32 | 2 |
| 308 | 14-3-3 protein zeta/delta | P63104 | 4.73/5.31 | 27.90/53.92 | 14 | 56 | 2.30E-42 | 3 |
| 309 | Eukaryotic translation initiation factor 5A-1 | P63241 | 5.08/5.63 | 17.05/29.02 | 6 | 45 | 6.00E-04 | 3 |
| 310 | Guanine nucleotide-binding protein subunit beta 2-like 1 |
P63244 | 7.56/7.56 | 35.51/63.97 | 18 | 70 | 4.30E-33 | 3 |
| 311 | Actin, cytoplasmic 2 | P63261 | 5.31/5.80 | 42.11/76.93 | 17 | 46 | 3.60E-57 | 1 |
| 312 | Ser/Thr protein phosphatase 2A catalytic subunit alpha isoform |
P67775 | 5.30/5.76 | 36.14/70.58 | 10 | 45 | 3.30E-06 | 3 |
| 313 | Elongation factor 1-alpha 1 | P68104 | 9.10/7.87 | 50.45/106.51 | 7 | 19 | 1.50E-19 | 6 |
| 314 | Tubulin alpha-ubiquitous chain | P68363 | 4.94/5.99 | 50.80/87.80 | 13 | 39 | 4.60E-07 | 2 |
| 315 | Tubulin beta-2C chain | P68371 | 4.79/5.59 | 50.26/95.91 | 18 | 38 | 1.50E-29 | 3 |
| 316 | Histone H3.1 | P68431 | 11.27/8.58 | 15.44/27.15 | 4 | 20 | 2.70E-07 | 3 |
| 317 | Hemoglobin subunit beta | P68871 | 6.75/7.05 | 16.10/19.88 | 13 | 89 | 4.30E-79 | 1 |
| 318 | Hemoglobin subunit alpha | P69905 | 8.72/8.49 | 15.31/233.09 | 6 | 62 | 2.70E-34 | 1 |
| 319 | T-complex protein 1 subunit beta | P78371 | 6.01 | 57.79 | 14 | 42 | 6.29E-10 | 6 |
| 320 | Glutathione transferase omega-1 | P78417 | 6.23/6.36 | 27.83/52.67 | 16 | 46 | 7.30E-35 | 1 |
| 321 | Neutrophil gelatinase-associated lipocalin Precursor |
P80188 | 9.02/8.04 | 22.74/41.77 | 12 | 63 | 1.20E-17 | 4 |
| 322 | Protein S100-A12 | P80511 | 5.83/6.30 | 10.57/11.87 | 6 | 45 | 1.70E-21 | 1 |
| 323 | Nuclear protein Hcc-1 | P82979 | 6.13/6.46 | 23.71/59.89 | 4 | 18 | 2.30E-03 | 3 |
| 324 | ADP-ribosylation factor 1 | P84077 | 6.32/6.31 | 20.70/29.64 | 11 | 69 | 8.50E-16 | 3 |
| 325 | Histone H3.3 | P84243 | 11.27/7.29 | 15.38/26.42 | 8 | 44 | 2.10E-14 | 3 |
| 326 | Sorbitol dehydrogenase | Q00796 | 8.25/9.59 | 38.90/63.57 | 12 | 36 | 2.80E-05 | 6 |
| 327 | Adenylyl cyclase-associated protein 1 | Q01518 | 8.12/6.70 | 52.22/109.94 | 8 | 27 | 6.80E-17 | 6 |
| 328 | Lactoylglutathione lyase | Q04760 | 5.12/5.38 | 20.99/35.44 | 6 | 32 | 1.00E-03 | 1 |
| 329 | Galectin-10 | Q05315 | 6.82/7.15 | 16.58/20.60 | 6 | 66 | 1.10E-11 | 1 |
| 330 | Proteasome activator complex subunit 1 | Q06323 | 5.78/6.14 | 28.88/59.02 | 15 | 61 | 1.80E-30 | 3 |
| 331 | Peroxiredoxin-1 | Q06830 | 8.27/8.26 | 22.32/40.87 | 18 | 75 | 2.30E-64 | 1 |
| 332 | Splicing factor, arginine/serine-rich 1 | Q07955 | 10.37/6.40 | 27.84/68.78 | 9 | 38 | 4.80E-05 | 3 |
| 333 | Rho-GTPase-activating protein 1 | Q07960 | 5.85/6.34 | 50.46/96.08 | 21 | 57 | 1.80E-25 | 3 |
| 334 | Secernin-1 | Q12765 | 4.66/5.14 | 46.80/10.21 | 12 | 29 | 9.20E-07 | 3 |
| 335 | Nuclear pore complex protein Nup160 | Q12769* | 5.41/9.17 | 151.13/83.21 | 12 | 12 | 4.60E-03 | 3 |
| 336 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase | Q13011 | 8.16 | 36.14 | 7 | 25 | 2.51E-06 | 6 |
| 337 | Ubiquitin-conjugating enzyme E2 variant 1 | Q13404 | 8.56/6.07 | 26.07/55.70 | 8 | 32 | 4.60E-07 | 4 |
| 338 | Dynactin subunit 2 | Q13561 | 5.10/5.30 | 44.19/52.24 | 10 | 35 | 1.70E-06 | 6 |
| 339 | NEDD8-activating enzyme E1 subunit | Q13564 | ND | ND | ND | ND | 1.20E-04 | 6 |
| 340 | Spectrin alpha chain, brain | Q13813 | 5.22/5.55 | 285.16/203.12 | 40 | 23 | 7.30E-18 | 2 |
| 341 | Spliceosome RNA helicase BAT1 | Q13838 | 5.44/5.99 | 49.42/95.04 | 15 | 35 | 9.20E-18 | 3 |
| 342 | Coactosin-like protein | Q14019 | 5.54/6.69 | 16.05/25.85 | 12 | 57 | 3.40E-19 | 4 |
| 343 | Heterogeneous nuclear ribonucleoprotein D0 |
Q14103 | 7.62/5.93 | 38.58/91.86 | 7 | 22 | 5.20E-05 | 4 |
| 344 | Septin-6 | Q14141 | 6.24 | 50.08 | 17 | 35 | 7.92E-08 | 6 |
| 345 | Keratin, type I cuticular Ha3-II | Q14525 | 4.81/5.77 | 47.33/192.32 | 14 | 44 | 1.50E-12 | 2 |
| 346 | Neutral alpha-glucosidase AB precursor | Q14697 | 5.74/6.21 | 107.26/196.40 | 24 | 28 | 9.20E-13 | 3 |
| 347 | Major vault protein | Q14764 | 5.34/5.86 | 99.55/174.43 | 19 | 22 | 1.70E-11 | 2 |
| 348 | LIM and SH3 domain protein 1 | Q14847 | 6.61/6.81 | 30.10/71.22 | 10 | 33 | 1.80E-11 | 3 |
| 349 | Orphan nuclear receptor NR1I3 | Q14994 | ND | ND | ND | ND | 2.50E-03 | 6 |
| 350 | Septin-2 | Q15019 | 6.15/6.48 | 41.69/72.94 | 7 | 26 | 1.60E-04 | 2 |
| 351 | Neutrophil cytosol factor 4 | Q15080 | 6.40/6.49 | 39.12/74.10 | 8 | 26 | 1.80E-11 | 2 |
| 352 | Protein disulfide-isomerase A6 precursor | Q15084 | 4.95/5.42 | 48.90/88.78 | 15 | 43 | 2.90E-15 | 3 |
| 353 | Poly(rC)-binding protein 1 | Q15365 | 6.66 | 37.99 | 7 | 30 | 3.97E-08 | 6 |
| 354 | Ras suppressor protein 1 | Q15404 | 8.57/8.54 | 31.52 | 16 | 56 | 1.10E-27 | 3 |
| 355 | Protein phosphatase 1 regulatory subunit 7 | Q15435 | 4.84/5.39 | 41.65 | 10 | 32 | 3.90E-06 | 3 |
| 356 | Syntaxin-binding protein 2 | Q15833 | 6.11/6.45 | 66.85 | 13 | 33 | 1.80E-09 | 3 |
| 357 | Ras-related protein Rab-11B | Q15907 | 5.65/5.80 | 24.59 | 9 | 40 | 2.70E-24 | 3 |
| 358 | Histone H2A type 2-C | Q16777 | 10.90/6.70 | 14.00/26.34 | 4 | 49 | 7.80E-04 | 3 |
| 359 | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor |
Q16836 | 8.88/5.86 | 34.21/27.24 | 10 | 38 | 1.80E-18 | 4 |
| 360 | Thioredoxin reductase 1, cytoplasmic precursor |
Q16881 | 6.07 | 55.47 | 11 | 27 | 1.58E-08 | 6 |
| 361 | Chaperonin containing TCP1, subunit 8 | Q53HU0 | ND | ND | ND | ND | 1.40E-05 | 6 |
| 362 | This TrEMBL entry deleted | Q5JV65 | ND | ND | ND | ND | 4.80E-26 | 6 |
| 363 | Histone H2B type 2-F | Q5QNW6 | 10.31/9.82 | 13.98/27.27 | 8 | 61 | 5.80E-20 | 3 |
| 364 | Twinfilin-2 | Q6IBS0 | 6.37/6.71 | 39.75/79.74 | 10 | 42 | 4.60E-07 | 3 |
| 365 | NAPRT Protein | Q6PJL1 | ND | ND | ND | ND | 5.50E-24 | 6 |
| 366 | Staphylococcal nuclease domain-containing protein 1 |
Q7KZF4 | 6.74/6.92 | 102.62/200.89 | 21 | 27 | 7.30E-14 | 3 |
| 367 | Histone H2A type 3 | Q7L7L0 | 11.05/5.96 | 14.10/25.95 | 5 | 35 | 8.50E-21 | 3 |
| 368 | Keratin, type II cytoskeletal 1b | Q7Z794 | ND | ND | ND | ND | 3.00E-11 | 6 |
| 369 | Unc-112-related protein 2 | Q86UX7 | 6.52/6.21 | 76.48/68.65 | 23 | 29 | 4.60E-22 | 3 |
| 370 | Ras-related protein Rab-43 | Q86YS6 | 5.44/5.10 | 23.55/56.43 | 7 | 45 | 5.80E-07 | 4 |
| 371 | Dedicator of cytokinesis protein 3 | Q8IZD9 | 6.52/9.18 | 235.01/33.43 | 12 | 7 | 8.00E-03 | 6 |
| 372 | Nesprin-1 | Q8NF91 | 5.38 | 1011.04 | 35 | 6 | 3.15E-03 | 6 |
| 373 | Gamma-glutamyl hydrolase precursor | Q92820 | 6.67/6.59 | 36.34/59.02 | 4 | 14 | 4.30E-04 | 1 |
| 374 | Probable ATP-dependent RNA helicase DDX17 |
Q92841 | 8.82/9.65 | 72.95/105.93 | 13 | 22 | 8.20E-03 | 6 |
| 375 | Histone H2A type 1-C | Q93077 | 11.05/3.49 | 14.10/18.34 | 5 | 23 | 6.80E-10 | 3 |
| 376 | Histone H2B type 1-H | Q93079 | 10.31/8.86 | 13.93/27.11 | 3 | 27 | 5.40E-10 | 3 |
| 377 | Uncharacterized protein C19orf10 precursor | Q969H8 | 6.20/6.16 | 18.90/53.14 | 4 | 27 | 2.20E-03 | 4 |
| 378 | Far upstream element-binding protein 1 | Q96AE4 | 7.18/8.36 | 67.69/60.93 | 19 | 39 | 2.90E-33 | 4 |
| 379 | EF-hand domain-containing protein 2 | Q96C19 | 5.15/5.37 | 26.79/57.61 | 23 | 62 | 3.60E-28 | 2 |
| 380 | Phosphoglucomutase-2 | Q96G03 | 6.28 | 68.75 | 19 | 35 | 1.26E-09 | 6 |
| 381 | ERO1-like protein alpha precursor | Q96HE7 | 5.48/5.87 | 55.21/127.76 | 11 | 27 | 2.90E-11 | 3 |
| 382 | Abhydrolase domain-containing protein 14B | Q96IU4 | 5.94/6.05 | 22.45/36.45 | 6 | 38 | 2.10E-03 | 1 |
| 383 | Histone H2A type 1-H | Q96KK5 | 10.88/7.06 | 13.93/20.48 | 5 | 38 | 2.70E-08 | 3 |
| 384 | Cytosolic nonspecific dipeptidase | Q96KP4 | 5.66 | 53.19 | 12 | 35 | 6.29E-04 | 6 |
| 385 | RNA-binding protein 14 | Q96PK6 | 9.68/9.71 | 69.62/102.34 | 14 | 25 | 8.50E-04 | 6 |
| 386 | Calponin-2 | Q99439 | 6.92/6.99 | 34.07/60.59 | 14 | 49 | 1.10E-11 | 2 |
| 387 | Synaptic vesicle membrane protein VAT-1 homolog |
Q99536 | 5.88/6.17 | 42.12/82.48 | 16 | 50 | 4.60E-14 | 3 |
| 388 | Translin-associated protein X | Q99598 | 6.10/6.38 | 33.21/60.00 | 10 | 50 | 1.50E-06 | 2 |
| 389 | Monoglyceride lipase | Q99685 | 6.49/6.18 | 33.47/67.37 | 6 | 24 | 7.40E-04 | 4 |
| 390 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Q99714 | 7.66/6.25 | 27.13/18.90 | 7 | 11 | 3.60E-04 | 4 |
| 391 | Aconitate hydratase, mitochondrial precursor | Q99798 | 7.36/6.76 | 86.11/52.51 | 13 | 20 | 9.20E-42 | 4 |
| 392 | Copine-1 | Q99829 | 5.52/5.92 | 59.65/119.95 | 13 | 24 | 2.30E-22 | 1 |
| 393 | Histone H2A type 1-J | Q99878 | 10.88/5.71 | 13.93/25.78 | 6 | 38 | 1.10E-15 | 3 |
| 394 | Uncharacterized protein C9orf142 | Q9BUH6 | 5.39/5.60 | 21.97/38.08 | 9 | 42 | 3.40E-09 | 2 |
| 395 | Transmembrane emp24 domain-containing protein 9 precursor |
Q9BVK6 | 6.67/6.57 | 25.20/40.69 | 11 | 55 | 2.70E-06 | 2 |
| 396 | Kinesin light chain 2 | Q9H0B6 | 6.72/6.36 | 69.29/79.92 | 14 | 29 | 8.20E-03 | 2 |
| 397 | Haloacid dehalogenase-like hydrolase domain-containing protein 2 |
Q9H0R4 | ND | ND | ND | ND | 1.70E-05 | 6 |
| 398 | Ras-related protein Rab-1B | Q9H0U4 | 5.55/5.64 | 22.33/41.56 | 4 | 20 | 2.10E-05 | 3 |
| 399 | EH domain-containing protein 1 | Q9H4M9 | 6.35 | 60.65 | 13 | 26 | 9.98E-04 | 6 |
| 400 | Sideroflexin-1 | Q9H9B4 | 9.22/5.90 | 35.88/27.94 | 10 | 34 | 1.30E-05 | 4 |
| 401 | Phosphopantothenate--cysteine ligase | Q9HAB8 | 6.25/6.74 | 33.98/65.20 | 8 | 24 | 2.00E-06 | 3 |
| 402 | Retinoid-inducible serine carboxypeptidase precursor |
Q9HB40 | 5.61/6.34 | 51.08/26.26 | 8 | 15 | 4.60E-08 | 1 |
| 403 | Adipocyte plasma membrane-associated Protein |
Q9HDC9 | 5.82/7.04 | 46.62/50.00 | 21 | 48 | 2.90E-38 | 4 |
| 404 | Exosome complex exonuclease RRP41 | Q9NPD3 | 6.08/6.39 | 26.65/48.50 | 9 | 40 | 6.60E-04 | 2 |
| 405 | Protein FAM49B | Q9NUQ9 | 5.76/6.03 | 37.01/60.98 | 16 | 49 | 4.60E-22 | 1 |
| 406 | Dipeptidyl-peptidase 3 | Q9NY33 | 5.02/5.37 | 82.88/153.84 | 9 | 12 | 9.20E-18 | 3 |
| 407 | Tropomodulin-3 | Q9NYL9 | ND | ND | ND | ND | 1.20E-29 | 6 |
| 408 | EH-domain-containing protein 3 | Q9NZN3 | 6.06/6.44 | 61.97/50.21 | 12 | 24 | 7.60E-04 | 2 |
| 409 | Vacuolar protein sorting 29 | Q9UBQ0 | 6.29/6.70 | 20.66/35.44 | 8 | 43 | 3.60E-13 | 2 |
| 410 | Fructose-1,6-bisphosphate aldolase A [Fragment] |
Q9UCN2 | ND | ND | ND | ND | 7.20E-05 | 6 |
| 411 | Protein NipSnap3A | Q9UFN0 | 9.21/8.65 | 28.56/37.28 | 6 | 25 | 5.20E-06 | 6 |
| 412 | Mitogen-activated protein kinase kinase 1 | Q9UHA4 | 6.73/6.59 | 13.67/24.23 | 3 | 43 | 3.60E-07 | 3 |
| 413 | N-acetylglucosamine kinase | Q9UJ70 | 5.82/6.11 | 37.69/70.84 | 14 | 44 | 5.40E-15 | 2 |
| 414 | DCC-interacting protein 13 alpha | Q9UKG1 | ND | ND | ND | ND | 6.90E-08 | 6 |
| 415 | Proteasome activator complex subunit 2 | Q9UL46 | 5.44/5.69 | 27.52/55.19 | 11 | 43 | 5.20E-06 | 2 |
| 416 | Apoptosis-associated speck-like protein containing a CARD |
Q9ULZ3 | 5.95/6.54 | 21.67/38.14 | 7 | 37 | 7.30E-09 | 1 |
| 417 | Protein-arginine deiminase type-4 | Q9UM07 | 6.15/6.58 | 75.13/116.76 | 9 | 19 | 4.60E-08 | 2 |
| 418 | NSFL1 cofactor p47 | Q9UNZ2 | ND | ND | ND | ND | 5.50E-17 | 6 |
| 419 | RuvB-like 2 | Q9Y230 | 5.49/5.73 | 51.30/87.80 | 18 | 43 | 5.40E-11 | 2 |
| 420 | Cofilin-2 | Q9Y281 | 7.66/7.49 | 18.84/29.00 | 5 | 28 | 1.50E-03 | 1 |
| 421 | Trafficking protein particle complex subunit 4 | Q9Y296 | 5.83/6.24 | 24.44/40.69 | 13 | 60 | 3.60E-14 | 2 |
| 422 | SH3 domain-binding protein 1 | Q9Y3L3 | 6.33/6.44 | 76.01/139.19 | 4 | 5 | 1.00E-03 | 2 |
| 423 | Talin-1 | Q9Y490 | 5.77/6.23 | 271.77/231.63 | 47 | 27 | 2.90E-45 | 1 |
| 424 | Heme-binding protein 2 | Q9Y5Z4 | 4.58/5.81 | 22.86/69.71 | 3 | 18 | 4.60E-12 | 3 |
| 425 | Actin, alpha cardiac muscle 1 | P68032 | 5.23 | 42.33 | 4 | 16 | 1.58E-03 | 6 |
| 426 | High mobility group protein B2 | P26583 | 5.62/7.16 | 24918.2/44.58 | 11 | 43 | 5.80E-28 | 4 |
Figure 2.
Comparison of eosinophil proteome maps focused at four different pH ranges: 3–10; 4–7; 5–8; and 6–11. Each gel was loaded with 200 µg of total eosinophil cell lysate.
Figure 3.
Gel-to-gel correlation of five replicate gels showing 2-DE reproducibility. Log normalized spot volumes for gel 1 were plotted pairwise versus gels 2 to 5 and the Pearson’s correlation coefficient shown in squares was calculated. Gels (11 cm) were focused over the pH range 3–10 and were stained with Sypro Ruby. Gel image analysis utilized Nonlinear SameSpots software.
Table 2.
Summary prefractionation protein ID’s
| Fractions analyzed |
Protein IDs | Non-redundant Protein IDsa |
|---|---|---|
|
Subcellular fractionsb | ||
| 1-cytoplasm | 625 | 95 |
| 2-organelle | 425 | 50 |
| 3-nucleus | 693 | 141 |
| 4-cytoskeleton | 345 | 52 |
| Total subcellular | 2088 | 338 |
| IEF fractionsc | ||
| pH 3.0–5.4 | 44 | 4 |
| pH 5.4–7.0 | 181 | 13 |
| pH 7.0–10.0 | 226 | 16 |
| Total IEF | 451 | 33 |
| Whole cell lysate | 480 | 63 |
| Total all fractions | 3019 | 434 |
Had Mascot expectation scores of 10−3 or less.
According to Calbiochem’s ProteoExtract® Subcellular Proteome Extraction kit.
Using ZOOM® IEF Fractionator (Invitrogen).
Figure 4.
Western blot analysis of eight randomly selected eosinophil proteins to demonstrate their distribution by differential solubility using a commercial kit (Calbiochem’s ProteoExtract® Subcellular Proteome Extraction kit). The kit employed four solubility fractions F1 to F4 as shown (see also Table 2). 50 µg of cell lysates were applied to each lane.
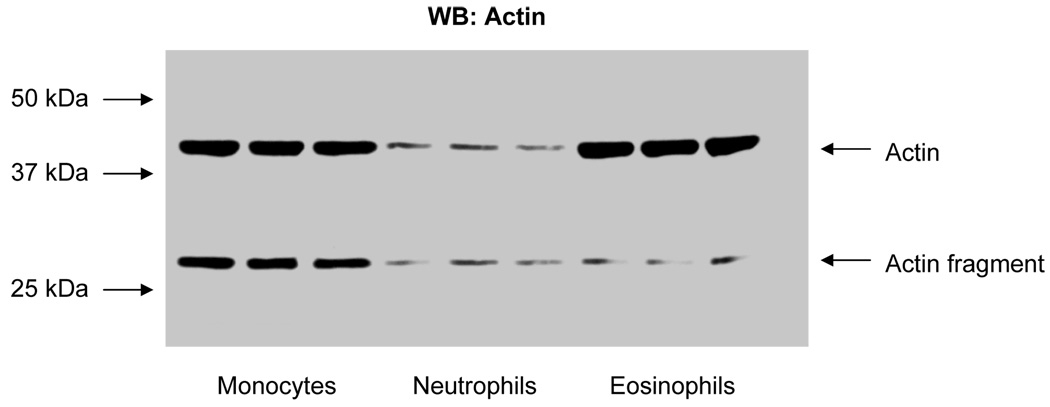
Actin was the most prominent protein expressed in eosinophils. Because of this fact, we chose to comparatively evaluate actin levels in other leukocytes. Fig. 5 shows the comparative distribution of actin and an actin proteolytic cleavage fragment by Western blot analysis of monocyte, neutrophil, and eosinophil cell lysates.
Figure 5.
Western blot analysis comparing actin levels in monocytes, neutrophils, and eosinophils in triplicate. 50 µg of cell lysate protein was loaded in each lane. Actin fragment was an N-terminal product as a result of proteolysis (see Discussion).
3.2 Ingenuity Pathway Analysis software application
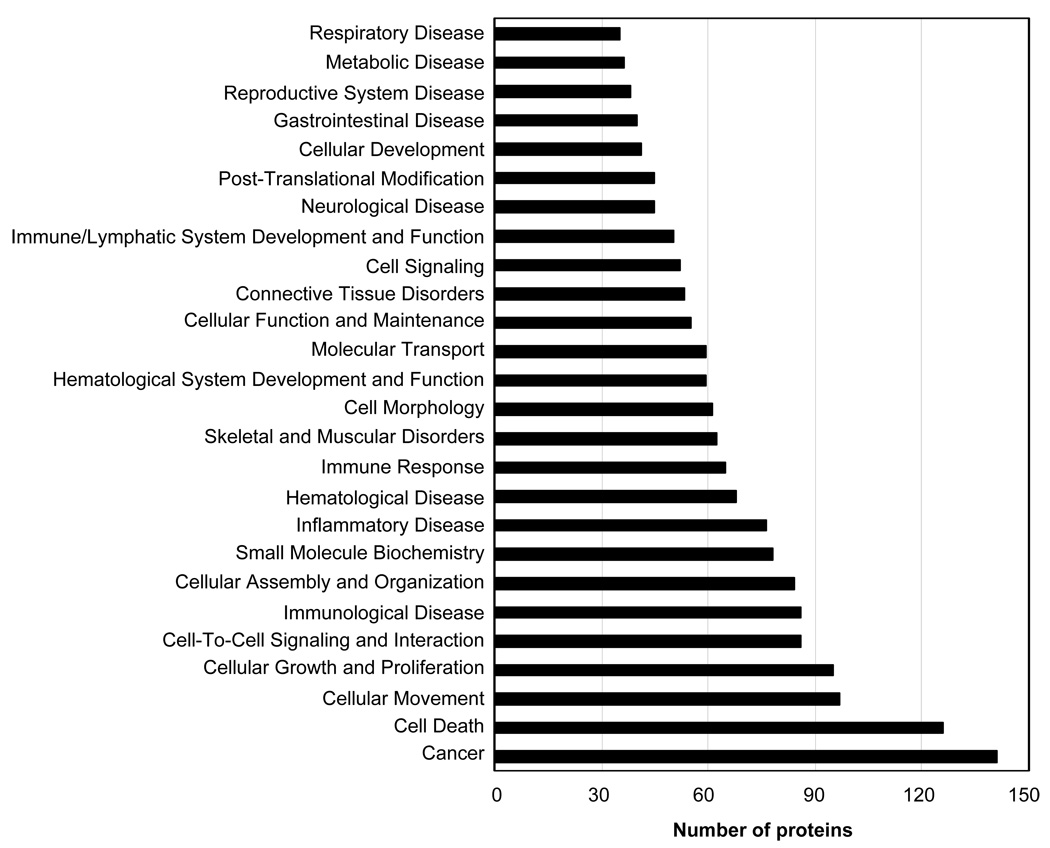
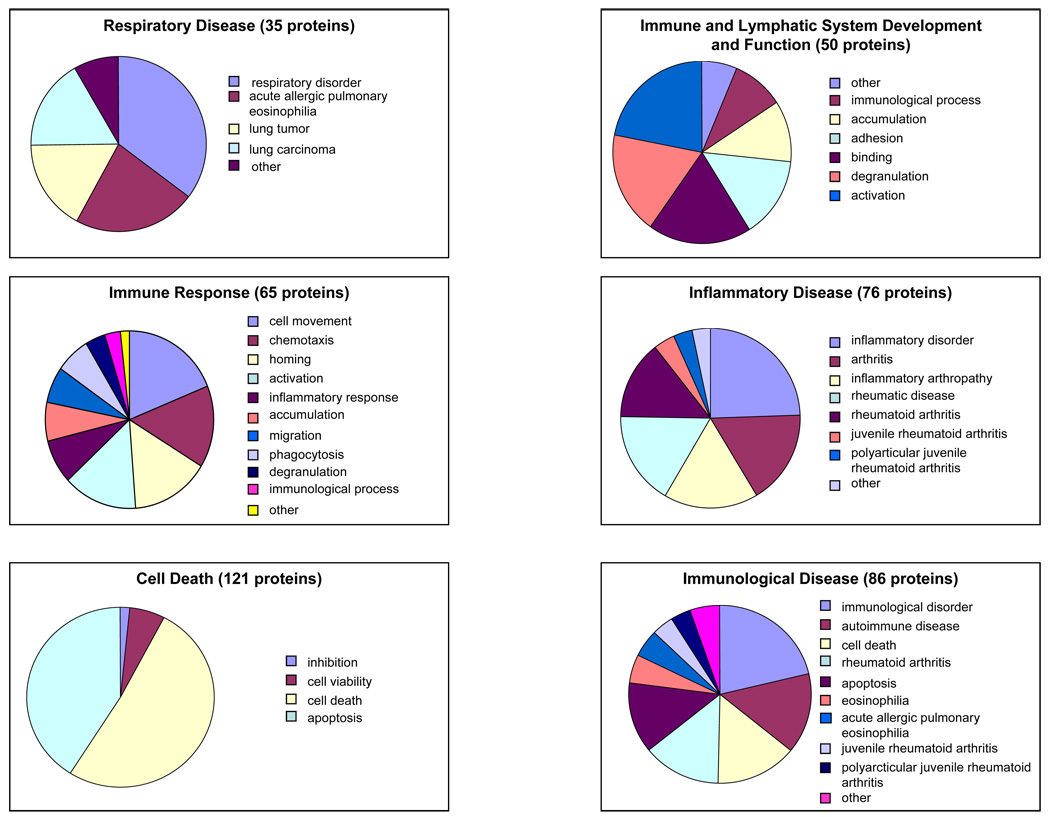
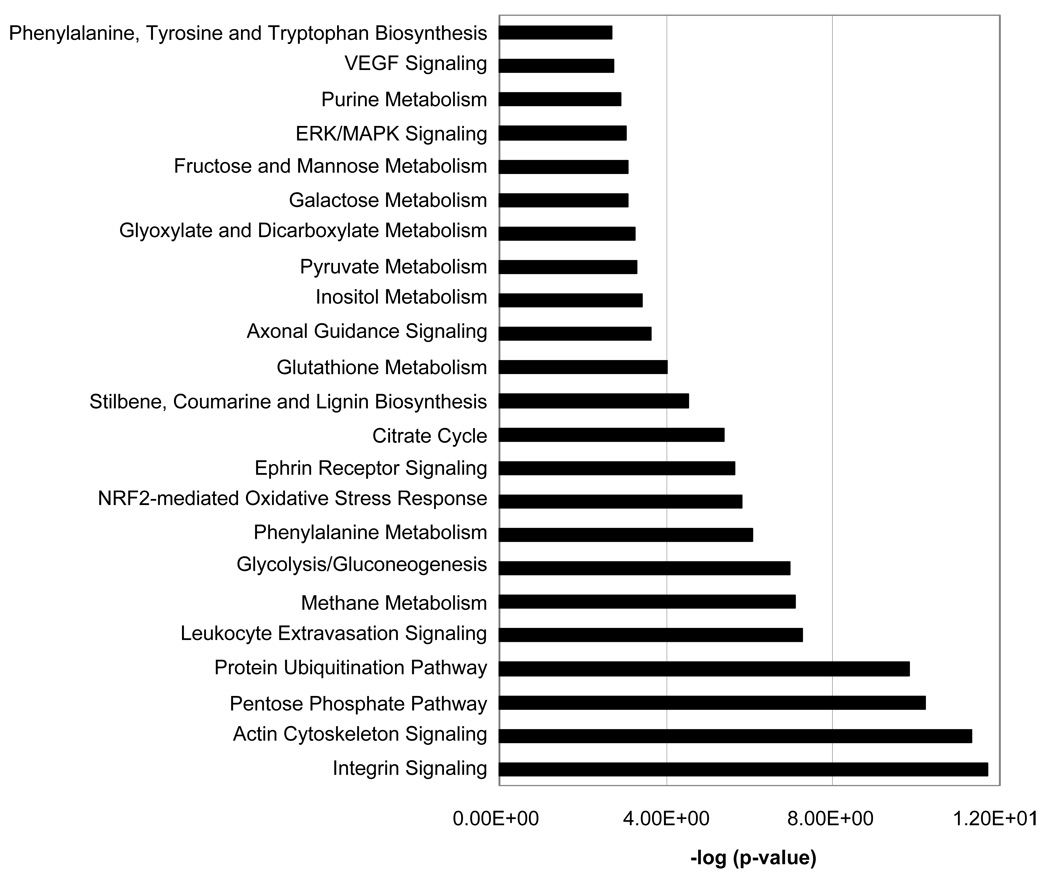
Ingenuity software was applied to the analysis of the eosinophil expression dataset to probe the relevant biological functions of the identified proteins in the dataset (Table 1). Some functions and diseases relevant to the dataset are shown in Fig. 6. Fig. 7 shows selected groups of proteins that highlight in more detail the proportion of the identified proteins whose function or impact are particularly relevant to eosinophil biological activity; namely, immunological disease, inflammatory disease, immune response, immune and lymphatic system development and function, and respiratory disease. Fig. 8 gives a histogram of the top canonical pathways associated with the dataset. A small p-value indicated a strong association between the dataset and the respective pathway. Proteins found in selected eosinophil disease functional and canonical pathway subsets (Figs. 6 and 8) are listed in supplemental Tables S1 and S2 (see www.proteomics-journal.com).
Figure 6.
Ingenuity Pathway Analysis showing the distribution of identified eosinophil proteins from Table 1 into disease and functional categories.
Figure 7.
Subclassification of eosinophilic proteins from Table 1 and Fig. 6 shows a distribution of eosinophilic proteins into diseases and functional categories highly consistent with what is general appreciated regarding eosinophilic biological activity.
Figure 8.
Ingenuity Pathway Analysis showing the distribution of identified eosinophil proteins (Table 1) into canonical pathways showing a strong emphasis on signaling pathways consistent with the dynamic nature of the eosinophil.
4 Discussion
We have identified 3,141 proteins which had Mascot expectation scores of 10−3 or less. Of these, 426 proteins were unique and non-redundant as identified using the SwissProt protein database. We did not attempt to distinguish differences between males and females nor did we address the extent of observed polymorphic variations between individuals since large numbers of donors would be required. However, further studies are planned to deal with these important issues. Significantly, of the 426 non-redundant proteins 231were novel proteins not previously reported to occur in eosinophils. Since only 8% of all proteins excised and analyzed from 2-D gels were among the unique, non-redundant dataset (426 proteins), the question arises as to the occurrence and nature of the redundant protein dataset (2715 proteins).
There are many explanations for protein redundancy including phosphorylation. We undertook a preliminary assessment of the eosinophil phosphoproteome using Pro-Q Diamond staining in order to evaluate the contribution of phosphorylation to redundancy (results not shown). The characterization of the eosinophil phosphoproteome by 2-DE will be separately reported. We found that many eosinophil proteins were phosphorylated; phosphorylation can be variable at a given site and may be variable as to the number of sites modified all of which contribute to redundancy. In addition, as shown in Fig. 1, 2-D gel analysis indicated a number of horizontal protein spots, identified as the same protein by MS, with the same Mr but having different pIs, indicating polymorphism and/or posttranslational modification. In addition to phosphorylation, a number of modifications can account for such variations; as for example, acetylation, sialylation, sulfation, and methylation. Furthermore, since many proteins have attached carbohydrate moieties, these can give rise to significant pI and/or Mr variations. Finally, proteolytic processing/modifications must be considered among the relevant causes of protein redundancy. Clearly, the above examples are not an exhaustive list of factors leading to protein redundancies. The observed high protein redundancy likely reflected the dynamic character of the eosinophil and underscores the fact that posttranslational modifications may be the result of various regulatory and signaling events.
This proteomic dataset is the largest comprehensive proteomic dataset of proteins expressed in normal peripheral blood eosinophils reported to date. There have been two reports of comparative proteomic studies. Waschnagg et al. [12] very recently evaluated protein expression differences induced by Birch pollen allergy and identified 97 unique non-redundant eosinophil proteins of which 90 occur in our list of 426 (Table 1) which is an excellent agreement. However, a comparative proteomic study of healthy versus atopic dermatitis patients identified 51 differentially expressed proteins of which only three are included in Table 1 [13].
In this study we have made some attempt at characterizing the less abundant proteins using ZOOM® pre-fractionation IEF and subcellular fractionation methods. Protein distribution into various fractions using a commercial subcellular fractionation method allowed for the reduction of protein complexity and increased the number of less abundant proteins observed. We found that this fractionation method performed better in reducing protein losses than many other subcellular fractionation methods that can incur appreciable protein loss. Furthermore, the differential solubilization method was amenable to small sample size, gave high protein recoveries, had relatively high throughput, and processing time was fairly short minimizing protein alterations. However, this method is not sufficiently adequate to predict protein localization to specific subcellular compartments.
Characterization of the dataset (Table 1) using Ingenuity Pathway Analysis revealed a number of interesting features. Especially worthy of note was that 312 of the 434 (72%) identified non-redundant proteins could be subdivided into categories (Fig. 6) which were related to known eosinophil biological activities directly; e.g., eosinophilia, cell movement, chemotaxis and activation, or indirectly; e.g., autoimmune diseases. We were able to detect and positively identify many proteins that were relevant to eosinophil functions involving survival and activation. Recent studies strongly suggest that tissue eosinophilia is more dependent on increased survival in peripheral tissues than increased de novo generation in the bone marrow followed by blood to tissue translocation [14]. Analysis of eosinophil turnover in vivo revealed their active recruitment to the peritoneal cavity and their prolonged survival there [14]. In this regard our Ingenuity Pathway analysis showed a considerable number of proteins (~125) involved in cell death and survival (Fig. 6). Most of these proteins have previously not been correlated with eosinophil survival processes. However, some of these proteins were shown to play roles in other aspects of eosinophil biology. These observations emphasized the need for more studies to investigate the pro- and anti-apoptotic proteins that regulate eosinophil survival in end organs to induce or prevent apoptosis in cells depending on whether the need is to protect against helminth parasites or ameliorate eosinophil-associated diseases.
Eosinophils are secretory cells that contain large amounts of granules occupying about one-fifth of the cytoplasm [1]. Four major populations of granules have been identified; namely, primary, secondary, small granules, and as well lipid bodies [1]. Our 2-DE studies identified four of the major proteins found in the secondary granules that included, ECP, EDN, EPO, and MBP as well as galectin-10 found in the primary granules. Numerous other proteins have also been reported to occur in the granules [1]. ECP is a secretory ribonuclease associated with host defense against nonphagocytosable pathogens, such as helminthic parasites. It also has antibacterial activity which is not shared by EDN, another closely-related neurotoxic eosinophil ribonuclease. The mechanism of action of ECP is thought to involve pore formation in target membranes which is apparently not dependent on its RNAse activity [15]. On the other hand, EDN which shares 70% homology with ECP has been implicated in antiviral activity against respiratory infections mainly due to its ribonuclease activity [16]. EPO is an eosinophil haloperoxidase that catalyzes the peroxidative oxidation of halides present in the plasma as well as hydrogen peroxide generated by dismutation of superoxide produced during respiratory burst. This reaction leads to the formation of bactericidal hypohalous acids [17]. MBP was traditionally associated with toxicity against helminthic worms and is at least partly responsible for tissue damage in bronchial mucosa in asthma. The mechanism of its action is believed to be increased membrane permeability through surface charge interactions leading to perturbation of the cell-surface lipid bi-layer. These granule proteins are actively released from activated eosinophils and little if any active transcription occurs in mature eosinophils. The role of eosinophils in the pathophysiology of bacterial and viral infections is still not well elucidated.
The second most abundant and notable protein observed by 2-D gel analysis of eosinophil cell lysates was galectin-10 (Table 1, Fig. 1; ID 329) which occurs mainly in the primary granules of eosinophils and for many years was referred to as lysophospholipase or Charcot-Layden crystal protein. However, new evidence gives strong indication that it belongs to the galectin superfamily of proteins and was designated as galectin-10 by Ackerman et al. [18, 19]. Previously galectin-10 was thought to occur only in eosinophils and basophils but recent work has also identified it in human CD4+CD25+ regulatory T cells (CD25+ Treg cells) where it is thought to function in maintaining immunological self-tolerance by suppressing autoaggressive T-cells [20]. Eosinophilic galectin-10 also appears to have lectin-like properties and can bind mannose (in the crystal) [18]. Further investigations are required to elucidate the biological function of this interesting protein. Gel analysis results from both 1-D and 2-D gels, including Western blot analysis using anti-galectin-10, showed that galectin-10 distributed in multiple gel locations. Repeated gel analysis by 1D SDS-PAGE and Western blotting of eosinophil cell lysates gave three bands of molecular weights ~ 17 KDa, ~ 25 KDa, and ~ 75 KDa. Galectin-10 has been reported to be unique in having a propensity to aggregate even in dissociating conditions [21]. Gel analysis by 2-DE was also anomalous with spots at ~ 17 KDa and ~ 25 KDa and pronounced vertical streaking likely due to precipitation at its pI in the first dimension of 2-DE (Fig. 1). Some dimer formation was also noted (Fig. 1). Western blot 2-DE analysis of galectin-10 also showed multiple horizontal spots at ~ 17 KDa indicating polymorphism and/or posttranslational modifications. N-terminal acetylation and isoforms for galectin-10 were also identified by 2-DE of CD25+ Treg-cell lysates and human eosinophils [20]. A separate study will be required to fully characterize the various isoforms associated with galectin-10.
The 2-D gels showed that eosinophils have especially high amounts of actin (Fig. 1, Table 1; ID 311) which contributed to the relatively lower abundance of other proteins in the cell lysate samples. Actin cytoskeletal structures and associated proteins likely play important roles in eosinophil functions; such as, signaling (Fig. 8), cell motility, degranulation, phagocytosis, and activation [22–24]. Some of the actin was proteolytically processed to smaller Mr forms (Figs. 1 and 5; Table 1; ID 275) and some actin was also phosphorylated (Pro-Q Diamond staining not shown). Actin phosphorylation in other cells was previously reported [25, 26] and plays an important role in actin polymerization [27, 28]. Protein MS identification did not distinguish between β- or α-actin since β- and α-actin have virtually identical primary structures. Although nonmuscle actin and actin-associated proteins are reported to occur in the nucleus of cells [29], our previous studies using FITC-phalliodin, anti-α-fodrin [24], and anti-actin (results not shown) did not indicate the occurrence of nuclear actin in eosinophils. However, these results could not rule out the occurrence of very low levels of nuclear actin that could not be detected under the experimental conditions employed or that the actin Ab was not reactive to nuclear actin due to some unique complex formation. Actin levels in eosinophils and monocytes were relatively high when compared with neutrophils with considerably more proteolytic processing occurring in monocytes (Fig. 5). This was also confirmed by 2-DE of cell lysates (results not shown). The observed actin fragment represented the N-terminal domain of actin, since the anti-actin antibody used was raised aganist an N-terminal fragment (actin 1–19). Numerous actin complex-associated proteins were identified; for example, actin-related protein 2/3 complex, gelsolin, vimentin, Rho GDP-dissociation inhibitor, transgelin, moesin, coactosin-like protein, tubulin, cofilin, L-plastin, calreticulin, myosin, F-actin capping proteins cap Z alpha-1 and beta, and macrophage protein G, α-actinin, profilin, dynactin, coronin, nesprin, kinesin, tropomodulin, talin, and spectrin. The facile identification of these proteins was undoubtedly related to the high level expression of cytoplasmic actin in eosinophils. The high actin level and abundant associated proteins underscore the importance of the cytoskeleton in the biological activity of eosinophils especially motility and activation
An important advantage of proteomic analysis by 2-DE is the visualization and potential identification of polymorphisms and/or posttranslational modifications. Fig. 1 shows a number of proteins that are likely to be posttranslationally modified as evidenced by repeated horizontal protein spots from the same protein identified by MS; for example, Fig. 1, Table 1; ID’s 31, 36, 45, 48, 49, 50, 53, 59, 61, 66, 107, 135, 167, 182, 193, 216, 257, 283, 304 and 329. Some of these proteins have not been previously reported to be posttranslationally modified; as for example, Table 1: 31, 59, 107, 135, and 257. We expect that the herein described protein expression results represent the largest.
In summary, the herein described protein expression results represent the largest comprehensive reporting of the human eosinophil proteome. The identification of proteins in any proteome study is somewhat asymptotic and probably not 100% achievable by current technologies. This proteome map will be especially valuable as a baseline to compare with eosinophils from disease and pharmacologically treated states.
Supplementary Material
Acknowledgements
Funding information
This study was supported by the National Institutes of Health, National Heart, Lung and Blood Institute’s Proteomics Initiative NO1-HV-28184 (to A. K.), the National Institutes of Health grants 1-R24 CA88317 (to A.K.), 1-P30 ES06676 (to C. Elferink) and 1-P01AI062885 (to A. Brasier).
Abbreviations
- HBSS
Hank’s balanced saline solution
- TCEP
tri-(2-carboxyethyl) phosphine
- IPG
immobilized pH gradient
- RT
room temperature
- GM-CSF
granulocyte-macrophage colony-stimulating factor
- RANTES
regulated upon activation, normal T cell expressed and secreted
- ID
identification
- EOS
eosinophil
- IPA
Ingenuity Pathway Analysis
- EPO
eosinophil peroxidase
- MBP
major basic protein
- ECP
eosinophil cationic protein
- EDN
eosinophil-derived neurotoxin
- CLC
Charcot-Leyden crystal protein
- IL
interleukin
Footnotes
Conflict of interest - The authors have no financial conflict of interest.
References
- 1.Giembycz MA, Lindsay MA. Pharmacology of the eosinophil. Pharmacol. Rev. 1999;51:213–339. [PubMed] [Google Scholar]
- 2.Rothenberg ME, Hogan SP. The eosinophil. Annu. Rev. Immunol. 2006;24:147–174. doi: 10.1146/annurev.immunol.24.021605.090720. [DOI] [PubMed] [Google Scholar]
- 3.Ganley-Leal LM, Mwinzi PN, Cetre-Sossah CB, Andove J, et al. Correlation between eosinophils and protection against reinfection with Schistosoma mansoni and the effect of human immunodeficiency virus type 1 coinfection in humans. Infect. Immun. 2006;74:2169–2176. doi: 10.1128/IAI.74.4.2169-2176.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melo RCN, Spencer LA, Dvorak AM, Weller PF. Mechanisms of eosinophil secretion: large vesiculotubular carriers mediate transport and release of granule-derived cytokines and other proteins. J. Leukoc. Biol. 2008;83:229–236. doi: 10.1189/jlb.0707503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pazdrak K, Young TW, Stafford S, Olszewska-Pazdrak B, Straub C, et al. Cross-Talk between ICAM-1 and granulocyte-macrophage colony-stimulating factor receptor signaling modulates eosinophil survival and activation. J. Immunol. 2008;180:4182–4190. doi: 10.4049/jimmunol.180.6.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsumoto K, Appiah-Pippim J, Schleimer RP, Bickel CA, et al. CD44 and CD69 represent different types of cell-surface activation markers for human eosinophils. Am. J. Respir. Cell Mol. Biol. 1998;18:860–866. doi: 10.1165/ajrcmb.18.6.3159. [DOI] [PubMed] [Google Scholar]
- 7.Wardlaw A. Eosinophil density: what does it mean? Clin. Exper. Allergy. 1995;25:1145–1149. doi: 10.1111/j.1365-2222.1995.tb03036.x. [DOI] [PubMed] [Google Scholar]
- 8.Gruart V, Balloul JM, Prin L, Tomassini M. Variations in protein expression related to human eosinophil heterogeneity. J. Immunol. 1989;142:4416–4421. [PubMed] [Google Scholar]
- 9.Fukuda T, Gleich GJ. Heterogeneity of human eosinophils. J. Allergy Clin. Immunol. 1989;83:369–373. doi: 10.1016/0091-6749(89)90120-6. [DOI] [PubMed] [Google Scholar]
- 10.Pleis JR, Lethbridge-Cejku M. Summary Health Stastistics for U.S. Adults: National Health Interview Survey, Vital Health Stat. 2007;10(235) [PubMed] [Google Scholar]
- 11.Matsumoto K, Appiah-Pippim J, Schleimer RP, Bickel CA, et al. CD44 and CD69 represent different types of cell-surface activation markers for human eosinophils. Am. J. Respir. Cell Mol. Biol. 1998;18:860–866. doi: 10.1165/ajrcmb.18.6.3159. [DOI] [PubMed] [Google Scholar]
- 12.Waschnagg C, Forsberg J, Engström Å, Odreman F, Venge P, Garcia R. The human eosinophil proteome. Changes induced by Birch pollen allergy. J. Proteome Res. 2009 doi: 10.1021/pr800984e. DO1: 10.1021/pr800984e. [DOI] [PubMed] [Google Scholar]
- 13.Yoon SW, Kim TY, Sung MH, Kim CJ, Poo H. Comparative proteomic analysis of peripheral blood eosiniophils from healthy donors and atopic dermatitis patients with eosinophilia. Proteomics. 2005;5:1987–1995. doi: 10.1002/pmic.200401086. [DOI] [PubMed] [Google Scholar]
- 14.Ohnmacht C, Pullner A, Van Rooijen N, Voehringer D.Analysis of eosinophil turnover in vivo reveals their active recruitment to and prolonged survival in the peritoneal cavity J. Immunol 200717947664774 [DOI] [PubMed] [Google Scholar]
- 15.Domachowske JB, Dyer KD, Adams AG, Leto TL, Rosenberg HF. Eosinophil cationic protein/RNase 3 is another RNase A-family ribonuclease with direct antiviral activity. Nucleic Acids Res. 1998;26:3358–3363. doi: 10.1093/nar/26.14.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenberg HF. Eosinophil-derived neurotoxin/RNase 2; connecting the past, the present and the future. Curr. Pharm. Biotechnol. 2008;9:135–140. doi: 10.2174/138920108784567236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jong EC, Henderson WR, Klebanoff SJ. Bactericidal activity of eosinophil peroxidase. J. Immunol. 1980;124:1378–1382. [PubMed] [Google Scholar]
- 18.Ackerman SJ, Liu L, Kwatia MA, Savage MP, et al. Charcot-Leyden crystal protein (galectin-10) is not a dual function galectin with lysophospholipase activity but binds a lysophospholipase inhibitor in a novel structural fashion. J. Biol. Chem. 2002;277:14859–14868. doi: 10.1074/jbc.M200221200. [DOI] [PubMed] [Google Scholar]
- 19.Ackerman SJ, Corrette SE, Rosenberg HF, Bennett JC, et al. Molecular cloning and characterization of human eosinophil Charcot-Leyden crystal protein (lysophospholipase) J. Immunol. 1993;150:456–468. [PubMed] [Google Scholar]
- 20.Kubach J, Lutter P, Bopp T, Stoll S, et al. Human CD4+CD25+ regulatory T cells: proteome analysis identifies galectin-10 as a novel marker essential for their energy and suppressive function. Blood. 2007;110:1550–1558. doi: 10.1182/blood-2007-01-069229. [DOI] [PubMed] [Google Scholar]
- 21.Ackerman SJ, Loegering DA, Gleich GJ. The human eosinophil Charcot-Leyden crystal protein: biochemical characteristics and measurement by radioimmunoassay. J. Immunol. 1980;125:2118–2126. [PubMed] [Google Scholar]
- 22.Boehme SA, Sullivan SK, Crowe PD, Santos M, et al. Activation of mitogen-activated protein kinase regulates eotaxin-induced eosinophil migration. J. Immunol. 1999;163:1611–1618. [PubMed] [Google Scholar]
- 23.Suzuki M, Kato M, Hanaka H, Izumi T, et al. Actin assembly is a crucial factor for superoxide anion generation from adherent human eosinophils. J. Allergy Clin. Immunol. 2003;112:126–133. doi: 10.1067/mai.2003.1515. [DOI] [PubMed] [Google Scholar]
- 24.Starosta V, Pazdrak K, Boldogh I, Svider T, Kurosky A. Lipoxin A4 counterregulates GM-CSF signaling in eosinophilic granulocytes. J. Immunol. 2008;181:8688–8699. doi: 10.4049/jimmunol.181.12.8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rush J, Moritz A, Lee KA, Guo A, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotech. 2004;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 26.Zahedi RP, Lewandrowski U, Wiesner J, Wortelkamp S, et al. Phosphoproteome of resting human platelets. J. Proteome Res. 2008;7:526–534. doi: 10.1021/pr0704130. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Shu S, Hong MSS, Levine RL, Korn ED. Phosphorylation of actin Tyr-53 inhibits filament nucleation and elongation and stabilizes filaments. Proc. Natl. Acad. Sci. USA. 2006;103:13694–13699. doi: 10.1073/pnas.0606321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Zheng Y, Yu Y, Xia L, et al. Phosphorylation of β-actin by protein kinase C-delta in camptothecin analog-induced leukemic cell apoptosis. Acta Pharmacol. Sinica. 2008;29:135–142. doi: 10.1111/j.1745-7254.2008.00753.x. [DOI] [PubMed] [Google Scholar]
- 29.Pederson T, Aebi U. Actin in the nucleus: what form and what for? J. Struct. Biol. 2003;140:3–9. doi: 10.1016/s1047-8477(02)00528-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.