Abstract
Traumatic brain injury (TBI) triggers a cascade of apoptotic-related events that include BCL2 expression, a pro-survival protein in the apoptosis pathway. The purpose of this study was to use tagging single nucleotide polymorphism (tSNP) genotypes to screen the BCL2 gene to determine if genetic variability in the BCL2 gene influences outcomes in 205 patients with severe TBI. Outcomes (Glasgow Outcome Scale [GOS], Disability Rating Scale [DRS], mortality, and Neurobehavioral Rating Scale–Revised [NRS-R]) were analyzed at 3, 6, 12, and 24 months. Multivariate analysis demonstrates that there were four tSNPs of significant interest: rs17759659, rs1801018, rs7236090, and rs949037. Presence of the variant allele for rs17759659 was associated with poorer outcomes (GOS p = 0.001; DRS p = 0.002), higher mortality (p = 0.02; OR = 4.23; CI 1.31,13.61), and worse NRS-R scores (p = 0.05). Presence of the variant allele for rs1801018 was associated with poorer outcomes (GOS p = 0.02; DRS p = 0.009), and mortality (p = 0.03; OR = 3.86; CI 1.18,12.59). Being homozygous for the wild-type allele for rs7236090 was associated with favorable outcomes on the NRS-R (p = 0.007), while homozygosity for the variant genotype was associated with favorable outcomes on the GOS (p = 0.007) and DRS (p = 0.006). The homozygous variant for rs949037 was associated with favorable outcomes (GOS p = 0.04; DRS p = 0.03), and the homozygous wild-type was associated with increased mortality at 3 months (p = 0.005; OR = 3.67; CI 1.08,12.49). The only finding that stood up to Bonferroni correction was rs17759659 for GOS. These data support the possibility that genetic variability for pro-survival proteins, particularly genetic variation in the BCL2 gene, impacts outcomes after severe TBI.
Key words: apoptosis, BCL2, genotype, outcomes, traumatic brain injury
Introduction
There are 1.4 million people in the United States who seek medical treatment each year for a new traumatic brain injury (TBI) (Langlois et al., 2006). Of these, 51,000 will die and 235,000 are hospitalized due to their injuries (Langlois et al., 2006). As a consequence of TBI, 5.3 million Americans live with long-term or lifelong disabilities, and 125,000 new cases are added each year (Thurman et al., 1999). These disabilities may adversely affect activities of daily living, as well as cognition and behavior. At 1 year post-injury, 40% of people who were hospitalized for TBI report having at least one unmet need for continued services (Corrigan et al., 2004). These needs are largely in the neurobehavioral domain, such as memory and problem solving, managing stress and emotions, controlling temper, and employment/job skills issues (Corrigan et al., 2004). The variability in how a person recovers from a severe TBI may in part be related to the primary injury, sequelae of secondary injury, and the genetic makeup of each individual, as well as the response to therapeutic interventions and rehabilitation.
There are two common forms of neuronal death following TBI: necrosis and apoptosis (Zhang et al., 2005). The focus of this study is related to apoptosis. TBI sets into motion a cascade of secondary pro- and anti-apoptotic events, and the BCL2 (B-cell lymphoma 2) proto-oncogene family plays a key role in this pathway (Bredesen, 2000; Garcia et al., 1992; Hockenbery et al., 1990; Kane et al., 1993; Mah et al., 1993; Myers et al., 1995; Nunez et al., 1990). Following TBI, apoptosis can occur within the site of injury and in distant regions days to weeks after the trauma (Clark et al., 1997).
BCL2 is a proto-oncogene protein that inhibits apoptosis. In experimental models, overexpression of BCL2 protein in the central nervous system can prevent apoptosis in the presence of neurons that respond to nerve growth factor. BCL2 protein limits anti-apoptotic effects in neurons that respond to ciliary growth factor (Graham et al., 2000). Damage to the mitochondria from cellular events related to injury results in the release of apoptosis promoters (i.e., BAX and BAD), survival promoters (i.e., BCL-x and BCL2), or both (Graham et al., 2000). The BCL2 family regulates apoptosis/cell death or survival by regulating the permeability of the mitochondrial outer membrane and permeability transition pore (Bahr, 2000).
To date there have been some BCL2 studies evaluating their role in TBI pathology; however, none have addressed this relationship using a genetic variation approach in humans. Of the studies conducted, there is empirical evidence that BCL2 levels are increased in human brain and/or cerebrospinal fluid (CSF) after TBI compared to patients without TBI or neurological injury (Clark et al., 1999, 2000; Yang and Xue, 2004). Minambres and colleagues (2008) reported that in vivo samples from the pericontusional zone of TBI victims had significantly higher BCL2 concentrations than non-head-injured subjects at autopsy. In contrast, pediatric TBI subjects who had an increase in BCL2 protein concentrations had a lower mortality rate and better Glasgow Outcome Scale (GOS) scores (Clark et al., 2000). While the role of BCL-2 levels in these clinical studies of TBI pathology may vary by study population, the work reviewed here demonstrates that BCL2 concentrations are increased after TBI, and that BCL2 levels are related to global functional outcomes and mortality after TBI. However, none of these studies examined molecular variation of the BCL2 gene in the TBI population in general, or in relationship to the outcomes seen after injury. The purpose of this study was to investigate if variation in the BCL2 gene contributes to variability in the outcomes attained after severe TBI.
Methods
Study design and subjects
This study utilized a descriptive, longitudinal, between-group, and within-subject design to examine BCL2 genotypes in 205 subjects with severe TBI. All severe TBI patients admitted to the University of Pittsburgh Medical Center (UPMC) Presbyterian Hospital, a level 1 trauma center in a tertiary care institution, from May 2000–April 2007, who met the inclusion criteria were eligible to participate in the study and were approached with proxy consent. As part of the inclusion criteria, (1) all subjects were between the ages of 16 and 75 years, (2) all had an initial Glasgow Coma Scale score (GCS) ≤8, (3) all had an external ventricular drain (EVD) for intracranial pressure (ICP) monitoring and CSF drainage and sampling, (4) all had a positive computed tomography (CT) scan for TBI (an abnormal CT scan due to post-traumatic lesions: a. depressed skull fracture, b. epidural hematoma, c. subdural hematoma, d. traumatic subarachnoid hemorrhage, e. cerebral contusion, f. diffuse axonal injury, g. any combination of the above), and (5) all had a signed written consent from next-of-kin. Subjects were excluded from the study if they had a penetrating/open TBI or had a pre-existing neurological condition. Protected populations such as prisoners and persons with mental retardation or developmental disabilities were also not recruited. Global and neurobehavioral outcomes were assessed at 3, 6, 12, and 24 months post-injury. DNA was extracted from CSF or blood specimens. This study was reviewed and approved by the University of Pittsburgh Institutional Review Board.
Sample characteristics
There were 205 subjects with demographic, genotype, and outcomes data available for neurobehavioral (Neurobehavioral Rating Scale–Revised [NRS-R]), and global functional outcomes (GOS, Disability Rating Scale [DRS], and mortality). Sample characteristics for these analyses are displayed in Table 1. While non-Caucasian were recruited for the overall study, they were not included in the analyses of genotype data because of insufficient representation in the sample available that had both genotype and outcomes data available. Limiting the analysis to non-Hispanic Caucasians was done in an attempt to control for population stratification (14 subjects were non-Caucasian [6.4%], therefore 205 subjects were non-Hispanic Caucasians). The NRS-R (n = 99) analyses utilized a smaller subsample of survivors and those who could actively participate in the interview procedures. The percentage of GCS scores between 6 and 8 were higher in subjects able to complete the NRS-R (76.2%), compared to the functional outcome group (60%), which is in part related to the requirement for direct patient interactions in a semi-structured interview for the NRS-R.
Table 1.
Summary of Sample Characteristics and Preliminary Mixed Effects Regression Modeling Covariate Analyses
| |
|
Global functional outcomes |
Neurobehavioral outcomes |
|||||
|---|---|---|---|---|---|---|---|---|
| |
|
GOS and DRS (n = 205) |
Mortality (n = 131) |
NRS-R (n = 101) |
||||
| Mean (SD) | GOS p | DRS p | Mean (SD) | p | Mean (SD) | p | ||
| Age, mean (SD) | 34.51 (14.74) | <0.0001* | <0.0001* | 35.24 (14.88) | <0.0001# | 31.21 (12.27) | 0.001* | |
| range 16–73 years | range 16–72 years | range 16–67 years | ||||||
| n(%) | p | p | n(%) | p | n(%) | p | ||
| Admission GCS score, n(%) | GCS 3–5 | 82 (40) | <0.0001* | <0.0001* | 48 (36.6) | <0.0001# | 23 (22.8) | 0.24 |
| GCS 6–8 | 123 (60) | 83 (63.4) | 77 (76.2) | |||||
| Gender, n(%) | Male | 163 (79.5) | 0.48 | 0.47 | 104 (79.4) | 0.36 | 81 (80.2) | 0.95 |
| Female | 42 (20.5) | 27 (20.6) | 20 (19.8) | |||||
| Hypothermia, n(%) | Yes | 38 (18.5) | 0.74 | 0.50 | 19 (14.5) | 0.46 | 21 (20.8) | 0.24 |
| No | 164 (80) | 112 (85.5) | 80 (79.2) | |||||
| Unknown | 3 (1.5) | 0 | 0 | |||||
| Hypotension, n(%) | Yes | 11 (5.4) | 0.43 | 0.31 | 10 (7.6) | 0.03# | 6 (5.9) | 0.55 |
| No | 140 (68.3) | 121 (92.4) | 60 (59.0) | |||||
| Unknown | 54 (26.3) | 0 | 35 (34.7) | |||||
| Hypoxia, n(%) | Yes | 25 (12.2) | 0.61 | 0.47 | 24 (18.3) | 0.54 | 13 (12.9) | 0.66 |
| No | 126 (61.5) | 107 (81.7) | 53 (52.5) | |||||
| Unknown | 54 (26.3) | 0 | 35 (34.7) | |||||
p ≤ 0.2 criterion for inclusion in the Primary Mixed Effects Regression Modeling.
p ≤ 0.3 criterion for inclusion in the Primary Binary Logistic Regression Cross-Sectional Analysis at 3 months; by chi square, based on n = 205.
Mortality and NRS-R sample statistics are based on the subsample size.
GOS, Glasgow Outcome Scale; DRS, Disability Rating Scale; NRS-R, Neurobehavioral Rating Scale–Revised; GCS, Glasgow Coma Scale; SD, standard deviation
Standard of care
All potential subjects were admitted to the level 1 trauma center with a diagnosis of severe TBI within 48 h of injury. Patients received aggressive medical treatment in accordance with guidelines for the management of severe head injury (Bullock et al., 1996; Povlishock, 2000, 2004). EVDs were placed in all subjects as part of routine care. Blood levels were drawn on admission for alcohol, drug, and other toxin levels. Participants were monitored continuously for physiological parameters, including ICP, cerebral perfusion pressure (CPP), arterial blood pressure, central venous pressure, pulse oximetry, respiratory rate, heart rate and rhythm, and core temperature. These physiological variables were documented in real time. Some of the patients (n = 38) received hypothermia treatment (target temperature 32.5–34.0°C) as part of a randomized, prospective clinical trial. The distribution of alleles in the hypothermic group versus the non-hypothermic group were assessed for expected distribution within a population using the Hardy-Weinberg equilibrium evaluation, and all distributions met the expected distribution patterns (data not shown). Hypothermia would not change a participant's BCL-2 genotypes; however, it may alter response to therapy in a gene-specific manner.
BCL2 genotyping
The use of tagging single nucleotide polymorphisms (tSNPs) for BCL2 allowed us to efficiently characterize all of the genetic variability within and around the gene and a subset of polymorphisms within the gene. This approach capitalizes on our knowledge that polymorphisms within a block of linkage disequilibrium in a region of DNA are often highly correlated, and therefore often redundant in the information they provide. Selecting tSNPs allows us to use one SNP per region of linkage disequilibrium without losing significant data about the variability of that region.
Based on HapMap data (April, 2007) there were 20 tSNPs for the BCL2 gene, with a minor allele frequency (MAF) of ≥30% and r2 ≥0.80 (rs1026825, rs12454712, rs12968517, rs1381548, rs1481031, rs17756073, rs17759659, rs1801018, rs1944419, rs3810027, rs4456611, rs4941185, rs7230970, rs7236090, rs8083946, rs899968, andrs949037). TaqMan-based assays could not be successfully manufactured for rs7231914, rs8089538, and rs2850762 at the time of genotyping. Therefore in this study we genotyped the remaining 17 tSNPs.
An ABI Prism® 7000 Sequence Detection System was used to conduct allele discrimination using commercially available assays and SDS 2.0 software (Applied Biosystems Inc., Carlsbad, CA). Quality controls consisted of (1) independently duplicating 10% of the genotypes; (2) independent double calls were made and compared for each sample; and (3) deviation from Hardy-Weinberg equilibrium was assessed for each tSNP. Lab technicians were blinded to outcome data.
Analysis included dichotomized genotype data. BCL2 genotype data were dichotomized for each tSNP by combining homozygous variants with heterozygotes and comparing them to homozygous wild-types, thereby analyzing the importance of harboring at least one copy of the variant allele. Dichotomization was based on the frequency of alleles in our study population.
Measures of outcomes
A battery of neurobehavioral and global outcome tests was conducted for each participant at 3, 6, 12, and 24 months post-TBI. The instruments were administered by trained technicians, under the direction of a neuropsychologist. The technician was blinded to patient genotypes. Face-to-face visits took place either at the outpatient neurosurgical clinic of the medical center, a rehabilitation setting, or the participant's home. Phone interviews were utilized to collect the GOS and DRS outcomes data for subjects who were unavailable or refused to complete a face-to-face visit.
Glasgow Outcome Scale
The GOS, a clinical observation scale, categorizes global functional outcomes into five levels. It is an ordinal level of measurement: 1 = death, 2 = persistent vegetative state, 3 = severe disability, 4 = moderate disability, and 5 = good recovery (Jennett, 1976a, 1976b; Jennett and Bond, 1975). A consensus conference in 1998 recommended evaluating basic functional assessment at 3, 6, and 12 months following a TBI (Wilson et al., 1998).
Disability Rating Scale
The DRS was designed to assess disability in patients recovering from severe TBI as they progress from severe coma to community reintegration (Rappaport et al., 1982). The total score ranges from 0 (no disability) to 30 (death). There are eight items in the measurement tool, which assess four areas: awareness/arousal, cognitive ability for self-care, level of physical dependence on others, and estimated ability for employment, school, or homemaking. This scale has been shown to be a useful predictor of outcome following TBI (McCauley et al., 2001).
The Neurobehavioral Rating Scale–Revised
The NRS-R was developed by Levin and colleagues (1990) to increase reliability and content validity of the original 1987 version, the Neurobehavioral Rating Scale (NRS), and to provide supplementary information to more global measures of functional outcome such as the GOS or DRS (McCauley et al., 2001). Both the NRS and the NRS-R are semi-structured interviews that require a trained examiner. When using the NRS-R, the examiner bases scores on a 15- to 20-min interview (two-thirds of the items), and patient observation during the interview (one-third of the items). The 29 items are scored on a 4-point Likert-type scale (absent, mild, moderate, and severe). NRS-R items address issues such as, but not limited to, mental flexibility, irritability, tension/anxiety, alertness, attention, decreased initiation/motivation, suspiciousness, mental fatigability, hallucinating behavior, and motor retardation (Lezek et al., 2004). The scores for each item are summed to yield a total score.
Statistical analysis
Bivariate mixed-models analysis was used to identify tSNPs of interest, and potential covariates (age, gender, admission GCS score, documented hypoxia and/or hypotension before admission, and the presence of hypothermia/cooling) for the GOS, DRS, and NRS-R, using a preliminary significant criterion cut-off of overall p value (p for type 3) (p ≤ 0.2). p Values for type 3 tests were utilized to determine overall significance when assessing all three genotypes (i.e., AA, GG, and AG) at the same time. Genotypes were also analyzed based on dichotomization of the presence or absence of the variant allele, and utilized a p value. From these findings, primary multivariate analyses were conducted using mixed-effects regression modeling with a significant p value set at ≤0.05. The majority of the deceased subjects had expired by the 3-month time point (n = 51 versus 24 months n = 65). Therefore we chose to cross-sectionally examine mortality at the 3-month outcome time point only. This was done to ascertain which tSNPs had the strongest association with early mortality. Mortality status at 3 months was run as a cross-sectional analysis using logistic regression to ascertain tSNPs and covariates of interest. A preliminary bivariate logistic regression analysis was conducted with variables exhibiting overall significance (p for type 3 of p ≤ 0.3), by genotype. The primary multivariate logistic analysis was conducted with a criterion of p ≤0.05. A priori, regardless of the preliminary significance, all of the models controlled for age, gender, and admission GCS score. No other covariates were included in the GOS, DRS, and NRS-R models, because they did not meet the significance criterion. Due to preliminary bivariate logistic regression analysis, hypotension (p = 0.03) was added as an additional covariate to the mortality model analysis. The conservative Bonferroni correction was used to generate an adjusted alpha for statistical importance (α = 0.001).
Haplotype analysis
Genotype data were analyzed using HaploView 4.2 (Barrett et al., 2008) for haplotype construction. We used the linkage format association tests. The data were coded to support a cohort of unrelated individuals. The affection status, coding for affected and non-affected individuals, was dichotomized into phenotype groups for GOS at 3, 6, and 12 months outcome (poor [GOS 1–3] versus good [GOS 4–5] outcomes), and mortality at 3 months for the chi-square association analysis.
Results
Identified covariates and data quality assessment
A summary of the sample characteristics and preliminary analyses of the covariates for functional and neurocognitive outcomes are reported in Table 1.
All of the tSNPs genotyped for the overall sample and each subsample were in Hardy-Weinberg equilibrium (data not shown), and were representative of the general population based on dbSNP data (www.ncbi.nlm.nih.gov/SNP). Table 2 provides a detailed genotype distribution for each of the 17 BCL2 tSNPs for the 205 subjects genotyped in the overall sample for this study.
Table 2.
BCL2 Genotype Information
| tSNP rs number | Alleles base Δ | Homozygous wild-type | Homozygous variant | Heterozygous allele | n | Study MAF | HapMap MAF | Location tSNP | Position | |
|---|---|---|---|---|---|---|---|---|---|---|
| 3′ end | rs17756073 | A/G | 97 (AA) | 26 (GG) | 74 (A/G) | 197 | 0.320 | 0.3 | intron | 58960763 |
| I | rs4456611 | C/T | 52 (CC) | 46 (TT) | 98 (C/T) | 196 | 0.485 | 0.467 | intron | 58961432 |
| I | rs1026825 | A/G | 59 (AA) | 48 (GG) | 98 (A/G) | 205 | 0.473 | 0.458 | intron | 58971255 |
| I | rs7230970 | C/T | 71 (TT) | 39 (CC) | 83 (C/T) | 193 | 0.417 | 0.473 | intron | 58972056 |
| I | rs899968 | A/C | 74 (CC) | 39 (AA) | 86 (A/C) | 199 | 0.412 | 0.425 | intron | 58973244 |
| I | rs4941185 | A/G | 66 (AA) | 42 (GG) | 91 (A/G) | 199 | 0.440 | 0.4 | intron | 58974534 |
| I | rs12454712 | C/T | 92 (TT) | 27 (CC) | 85 (C/T) | 204 | 0.341 | 0.362 | intron | 58996864 |
| I | rs1481031 | A/G | 91 (AA) | 20 (GG) | 91 (A/G) | 202 | 0.324 | 0.325 | intron | 59003065 |
| I | rs3810027 | C/G | 83 (CC) | 25 (GG) | 92 (C/G) | 200 | 0.355 | 0.331 | intron | 59054958 |
| I | rs8083946 | A/G | 80 (GG) | 37 (AA) | 87 (A/G) | 204 | 0.395 | 0.333 | intron | 59056901 |
| I | rs1944419 | A/T | 50 (AA) | 47 (TT) | 99 (A/T) | 196 | 0.492 | 0.458 | intron | 59075593 |
| I | rs12968517 | C/T | 84 (CC) | 33 (TT) | 87 (C/T) | 205 | 0.373 | 0.292 | intron | 59092951 |
| I | rs7236090 | C/T | 61 (CC) | 40 (TT) | 96 (C/T) | 197 | 0.421 | 0.45 | intron | 59098091 |
| I | rs1381548 | A/G | 70 (GG) | 37 (AA) | 90 (A/G) | 197 | 0.416 | 0.392 | intron | 59108376 |
| I | rs17759659 | A/G | 72 (AA) | 29 (GG) | 100 (A/G) | 201 | 0.393 | 0.483 | intron | 59109624 |
| I | rs949037 | C/T | 64 (TT) | 46 (CC) | 88 (C/T) | 198 | 0.455 | 0.4 | intron | 59129993 |
| 5′ end | rs1801018 | A/G | 68 (GG) | 43 (AA) | 92 (A/G) | 203 | 0.438 | 0.475 | exon 2 synonymous | 59136859 |
tSNP, tagging single nucleotide polymorphism; MAF, minor allele frequency.
Haplotype analysis
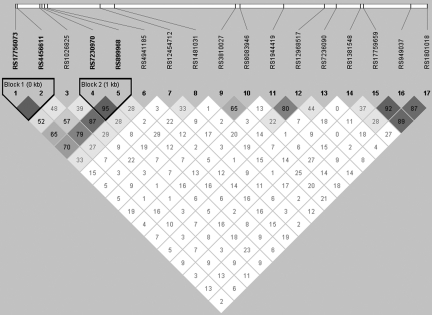
HaploView 4.2 (Barrett et al., 2008) constructed two haplotype blocks covering four tSNPs (Fig. 1). The chi-square analyses were not significant at any time point (data not shown).
FIG. 1.
This LD plot for the BCL2 gene was generated using HaploView 4.2 (Barrett et al., 2008) for haplotype construction. Two haplotype blocks were constructed covering four tSNPs. The chi-square was not significant at any time point for either of the haplotype blocks (tSNP, tagging single nucleotide polymorphism; LD, linkage disequilibrium).
Preliminary analyses for outcomes variables
The preliminary analyses for BCL2 genotypes and GOS, DRS, mortality, and NRS-R outcomes are presented in Supplementary Tables 1 and 2 (see online supplementary material at http://www.liebertonline.com).
GOS and DRS
Table 3 gives detailed formation on the statistically significant and trending tSNPs for GOS and DRS outcomes. The statistically significant tSNPs (rs12968517, rs17759659, rs1801018, rs7236090, and rs940370 are clustered in the 5′ end of the gene, and are also implicated in the mortality analyses (Fig. 2). tSNP rs12968517 (TT) was associated with lower GOS scores (p = 0.04), and higher DRS scores (p = 0.04), both indicating poorer outcomes. tSNP rs17759659 (AA) was associated with favorable outcomes: higher GOS scores (p = 0.003) and lower DRS scores (p = 0.003). In the dichotomized analysis (combination of the homozygous variants with heterozygotes, and comparing them to homozygous wild-types), the presence of the variant allele (G) was associated with significantly poorer outcomes (GOS p = 0.001; DRS p = 0.002). The GOS analysis met significance under the Bonferroni correction (p ≤ 0.001). tSNP rs1801018 (GG) was associated with higher GOS scores (p = 0.007) and lower DRS scores (p = 0.002), both indicating better outcomes. In the dichotomized analysis, the presence of the variant allele (A) was associated with significantly poorer outcomes (lower GOS scores p = 0.02; higher DRS scores p = 0.009). tSNP rs7236090 (TT) was associated with favorable outcomes: higher GOS scores (p = 0.007; overall p value for type 3 tests p = 0.03), and lower DRS scores (p = 0.006; p for type 3 p = 0.02). tSNP rs949037 (CC) was associated with higher GOS scores (p = 0.04; p for type 3 p = 0.02), and lower DRS scores (p = 0.03; p for type 3 p = 0.04), both indicating better outcomes.
Table 3.
Primary Mixed Effects Regression Modeling of Global Functional Outcomes: GOS, DRS, and BCL2 tSNPs
| |
|
|
GOS |
|
DRS |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
|
Genotype |
|
Dichotomized |
|
Genotype |
|
Dichotomized |
||||
| tSNP | Variable | coefff | p | coefff | p | coefff | p | coefff | p | ||||
| rs12454712 | Genotype | CC | −0.08 | 0.77 | CC & CT | −0.12 | 0.47 | CC | −0.20 | 0.94 | CC & CT | 0.33 | 0.82 |
| (n = 204) | TT | 0.10 | 0.58 | TT | reference | TT | −0.38 | 0.81 | TT | reference | |||
| CT | reference | CT | reference | ||||||||||
| overall | 0.74 | overall | 0.97 | ||||||||||
| Month | 3 | −0.34 | < 0.0001*** | 3 | −0.34 | < 0.0001*** | 3 | 1.99 | < 0.0001*** | 3 | 1.99 | < 0.0001*** | |
| 6 | −0.16 | 0.002* | 6 | −0.16 | 0.002* | 6 | 0.58 | 0.03 | 6 | 0.58 | 0.03* | ||
| 12 | −0.09 | 0.09† | 12 | −0.09 | 0.09† | 12 | 0.03 | 0.09† | 12 | 0.03 | 0.90 | ||
| 24 | reference | 24 | reference | 24 | reference | 24 | reference | ||||||
| overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | ||||||
| Age | −0.04 | <0.0001*** | −0.04 | <0.0001*** | 0.34 | <0.0001*** | 0.34 | <0.0001*** | |||||
| GCS | 1.09 | <0.0001*** | 1.10 | <0.0001*** | −10.66 | <0.0001*** | −10.66 | <0.0001*** | |||||
| Gender | −0.09 | 0.64 | −0.09 | 0.63 | 0.94 | 0.61 | 0.94 | 0.61 | |||||
| rs12968517 | Genotype | CC | −0.11 | 0.54 | TT & CT | −0.02 | 0.89 | CC | 1.11 | 0.53 | TT & CT | 0.07 | 0.97 |
| (n = 204) | TT | −0.48 | 0.04* | CC | reference | TT | 4.14 | 0.04* | CC | reference | |||
| CT | reference | CT | reference | ||||||||||
| overall | 0.13 | overall | 0.16 | ||||||||||
| Month | 3 | −0.34 | <0.0001*** | 3 | −0.34 | <0.0001*** | 3 | 2.01 | <0.0001*** | 3 | 2.01 | <0.0001*** | |
| 6 | −0.16 | 0.001*** | 6 | −0.16 | 0.001*** | 6 | 0.61 | 0.03* | 6 | 0.61 | 0.02* | ||
| 12 | −0.09 | 0.08 | 12 | −0.09 | 0.08† | 12 | 0.05 | 0.85 | 12 | 0.05 | 0.85 | ||
| 24 | reference | 24 | reference | 24 | reference | 24 | reference | ||||||
| overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | ||||||
| Age | −0.04 | <0.0001*** | −0.04 | <0.0001*** | 0.33 | <0.0001*** | 0.33 | <0.0001*** | |||||
| GCS | 1.05 | <0.0001*** | 1.06 | <0.0001*** | −10.29 | <0.0001*** | −10.32 | <0.0001*** | |||||
| Gender | −0.08 | 0.68 | −0.10 | 0.64 | 0.73 | 0.70 | 0.89 | 0.64 | |||||
| rs1481031 | Genotype | AA | −0.12 | 0.49 | GG &AG | 0.17 | 0.29 | AA | 1.11 | 0.49 | GG &AG | −1.47 | 0.33 |
| (n = 202) | GG | 0.30 | 0.29 | AA | reference | GG | −2.00 | 0.46 | AA | reference | |||
| AG | reference | AG | reference | ||||||||||
| overall | 0.33 | overall | 0.48 | ||||||||||
| Month | 3 | −0.34 | <0.0001*** | 3 | −0.34 | <0.0001*** | 3 | 2.00 | <0.0001*** | 3 | 2.00 | <0.0001*** | |
| 6 | −0.16 | 0.002* | 6 | −0.16 | 0.002* | 6 | 0.57 | 0.03* | 6 | 0.57 | 0.03* | ||
| 12 | −0.09 | 0.08† | 12 | −0.09 | 0.08† | 12 | 0.05 | 0.86 | 12 | 0.05 | 0.86 | ||
| 24 | reference | 24 | reference | 24 | reference | 24 | reference | ||||||
| overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | ||||||
| Age | −0.04 | <0.0001*** | −0.04 | <0.0001*** | 0.33 | <0.0001*** | 0.33 | <0.0001*** | |||||
| GCS | 1.03 | <0.0001*** | 1.03 | <0.0001*** | −10.12 | <0.0001*** | −10.13 | <0.0001*** | |||||
| Gender | −0.08 | 0.69 | −0.08 | 0.69 | 0.68 | 0.72 | 0.70 | 0.71 | |||||
| rs17756073 | Genotype | AA | 0.05 | 0.77 | GG & AG | −0.15 | 0.37 | AA | 0.35 | 0.83 | GG & AG | 0.77 | 0.62 |
| (n = 197) | GG | −0.36 | 0.17 | AA | reference | GG | 4.08 | 0.09† | AA | reference | |||
| AG | reference | AG | reference | ||||||||||
| overall | 0.27 | overall | 0.21 | ||||||||||
| Month | 3 | −0.34 | <0.0001*** | 3 | −0.34 | <0.0001*** | 3 | 1.99 | <0.0001*** | 3 | 1.99 | <0.0001*** | |
| 6 | −0.16 | 0.002* | 6 | −0.16 | 0.002* | 6 | 0.57 | 0.03* | 6 | 0.57 | 0.03* | ||
| 12 | −0.08 | 0.12 | 12 | −0.08 | 0.12 | 12 | 0.01 | 0.97 | 12 | 0.01 | 0.97 | ||
| 24 | reference | 24 | reference | 24 | reference | 24 | reference | ||||||
| overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | ||||||
| Age | −0.04 | <0.0001*** | −0.04 | <0.0001*** | 0.34 | <0.0001*** | 0.34 | <0.0001*** | |||||
| GCS | 1.06 | <0.0001*** | 1.09 | <0.0001*** | −10.19 | <0.0001*** | −10.54 | <0.0001*** | |||||
| Gender | −0.09 | 0.67 | −0.08 | 0.71 | 0.86 | 0.65 | 0.72 | 0.70 | |||||
| rs17759659 | Genotype | AA | 0.52 | 0.003* | GG & AG | −0.53 | 0.001*** | AA | −4.78 | 0.003* | GG & AG | 4.71 | 0.002* |
| (n = 201) | GG | −0.08 | 0.74 | AA | reference | GG | −0.29 | 0.89 | AA | reference | |||
| AG | reference | AG | reference | ||||||||||
| overall | 0.005* | overall | 0.01* | ||||||||||
| Month | 3 | −0.35 | <0.0001*** | 3 | −0.35 | <0.0001*** | 3 | 2.04 | <0.0001*** | 3 | 2.04 | <0.0001*** | |
| 6 | −0.16 | 0.002* | 6 | −0.16 | 0.002* | 6 | 0.58 | 0.03* | 6 | 0.58 | 0.03* | ||
| 12 | −0.09 | 0.09† | 12 | −0.09 | 0.09† | 12 | 0.03 | 0.93 | 12 | 0.03 | 0.93 | ||
| 24 | reference | 24 | reference | 24 | reference | 24 | reference | ||||||
| overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | ||||||
| Age | −0.04 | <0.0001*** | −0.04 | <0.0001*** | 0.37 | <0.0001*** | 0.37 | <0.0001*** | |||||
| GCS | 1.08 | <0.0001*** | 1.08 | <0.0001*** | −10.44 | <0.0001*** | −10.44 | <0.0001*** | |||||
| Gender | −0.01 | 0.96 | 0.01 | 0.97 | −0.22 | 0.91 | −0.23 | 0.90 | |||||
| rs1801018 | Genotype | AA | 0.30 | 0.16 | AA & AG | −0.41 | 0.02* | AA | −3.65 | 0.06† | AA & AG | 4.21 | 0.009* |
| (n = 203) | GG | 0.50 | 0.007* | GG | reference | GG | −5.32 | 0.002* | GG | reference | |||
| AG | reference | AG | reference | ||||||||||
| overall | 0.02* | overall | 0.006** | ||||||||||
| Month | 3 | −0.33 | <0.0001*** | 3 | −0.34 | <0.0001*** | 3 | 1.97 | <0.0001*** | 3 | 1.97 | <0.0001*** | |
| 6 | −0.15 | 0.003* | 6 | −0.15 | 0.003* | 6 | 0.57 | 0.03* | 6 | 0.57 | 0.03* | ||
| 12 | −0.09 | 0.09† | 12 | −0.09 | 0.09† | 12 | 0.03 | 0.91 | 12 | 0.03 | 0.91 | ||
| 24 | reference | 24 | reference | 24 | reference | 24 | reference | ||||||
| overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | ||||||
| Age | −0.04 | <0.0001*** | −0.04 | <0.0001*** | 0.33 | <0.0001*** | 0.34 | <0.0001*** | |||||
| GCS | 1.08 | <0.0001*** | 1.07 | <0.0001*** | −10.47 | <0.0001*** | −10.45 | <0.0001*** | |||||
| Gender | −0.08 | 0.69 | −0.05 | 0.81 | 0.64 | 0.73 | 0.25 | 0.89 | |||||
| rs1944419 | Genotype | AA | 0.32 | 0.26 | AA & AG | −0.22 | 0.25 | AA | 2.28 | 0.22 | AA & AG | −2.01 | 0.25 |
| (n = 196) | GG | 0.52 | 0.94 | GG | reference | GG | 0.83 | 0.66 | GG | reference | |||
| AG | reference | AG | reference | ||||||||||
| overall | 0.51 | overall | 0.47 | ||||||||||
| Month | 3 | −0.33 | <0.0001*** | 3 | −0.33 | <0.0001*** | 3 | 1.96 | <0.0001*** | 3 | 1.96 | <0.0001*** | |
| 6 | −0.14 | 0.006* | 6 | −0.14 | 0.006* | 6 | 0.56 | 0.04* | 6 | 0.56 | 0.04* | ||
| 12 | −0.08 | 0.11 | 12 | −0.08 | 0.11 | 12 | 0.00 | 1.00 | 12 | 0.00 | 1.00 | ||
| 24 | reference | 24 | reference | 24 | reference | 24 | reference | ||||||
| overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | ||||||
| Age | −0.04 | <0.0001*** | −0.04 | <0.0001*** | 0.33 | <0.0001*** | 0.33 | <0.0001*** | |||||
| GCS | 1.03 | <0.0001*** | 1.03 | <0.0001*** | −10.23 | <0.0001*** | −10.22 | <0.0001*** | |||||
| Gender | −0.04 | 0.83 | 0.05 | 0.83 | −0.61 | 0.76 | −0.68 | 0.73 | |||||
| rs4456611 | Genotype | CC | −0.003 | 0.99 | TT & CT | −0.10 | 0.58 | CC | 0.94 | 0.61 | TT & CT | 0.19 | 0.91 |
| (n = 196) | TT | −0.34 | 0.09† | CC | reference | TT | 3.44 | 0.06† | CC | reference | |||
| CT | reference | CT | reference | ||||||||||
| overall | 0.21 | overall | 0.17 | ||||||||||
| Month | 3 | −0.33 | <0.0001*** | 3 | −0.33 | <0.0001*** | 3 | 1.81 | <0.0001*** | 3 | 1.81 | <0.0001*** | |
| 6 | −0.14 | 0.005* | 6 | −0.14 | 0.006* | 6 | 0.46 | 0.09† | 6 | 0.46 | |||
| 12 | −0.09 | 0.08† | 12 | −0.09 | 0.08† | 12 | −0.01 | 0.98 | 12 | −0.01 | |||
| 24 | reference | 24 | reference | 24 | reference | 24 | reference | ||||||
| overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | ||||||
| Age | −0.04 | <0.0001*** | −0.04 | <0.0001*** | 0.36 | <0.0001*** | 0.36 | <0.0001*** | |||||
| GCS | 1.13 | <0.0001*** | 1.14 | <0.0001*** | −10.95 | <0.0001*** | −10.93 | <0.0001*** | |||||
| Gender | −0.06 | 0.74 | −0.06 | 0.77 | 0.64 | 0.73 | 0.58 | 0.76 | |||||
| rs4941185 | Genotype | AA | −0.30 | 0.11 | GG & AG | 0.15 | 0.39 | AA | 2.45 | 0.16 | GG & AG | −0.91 | 0.58 |
| (n = 199) | GG | −0.48 | 0.03* | AA | reference | GG | 4.80 | 0.02* | AA | reference | |||
| AG | reference | AG | reference | ||||||||||
| overall | 0.06† | overall | 0.05* | ||||||||||
| Month | 3 | −0.34 | <0.0001*** | 3 | −0.34 | <0.0001*** | 3 | 1.99 | <0.0001*** | 3 | 1.99 | <0.0001*** | |
| 6 | −0.16 | 0.002* | 6 | −0.16 | 0.002* | 6 | 0.59 | 0.03* | 6 | 0.59 | 0.03* | ||
| 12 | −0.10 | 0.05† | 12 | −0.10 | 0.05† | 12 | 0.08 | 0.77 | 12 | 0.08 | 0.78 | ||
| 24 | reference | 24 | reference | 24 | reference | 24 | reference | ||||||
| overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | ||||||
| Age | −0.04 | <0.0001*** | −0.04 | <0.0001*** | 0.34 | <0.0001*** | 0.34 | <0.0001*** | |||||
| GCS | 0.97 | <0.0001*** | 1.01 | <0.0001*** | −9.50 | <0.0001*** | −9.95 | <0.0001*** | |||||
| Gender | −0.07 | 0.73 | −0.08 | 0.69 | 0.79 | 0.67 | 0.82 | 0.66 | |||||
| rs7236090 | Genotype | CC | 0.12 | 0.52 | TT & CT | 0.05 | 0.77 | CC | −0.10 | 0.95 | TT & CT | −1.47 | 0.37 |
| (n = 197) | TT | 0.59 | 0.007* | CC | reference | TT | −5.56 | 0.006* | CC | reference | |||
| CT | reference | CT | reference | ||||||||||
| overall | 0.03* | overall | 0.02* | ||||||||||
| Month | 3 | −0.34 | <0.0001*** | 3 | −0.34 | <0.0001*** | 3 | 2.03 | <0.0001*** | 3 | 2.03 | <0.0001*** | |
| 6 | −0.16 | 0.002* | 6 | −0.16 | 0.002* | 6 | 0.57 | 0.04* | 6 | 0.56 | 0.04* | ||
| 12 | −0.08 | 0.11 | 12 | −0.08 | 0.11 | 12 | 0.04 | 0.89 | 12 | 0.04 | 0.89 | ||
| 24 | reference | 24 | reference | 24 | reference | 24 | reference | ||||||
| overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | ||||||
| Age | −0.04 | <0.0001*** | −0.04 | <0.0001*** | 0.35 | <0.0001*** | 0.35 | <0.0001*** | |||||
| GCS | 1.03 | <0.0001*** | 1.05 | <0.0001*** | −9.75 | <0.0001*** | −10.02 | <0.0001*** | |||||
| Gender | −0.12 | 0.56 | −0.10 | 0.61 | 0.99 | 0.59 | 1.08 | 0.57 | |||||
| rs949037 | Genotype | CC | 0.43 | 0.04* | CC & CT | 0.33 | 0.07† | CC | −4.23 | 0.03* | CC & CT | −2.23 | 0.18 |
| (n = 198) | TT | −0.18 | 0.35 | TT | reference | TT | 0.74 | 0.68 | TT | reference | |||
| CT | reference | CT | reference | ||||||||||
| overall | 0.02* | overall | 0.04* | ||||||||||
| Month | 3 | −0.34 | <0.0001*** | 3 | −0.34 | <0.0001*** | 3 | 2.04 | <0.0001*** | 3 | 2.03 | <0.0001*** | |
| 6 | −0.16 | 0.002* | 6 | −0.16 | 0.002* | 6 | 0.58 | 0.03* | 6 | 0.58 | 0.03* | ||
| 12 | −0.08 | 0.10 | 12 | −0.08 | 0.10 | 12 | 0.04 | 0.89 | 12 | 0.04 | 0.89 | ||
| 24 | reference | 24 | reference | 24 | reference | 24 | reference | ||||||
| overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | overall | <0.0001*** | ||||||
| Age | −0.04 | <0.0001*** | −0.04 | <0.0001*** | 0.34 | <0.0001*** | 0.35 | <0.0001*** | |||||
| GCS | 1.10 | <0.0001*** | 1.08 | <0.0001*** | −10.79 | <0.0001*** | −10.62 | <0.0001*** | |||||
| Gender | −0.03 | 0.86 | −0.04 | 0.84 | 0.26 | 0.89 | 0.44 | 0.82 | |||||
p ≤ 0.05; ***p ≤ 0.001; †trend, ***p ≤ 0.001 meets Bonferroni correction.
Month = time point of the outcome assessment post-injury (3, 6, 12, and 24 months).
Overall = p for type 3 test.
tSNP, tagging single nucleotide polymorphism; GOS, Glasgow Outcome Scale; DRS, Disability Rating Scale; GCS, Glasgow Coma Scale.
FIG. 2.
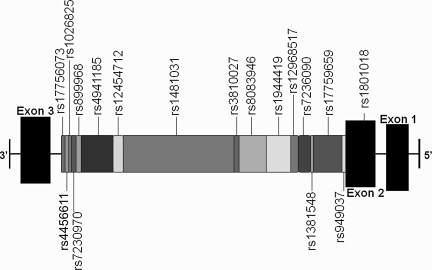
BCL2 gene with tSNPs on chromosome 18q21.3. Based on HapMap data (April 2007) and the National Center for Biotechnology Information SNP database information (April 2007). This illustration was created to display the location of the 17 tSNPs examined in this study, as well as the position of the exons on the gene (tSNP, tagging single nucleotide polymorphism).
For all of the tSNPs studied, the following covariates were associated with poorer outcomes with the Bonferroni correction: older age (p < 0.0001) and more severe GCS score on admission (p < 0.0001).
Mortality
Table 4 gives detailed formation on the statistically significant and trending tSNPs for the mortality outcome. tSNP rs12968517 (TT) was associated with higher mortality (p = 0.02; OR = 4.84; CI 1.08,21.65). tSNP rs17759659 (AA) was associated with lower mortality (p = 0.02; OR = 0.23; CI 0.07,0.78). In the dichotomized analysis, the presence of the rs17759659 variant allele (G) was associated with significantly higher mortality by 3 months (p = 0.02; OR = 4.23; CI 1.3,13.61). tSNP rs1801018 (GG) was associated with lower mortality (p = 0.03; OR = 0.26; CI 0.08,0.87; overall p value for type 3 tests p = 0.08), indicating better outcomes. In the dichotomized analysis, the presence of the variant allele (A) was associated with significantly poorer outcomes (p = 0.03; OR = 3.86; CI 1.18,12.59). tSNP rs7236090 (TT) showed a trend toward lower mortality (p = 0.09; OR = 0.32; CI 0.08,1.30), indicating better outcome. tSNP rs949037 (TT) was associated with higher mortality (p = 0.005; OR = 3.67; CI 1.08,12.49), indicating a worse outcome. For all of the tSNPs analyzed, older age (p-value range <0.0001–0.0004), female gender (p-value range 0.04–0.79), more severe GCS score on admission (p < 0.0001), and hypotension (p-value range 0.004–0.03), were all statistically associated with or trending toward poorer outcomes.
Table 4.
Primary Binary Logistic Regression Model Analyses of 3-Month Mortality Outcome and BCL2 tSNPs
| |
|
|
Genotypes |
|
Dichotomized genotypes |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tSNP | Variable | Genotype | df | coefff | Odds ratio | 95% Wald confidence limits | p | Genotype | df | coefff | Odds ratio | 95% Wald confidence limits | p | ||
| rs12968517 | Genotype | CC | 2 | −0.76 | 0.71 | 0.22 | 2.22 | 0.05* | TT & CT | 1 | 0.38 | 2.13 | 0.73 | 6.21 | 0.17 |
| (n = 130) | TT | 1.17 | 4.84 | 1.08 | 21.65 | 0.02* | CC | reference | |||||||
| CT | reference | ||||||||||||||
| overall | 0.05* | ||||||||||||||
| Age | 1 | 0.07 | 1.07 | 1.03 | 1.12 | 0.0004*** | 1 | 0.07 | 1.07 | 1.03 | 1.11 | 0.0003*** | |||
| GCS | 1 | −1.11 | 0.11 | 0.04 | 0.31 | <0.0001*** | 1 | −1.12 | 0.11 | 0.04 | 0.30 | <0.0001*** | |||
| Gender | 1 | 0.59 | 3.28 | 1.06 | 10.18 | 0.04* | 1 | 0.59 | 3.28 | 1.05 | 10.26 | 0.41 | |||
| Hypotension | 1 | −1.46 | 0.05 | 0.01 | 0.40 | 0.005* | 1 | 1.47 | 0.05 | 0.01 | 0.38 | 0.004* | |||
| rs17756073 | Genotype | AA | 2 | −0.20 | 1.22 | 0.39 | 3.79 | 0.58 | GG & AG | 1 | 0.05 | 1.11 | 0.41 | 3.01 | 0.84 |
| (n = 125) | GG | 0.60 | 2.70 | 0.53 | 13.71 | 0.23 | AA | reference | |||||||
| AG | reference | ||||||||||||||
| overall | 0.47 | ||||||||||||||
| Age | 1 | 1.07 | 1.04 | 1.12 | 0.0002*** | 1 | 0.07 | 1.07 | 1.03 | 1.11 | 0.0002*** | ||||
| GCS | 1 | 0.12 | 0.04 | 0.34 | <0.0001*** | 1 | −1.09 | 0.11 | 0.04 | 0.32 | <0.0001*** | ||||
| Gender | 1 | 2.77 | 0.88 | 8.68 | 0.08† | 1 | 0.45 | 2.44 | 0.81 | 7.37 | 0.11 | ||||
| Hypotension | 1 | 0.12 | 0.02 | 0.70 | 0.02* | 1 | −1.09 | 0.11 | 0.02 | 0.62 | 0.01* | ||||
| rs17759659 | Genotype | AA | 2 | −0.94 | 0.23 | 0.07 | 0.78 | 0.02* | GG & AG | 1 | 0.72 | 4.23 | 1.31 | 13.61 | 0.02* |
| (n = 128) | GG | 0.43 | 0.92 | 0.22 | 3.84 | 0.37 | AA | reference | |||||||
| AG | reference | ||||||||||||||
| overall | 0.05* | ||||||||||||||
| Age | 1 | 0.09 | 1.10 | 1.05 | 1.14 | <0.0001*** | 1 | 0.09 | 1.10 | 1.05 | 1.14 | <0.0001*** | |||
| GCS | 1 | −1.21 | 0.09 | 0.03 | 0.28 | <0.0001*** | 1 | −1.21 | 0.09 | 0.03 | 0.28 | <0.0001*** | |||
| Gender | 1 | 0.58 | 3.17 | 0.96 | 10.39 | 0.06† | 1 | 0.57 | 3.14 | 0.97 | 10.18 | 0.06† | |||
| Hypotension | 1 | −1.18 | 0.09 | 0.02 | 0.61 | 0.01* | 1 | −1.19 | 0.09 | 0.01 | 0.59 | 0.01* | |||
| rs1801018 | Genotype | AA | 2 | 0.41 | 0.93 | 0.25 | 3.54 | 0.36 | AA & AG | 1 | 0.68 | 3.86 | 1.18 | 12.59 | 0.03* |
| (n = 130) | GG | −0.89 | 0.26 | 0.08 | 0.87 | 0.03* | GG | reference | |||||||
| AG | reference | ||||||||||||||
| overall | 0.08† | ||||||||||||||
| Age | 1 | 0.08 | 1.09 | 1.04 | 1.13 | 0.0001*** | 1 | 0.08 | 1.09 | 1.04 | 1.13 | 0.0001*** | |||
| GCS | 1 | −1.14 | 0.10 | 0.04 | 0.30 | <0.0001*** | 1 | −1.14 | 0.10 | 0.04 | 0.30 | <0.0001*** | |||
| Gender | 1 | 0.52 | 2.82 | 0.91 | 8.75 | 0.07† | 1 | 0.51 | 2.80 | 0.91 | 8.57 | 0.07† | |||
| Hypotension | 1 | −1.13 | 0.10 | 0.02 | 0.59 | 0.01* | 1 | −1.14 | 0.10 | 0.02 | 0.57 | 0.01* | |||
| rs4456611 | Genotype | CC | 2 | −0.59 | 0.70 | 0.21 | 2.33 | 0.13 | TT & CT | 1 | 0.35 | 2.00 | 0.66 | 6.11 | 0.22 |
| (n = 128) | TT | 0.83 | 2.92 | 0.84 | 10.13 | 0.04* | CC | reference | |||||||
| CT | reference | ||||||||||||||
| overall | 0.12 | ||||||||||||||
| Age | 1 | 0.08 | 1.09 | 1.04 | 1.13 | <0.0001*** | 1 | 0.08 | 1.08 | 1.04 | 1.13 | <0.0001*** | |||
| GCS | 1 | −1.14 | 0.10 | 0.04 | 0.30 | <0.0001*** | 1 | −1.16 | 0.10 | 0.03 | 0.29 | <0.0001*** | |||
| Gender | 1 | 0.57 | 3.14 | 0.99 | 9.97 | 0.05* | 1 | 0.49 | 2.68 | 0.90 | 8.07 | 0.08† | |||
| Hypotension | 1 | −1.19 | 0.09 | 0.02 | 0.59 | 0.01* | 1 | −1.15 | 0.10 | 0.02 | 0.57 | 0.01 | |||
| rs4941185 | Genotype | AA | 2 | 0.07 | 1.04 | 0.35 | 3.03 | 0.83 | GG & AG | 1 | −0.04 | 0.92 | 0.35 | 2.44 | 0.87 |
| (n = 129) | GG | −0.11 | 0.86 | 0.23 | 3.32 | 0.79 | AA | reference | |||||||
| AG | reference | ||||||||||||||
| overall | 0.96 | ||||||||||||||
| Age | 1 | 0.07 | 1.07 | 1.04 | 1.12 | 0.0001*** | 1 | 0.07 | 1.07 | 1.04 | 1.12 | 0.0001*** | |||
| GCS | 1 | −1.15 | 0.10 | 0.04 | 0.29 | <0.0001*** | 1 | −1.14 | 0.10 | 0.04 | 0.29 | <0.0001*** | |||
| Gender | 1 | 0.49 | 2.68 | 0.90 | 8.02 | 0.77 | 1 | 0.48 | 2.63 | 0.90 | 7.73 | 0.79 | |||
| Hypotension | 1 | −1.37 | 0.07 | 0.01 | 0.42 | 0.004* | 1 | −1.36 | 0.07 | 0.01 | 0.43 | 0.004* | |||
| rs7230970 | Genotype | CC | 2 | −0.37 | 0.30 | 0.07 | 1.41 | 0.45 | CC & TT | 1 | 0.47 | 2.54 | 0.86 | 7.47 | 0.09† |
| (n = 126) | TT | −0.45 | 0.28 | 0.09 | 0.90 | 0.25 | TT | reference | |||||||
| CT | reference | ||||||||||||||
| overall | 0.07† | ||||||||||||||
| Age | 1 | 0.08 | 1.08 | 1.04 | 1.13 | 0.0001*** | 1 | 0.08 | 1.09 | 1.04 | 1.13 | <0.0001*** | |||
| GCS | 1 | −1.30 | 0.08 | 0.02 | 0.24 | <0.0001*** | 1 | −1.24 | 0.08 | 0.03 | 0.26 | <0.0001*** | |||
| Gender | 1 | 0.52 | 2.83 | 0.90 | 8.92 | 0.08† | 1 | 0.50 | 2.74 | 0.88 | 8.51 | 0.08† | |||
| Hypotension | 1 | −1.04 | 0.12 | 0.02 | 0.78 | 0.03* | 1 | −1.20 | 0.09 | 0.02 | 0.56 | 0.01* | |||
| rs7236090 | Genotype | CC | 2 | 0.39 | 1.03 | 0.34 | 3.09 | 0.28 | TT & CT | 1 | −0.19 | 0.68 | 0.25 | 1.87 | 0.45 |
| (n = 128) | TT | −0.76 | 0.32 | 0.08 | 1.30 | 0.09† | CC | reference | |||||||
| CT | reference | ||||||||||||||
| overall | 0.23 | ||||||||||||||
| Age | 1 | 0.07 | 1.08 | 1.04 | 1.12 | 0.0001*** | 1 | 0.07 | 1.07 | 1.03 | 1.11 | 0.0002*** | |||
| GCS | 1 | −1.11 | 0.11 | 0.04 | 0.32 | <0.0001*** | 1 | −1.08 | 0.12 | 0.04 | 0.33 | <0.0001*** | |||
| Gender | 1 | 0.57 | 3.10 | 1.02 | 9.45 | 0.05* | 1 | 0.52 | 2.85 | 0.97 | 8.36 | 0.06† | |||
| Hypotension | 1 | −1.08 | 0.12 | 0.02 | 0.69 | 0.02* | 1 | −1.06 | 0.12 | 0.02 | 0.68 | 0.02* | |||
| rs899968 | Genotype | AA | 2 | −0.42 | 0.70 | 0.16 | 3.12 | 0.38 | AA & AC | 1 | −0.32 | 0.52 | 0.19 | 1.42 | 0.20 |
| (n = 128) | CC | 0.49 | 1758.00 | 0.61 | 5.06 | 0.18 | CC | reference | |||||||
| AC | reference | ||||||||||||||
| overall | 0.40 | ||||||||||||||
| Age | 1 | 0.08 | 1.08 | 1.04 | 1.13 | 0.0001*** | 1 | 0.08 | 1.08 | 1.04 | 1.12 | 0.0001*** | |||
| GCS | 1 | −1.11 | 0.11 | 0.04 | 0.31 | <0.0001*** | 1 | −1.11 | 0.11 | 0.04 | 0.31 | <0.0001*** | |||
| Gender | 1 | 0.56 | 3.08 | 1.01 | 9.38 | 0.05* | 1 | 0.58 | 3.21 | 1.07 | 9.63 | 0.04* | |||
| Hypotension | 1 | −1.32 | 0.07 | 0.01 | 0.45 | 0.005* | 1 | −1.31 | 0.07 | 0.01 | 0.47 | 0.006* | |||
| rs949037 | Genotype | CC | 2 | −1.13 | 0.35 | 0.08 | 1.53 | 0.02* | CC & CT | 1 | −0.79 | 0.21 | 0.06 | 0.67 | 0.01* |
| (n = 126) | TT | 1.21 | 3.67 | 1.08 | 12.49 | 0.005* | TT | reference | |||||||
| CT | reference | ||||||||||||||
| overall | 0.02* | ||||||||||||||
| Age | 1 | 0.09 | 1.09 | 1.05 | 1.14 | <0.0001*** | 1 | 0.09 | 1.09 | 1.05 | 1.14 | <0.0001*** | |||
| GCS | 1 | −1.05 | 0.12 | 0.04 | 0.35 | 0.0001*** | 1 | −1.04 | 0.13 | 0.04 | 0.36 | 0.0001*** | |||
| Gender | 1 | 0.51 | 2.78 | 0.85 | 9.05 | 0.09† | 1 | 0.51 | 2.76 | 0.88 | 8.64 | 0.08† | |||
| Hypotension | 1 | −1.24 | 0.08 | 0.01 | 0.57 | 0.01* | 1 | −1.35 | 0.07 | 0.01 | 0.46 | 0.01* | |||
p ≤ 0.05; ***p ≤ 0.001; †trend; ***p ≤ 0.001 meets Bonferroni correction.
Overall = p for type 3 test.
tSNP, tagging single nucleotide polymorphism.
NRS-R
Table 5 gives detailed formation on the statically significant and trending tSNPs for the neurocognitive outcomes as measured by the NRS-R. tSNP rs12454712 (TT) was associated with lower NRS-R scores (p = 0.02; p for type 3 p = 0.05), indicating better outcomes. In the dichotomized analysis, the presence of the rs12454712 variant allele (C) was associated with significantly higher NRS-R scores, which are associated with poorer outcomes (p = 0.01). tSNP rs17759659 (GG) was associated with higher NRS-R scores (p = 0.03; p for type 3 p = 0.02), indicating poorer outcome. In the dichotomized analysis, the presence of the rs17759659 variant allele (G) was associated with significantly higher NRS-R scores, or poorer neurobehavioral outcomes (p = 0.05). Due to equal AA and TT groups among our sample, two dichotomized analyses were completed for rs1944419, featuring either AA or TT as the wild-type. When AA was analyzed as the wild-type, the presence of the rs1944419 variant allele (T) was associated with lower NRS-R scores, and thus more favorable outcomes (p = 0.05). For tSNP rs4456611, the dichotomized analysis showed that the presence of the variant allele (C) was associated with a trend toward lower NRS-R scores, or more favorable outcomes (p = 0.06). The homozygous variant of rs7236090 (CC) was associated with lower NRS-R scores, also a favorable outcome (p = 0.007; p for type 3 p = 0.02). In the dichotomized analysis the presence of the rs7236090 variant allele (C) was associated with significantly lower NRS-R scores, and thus more favorable outcomes (p = 0.005). For all of the tSNPs studied, older age (p-value range = 0.0002–0.01) was associated with poorer outcomes.
Table 5.
Primary Mixed Effects Regression Modeling Analyses of Neurobehavioral Outcomes: NRS-R and BCL2 tSNPs
| |
|
Genotypes |
Dichotomized genotypes |
||||
|---|---|---|---|---|---|---|---|
| tSNP | Variables | coefff | p | coefff | p | ||
| rs1026825 | Genotype | AA | −2.68 | 0.23 | GG & AG | 2.63 | 0.20 |
| (n = 99) | GG | −0.11 | 0.96 | AA | reference | ||
| AG | reference | ||||||
| overall | 0.45 | ||||||
| Months | 3 | 0.07 | 0.96 | 3 | 0.05 | 0.97 | |
| 6 | −0.54 | 0.69 | 6 | −0.55 | 0.69 | ||
| 12 | reference | 12 | reference | ||||
| overall | 0.88 | overall | 0.88 | ||||
| Age | 0.24 | 0.002* | 0.24 | 0.002* | |||
| GCS | 2.67 | 0.23 | 2.68 | 0.23 | |||
| Gender | −0.55 | 0.82 | −0.56 | 0.81 | |||
| rs12454712 | Genotype | CC | 0.99 | 0.77 | CC & CT | 4.44 | 0.01* |
| (n = 99) | TT | −4.28 | 0.02* | TT | reference | ||
| CT | reference | ||||||
| overall | 0.05* | ||||||
| Months | 3 | −0.16 | 0.91 | 3 | −0.19 | 0.90 | |
| 6 | −0.68 | 0.62 | 6 | −0.68 | 0.62 | ||
| 12 | reference | 12 | reference | ||||
| overall | 0.87 | overall | 0.87 | ||||
| Age | 0.25 | 0.001*** | 0.25 | 0.001*** | |||
| GCS | 2.05 | 0.34 | 2.04 | 0.34 | |||
| Gender | −2.08 | 0.36 | −2.00 | 0.37 | |||
| rs1481031 | Genotype | AA | −0.60 | 0.77 | GG & AG | 1.42 | 0.47 |
| (n = 97) | GG | 4.57 | 0.14 | AA | reference | ||
| AG | reference | ||||||
| overall | 0.26 | ||||||
| Months | 3 | 0.87 | 0.54 | 3 | 0.81 | 0.57 | |
| 6 | 0.04 | 0.97 | 6 | −0.03 | 0.98 | ||
| 12 | reference | 12 | reference | ||||
| overall | 0.78 | overall | 0.80 | ||||
| Age | 0.23 | 0.004* | 0.25 | 0.002* | |||
| GCS | 1.71 | 0.46 | 2.02 | 0.38 | |||
| Gender | −0.77 | 0.74 | −1.05 | 0.66 | |||
| rs17756073 | Genotype | AA | −3.26 | 0.10 | GG & AG | 3.07 | 0.10 |
| (n = 96) | GG | −0.75 | 0.80 | AA | reference | ||
| AG | reference | ||||||
| overall | 0.25 | ||||||
| Months | 3 | 0.09 | 0.95 | 3 | 0.08 | 0.95 | |
| 6 | −0.49 | 0.73 | 6 | −0.50 | 0.72 | ||
| 12 | reference | 12 | reference | ||||
| overall | 0.90 | overall | 0.90 | ||||
| Age | 0.24 | 0.002* | 0.24 | 0.002* | |||
| GCS | 2.47 | 0.27 | 2.49 | 0.26 | |||
| Gender | −0.71 | 0.76 | −0.72 | 0.76 | |||
| rs17759659 | Genotype | AA | −2.13 | 0.28 | GG & AG | 3.59 | 0.05* |
| (n = 98) | GG | 5.51 | 0.03* | AA | reference | ||
| AG | reference | ||||||
| overall | 0.02* | ||||||
| Months | 3 | −0.01 | 1.00 | 3 | −0.09 | 0.95 | |
| 6 | −0.63 | 0.65 | 6 | −0.71 | 0.60 | ||
| 12 | reference | 12 | reference | ||||
| overall | 0.87 | overall | 0.85 | ||||
| Age | 0.28 | 0.0002*** | 0.28 | 0.0002*** | |||
| GCS | 2.62 | 0.22 | 2.79 | 0.20 | |||
| Gender | −1.30 | 0.55 | −1.21 | 0.59 | |||
| rs1944419 | Genotype | AA | 3.88 | 0.10 | TT & AT | −4.46 | 0.05* |
| (n = 96) | TT | −2.03 | 0.38 | AA | reference | ||
| AT | reference | ||||||
| overall | 0.10 | ||||||
| Months | 3 | 0.07 | 0.96 | 3 | 0.03 | 0.98 | |
| 6 | −0.88 | 0.54 | 6 | −0.93 | 0.52 | ||
| 12 | reference | 12 | reference | ||||
| overall | 0.75 | overall | 0.74 | ||||
| Age | 0.24 | 0.002* | 0.25 | 0.001*** | |||
| GCS | 3.56 | 0.12 | 3.41 | 0.13 | |||
| Gender | −1.50 | 0.52 | −1.38 | 0.55 | |||
| Genotype | AA & AT | 3.13 | 0.16 | ||||
| TT | reference | ||||||
| Months | 3 | 0.01 | 0.99 | ||||
| 6 | −0.87 | 0.54 | |||||
| 12 | reference | ||||||
| overall | 0.77 | ||||||
| Age | 0.24 | 0.003* | |||||
| GCS | 2.84 | 0.21 | |||||
| Gender | −1.34 | 0.56 | |||||
| rs4456611 | Genotype | CC | −2.31 | 0.36 | CC & CT | −4.14 | 0.06† |
| (n = 94) | TT | 3.48 | 0.14 | TT | reference | ||
| CT | reference | ||||||
| overall | 0.12 | ||||||
| Months | 3 | 0.98 | 0.50 | 3 | 0.98 | 0.50 | |
| 6 | 0.09 | 0.95 | 6 | 0.08 | 0.95 | ||
| 12 | reference | 12 | reference | ||||
| overall | 0.75 | overall | 0.75 | ||||
| Age | 0.26 | 0.001*** | 0.26 | 0.001*** | |||
| GCS | 1.70 | 0.47 | 1.06 | 0.64 | |||
| Gender | −2.71 | 0.25 | −2.51 | 0.28 | |||
| rs7236090 | Genotype | CC | −6.02 | 0.007* | CC & CT | 5.69 | 0.005* |
| (n = 95) | TT | −0.93 | 0.68 | TT | reference | ||
| CT | reference | ||||||
| overall | 0.02* | ||||||
| Months | 3 | 1.05 | 0.47 | 3 | 0.97 | 0.50 | |
| 6 | 0.25 | 0.85 | 6 | 0.19 | 0.89 | ||
| 12 | reference | 12 | reference | ||||
| overall | 0.75 | overall | 0.77 | ||||
| Age | 0.20 | 0.01* | 0.20 | 0.01* | |||
| GCS | 1.53 | 0.48 | 1.53 | 0.48 | |||
| Gender | −0.73 | 0.76 | −0.76 | 0.74 | |||
p ≤ 0.05; ***p ≤ 0.001; †trend; ***p ≤ 0.001 meets Bonferroni correction.
Overall = p for type 3 test.
tSNP, tagging single nucleotide polymorphism; NRS-R, Neurobehavioral Rating Scale–Revised.
Month = time point of the outcome assessment post-injury (3, 6, and 12 months).
Discussion
To our knowledge this is the first study to examine the relationship between genetic variability in the BCL2 gene and outcomes after TBI. The results of this study show that there is a relationship between BCL2 genotype and global functional outcomes. Our pilot study was exploratory in nature and provides novel information regarding BCL2 genotype and TBI to inform future studies.
Outcomes
Our study found that for tSNP rs17759659, the presence of the variant allele (G) was associated with significantly poorer GOS scores with the conservative Bonferroni correction. This tSNP is in the large intron II region towards the 5′ end of the gene. The function of this tSNP is unknown. There were additional tSNPs representing the intron II region of the gene that were significant (using a traditional alpha of α = 0.05) in the global function outcomes analyses (rs12968517, rs7236090, and rs949037), indicating that this region may play a role in outcomes after TBI. The size of intron II may be an indication that it has a functional role (Seto et al., 1988), potentially related to mRNA stability or processing.
Our study found that tSNP rs1801018, variant present genotypes (AA or AG), was associated with poor outcomes using a traditional alpha (low GOS scores, high DRS scores, and increased rate of mortality). In addition, the homozygous wild-type GG was associated with favorable outcomes (high GOS scores, low DRS scores, and lower mortality). The continuity of the findings for this tSNP indicates that the presence of one or two copies of the A allele is associated with poorer outcomes, therefore allele dosing is not an issue; simply the presence of the variant allele drives a tendency toward susceptibility to poor outcomes.
While rs1801018 did not reach significance under the Bonferroni correction, there are some trending associations that make it a candidate for subsequent studies, particularly since it has the potential to tag all of the activity in the promoter region of the gene. The BCL2 gene is believed to have two promoter regions, P1 and P2. P2 is located 1.3 kb downstream from P1 and is associated with exon 2 (the location of rs1801018) (Bredow et al., 2007; Seto et al., 1988; Young and Korsmeyer, 1993). P2 has two discrete transcription initiation sites and decreases the activity of the P1 promoter, thus functioning as a negative regulatory element (Young and Korsmeyer, 1993). This entire region encompassing both promoter regions appears to be characterized by tSNP rs1801018, leading us to hypothesize that a potential mechanism to explain the trending association of this tSNP with outcomes after TBI may be related to gene regulation.
In addition, tSNP rs1801018 has also been investigated in other pathologies. In the oncology literature tSNP rs1801018 is significantly associated with a greater susceptibility to chronic myeloid leukemia (CML; Kim et al., 2009). In allelic analyses of rs1801018, the presence of the A allele is associated with increased risk for CML, which was inferred to mean decreased apoptosis and more cell survival, and hence oncogenesis. While our studies differ in methodology and population, our findings in light of the CML-based findings would lead us to believe that decreased apoptosis may play a role in poorer outcomes post-TBI, potentially though the survival of dysfunctional cells that would otherwise undergo cell death. Harboring dysfunctional neurons may contribute to glial scar formation and act as a barrier to neuronal growth and prevent neuronal regeneration, as suggested by BCL2 transgenetic mouse model studies in which this was associated with poor outcomes (Nakamura et al., 1999).
The mortality analysis had no tSNP findings that were significant after Bonferroni correction. This may be related to the cross-sectional nature of the analysis, that limited the outcome to mortality at 3 months. In addition, mortality was unique in that hypotension was only a covariate of interest in the preliminary analysis for mortality at 3 months (which was then grounds for inclusion in the primary model). We speculate that hypotension was included in the mortality model because it was a cross-sectional analysis, and it may impact 3-month mortality; however, when analyzing GOS and DRS scores at the 3-, 6-, 12-, and 24-month follow-up points hypotension was not statistically significant.
None of our neurobehavioral findings met significance under the Bonferroni correction.
Therapeutic implications
This study was a first step in exploring BCL2 genotypes and how they affect long-term neurobehavioral outcomes after severe TBI in adults. Replication of these findings is needed, however, it does add to the body of knowledge that implicates a role for BCL2 in post–brain injury cellular events with potential for intervention. BCL2 gene-based interventions have significantly impacted TBI lesion size in animal models. A BCL2 fusion protein-recombinant adenovirus gene therapy intervention after TBI in rats was shown to significantly reduce apoptosis at 3 days post-injury (p < 0.01; Yang et al., 2006). Given the success seen with BCL2-based therapies in animal models, the development of BCL2 genotype-based therapies for humans may be possible.
Though there are several interventions that have successfully altered BCL2/BCL2 expression, including pharmaceutical BCL2 protein derivative (Panizzon et al., 1998), recombinant human erythropoietin (Liao et al., 2008), and hyperbaric oxygenation (Liu et al., 2006; Vlodavsky et al., 2005), none of the results have been duplicated nor did they explore outcomes. More research needs to be conducted in this area with outcomes added to the study designs. The full implications of BCL2 genotypes and their effects on outcomes after severe TBI have yet to be elucidated; however, the findings detailed here form the basis for better understanding of the physiologic implications of BCL2 in outcomes after TBI.
Limitations
One limitation is external validity beyond male Caucasians. While the demographics of the overall sample were representative of non-penetrating TBI in the greater Pittsburgh area, the generalizability of the data is limited due to the lack of diversity in ancestry and gender in the sample. An additional limitation involves sample size, particularly for analyses of outcomes at time points further out from the injury, and those that require survival such as the NRS. While our study is adequately powered to assess outcomes for which death is a variable, other subgroup analyses require further confirmation using larger sample sizes. Due to technological difficulties in the manufacturing of three tSNP assays (rs2850762, rs7231914, and rs8089538), we were unable to characterize the entire gene at MAF of ≥30% and r2 ≥0.80; however, this would not alter the findings presented in this paper.
Supplementary Material
Acknowledgments
This study utilized biological samples and data collected from federally-funded studies that examined genes and gene products in TBI patients, and how they related to outcomes attained by TBI patients (R01NR04801, NR008424, 5P50NS30318, and R49/CCR 323155-03). We thank the nurses of the neurotrauma intensive care unit for their assistance with sample collection and support of our work. Genotype data collection was supported by the Leslie A. Hoffman Endowed Research Award at the University of Pittsburgh School of Nursing, and the Sigma Theta Tau Eta Chapter at the University of Pittsburgh. Postdoctoral fellowship is funded by the University of Pittsburgh School of Nursing, Targeted Research and Academic Training Program for Nurses in Genomics (T32 NR009759).
Author Disclosure Statement
No competing financial interests exist.
References
- Bahr M. Live or let die—retinal ganglion cell death and survival during development and in the lesioned adult CNS. Trends Neurosci. 2000;23:483–490. doi: 10.1016/s0166-2236(00)01637-4. [DOI] [PubMed] [Google Scholar]
- Barrett J.C. Fry B. Maller J. Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15; doi: 10.1093/bioinformatics/bth457. [PubMed ID:15297300]. [DOI] [PubMed] [Google Scholar]
- Bredesen D.E. Apoptosis: overview and signal transduction pathways. J. Neurotrauma. 2000;17:801–810. doi: 10.1089/neu.2000.17.801. [DOI] [PubMed] [Google Scholar]
- Bredow S. Juri D.E. Cardon K. Tesfaigzi Y. Identification of a novel Bcl-2 promoter region that counteracts in a p53-dependent manner the inhibitory P2 region. Gene. 2007;404:110–116. doi: 10.1016/j.gene.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock R. Chesnut R.M. Clifton G. Ghajar J. Marion D.W. Narayan R.K. Newell D.W. Pitts L.H. Rosner M.J. Wilberger J.W. Guidelines for the management of severe head injury. Brain Trauma Foundation. Eur. J. Emerg. Med. 1996;3:109–127. doi: 10.1097/00063110-199606000-00010. [DOI] [PubMed] [Google Scholar]
- Clark R.S. Chen J. Watkins S.C. Kochanek P.M. Chen M. Stetler R.A. Loeffert J.E. Graham S.H. Apoptosis-suppressor gene bcl-2 expression after traumatic brain injury in rats. J. Neurosci. 1997;17:9172–9182. doi: 10.1523/JNEUROSCI.17-23-09172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R.S. Kochanek P.M. Adelson P.D. Bell M.J. Carcillo J.A. Chen M. Wisniewski S.R. Janesko K. Whalen M.J. Graham S.H. Increases in bcl-2 protein in cerebrospinal fluid and evidence for programmed cell death in infants and children after severe traumatic brain injury. J. Pediatr. 2000;137:197–204. doi: 10.1067/mpd.2000.106903. [DOI] [PubMed] [Google Scholar]
- Clark R.S. Kochanek P.M. Chen M. Watkins S.C. Marion D.W. Chen J. Hamilton R.L. Loeffert J.E. Graham S.H. Increases in Bcl-2 and cleavage of caspase-1 and caspase-3 in human brain after head injury. FASEB J. 1999;13:813–821. doi: 10.1096/fasebj.13.8.813. [DOI] [PubMed] [Google Scholar]
- Corrigan J.D. Whiteneck G. Mellick D. Perceived needs following traumatic brain injury. J. Head Trauma Rehabil. 2004;19:205–216. doi: 10.1097/00001199-200405000-00002. [DOI] [PubMed] [Google Scholar]
- Garcia I. Martinou I. Tsujimoto Y. Martinou J.C. Prevention of programmed cell death of sympathetic neurons by the bcl-2 proto-oncogene. Science. 1992;258:302–304. doi: 10.1126/science.1411528. [DOI] [PubMed] [Google Scholar]
- Graham S.H. Chen J. Clark R.S. Bcl-2 family gene products in cerebral ischemia and traumatic brain injury. J. Neurotrauma. 2000;17:831–841. doi: 10.1089/neu.2000.17.831. [DOI] [PubMed] [Google Scholar]
- Hockenbery D. Nunez G. Milliman C. Schreiber R.D. Korsmeyer S.J. Bcl-2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature. 1990;348:334–336. doi: 10.1038/348334a0. [DOI] [PubMed] [Google Scholar]
- Jennett B. Bond M. Assessment of outcome after severe brain damage. Lancet. 1975;1:480–484. doi: 10.1016/s0140-6736(75)92830-5. [DOI] [PubMed] [Google Scholar]
- Jennett B. Outcome after severe head injury: definitions and predictions. Med. J. Aust. 1976a;2:475–477. [PubMed] [Google Scholar]
- Jennett B. Predicting outcome after head injury. Proc. R. Soc. Med. 1976b;69:140–141. [PMC free article] [PubMed] [Google Scholar]
- Kane D.J. Sarafian T.A. Anton R. Hahn H. Gralla E.B. Valentine J.S. Ord T. Bredesen D.E. Bcl-2 inhibition of neural death: decreased generation of reactive oxygen species. Science. 1993;262:1274–1277. doi: 10.1126/science.8235659. [DOI] [PubMed] [Google Scholar]
- Kim D.H. Xu W. Ma C. Liu X. Siminovitch K. Messner H.A. Lipton J.H. Genetic variants in the candidate genes of the apoptosis pathway and susceptibility to chronic myeloid leukemia. Blood. 2009;113:2517–2525. doi: 10.1182/blood-2008-07-169110. [DOI] [PubMed] [Google Scholar]
- Langlois J.A. Rutland-Brown W. Thomas K.E. Traumatic brain injury in the United States: Emergency department visits, hospitalizations, and deaths. Centers for Disease Control and Prevention, National Center for Injury Prevention and Control; Atlanta, GA: 2006. [Google Scholar]
- Levin H.S. Gary H.E., Jr. Eisenberg H.M. Ruff R.M. Barth J.T. Kreutzer J. High W.M., Jr. Portman S. Foulkes M.A. Jane J.A., et al. Neurobehavioral outcome 1 year after severe head injury. Experience of the Traumatic Coma Data Bank. J. Neurosurg. 1990;73:699–709. doi: 10.3171/jns.1990.73.5.0699. [DOI] [PubMed] [Google Scholar]
- Lezek M.D. In: Neuropsychological Assessment. 4th. Howieson D.B., editor; Loring D.W., editor. Oxford University Press; New York: 2004. [Google Scholar]
- Liao Z.B. Zhi X.G. Shi Q.H. He Z.H. Recombinant human erythropoietin administration protects cortical neurons from traumatic brain injury in rats. Eur. J. Neurol. 2008;15:140–149. doi: 10.1111/j.1468-1331.2007.02013.x. [DOI] [PubMed] [Google Scholar]
- Liu Z. Jiao Q.F. You C. Che Y.J. Su F.Z. Effect of hyperbaric oxygen on cytochrome C, Bcl-2 and Bax expression after experimental traumatic brain injury in rats. Chin. J. Traumatol. 2006;9:168–174. [PubMed] [Google Scholar]
- Mah S.P. Zhong L.T. Liu Y. Roghani A. Edwards R.H. Bredesen D.E. The protooncogene bcl-2 inhibits apoptosis in PC12 cells. J. Neurochem. 1993;60:1183–1186. doi: 10.1111/j.1471-4159.1993.tb03275.x. [DOI] [PubMed] [Google Scholar]
- McCauley S.R. Levin H.S. Vanier M. Mazaux J.M. Boake C. Goldfader P.R. Rockers D. Butters M. Kareken D.A. Lambert J., et al. The neurobehavioural rating scale-revised: sensitivity and validity in closed head injury assessment. J. Neurol. Neurosurg. Psychiatry. 2001;71:643–651. doi: 10.1136/jnnp.71.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minambres E. Ballesteros M.A. Mayorga M. Marin M.J. Munoz P. Figols J. Lopez-Hoyos M. Cerebral apoptosis in severe traumatic brain injury patients: an in vitro, in vivo, and postmortem study. J. Neurotrauma. 2008;25:581–591. doi: 10.1089/neu.2007.0398. [DOI] [PubMed] [Google Scholar]
- Myers K.M. Fiskum G. Liu Y. Simmens S.J. Bredesen D.E. Murphy A.N. Bcl-2 protects neural cells from cyanide/aglycemia-induced lipid oxidation, mitochondrial injury, and loss of viability. J. Neurochem. 1995;65:2432–2440. doi: 10.1046/j.1471-4159.1995.65062432.x. [DOI] [PubMed] [Google Scholar]
- Nakamura M. Raghupathi R. Merry D.E. Scherbel U. Saatman K.E. McIntosh T.K. Overexpression of Bcl-2 is neuroprotective after experimental brain injury in transgenic mice. J. Comp. Neurol. 1999;412:681–692. doi: 10.1002/(sici)1096-9861(19991004)412:4<681::aid-cne9>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Nunez G. London L. Hockenbery D. Alexander M. McKearn J.P. Korsmeyer S.J. Deregulated Bcl-2 gene expression selectively prolongs survival of growth factor-deprived hematopoietic cell lines. J. Immunol. 1990;144:3602–3610. [PubMed] [Google Scholar]
- Panizzon K.L. Shin D. Frautschy S. Wallis R.A. Neuroprotection with Bcl-2(20-34) peptide against trauma. Neuroreport. 1998;9:4131–4136. doi: 10.1097/00001756-199812210-00024. [DOI] [PubMed] [Google Scholar]
- Povlishock E.J.T. Guidelines for the management of severe head injury. J. Neurotrauma. 2000;17:451–627. [Google Scholar]
- Polvishock J.T.E. Guidelines for the management of severe TBI [Special Issue] J. Neurotrauma. 2004;24:s1–s106. doi: 10.1089/neu.2007.9999. [DOI] [PubMed] [Google Scholar]
- Rappaport M. Hall K.M. Hopkins K. Belleza T. Cope D.N. Disability rating scale for severe head trauma: coma to community. Arch. Phys. Med. Rehabil. 1982;63:118–123. [PubMed] [Google Scholar]
- Seto M. Jaeger U. Hockett R.D. Graninger W. Bennett S. Goldman P. Korsmeyer S.J. Alternative promoters and exons, somatic mutation and deregulation of the Bcl-2-Ig fusion gene in lymphoma. EMBO J. 1988;7:123–131. doi: 10.1002/j.1460-2075.1988.tb02791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman D.J. Alverson C. Dunn K.A. Guerrero J. Sniezek J.E. Traumatic brain injury in the United States: A public health perspective. J. Head Trauma Rehabil. 1999;14:602–615. doi: 10.1097/00001199-199912000-00009. [DOI] [PubMed] [Google Scholar]
- Vlodavsky E. Palzur E. Feinsod M. Soustiel J.F. Evaluation of the apoptosis-related proteins of the BCL-2 family in the traumatic penumbra area of the rat model of cerebral contusion, treated by hyperbaric oxygen therapy: a quantitative immunohistochemical study. Acta Neuropathol. 2005;110:120–126. doi: 10.1007/s00401-004-0946-8. [DOI] [PubMed] [Google Scholar]
- Wilson J.T. Pettigrew L.E. Teasdale G.M. Structured interviews for the Glasgow Outcome Scale and the extended Glasgow Outcome Scale: guidelines for their use. J. Neurotrauma. 1998;15:573–585. doi: 10.1089/neu.1998.15.573. [DOI] [PubMed] [Google Scholar]
- Yang S.Y. Xue L. Human neuronal apoptosis secondary to traumatic brain injury and the regulative role of apoptosis-related genes. Chin. J. Traumatol. 2004;7:159–164. [PubMed] [Google Scholar]
- Yang X.F. Zheng X.S. Liu W.G. Feng J.F. Bcl-2 gene therapy for apoptosis following traumatic brain injury. Chin. J. Traumatol. 2006;9:276–281. [PubMed] [Google Scholar]
- Young R.L. Korsmeyer S.J. A negative regulatory element in the bcl-2 5′-untranslated region inhibits expression from an upstream promoter. Mol. Cell Biol. 1993;13:3686–3697. doi: 10.1128/mcb.13.6.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Chen Y. Jenkins L.W. Kochanek P.M. Clark R.S. Bench-to-bedside review: Apoptosis/programmed cell death triggered by traumatic brain injury. Crit. Care. 2005;9:66–75. doi: 10.1186/cc2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.