Abstract
Satellite cells, the adult stem cells responsible for skeletal muscle regeneration, are defined by their location between the basal lamina and the fiber sarcolemma. Increasing evidence suggests that satellite cells represent a heterogeneous population of cells with distinct embryological origin and multiple levels of biochemical and functional diversity. This review focuses on the rich diversity of the satellite cell population based on studies across species. Ultimately, a more complete characterization of the heterogeneity of satellite cells will be essential to understand the functional significance in terms of muscle growth, homeostasis, tissue repair, and aging.
Keywords: Satellite cell, heterogeneity, muscle, regeneration, lineage, stem cell
1. Introduction
Almost 50 years ago, Alex Mauro observed by electron microscopy the presence of mononucleated cells, which he termed “satellite cells”, that were intimately associated with skeletal muscle fibers of the frog [1]. Mauro noted that satellite cells (SCs) were “wedged” between the muscle fiber membrane and the basal lamina, and hypothesized that they could be involved in muscle regeneration.
SCs were subsequently identified in skeletal muscles of other vertebrates, including humans, and their involvement in muscle regeneration became unquestionable [2]. During regeneration, SCs break quiescence, undergo proliferative expansion, generate myoblasts and terminally differentiate by fusing to each other or with damaged fibers [3]. In growing muscles, SC progeny similarly fuse with existing fibers [4]. Importantly, a fraction of SCs do not terminally differentiate, but rather replenish the SC pool by self-renewal and reoccupy a position under the basal lamina [5]. SCs are rare in uninjured muscles, typically accounting for <2% of the nuclear content of muscle, but with higher estimates noted in specific muscles and species [6-7]. Nevertheless, they have remarkable proliferative potential and can efficiently repair even severely damaged muscles [8].
This historical, purely anatomical definition of SCs does not suggest any heterogeneity within the population. However, an increasing number of studies have indicated that the SC pool is more heterogeneous than originally anticipated. In this review we will provide a summary of the evidence supporting this idea, identifying different levels of heterogeneity.
2. Heterogeneity based on embryological origin
Different muscles exhibit distinct characteristics, including anatomical structure, contractile and metabolic properties, fiber composition, blood supply, pattern of innervation and embryonic origin. Moreover, they have different regenerative capacities [9] and are differentially affected in genetic disorders [10]. Numerous studies have begun to clarify the developmental, cellular and molecular bases of this diversification [11-13]. All the muscles in the trunk and limbs develop from the somites [14], whereas, in the head, only the muscles of the tongue,and some muscles of larynx and neck are believed to be somitic in origin [11]. Extraocular muscles (EOM) are derived from the prechordal and cranial paraxial mesoderm. The remaining head muscles (the branchiomeric muscles) control facial expression, pharyngeal function and jaw movements and originate from the paraxial unsegmented mesoderm and the lateral splanchnic mesoderm [11, 15]. Molecular and biochemical properties distinguish head from body muscles, and there are even biochemical distinctions among muscles of the head [16]. Similarly body muscles are heterogeneous. Based on developmental and innervation pattern, it is possible to distinguish epaxial muscles of the back from hypaxial muscles, such as limb muscles and diaphragm [12, 17] (Table 1). Lineage-tracing studies both in chick and mouse have disclosed differences in the origin of these different muscles groups. Importantly, extraocular, branchiomeric and somitic muscles (both in the head and in the body) follow distinct genetic programs during development [18-25].
Table 1. Heterogeneity of SCs based on lineage history and muscle of origin.
SCs resident in different muscle groups originate from different embryonic tissues and can be discriminated according to their lineage history, based on Cre/loxP-mediated cell tracing experiments. Indicated Cre driver mice express Cre recombinase under the control of the regulatory sequences of specific genes and can be used for genetic labeling of precursor cells in vivo. A “+” indicates that SCs of the individual muscle are derived from, or induced to have been derived from (see notes), progenitors of the lineage that his mapped by the specific Cre driver.
| Muscle group | Developmental origin | Lineage (Cre-drivers) | References | |||||
|---|---|---|---|---|---|---|---|---|
| Myf5Cre | MyoDCre | Pax3Cre | MCre2 | Mesp1Cre | Isl1Cre | |||
| EOM | Cranial unsegmented mesoderm |
+ | + | − | − | + | − | [18-19, 127] |
| Branchiomeric | Cranial unsegmented mesoderm |
+ | +1 | − | − | + | + | [18-19] |
| Tongue (hypaxial) |
Occipital somites | + | +1 | + | +1 | + | − | [19] |
| Limbs (hypaxial) |
Somitic dermomyotome |
+ | + | + | + | − | − | [18-19, 31, 87, 127] |
| Inter-limb (hypaxial) |
Somitic dermomyotome |
+1 | + | +3 | +1 | − 1 | − 1 | [127] |
| Back (epaxial) | Somitic dermomyotome |
+1 | +1 | + | − | − 1 | − 1 | [30-31] |
Not formally proven, but deduced from lineage studies on the corresponding muscle.
Hypaxial Pax3-enhancer [31].
Pax3nlacz/Pax3GFP are transcriptionally active in interlimb SCs (see text).
Given the enormous diversity among muscles, it is not surprising that distinctions between the SCs residing within them would be found. In amniotes, the presence of SCs seems to be a common feature of all adult skeletal muscles, although the density of SCs differs among different muscle groups [26-28]. Beyond the simple property of number or density, evidence suggests that SCs resident in different muscle compartments are not identical but differ in terms of embryonic origin, lineage history, gene expression pattern and functional behavior in vitro and in vivo (see below).
Chick/quail chimera experiments, together with molecular and retroviral labeling experiments, showed that SCs of the limb (hypaxial) and back (epaxial) muscles are somitic in origin [29-31]. It has been proposed that cells expressing Pax3 and Pax7, which are resident in the central portion of the somitic dermomyotome, could be their embryonic ancestors [32-33]. SCs of the limb are believed to derive from Pax3+ cells, which migrate from the lateral lip of the dermomyotome [31] and are initially negative for Pax7 but subsequently become dependent on Pax7 expression [32-36].The developmental origin of the SCs of other hypaxial muscles has not been evaluated carefully but is believed to be similar to that proposed for the hindlimbs. These observations suggest that the SCs of the body muscle derive from the same embryonic precursors as the muscles in which they reside.
Observations obtained with chick/quail transplantation experiments suggest that SCs of non-somitic head muscles also share a developmental origin with the muscles in which reside [19]. Homotopic transplantation of unsegmented quail mesoderm into the head of chick embryos revealed SCs of quail origin in newborn eye and branchiomeric muscles [19]. Additionally, a comparative lineage analysis has recently been performed by crossing several tissue-specific Cre-expressing mice with reporter lines. These analyses revealed that SCs in different compartments express different combinations of markers, suggesting distinct embryonic lineages [19]. Similar to the transplantation experiments, a strict correlation between SCs and the muscles in which they reside is evident from these lineage studies [19] (Table 1).
2.1 Molecular characterization
Muscles of different developmental origin or fiber type composition present important differences in term of function and gene expression, identifying specific muscle “allotypes”. The categorization into different allotypes is supported by genome-wide expression analyses of whole muscles [37-38]. SCs in different muscles also differ in terms of gene expression. SCs sorted by FACS from mouse EOM express higher levels of Alx4, Pitx1, Pitx2 and Tcf21 compared to limb SCs [18]. Those from branchiomeric muscles also express higher levels of Tcf21 [18-19]. Pax3 and Lbx1 expression is lower in head SCs compared to those in limb [18]. Remarkably, EOMs are lost in mice in which the expression of Pitx2 is abolished [24-25]. In embryos in which both musculin and Tcf21 are genetically ablated, a subset of branchial muscles fails to develop [22], whereas in Pax3 and Lbx1 knock-out mice, limb myogenesis is affected [39]. Thus the differences in gene expression reflect the developmental ontology of SCs, indicating that the genetic requirements for SC generation could reflect the genetic programs governing the formation of the corresponding muscles.
Differences in gene expression have also been reported for SCs sorted from the diaphragm compared to limb muscles. In particular, SCs from the diaphragm express higher levels of Pax3 [40]. This observation correlates with evidence from mice in which the Pax3 locus drives the expression of reporter genes [34, 41]. In these mice, only the SCs of the diaphragm, a subset of the forelimb and intercostal muscles, and the hindlimb gracilis muscle express the reporter [34].
2.2 Characterization in vitro and after transplantation in vivo
The finding that the molecular signature of SCs is different in different muscles raises the question whether this signature is intrinsic to the cells or depends on the environment. When SCs were sorted from adult mouse EOM, they failed to express EOM-specific markers such as Myh13 and Myh15 after differentiation in vitro [18]. Similarly, when sorted EOM SCs were transplanted into tibialis anterior (TA) muscles, they efficiently engrafted but failed to express EOM-specific markers [18]. These data suggest that the EOM SC phenotype is not dictated by intrinsic cues. However, this conclusion is challenged by the identification of a different pattern of gene expression in myogenic cell lines obtained from EOM and limb muscles [42], suggesting a hereditable EOM-specific signature.
SCs from mouse extensor digitorum longus (EDL) and masseter present differences in gene expression (i.e. Pax3, Lbx1, Tcf21) when freshly isolated and immediately assayed, and those differences are maintained when the cells are propagated in culture, again, indicating a heritable, muscle-specific signature [18-19, 28]. When myoblasts isolated from feline masseter were isolated and induced to differentiate in vitro, expression of masticatory-specific genes became apparent [43]. Furthermore, when SC cultures were treated with BMP4, a greater expression of cardiac genes, such as Isl1 and Tbx20, was observed in branchiomeric compared to the limb cultures [19]. Finally, when feline (fast) masticatory muscles were transplanted into a fast limb muscle bed and allowed to regenerate and become innervated, the regenerated muscles still expressed masticatory MyHC [44]. This seems to be independent of innervation [45] and donor basal lamina [46]. Intriguingly, when the same muscles were transplanted in the slow soleus bed, only a few transplanted fibers expressed the masticatory MyHC [44]. These studies suggest that branchiomeric SCs are programmed to express a masticatory-specific set of myogenic genes, but that this program can be overridden by a slow, but not by a fast, muscle environment. A cell-autonomous signature characterizes not only the branchiomeric SCs, but also those of the diaphragm. Indeed, GFP+ SCs sorted from the diaphragm of Pax3GFP/+ mice and engrafted into the GFP− TA muscle retain their initial phenotype [41].
3. Heterogeneity based on fiber association
The multinucleated myofibers that comprise each muscle are, themselves, heterogeneous. On the basis of anatomical and functional properties, two major groups of fibers can be identified: extrafusal and intrafusal. Extrafusal fibers are far more numerous, are innervated by motor neurons, and are responsible for the maintenance of posture and locomotion. Within the extrafusal fiber population, further heterogeneity exists (see below). Intrafusal fibers are important for the control of muscle contraction by monitoring changes in muscle length, which are then transduced into proprioceptive signals by sensory neurons [47]. Below we will discuss heterogeneity of SCs based on their association with either extrafusal or intrafusal fibers.
3.1 Extrafusal fibers: fast vs. slow
Adult extrafusal fibers are heterogeneous based on their speed of contraction, which depends mainly on the ATPase activity of the predominant myosin isoform that is expressed, and are categorized broadly as being either “fast” or “slow” contracting fibers. In rodents, a single slow and three adult fast myosin heavy chain (MyHC) genes have been identified [48].
The characteristics of the mature fibers are indisputably influenced by extrinsic factors such as functional demands, electrical stimulation, and hormones [49]. Nevertheless, work of several investigators also suggests that fiber type is dependent, at least in part, on the intrinsic properties of the progenitors which contributed to their generation [12].
Several differences in SCs associated with fast or slow fibers/muscles have been reported, including differences in the proliferation rate and differentiation capacity of their progeny in culture [50-52]. Moreover, a higher adipogenic potential was observed in cells isolated from the slow soleus than from predominantly fast muscles in the rat [53]. Myoblasts from fast or slow muscles differ in the levels of expression of many genes, such as AChE [54], FGF-receptors [55], and Pax7 [56]. Notably, all these differences were observed in vitro and thus in absence of innervation or a specific milieu in vivo, further suggesting an intrinsic diversity of “slow” and “fast” SCs.
Many studies have evaluated the pattern of myosin expression in cultures of myotubes derived from SCs associated with fast or slow contracting fibers. Myogenic cells from avian [51, 57] and rodent [50, 58-59] fast muscles or fast fibers clearly give rise to myotubes expressing exclusively fast MyHC whereas cells from slow muscles/fibers give rise to myotubes expressing slow MyHC and fast MyHC. Additionally, by engineering three-dimensional muscle constructs from myogenic cells isolated from rat TA and soleus, differences in the dynamics of contraction were reported, indicating that the cells from fast and slow muscles give rise to functionally distinct differentiated muscle cells in vitro [60]. Intriguingly, it has been reported that, in contrast to birds and rodents, human fast and slow muscles/fibers give rise to myogenic clones almost all of which co-express fast and slow MyHC [61-62].
The capacity of different subpopulation of SCs to maintain a specific phenotype for multiple cycles of replication suggests an epigenetic control of SC heterogeneity. Although hereditable, epigenetic modifications can be altered under specific stimuli; the observation that quail SC-derived myoblasts can change over time in culture, giving rise to myotubes with different patterns of MyHC expression [51], and that the in vitro conditions can affect the differentiation program in rodent cells [59], reveal a certain degree of plasticity in the SCs. Indeed, transplantation experiments indicate that myoblasts can express their intrinsic program if they differentiate alone, but that this program is overridden upon fusion with a fiber that expresses a different phenotype [63].
Nevertheless, there is considerable evidence of intrinsic differences between SCs from fast and slow muscles [64-66]. When clones of SC-derived myoblasts with the capacity to differentiate into both fast and slow myotubes in vitro were transplanted into the limb buds of developing embryos, they generated fibers co-expressing fast and slow MyHC [66]. Moreover, in experiments in which the rat soleus was injured and transplanted to the bed of the EDL, the signature of the SCs of the soleus was revealed by the presence of fibers expressing slow MyHC when analyzed after 3 months [65]. However, this signature could not be observed after 6 months, suggesting that the slow phenotype was stable, but not irreversible, and that extrinsic influences such as the innervation pattern could eventually override the intrinsic program [67]. These observations also suggest that innervation can efficiently reprogram mature regenerated fibers but not SCs themselves. Indeed, when fast or slow muscles were stimulated by slow or fast innervation patterns, respectively, a large shift of fiber-type was observed in the muscles, but the in vitro differentiation predisposition of the SCs was not changed [68].
3.2 Intrafusal fibers
Intrafusal fibers express genes specific to embryonic myogenesis [47]. They are thus believed to persist in a relatively immature state. Studies suggest that this peculiarity could reflect the existence of unique types of intrafusal myogenic progenitors with distinct differentiation programs [47]. Nevertheless, it is not known to what extent spindle fiber SCs are intrinsically predetermined. Indeed, sensory innervation seems to be important for proper spindle fiber formation and studies involving the transplantation of growing muscles indicate that intrafusal SCs show great plasticity, as their MyHC expression can be respecified towards the extrafusal muscle fiber phenotype by foreign motor innervation [69-70].
The idea of the existence of intrafusal SCs with specific characteristics is corroborated by the report of the expression of Pax3 in a subpopulation of intrafusal fiber-associated SCs [71]. It remains undetermined if this difference in Pax3 expression is intrinsic or due to the innervation of the intrafusal fibers. Similar to the SCs in the EOM [72], a small fraction of the SCs in the spindle fibers could also be mitotically active in absence of external insults. In this sense, it is interesting to note that both EOM and spindle fibers contain a particularly high density of SCs [73-74].
4. Heterogeneity based on postnatal stage
Prenatally, myogenic progenitors establish the patterning of skeletal muscle [12]. After birth, skeletal muscle undergoes robust growth associated with the general growth of the organism. During this phase, muscle fibers increase in size by the fusion of post-natal progenitors. During this phase, it is difficult to equate the sublaminar cells, which are proliferative, with the entirely quiescent SCs of mature muscle. In mature, adult muscle, SCs can be activated in response to specific stimuli and are involved in the maintenance of muscle homeostasis and in the response to external insults or changes of functional demand [75]. Although relatively stable, skeletal muscle progressively changes with age, and those changes are accompanied also by changes in the SC population [76]. Characterizations of SCs in the young (growing), adult and aged muscle environment are considered below.
4.1 Postnatal growing muscle
Sublaminar cells can first be detected during late fetal stage [77]. It is generally believed that SCs and their progeny are responsible for the postnatal growth of the muscles. This idea is supported by the observation that myogenic cells, located beneath the basal lamina, can be frequently observed during postnatal growth. Moreover, newborn myoblasts give rise to functional SCs when transplanted in the adult muscle [78]. As in the adult (see below), distinct subsets of SCs have been proposed for growing muscle [79-80].
With regard to the presence of SCs in the neonatal period, clear differences have been observed between newborn and adult myogenic cells. Newborn myoblasts engraft less efficiently than their adult counterparts [81]. However, they regenerate fibers that are largely peripherally nucleated, whereas regenerated adult mouse skeletal muscles are normally centrally nucleated [82]. Moreover, the activation of myoblasts during neonatal growth may be different from that occurring during adult muscle regeneration [83]. Additionally, a cell-intrinsic difference between neonatal progenitors and adult SCs in their Pax7-dependency has been recently demonstrated [35].
Importantly, studies indicate that the pool of myogenic cells present in juvenile muscles is more complex than initially predicted. First, until approximately 3 weeks of age in the mouse, a subset of the myogenic progenitors in growing muscles are actively proliferating or committed to terminal differentiation [84]. This is distinct from the adult where essentially all the progenitors are quiescent, undifferentiated SCs in absence of an external injury or stimuli. Furthermore, although evidence suggests that immediately after birth the majority of myoblasts exhibit characteristics distinct from fetal myoblasts in vitro [85], it is not known when prenatal progenitors are no longer present in muscles. Finally, the recent description of myogenic Pw1+/Pax7− interstitial cells (“PICs”) in postnatal growing muscle further increases the level of complexity [86].
4.2 Biochemical and functional heterogeneity of adult SCs
There is increasing evidence that there is SC heterogeneity beyond that conferred by the muscle in which they reside or the fiber type with which they are associated. Indeed, SCs present in the same muscle and even on the same fiber can express different markers and exhibit different functions, as described below.
4.2.1 Markers
For years, electron microscopy remained the only way to identify SCs. Over the past decades, molecular markers have been found which are sufficiently specific to allow SC identification at the light microscope level in muscle sections or single fiber preparations. Some of the markers are membrane proteins, which therefore can be used to isolate SCs with FACS, although in most cases, combinations of positive and negative markers are required to obtain a high purity. Mutant mice in which the expression of reporter genes is under the control of SC-specific genes also represent a way to identify and isolate SCs. A list of recognized SC markers is provided in Table 2.
Table 2. Quiescent SC markers.
A list of markers expressed in quiescent SCs is provided together with information regarding their expression after activation and their usage for prospective isolation by FACS.
| Gene/Protein | Active SC1 |
FACS | Comments | References |
|---|---|---|---|---|
| Membrane | ||||
| Syndecan 3-4 | + | yes (7% are CD31+) | nearly all SCs | [90, 156] |
| Integrin-α7 | + | with Itgβ1+/Lin−2 or CD34+ /Lin−2 or Syn4+/Lin−2 |
nearly all SCs | [87, 91, 125- 126, 157] |
| Integrin-β1 | + | with Itga 7+/ Lin−2 or CXCR4+/Lin−2 |
~90% of SCs | [87, 89] |
| CXCR4 | + | with1 Itgβ+ /Lin−2 | ~80% of SCs | [89, 94] |
| CD34 | +/− | with Itgα7+/ Lin−2 | majority of SCs | [88, 91, 126, 158] |
| Vcam1 | + | with Lin−2,3 | nearly all SCs | [159-160] |
| SM/C2.6 | + | yes | [161] | |
| NCAM1 (CD56) | + | yes | used for FACS in human | [162] |
| Calcitonin receptor | ~ | n.t.5 | great majority of SCs | [157, 163] |
| Nrn1 | + | n.t.5 | [160] | |
| Igsf4a | + | n.t.5 | [160] | |
| Cadherin 15 (Mcad) | + | n.t.5 | majority of SCs | [88] |
| c-Met | + | n.t.5 | nearly all SCs | [99] |
| Caveolin-1 | + | n.t.5 | nearly all SCs | [157] |
| Nucleus | ||||
| Pax7 | + | Pax7GFP | nearly all SCs | [36, 93] |
| Pax3 | + | Pax3GFP | subsets of muscles | [41] |
| Myf5 | + | Myf5Cre/R26RSTOP-YFP | 80-90% of SCs4 | [87-88] |
| Foxk1 (Mnf) | + | switch of isoform during activation |
[164] | |
| Msx1 | − | [92] | ||
| Sox8-9 | + | [165] | ||
| Cytoplasm | ||||
| Nestin | − | NestinGFP | endogenous protein differs from NestinGFP |
[40] |
| Desmin | + | higher in differentiation | [40, 166] | |
| Secreted | ||||
| Myostatin | ~ | [92, 167] | ||
| Bdnf | + | 64% of diaphragm SCs | [168] |
Expression after activation: + present; − not expressed; ~ expressed at low level or unclear;+/− switch of isoform [88] before down-regulation [158].
Lin−: negative selection for CD45 (hematopoietic); CD31 (endothelial) and in certain cases Sca1 and/or CD11b.
Unpublished.
Data obtained with Myf5nlacz and Myf5Cre mice.
Not tested.
Some markers characterize specific subpopulations of SCs. For example, not all the SCs express CD34 or M-cadherin, have an active Myf5 locus, or have previously expressed Myf5 [87-88]. Only specific subsets of quiescent SCs express Itgß1 or CXCR4 [89] or the “side population” (SP) cell marker ABCG2 [90]. Single cell expression studies also suggest variability in the SC pool [91-92]. Importantly, many markers are expressed at low level and therefore some of the heterogeneity might arise due to detection problems. Indeed, different results have been obtained when different techniques have been used, as exemplified by studies of Myf5, M-cadherin and Sca1 [40, 88, 90, 92-94]. Additionally, only in rare cases have differences in gene expression been linked with functional heterogeneity [87, 89-90]. Finally, due to the complexity of the SC niche [76], some of the differences in gene expression could simply reflect transient states of SCs under the influence of local cues.
4.2.2 Activation, proliferation and lineage progression
In early studies of injured rat muscles, desmin+, MyoD+ and myogenin+ myoblasts were observed within 12 hours after injury, but SC proliferation was not detected until 24 hours [95]. This temporal pattern suggested the existence of two populations of precursor cells in adult muscles: more committed SCs, which are ready for immediate differentiation without preceding through cell division, and more immature SCs, which undergo mitosis before generating daughter cells capable of differentiating [95]. Similar to embryonic myogenic progenitors [96-97], SCs have been shown to enter the myogenic program by inducing either MyoD or Myf5 [92, 98-99] (Figure 1). It is not known if SCs follow a different developmental program depending on the predominant expression of MyoD or Myf5, although the delayed differentiation reported for MyoD−/− myoblasts [100] and the precocious differentiation observed in Myf5−/− myoblasts [101] may suggest that this is the case. Several studies also indicate a variability in terms of morphology, adhesion properties, proliferation, clonogenic capacity, survival, differentiation and fusion ability in the myogenic precursors isolated by enzymatic digestion from single muscles [93, 102-105].
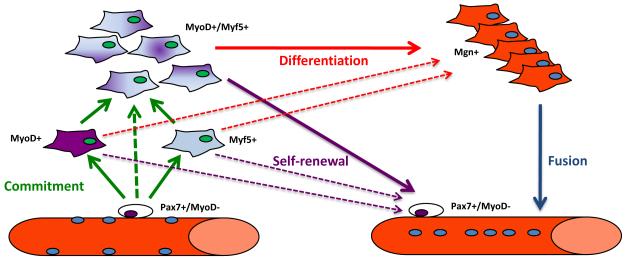
Figure 1. Schematic representation of SC myogenic progression.
Upon activation, SCs (initially Pax7+/MyoD−) can enter the myogenic program by either inducing Myf5 or MyoD expression. Most of the cells subsequently express both determination genes and terminally differentiate by inducing Myogenin (Mgn+) expression and then fusing to form multinucleated fibers. A fraction of the SCs self-renew by down-regulating MyoD. Dashed lines represent possible but not proven connections. A fraction of SCs possibly differentiate without entering the cell cycle (see text). The newly formed fiber on the right is depicted as having centrally placed nuclei to reflect the process that occurs during muscle regeneration as opposed to homeostatic turnover of mature myonuclei by SCs which occurs normally at a very low level.
The capacity of SCs to differentiate into alternative lineages, such as fibrogenic, adipogenic and osteogenic lineages, has been suggested by in vitro analyses of cells associated with muscle fibers [106-112]. Only in the case of the fibrotic conversion has the SC origin been confirmed by lineage tracing, thus ruling out the possibility of contaminating cells associated to the fiber [106]. Intriguingly, recent work with mouse SCs suggests that only a fraction of SCs possess a transdifferentiation potential, thus linking intrinsic SC heterogeneity to multipotentiality [107, 113]. The recent observation that in human muscles a subpopulation of cells expressing the myogenic marker CD56 and co-expressing CD34 contains cells with adipogenic potential further supports this possibility [114].
4.2.3 Self-renewal and quiescence
The idea that SCs can both terminally differentiate into multinucleated fibers and self-renew was definitively confirmed only recently by experiments in which few SCs were transplanted and shown to contribute both to muscle regeneration and to the replenishment of the undifferentiated SC pool [27]. Different models, not necessarily mutually exclusive, implying stochastic events and/or asymmetric cell divisions, can explain how the progeny of SCs can adopt these divergent fates [115].
Undifferentiated subpopulations of cells that can self-renew and give rise to terminally differentiated myotubes have been identified in cultures of single muscle precursors established from adult muscles [116-117]. Further studies on single fiber-associated SCs in suspension also confirmed that a subset of activated SCs will retain Pax7 expression, turn off the expression of MyoD, return to quiescence and thus renew the stem cell pool [118-119] (Figure 1). These studies indicate that SCs can adopt alternative fates but do not discriminate whether all SCs have this capacity or if stem-cell like properties reside only in a subset of cells.
The hypothesis that SCs are heterogeneous in terms of self-renewal is supported by evidence from transplantation experiments. Only a small subpopulation of cultured myoblasts was shown to efficiently survive transplantation and contribute to regeneration, exhibiting stem cell-like properties [120]. This subpopulation is characterized by a slow rate of replication in vitro but efficiently expands in vivo [120]. Experiments involving freshly sorted SCs or fiber associated SCs showed a high rate of successful engraftment, suggesting that the expansion of these cells in culture before transplantation reduces their regenerative capacity, possibly selecting against cells with stem-cell properties [41]. Moreover, when single sorted SCs are transplanted, only a small percentage engraft [91]. It is not known if these latter observations simply reflect a stochastic survival of the cells after transplantation or a true heterogeneity.
The behavior of muscle stem cells under specific experimental conditions further suggests the presence of subsets of SCs with different self-renewal ability. Myogenic cells that present different proliferative/regenerative potentials and that can be differentially activated by distinct types of injury were identified on the basis of their sensitivity to irradiation [121]. The finding suggests that an altered environment possibly selects for specific subpopulations of SCs with different self-renewing ability [121].This idea was supported by the observation that the aging process may select for subpopulations of SCs with high self-renewal ability [122].
Recently, non-random DNA segregation was shown to be strongly associated with divergent fates of self-renewing SCs: in the great majority of the cases, the daughter cell inheriting the older templates retained the more immature phenotype, whereas the daughter inheriting the newer templates exhibited signs of differentiation [123-124]. The observation that asymmetric segregation of parental DNA occurs in a fraction of SCs or their progeny with frequencies ranging from as low as 7% [123] to as high as 50% [124], albeit under different experimental conditions, suggests that the ability to segregate parental DNA non-randomly may be a property of a subset of SCs.
Cd45−/Sca1−/Mac1−/ß1integrin+/CXCR4+ SCs represent a subset of fiber-associated cells with high myogenic properties and self-renewal capacity after transplantation [89, 94]. Transplanted cells not only self-renew, but also give rise to CXCR4−/ß1integrin− and CXCR4−/ß1integrin+ cells, suggesting that ß1integrin+/CXCR4+ may be the more primitive stem cells [89]. In this sense, as the majority of the Pax7+ cells associated with the fiber are CXCR4+ and ß1integrin+ [89], it would be interesting to compare their ability to self-renew with the smaller CXCR4− and/or ß1integrin− fractions. Similarly, a small fraction of SCs expressing the side population marker ABCG2 and Sca1 were identified [90]. They represent 3-10% of the Syndecan-4+ cells, co-express the SC marker Pax7, and apparently engraft more efficiently than the rest of the SC pool [90]. The requirement of Sprouty1 for the return to quiescence of a subset of SCs after injury further suggest the presence of distinct fractions in the pool of self-renewing SCs [125].
Based on studies using mice expressing Cre under the control of the regulatory regions of Myf5 (Myf5Cre) intercrossed with Rosa26YFP reporter mice, it was concluded that approximately 10% of the Pax7+ SCs have never expressed Myf5 by virtue of being YFP− [87]. Transplantation revealed that these cells extensively contributed to the SC reservoir, whereas YFP+ cells only occasionally gave rise to SCs. Both YFP+ and YFP− SCs efficiently contributed to fiber regeneration [87]. Importantly, exogenous Wnt7a stimulated the expansion of YFP− “satellite stem cells” [126]. The genetic strategy, such as using Myf5Cre, represents a powerful approach to disclose SC heterogeneity, but there are several caveats that need to be borne in mind. First, YFP− cells could result from inefficient recombination. Additionally, the presence of YFP does not imply that cells co-express Myf5. It is possible that a subset of the YFP+ SCs no longer expresses Myf5 and thus may be functionally equivalent to the YFP− cells. It is also important to note that a subset of the YFP+ SCs in vivo could be derived from pre- or perinatal myoblasts and not from YFP− SCs. Similar lineage studies with MyoDCre mice revealed that almost all the SCs are derived from myoblasts expressing MyoD pre- or perinatally [127]. These observations raise some questions about the homogeneity of the (Myf5Cre)YFP+ population of SCs. Future studies will determine the developmental history of SCs with regard to Myf5 and MyoD expression and whether a distinct subset is more stem-like.
4.3 Aging muscle
In aged skeletal muscles, many aspects of the regenerative response are impaired [106, 128-130]. This correlates with an age-dependent decrease in the functionality of the SCs, and possibly with a decrease in SCs numbers, although there is debate about the magnitude and functional significance of any observed changes in SC number with age [131]. The niche in which the SCs reside is profoundly modified during aging [76]. Alteration both in the local and systemic environment of SCs impacts their function, accounting for cell-extrinsic changes in addition to any cell-intrinsic changes that accompany aging and contribute to impaired SC functionality [131]. Once established, cell-intrinsic changes may be irreversible or may in fact be reversible [132], depending on the nature of those changes. Either way, these changes could represent an additional source of SCs heterogeneity.
The transcriptional profile of SCs (or their progeny) changes with age [133]. Myogenic cells from aged muscles showed a delayed response to activation cues and an initially reduced proliferative expansion in vitro [103, 130, 134-137]. Intriguingly in the aged muscle, a reduced expression of the Notch-ligand Delta affects Notch signaling and results in a reduced proliferation of myogenic progenitors and impaired regeneration [130]. TGFβ, Wnt, and IGF pathways have also implicated in the impaired SCs proliferation and myogenic progression associated with the aging process [138-141]. Furthermore, impaired differentiation ability in vitro has been reported for aged SCs [122, 142]. Evidence suggests an increased tendency of aged SCs to enter alternative differentiation programs by adopting an adipogenic and fibroblastic fate [106, 143], and SCs may be more prone to undergo senescence or apoptosis with aging [144]. Finally, besides affecting the functionality of SCs, the aged environment possibly selects for specific subpopulations of SCs with distinct regenerative potential [122]. Collectively, these observations disclose multiple levels of diversification between muscle stem cells present in the young and old muscle.
5. Concluding remarks
An increasing amount of evidence indicates, as described herein, that SCs are heterogeneous by many criteria. Nevertheless many unanswered questions remain. How are different subpopulation of SCs related to one another from lineage and functional perspectives? Which roles do different subpopulations exert under normal conditions of growth, homeostasis and regeneration? How are the properties and functionality of SCs or specific subsets of SCs affected by pathological conditions? In the setting of diseases, extrinsic influences, different from those acting in healthy adult muscle, may impart both reversible and irreversible changes to SCs, as recently described for aging muscle [131]. These processes remain poorly understood and require further investigation.
Muscle regeneration depends primarily, if not exclusively, upon SCs. However, other cell types have been shown to have myogenic potential [145-146], although in most cases the physiological relevance remains unclear. Even more uncertain is the relationship of these cells with SCs. Intriguingly, bone marrow derived cells [147-148], SP cells [149], mesenchymal cells from the synovia [150], AC133+ cells [151], pericytes [152], mesoangioblasts [153] and even embryonic stem cells [154] and induced pluripotent stem cells (iPS) [155] (or the progeny of all of these populations) have been found in the SC position or to express SC markers after transplantation. Although in certain cases (for example for ES cells) the capacity to generate SC-like cells is not meant to model a natural process, the physiological contribution to the SC pool by other cell types is worthy of future investigation. A better understanding of the origin and heterogeneity of SCs, together with further studies of the mechanisms driving their self-renewal, quiescence, proliferation and differentiation, will all be important steps for harnessing the potential of adult stem cells in order to apply them, ultimately, to regenerative medicine.
Acknowledgements
We thank Suchitra Gopinath for discussions. This work was supported by grants from the Department of Veterans Affairs and the NIH (AG23806, AR056849, and an NIH Director’s Pioneer Award (OD000392)) to TAR.
Abbreviations
- SC
satellite cell
- EDL
extensor digitorum longus
- TA
tibialis anterior
- EOM
extra-ocular muscle
- MyHC
myosin heavy chain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Mauro A. Satellite cell of skeletal muscle fibers. The Journal of Biophysical and Biochemical Cytology. 1961;9:493–5. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kuang S, Rudnicki MA. The emerging biology of satellite cells and their therapeutic potential. Trends in Molecular Medicine. 2008;14:82–91. doi: 10.1016/j.molmed.2007.12.004. [DOI] [PubMed] [Google Scholar]
- [3].Reznik M. Thymidine-3H uptake by satellite cells of regenerating skeletal muscle. The Journal of Cell Biology. 1969;40:568–71. doi: 10.1083/jcb.40.2.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Moss FP, Leblond CP. Nature of dividing nuclei in skeletal muscle of growing rats. The Journal of Cell Biology. 1970;44:459–61. doi: 10.1083/jcb.44.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Moss FP, Leblond CP. Satellite cells as the source of nuclei in muscles of growing rats. The Anatomical Record. 1971;170:421–35. doi: 10.1002/ar.1091700405. [DOI] [PubMed] [Google Scholar]
- [6].Allbrook DB, Han MF, Hellmuth AE. Population of Muscle Satellite Cells in Relation to Age and Mitotic Activity. Pathology. 1971;3:233–43. doi: 10.3109/00313027109073739. [DOI] [PubMed] [Google Scholar]
- [7].Schultz E. A quantitative study of the satellite cell population in postnatal mouse lumbrical muscle. The Anatomical Record. 1974;180:589–95. doi: 10.1002/ar.1091800405. [DOI] [PubMed] [Google Scholar]
- [8].Zammit PS, Heslop L, Hudon V, Rosenblatt JD, Tajbakhsh S, Buckingham ME, et al. Kinetics of Myoblast Proliferation Show That Resident Satellite Cells Are Competent to Fully Regenerate Skeletal Muscle Fibers. Experimental Cell Research. 2002;281:39–49. doi: 10.1006/excr.2002.5653. [DOI] [PubMed] [Google Scholar]
- [9].Pavlath GK, Thaloor D, Rando TA, Cheong M, English AW, Zheng B. Heterogeneity among muscle precursor cells in adult skeletal muscles with differing regenerative capacities. Developmental Dynamics. 1998;212:495–508. doi: 10.1002/(SICI)1097-0177(199808)212:4<495::AID-AJA3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- [10].Emery AEH. The muscular dystrophies. The Lancet. 2002;359:687–95. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- [11].Noden DM, Francis-West P. The differentiation and morphogenesis of craniofacial muscles. Developmental Dynamics. 2006;235:1194–218. doi: 10.1002/dvdy.20697. [DOI] [PubMed] [Google Scholar]
- [12].Biressi S, Molinaro M, Cossu G. Cellular heterogeneity during vertebrate skeletal muscle development. Developmental Biology. 2007;308:281–93. doi: 10.1016/j.ydbio.2007.06.006. [DOI] [PubMed] [Google Scholar]
- [13].Sambasivan R, Tajbakhsh S. Skeletal muscle stem cell birth and properties. Seminars in Cell & Developmental Biology. 2007;18:870–82. doi: 10.1016/j.semcdb.2007.09.013. [DOI] [PubMed] [Google Scholar]
- [14].Christ B, Ordahl CP. Early stages of chick somite development. Anatomy and Embryology. 1995;191:381–96. doi: 10.1007/BF00304424. [DOI] [PubMed] [Google Scholar]
- [15].Nathan E, Monovich A, Tirosh-Finkel L, Harrelson Z, Rousso T, Rinon A, et al. The contribution of Islet1-expressing splanchnic mesoderm cells to distinct branchiomeric muscles reveals significant heterogeneity in head muscle development. Development. 2008;135:647–57. doi: 10.1242/dev.007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mootoosamy RC, Dietrich S. Distinct regulatory cascades for head and trunk myogenesis. Development. 2002;129:573–83. doi: 10.1242/dev.129.3.573. [DOI] [PubMed] [Google Scholar]
- [17].Buckingham M, Vincent SD. Distinct and dynamic myogenic populations in the vertebrate embryo. Current Opinion in Genetics & Development. 2009;19:444–53. doi: 10.1016/j.gde.2009.08.001. [DOI] [PubMed] [Google Scholar]
- [18].Sambasivan R, Gayraud-Morel B, Dumas G, Cimper C, Paisant S, Kelly RG, et al. Distinct Regulatory Cascades Govern Extraocular and Pharyngeal Arch Muscle Progenitor Cell Fates. Developmental Cell. 2009;16:810–21. doi: 10.1016/j.devcel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- [19].Harel I, Nathan E, Tirosh-Finkel L, Zigdon H, Guimarães-Camboa N, Evans SM, et al. Distinct Origins and Genetic Programs of Head Muscle Satellite Cells. Developmental Cell. 2009;16:822–32. doi: 10.1016/j.devcel.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the Genetic Hierarchies Controlling Skeletal Myogenesis: Pax-3 and Myf-5 Act Upstream of MyoD. Cell. 1997;89:127–38. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- [21].Kassar-Duchossoy L, Gayraud-Morel B, Gomès D, Rocancourt D, Buckingham M, Shinin V, et al. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–71. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- [22].Lu J-r, Bassel-Duby R, Hawkins A, Chang P, Valdez R, Wu H, et al. Control of Facial Muscle Development by MyoR and Capsulin. Science. 2002;298:2378–81. doi: 10.1126/science.1078273. [DOI] [PubMed] [Google Scholar]
- [23].Kelly RG, Jerome-Majewska LA, Papaioannou VE. The del22q11.2 candidate gene Tbx1 regulates branchiomeric myogenesis. Hum Mol Genet. 2004;13:2829–40. doi: 10.1093/hmg/ddh304. [DOI] [PubMed] [Google Scholar]
- [24].Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–51. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- [25].Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R, et al. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development. 1999;126:5749–58. doi: 10.1242/dev.126.24.5749. [DOI] [PubMed] [Google Scholar]
- [26].Schmalbruch H, Hellhammer U. The number of nuclei in adult rat muscles with special reference to satellite cells. The Anatomical Record. 1977;189:169–75. doi: 10.1002/ar.1091890204. [DOI] [PubMed] [Google Scholar]
- [27].Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, et al. Stem Cell Function, Self-Renewal, and Behavioral Heterogeneity of Cells from the Adult Muscle Satellite Cell Niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- [28].Ono Y, Boldrin L, Knopp P, Morgan JE, Zammit PS. Muscle satellite cells are a functionally heterogeneous population in both somite-derived and branchiomeric muscles. Developmental Biology. 2010;337:29–41. doi: 10.1016/j.ydbio.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Armand O, Boutineau AM, Mauger A, Pautou MP, Kieny M. Origin of satellite cells in avian skeletal muscles. Archives d’anatomie microscopique et de morphologie expérimentale. 1983;72:163–81. [PubMed] [Google Scholar]
- [30].Gros J, Manceau M, Thomé V, Marcelle C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–8. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- [31].Schienda J, Engleka KA, Jun S, Hansen MS, Epstein JA, Tabin CJ, et al. Somitic origin of limb muscle satellite and side population cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:945–50. doi: 10.1073/pnas.0510164103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomès D, Tajbakhsh S. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes & Development. 2005;19:1426–31. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–53. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- [34].Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, et al. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. The Journal of Cell Biology. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lepper C, Conway SJ, Fan C-M. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–31. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 Is Required for the Specification of Myogenic Satellite Cells. Cell. 2000;102:777–86. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- [37].Porter JD, Khanna S, Kaminski HJ, Rao JS, Merriam AP, Richmonds CR, et al. Extraocular muscle is defined by a fundamentally distinct gene expression profile. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12062–7. doi: 10.1073/pnas.211257298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Porter JD, Merriam AP, Leahy P, Gong B, Feuerman J, Cheng G, et al. Temporal gene expression profiling of dystrophin-deficient (mdx) mouse diaphragm identifies conserved and muscle group-specific mechanisms in the pathogenesis of muscular dystrophy. Hum Mol Genet. 2004;13:257–69. doi: 10.1093/hmg/ddh033. [DOI] [PubMed] [Google Scholar]
- [39].Birchmeier C, Brohmann H. Genes that control the development of migrating muscle precursor cells. Current Opinion in Cell Biology. 2000;12:725–30. doi: 10.1016/s0955-0674(00)00159-9. [DOI] [PubMed] [Google Scholar]
- [40].Day K, Shefer G, Richardson JB, Enikolopov G, Yablonka-Reuveni Z. Nestin-GFP reporter expression defines the quiescent state of skeletal muscle satellite cells. Developmental Biology. 2007;304:246–59. doi: 10.1016/j.ydbio.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, et al. Direct Isolation of Satellite Cells for Skeletal Muscle Regeneration. Science. 2005;309:2064–7. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- [42].Porter JD, Israel S, Gong B, Merriam AP, Feuerman J, Khanna S, et al. Distinctive morphological and gene/protein expression signatures during myogenesis in novel cell lines from extraocular and hindlimb muscle. Physiol Genomics. 2006;24:264–75. doi: 10.1152/physiolgenomics.00234.2004. [DOI] [PubMed] [Google Scholar]
- [43].Kang LHD, Rughani A, Walker ML, Bestak R, Hoh JFY. Expression of Masticatory-specific Isoforms of Myosin Heavy Chain, Myosin Binding Protein-C, and Tropomyosin in Muscle Fibers and Satellite Cell Cultures of Cat Masticatory Muscle. J Histochem Cytochem. 2010 doi: 10.1369/jhc.2010.955419. jhc.2010.955419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hoh JFY, Hughes S. Myogenic and neurogenic regulation of myosin gene expression in cat jaw-closing muscles regenerating in fast and slow limb muscle beds. Journal of Muscle Research and Cell Motility. 1988;9:59–72. doi: 10.1007/BF01682148. [DOI] [PubMed] [Google Scholar]
- [45].Hoh JFY, Hughes S. Expression of superfast myosin in aneural regenerates of cat jaw muscle. Muscle & Nerve. 1991;14:316–25. doi: 10.1002/mus.880140405. [DOI] [PubMed] [Google Scholar]
- [46].Hoh JFY, Hughes S. Basal lamina and superfast myosin expression in regenerating cat jaw muscle. Muscle & Nerve. 1991;14:398–406. doi: 10.1002/mus.880140503. [DOI] [PubMed] [Google Scholar]
- [47].Walro JM, Kucera J. Why adult mammalian intrafusal and extrafusal fibers contain different myosin heavy-chain isoforms. Trends in Neurosciences. 1999;22:180–4. doi: 10.1016/s0166-2236(98)01339-3. [DOI] [PubMed] [Google Scholar]
- [48].Schiaffino S, Reggiani C. Myosin isoforms in mammalian skeletal muscle. J Appl Physiol. 1994;77:493–501. doi: 10.1152/jappl.1994.77.2.493. [DOI] [PubMed] [Google Scholar]
- [49].Pette D, Staron RS. Mammalian Skeletal Muscle Fiber Type Transitions. In: Kwang WJ, editor. International Review of Cytology. Academic Press; 1997. pp. 143–223. [DOI] [PubMed] [Google Scholar]
- [50].Barjot C, Cotten M-L, Goblet C, Whalen RG, Bacou F. Expression of myosin heavy chain and of myogenic regulatory factor genes in fast or slow rabbit muscle satellite cell cultures. Journal of Muscle Research and Cell Motility. 1995;16:619–28. doi: 10.1007/BF00130243. [DOI] [PubMed] [Google Scholar]
- [51].Feldman JL, Stockdale FE. Skeletal muscle satellite cell diversity: Satellite cells form fibers of different types in cell culture. Developmental Biology. 1991;143:320–34. doi: 10.1016/0012-1606(91)90083-f. [DOI] [PubMed] [Google Scholar]
- [52].Lagord C, Soulet L, Bonavaud S, Bassaglia Y, Rey C, Barlovatz-Meimon G, et al. Differential myogenicity of satellite cells isolated from extensor digitorum longus (EDL) and soleus rat muscles revealed in vitro. Cell and Tissue Research. 1998;291:455–68. doi: 10.1007/s004410051015. [DOI] [PubMed] [Google Scholar]
- [53].Yada E YK, Nishihara M. Adipogenic potential of satellite cells from distinct skeletal muscle origins in the rat. J Vet Med Sci. 2006;68:479–86. doi: 10.1292/jvms.68.479. [DOI] [PubMed] [Google Scholar]
- [54].Barjot C, Jbilo O, Chatonnet A, Bacou F. Expression of acetylcholinesterase gene during in vitro differentiation of rabbit muscle satellite cells. Neuromuscular Disorders. 1993;3:443–6. doi: 10.1016/0960-8966(93)90093-y. [DOI] [PubMed] [Google Scholar]
- [55].Martelly I, Soulet L, Bonnavaud S, Cebrian J, Gautron J, Barritault D. Differential expression of FGF receptors and of myogenic regulatory factors in primary cultures of satellite cells originating from fast (EDL) and slow (Soleus) twitch rat muscles. Cellular and molecular biology. 2000;46:1239–48. [PubMed] [Google Scholar]
- [56].Brzóska E, Przewozniak M, Grabowska I, Janczyk-Ilach K, Moraczewski J. Pax3 and Pax7 expression during myoblast differentiation in vitro and fast and slow muscle regeneration in vivo. Cell Biology International. 2009;33:483–92. doi: 10.1016/j.cellbi.2008.11.015. [DOI] [PubMed] [Google Scholar]
- [57].Matsuda R, Spector DH, Strohman RC. Regenerating adult chicken skeletal muscle and satellite cell cultures express embryonic patterns of myosin and tropomyosin isoforms. Developmental Biology. 1983;100:478–88. doi: 10.1016/0012-1606(83)90240-3. [DOI] [PubMed] [Google Scholar]
- [58].Rosenblatt JD, Parry DJ, Partridge TA. Phenotype of adult mouse muscle myoblasts reflects their fiber type of origin. Differentiation. 1996;60:39–45. doi: 10.1046/j.1432-0436.1996.6010039.x. [DOI] [PubMed] [Google Scholar]
- [59].Dusterhoft S, Pette D. Satellite cells from slow rat muscle express slow myosin under appropriate culture conditions. Differentiation. 1993;53:25–33. doi: 10.1111/j.1432-0436.1993.tb00642.x. [DOI] [PubMed] [Google Scholar]
- [60].Huang Y-C, Dennis RG, Baar K. Cultured slow vs. fast skeletal muscle cells differ in physiology and responsiveness to stimulation. Am J Physiol Cell Physiol. 2006;291:C11–7. doi: 10.1152/ajpcell.00366.2005. [DOI] [PubMed] [Google Scholar]
- [61].Edom F, Mouly V, Barbet JP, Fiszman MY, Butler-Browne GS. Clones of Human Satellite Cells Can Express in Vitro both Fast and Slow Myosin Heavy Chains. Developmental Biology. 1994;164:219–29. doi: 10.1006/dbio.1994.1193. [DOI] [PubMed] [Google Scholar]
- [62].Bonavaud S, Agbulut O, Nizard R, D’honneur G, Mouly V, Butler-Browne G. A discrepancy resolved: human satellite cells are not preprogrammed to fast and slow lineages. Neuromuscular disorders : NMD. 2001;11:747–52. doi: 10.1016/s0960-8966(01)00222-x. [DOI] [PubMed] [Google Scholar]
- [63].Hughes SM, Blau HM. Muscle fiber pattern is independent of cell lineage in postnatal rodent development. Cell. 1992;68:659–71. doi: 10.1016/0092-8674(92)90142-y. [DOI] [PubMed] [Google Scholar]
- [64].Kalhovde JM, Jerkovic R, Sefland I, Cordonnier C, Calabria E, Schiaffino S, et al. ‘Fast’ and ‘slow’ muscle fibres in hindlimb muscles of adult rats regenerate from intrinsically different satellite cells. The Journal of Physiology. 2005;562:847–57. doi: 10.1113/jphysiol.2004.073684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Snoj-Cvetko E, Smerdu V, Sketelj J, Dolenc I, D’Albis A, Janmot C, et al. Adaptive range of myosin heavy chain expression in regenerating soleus is broader than in mature muscle. Journal of Muscle Research and Cell Motility. 1996;17:401–9. doi: 10.1007/BF00123357. [DOI] [PubMed] [Google Scholar]
- [66].DiMario JX, Fernyak SE, Stockdale FE. Myoblasts transferred to the limbs of embryos are committed to specific fibre fates. Nature. 1993;362:165–7. doi: 10.1038/362165a0. [DOI] [PubMed] [Google Scholar]
- [67].Eržen I, Primc M, Janmot C, Cvetko E, Sketelj J, d‘Albis A. Myosin Heavy Chain Profiles in Regenerated Fast and Slow Muscles Innervated by the Same Motor Nerve Become Nearly Identical. The Histochemical Journal. 1999;31:277–83. doi: 10.1023/a:1003709700270. [DOI] [PubMed] [Google Scholar]
- [68].Barjot C, Rouanet P, Vigneron P, Janmot C, D’Albis A, Bacou F. Transformation of slow- or fast-twitch rabbit muscles after cross-reinnervation or low frequency stimulation does not alter the in vitro properties of their satellite cells. Journal of Muscle Research and Cell Motility. 1998;19:25–32. doi: 10.1023/a:1005396125746. [DOI] [PubMed] [Google Scholar]
- [69].Soukup T, Jirmanov I, Mrckov K, Zacharov G, Thornell LE. Expression of myosin heavy chain (MyHC) isoforms in rat intrafusal muscle fibres after neonatal deefferentation and subsequent denervation. General physiology and biophysics. 1999;18(Suppl 1):81–3. [PubMed] [Google Scholar]
- [70].Soukup T, Thornell L-E. Expression of myosin heavy chain isoforms in regenerated muscle spindle fibres after muscle grafting in young and adult rats--plasticity of intrafusal satellite cells. Differentiation. 1998;62:179–86. doi: 10.1046/j.1432-0436.1998.6240179.x. [DOI] [PubMed] [Google Scholar]
- [71].Kirkpatrick LJ, Yablonka-Reuveni Z, Rosser BWC. Retention of Pax3 Expression in Satellite Cells of Muscle Spindles. J Histochem Cytochem. 2010;58:317–27. doi: 10.1369/jhc.2009.954792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].McLoon LK, Wirtschafter JD. Continuous myonuclear addition to single extraocular myofibers in uninjured adult rabbits. Muscle & Nerve. 2002;25:348–58. doi: 10.1002/mus.10056. [DOI] [PubMed] [Google Scholar]
- [73].Kirkpatrick LJ, Allouh MZ, Nightingale CN, Devon HG, Yablonka-Reuveni Z, Rosser BWC. Pax7 Shows Higher Satellite Cell Frequencies and Concentrations Within Intrafusal Fibers of Muscle Spindles. J Histochem Cytochem. 2008;56:831–40. doi: 10.1369/jhc.2008.951608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Pacheco-Pinedo EC, Budak MT, Zeiger U, Jorgensen LH, Bogdanovich S, Schroder HD, et al. Transcriptional and functional differences in stem cell populations isolated from extraocular and limb muscles. Physiol Genomics. 2009;37:35–42. doi: 10.1152/physiolgenomics.00051.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Cassano M, Quattrocelli M, Crippa S, Perini I, Ronzoni F, Sampaolesi M. Cellular mechanisms and local progenitor activation to regulate skeletal muscle mass. Journal of Muscle Research and Cell Motility. 2009;30:243–53. doi: 10.1007/s10974-010-9204-y. [DOI] [PubMed] [Google Scholar]
- [76].Gopinath SD, Rando TA. Stem Cell Review Series: Aging of the skeletal muscle stem cell niche. Aging Cell. 2008;7:590–8. doi: 10.1111/j.1474-9726.2008.00399.x. [DOI] [PubMed] [Google Scholar]
- [77].Feldman JL, Stockdale FE. Temporal appearance of satellite cells during myogenesis. Developmental Biology. 1992;153:217–26. doi: 10.1016/0012-1606(92)90107-r. [DOI] [PubMed] [Google Scholar]
- [78].Heslop L, Beauchamp JR, Tajbakhsh S, Buckingham ME, Partridge TA, Zammit PS. Transplanted primary neonatal myoblasts can give rise to functional satellite cells as identified using the Myf5nlacZl+mouse. Gene Therapy. 2001;8:778. doi: 10.1038/sj.gt.3301463. [DOI] [PubMed] [Google Scholar]
- [79].Schultz E. Satellite Cell Proliferative Compartments in Growing Skeletal Muscles. Developmental Biology. 1996;175:84–94. doi: 10.1006/dbio.1996.0097. [DOI] [PubMed] [Google Scholar]
- [80].Molnar G, Ho ML, Schroedl NA. Evidence for multiple satellite cell populations and a non-myogenic cell type that is regulated differently in regenerating and growing skeletal muscle. Tissue and Cell. 1996;28:547–56. doi: 10.1016/s0040-8166(96)80057-7. [DOI] [PubMed] [Google Scholar]
- [81].Lee-Pullen T, Bennett A, Beilharz M, Grounds M, Sammels L. Superior Survival and Proliferation after Transplantation of Myoblasts Obtained from Adult Mice Compared with Neonatal Mice. Transplantation. 2004;78:1172–6. doi: 10.1097/01.tp.0000137936.75203.b4. [DOI] [PubMed] [Google Scholar]
- [82].Morgan JE, Hoffman EP, Partridge TA. Normal myogenic cells from newborn mice restore normal histology to degenerating muscles of the mdx mouse. The Journal of Cell Biology. 1990;111:2437–49. doi: 10.1083/jcb.111.6.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Creuzet S, Lescaudron L, Li Z, Fontaine-Pérus J. MyoD, Myogenin, and Desmin-nls-lacZ Transgene Emphasize the Distinct Patterns of Satellite Cell Activation in Growth and Regeneration. Experimental Cell Research. 1998;243:241–53. doi: 10.1006/excr.1998.4100. [DOI] [PubMed] [Google Scholar]
- [84].White RB, Biérinx AS, Gnocchi VF, Zammit PS. Dynamics of muscle fibre growth during postnatal mouse development. BMC Developmental Biology. 2010;10:1–11. doi: 10.1186/1471-213X-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Cossu G, Molinaro M, Pacifici M. Differential response of satellite cells and embryonic myoblasts to a tumor promoter. Developmental Biology. 1983;98:520–4. doi: 10.1016/0012-1606(83)90382-2. [DOI] [PubMed] [Google Scholar]
- [86].Mitchell KJ, Pannerec A, Cadot B, Parlakian A, Besson V, Gomes ER, et al. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nat Cell Biol. 2010;12:257–66. doi: 10.1038/ncb2025. [DOI] [PubMed] [Google Scholar]
- [87].Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric Self-Renewal and Commitment of Satellite Stem Cells in Muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Beauchamp JR, Heslop L, Yu DSW, Tajbakhsh S, Kelly RG, Wernig A, et al. Expression of Cd34 and Myf5 Defines the Majority of Quiescent Adult Skeletal Muscle Satellite Cells. The Journal of Cell Biology. 2000;151:1221–34. doi: 10.1083/jcb.151.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Cerletti M, Jurga S, Witczak CA, Hirshman MF, Shadrach JL, Goodyear LJ, et al. Highly Efficient, Functional Engraftment of Skeletal Muscle Stem Cells in Dystrophic Muscles. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Tanaka KK, Hall JK, Troy AA, Cornelison DDW, Majka SM, Olwin BB. Syndecan-4-Expressing Muscle Progenitor Cells in the SP Engraft as Satellite Cells during Muscle Regeneration. Cell Stem Cell. 2009;4:217–25. doi: 10.1016/j.stem.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–6. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Cornelison DDW, Olwin BB, Rudnicki MA, Wold BJ. MyoD−/− Satellite Cells in Single-Fiber Culture Are Differentiation Defective and MRF4 Deficient. Developmental Biology. 2000;224:122–37. doi: 10.1006/dbio.2000.9682. [DOI] [PubMed] [Google Scholar]
- [93].Bosnakovski D, Xu Z, Li W, Thet S, Cleaver O, Perlingeiro RCR, et al. Prospective Isolation of Skeletal Muscle Stem Cells with a Pax7 Reporter. Stem Cells. 2008;26:3194–204. doi: 10.1634/stemcells.2007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Sherwood RI, Christensen JL, Conboy IM, Conboy MJ, Rando TA, Weissman IL, et al. Isolation of Adult Mouse Myogenic Progenitors: Functional Heterogeneity of Cells within and Engrafting Skeletal Muscle. 2004;119:543–54. doi: 10.1016/j.cell.2004.10.021. [DOI] [PubMed] [Google Scholar]
- [95].Rantanen J, Hurme T, Lukka R, Heino J, Kalimo H. Satellite cell proliferation and the expression of myogenin and desmin in regenerating skeletal muscle: evidence for two different populations of satellite cells. Laboratory Investigation. 1995;72:341–7. [PubMed] [Google Scholar]
- [96].Kablar B, Krastel K, Tajbakhsh S, Rudnicki MA. Myf5 and MyoD activation define independent myogenic compartments during embryonic development. Developmental Biology. 2003;258:307–18. doi: 10.1016/s0012-1606(03)00139-8. [DOI] [PubMed] [Google Scholar]
- [97].Cossu G, Kelly R, Tajbakhsh S, Di Donna S, Vivarelli E, Buckingham M. Activation of different myogenic pathways: myf-5 is induced by the neural tube and MyoD by the dorsal ectoderm in mouse paraxial mesoderm. Development. 1996;122:429–37. doi: 10.1242/dev.122.2.429. [DOI] [PubMed] [Google Scholar]
- [98].Cooper R, Tajbakhsh S, Mouly V, Cossu G, Buckingham M, Butler-Browne G. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J Cell Sci. 1999;112:2895–901. doi: 10.1242/jcs.112.17.2895. [DOI] [PubMed] [Google Scholar]
- [99].Cornelison DDW, Wold BJ. Single-Cell Analysis of Regulatory Gene Expression in Quiescent and Activated Mouse Skeletal Muscle Satellite Cells. Developmental Biology. 1997;191:270–83. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- [100].Sabourin LA, Girgis-Gabardo A, Seale P, Asakura A, Rudnicki MA. Reduced Differentiation Potential of Primary MyoD−/− Myogenic Cells Derived from Adult Skeletal Muscle. The Journal of Cell Biology. 1999;144:631–43. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Montarras D, Lindon C, Pinset C, Domeyne P. Cultured myf5 null and myoD null muscle precursor cells display distinct growth defects. Biol Cell. 2000;92:565–72. doi: 10.1016/s0248-4900(00)01110-2. [DOI] [PubMed] [Google Scholar]
- [102].Rouger K, Brault M, Daval N, Leroux I, Guigand L, Lesoeur J, et al. Muscle satellite cell heterogeneity: in vitro and in vivo evidences for populations that fuse differently. Cell and Tissue Research. 2004;317:319–26. doi: 10.1007/s00441-004-0911-9. [DOI] [PubMed] [Google Scholar]
- [103].Schultz E, Lipton BH. Skeletal muscle satellite cells: Changes in proliferation potential as a function of age. Mechanisms of Ageing and Development. 1982;20:377–83. doi: 10.1016/0047-6374(82)90105-1. [DOI] [PubMed] [Google Scholar]
- [104].Schultz E, Jaryszak DL. Effects of skeletal muscle regeneration on the proliferation potential of satellite cells. Mechanisms of Ageing and Development. 1985;30:63–72. doi: 10.1016/0047-6374(85)90059-4. [DOI] [PubMed] [Google Scholar]
- [105].Yablonka-Reuveni Z, Quinn LS, Nameroff M. Isolation and clonal analysis of satellite cells from chicken pectoralis muscle. Developmental Biology. 1987;119:252–9. doi: 10.1016/0012-1606(87)90226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, et al. Increased Wnt Signaling During Aging Alters Muscle Stem Cell Fate and Increases Fibrosis. Science. 2007;317:807–10. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- [107].Rossi CA, Pozzobon M, Ditadi A, Archacka K, Gastaldello A, Sanna M, et al. Clonal Characterization of Rat Muscle Satellite Cells: Proliferation, Metabolism and Differentiation Define an Intrinsic Heterogeneity. PLoS ONE. 2010;5:e8523. doi: 10.1371/journal.pone.0008523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Asakura A, Rudnicki MA, Komaki M. Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation. 2001;68:245–53. doi: 10.1046/j.1432-0436.2001.680412.x. [DOI] [PubMed] [Google Scholar]
- [109].Shefer G, Wleklinski-Lee M, Yablonka-Reuveni Z. Skeletal muscle satellite cells can spontaneously enter an alternative mesenchymal pathway. J Cell Sci. 2004;117:5393–404. doi: 10.1242/jcs.01419. [DOI] [PubMed] [Google Scholar]
- [110].Csete M, Walikonis J, Slawny N, Wei Y, Korsnes S, Doyle JC, et al. Oxygen-mediated regulation of skeletal muscle satellite cell proliferation and adipogenesis in culture. Journal of Cellular Physiology. 2001;189:189–96. doi: 10.1002/jcp.10016. [DOI] [PubMed] [Google Scholar]
- [111].Wada MR, Inagawa-Ogashiwa M, Shimizu S, Yasumoto S, Hashimoto N. Generation of different fates from multipotent muscle stem cells. Development. 2002;129:2987–95. doi: 10.1242/dev.129.12.2987. [DOI] [PubMed] [Google Scholar]
- [112].Hashimoto N, Kiyono T, Wada MR, Umeda R, Goto Y-i, Nonaka I, et al. Osteogenic properties of human myogenic progenitor cells. Mechanisms of Development. 125:257–69. doi: 10.1016/j.mod.2007.11.004. [DOI] [PubMed] [Google Scholar]
- [113].Hashimoto N, Murase T, Kondo S, Okuda A, Inagawa-Ogashiwa M. Muscle reconstitution by muscle satellite cell descendants with stem cell-like properties. Development. 2004;131:5481–90. doi: 10.1242/dev.01395. [DOI] [PubMed] [Google Scholar]
- [114].Pisani DF, Dechesne CA, Sacconi S, Delplace S, Belmonte N, Cochet O, et al. Isolation of a Highly Myogenic CD34-Negative Subset of Human Skeletal Muscle Cells Free of Adipogenic Potential. Stem Cells. 2010;28:753–64. doi: 10.1002/stem.317. [DOI] [PubMed] [Google Scholar]
- [115].Dhawan J, Rando TA. Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends in Cell Biology. 2005;15:666–73. doi: 10.1016/j.tcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- [116].Baroffio A, Hamann M, Bernheim L, Bochaton-Piallat M-L, Gabbiani G, Bader CR. Identification of self-renewing myoblasts in the progeny of single human muscle satellite cells. Differentiation. 1996;60:47–57. doi: 10.1046/j.1432-0436.1996.6010047.x. [DOI] [PubMed] [Google Scholar]
- [117].Yoshida N, Yoshida S, Koishi K, Masuda K, Nabeshima Y. Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates ‘reserve cells’. J Cell Sci. 1998;111:769–79. doi: 10.1242/jcs.111.6.769. [DOI] [PubMed] [Google Scholar]
- [118].Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates. The Journal of Cell Biology. 2004;166:347–57. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Olguin HC, Olwin BB. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Developmental Biology. 2004;275:375–88. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Beauchamp JR, Morgan JE, Pagel CN, Partridge TA. Dynamics of Myoblast Transplantation Reveal a Discrete Minority of Precursors with Stem Cell–like Properties as the Myogenic Source. The Journal of Cell Biology. 1999;144:1113–22. doi: 10.1083/jcb.144.6.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Heslop L, Morgan J, Partridge T. Evidence for a myogenic stem cell that is exhausted in dystrophic muscle. J Cell Sci. 2000;113:2299–308. doi: 10.1242/jcs.113.12.2299. [DOI] [PubMed] [Google Scholar]
- [122].Collins CA, Zammit PS, Ruiz AP, Morgan JE, Partridge TA. A Population of Myogenic Stem Cells That Survives Skeletal Muscle Aging. Stem Cells. 2007;25:885–94. doi: 10.1634/stemcells.2006-0372. [DOI] [PubMed] [Google Scholar]
- [123].Shinin V, Gayraud-Morel B, Gomès D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nature Cell Biology. 2006;8:677–82. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- [124].Conboy MJ, Karasov AO, Rando TA. High Incidence of Non-Random Template Strand Segregation and Asymmetric Fate Determination In Dividing Stem Cells and their Progeny. PLoS Biology. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Shea KL, Xiang W, LaPorta VS, Licht JD, Keller C, Basson MA, et al. Sprouty1 Regulates Reversible Quiescence of a Self-Renewing Adult Muscle Stem Cell Pool during Regeneration. Cell Stem Cell. 2010;6:117–29. doi: 10.1016/j.stem.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Le Grand F, Jones AE, Seale V, Scimè A, Rudnicki MA. Wnt7a Activates the Planar Cell Polarity Pathway to Drive the Symmetric Expansion of Satellite Stem Cells. 2009;4:535–47. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kanisicak O, Mendez JJ, Yamamoto S, Yamamoto M, Goldhamer DJ. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Developmental Biology. 2009;332:131–41. doi: 10.1016/j.ydbio.2009.05.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Carlson BM, Faulkner JA. Muscle transplantation between young and old rats: age of host determines recovery. Am J Physiol Cell Physiol. 1989;256:C1262–6. doi: 10.1152/ajpcell.1989.256.6.C1262. [DOI] [PubMed] [Google Scholar]
- [129].McGeachie JK, Grounds MD. Retarded myogenic cell replication in regenerating skeletal muscles of old mice: an autoradiographic study in young and old BALBc and SJL/J mice. Cell and Tissue Research. 1995;280:277–82. doi: 10.1007/BF00307799. [DOI] [PubMed] [Google Scholar]
- [130].Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-Mediated Restoration of Regenerative Potential to Aged Muscle. Science. 2003;302:1575–7. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- [131].Brack A, Rando T. Intrinsic Changes and Extrinsic Influences of Myogenic Stem Cell Function During Aging. Stem Cell Reviews and Reports. 2007;3:226–37. doi: 10.1007/s12015-007-9000-2. [DOI] [PubMed] [Google Scholar]
- [132].Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–4. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- [133].Bortoli S, Renault V, Eveno E, Auffray C, Butler-Browne G, Piétu G. Gene expression profiling of human satellite cells during muscular aging using cDNA arrays. Gene. 2003;321:145–54. doi: 10.1016/j.gene.2003.08.025. [DOI] [PubMed] [Google Scholar]
- [134].Bockhold KJ, Rosenblatt JD, Partridge TA. Aging normal and dystrophic mouse muscle: Analysis of myogenicity in cultures of living single fibers. Muscle & Nerve. 1998;21:173–83. doi: 10.1002/(sici)1097-4598(199802)21:2<173::aid-mus4>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- [135].Barani AE, Durieux A-C, Sabido O, Freyssenet D. Age-related changes in the mitotic and metabolic characteristics of muscle-derived cells. J Appl Physiol. 2003;95:2089–98. doi: 10.1152/japplphysiol.00437.2003. [DOI] [PubMed] [Google Scholar]
- [136].Shefer G, Van de Mark DP, Richardson JB, Yablonka-Reuveni Z. Satellite-cell pool size does matter: Defining the myogenic potency of aging skeletal muscle. Developmental Biology. 2006;294:50–66. doi: 10.1016/j.ydbio.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Chakravarthy MV, Davis BS, Booth FW. IGF-I restores satellite cell proliferative potential in immobilized old skeletal muscle. J Appl Physiol. 2000;89:1365–79. doi: 10.1152/jappl.2000.89.4.1365. [DOI] [PubMed] [Google Scholar]
- [138].Barton-Davis ER, Shoturma DI, Musaro A, Rosenthal N, Sweeney HL. Viral mediated expression of insulin-like growth factor I blocks the aging-related loss of skeletal muscle function. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15603–7. doi: 10.1073/pnas.95.26.15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Carlson ME, Hsu M, Conboy IM. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature. 2008;454:528–32. doi: 10.1038/nature07034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Carlson ME, Conboy MJ, Hsu M, Barchas L, Jeong J, Agrawal A, et al. Relative roles of TGF-beta1 and Wnt in the systemic regulation and aging of satellite cell responses. Aging Cell. 2009;8:676–89. doi: 10.1111/j.1474-9726.2009.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Musarò A, McCullagh K, Paul A, Houghton L, Dobrowolny G, Molinaro M, et al. Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nature Genetics. 2001;27:195. doi: 10.1038/84839. [DOI] [PubMed] [Google Scholar]
- [142].Charge SBP, Brack AS, Hughes SM. Aging-related satellite cell differentiation defect occurs prematurely after Ski-induced muscle hypertrophy. Am J Physiol Cell Physiol. 2002;283:C1228–41. doi: 10.1152/ajpcell.00206.2002. [DOI] [PubMed] [Google Scholar]
- [143].Taylor-Jones JM, McGehee RE, Rando TA, Lecka-Czernik B, Lipschitz DA, Peterson CA. Activation of an adipogenic program in adult myoblasts with age. Mechanisms of Ageing and Development. 2002;123:649–61. doi: 10.1016/s0047-6374(01)00411-0. [DOI] [PubMed] [Google Scholar]
- [144].Jejurikar SS, Henkelman EA, Cederna PS, Marcelo CL, Urbanchek MG, Kuzon JWM. Aging increases the susceptibility of skeletal muscle derived satellite cells to apoptosis. Experimental Gerontology. 2006;41:828–36. doi: 10.1016/j.exger.2006.06.053. [DOI] [PubMed] [Google Scholar]
- [145].Cossu G, Biressi S. Satellite cells, myoblasts and other occasional myogenic progenitors: Possible origin, phenotypic features and role in muscle regeneration. Seminars in Cell & Developmental Biology. 2005;16:623–31. doi: 10.1016/j.semcdb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- [146].Tedesco FS, Dellavalle A, Diaz-Manera J, Messina G, Cossu G. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. The Journal of Clinical Investigation. 2010;120:11–9. doi: 10.1172/JCI40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].LaBarge MA, Blau HM. Biological Progression from Adult Bone Marrow to Mononucleate Muscle Stem Cell to Multinucleate Muscle Fiber in Response to Injury. 2002;111:589–601. doi: 10.1016/s0092-8674(02)01078-4. [DOI] [PubMed] [Google Scholar]
- [148].Dreyfus PA, Chretien F, Chazaud B, Kirova Y, Caramelle P, Garcia L, et al. Adult Bone Marrow-Derived Stem Cells in Muscle Connective Tissue and Satellite Cell Niches. Am J Pathol. 2004;164:773–9. doi: 10.1016/S0002-9440(10)63165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Asakura A, Seale P, Girgis-Gabardo A, Rudnicki MA. Myogenic specification of side population cells in skeletal muscle. The Journal of Cell Biology. 2002;159:123–34. doi: 10.1083/jcb.200202092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].De Bari C, Dell’Accio F, Vandenabeele F, Vermeesch JR, Raymackers J-M, Luyten FP. Skeletal muscle repair by adult human mesenchymal stem cells from synovial membrane. The Journal of Cell Biology. 2003;160:909–18. doi: 10.1083/jcb.200212064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Torrente Y, Belicchi M, Sampaolesi M, Pisati F, Meregalli M, D’Antona G, et al. Human circulating AC133+ stem cells restore dystrophin expression and ameliorate function in dystrophic skeletal muscle. The Journal of Clinical Investigation. 2004;114:182–95. doi: 10.1172/JCI20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Dellavalle A, Sampaolesi M, Tonlorenzi R, Tagliafico E, Sacchetti B, Perani L, et al. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat Cell Biol. 2007;9:255–67. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- [153].Sampaolesi M, Torrente Y, Innocenzi A, Tonlorenzi R, D’Antona G, Pellegrino MA, et al. Cell Therapy of {alpha}-Sarcoglycan Null Dystrophic Mice Through Intra-Arterial Delivery of Mesoangioblasts. Science. 2003;301:487–92. doi: 10.1126/science.1082254. [DOI] [PubMed] [Google Scholar]
- [154].Chang H, Yoshimoto M, Umeda K, Iwasa T, Mizuno Y, Fukada S-i, et al. Generation of transplantable, functional satellite-like cells from mouse embryonic stem cells. FASEB J. 2009;23:1907–19. doi: 10.1096/fj.08-123661. [DOI] [PubMed] [Google Scholar]
- [155].Mizuno Y, Chang H, Umeda K, Niwa A, Iwasa T, Awaya T, et al. Generation of skeletal muscle stem/progenitor cells from murine induced pluripotent stem cells. FASEB J. 2010 doi: 10.1096/fj.09-137174. fj.09-137174. [DOI] [PubMed] [Google Scholar]
- [156].Cornelison DDW, Filla MS, Stanley HM, Rapraeger AC, Olwin BB. Syndecan-3 and Syndecan-4 Specifically Mark Skeletal Muscle Satellite Cells and Are Implicated in Satellite Cell Maintenance and Muscle Regeneration. Developmental Biology. 2001;239:79–94. doi: 10.1006/dbio.2001.0416. [DOI] [PubMed] [Google Scholar]
- [157].Gnocchi VF, White RB, Ono Y, Ellis JA, Zammit PS. Further Characterisation of the Molecular Signature of Quiescent and Activated Mouse Muscle Satellite Cells. PLoS ONE. 2009;4:e5205. doi: 10.1371/journal.pone.0005205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Ieronimakis N, Balasundaram G, Rainey S, Srirangam K, Yablonka-Reuveni Z, Reyes M. Absence of CD34 on Murine Skeletal Muscle Satellite Cells Marks a Reversible State of Activation during Acute Injury. PLoS ONE. 2010;5:e10920. doi: 10.1371/journal.pone.0010920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Rosen GD, Sanes JR, LaChance R, Cunningham JM, Roman J, Dean DC. Roles for the integrin VLA-4 and its counter receptor VCAM-1 in myogenesis. Cell. 1992;69:1107–19. doi: 10.1016/0092-8674(92)90633-n. [DOI] [PubMed] [Google Scholar]
- [160].Seale P, Ishibashi J, Holterman C, Rudnicki MA. Muscle satellite cell-specific genes identified by genetic profiling of MyoD-deficient myogenic cell. Developmental Biology. 2004;275:287–300. doi: 10.1016/j.ydbio.2004.07.034. [DOI] [PubMed] [Google Scholar]
- [161].Fukada S-i, Higuchi S, Segawa M, Koda K-i, Yamamoto Y, Tsujikawa K, et al. Purification and cell-surface marker characterization of quiescent satellite cells from murine skeletal muscle by a novel monoclonal antibody. Experimental Cell Research. 2004;296:245–55. doi: 10.1016/j.yexcr.2004.02.018. [DOI] [PubMed] [Google Scholar]
- [162].Illa I, Leon-Monzon M, Dalakas MC. Regenerating and denervated human muscle fibers and satellite cells express neural cell adhesion molecule recognized by monoclonal antibodies to natural killer cells. Annals of Neurology. 1992;31:46–52. doi: 10.1002/ana.410310109. [DOI] [PubMed] [Google Scholar]
- [163].Fukada S-i, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, et al. Molecular Signature of Quiescent Satellite Cells in Adult Skeletal Muscle. Stem Cells. 2007;25:2448–59. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- [164].Garry DJ, Meeson A, Elterman J, Zhao Y, Yang P, Bassel-Duby R, et al. Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5416–21. doi: 10.1073/pnas.100501197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Schmidt K, Glaser G, Wernig A, Wegner M, Rosorius O. Sox8 Is a Specific Marker for Muscle Satellite Cells and Inhibits Myogenesis. Journal of Biological Chemistry. 2003;278:29769–75. doi: 10.1074/jbc.M301539200. [DOI] [PubMed] [Google Scholar]
- [166].Kaufman SJ, Foster RF. Replicating myoblasts express a muscle-specific phenotype. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:9606–10. doi: 10.1073/pnas.85.24.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].McCroskery S, Thomas M, Maxwell L, Sharma M, Kambadur R. Myostatin negatively regulates satellite cell activation and self-renewal. The Journal of Cell Biology. 2003;162:1135–47. doi: 10.1083/jcb.200207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Mousavi K, Jasmin BJ. BDNF Is Expressed in Skeletal Muscle Satellite Cells and Inhibits Myogenic Differentiation. J Neurosci. 2006;26:5739–49. doi: 10.1523/JNEUROSCI.5398-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]