Abstract
Stable integration of foreign DNA into the frog genome has been the purpose of several studies aimed at generating transgenic animals or producing mutations of endogenous genes. Inserting DNA into a host genome can be achieved in a number of ways. In Xenopus, different strategies have been developed which exhibit specific molecular and technical features. Although several of these technologies were also applied in various model organizms, the attributes of each method have rarely been experimentally compared. Investigators are thus confronted with a difficult choice to discriminate which method would be best suited for their applications. To gain better understanding, a transgenesis workshop was organized by the X-omics consortium. Three procedures were assessed side-by-side, and the results obtained are used to illustrate this review. In addition, a number of reagents and tools have been set up for the purpose of gene expression and functional gene analyses. This not only improves the status of Xenopus as a powerful model for developmental studies, but also renders it suitable for sophisticated genetic approaches. Twenty years after the first reported transgenic Xenopus, we review the state of the art of transgenic research, focusing on the new perspectives in performing genetic studies in this species.
Keywords: genetic modification, phi-C31 integrase, I-SceI meganuclease, restriction-enzyme-mediated integration (REMI), transgenesis workshop, Xenopus
Introduction
Studies using the amphibian Xenopus laevis have contributed to a better understanding of the mechanisms of early development for over a century. Several aspects of its physiology promote this species as an ideal laboratory animal for many biological studies (Danilchick et al., 1991). However, X. laevis was left aside as a model for genetic manipulations, due to the difficulty in producing and analysing inheritable genomic modifications. The first limitation occurs due to the genomic duplication that has occurred several times in most fishes and amphibians (Ohno, 1999). The genus Xenopus contains nearly 20 species that are all, except one, polyploid (Graf and Kobel, 1991). X. laevis is considered as a tetraploid and has a generation time of 1–2 years, which together make traditional genetic studies impractical (Bisbee et al., 1977; Evans et al., 2004, 2005; Morin et al., 2006). In addition, there have been few available tools to modify its genetic information for many years. However, technical innovations that allow classical experimental procedures to be combined with sophisticated genetic studies may reinvigorate the use of Xenopus as a model system.
Two key factors have contributed to this evolution. The first one involves the use of Xenopus tropicalis. X. tropicalis is the only diploid species of the Xenopus genus and has a 6–9 months generation time. X. tropicalis retains nearly all the technical advantages of X. laevis, with the added potential of being amenable to genetic studies (Amaya et al., 1998; Khokha et al., 2002). Importantly, recent results show that cross-species experiments are likely to be successful, due to a high sequence conservation in a number of genes (Chalmers et al., 2005). The research community can therefore build on knowledge, technical expertise and tools developed using X. laevis for their applications in X. tropicalis. In addition, the genome sequence is now available and a variety of genomic resources are accessible and organized via the Internet resource Xenbase (http://www.xenbase.org) (Richardson and Chapman, 2003; Bowes et al., 2008).
The second factor was the development of an efficient method of transgenesis that directly produces non-mosaic transgenic animals. On the basis of the REMI (restriction-enzyme-mediated integration) strategy, this method provided a first step towards the engineering of tools and reagents which are compatible with genetic approaches (Kroll and Amaya, 1996). This transgenic procedure offered an unequalled way to perform large-scale gene expression regulation studies in whole developing embryos. Besides, the method could greatly improve the ability to manipulate gene expression in functional assays. Gain- and loss-of-function experiments have been traditionally performed via microinjections of RNA, DNA, morpholinos or antibodies at a given stage of development. Because of the relative short half-life of injected products, their dilution and mosaic distribution as development proceeds, these approaches are often limited to the analysis of certain genes that act early in development (Amaya, 2005). The REMI procedure makes overexpression of cloned cDNA possible with high precision in time and space, even at late developmental stages, and allows heritable changes in gene expression (Figure 1). Finally, the generation of individuals lacking endogenous gene function using random insertion mutagenesis could easily be envisaged (Bronchain et al., 1999). The transfer of this technology to X. tropicalis opened ways to manipulate the amphibian genome in a diploid species (Offield et al., 2000). However, as promising as these additions to experimental approaches are, broad use and adaptation have been slow.
Figure 1. REMI transgenesis procedure.
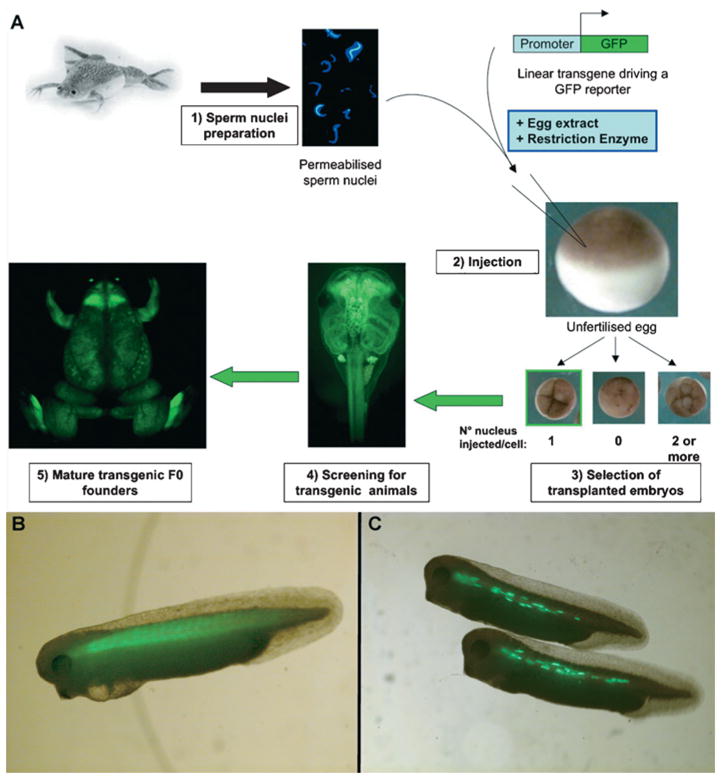
(A) The REMI method has been successfully used to generate transgenic X. laevis tadpoles and lines. Sperm nuclei were isolated, as described by Murray (1991), with the modifications by Kroll and Amaya (1996). The quality and concentration of the sperm solution are determined by Hoechst staining in a haemocytometer (1). The cells are then incubated with the linear transgene along with egg extract and RE. Methods that aimed at improving the REMI method have discarded the use of egg extract and RE (blue box), which made the method very similar to an ICSI protocol (Sparrow et al., 2000). The mixture is then injected into unfertilized eggs (2) and the transplanted embryos are selected at the four-cell stage (3). All eggs are activated by the injection, but only the embryos that cleaved normally are isolated for further analyses. Indeed, as the nuclei are injected at a constant flow rate, at best a third of the eggs are expected to receive a single nucleus. As development proceeds, embryos are scored for the expression of the transgene (4) and placed into an husbandry facility to obtain mature F0 founder animals to derive transgenic lines. Here the generation of an F0 expressing GFP under the control of the ubiquitous CMV (cytomegalovirus) promoter is shown. (B) Tadpole generated using the REMI method expressing a GFP reporter under the control of the cardiac actin promoter. Striated muscle cells of the somites express GFP in an homogenous fashion. (C) Tadpoles expressing a GFP reporter under the control of the cardiac actin promoter generated by microinjection. Microinjected embryos exhibit a punctuated GFP signal in few cells within the somites, reminiscent of a mosaic expression.
Here, we review some of the innovations in the field of transgenesis in Xenopus. In particular, we attempt to highlight and discuss technical aspects that may have interfered with its wider use. In this review, we report on results obtained during a transgenesis workshop organized by the X-omics consortium. The aim of the workshop was primarily to learn different methods of producing transgenic frogs. In addition, we were able to evaluate the panel of applications and to estimate the potential benefits and drawbacks of various procedures, namely the phi-C31 integrase, I-SceI meganuclease and REMI approaches in X. laevis. Finally, we provide perspectives that may stimulate new research using transgenesis in Xenopus species.
Two decades of transgenesis
Inheritable genetic modifications using DNA microinjections
Transgenesis is not new in amphibians. Several reports in the early 1980s, showed that exogenous DNA could persist to adulthood in the frog tissues following DNA microinjection into fertilized eggs, but exhibited a mosaic pattern of distribution (Rusconi and Schaffner, 1981; Etkin and Roberts, 1983; Andres et al., 1984; Bendig and Williams, 1984; Etkin et al., 1984). In 1987, L.D. Etkin and B. Pearman first reported and characterized transgenic X. laevis animals, showing that some microinjected transformants could transmit the exogenous DNA to their offspring (Etkin and Pearman, 1987). In addition, they estimated that in 5–10% of the transformants in which the exogenous DNA persisted to adulthood, one or more integrations had occurred. They concluded that, although the initial transformants were highly mosaic, bringing the animals to further filial generations (F), F1 or F2, could be very useful in assessing tissue-specific expression of injected DNA.
Germinal transgenesis by REMI
The mosaic distribution of exogenous DNA can, indeed, obscure the activity of a given transgene, making promoter/enhancer analyses difficult to interpret after microinjections. Ten years ago, K.L. Kroll and E. Amaya published a protocol for producing transgenic Xenopus using a REMI strategy (Kroll and Amaya, 1996; Amaya and Kroll, 1999). The aim of this method was to reduce mosaic expression of the transgene through an integration step into the host genome occurring in vitro within isolated sperm nuclei.
The REMI method relies on the RE (restriction enzyme)-mediated incorporation of linear DNA template into the host genome before the first cell division occurs. This ensures that all the cells of the embryo carry the foreign DNA, and that germline transmission from mature founders is possible (Kroll and Kirschner, 1999). Briefly, the procedure involves the incubation of a linearized transgene and isolated sperm nuclei along with the RE to favour the generation of double-stranded breaks and egg extracts to promote genomic DNA decondensation. The foreign DNA is thought to incorporate randomly into the genomic DNA during a process of DNA repair. The manipulated nuclei are then transplanted into unfertilized eggs, thus generating transgenic embryos (Figure 1).
The REMI transgenesis method compares favourably to transgenesis procedures used in other vertebrates. A large number of transgenic embryos are obtained within a short period of time (1 day) at relatively low cost, and the rate of transgenesis ranges from 30 to 70%, depending on the transgene. By using a transgene coupled to a fluorescent reporter, screening transgenic embryos can be rapidly accomplished and reporter gene expression or phenotypes can be studied directly in developing embryos. In addition, the transgenic embryos are non-mosaic at the F0 (Bronchain et al., 1999; Marsh-Armstrong et al., 1999). This implies that only one generation is required to obtained homozygous animals when using a gynogenesis procedure or two generations in a full-sibling mating scheme (see below and Figure 5). These features of the REMI transgenesis technique make it a powerful tool to manipulate gene function and to study gene expression in whole embryos.
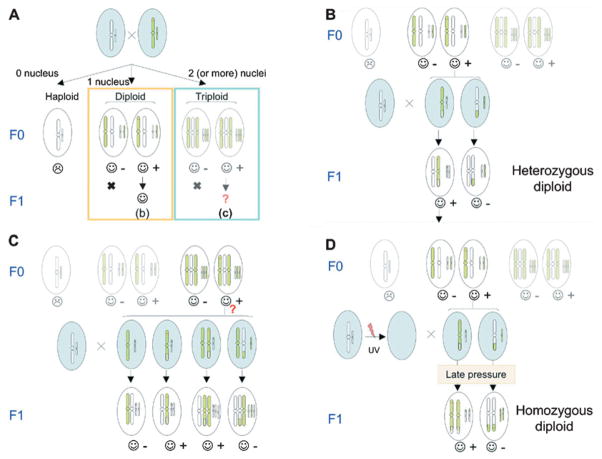
Figure 5. Generation of diploid and triploid transgenic lines.
(A) Haploid, (B) diploid and (C) triploid individuals can be generated following the transplantation of a random number of male nuclei during the REMI procedure. If the egg does not receive a nucleus, the embryo will be haploid and will not develop beyond the neurula stage (☹), unlike diploid and triploid animals that will give rise to founders (☺). (B) If the egg received a single nucleus, then the embryo generated will produce an heterozygous diploid F0 founder and lines can be derived as shown. Transgenic embryos can be selected by observing the expression of a marker gene, such as GFP, and individuals carrying the transgene (+) are isolated from non-carriers (−). (C) In the case of multiple nuclei transplantation, the embryos generated will be polyploid. It is not known how many nuclei can be maintained in Xenopus. Multiploid cells have been shown to suffer from chromosomal instability and degenerate. However, triploid individuals that result from transplantation of two male nuclei are stable and develop to adulthood (Smith, 1958; Kawahara, 1978; Tompkins, 1978; Tompkins and Reinschmidt, 1991; Noramly et al., 2005). It is not known if these animals are fertile (?) and whether lines can be derived form them. However, triploid embryos could be generated by REMI and maintained in animal facilities. (D) Finally, F0 diploid transgenic female founders can be made homozygous at the F1 generation by using a gynogenesis procedure. In this case, the genomic contribution of the male DNA is eliminated by UV treatment. The UV-treated spermatozoids are used for in vitro fertilization of the transgenic eggs. Inhibition of the first cleavage by pressure re-establishes the diploid state and development can proceed normally.
Although the majority of integration events occur before the first cleavage, in practice additional integrations may arise later in development. Indeed, half of the transgenic embryos that are probably the result of an integration event in one blastomere at the two-cell stage can be obtained using REMI (Figure 2A and see Supplementary Figure 2B at http://www.biolcell.org/boc/100/boc1000503add.htm). In very few cases, the transgene expression pattern suggests even later integration events, and large clones can be observed (Figure 2B). These types of integration events are apparently rare, and are often undetectable in embryos that have already integrated DNA before the first division. Thus, although most of the embryos generated during the REMI procedure have a single integration pattern, some transformants are insertionally mosaic.
Figure 2. Transgenic animals generated by the REMI method: diversity within F0 founders and possible mosaicism.
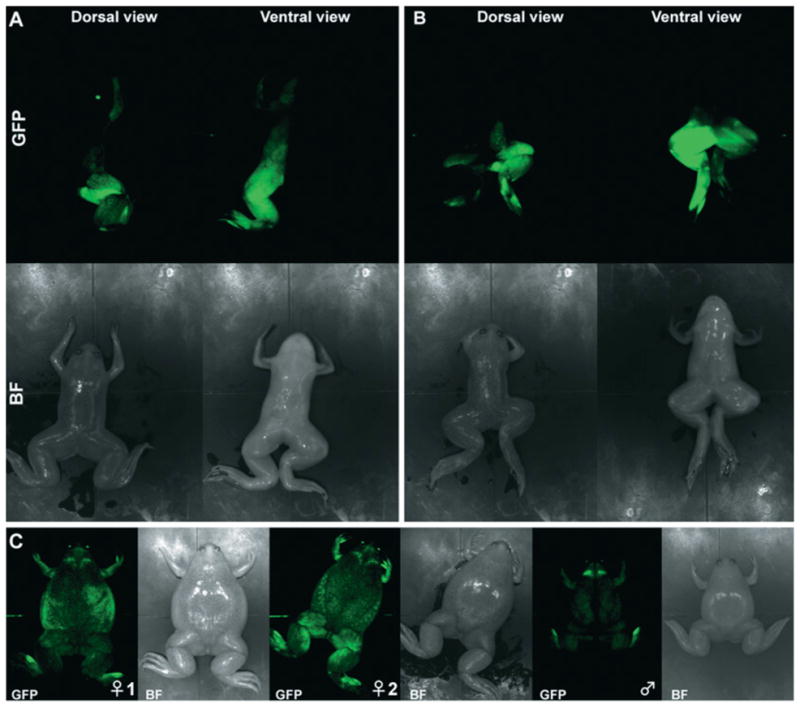
(A) Hemi-transgenic individual showing a left–right restricted distribution of GFP expression using a CMV (cytomegalovirus)–GFP reporter transgene. (B) Hemi-transgenic individual showing an antero–posterior-restricted distribution of GFP expression from a CMV–GFP reporter transgene. (C) Non-mosaic F0 founders showing different level of GFP expression from the same CMV–GFP reporter construct. In all panels, bright-field (BF) images are shown along the GFP pictures. Pictures were taken using the Aequoria system (Hamamatsu).
What are the molecular mechanisms of DNA integration using REMI in Xenopus?
To date, it is still unclear when and how transgene integrations occur. Published protocols have simplified the original procedure, discarding the use of egg extracts and RE (Figure 1). The modified procedure is now similar to the ICSI (intracytoplasmic sperm injection) used to generate transgenic mice (Perry et al., 1999; Sparrow et al., 2000; Smith et al., 2006). This challenges the idea that transgenic nuclei are being generated in vitro during the REMI procedure. If, indeed, most of the events triggering transgene integration occur after transplantation of the male pro-nucleus, then why was the REMI method successful at generating useful transgenic when conventional microinjections of linearized DNA into fertilized eggs is not (see respectively Figure 1B and 1C)?
Impact of injection timing and location
Two factors that may affect the efficiency of integration are: (i) the location of the exogenous DNA with respect to the pronuclei, and (ii) when it contacts genomic DNA. Using the REMI procedure, proximity and exposure time are not an issue, since it ensures that the exogenous DNA is present right after fertilization within the zygote as a linear template, thus during the entire course of the first cell cycle, unlike when performing microinjection experiments (Figure 1).
Impact of the amount of exogenous DNA and enzyme
Despite the large amounts of exogenous DNA delivered during microinjection experiments (ranging from the order of 10 to a few 100 pg per cell), it is often maintained as extrachromosomal structures and randomly segregates in daughter cells, resulting in a mosaic distribution (Etkin and Pearman, 1987; Etkin, 1991). Integrations are possible, but the efficiency probably depends on the generation of genomic DNA breaks as development proceeds, and on the size of the transgene concatemers initially formed. In fact, adding RE was shown to increase the efficiency of transgenesis, perhaps by limiting the extent of concatenation, thus making the exogenous DNA more amenable for integration. In the REMI procedure, the linear DNA ends up being injected at a concentration 10–100 times below what is normally used for conventional microinjections (1–10 pg). However, its distribution within the cell is localized to the site of the forming nucleus and little diffusion is expected. Indeed, following microinjection into fertilized eggs, the exogenous DNA appears to ‘stick’ to chromosomes during mitosis (Etkin and Pearman, 1987).
Increasing the amount of linear DNA in the REMI protocol does not dramatically improve the efficiency of early integration events. In fact, it results in increasing the number of embryos with detectable mosaic expression (often seen in the somites). One interpretation for these data is that part of the exogenous DNA inserts into the genome before the first cell cycle and that the remaining DNA is maintained as extrachromosomal structures (Smith et al., 2005).
Impact of chromosomal breaks
Various mechanisms have been suggested to underlie DNA integration following microinjection experiments in the mouse. Chromosomal breaks is considered to be an important limiting factor, resulting, in most cases, in DNA integration at a single site (Brinster et al., 1985). In contrast, F0 Xenopus transgenic animals generated by REMI often carry more than one integration event, as estimated by the ratio of transgenic compared with non-transgenic F1 siblings (Bronchain et al., 1999; Kroll and Kirschner, 1999; Marsh-Armstrong et al., 1999). In the REMI procedure, the sperm nuclei are often maintained as a frozen preparation. Nuclei isolation and/or storing procedure may favour the mechanical formation of numerous genomic DNA breaks that must be repaired before the cell initiates the first division. Importantly, repair mechanisms that would result in major chromosomal rearrangements cannot be excluded with this method.
Taken together, these observations partly explain the predominance of early integration events using the REMI method in Xenopus, the transgene inserting predominantly as a concatemer and often in multiple sites. The time and place at which the exogenous DNA is brought into contact with the host genome are likely effectors of transgenic efficiency. It also points to the requirement for maternal components that promote exogenous DNA incorporation. A better understanding of the early mechanisms of DNA integration would help narrow down the critical steps of the REMI procedure and would greatly benefit future improvements of transgenic methods in Xenopus.
Specific features of transgenic animals generated by the REMI method
The molecular nature of exogenous DNA insertions mediated by REMI, which includes numerous independent events, random integration sites and transgene concatenation, brings some important experimental considerations when working with founder animals. The mutagenic aspects of the method can be regarded as a source of major drawbacks for some applications. However, the high efficiency and the majority of non-mosaic events provide the researcher with an almost guaranteed germline transmission from few founder animals.
Limitations when working with F0 transgenic animals
Transgenic animals can be studied at the F0, but the genetic variability among founders may complicate data interpretation. Each F0 arises from an independent event with regard to the site of integrations and copy number. The transgene expression level can therefore be variable among individuals (Figure 2C). In addition, the fragile sperm nuclei can easily be damaged during the procedure, impinging on genomic DNA integrity and making the method very mutagenic (Bronchain et al., 1999). The potential generation of genetic abnormalities using REMI has been correlated with the resulting embryos exhibiting numerous developmental defects. These considerations, in part, explain the overall low survival rate of the embryos generated by REMI compared with standard microinjection techniques (Sparrow et al., 2000; Smith and Mohun, 2005).
There are two significant drawbacks to abnormal development and low survival rates. First, phenotypic analyses following mis-expression studies can be difficult to interpret in such a background. Secondly, the small number of surviving animals makes studies at late stages of development tedious. The occurrence of genetic variability at the F0 can be overcome by establishing transgenic lines, most practically by using X. tropicalis. Both Xenopus species are amenable to methods such as microinjections, grafting and dissection, as well as to REMI and gynogenesis procedures (Offield et al., 2000; Sparrow et al., 2000; Noramly et al., 2005). The diploid genome and shorter generation time of X. tropicalis confer a significant advantage and render generation of homozygous transgenic lines a feasible task in a relatively short period of time when using gynogenesis (see Figure 5D).
Genome ploidy issues
In the REMI protocol, the sperm nuclei are injected using an infusion pump. On average, at best, a third of the injected embryos are expected to receive a single nucleus and to develop properly (Figure 1 and see Figure 5B). Triploid and some hyperdiploid cells can easily be identified by virtue of the large excess of genomic DNA. This usually leads to the formation of bigger cells and can be directly quantified by FACS analyses (Flajnik et al., 1984). Alternatively, counting the proportion of nucleoli in cells will give a good estimation of the ploidy status of the individuals, as deviation from the expected frequencies is indicative of chromosome number mosaics (Smith, 1958; Muller et al., 1978). A simple karyotype will provide a lot of information, but the use of FISH (fluorescent in situ hybridization) analyses on chromosomes from transgenic individuals would be greatly beneficial. A FISH protocol has been developed for the MHC (myosin heavy chain) and Ig loci of Xenopus (Courtet et al., 2001). Initially, the limit of detection required at least three copies per genome. In the REMI transgenic procedure, the transgene integrates in multiple copies per insertion site, providing suitable circumstances for this analysis. Recently, a protocol to improve the sensitivity of the method was developed that coupled FISH to TSA (tyramide signal amplification). This strategy was successfully used to localize single-copy genes in both Xenopus species (Krylov et al., 2003; Tlapakova et al., 2005; Krylov et al., 2007). A workshop on cytogenetics of X. tropicalis (Workshop on Cytogenetics of Xenopus tropicalis, 17–21 September 2007, Charles University, Prague, Czech Republic), showed that this approach offers a very powerful tool to rapidly perform genetic mapping.
SMGT (sperm-mediated gene transfer)
To date, the REMI method remains the most broadly used procedure for making transgenic Xenopus. However, additional strategies have been developed. Reports have highlighted the use of transfected spermatozoa from different species to generate transgenic animals, including Xenopus (Habrova et al., 1996; Takac et al., 1998; Smith and Spadafora, 2005). Different strategies have been employed to transform sperm DNA, such as liposome transfection, electroporation and dipping cells in a solution of naked DNA. SMGT has been combined with other methods to enhance the performance of the technique. Interestingly, SMGT in conjunction with REMI was shown to be very efficient in cattle (Shemesh et al., 2000).
The field of SMGT initially generated considerable interest (Brackett et al., 1971; Lavitrano et al., 1989). Indeed, the straightforwardness of the method is appealing, as it relies on the use of sperm for artificial insemination. Sperm cells are used as vectors for transmitting their own, but also the exogenous DNA, to the zygote. In species refractory to microinjections, such as farm animals, SMGT was used as an alternative for transgenesis (Lavitrano et al., 2006). In X. laevis, transgenic animals were obtained by SMGT (Habrova et al., 1996; Takac et al., 1998). However, the integrity of the transgene appeared compromised in these animals and their progeny. In fact, stable transgene integrations have rarely been detected following SMGT in many species (Smith and Spadafora, 2005).
The field of SMGT has been controversial and very few investigators are exploiting the use of this technology for making transgenic animals (Brinster et al., 1989; Smith, 1999; Smith and Spadafora, 2005). However, it would be worth exploring the mechanisms further by which SMGT is controlled in the frog. If issues such as transgene integrity and stability are solved, SMGT may result in being a method of choice, as being efficient, rapid, inexpensive and amenable to most laboratories. Developing strategies to transform sperm cells (or oocytes) for use in in vitro fertilization would clearly improve transgenic procedures in frogs.
TEs (transposable elements)
A variety of methods based on specific enzymatic activities have been tested in Xenopus, in addition to REs to promote insertion of new DNA fragments into the genome. These include mobile elements in the form of TEs. Different families of transposable systems have been tested in Xenopus such as Sleeping Beauty, a Tc1/mariner family of TEs (Ivics and Izsvak, 2004). The expectation was to identify a system that would compare favourably with the REMI method. So far, the data obtained in embryos has not reached the expected efficiency, with a rate of transgenesis below 6% (Sinzelle et al., 2006). The F0 transformants obtained exhibited a high degree of mosaic distribution. However, each family of TEs has its own mechanisms of action, which can affect how well it performs in a given host. Recently, a Tol2 TE system has been described in Xenopus with promising features (Hamlet et al., 2006).
I-SceI meganuclease procedure for making transgenic Xenopus
The I-SceI meganuclease (meganuclease) is an endonuclease which recognises an 18 bp sequence and was originally isolated from Saccharomyces cerevisiae. Initially developed in the fish medaka, a transgenic method based on the meganuclease was subsequently adapted in X. laevis and X. tropicalis (Ogino et al., 2006a, 2006b; Pan et al., 2006). Importantly, germline transmission has been demonstrated for this method that makes it compatible with multi-generation experiments (Ogino et al., 2006b; Pan et al., 2006).
The meganuclease procedure requires only the I-SceI endonuclease, a commercially available meganuclease, and a transgene construct containing the 18 bp recognition site. The construct is digested with the meganuclease and the mixture is directly microinjected into fertilized eggs. Efficient integration requires an injection between the animal pole and the sperm entry site, thus in close proximity to the forming nucleus. Using the meganuclease method, as many as 20% in X. laevis or 30% in X. tropicalis express the transgene at Stage 44–46, with an excellent survival rate (Ogino et al., 2006b; Pan et al., 2006). The number of transgene copies ranges from one to eight per transgenic embryos, in one to two insertion sites. These results are, to some extent, comparable with what was previously reported by Etkin and Pearman (1987) using a standard microinjection strategy. However, using the meganuclease method, a substantial number of non-mosaic embryos is generated, from 2 to 14%, depending on the promoter–reporter construct used (Pan et al., 2006).
It is likely that the meganuclease, along with the REMI, ICSI and microinjection, procedures share at least some of the same molecular mechanisms for transgene integration. Using these methods, the insertion events are random and multiple, and the exogenous DNA insert as concatemers. Significant advantages of the meganuclease method over the REMI (or ICSI) procedure are the ease of manipulation, a good survival rate and efficiency in both Xenopus species (Table 1).
Table 1.
Principal features of transgenic methods performed in Xenopus
| Procedure | Transgenic method |
||||
|---|---|---|---|---|---|
| Micro-injection | REMI | Meganuclease | Integrase | TE | |
| Injection setups | Conventional | Infusion pump | Conventional | Conventional | Conventional |
| Eggs | In vitro fertilized | Unfertilized | In vitro fertilized | In vitro fertilized | In vitro fertilized |
| Specific reagents | Sperm nuclei | Integrase mRNA | Transposase mRNA | ||
| Exogenous DNA | Linear plasmids | Linear plasmids/BACs | Circular plasmids | Plasmids* | Plasmids* |
| Type of insertion | |||||
| Concatemer formation | Yes | Yes | Probable?(1 to 8 copy) | No (single copy) | ? |
| Sites | Multiple | Multiple | Multiple | Single | Multiple? |
| Location | Random? | Random? | Random? | No? | TE sites? |
| Transgene distribution | |||||
| Mosaic | Yes | ? | Yes | Yes | Yes |
| Non-mosaic | None | Almost all | 2–12% | ? | None |
| Other features | |||||
| Efficiency | 5–10% | Over 20% | Up to 14% | 25–35% | 6.5% for SB; 30% for Tol2 |
| Germline transmission | Yes | Yes | Yes | Not tested | Yes |
| X. tropicalis | Not tested | Yes | Yes | Not tested | Yes |
| Reference | Etkin and Pearman (1987) | Kroll and Amaya (1996) | Ogino et al. (2006b); Pan et al. (2006) | Allen and Weeks (2005) | Tol2, Hamlet et al. (2006); SB, Sinzelle et al. (2006) |
RNAse-free sample.
?, unknown or uncertain.
phi-C31-integrase-mediated transgenesis
The methods that we have described so far depend on mechanisms of DNA insertion that are nearly random. Researchers have taken advantage of the mechanism by which the bacteriophage phi-C31 infects Streptomyces (Groth et al., 2000; Allen and Weeks, 2005, 2006). The phage DNA encodes the phi-C31 integrase that catalyses a site-specific recombination into the host genome. Two different short DNA sequences are needed to sustain integration by this integrase. The attP site in the phage DNA and the attB site in the bacterium genome are recombined to generate two new sequences (attR and attL). The integrase does not require accessory proteins to mediate integration, but cannot catalyse the reverse reaction (excision) in this context. This feature ensures that the integration events are unidirectional when the integrase is provided alone. This transgenic approach involves the co-injection of mRNA encoding the integrase with a plasmid containing an attB site, using standard microinjection techniques, into fertilized eggs. As the sequence requirements for attP sites can be relatively promiscuous, the integration event relies on the existence of pseudo-attP sites in the frog genome.
The integrase method offers a unique way to promote single-copy transgene insertion in a sequence-restricted manner. A major advantage lies in the ease of the manipulation itself that is based on microinjection methodologies in fertilized eggs. Of note, transgene germline transmission and efficiency in X. tropicalis have not yet been demonstrated (Table 1).
Use of transgenesis in promoter/enhancer studies: where do we stand?
If one assesses the number of PubMed entries using the following search criteria, ‘genetically modified animals’ and ‘Xenopus’, 195 publications are selected (Figure 3A). Among these publications, a majority also refers to ‘gene expression regulation’, including promoter and enhancer studies (Figure 3C).
Figure 3. Publication status on transgenesis in Xenopus.
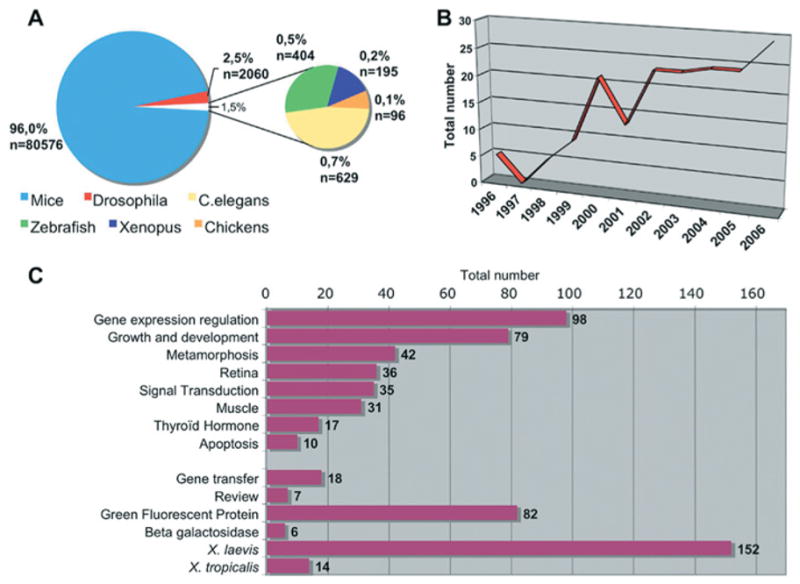
(A) Proportion of PubMed entries taking into account a given model organism and genetically modified animals. The data were obtained by using the following search criteria. For Xenopus, zebrafish, chickens and Caernorhabditis elegans: “Name of the organism”[MeSH] AND Animals, Genetically Modified[MeSH] NOT Mice[MeSH] NOT Drosophila[MeSH]; for mice: Mice[MeSH] AND Animals, Genetically Modified[MeSH] NOT Drosophila[MeSH]; and for Drosophila: Drosophila[MeSH] AND Animals, Genetically Modified[MeSH] NOT Mice[MeSH]. (B) Evolution over years of the number of PubMed entries associated to Xenopus and genetically modified animals. (C) Topic distribution of the PubMed entries referring to both ‘Xenopus’ and ‘Genetically modified animals’. The data were obtained using the topics mentioned on the histogram and the search criteria described above: Xenopus[MeSH] AND Animals, Genetically Modified[MeSH] NOT Mice[MeSH] NOT Drosophila[MeSH]. These searches were performed in October 2007.
Looking for the ideal reporter gene
The transgenesis methods described above offer a unique way to perform high-throughput promoter/enhancer assays. Hundreds of transformants can be obtained within hours and, in combination with the use of fluorescent markers, such as the GFP (green fluorescent protein), the activities of a transgene can be monitored in whole living embryos as development proceeds. In most assays, investigators favoured GFP as a reporter gene (Figure 3C). GFP expression can easily be detected in living embryos at many stages of development. However, in practice, the development of pigmented cells, and the autofluorescence of the vitellus and internal organs, can make the observation of GFP difficult in wild-type animals. Performing experiments in X. laevis mutants, such as the periodic and reticulated albinos, may facilitate monitoring GFP expression (Figure 4). To date, no such mutants have been isolated in X. tropicalis.
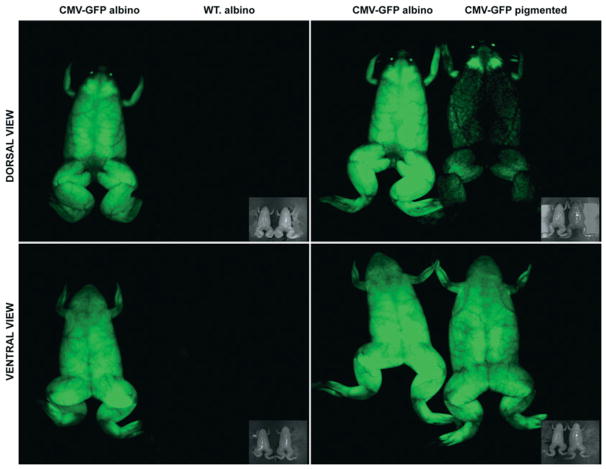
Figure 4. Glow in pigmented frogs: the advantage of the albino mutants.
GFP fluorescence was analysed in an adult albino frog transgenic for a CMV (cytomegalovirus)–GFP reporter transgene that drives GFP expression ubiquitously. In the left-hand panels, the transgenic albino frog is shown next to a wild-type albino animal, and in the right-hand panels next to a pigmented frog transgenic for the same construct. The pigments obscure the analyses of GFP expression on the dorsal side of the pigmented animal (upper panels). However, when observed on the ventral side, both the albino and pigmented animal express GFP to a similar extent (lower panels). Bright-field images are provided as insets in the main pictures. Pictures were taken using the Aequoria system (Hamamatsu).
Despite its obvious advantages, the use of GFP to monitor expression has some drawbacks. Its biochemical features, such as stability and threshold levels required for detection, makes it inappropriate to assess dynamic or low levels of gene expression. In numerous reports, GFP expression was assessed by fluorescence microscopy and/or ISH (in situ hybridization) studies. A discrepancy was occasionally noticed between the expression domains of GFP mRNA and the observed fluorescence. This effect was attributed to a lag in the accumulation and processing of the GFP protein, as well as differences in mRNA compared with protein stabilities (Hartley et al., 2001; Polli and Amaya, 2002; Hutcheson et al., 2005). However, the engineering of new GFP variants, as well as the isolation of novel FPs (fluorescent peptides), have contributed to establishing promising sources of FPs to be used in Xenopus. Most of these FPs remain untested in Xenopus. Finally, an interesting alternative to investigate the control of gene expression is the use of a cross-species approach. In this case, the identification of the transgene activity can be monitored by the use of species-specific ISH probes. In this context, the dynamic nature of the transgene expression is expected to closely resemble the endogenous one (Polli and Amaya, 2002).
Transgenesis and position effects
One expectation when isolating promoter/enhancer elements is that the transgene expression resembles the in vivo state as much as possible. A limitation that persists when using transgenesis results from the random nature of integration events into the host genome. Indeed, it is accepted that the sequences surrounding the transgene integration site can modify the expected expression pattern, a phenomena known as CPEs (chromosomal position effects) (Wilson et al., 1990). In transgenesis experiments, it is impossible to target a specific site and, with the exception of the integrase method, to control the transgene copy number. Furthermore, the transgene can insert as a concatemer, resulting in new arrangements of DNA fragments. In this context, the nature of the transgenic construct itself may play an unpredictable role in the final pattern of expression. Some investigators remove unnecessary sequences from the vector in order to prevent interference from potential enhancers found in selection cassettes (Hirsch et al., 2002). These potential problems must be considered when interpreting promoter/enhancer studies in F0 animals.
A number of strategies have been proposed to overcome CPEs. Targeting the site of integration by homologous recombination (knock-in) would solve most problems, however, this technology is yet to be developed in Xenopus. An alternative strategy relies on the generation of transgenic lines carrying preferred sites for transgene integration. The Cre/Lox-mediated site-specific integration strategy has been successfully used to minimize variability among transgenic lines in various species. Interestingly, the Cre and FLP recombinases have been shown to be active in Xenopus, and an inducible system based on the Cre recombinase has been developed in transgenic Xenopus (Werdien et al., 2001; Ryffel et al., 2003; Waldner et al., 2006).
The use of insulator elements within transgenic vectors has proven to be very useful to provide an efficient relief from CPEs and unintended activation by enhancer elements (Allen and Weeks, 2005). In particular, it has been shown that ‘bracketing’ the reporter construct with tandem copies of the chicken β-globin 5′-HS4 insulator was sufficient to block the spread of chromatin silencing (Chung et al., 1993). The development of vectors containing multiple insulator elements, in which genes of interest and adequate reporters could be inserted, may be of great interest to the community.
The occurrence of CPEs suggests that genes are organized as transcription units that remain independent from one another as contiguous fragments on chromosomes through insulation from neighbouring sequences (Laemmli et al., 1992; Dillon and Grosveld, 1994). In this context, it is not surprising that standard transgenic constructs, which often contain a few kb of upstream sequences, display CPE and unfaithful expression. The transfer of large genomic DNA fragments, such as BACs/YACs (bacterial artificial chromosomes/yeast artificial chromosomes) has become a method of choice to achieve optimal expression from transgenes in various species (Giraldo and Montoliu, 2001). Indeed, BACs (or YACs) are in a range of size compatible with the maintenance of a genome region that could behave as a functionally independent gene regulation unit (Giraldo and Montoliu, 2001). Large genomic DNA fragments have not been extensively used as transgenic vectors in Xenopus. However, using REMI, the fact that transgenes over 10 kb long insert into the host genome as concatemers, as well as the report of a successful example of BAC transgenesis, suggests that this method is well suited for this type of gene transfer (Kelly et al., 2005).
Transgenesis and functional studies: targeted and inducible expression systems
In a relatively short period of time and with the completion of the X. tropicalis genome sequence, an impressive amount of nucleotide sequence information has accumulated. This wealth of data now calls for functional studies. One striking aspect of the Xenopus publications in which a transgenic method has been used is the representation of relatively few topics of research areas. Among these publications, 68% relate to ‘retina’ and ‘muscle’ development, and ‘metamorphosis’, including thyroid hormone metabolism, and finally signal transduction that includes predominantly the Wnt, BMP (bone morphogenetic protein), HH (Hedgehog) and FGF (fibroblast growth factor) signalling pathways (Figure 3C). An explanation for this may come from the original research interests of the investigators that have participated in the continuous improvement of the transgenic methods. Another explanation lies in the experiment itself. Manipulating the function of genes involved in limb formation, naturally induced apoptosis or thyroid hormone signalling involves reaching metamorphosis. In addition, many experiments require the gene to be expressed in all cells of a tissue of interest. This is particularly relevant when assessing gene functions in a given cell-signalling pathway. For these purposes, microinjection of DNA that is not integrated into the genome fails to meet the needs of the investigator (Kroll and Amaya, 1996).
For studying genes involved in relatively late developmental stages, or displaying lethality when misexpressed, the development of targeted/conditional gene expression systems is critical. A variety of regulatable approaches have been reported, including the use of the Gal4-UAS binary system, the modified progesterone RU-486 or tetracycline inducible systems, the Cre/FLP site-specific recombinases and finally heat shock or metallothionein inducible promoters, (Marsh-Armstrong et al., 1999; Wheeler et al., 2000; Werdien et al., 2001; Chae et al., 2002; Hartley et al., 2002; Ryffel et al., 2003; Das and Brown, 2004; Waldner et al., 2006). Optimization of the various systems is now underway, and a plausible way to achieve this goal is to combine binary and hormone-inducible systems (Chae et al., 2002). An attractive aspect of this strategy lies in the generation of numerous activator and effector lines, the combination of which by simple cross-fertilization result in a variety of transgenic animals expressing a gene of interest in restricted domains in time and space. In this scheme, the characterization and annotation of a range of tissue-specific promoters, as well as the construction of compatible effector lines, should expand interest in the approach.
Tracing biological activities in vivo
Protein localization and interactions
FPs have been exploited to determine the subcellular localization and dynamics of a given protein as a first step to establish functional networks. Transgenesis makes this type of approach possible in living cells, at a high spatial and temporal resolution when combined with advanced imaging tools (Coen et al., 2001; Stubbs et al., 2006).
Transgenic sensor lines
Not only might FPs be used to monitor protein interactions, they may also be useful to image signalling networks and to monitor environmental conditions. In this scheme, transgenic Xenopus lines have been established to visualize the in vivo activity of the retinoic acid and Wnt/β-catenin signalling networks during development (Geng et al., 2003; Luria and Furlow, 2004). These sensor lines are based on the induction of a reporter gene placed under the control of an artificial promoter that mediates transcriptional responsiveness provided by specific transcription-factor-binding sites that are fused to a minimal promoter. This strategy was further exploited to monitor the global inhibition of the Lef1/Tcf (lymphoid enhancer-binding factor 1/T-cell-specific factor)-dependent signalling pathway (Deroo et al., 2004; Denayer et al., 2006).
Most research dedicated to environmental monitoring has focused on plants. However, environmental contamination by toxins alters numerous physiological functions in nearly all classes of vertebrates, in particular, the endocrine system (Kloas et al., 1999). The use of transgenic Xenopus as detectors of environmental changes offers an attractive approach to assess the impact of a variety of environmental conditions on vertebrate development and physiology. Oofusa et al. (2003) fused a metallothionein promoter to GFP to assess the effect of zinc and cadmium concentrations in water. The expectation was to obtain a readout of the activation of genes used for protection from heavy metals. Furlow and Brown (1999) showed that some transcriptional regulatory elements of the TH/bZip gene behaved as a thyroid-hormone-responsive construct. Thus alteration of its transcriptional regulation may be used to reflect the presence of endocrine disruptors in the environment (Turque et al., 2005).
Amphibians have long been used as indicator species of environmental changes or contamination. Transgenic approaches open new fields of research for the development of monitoring programmes. In fact, the development of biotechnological tools to perform environmental risk assessment using transgenic Xenopus is ongoing (WatchFrog company; http://www.watchfrog.fr/).
Cell selection and ablation
The development of selection procedures through exposure to toxic proteins has been initiated by making transgenic animals carrying the aphA-2 gene that confers resistance to the antibiotic G418 (Moritz et al., 2002). This strategy ultimately aims at developing tools to select transgenic animals expressing non-fluorescent transgene products. However, long-term effects of G418 exposure have not been determined.
Methods to perform targeted cell ablation experiments have been tested in transgenic Xenopus. Activation of the mitochondrial apoptotic pathway was shown to provide an efficient means to induce genetic cell ablation (Du Pasquier et al., 2007). Alternatively, the expression of the M2(H37A) toxic ion channel was also shown to be cytotoxic in embryos and to promote cell death (Smith et al., 2007). The combination of these cell ablation strategies with inducible expression systems would provide a way to monitor the timing and level of cytotoxic effects. Such methods not only open new ways to assess cell fates and functions during development, but also give the opportunity to challenge regeneration processes.
Conclusions and perspectives
We have provided an historical survey and a review of the status of Xenopus transgenesis. To summarize, the transgenic techniques have contributed enormously to promote Xenopus as a model organism that is compatible with genetic studies. Transgenic experiments were primarily designed to investigate how gene expression is controlled in vivo. In addition, an effort has been made to develop suitable reagents for sophisticated functional gene analyses and tracking biological activities in vivo. The generation of transgenic lines, as well as genetically defined strains in various laboratories, now calls for the development of structures to maintain and propagate these resources. Of particular interest are lines that would allow the rapid allocation of gene expression domains to specific tissues or subcellular compartments, as well as sensor lines. In addition, a collection of transgenic lines adapted to functional gene analyses that rely on the use of hormone and/or binary inducible systems appears crucial.
Transgenesis procedures and large-scale screens
Despite numerous technical advances, transgenesis is still not broadly used in Xenopus and remains mastered in a limited number of laboratories (Figure 3). This status is in sharp contrast with the work that has been conducted using other model organisms, as exemplified by the data obtained in fish (Amsterdam and Becker, 2005). Two prominent examples are insertional mutagenesis screens that take advantage of retroviral vectors to promote transgene insertions, and gene/enhancer trap screens that use both transposons and retroviruses (Amsterdam and Hopkins, 2004; Balciunas et al., 2004; Ellingsen et al., 2005; Kotani et al., 2006). Both fish and Xenopus share significant advantages that make them convenient vertebrates in which to perform genetic approaches to development, namely transparency of the embryos, external/rapid development and fecundity. The ability to directly obtained non-mosaic transgenic founder animals using the REMI method, thus saving one generation in a breeding scheme to unravel mutations, should have boosted Xenopus as a model to perform high-throughput studies using transgenesis. However, large-scale screens based on transgenic approaches have not been reported yet in Xenopus.
So far, the REMI procedure has been the most broadly used method to generate transgenic Xenopus. Thus, although very efficient, the REMI method as it stands may not be ideally suited for these investigations. One explanation is that the REMI method is difficult to initiate and investigators are confronted with numerous technical challenges. Most investigators working on Xenopus are familiar with microinjection techniques and are thus faced with the technical difficulty of injecting large nuclei into unfertilized eggs, which are softer than one-cell-stage embryos. Furthermore, the REMI method often leads to developmental defects that limit the ability to study late stages of development and the generation of transgenic founders to make lines. New transgenic methods have been developed which display advantageous technical features that overcome most of the difficulties encountered using REMI. In particular, methods that are based on microinjection techniques, such as the integrase or meganuclease methods, can circumvent most of the undesirable side effects on genetic integrity generated by the REMI method.
Choosing procedures to perform transgenesis in Xenopus
The message that emerged from the recent technical developments described in the present review is that at least three efficient methods of transgenesis are currently available, the REMI, the I-SceI meganuclase and the phi-C31 integrase strategies. Each of the methods gives the opportunity to manipulate gene expression and generate Xenopus transgenic lines. However, these transgenic procedures utilize different molecular mechanisms to mediate DNA integration and may not be fully interchangeable, as the resulting transgenic animals are intrinsically different (Table 1). The choice of the method relies on the type of experiments to be performed and the technical expertise available in a given laboratory. To make investigators familiar with the various transgenic procedures, a workshop was organized by the X-omics consortium in June 2006. An evaluation of the panel of best-suited applications, as well as interesting insights into the potential advantages and drawbacks of each method, were assessed (see Supplementary material at http://www.biolcell.org/boc/100/boc1000503add.htm). Briefly, the experiments performed during the workshop clearly demonstrated that we have tools that provide answers to most experimental requirements. We suggest that resources to facilitate further the development of transgenesis as a routine procedure in Xenopus would be valuable. These resources include the development of common specialized stock centres, cryopreservation of stocks of transgenic sperm and technical training centres.
Is there a need for improving transgenic methodologies in Xenopus?
The observation that transgenic procedures are largely underused is particularly striking with respect to the evolution of transgenesis approaches in other model organisms, such as zebrafish. Numerous large-scale investigations have been performed in these species using transgenic approaches. In Xenopus, a single example of large-scale screening based on chemical mutagenesis has been reported in the literature, demonstrating that these investigations are technically feasible using this model (Noramly et al., 2005). One consideration is that the initial REMI method was not well suited for high-throughput studies in Xenopus and that more recently developed methods await to be challenged to fully exploit transgenesis in Xenopus. What is lacking in the field are transgenic methodologies that could put forward the major advantages of the Xenopus model, in particular, its external development and the ability to produce a large number of eggs with the use of a diploid species whose genome has been sequenced. In other words, future developments concern the improvement or development of efficient imaging systems dedicated to Xenopus, as well as the ability to generate large numbers of non-mosaic F0 transgenic animals in X. tropicalis.
Supplementary Material
Acknowledgments
We thank Chantal Ballagny and Annie Legal for providing us with their support in the administration and management of the workshop. We would also like to thank Christophe De Medeiros, Johanna Hamdache and Karine Parain for their tremendous technical support during the workshop. We are very grateful to Raphaël Thuret, for his help in supervising the training sessions, and to Daniel Roche for his contribution in the REMI experiment (REMI2). We are particularly grateful to the workshop participants for their wonderful contribution, namely: E.J. Bellefroid, C. Ben, J.F. Bodart, F. Broders, E. M. Callery, T. Carle, L. Coen, D. Du Pasquier, L. Fairclough, C. Jäckh, M. Locker, S. Menoret, A. H. Monsoro-Burq, O. Nentwich, M. Nichane, M. Perron, A. Rana, J.F. Riou, D. Roche, A. Sebillot, S. Smith, V. Thomé, R. Thuret, K. Treguer, M. Umbhauer, C. Van Campenhout and Q. Ymlahi-Ouazzani. We acknowledge Anne-Hélène Monsoro-Burq, Jean François Riou, Muriel Umbhauer, David Du Pasquier and the Watchfrog company for sharing their material and expertise during the workshop. We thank Leica microsystems and Micro Mecanique SAS companies for providing us with fluorescent and light microscopes respectively during the course of the workshop. Finally, we thank Morgane Locker for fruitful discussions and critical comments on the preparation of the manuscript. The workshop was supported by the European Community FP6 (X-omics coordinated action no. 512065), Centre National de la Recherche Scientifique, Université Paris-Sud XI, L’Association pour la Recherche contre le Cancer and le Ministère de l’Education Nationale, de la Recherche et de la Technologie.
Abbreviations
- BAC
bacterial artificial chromosome
- CPE
chromosomal position effect
- F
filial generation
- FISH
fluorescent in situ hybridization
- FP
fluorescent peptides
- GFP
green fluorescent protein
- ICSI
intracytoplasmic sperm injection
- ISH
in situ hybridization
- RE
restriction enzyme
- REMI
restriction-enzyme-mediated integration
- SMGT
sperm-mediated gene transfer
- TE
transposable element
- YAC
yeast artificial chromosome
- Xenopus
The pipid frog Xenopus is one of the favourite amphibian models of biologists, especially embryologists. The genus of Xenopus laevis (the African-clawed toad) is widely used in developmental biology and was formerly used in pregnancy diagnosis. It can be induced to ovulate and mate any time of the year, following a simple injection of gonadotropic hormones. Xenopus embryos are large and easily manipulated. The function of various macromolecules, such as RNA and protein, can be assayed by microinjection into living embryos
- Xenopus tropicalis
Xenopus (Silurana) tropicalis forms a separate, but evolutionarily related, lineage from X. laevis. Both species are highly similar in morphology, and share the same advantages with respect to embryological manipulation
- Transgenesis
This is a procedure that introduces an exogenous DNA, called a transgene, into the genome of a living organism. Alternatively, the transgene can be introduced into germ cells that are used for fecundation. The transgenic organism will exhibit a new property and will transmit it to its offspring
- Non-mosaic transgenic animal
An individual made up exclusively of cells that are genetically identical for the transgene insertion events
- Morpholino
These are oligounucleotides made of short chains of approx. 25 subunits, each containing a nucleic acid base, a morpholine ring and a non-ionic phosphorodiamidate intersubunit linkage. Antisense morpholinos are used to knockdown gene expression, redirect splicing or block miRNA (microRNA) maturation
- Mosaic distribution or pattern
The variegated level and/or expression of a molecule within cells of a single individual
- Integrase
Enzyme that facilitates prophage integration into or excision from a bacterial DNA
- Meganuclease
Sequence-specific endonuclease with large (>12 bp) cleavage sites
- Transformant
An organism that has been genetically altered through the uptake of foreign DNA
- Transgene
A genetic material that has been transferred by genetic engineering techniques from one organism to another. In practice, it describes a segment of DNA containing a sequence that has been isolated from one organism and is introduced into a different organism
- Germline transmission
A process referring to the passage of a transgene that has been incorporated into the germline of an organism to its offspring
- Founders
Individuals with a genetic trait of interest that will be used to derive lines. The F0s are animals produced during the transgenesis procedure, for which the primary genetic trait of interest is the transgene
- Gynogenesis
Development of an egg that is stimulated by a sperm in the absence of any participation of the sperm nucleus. In Xenopus this is achieved by using UV-irradiated sperm. The diploid status can be re-established either by blocking polar body extrusion, or through inhibition of the first division. The resulting embryos are called gynogenic animals
- Blastomeres
Cells formed by division of the fertilized egg. The first division establishes the left–right axis in Xenopus
- Extrachromosomal structures
Extrachromosomal DNA that is maintained in a cell distinct from the chromosomes. These structures have been shown to replicate and to segregate independently from the genomic DNA in a random fashion
- Concatemers
DNA arrays of covalently joined sequences generated by concatenation. Concatenation has been observed when linear transgenes are microinjected into cells and during the REMI procedure
- Hyperdiploid
An organism exhibiting a chromosome number greater than the diploid number
- Transposable element
A DNA sequence that can move from one location to another, for example, from a transgene to the host genome
- Stage 44–46
Stages of Xenopus embryonic development according to Nieuwkoop and Faber (1994). These stages will correspond to swimming tadpoles
References
*Articles of special interest
- Allen BG, Weeks DL. Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat Methods. 2005;2:975–979. doi: 10.1038/nmeth814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BG, Weeks DL. Using phiC31 integrase to make transgenic Xenopus laevis embryos. Nat Protoc. 2006;1:1248–1257. doi: 10.1038/nprot.2006.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaya E. Xenomics. Genome Res. 2005;15:1683–1691. doi: 10.1101/gr.3801805. [DOI] [PubMed] [Google Scholar]
- Amaya E, Kroll KL. A method for generating transgenic frog embryos. Methods Mol Biol. 1999;97:393–414. doi: 10.1385/1-59259-270-8:393. [DOI] [PubMed] [Google Scholar]
- Amaya E, Offield MF, Grainger RM. Frog genetics: Xenopus tropicalis jumps into the future. Trends Genet. 1998;14:253–255. doi: 10.1016/s0168-9525(98)01506-6. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Becker TS. Transgenes as screening tools to probe and manipulate the zebrafish genome. Dev Dyn. 2005;234:255–268. doi: 10.1002/dvdy.20541. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Hopkins N. Retroviral-mediated insertional mutagenesis in zebrafish. Methods Cell Biol. 2004;77:3–20. doi: 10.1016/s0091-679x(04)77001-6. [DOI] [PubMed] [Google Scholar]
- Andres AC, Muellener DB, Ryffel GU. Persistence, methylation and expression of vitellogenin gene derivatives after injection into fertilized eggs of Xenopus laevis. Nucleic Acids Res. 1984;12:2283–2302. doi: 10.1093/nar/12.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D, Davidson AE, Sivasubbu S, Hermanson SB, Welle Z, Ekker SC. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics. 2004;5:62. doi: 10.1186/1471-2164-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendig MM, Williams JG. Differential expression of the Xenopus laevis tadpole and adult β-globin genes when injected into fertilized Xenopus laevis eggs. Mol Cell Biol. 1984;4:567–570. doi: 10.1128/mcb.4.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisbee CA, Baker MA, Wilson AC, Haji-Azimi I, Fischberg M. Albumin phylogeny for clawed frogs (Xenopus) Science. 1977;195:785–787. doi: 10.1126/science.65013. [DOI] [PubMed] [Google Scholar]
- Bowes JB, Snyder KA, Segerdell E, Gibb R, Jarabek C, Noumen E, Pollet N, Vize PD. Xenbase: a Xenopus biology and genomics resource. Nucleic Acids Res. 2008;36:D761–D767. doi: 10.1093/nar/gkm826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackett BG, Baranska W, Sawicki W, Koprowski H. Uptake of heterologous genome by mammalian spermatozoa and its transfer to ova through fertilization. Proc Natl Acad Sci USA. 1971;68:353–357. doi: 10.1073/pnas.68.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Chen HY, Trumbauer ME, Yagle MK, Palmiter RD. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc Natl Acad Sci USA. 1985;82:4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Sandgren EP, Behringer RR, Palmiter RD. No simple solution for making transgenic mice. Cell. 1989;59:239–241. doi: 10.1016/0092-8674(89)90282-1. [DOI] [PubMed] [Google Scholar]
- Bronchain OJ, Hartley KO, Amaya E. A gene trap approach in Xenopus. Curr Biol. 1999;9:1195–1198. doi: 10.1016/S0960-9822(00)80025-1. [DOI] [PubMed] [Google Scholar]
- Chae J, Zimmerman LB, Grainger RM. Inducible control of tissue-specific transgene expression in Xenopus tropicalis transgenic lines. Mech Dev. 2002;117:235–241. doi: 10.1016/s0925-4773(02)00219-8. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Goldstone K, Smith JC, Gilchrist M, Amaya E, Papalopulu N. A Xenopus tropicalis oligonucleotide microarray works across species using RNA from Xenopus laevis. Mech Dev. 2005;122:355–363. doi: 10.1016/j.mod.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- Coen L, du Pasquier D, Le Mevel S, Brown S, Tata J, Mazabraud A, Demeneix BA. Xenopus Bcl-X(L) selectively protects Rohon–Beard neurons from metamorphic degeneration. Proc Natl Acad Sci USA. 2001;98:7869–7874. doi: 10.1073/pnas.141226798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtet M, Flajnik M, Du Pasquier L. Major histocompatibility complex and immunoglobulin loci visualized by in situ hybridization on Xenopus chromosomes. Dev Comp Immunol. 2001;25:149–157. doi: 10.1016/s0145-305x(00)00045-8. [DOI] [PubMed] [Google Scholar]
- Danilchick M, Peng HB, Kay BK. Xenopus laevis: practical uses in cell and molecular biology. Pictorial collage of embryonic stages. Methods Cell Biol. 1991;36:679–681. [PubMed] [Google Scholar]
- Das B, Brown DD. Controlling transgene expression to study Xenopus laevis metamorphosis. Proc Natl Acad Sci USA. 2004;101:4839–4842. doi: 10.1073/pnas.0401011101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denayer T, Van Roy F, Vleminckx K. In vivo tracing of canonical Wnt signaling in Xenopus tadpoles by means of an inducible transgenic reporter tool. FEBS Lett. 2006;580:393–398. doi: 10.1016/j.febslet.2005.11.084. [DOI] [PubMed] [Google Scholar]
- Deroo T, Denayer T, Van Roy F, Vleminckx K. Global inhibition of Lef1/Tcf-dependent Wnt signaling at its nuclear end point abrogates development in transgenic Xenopus embryos. J Biol Chem. 2004;279:50670–50675. doi: 10.1074/jbc.M408969200. [DOI] [PubMed] [Google Scholar]
- Dillon N, Grosveld F. Chromatin domains as potential units of eukaryotic gene function. Curr Opin Genet Dev. 1994;4:260–264. doi: 10.1016/s0959-437x(05)80053-x. [DOI] [PubMed] [Google Scholar]
- Du Pasquier D, Chesneau A, Ymlahi-Ouazzani Q, Boistel R, Pollet N, Ballagny C, Sachs LM, Demeneix B, Mazabraud A. tBid mediated activation of the mitochondrial death pathway leads to genetic ablation of the lens in Xenopus laevis. Genesis. 2007;45:1–10. doi: 10.1002/dvg.20252. [DOI] [PubMed] [Google Scholar]
- Ellingsen S, Laplante MA, Konig M, Kikuta H, Furmanek T, Hoivik EA, Becker TS. Large-scale enhancer detection in the zebrafish genome. Development. 2005;132:3799–3811. doi: 10.1242/dev.01951. [DOI] [PubMed] [Google Scholar]
- Etkin LD. Persistence of bovine papillomavirus (pBPV-1) as an extrachromosomal element in tissues of microinjected Xenopus laevis. Nucleic Acids Res. 1991;19:191–192. doi: 10.1093/nar/19.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etkin LD, Roberts M. Transmission of integrated sea urchin histone genes by nuclear transplantation in Xenopus laevis. Science. 1983;221:67–69. doi: 10.1126/science.6857265. [DOI] [PubMed] [Google Scholar]
- Etkin LD, Pearman B. Distribution, expression and germ line transmission of exogenous DNA sequences following microinjection into Xenopus laevis eggs. Development. 1987;99:15–23. doi: 10.1242/dev.99.1.15. [DOI] [PubMed] [Google Scholar]
- Etkin L, Pearman B, Roberts M, Bektesh SL. Replication, integration and expression of exogenous DNA injected into fertilized eggs of Xenopus laevis. Differentiation. 1984;26:194–202. doi: 10.1111/j.1432-0436.1984.tb01395.x. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Kelley DB, Tinsley RC, Melnick DJ, Cannatella DC. A mitochondrial DNA phylogeny of African clawed frogs: phylogeography and implications for polyploid evolution. Mol Phylogenet Evol. 2004;33:197–213. doi: 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Kelley DB, Melnick DJ, Cannatella DC. Evolution of RAG-1 in polyploid clawed frogs. Mol Biol Evol. 2005;22:1193–1207. doi: 10.1093/molbev/msi104. [DOI] [PubMed] [Google Scholar]
- Flajnik MF, Horan PK, Cohen N. A flow cytometric analysis of the embryonic origin of lymphocytes in diploid/triploid chimeric Xenopus laevis. Dev Biol. 1984;104:247–254. doi: 10.1016/0012-1606(84)90052-6. [DOI] [PubMed] [Google Scholar]
- Furlow JD, Brown DD. In vitro and in vivo analysis of the regulation of a transcription factor gene by thyroid hormone during Xenopus laevis metamorphosis. Mol Endocrinol. 1999;13:2076–2089. doi: 10.1210/mend.13.12.0383. [DOI] [PubMed] [Google Scholar]
- Geng X, Xiao L, Lin GF, Hu R, Wang JH, Rupp RA, Ding X. Lef/Tcf-dependent Wnt/β-catenin signaling during Xenopus axis specification. FEBS Lett. 2003;547:1–6. doi: 10.1016/s0014-5793(03)00639-2. [DOI] [PubMed] [Google Scholar]
- Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- Graf JD, Kobel HR. Genetics of Xenopus laevis. Methods Cell Biol. 1991;36:19–34. doi: 10.1016/s0091-679x(08)60270-8. [DOI] [PubMed] [Google Scholar]
- Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habrova V, Takac M, Navratil J, Macha J, Ceskova N, Jonak J. Association of Rous sarcoma virus DNA with Xenopus laevis spermatozoa and its transfer to ova through fertilization. Mol Reprod Dev. 1996;44:332–342. doi: 10.1002/(SICI)1098-2795(199607)44:3<332::AID-MRD7>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Hamlet MR, Yergeau DA, Kuliyev E, Takeda M, Taira M, Kawakami K, Mead PE. Tol2 transposon-mediated transgenesis in Xenopus tropicalis. Genesis. 2006;44:438–445. doi: 10.1002/dvg.20234. [DOI] [PubMed] [Google Scholar]
- Hartley KO, Hardcastle Z, Friday RV, Amaya E, Papalopulu N. Transgenic Xenopus embryos reveal that anterior neural development requires continued suppression of BMP signaling after gastrulation. Dev Biol. 2001;238:168–184. doi: 10.1006/dbio.2001.0398. [DOI] [PubMed] [Google Scholar]
- Hartley KO, Nutt SL, Amaya E. Targeted gene expression in transgenic Xenopus using the binary Gal4-UAS system. Proc Natl Acad Sci USA. 2002;99:1377–1382. doi: 10.1073/pnas.022646899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch N, Zimmerman LB, Gray J, Chae J, Curran KL, Fisher M, Ogino H, Grainger RM. Xenopus tropicalis transgenic lines and their use in the study of embryonic induction. Dev Dyn. 2002;225:522–535. doi: 10.1002/dvdy.10188. [DOI] [PubMed] [Google Scholar]
- Hutcheson DA, Hanson MI, Moore KB, Le TT, Brown NL, Vetter ML. bHLH-dependent and -independent modes of Ath5 gene regulation during retinal development. Development. 2005;132:829–839. doi: 10.1242/dev.01653. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Izsvak Z. Transposable elements for transgenesis and insertional mutagenesis in vertebrates: a contemporary review of experimental strategies. Methods Mol Biol. 2004;260:255–276. doi: 10.1385/1-59259-755-6:255. [DOI] [PubMed] [Google Scholar]
- Kawahara H. Production of triploid and gynogenetic diploid Xenopus by cold treatment. Dev Growth Differ. 1978;20:227–236. doi: 10.1111/j.1440-169X.1978.00227.x. [DOI] [PubMed] [Google Scholar]
- Kelly LE, Davy BE, Berbari NF, Robinson ML, El-Hodiri HM. Recombineered Xenopus tropicalis BAC expresses a GFP reporter under the control of Arx transcriptional regulatory elements in transgenic Xenopus laevis embryos. Genesis. 2005;41:185–191. doi: 10.1002/gene.20113. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, Papalopulu N, Smith JC, Zorn AM, Harland RM, Grammer TC. Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn. 2002;225:499–510. doi: 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- Kloas W, Lutz I, Einspanier R. Amphibians as a model to study endocrine disruptors: II. Estrogenic activity of environmental chemicals in vitro and in vivo. Sci Total Environ. 1999;225:59–68. doi: 10.1016/s0048-9697(99)80017-5. [DOI] [PubMed] [Google Scholar]
- Kotani T, Nagayoshi S, Urasaki A, Kawakami K. Transposon-mediated gene trapping in zebrafish. Methods. 2006;39:199–206. doi: 10.1016/j.ymeth.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Kirschner MW. Easy passage: germline transgenesis in frogs. Proc Natl Acad Sci USA. 1999;96:14189–14190. doi: 10.1073/pnas.96.25.14189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov V, Macha J, Tlapakova T, Takac M, Jonak J. The c-SRC1 gene visualized by in situ hybridization on Xenopus laevis chromosomes. Cytogenet Genome Res. 2003;103:169–172. doi: 10.1159/000076307. [DOI] [PubMed] [Google Scholar]
- Krylov V, Tlapakova T, Macha J. Localization of the single copy gene Mdh2 on Xenopus tropicalis chromosomes by FISH-TSA. Cytogenet Genome Res. 2007;116:110–112. doi: 10.1159/000097427. [DOI] [PubMed] [Google Scholar]
- Laemmli UK, Kas E, Poljak L, Adachi Y. Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr Opin Genet Dev. 1992;2:275–285. doi: 10.1016/s0959-437x(05)80285-0. [DOI] [PubMed] [Google Scholar]
- Lavitrano M, Camaioni A, Fazio VM, Dolci S, Farace MG, Spadafora C. Sperm cells as vectors for introducing foreign DNA into eggs: genetic transformation of mice. Cell. 1989;57:717–723. doi: 10.1016/0092-8674(89)90787-3. [DOI] [PubMed] [Google Scholar]
- Lavitrano M, Busnelli M, Cerrito MG, Giovannoni R, Manzini S, Vargiolu A. Sperm-mediated gene transfer. Reprod Fertil Dev. 2006;18:19–23. doi: 10.1071/rd05124. [DOI] [PubMed] [Google Scholar]
- Luria A, Furlow JD. Spatiotemporal retinoid-X receptor activation detected in live vertebrate embryos. Proc Natl Acad Sci USA. 2004;101:8987–8992. doi: 10.1073/pnas.0307053101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh-Armstrong N, Huang H, Berry DL, Brown DD. Germ-line transmission of transgenes in Xenopus laevis. Proc Natl Acad Sci USA. 1999;96:14389–14393. doi: 10.1073/pnas.96.25.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Chang E, Petrescu A, Liao N, Griffith M, Chow W, Kirkpatrick R, Butterfield YS, Young AC, Stott J, et al. Sequencing and analysis of 10,967 full-length cDNA clones from Xenopus laevis and Xenopus tropicalis reveals post-tetraploidization transcriptome remodeling. Genome Res. 2006;16:796–803. doi: 10.1101/gr.4871006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz OL, Biddle KE, Tam BM. Selection of transgenic Xenopus laevis using antibiotic resistance. Transgenic Res. 2002;11:315–319. doi: 10.1023/a:1015612022976. [DOI] [PubMed] [Google Scholar]
- Muller WP, Thiebaud CH, Ricard L, Fischberg M. The induction of triploidy by pressure in Xenopus laevis. Rev Suisse Zool. 1978;85:20–26. doi: 10.5962/bhl.part.82213. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin): a systemical and chronological survey of the development from the fertilized egg till the end of metamorphosis. Garland, New York: 1994. [Google Scholar]
- Noramly S, Zimmerman L, Cox A, Aloise R, Fisher M, Grainger RM. A gynogenetic screen to isolate naturally occurring recessive mutations in Xenopus tropicalis. Mech Dev. 2005;122:273–287. doi: 10.1016/j.mod.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Offield MF, Hirsch N, Grainger RM. The development of Xenopus tropicalis transgenic lines and their use in studying lens developmental timing in living embryos. Development. 2000;127:1789–1797. doi: 10.1242/dev.127.9.1789. [DOI] [PubMed] [Google Scholar]
- Ogino H, McConnell WB, Grainger RM. High-throughput transgenesis in Xenopus using I-SceI meganuclease. Nat Protoc. 2006a;1:1703–1710. doi: 10.1038/nprot.2006.208. [DOI] [PubMed] [Google Scholar]
- Ogino H, McConnell WB, Grainger RM. Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease. Mech Dev. 2006b;123:103–113. doi: 10.1016/j.mod.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Ohno S. Gene duplication and the uniqueness of vertebrate genomes circa 1970–1999. Semin Cell Dev Biol. 1999;10:517–522. doi: 10.1006/scdb.1999.0332. [DOI] [PubMed] [Google Scholar]
- Oofusa K, Tooi O, Kashiwagi A, Kashiwagi K, Kondo Y, Obara M, Yoshizato K. Metal ion-responsive transgenic Xenopus laevis as an environmental monitoring animal. Environ Toxicol Pharmacol. 2003;13:153–159. doi: 10.1016/S1382-6689(03)00003-6. [DOI] [PubMed] [Google Scholar]
- Pan FC, Chen Y, Loeber J, Henningfeld K, Pieler T. I-SceI meganuclease-mediated transgenesis in Xenopus. Dev Dyn. 2006;235:247–252. doi: 10.1002/dvdy.20608. [DOI] [PubMed] [Google Scholar]
- Perry AC, Wakayama T, Kishikawa H, Kasai T, Okabe M, Toyoda Y, Yanagimachi R. Mammalian transgenesis by intracytoplasmic sperm injection. Science. 1999;284:1180–1183. doi: 10.1126/science.284.5417.1180. [DOI] [PubMed] [Google Scholar]
- Polli M, Amaya E. A study of mesoderm patterning through the analysis of the regulation of Xmyf-5 expression. Development. 2002;129:2917–2927. doi: 10.1242/dev.129.12.2917. [DOI] [PubMed] [Google Scholar]
- Richardson PM, Chapman J. The Xenopus tropicalis genome project. Current Genomics. 2003;4:645–652. [Google Scholar]
- Rusconi S, Schaffner W. Transformation of frog embryos with a rabbit beta-globin gene. Proc Natl Acad Sci USA. 1981;78:5051–5055. doi: 10.1073/pnas.78.8.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryffel GU, Werdien D, Turan G, Gerhards A, Goosses S, Senkel S. Tagging muscle cell lineages in development and tail regeneration using Cre recombinase in transgenic Xenopus. Nucleic Acids Res. 2003;31:e44. doi: 10.1093/nar/gng044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh M, Gurevich M, Harel-Markowitz E, Benvenisti L, Shore LS, Stram Y. Gene integration into bovine sperm genome and its expression in transgenic offspring. Mol Reprod Dev. 2000;56:306–308. doi: 10.1002/(SICI)1098-2795(200006)56:2+<306::AID-MRD21>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Sinzelle L, Vallin J, Coen L, Chesneau A, Pasquier DD, Pollet N, Demeneix B, Mazabraud A. Generation of trangenic Xenopus laevis using the Sleeping Beauty transposon system. Transgenic Res. 2006;15:751–760. doi: 10.1007/s11248-006-9014-6. [DOI] [PubMed] [Google Scholar]
- Smith S. Induction of triploidy in the South African clawed frog, Xenopus laevis (Daudin) Nature. 1958;181:290. doi: 10.1038/181290a0. [DOI] [PubMed] [Google Scholar]
- Smith KR. Sperm cell mediated transgenesis: a review. Anim Biotechnol. 1999;10:1–13. doi: 10.1080/10495399909525917. [DOI] [PubMed] [Google Scholar]
- Smith SJ, Mohun TJ. Frog transgenesis made simple. Nat Methods. 2005;2:897–898. doi: 10.1038/nmeth1205-897. [DOI] [PubMed] [Google Scholar]
- Smith K, Spadafora C. Sperm-mediated gene transfer: applications and implications. Bioessays. 2005;27:551–562. doi: 10.1002/bies.20211. [DOI] [PubMed] [Google Scholar]
- Smith SJ, Ataliotis P, Kotecha S, Towers N, Sparrow DB, Mohun TJ. The MLC1v gene provides a transgenic marker of myocardium formation within developing chambers of the Xenopus heart. Dev Dyn. 2005;232:1003–1012. doi: 10.1002/dvdy.20274. [DOI] [PubMed] [Google Scholar]
- Smith SJ, Fairclough L, Latinkic BV, Sparrow DB, Mohun TJ. Xenopus laevis transgenesis by sperm nuclear injection. Nat Protoc. 2006;1:2195–2203. doi: 10.1038/nprot.2006.325. [DOI] [PubMed] [Google Scholar]
- Smith SJ, Kotecha S, Towers N, Mohun TJ. Targeted cell-ablation in Xenopus embryos using the conditional, toxic viral protein M2(H37A) Dev Dyn. 2007;236:2159–2171. doi: 10.1002/dvdy.21233. [DOI] [PubMed] [Google Scholar]
- Sparrow DB, Latinkic B, Mohun TJ. A simplified method of generating transgenic Xenopus. Nucleic Acids Res. 2000;28:E12. doi: 10.1093/nar/28.4.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Davidson L, Keller R, Kintner C. Radial intercalation of ciliated cells during Xenopus skin development. Development. 2006;133:2507–2515. doi: 10.1242/dev.02417. [DOI] [PubMed] [Google Scholar]
- Takac M, Habrova V, Macha J, Ceskova N, Jonak J. Development of transgenic Xenopus laevis with a high C-Src gene expression. Mol Reprod Dev. 1998;50:410–419. doi: 10.1002/(SICI)1098-2795(199808)50:4<410::AID-MRD5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Tlapakova T, Krylov V, Macha J. Localization, structure and polymorphism of two paralogous Xenopus laevis mitochondrial malate dehydrogenase genes. Chromosome Res. 2005;13:699–706. doi: 10.1007/s10577-005-0987-4. [DOI] [PubMed] [Google Scholar]
- Tompkins R. Triploid and gynogenic diploid Xenopus laevis. J Exp Zool. 1978;203:251–256. [Google Scholar]
- Tompkins R, Reinschmidt D. Experimentally induced homozygosity in Xenopus laevis. Methods Cell Biol. 1991;36:35–44. doi: 10.1016/s0091-679x(08)60271-x. [DOI] [PubMed] [Google Scholar]
- Turque N, Palmier K, Le Mevel S, Alliot C, Demeneix BA. A rapid, physiologic protocol for testing transcriptional effects of thyroid-disrupting agents in premetamorphic Xenopus tadpoles. Environ Health Perspect. 2005;113:1588–1593. doi: 10.1289/ehp.7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldner C, Sakamaki K, Ueno N, Turan G, Ryffel GU. Transgenic Xenopus laevis strain expressing cre recombinase in muscle cells. Dev Dyn. 2006;235:2220–2228. doi: 10.1002/dvdy.20880. [DOI] [PubMed] [Google Scholar]
- Werdien D, Peiler G, Ryffel GU. FLP and Cre recombinase function in Xenopus embryos. Nucleic Acids Res. 2001;29:E53–E53. doi: 10.1093/nar/29.11.e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GN, Hamilton FS, Hoppler S. Inducible gene expression in transgenic Xenopus embryos. Curr Biol. 2000;10:849–852. doi: 10.1016/s0960-9822(00)00596-0. [DOI] [PubMed] [Google Scholar]
- Wilson C, Bellen HJ, Gehring WJ. Position effects on eukaryotic gene expression. Annu Rev Cell Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.