Abstract
The yeast vacuolar membrane protein Ycf1p and its mammalian counterpart, MRP1, belong to the ABCC subfamily of ATP-binding cassette (ABC) transporters. Genetic evidence suggests that the yeast casein kinase 2α, Cka1p, negatively regulates Ycf1p function via phosphorylation of Ser251 within the N-terminus. In this study we provide strong evidence that Cka1p regulates Ycf1p function via phosphorylation of Ser251. We show that the CK2 holoenzyme interacts with Ycf1p. However, genetic analysis suggests that only Cka1p is required for Ser251 phosphorylation; as deletion of CKA1 significantly reduces Ser251 phosphorylation in vivo. Furthermore, purified recombinant Cka1p phosphorylates an Ycf1p-derived peptide containing Ser251. We also demonstrate that Ycf1p function is induced in response to high salt stress. Induction of Ycf1p function strongly correlates with reduced phosphorylation of Ser251. Importantly, Cka1p activity in vivo is similarly reduced in response to salt stress, consistent with our finding that Cka1p directly phosphorylates Ser251 of Ycf1p. We provide genetic and biochemical evidence that strongly suggests that the induction of Ycf1p function is the result of decreased phosphorylation of Ser251. In conclusion, our work demonstrates a novel biochemical role for Cka1p regulation of Ycf1p function in the cellular response of yeast to salt stress.
Keywords: Saccharomyces cerevisiae, Ycf1p, ABC Transporter, casein kinase 2, CK2, Cka1p, phosphorylation, regulation, salt stress
Introduction
Transporters of the ATP-Binding Cassette (ABC) superfamily are expressed in all organisms from microbes to mammals, and function in the transport of chemically diverse compounds across cellular membranes (Gottesman & Ambudkar, 2001, Dean, 2005). Recent interest has focused on the ABCC subfamily of ABC transporters, whose prototype member is the mammalian multidrug resistance-associated protein 1 (MRP1) also called ABCC1. Mutations in several members of the ABCC subfamily cause human diseases, including cystic fibrosis, pseudoxanthoma elasticum, and Dubin Johnson Syndrome, resulting from mutations in ABCC7 (CFTR), ABCC6 (MRP6), and ABCC2 (MRP2), respectively (Gottesman & Ambudkar, 2001, Dean, 2005, Cole & Deeley, 2006). In addition, overexpression of MRP1 and other ABCC proteins is associated with multidrug resistance in a variety of tumor cell lines (Cole, et al., 1992, Gottesman & Ambudkar, 2001, Haimeur, et al., 2004, Dean, 2005).
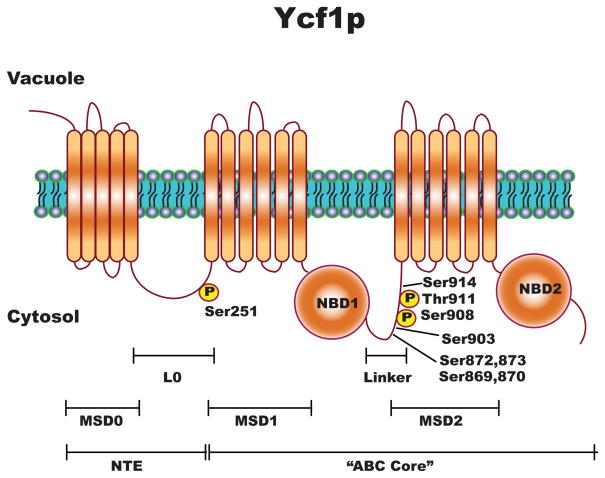
The ABCC transporters are distinguished from the other subfamily of ABC transporters by two unique features. First, several transport substrates in the form of glutathione conjugates or complexes (Borst & Elferink, 2002, Haimeur, et al., 2004). Second, in addition to the ABC “core” domain composed of two membrane spanning domains (MSDs) and two nucleotide binding domains (NBDs) (Fig. 1), the ABCCs have a unique and large N-terminal extension (NTE). The NTE consists of either five additional membrane spans and a large intracellular loop (“long” ABCC’s) or an extended N-terminal cytosolic domain (“short” ABCC’s) (Borst & Elferink, 2002, Haimeur, et al., 2004, Paumi, et al., 2009) (Fig. 1). Functionally, the NTE plays a role in protein localization and substrate binding (Bakos, et al., 2000, Ren, et al., 2001, Fernandez, et al., 2002, Mason & Michaelis, 2002, Qian, et al., 2002, Westlake, et al., 2003, Westlake, et al., 2005, Ren, et al., 2006, Yang, et al., 2007).
Figure 1.
Substrate specificity, the biochemical properties, and the transcriptional regulation of ABCC transporters have been relatively well-studied (Wemmie, et al., 1994, Borst & Elferink, 2002, Sharma, et al., 2002, Haimeur, et al., 2004). In contrast, the post-translational regulation of the ABCCs, has, in general, not been well-characterized. To date, ABCC2 (MRP2) (Minami, et al., 2009), ABCC7 (CFTR) (Cheng, et al., 1991, Chappe, et al., 2003, Howell, et al., 2004, Chappe, et al., 2005), and the yeast ABCC, Ycf1p (Li, et al., 2007, Smolka, et al., 2007, Albuquerque, et al., 2008) have been shown to be post-translationally modified. Ycf1p, a typical ABCC, contains an NTE, and has been shown to be positively and negatively regulated via phosphorylation in both the linker region of the ABC core and the NTE (Eraso, et al., 2004, Li, et al., 2007, Smolka, et al., 2007, Albuquerque, et al., 2008, Paumi, et al., 2008).
To examine the role of phosphorylation in the regulation of ABCC transporter function, we are using Saccharomyces cerevisiae. Yeast are a highly tractable system for genetic and biochemical analysis. The best-studied full-length ABCC subfamily member in yeast is Ycf1p, a homologue of human ABCC1 (MRP1) (Fig. 1) (Szczypka, et al., 1994, Li, et al., 1996, Li, et al., 1997, Wemmie & Moye-Rowley, 1997, Mason & Michaelis, 2002, Mason, et al., 2003). Ycf1p substrate specificity and function has been shown to be highly homologous to human MRP1 (ABCC1) (Tommasini, et al., 1996). Unlike MRP1, which is localized to the plasma membrane and effluxes toxins from the cytosol to the extracellular space, Ycf1p is localized to the vacuolar membrane and transports substrates into the vacuole lumen thereby effectively sequestering them from cytosolic targets (Szczypka, et al., 1994, Li, et al., 1996, Paumi, et al., 2009). Ycf1p was initially characterized as a protein required for cadmium resistance in yeast and was later shown to be required for the detoxification of a broad range of cellular toxins including heavy metal complexes (cadmium, lead, mercury, arsenic), oxidants (diamide), and endogenous toxins (Szczypka, et al., 1994, Li, et al., 1996, Chaudhuri, et al., 1997, Li, et al., 1997, Ghosh, et al., 1999, Pascolo, et al., 2001, Gueldry, et al., 2003, Song, et al., 2003, Jungwirth & Kuchler, 2006). Recent evidence, from multiple laboratories, suggests that post-transcriptional regulation of Ycf1p is an important regulatory mechanism of transporter function (Wemmie, et al., 1994, Sharma, et al., 2002, Eraso, et al., 2004, Paumi, et al., 2007, Paumi, et al., 2008).
Several phosphoproteomic studies within yeast have shown that Ycf1p is phosphorylated (Chi, et al., 2007, Smolka, et al., 2007, Albuquerque, et al., 2008). The goal of these previous studies was to identify phosphorylation changes within the context of the entire yeast proteome. Three of the aforementioned studies identified Ser251, which is within the L0 (Fig. 1), as a major phosphorylation site. In addition, multiple phosphorylation sites within the linker domain were identified (Li, et al., 2007, Smolka, et al., 2007, Albuquerque, et al., 2008). Genetic evidence suggests that Cka1p, which is the yeast homolog of human CK2α, negatively regulates Ycf1p function via phosphorylation of Ser251 (Paumi, et al., 2008). However, the functional significance of Cka1p-dependent phosphorylation and thus regulation of Ycf1p remains unclear. Here we demonstrate that Ycf1p function is induced in response to salt stress via the attenuation of Cka1p-dependnet phosphorylation of Ycf1p at Ser251.
In this study, we examine the mechanism and functional significance of the negative regulation of Ycf1p as mediated by Cka1p.. Our results suggest that Cka1p negatively regulates Ycf1p function solely through the direct phosphorylation of Ser251. To this end, we show that each of the subunits of CK2 (Cka1p, Cka2p, Ckb1p, and Ckb2p) physically interact with Ycf1p. However, genetic and biochemical studies suggest that phosphorylation of Ycf1p at Ser251 requires only Cka1p. Taken together these results support the previous findings from Paumi et. al. 2008 that originally demonstrated the negative regulation of Ycf1p by Cka1p via direct phosphorylation of Ser251 (Paumi, et al., 2008). In extension, we now provide novel evidence for a role of Ycf1p in the cellular response to salt stress. Cka1p activity is shown to be attenuated in response to high salt, which results in the decline in Cka1p-dependent phosphorylation of Ser251 within Ycf1p and thus increases Ycf1p function. Further, we provide genetic and biochemical evidence that strongly suggests that induction of Ycf1p function in response to salt stress is indeed the result of decreased phosphorylation of Ser251. In conclusion, this work provides evidence that regulation of Ycf1p function via Cka1p plays an important role in cell stress response and implicates a similar role of human CK2α in regulating human MRP1 function in response to salt or other possible cell stresses.
Materials and Methods
Materials
All materials for Fmoc solid phase synthesis were purchased from Advanced Chemtech (Louisville, KY) and used without additional purification. The ATPγS was purchased from Roche Applied Science (Indianapolis, IN), and the [γ-32P] adenosine triphosphate was purchased from Perkin Elmer (Wellesley, MA). All additional chemicals were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO) and used without further purification.
Yeast Strains, Media, and Growth Conditions
Yeast strains used in this study are listed in Table 1. Standard drop-out media (SC) was prepared as previously described (Burke, 2000). The plates used for the cadmium spot tests were prepared by adding cadmium (CdSO4) at the indicated concentrations in SC medium immediately before pouring plates as described (Mason & Michaelis, 2002, Mason, et al., 2003). Cultures were grown at 30°C unless otherwise noted.
Table I.
Yeast strains used in this study
| Straina | Relevant Genotype | Reference |
|---|---|---|
| CP59b | met3Δ leu2Δ ura3Δ his3Δ | Open Biosystems |
| CP60 | met3Δ leu2Δ ura3Δ his3Δ ycf1::KanMX | Open Biosystems |
| CP105 | ura3Δ ade2Δ LEU2 TRP1 YCF1 | This Study |
| CP106 | ura3Δ ADE2 LEU2 TRP1 YCF1 | This Study |
| CP135 | met15Δ leu2Δ ura3Δ his3Δ ycf1Δ::KanMx [2μ YCF1-GFP URA3] | This Study |
| CP136 | met15Δ leu2Δ ura3Δ his3Δ ycf1Δ::KanMx [2μ YCF1-Ser251Ala -GFP URA3] | This Study |
| CP147 | met15Δ leu2Δ ura3Δ his3Δ cka1Δ::KanMx | Open Biosystems |
| CP151 | met15Δ leu2Δ ura3Δ his3Δ cka1Δ::KanMX [2μ YCF1-GFP URA3] | This Study |
| CP156 | met15Δ leu2Δ ura3Δ his3Δ YCF1:TAP-HIS3 | Open Biosystems |
| CP157 | met15Δ leu2Δ ura3Δ his3Δ ycf11Δ::KanMX cka1Δ::URA3 | Paumi et. al. 2009 |
| CP159 | met15Δ leu2Δ ura3Δ his3Δ cka2Δ::KanMX | Open Biosystems |
| CP164 | met15Δ leu2Δ ura3Δ his3Δ YCF1:TAP-HIS3 KanMx-PGAL-GST-CKA1 | This Study |
| CP165 | met15Δ leu2Δ ura3Δ his3Δ ycf1Δ::KanMX [2μ YCF1-S869A -GFP URA3] | This Study |
| CP166 | met15Δ leu2Δ ura3Δ his3Δ ycf1Δ::KanMX [2μ YCF1-S870A -GFP URA3] | This Study |
| CP167 | met15Δ leu2Δ ura3Δ his3Δ ycf1Δ::KanMX [2μ YCF1-S872A -GFP URA3] | This Study |
| CP168 | met15Δ leu2Δ ura3Δ his3Δ ycf1Δ::KanMX [2μ YCF1-S873A -GFP URA3] | This Study |
| CP169 | met15Δ leu2Δ ura3Δ his3Δ ycf1Δ::KanMX [2μ YCF1-S903A -GFP URA3] | This Study |
| CP170 | met15Δ leu2Δ ura3Δ his3Δ ycf1Δ::KanMX [2μ YCF1-S914A -GFP URA3] | This Study |
| CP174 | PYES2/CT::cka1::v5::6his | Kubinski et. al. 2007 |
| CP176 | met15Δ leu2Δ ura3Δ his3Δ cka2Δ::KanMX [2μ CKA2:MYC:6xHis LEU2] | This Study |
| CP177 | met15Δ leu2Δ ura3Δ his3Δ pepΔ::KanMX [2μ YCF1-GFP URA3] | This Study |
| CP178 | met15Δ leu2Δ ura3Δ his3Δ cka2Δ::KanMX [2μ YCF1-GFP URA3] | This Study |
| CP179 | met15Δ leu2Δ ura3Δ his3Δ KanMx- PGAL-GST-CKA1 | This Study |
| CP180 | met15Δ leu2Δ ura3Δ his3Δ pepΔ::KanMX [2μ YCF1-Ser251Ala-GFP URA3] | This Study |
| CP187 | met15Δ leu2Δ ura3Δ his3Δ cka1Δ::LEU2 pep4Δ::KanMX [2μ YCF1-TAP URA3] | This Study |
| CP188 | met15Δ leu2Δ ura3Δ his3Δ pep4Δ::KanMX [2μ YCF1-TAP URA3] | This Study |
| CP189 | met15Δ leu2Δ ura3Δ his3Δ pep4Δ::KanMX [2μ YCF1-Ser869Ala-GFP URA3] | This Study |
| CP190 | met15Δ leu2Δ ura3Δ his3Δ pep4Δ::KanMX [2μ YCF1-Ser870Ala-GFP URA3] | This Study |
| CP191 | met15Δ leu2Δ ura3Δ his3Δ pep4Δ::KanMX [2μ YCF1-Ser872Ala-GFP URA3] | This Study |
| CP192 | met15Δ leu2Δ ura3Δ his3Δ pep4Δ::KanMX [2μ YCF1-Ser873Ala-GFP URA3] | This Study |
| CP193 | met15Δ leu2Δ ura3Δ his3Δ pep4Δ::KanMX [2μ YCF1-Ser903Ala-GFP URA3] | This Study |
| CP194 | met15Δ leu2Δ ura3Δ his3Δ pep4Δ::KanMX [2μ YCF1-Ser914Ala-GFP URA3] | This Study |
| CP195 | met15Δ leu2Δ ura3Δ his3Δ CKA1-V5-His::KanMx | Open Biosystems |
| CP200 | met15Δ leu2Δ ura3Δ his3Δ pep4Δ::KanMX [2μ YCF1-Ser908Ala,Thr911Ala-GFP URA3] | This Study |
| CP224 | met15Δ leu2Δ ura3Δ his3Δ ckb1Δ::KanMX | Open Biosystems |
| CP225 | met15Δ leu2Δ ura3Δ his3Δ ckb2Δ::KanMX | Open Biosystems |
| CP226 | met15Δ leu2Δ ura3Δ his3Δ YCF1:TAP-HIS3 [2μ PGAL-GST-CKA2 URA3] | This Study |
| CP227 | met15Δ leu2Δ ura3Δ his3Δ YCF1:TAP-HIS3 [2μ PGAL-GST-CKB1 URA3] | This Study |
| CP228 | met15Δ leu2Δ ura3Δ his3Δ YCF1:TAP-HIS3 [2μ PGAL-GST-CKB2 URA3] | This Study |
| CP229 | met15Δ leu2Δ ura3Δ his3Δ ckb1Δ::KanMX [2μ YCF1--GFP URA3] | This Study |
| CP230 | met15Δ leu2Δ ura3Δ his3Δ ckb2Δ::KanMX [2μ YCF1-GFP URA3] | This Study |
| CP239 | met3Δ leu2Δ ura3Δ his3Δ ycf1::KanMX [2μ YCF1 - GFP URA3] | This Study |
| CP240 | met3Δ leu2Δ ura3Δ his3Δ ycf1::KanMX [2μ YCF1 – Ser251Ala-GFP URA3] | This Study |
| CP241 | met3Δ leu2Δ ura3Δ his3Δ ycf1::KanMX [2μ YCF1 – Ser251Glu-GFP URA3] | This Study |
All CP strains listed here are isogenic to CP59
CP59 is the Paumi lab isolate of BY4741b CP59 is the Paumi lab isolate of BY4741
The GST-Cka1p expressing strains, CP164 and CP181, were created by standard homologous recombination as described by Longtine et. al (Longtine, et al., 1998). In brief, the YCF1-TAP expressing strain (CP156) and non-expressing control strain (CP59) was purchased from Open Biosystems and confirmed by colony PCR analysis. The KanMX6-PGALGST fusion sequence was PCR amplified (using primers with 60 base pairs of homology to either the promoter region up to the ATG start codon (forward primer) or the ATG start codon + 57 bp downstream (reverse primer) of CKA1) from pFA6a-KanMX6-PGAL-GST and was subsequently gel purified and transformed into CP156 and CP59.
Plasmids
Plasmids used in this study are listed in Table 2. Phosphorylation site mutants of Ycf1p were created in the starting plasmid pCP58 [2μ YCF1-GFP URA3] using standard PCR-mediated mutagenesis followed by homologous recombination as described previously (Burke, 2000). Briefly primers were designed to make site specific mutations that would result in the phosphorylation site (almost exclusively serines) being changed to an alanine. Using a standard two-step PCR-mediated mutagenesis protocol for yeast (Burke, 2000), PCR products were generated containing the site specific mutations flanked by 200–300 base pairs of YCF1 homology on both the 5′ and 3′ ends. These PCR products and linearized/gapped pCP58 were gel purified and subsequently transformed into CP60. Single colonies containing mutant “healed” pCP58were selected on SC-URA plates. Plasmid were purified from yeast and sequenced to confirm the correctness of each mutation. PCR-mediated mutagenesis followed by homologous recombination resulted in the creation of pCP83, pCP84, pCP85, pCP94, pCP95, and pCP96 (YCF1 mutant plasmids Ser869Ala, Ser903Ala, Ser914Ala, Ser870Ala, Ser872Ala, and Ser873Ala, respectively).
Table II.
Plasmid used in this study
| Plasmid | Relevant Genotype | Reference |
|---|---|---|
| pCP25 (pFA6-) | KanMx6-PGAL1-GST | Longtine et. al. 1998 |
| pCP50 (pSM2243) | [2μ YCF1-Ser251Aala-GFP URA3] | Paumi et. al. 2008 |
| pCP58 (pSM1753) | [2μ YCF1-GFP URA3] | Mason et. al. 2002 |
| pCP74 | [2μ CKA2:MYC:6xHIS LEU2] | Schaerer-Brodbeck et. al. 2003 |
| pCP83 | [2μ YCF1-Ser869Ala-GFP URA3] | This Study |
| pCP84 | [2μ YCF1-Ser903Ala-GFP URA3] | This Study |
| pCP85 | [2μ YCF1-Ser914Ala-GFP URA3] | This Study |
| pCP94 | [2μ YCF1-Ser870Ala-GFP URA3] | This Study |
| pCP95 | [2μ YCF1-Ser872Ala-GFP URA3] | This Study |
| pCP96 | [2μ YCF1-Ser873Ala-GFP URA3] | This Study |
| pCP109 | [2μ PGAL-GST-CKA2 URA3] | Open Biosystems |
| pCP110 | [2μ PGAL-GST-CKB1 URA3] | Open Biosystems |
| pCP111 | [2μ PGAL-GST-CKB2 URA3] | Open Biosystems |
Growth Inhibition by Cadmium
Growth inhibition by CdSO4 and NaCl was monitored by spot tests as previously described (Mason & Michaelis, 2002, Mason, et al., 2003, Paumi, et al., 2007, Paumi, et al., 2008). Briefly, cells were grown overnight to saturation in SC-dropout media, subcultured at a 1:5000 dilution in the same medium and grown overnight to an OD600 of ~1.0. The overnight culture was diluted to an OD600 of 0.1, which in turn was diluted in 10-fold increments. Aliquots (4 μL) of each 10-fold dilution were spotted onto SC-dropout plates or S-GAL plates containing no drug (vehicle alone), 30 μM, 50 μM, or 200 μM CdSO4 and incubated for 2 or 4 days, respectively.
Synthesis and Purification of the Ser251 phosphospecific antibody, Ycf1p-Ser251-P
The synthesis and purification of the Ycf1p-Ser251-P phosphospecific antibody was created by Open Biosystems. In brief, a small peptide containing the Ycf1p-Ser251 CK2 consensus site, in a non-phosphorylated and a non-hydrolizable form (CKLPRNFSSEELSQ and CKLPRNF(pS)SEELSQ, respectively), were synthesized using standard techniques, and conjugated to KLH. Two rabbits were injected with the phosphorylated version of the synthetic peptide over a 90 day period (Open Biosystems 90 day protocol). Serum was collected at 3 separate times (Day 0, Day 45, and Day 90) and shipped. The specificity of each aliquot was tested by immunoprecipitation and western blotting for Ycf1p. One rabbit was found to produce Ycf1p specific antibodies. Serum containing Ycf1p-specific antibody was then subjected to negative affinity chromatography as per Open Biosystems standard protocol, and the resulting purified eluent was shipped. The eluent was tested for specificity against the Ycf1p WT protein and the Ser251Ala mutant at various dilutions (a test of linearity against serial dilutions of WT and the Ser251Ala mutant) and found to be specific for phosphorylation at Ser251 at a 1:500 dilution. Flow through from the negative selection by affinity chromatography was retained and resulted in an Ycf1p specific antibody (α-Ycf1p) that is not sensitive to the phosphorylation state of Ser251.
Western blotting and Co-immunoprecipitation Analysis of V5, GFP, and TAP- tagged proteins
Cell extracts were prepared and western blots of WT and mutant Ycf1p were performed as previously described (Wemmie & Moye-Rowley, 1997) with the following modifications: 50 mL of cells grown in log phase to an OD600 of 0.8 to 1.0 OD600 units were harvested and resuspended in 100 μL of RIPA buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40, and 0.5% deoxycholate) containing 1% SDS and fresh protease inhibitors (Roche Complete protease inhibitor cocktail, PMSF (1μM), and pepstatin A (5 μg/ml)) and broken with glass beads (10× 1 min vortexing and 1 min on ice). Following bead-beating, 900 μL of RIPA buffer plus protease inhibitors without SDS was added and the broken cells were microfuged at 2000 rpm for 5 minutes at 4°C to remove debris. The protein concentration in the supernatant was measured by Bradford Assay. For standard western blot analysis, 12.5 μg of total protein was solubilized in an equal volume of 2x SDS-PAGE sample buffer (50 mM Tris-HCl pH 6.8, 8 M urea, 0.1 mM EDTA, 15% SDS, 0.01% bromophenol blue, and 150 mM DTT added fresh), frozen overnight, and subsequently incubated at 37°C for 1 hour prior to SDS-PAGE and transfer to nitrocellulose. For western blot analysis of GFP-tagged Ycf1p from total protein extracts, membranes were blocked with 1% BSA for 1 hour and then incubated overnight with primary monoclonal mouse anti-GFP (1:1000) (Roche), monoclonal anti-V5 (1:5000) (Invitrogen) or monoclonal mouse anti-beta actin (1:5000) (Abcam), washed 3x times with TBST, and then incubated with the respective secondary antibody, sheep anti-mouse (1:2000). For TAP tagged Ycf1p, blots were probed with 1:1000 α-TAP (Open Biosystems) and for immunoprecipitations, cell lysates in RIPA containing Ycf1p-TAP were incubated with human α-IgG beads directly overnight. Alternatively, immunoprecipitations of Ycf1p were probed with the anti-Ycf1p-Ser251-P phosphospecific antibody (1:500) and an anti-rabbit secondary (1:5000). Protein was visualized using the chemiluminescent reagent (Denville Scientific) and quantitated using Adobe Photoshop (Miller, 2007, Luhtala & Parker, 2009).
Co-immunoprecipitation were performed as described previously (Paumi, et al., 2007) with the following modifications. Strains were grown overnight in a 500 mL volume to an OD600 of 0.8. Cells were collected by centrifugation, washed once with dH2O, and subjected to lysis by bead beating as previously described (Paumi, et al., 2007). One mg of protein was immunoprecipitated with 25 μL of anti-IgG agarose beads (Sigma) overnight at 4°C or alternatively overnight with 30 μL of anti-Ycf1p in combination with 30 μL of Protein A/G beads (Santa Cruz Biotechnology). After overnight incubation, beads were washed 4x with ice cold IP Lysis Buffer containing a complete set of protease inhibitors. The washed beads were then resuspended in an equal volume of SDS-PAGE Sample buffer, heated at 65°C for 10 minutes, and then subjected to SDS-PAGE and western blot analysis. For western blotting, membranes were blocked with 3% BSA for 1 hour, incubated overnight with anti-TAP (1:1000) (Open Biosystems), anti-GST (1:200) (Open Biosystems), anti-V5 (1:5000) (Invitrogen), anti-Ycf1p (1:500), or anti-Ycf1p-Ser251-P (1:500) and subsequently probed with anti-mouse or anti-rabbit secondary (1:5000) for 1 hour. Chemiluminescence (Denville Scientific) was used for protein detection.
In vitro Ycf1p-mediated transport assay
Yeast vacuoles were isolated and purified essentially as previously described (Roberts, et al., 1991, Paumi, et al., 2007), and Ycf1p-dependent transport was assayed by measuring the amount of the radiolabeled Ycf1p substrate, [3H]-estradiol-β-17-glucuronide ([3H] E2β17G), transported from outside to inside the vesiculated vacuoles. Transport assays were conducted as previously described (Paumi, et al., 2007) with minor modification as follows: 100 μL reaction volumes containing 4 mM of either NaATP or NaAMP-PCP; 10 mM MgCl2; 5 μM gramicidin-D; 10 mM creatine phosphate and 16 units/mL creatine kinase; 50 mM KCl; 400 mM Sorbitol; 25 mM Tris-Mes (pH 8.0); and the Ycf1p substrate [3H]-estradiol-17-glucaronide (Amersham, Piscataway, NJ) at a concentration of 400 μM. Reactions were initiated by the addition of 1–2 μg of purified vacuoles, incubated at 25°C for 5 or 10 minutes, and stopped by the addition of 1 mL of ice cold stop buffer (400 mM sorbitol and 3 mM Tris/Mes (pH 8.0)). Vacuoles were collected on 0.22 μm GV filters (Millipore) via vacuum manifold and washed. Disintegration per minute was measured (,the amount of Ycf1p levels were determined by western blotting (to normalize to protein level), and transport was reported as nmoles of substrate ([3H]-E217G) sequestered within the vacuoles per mg of total protein over 10 minutes (Paumi, et al., 2007).
Synthesis of Synthetic Peptides
All peptides were assembled by Fmoc solid phase peptide synthesis (SPPS) on a TETRAS peptide synthesizer (CreoSalus; Louisville, KY). In brief, the peptides were synthesized using Wang resins substituted with the first amino acid. Amino acid deprotection and coupling then followed standard methods to extend the peptide sequence as designed. Biotin was incorporated at the N-terminus via standard coupling with the exposed carboxylic acid functionality. Upon completion of assembly, the resin was washed with DMF, MeOH and CH2Cl2, and then dried. The peptide was cleaved from the resin by gentle mixing of the resin with a mixture of 6% phenol, 4% ddH2O, 90% trifluoroacetic acid for 5 h and the resin was filtered away. The peptides were precipitated by the addition of cold diethyl ether and dried under vacuum. The crude peptide was purified by reversed phase HPLC (ddH2O/AcCN, 0.5% TFA) and MALDI-TOF confirmed the mass of each of the peptides: (PKA-AcBr, [M+H], calc 2082.19, found 2082.62, calculated mass and experimentally determined mass match and confirm the synthesis of the crude peptide). Peptide synthesis yielded 5 peptides for use in the recombinant Cka1p, Cka2p, and hCK2α kinase assays. The 3 peptides synthesized were biotin-Ahx-RRRADDSDDDDDK (Control), biotin-Ahx-LPRNFSSEELS (Ycf1p-Ser251 peptide, Ser251 is underlined), and biotin-Ahx-LPRNFASEELS (Ycf1p-Ser251Ala peptide, Ala251 is underlined). Electrospray mass spectrometry confirmed the mass of each peptide.
Recombinant purification of Cka1p and Cka2p and Kinase Assays
In vitro kinase assays were used to examine the ability of recombinant Cka1p and Cka2p to phosphorylate synthetic short peptides derived from Ycf1p containing the Ser251 consensus sequence. Yeast strains expressing His tagged Cka1p and Cka2p (CP174 and CP176, respectively) were gifts from M.S. Szczypka and H. Riezman (Schaerer-Brodbeck & Riezman, 2003, Kubinski, et al., 2007). Recombinant Cka1p and Cka2p were purified by single step affinity chromatography or two step affinity chromatography as previously described (Kubinski, et al., 2007). In brief, cells were grown to an OD600nm of 0.8–1.0 and cells were harvested by centrifugation at 3000 × g. Cell pellets were washed 2x with ice cold ddH2O and subsequently resuspended in lysis buffer (10 mM HEPES pH 7.6, 150 mM NaCl, 5 mM MgCl2, 1 % Triton X-100, and PI’s (Protease Inhibitor cocktail complete from Roche, Pepstatin A, and Fresh PMSF). Cells were lysed by repeated bead beating. Cell lysates were spun at 500 × g for 10 min to precipitate the cellular debris. The supernatant was incubated overnight at 4°C with Bio-Rad profinity nickel affinity chromatography resin (Bio-Rad, Hercules, CA). Post overnight incubation, resin was washed 4x with lysis buffer and kinases were eluted (in 0.5 mL aliquots) by addition of lysis buffer containing 500 mM immidazole. Eluates were subjected to SDS-PAGE and coomassie staining was used to evaluate protein purification. Eluates containing the highly enriched recombinant kinases were then either subject to dialysis buffer (50 mM Tris HCl pH 7.5, 200 mM NaCl) in the case of Cka1p or subject to further purification by size-exclusion chromatography and subsequent dialysis in the case of Cka2p (Kubinski, et al., 2007).
Kinase assays were carried-out using a modification of the Sigma-Aldrich Biotin-substrate tagged method. In brief, radiometric assays were performed in 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 0.5 mM DTT, 100 ng/mL BSA, 100 μM ATP, 2 μL of [32P]-γ-ATP (10 Ci/mmole) (Perkin Elmer, Waltham, MA), and 200 mM NaCl using 250 μM biotinylated synthetic peptide (varying concentration of peptide for Michaelis-Menten kinetic analysis) as the substrate in a 100 μL reaction volume (This sentence needs restructured and clarified). Reactions were initiated by the addition of 1.6 μg of Cka1p enzyme and carried-out for varying amounts of time (0, 1, 2, 5, and 10 min) or a fixed time of 5 minutes at 30°C. Reactions were stopped by the addition of 4 μL of 0.5 M EDTA and 4 μL of 250 mg/mL avidin (Thermo Scientific-Pierce Protein Division, Rockford, Il). After addition of the stop solution the reactions were incubated for 5 minutes. After the 5 minute incubation, the terminated reaction aliquots were added to 30 kDa spin column (Pall Scientific, East Hills, NY), spun at 14,000 rpm for 5 min, and subsequently washed 2x with 100 μL wash solution (0.5 M Phosphate, 0.5 M NaCl, pH 8.5). Spin columns were submerged directly into scintillation vials containing 12 mL of scintillation cocktail and cpms of the phosphorylated peptides were counted via a Perkin Elmer scintillation counter. All experiments were normalized to a zero time point, performed in triplicate, and results were reported as nmoles of phosphorylated peptide formed per minute (or per 5 minutes) of reaction per milligram of recombinant protein.
Measurement of Ycf1p-dependent vacuole sequestration of MCB-SG and ade2Δ-associated red pigment accumulation
To measure Ycf1p function in vivo, Ycf1p-dependent formation of the ade2Δ-associated red pigment was measured as previously described (Paumi, et al., 2007). Red pigment accumulation was measured on a 730 UV/VIS Spectrometer (Beckman-Coultier, Brea, CA, USA) and normalized to total protein. Results are reported as Relative Red Pigment accumulation per mg of total protein. The red pigment accumulation assay was performed in triplicate. Similarly, monochlorobimane (MCB)-glutathione formation and Ycf1p-dependent sequestration was analyzed as previously described (Gulshan, et al., 2009). MCB is not itself fluorescent until conjugated to GSH, upon which it is immediately sequestered into the vacuole by Ycf1p. Cells were treated with MCB (Sigma-Aldrich, St. Louis, MO) as previously reported and analyzed by fluorescent microscopy (Li, et al., 1996, Gulshan, et al., 2009). Analysis was done on three separate samples and pooled until a total of 200 cells were counted.
Fluorescence Microscopy
To examine the localization of Ycf1p-GFP fusion proteins, cells were grown overnight to saturation in minimal media and then subcultured at a 1:1000 dilution in minimal medium and grown overnight to an OD600 of ~0.7. Cells were examined at 100X magnification on poly-lysine-coated slides using an Olympus BX512F equipped with fluorescence (EXFO X-cite 120 Fluorescence Illuminating System) and Nomarski optics. Images were captured with a QImaging Qicam Fast 1394 camera and digitally documented using Q-Capture Pro software (Paumi, et al., 2007, Paumi, et al., 2008).
Results
Cka1p-dependent phosphorylation at Ser251 exclusively regulates Ycf1p function
Genetic evidence suggests that Cka1p negatively regulates Ycf1p function via phosphorylation at Ser251 (Paumi, et al., 2008). However, it is possible that Cka1p may phosphorylate and thus regulate Ycf1p function through additional sites. Recently, three different groups have conducted phosphopeptide specific LC\MS\MS analysis of the entire yeast proteome in an effort to identify global phosphorylation changes under a variety of stresses (Li, et al., 2007, Smolka, et al., 2007, Albuquerque, et al., 2008). Each group identified multiple phosphorylated peptides derived from Ycf1p within their initial control screens. A phosphopeptide containing Ser251 was found and other phosphopeptides identified eight additional sites (Ser869, Ser870, Ser872, Ser873, Ser903, Ser908, Thr911, and Ser914) within the linker domain of Ycf1p (Li, et al., 2007, Smolka, et al., 2007, Albuquerque, et al., 2008). The phosphorylation sites at Ser908 and Thr911 were previously identified and characterized to be required for complete Ycf1p function (Li, et al., 1996, Li, et al., 1997, Eraso, et al., 2004).
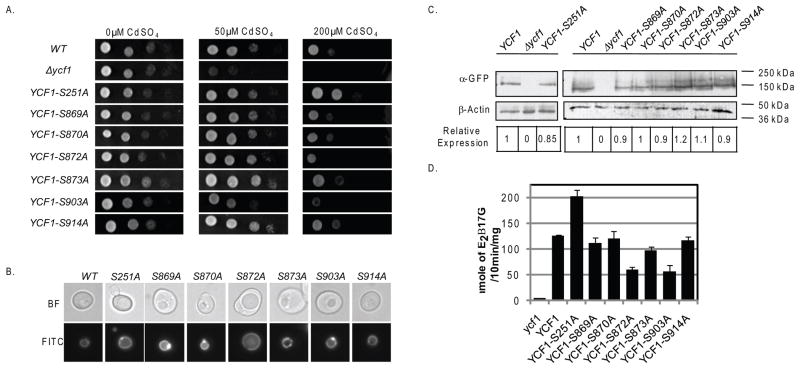
To rule out the possibility that Cka1p is regulating Ycf1p function through the phosphorylation of Ser251 and/or one or more sites within the linker domain, we compared the function of Ycf1p phospho-blocking alanine mutations (at Ser251 and the potential phosphorylation sites within the linker domain) to wild type (WT). We measured the activity of each mutant by cellular growth and biochemical Ycf1p-dependent transport assays (Fig. 2A and 2D). Previously, Eraso et. al. has shown that mutation of Ser908 and Thr911 to alanine resulted in decreased growth on cadmium and decreased Ycf1p-dependent transport activity (Eraso, et al., 2004). Therefore, we restricted our work to the other six uncharacterized phosphorylation sites, Ser869, Ser870, Ser872, Ser873, Ser903, and Ser914.
Figure 2.
To determine the ability of linker domain phosphorylation site mutants to grow on cadmium, we spotted 10-fold dilutions of each strain onto agar plates containing a range of cadmium concentrations (0, 50, and 200 μM) and allowed the strains to grow for varying lengths of time. We expected that if a phosphorylation site plays a role in Cka1p-dependent Ycf1p regulation then the corresponding alanine mutant would grow similar to the Ser251Ala Ycf1p mutant, which displays increased growth on cadmium compared to WT (Fig. 2A, compare rows 1 and 3). Our results showed that none of the strains expressing the 6 phosphorylation site mutants had increased growth as compared to WT (Fig. 2A, compare rows 1 to 4–9). Additionally, we found the strain expressing Ycf1p-Ser903Ala has a decreased ability to grow on cadmium (Fig. 2A, compare rows 1 and 8), a phenotype similar to that of the Ycf1p-Ser908Ala, Ycf1p-Thr911Ala, and the Ycf1p-Ser908Ala,Thr911Ala double mutant(Eraso, et al., 2004). It is worth noting that Ycf1p-Ser873Ala and Ycf1p-Ser914Ala have a very minimal, yet notable, increased resistance to the low concentration of cadmium (50μM) (Fig. 2A, 2nd column, compare rows 1 and 7 and 1 and 9). If phosphorylation of Ycf1p-Ser873 and Ycf1p-Ser914 play role in negatively regulating Ycf1p function similar to Ycf1p-Ser251, then we would expect that the alanine mutants would have increased resistance to high concentration of cadmium (200μM) as compared to WT, similar to Ycf1p-Ser251Ala (Fig. 2A, 3rd column, compare rows 1 and 7 and 1 and 9). However this is not the case and therefore suggests a more subtle phenotype that may be important under conditions not tested here. To confirm that our growth assay results were not due to mislocalization of the transporter, we tagged each mutant at the C-terminus with GFP and confirmed their correct localization to the vacuole rim similar to WT Ycf1p (Fig. 2B, compare panel 1 to panels 2–7).
In order to determine the effect that each phosphorylation site mutation has on Ycf1p function, we conducted in vitro transport assays as described in the Materials and Methods (Fig. 2D). Transport activity was normalized to protein expression (Fig. 2C). We reasoned that if a phosphorylation site plays a role in Cka1p-dependent regulation of Ycf1p function then mutation of the site to alanine would result in increased transporter activity as compared to WT, an effect similar to that of the Ycf1p-Ser251Ala mutant (Fig. 2D, compare lane 2 to 3) (Paumi, et al., 2008). In comparison, vesiculated vacuoles derived from strains expressing the linker domain phosphorylation site mutants did not have increased Ycf1p-dependent transporter activity similar to the Ycf1p-Ser251Ala mutant vacuoles (Fig. 2D, compare lane 3 to lanes 4–9). The transport activity of vacuoles derived from strains expressing the linker domain phosphorylation mutants, Ycf1p-Ser869Ala, Ycf1p-Ser870Ala, Ycf1p-Ser873Ala, and Ycf1p-S914Ala, are, in fact, similar to that of vacuoles derived from a WT strain (Fig. 2D, compare lane 2 to lanes 4,5,7, and 9). The activity of Ycf1p-Ser872Ala and Ycf1p-Ser903Ala, is less than that of WT (Fig. 2D, compare lane 2 to lanes 6 and 8). The phenotype of Ycf1p-Ser872Ala and Ycf1p-Ser903Ala is similar to the phenotype shown here for the double mutant Ycf1p-Ser908Ala,Thr911Ala and the phenotype previously reported for the single mutants Ycf1p-Ser908Ala and Ycf1p-Thr911Ala, and the double mutant Ycf1p-Ser908Ala,Thr911Ala (Eraso, et al., 2004). Together these results suggest that Ser869, Ser870, Ser872, Ser873, Ser903, and Ser914, within the linker region, do not play a role in Cka1p-dependent negative regulation of Ycf1p function.
Cka1p is required for the phosphorylation of Ycf1p-Ser251 in vivo
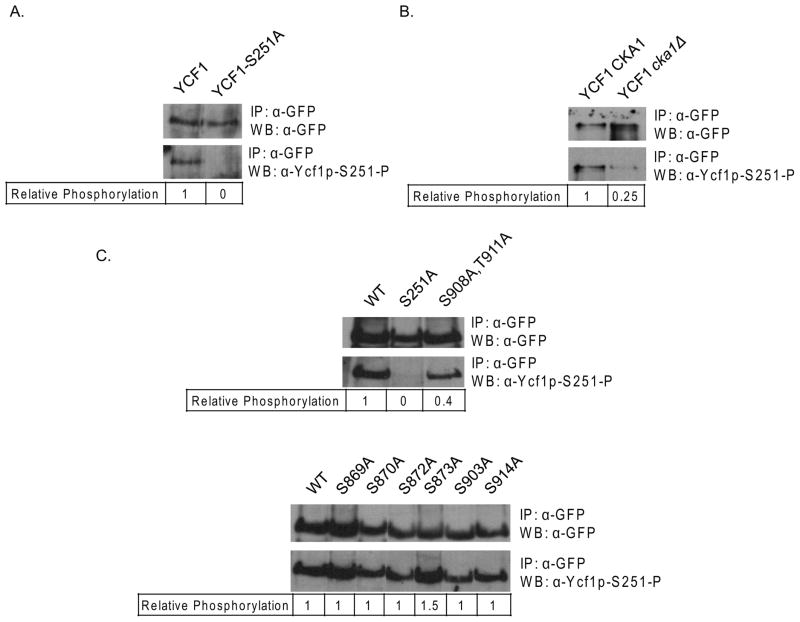
Although the study by Paumi et. al. 2008 provides substantial support for direct phosphorylation of Ycf1p by Cka1p, the direct phosphorylation of Ser251 by Cka1p has yet to be confirmed. To determine if indeed Cka1p does regulate Ycf1p function via direct phosphorylation of Ser251, a phosphospecific antibody to Ser251 was synthesized, Ycf1p-Ser251-P (see Materials and Methods). To test the specificity of our antibody, WT Ycf1p and Ycf1p-Ser251A were immunoprecipitated and subjected to western blot analysis with anti-Ycf1p-Ser251-P (Fig. 3A). Anti-Ycf1p-Ser251-P clearly recognized WT and was unable to recognize the band corresponding to Ycf1p-Ser251A. This result suggests that the α-Ycf1p-Ser251-P phosphospecific antibody is specific for phosphorylated Ycf1p-Ser251. To determine if deletion of the Cka1p gene eliminates phosphorylation at Ser251, as suggested by Paumi et. al. 2008, immunoprecipitations of Ycf1p from both a WT and cka1Δ strain were subjected to western blot analysis with anti-Ycf1p-Ser251-P (Fig. 3B). Our experiments revealed that deletion of cka1Δ results in near complete elimination of Ser251 phosphorylation in vivo. These results suggest phosphorylation of Ser251 plays an important role in the mechanism by which Cka1p-negatively regulates Ycf1p function.
Figure 3.
To determine if Cka1p-dependent phosphorylation of Ser251 is regulated in part by phosphorylation sites within the linker region of Ycf1p immunoprecipitations of each of the phosphorylation mutants (Fig. 2) were subjected to western blot analysis with the anti-Ycf1p-Ser251-P antibody (Fig. 3C). We reasoned that if the linker domain phosphorylation sites are involved in Cka1p-dependent regulation of Ycf1p, then mutation of the site(s) should result in altered phosphorylation of Ser251. To this end, our experiment revealed that phosphorylation of Ser251 is not altered by mutation of phosphorylation sites within the linker domain. However, we found one exception, the double mutant Ser908Ala,Thr911Ala. It has been previously shown that phosphorylation of Ycf1p-Ser908,Thr911 is required for Ycf1p transport activity and an Ycf1p-Ser908Ala,Thr911Ala mutant is a dominant mutation over the Ser251Ala mutation (Eraso, et al., 2004, Paumi, et al., 2008). Our finding suggests that the phosphorylation at Ser908 and Thr911 may be required for complete Cka1p-dependent phosphorylation of Ser251 and Cka1p regulation of Ycf1p function.
Cka1p, Cka2p, Ckb1p, and Ckb2p directly interact with Ycf1p
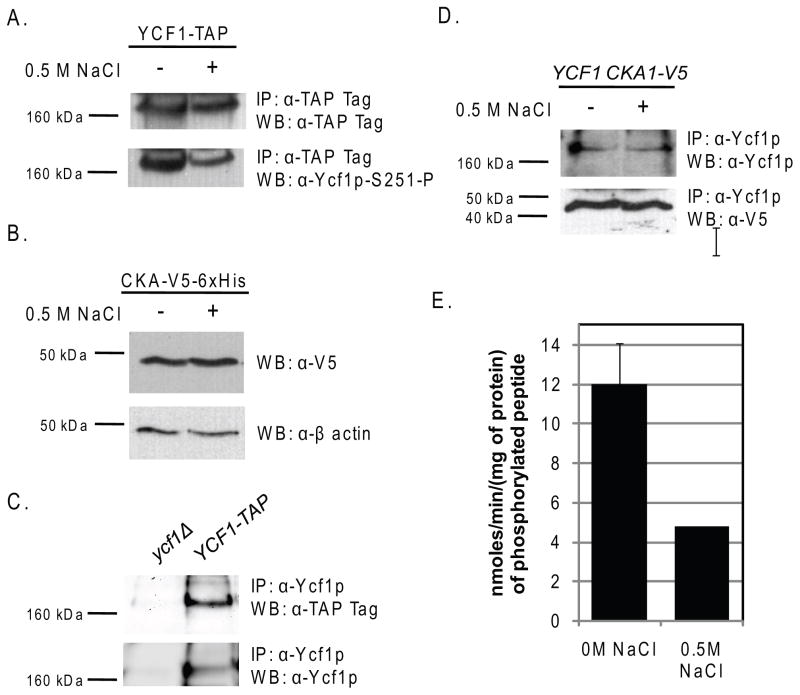
Cka1p was first identified as an interacting partner of Ycf1p using the integrated membrane yeast two hybrid (iMYTH) assay. This result was never confirmed but was supported by a number of in vivo and in vitro experiments that showed that Cka1p functionally interacted with Ycf1p (Paumi, et al., 2008). As is the case for most kinases, the interaction of a kinase with a substrate is very transient. The transient nature of kinase/substrate interactions, in combination with the relatively low cellular levels of many kinases, makes confirmation of kinase-protein interactions by traditional co-immunoprecipitations without overexpression of the kinase quite difficult.
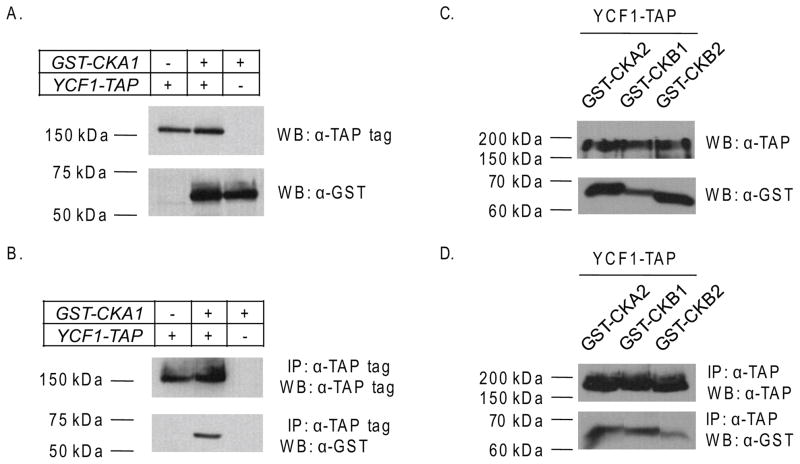
For a secondary analysis of the Cka1p and Ycf1p physical interaction, we performed a co-immunoprecipitation analysis. To increase our chances of seeing a possible interaction we utilized a chromosomally TAP-(Tandem Affinity Purification)tagged version of Ycf1p available through Open Biosystems. We used this strain to create a YCF1-TAP, GST-CKA1 double tag strain for subsequent co-immunoprecipitation analysis (described in Materials and Methods), a similar strain was made expressing GST-Cka1p alone for use as a control. Cka1p was N-terminally fused to GST and expressed under the control of a galactose inducible promoter to increase the overall expression levels. We confirmed that both Cka1p and Ycf1p were properly tagged and expressed by western blot analysis (Fig. 4A). Co-immunoprecipitation experiments were carried-out as described in the Materials and Methods. We treated equal amounts of cell lysates from all three strains with IgG agarose beads to capture Ycf1p-TAP and any binding partners. Analysis of lysates prepared by this method revealed that in the strain expressing both the Ycf1p-TAP and GST-Cka1p tagged proteins, Cka1p was present after immunoprecipitation of Ycf1p (Fig. 4B, lane 2). This result was not due to non-specific binding as shown by the co-immunoprecipitation of lysates derived from a GST-Cka1p alone or an Ycf1p-TAP alone strain (Fig. 4B, lane 1 and 3). Together with the iMYTH results, our data provide further supporting evidence that Ycf1p and Cka1p interact in vivo.
Figure 4.
It has long been appreciated that CK2 has both a regulatory subunit and a catalytic subunit (Poole, et al., 2005). The catalytic subunit can exist as a heterodimer of CK2α (Cka1p) and CK2α′ (Cka2p) or as a homodimer of either protein bound to a regulatory dimer, CK2β (Ckb1p) and CK2β′ (Ckb2p) in yeast (Domanska, et al., 2005, Poole, et al., 2005, Kubinski, et al., 2007). A number of recent studies have shown that CK2 specificity is in part dictated by the composition of the catalytic dimer (Domanska, et al., 2005, Berkey & Carlson, 2006, Kubinski, et al., 2007). These studies suggest that CK2α and CK2α′ substrate specificity is overlapping yet distinct (Domanska, et al., 2005, Berkey & Carlson, 2006, Kubinski, et al., 2007). Therefore, the composition of the catalytic dimer and the ratio of the different possible CK2 dimers in the cytosol are likely to dictate what substrates are phosphorylated.
To determine if the other subunits of the CK2 holoenzyme interact with Ycf1p, we performed a similar set of co-immunoprecipitation experiments for Cka2p, Ckb1p, and Cbk2p as we did for Cka1p (described above), however it is important to note that the Cka2p, Ckb1p, and the Ckb2p proteins were expressed from a plasmid rather than from the chromosome. This was done to ensure that we would have the best chances of identifying an interaction with Ycf1p. Total protein expression analysis confirmed that indeed all three proteins are expressed (Fig. 4C). Our results showed that all three of the CK2 holoenzyme subunits, Cka2p, Ckb1p, and Ckb2p, interact with Ycf1p (Fig. 4D). These results suggest that the CK2 holoenzyme does indeed interact with Ycf1p.
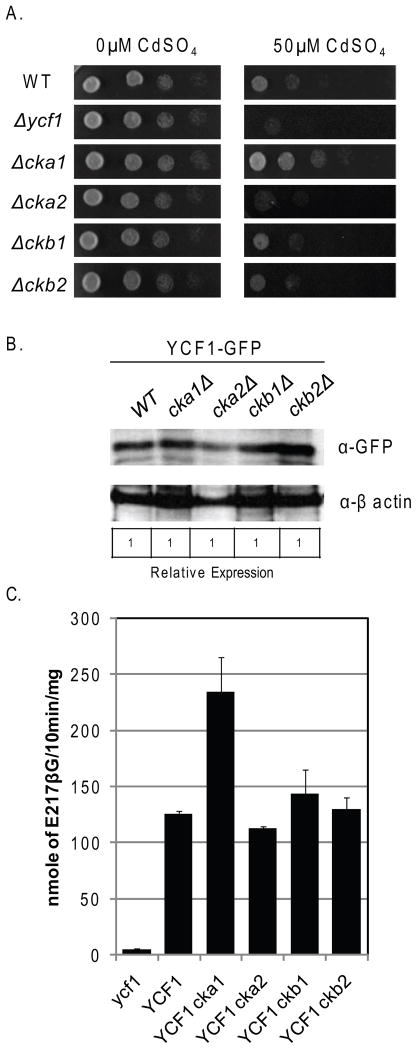
Suppression of Ycf1p function by Cka1p does not require Cka2p, Ckb1p, or Ckb2p
Interestingly, Cka2p, Ckb1, or Ckb2 were not identified as interacting partners for Ycf1p using iMYTH (Paumi, et al., 2008). However, our results above suggest the possibility that the intact CK2 holoenzyme is required for Cka1p-dependent regulation of Ycf1p. To determine if Cka2p, Ckb1p, and Ckb2p functionally interact with Ycf1p, we examined how deletion of cka2Δ, ckb1Δ, and ckb2Δ affects Ycf1p-dependent resistance to cadmium in vivo by dilution spot test growth assays and by Ycf1p-dependent in vitro transport activity assays (Fig. 5).We reasoned that if Cka2p, Ckb1p, or Ckb2p play a role in the negative regulation of Ycf1p by Cka1p then a strain deleted for any of the three proteins would phenotypically be similar to a cka1Δ strain and would have increased resistance to cadmium compared to WT (Fig. 5A, compare row 1 and 2, and 1 and 3), however this was not the case. Deletion of cka2Δ increases strain sensitivity to cadmium over that of a WT strain and deletion of ckb1Δ and ckb2Δ has no effect on strain sensitivity to cadmium (Fig. 5A, compare row 1 and 4, row 1 and 5, and row 1 and 6). This result suggests that Cka1p does not require Cka2p, Ckb1p, or Ckb2p function to regulate Ycf1p activity in vivo. Further, since CK2 holoenzyme assembly requires both Ckb1p and Ckb2p (Kubinski, et al., 2007), this result suggests that Cka1p alone is sufficient to negatively regulate Ycf1p function.
Figure 5.
To determine if deletion of cka2Δ, ckb1Δ, and ckb2Δ directly affects the transport activity of Ycf1p we carried-out in vitro transport assays as described above and in the Materials and Methods. Transport activity was normalized to Ycf1p expression (Fig. 5B). We reasoned that if Cka2p, Ckb1p, or Ckb2p played a role in Cka1p-dependent regulation of Ycf1p activity, then vesiculated vacuoles derived from a cka2Δ, ckb1Δ, and ckb2Δ strain would have increased Ycf1p-dependent transport activity as compared to WT, similar to that of vacuoles derived from a cka1Δ strain (Fig. 5C, compare lane 2 and 3). However, similar to our growth assay findings, this is not the case. Vacuoles derived from a cka2Δ strain have slightly less activity than vacuoles derived from a WT strain and vesiculated vacuoles from ckb1Δ and ckb2Δ had similar transport as WT (Fig. 5C, compare lanes 2 and 4, 2 and 5, and 2 and 6, respectively). This result is in agreement with the results from our growth analysis and strongly suggests that Cka2p, Ckb1p, and Ckb2p are not required for the suppression of Ycf1p function by Cka1p.
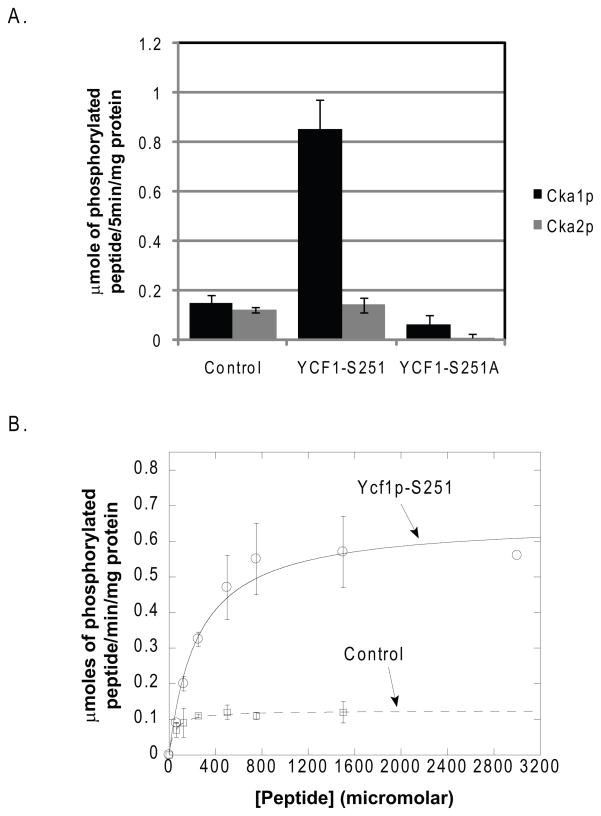
Cka1p is sufficient to phosphorylate Ser251 in vitro
Our results confirm the physical interaction of Cka1p with Ycf1p and support the claim that Cka1p negatively regulates Ycf1p function via the direct phosphorylation of Ser251. Mutation of all the identified phosphorylation sites in the Ycf1p linker region did not result in increased growth on cadmium or increased Ycf1p-dependent transport activity as is the case for the Ser251Ala or cka1Δ strains. Together, these studies suggest that Cka1p negatively regulates Ycf1p function solely via the direct phosphorylation of Ser251. To determine if this is the case, we carried-out a series of kinase assays with recombinant Cka1p and Cka2p as described in the Materials and Methods. Kinase assays were performed in the presence of a known substrate that has been optimized for human CK2 phosphorylation, a peptide derived from Ycf1p containing Ser251, and a control Ycf1p peptide containing the Ser251Ala mutation. Results of these experiments are shown in Fig. 6A. Incubation of Cka1p and Cka2p in the presence of radiolabeled ATP resulted in the phosphorylation of the control peptide (Fig. 6A, lanes 1 and 2) suggesting that our recombinant proteins are functional. Incubation of each kinase with the peptide derived from Ycf1p containing the Ser251 phosphorylation site revealed that both Cka1p and Cka2p can phosphorylate Ser251 but Cka1p phosphorylates the peptide about 9 times more efficiently (Fig. 6A, lanes 3 and 4, respectively). Kinase assays conducted in the presence of the Ycf1p-Ser251Ala-derived peptide showed no Cka2p-dependent phosphorylation and negligible Cka1p activity (Fig. 6A, lanes 6 and 5, respectively). Together these results suggest Cka1p negatively regulates Ycf1p function via the direct phosphorylation of Ser251 and support our previous results that show that Cka2p is not required for Cka1p-dependent Ycf1p regulation.
Figure 6.
To determine the kinetics of Cka1p-mediated Ycf1p-Ser251 peptide phosphorylation we conducted kinase assays with recombinant Cka1p in the presence of varying concentrations of either the Ycf1p-Ser251 peptide or the CK2 control optimized peptide substrate. Michaelis-Menten analysis showed that Cka1p has a Km of 46 μM and a Vmax of 0.12 μmoles/min/mg for the control peptide (Fig. 6B, dashed line) and a Km of 245 μM and a Vmax 0.65 μmoles/min/mg for the Ycf1p-Ser251 peptide (Fig. 6B, solid line). The Km and Vmax for Cka1p with the control peptide are in agreement with previously published CK2α kinase assays and confirms that Cka1p is functioning properly an in accordance with previous study findings (Marin, et al., 1994). Collectively, our results indicate that Cka1p phosphorylates Ser251 in vivo.
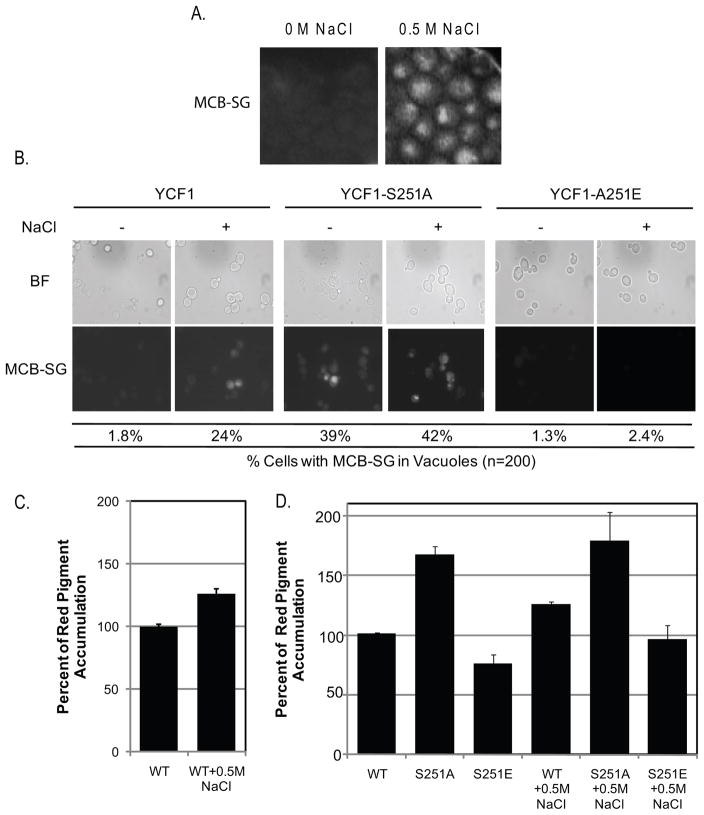
Cka1p regulation of Ycf1p is attenuated in response to salt stress
In a recent study, Soufi et. al. conducted a global analysis of the yeast osmotic stress response by quantitative proteomics and found peptides corresponding to Ycf1p-Ser251 (Soufi, et al., 2009). A comparison of the control, untreated sample with that of the high salt-treated sample suggested that phosphorylation of Ycf1p-Ser251 is decreased in response to salt stress (Soufi, et al., 2009). To determine the cellular role of the suppression of Ycf1p by Cka1p in response to salt stress, we characterized the effect of salt stress on Ycf1p-Ser251 phosphorylation status and Ycf1p function (Fig. 7 and 8). Interestingly, previous studies have reported a role for CK2 in the regulation of a cellular response(s) and the regulation of cellular resistance to salt stress (Kanhonou, et al., 2001, Hermosilla, et al., 2005, Chiang, et al., 2007). Therefore, we determined whether Ycf1p function is upregulated in response to salt stress via attenuation of Cka1p-dependent phosphorylation of Ser251.
Figure 7.
Figure 8.
To begin, we determined the phosphorylation status of Ycf1p-Ser251 in cells treated with 0.5 M NaCl as compare to untreated control cells (Fig. 7A) as described in the Materials and Methods. In agreement with the results obtained from Soufi et. al., phosphorylation of Ser251 is dramatically reduced (within 20 minutes) in response to high concentrations of salt as compared to untreated controls (Fig. 7A) (Soufi, et al., 2009). The decrease in Ycf1p-Ser251 phosphorylation in response to salt stress is responsive to the concentration of salt as phosphorylation decreases from 0M to 0.25M NaCl (Supp. Fig. 1) at which salt stress mediated reduction in the phosphorylation of Ser251 appears to hit a maximum. This result suggested that Ycf1p-Ser251 phosphorylation may be reduced via a change in Cka1p expression, localization, and/or function. To determine if phosphorylation of Ycf1p-Ser251 plays a role in the general regulation of Ycf1p in response to cell stress, we examined Ycf1p-Ser251 phosphorylation after treatment with 50μM cadmium for 1hr (described in detail in the Materials and Methods) (Supp. Fig. 2). These results showed that cadmium treatment does not alter Ycf1p-Ser251 phosphorylation status, but does increase Ycf1p expression. This change in expression of Ycf1p post cadmium treatment is in agreement with what has been previously reported (Li, et al., 1997) and suggests that regulation of Ycf1p-Ser251 via phosphorylation by Cka1p is not a general response to all cell stressors. In future experiments aimed at determining the cellular signals that are upstream of Cka1p we will examine more closely the role of Cka1p-mediated regulation of Ycf1p function via phosphorylation of Ser251 under multiple combinations of cellular insults. Therefore, we examined Cka1p expression in response to salt stress and found that expression of Cka1p was not altered (Fig. 7B). To determine if a change if Ycf1p-Cka1p protein interaction is responsible for the reduction in Ser251 phosphorylation, we carried-out Ycf1p co-immunoprecipitation experiments. To ensure interaction specificity, we immunoprecipitated endogenously expressed Ycf1p from a yeast strain expressing a V5 tagged version of Cka1p from its endogenous promoter with an Ycf1p specific antibody (Fig. 7C). Subsequently, as shown in Fig. 7D, we determined that the Ycf1p-Cka1p interaction was identical in both untreated and salt-treated cells. Therefore, we examined Cka1p function in response to salt stress (Fig. 7E). Our in vitro kinase assays that were carried-out with extracts derived from both untreated and 0.5 M NaCl-treated cells shows that Cka1p activity is reduced by about 60% in response to salt stress (Fig. 7E). Together these results suggest that Ycf1p-Ser251 phosphorylation is decreased in response to salt stress due to attenuated Cka1p function.
Importantly, our findings that the activity of Cka1p is reduced, not only correlated with a decrease in Ycf1p-Ser251 phosphorylation but also with an increase in Ycf1p function (Fig. 8A and 8C). Here we show, by microscopy, that Ycf1p-dependent vacuole accumulation of a known substrate, monochlorobimane-glutathione (MCB-SG), is increased (evident by an increase in fluorescence within the vacuoles of treated cells versus control untreated cells, Fig. 8A). In addition, the Ycf1p-dependent red pigment accumulation associated with the ade2Δ mutant is similarly increased in response to salt stress (Fig. 8C). It is important to note that although Ycf1p is responsible for a majority of ade2Δ-associated red pigment accumulation, the vacuole ABCC transporter, Bpt1p, plays a minor role in red pigment accumulation (Sharma, et al., 2002) and may account for a small fraction of the increased red pigment accumulation after salt treatment. Taken together these results suggest thatYcf1p function is induced in response to high salt concentrations in a Cka1p-dependent manner. To determine if indeed the increase in red pigment accumulation and the increased in vacuolar sequestration of MCB-SG was indeed due to decreased Ycf1p function due to decreased Ser251 phosphorylation we expressed WT Ycf1p, Ycf1p-Ser251Ala, and Ycf1p-Ser251Glu mutants in strains deleted for ycf1Δ ade2Δ and ycf1Δ respectively. We reasoned that if the increase in red pigment and MCB-SG in the vacuole is due to increased Ycf1p function via decreased phosphorylation of Ser251, and not another mechanism, then cells expressing Ycf1p-Ser251Glu should not have increased accumulation of either compound in response to salt stress and should mimic untreated WT cells (Fig. 8B and 8D), indeed this is the case for Ycf1p-Ser251Glu expressing cells. Further, expression of Ycf1p-Ser251Ala should mimic salt treated WT cells in both salt treated and untreated culture (Fig, 8B and 8D), and here too, this is the case for Ycf1p-Ser251Ala. These results suggest that Ycf1p function is induced in response to salt stress via decreased phosphorylation of Ser251, a result of decreased Cka1p activity. Based our results we expected an ycf1Δ strain to be more sensitive to salt stress than a WT strain and similarly a cka1Δ and an ycf1Δ cka1Δ would all have increased sensitivity to salt stress. However this turned out to not be the case, as spot tests for cell growth of WT, ycf1Δ, cka1Δ, and ycf1Δ cka1Δ strains showed no differences at high concentrations of salt (Supp. Fig. 3). Although our results do not show a direct role for Ycf1p in cellular resistance to salt stress, the results implicate an underlying role for Ycf1p in the cellular response of yeast to high salt.
Discussion
The ABCC transporter Ycf1p is critical for the detoxification of heavy metals in yeast. One of our long-term goals is to understand how Ycf1p activity is regulated. Recent studies with Ycf1p have shown that Ser251 in L0 is a site of phosphorylation and mutation of Ser251 to alanine results in increased transport function in vivo and in vitro (Chi, et al., 2007, Smolka, et al., 2007, Paumi, et al., 2008, Chi, 2007 #8). Further genetic evidence suggests that Ycf1p function is negatively regulated by the yeast CK2α subunit, Cka1p, via phosphorylation of Ser251 (Paumi, et al., 2008). However, it remains unclear what the role of Cka1p-dependent regulation of Ycf1p plays in cellular metabolism and response to cell stressors. In the work presented here, we address this key issue and present evidence that strongly suggests that Cka1p regulation of Ycf1p function is important in the cellular response to salt stress suggesting a new underlying role for Ycf1p in cellular resistance to high salt concentration.
In the present study we provide evidence that Cka1p-dependent regulation of Ycf1p function does not require or is regulated via phosphorylation sites within the linker domain of Ycf1p. Indeed if phosphorylation sites within the linker domain played a role in regulating Cka1p-mediated attenuation of Ycf1p function then individual alanine mutants would behave similarly to the Ser251Ala mutant phenotypically, but this is not the case (Fig. 2). Expression of alanine mutants for each of the linker domain (Fig. 1) phosphorylation sites did not result in increased strain resistance to high concentrations of cadmium or increased Ycf1p-dependent transport activity (Fig. 2B and D). It is important to note however that Ycf1p-Ser873Ala and Ycf1p-Ser914Ala have a subtle increase in resistance over that of WT at low concentration of cadmium. This subtle increased resistance to cadmium is not recapitulated at the higher concentration of cadmium and does not correlate with an increase in transporter function, which suggests that it may be the result of a nonspecific effect or that the differences we see at the low concentration is an experimental artifact. This subtle difference will be more closely examined in future studies. In addition we examined the possibility that phosphorylation within the linker region regulated Cka1p phosphorylation of Ser251. To this end, we synthesized a phosphospecific antibody for Ycf1p-Ser251 and showed that linker domain phosphorylation sites do not alter Ycf1p-Ser251 phosphorylation status with the exception of the Ycf1p-Ser908Ala,Thr911Ala double mutant (discussed below). Importantly, we show that deletion of cka1Δ almost completely eliminates Ser251 phosphorylation in vivo. Thus, Cka1p does appear to negatively regulate Ycf1p function solely through the phosphorylation of Ser251, as indicated by Paumi et. al. in JBC, 2008. We acknowledge the possibility that some level of control of Ycf1p function may be mediated by Cka1p-dependent phosphorylation of additional unidentified sites. However, this seems unlikely since the Ser251Ala mutant results in a remarkably similar phenotype to the cka1Δ deletion which almost completely eliminates Ser251 phosphorylation (Fig. 3A and B).
Interestingly, our analysis of Ser251 phosphorylation status in the context of linker domain phosphorylation site mutants revealed that the double mutant of Ser908 and Thr911 does have an effect on Ser251 phosphorylation status (Fig. 3C); mutation of Ser908 and Thr911 to alanine results in a 60% reduction in Ser251 phosphorylation. This result is consistent with the fact that a triple mutant of Ser251Ala, Ser908Ala, and Thr911Ala has the identical phenotype as the Ser908Ala,Thr911Ala double mutant (Paumi, et al., 2008) and supports the previous finding that the phenotype resulting from the Ser908Ala,Thr911Ala double mutant is dominant over the increased growth phenotype associated with the Ser251Ala mutant. One explanation of this finding is that phosphorylation of Ser908 and Thr911 play two roles in regulating Ycf1p function. First, phosphorylation of Ser908 and Thr911 is required for Ycf1p activity, and second, phosphorylation of Ser908 and Thr911 may play a role in the recruitment of Cka1p to Ycf1p. Thus, decreased phosphorylation at Ser908 and Thr911 would result in a reduced ability for Cka1p to associate with Ycf1p. Ycf1p-Ser872Ala and Ycf1p-Ser903Ala mutants also show decreased activity both in vivo and in vitro (Fig. 2A and 2D) similar to the Ycf1p-Ser908Ala,Thr911Ala mutant. However, the Ycf1p-Ser872Ala and Ycf1p-Ser903Ala mutants do not alter the phosphorylation of Ser251. These results suggest that phosphorylation of Ser872 and Ser903 may regulate Ycf1p function and Cka1p-mediated regulation of Ycf1p function via a different mechanism than Ser908 and Thr911. One possible mechanism is via the regulation of substrate recognition, a mutation that would be most likely dominant to a Ser251 mutation. This possibility could be quite important to all ABCC transporters as these sites are semi-conserved. The potential role of the phosphorylation sites within the linker domain is one of the major focuses of our current studies. Interestingly, many of the identified Ycf1p phosphorylation sites have MRP homologues (Paumi, et al., 2008). This may suggest that the regulation of the human and yeast MRPs is very complex involving multiple phosphorylation sites and multiple kinases.
To confirm the previously reported iMYTH results suggesting that Cka1p directly interacts with Ycf1p and to ask if all the subunits of the CK2 holoenzyme interact with Ycf1p, we conducted a number of co-immunoprecipitation experiments with endogenously-expressed Ycf1p-TAP. It is reasonable to believe that if Cka1p interacts with Ycf1p, then the entire holoenzyme complex also interacts with Ycf1p. Our studies showed that indeed Ycf1p interacts with all the subunits of the CK2 holoenzyme (Fig. 4). These results in combination with those previously reported by Paumi et. al. strongly suggests that Ycf1p and Cka1p do physically interact in vivo and that the interaction is not specific to Cka1p, but rather suggests the possibility that Cka1p can interact with Ycf1p as a member of the CK2 holoenzyme. CK2 holoenzyme formation is dependent upon the formation of a Ckb1p and Ckb2p dimer (Kubinski, et al., 2007). In the absence of Ckb1p and Ckb2p dimer formation the holoenzyme is inhibited (Kubinski, et al., 2007). The fact that Ycf1p is negatively regulated in the absence of CK2 holoenzyme formation suggests that Cka1p can phosphorylate Ycf1p-Ser251 as a monomer, although our data suggests the possibility that the Ycf1p-Cka1p interaction is strong enough to cause interaction of CK2 and Ycf1p. However, it is important to note that our CK2 holoenzyme-Ycf1p interaction analysis is based on overexpression of the individual subunits of the CK2 holoenzyme, and thus, the endogenous interaction requires confirmation.
One potential and very interesting explanation for our CK2 results, especially the appearance that deletion of Cka2p decreases resistance to cadmium rather than increases resistance to cadmium similar to deletion of Cka1p (Fig. 5A), is that the Cka1p/Cka2p heterodimer formation is in competition with Cka1p homodimer formation and the deletion of Cka2p results in increased Cka1p homodimer formation. The increase in Cka1p homodimer formation favors Cka1p specific substrates and hence increased Cka1p regulation of Ycf1p function (decrease growth on cadmium and decreased transporter activity). This model, for the role of Cka2p in Cka1p-dependent Ycf1p regulation, explains our results and suggests a role for Cka2p in regulating Cka1p specific activities of CK2.
To determine if Cka1p alone is truly sufficient to phosphorylate Ycf1p-Ser251 we conducted a series of kinase assays with short peptides derived from Ycf1p containing the Ser251 CK2 consensus site. Our results clearly indicate that recombinant Cka1p can interact with and phosphorylate Ser251 which supports the model that Cka1p negatively regulates Ycf1p function via phosphorylation of Ser251. However, it is important to note that in vitro kinase assays are not always a clear indicator of the ability of a kinase to phosphorylate a site in vivo; however, we have also shown that full phosphorylation of Ser251 in vivo requires Cka1p (Fig. 3). Therefore, together these results suggest that Cka1p negatively regulates Ycf1p function via direct phosphorylation of Ser251.
The most important finding of this work is the discovery of the potentially new and exciting role of Ycf1p in the cellular response to high salt. Our work suggests that Ycf1p function is upregulated in response to high salt concentration in a Cka1p-dependent manner via decreased phosphorylation of Ser251 within the NTE of Ycf1p. This finding is supported by previous studies that have demonstrated a role for CK2 in cellular response to salt stress (Kanhonou, et al., 2001, Hermosilla, et al., 2005). Furthermore, our findings provide important insight into the possible role for CK2 in regulating the function of the human ABCC transporters in normal cells and under stressed or disease conditions. For example, ABCC7 (CFTR) is regulated by CK2 in vivo and there is evidence to suggest that ABCC7 (a known Cl− channel) is regulated via a CK2α (Cka1p homologue)-dependent mechanism involving the protein salt channel, epithelium sodium channel 1 (ENaC1) (Mehta, 2008). Further, CK2 has been shown to modulate ABCA1 activity (Roosbeek, et al., 2004). Therefore it is reasonable to believe that CK2 may have a role in regulating the function of many of the ABC transporters.
In conclusion, our results presented here support the important mechanistic role of phosphorylation in both negatively and positively regulating Ycf1p transporter function and define a novel stress condition and the mechanism by which the CK2α kinase (Cka1p) regulates Ycf1p transporter function. Our future studies will focus on determining the cellular conditions and signaling pathways necessary for Cka1p-dependent Ycf1p regulation. In addition, we will define the role of Cka2p in regulating Cka1p activity, and determine if the human CK2α homologue, CSNK2A1, plays a similar role in regulating human MRP function in response to cellular stress.
Supplementary Material
Acknowledgments
This work is supported by P20RR020171 NIH COBRE (to CMP). We thank Dr. John D’Orazio for allowing us use of his fluorescent microscope, Dr. Andrew Pierce for providing the lab with multiple restriction enzymes during our cloning work, Dr. Susan Michaelis for her insightful comments at the onset of this work, Dr. Mary Vore for her insight and help in the initial preparation of this manuscript, and Dr. Nathan Vanderford for his help in the final preparation of this manuscript.
References
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakos E, Evers R, Calenda G, Tusnady GE, Szakacs G, Varadi A, Sarkadi B. Characterization of the amino-terminal regions in the human multidrug resistance protein (MRP1) J Cell Sci. 2000;113:4451–4461. doi: 10.1242/jcs.113.24.4451. [DOI] [PubMed] [Google Scholar]
- Berkey CD, Carlson M. A specific catalytic subunit isoform of protein kinase CK2 is required for phosphorylation of the repressor Nrg1 in Saccharomyces cerevisiae. Curr Genet. 2006;50:1–10. doi: 10.1007/s00294-006-0070-5. [DOI] [PubMed] [Google Scholar]
- Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–592. doi: 10.1146/annurev.biochem.71.102301.093055. Epub 2001 Nov 2009. [DOI] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T. Methods in Yeast Genetics. CSHL; Cold Spring Harbor: 2000. [Google Scholar]
- Chappe V, Irvine T, Liao J, Evagelidis A, Hanrahan JW. Phosphorylation of CFTR by PKA promotes binding of the regulatory domain. Embo J. 2005;24:2730–2740. doi: 10.1038/sj.emboj.7600747. Epub 2005 Jul 2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappe V, Hinkson DA, Zhu T, Chang XB, Riordan JR, Hanrahan JW. Phosphorylation of protein kinase C sites in NBD1 and the R domain control CFTR channel activation by PKA. J Physiol. 2003;548:39–52. doi: 10.1113/jphysiol.2002.035790. Epub 2003 Feb 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri B, Ingavale S, Bachhawat AK. apd1+, a gene required for red pigment formation in ade6 mutants of Schizosaccharomyces pombe, encodes an enzyme required for glutathione biosynthesis: a role for glutathione and a glutathione-conjugate pump. Genetics. 1997;145:75–83. doi: 10.1093/genetics/145.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Rich DP, Marshall J, Gregory RJ, Welsh MJ, Smith AE. Phosphorylation of the R domain by cAMP-dependent protein kinase regulates the CFTR chloride channel. Cell. 1991;66:1027–1036. doi: 10.1016/0092-8674(91)90446-6. [DOI] [PubMed] [Google Scholar]
- Chi A, Huttenhower C, Geer LY, et al. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci U S A. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. Epub 2007 Feb 2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang LY, Sheppard DC, Bruno VM, Mitchell AP, Edwards JE, Jr, Filler SG. Candida albicans protein kinase CK2 governs virulence during oropharyngeal candidiasis. Cell Microbiol. 2007;9:233–245. doi: 10.1111/j.1462-5822.2006.00784.x. [DOI] [PubMed] [Google Scholar]
- Cole SP, Deeley RG. Transport of glutathione and glutathione conjugates by MRP1. Trends Pharmacol Sci. 2006;27:438–446. doi: 10.1016/j.tips.2006.06.008. Epub 2006 Jul 2003. [DOI] [PubMed] [Google Scholar]
- Cole SP, Bhardwaj G, Gerlach JH, et al. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- Dean M. The genetics of ATP-binding cassette transporters. Methods Enzymol. 2005;400:409–429. doi: 10.1016/S0076-6879(05)00024-8. [DOI] [PubMed] [Google Scholar]
- Domanska K, Zielinski R, Kubinski K, Sajnaga E, Maslyk M, Bretner M, Szyszka R. Different properties of four molecular forms of protein kinase CK2 from Saccharomyces cerevisiae. Acta Biochim Pol. 2005;52:947–951. [PubMed] [Google Scholar]
- Eraso P, Martinez-Burgos M, Falcon-Perez JM, Portillo F, Mazon MJ. Ycf1-dependent cadmium detoxification by yeast requires phosphorylation of residues Ser908 and Thr911. FEBS Lett. 2004;577:322–326. doi: 10.1016/j.febslet.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Fernandez SB, Hollo Z, Kern A, Bakos E, Fischer PA, Borst P, Evers R. Role of the N-terminal transmembrane region of the multidrug resistance protein MRP2 in routing to the apical membrane in MDCKII cells. J Biol Chem. 2002;277:31048–31055. doi: 10.1074/jbc.M204267200. Epub 32002 Jun 31011. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Shen J, Rosen BP. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999;96:5001–5006. doi: 10.1073/pnas.96.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman MM, Ambudkar SV. Overview: ABC transporters and human disease. J Bioenerg Biomembr. 2001;33:453–458. doi: 10.1023/a:1012866803188. [DOI] [PubMed] [Google Scholar]
- Gueldry O, Lazard M, Delort F, Dauplais M, Grigoras I, Blanquet S, Plateau P. Ycf1p-dependent Hg(II) detoxification in Saccharomyces cerevisiae. Eur J Biochem. 2003;270:2486–2496. doi: 10.1046/j.1432-1033.2003.03620.x. [DOI] [PubMed] [Google Scholar]
- Gulshan K, Shahi P, Moye-Rowley WS. Compartment-specific Synthesis of Phosphatidylethanolamine Is Required for Normal Heavy Metal Resistance. Mol Biol Cell. 2009 doi: 10.1091/mbc.E09-06-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimeur A, Conseil G, Deeley RG, Cole SP. The MRP-related and BCRP/ABCG2 multidrug resistance proteins: biology, substrate specificity and regulation. Curr Drug Metab. 2004;5:21–53. doi: 10.2174/1389200043489199. [DOI] [PubMed] [Google Scholar]
- Hermosilla GH, Tapia JC, Allende JE. Minimal CK2 activity required for yeast growth. Mol Cell Biochem. 2005;274:39–46. doi: 10.1007/s11010-005-3112-2. [DOI] [PubMed] [Google Scholar]
- Howell LD, Borchardt R, Kole J, Kaz AM, Randak C, Cohn JA. Protein kinase A regulates ATP hydrolysis and dimerization by a CFTR (cystic fibrosis transmembrane conductance regulator) domain. Biochem J. 2004;378:151–159. doi: 10.1042/BJ20021428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungwirth H, Kuchler K. Yeast ABC transporters-- a tale of sex, stress, drugs and aging. FEBS Lett. 2006;580:1131–1138. doi: 10.1016/j.febslet.2005.12.050. Epub 2005 Dec 1122. [DOI] [PubMed] [Google Scholar]
- Kanhonou R, Serrano R, Palau RR. A catalytic subunit of the sugar beet protein kinase CK2 is induced by salt stress and increases NaCl tolerance in Saccharomyces cerevisiae. Plant Mol Biol. 2001;47:571–579. doi: 10.1023/a:1012227913356. [DOI] [PubMed] [Google Scholar]
- Kubinski K, Domanska K, Sajnaga E, Mazur E, Zielinski R, Szyszka R. Yeast holoenzyme of protein kinase CK2 requires both beta and beta′ regulatory subunits for its activity. Mol Cell Biochem. 2007;295:229–236. doi: 10.1007/s11010-006-9292-6. [DOI] [PubMed] [Google Scholar]
- Li X, Gerber SA, Rudner AD, et al. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- Li ZS, Szczypka M, Lu YP, Thiele DJ, Rea PA. The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J Biol Chem. 1996;271:6509–6517. doi: 10.1074/jbc.271.11.6509. [DOI] [PubMed] [Google Scholar]
- Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc Natl Acad Sci U S A. 1997;94:42–47. doi: 10.1073/pnas.94.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Luhtala N, Parker R. LSM1 over-expression in Saccharomyces cerevisiae depletes U6 snRNA levels. Nucleic Acids Res. 2009;37:5529–5536. doi: 10.1093/nar/gkp572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Meggio F, Sarno S, Andretta M, Pinna LA. Phosphorylation of synthetic fragments of inhibitor-2 of protein phosphatase-1 by casein kinase-1 and -2. Evidence that phosphorylated residues are not strictly required for efficient targeting by casein kinase-1. Eur J Biochem. 1994;223:647–653. doi: 10.1111/j.1432-1033.1994.tb19037.x. [DOI] [PubMed] [Google Scholar]
- Mason DL, Michaelis S. Requirement of the N-terminal extension for vacuolar trafficking and transport activity of yeast Ycf1p, an ATP-binding cassette transporter. Mol Biol Cell. 2002;13:4443–4455. doi: 10.1091/mbc.E02-07-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason DL, Mallampalli MP, Huyer G, Michaelis S. A region within a lumenal loop of Saccharomyces cerevisiae Ycf1p directs proteolytic processing and substrate specificity. Eukaryot Cell. 2003;2:588–598. doi: 10.1128/EC.2.3.588-598.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A. Cystic fibrosis as a bowel cancer syndrome and the potential role of CK2. Mol Cell Biochem. 2008;316:169–175. doi: 10.1007/s11010-008-9815-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. Quantifying western blots without expensive commercial quantification software. 2007. [Google Scholar]
- Minami S, Ito K, Honma M, et al. Posttranslational regulation of Abcc2 expression by SUMOylation system. Am J Physiol Gastrointest Liver Physiol. 2009;296:G406–413. doi: 10.1152/ajpgi.90309.2008. [DOI] [PubMed] [Google Scholar]
- Pascolo L, Petrovic S, Cupelli F, et al. Abc protein transport of MRI contrast agents in canalicular rat liver plasma vesicles and yeast vacuoles. Biochem Biophys Res Commun. 2001;282:60–66. doi: 10.1006/bbrc.2001.4318. [DOI] [PubMed] [Google Scholar]
- Paumi CM, Chuk M, Chevelev I, Stagljar I, Michaelis S. Negative regulation of the yeast ABC transporter Ycf1p by phosphorylation within its N-terminal extension. J Biol Chem. 2008;283:27079–27088. doi: 10.1074/jbc.M802569200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumi CM, Chuk M, Snider J, Stagljar I, Michaelis S. ABC transporters in Saccharomyces cerevisiae and their interactors: new technology advances the biology of the ABCC (MRP) subfamily. Microbiol Mol Biol Rev. 2009;73:577–593. doi: 10.1128/MMBR.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumi CM, Menendez J, Arnoldo A, et al. Mapping protein-protein interactions for the yeast ABC transporter Ycf1p by integrated split-ubiquitin membrane yeast two-hybrid analysis. Mol Cell. 2007;26:15–25. doi: 10.1016/j.molcel.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Poole A, Poore T, Bandhakavi S, McCann RO, Hanna DE, Glover CV. A global view of CK2 function and regulation. Mol Cell Biochem. 2005;274:163–170. doi: 10.1007/s11010-005-2945-z. [DOI] [PubMed] [Google Scholar]
- Qian YM, Grant CE, Westlake CJ, et al. Photolabeling of human and murine multidrug resistance protein 1 with the high affinity inhibitor [125I] LY475776 and azidophenacyl-[35S]glutathione. J Biol Chem. 2002;277:35225–35231. doi: 10.1074/jbc.M206058200. Epub 32002 Jul 35222. [DOI] [PubMed] [Google Scholar]
- Ren XQ, Furukawa T, Yamamoto M, Aoki S, Kobayashi M, Nakagawa M, Akiyama S. A functional role of intracellular loops of human multidrug resistance protein 1. J Biochem. 2006;140:313–318. doi: 10.1093/jb/mvj155. Epub 2006 Jul 2021. [DOI] [PubMed] [Google Scholar]
- Ren XQ, Furukawa T, Aoki S, et al. Glutathione-dependent binding of a photoaffinity analog of agosterol A to the C-terminal half of human multidrug resistance protein. J Biol Chem. 2001;276:23197–23206. doi: 10.1074/jbc.M101554200. Epub 22001 Apr 23111. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Roosbeek S, Peelman F, Verhee A, et al. Phosphorylation by protein kinase CK2 modulates the activity of the ATP binding cassette A1 transporter. J Biol Chem. 2004;279:37779–37788. doi: 10.1074/jbc.M401821200. Epub 32004 Jun 37724. [DOI] [PubMed] [Google Scholar]
- Schaerer-Brodbeck C, Riezman H. Genetic and biochemical interactions between the Arp2/3 complex, Cmd1p, casein kinase II, and Tub4p in yeast. FEMS Yeast Res. 2003;4:37–49. doi: 10.1016/S1567-1356(03)00110-7. [DOI] [PubMed] [Google Scholar]
- Sharma KG, Mason DL, Liu G, Rea PA, Bachhawat AK, Michaelis S. Localization, regulation, and substrate transport properties of Bpt1p, a Saccharomyces cerevisiae MRP-type ABC transporter. Eukaryot Cell. 2002;1:391–400. doi: 10.1128/EC.1.3.391-400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. Epub 12007 Jun 10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Sohn EJ, Martinoia E, et al. Engineering tolerance and accumulation of lead and cadmium in transgenic plants. Nat Biotechnol. 2003;21:914–919. doi: 10.1038/nbt850. Epub 2003 Jul 2020. [DOI] [PubMed] [Google Scholar]
- Soufi B, Kelstrup CD, Stoehr G, Frohlich F, Walther TC, Olsen JV. Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol Biosyst. 2009;5:1337–1346. doi: 10.1039/b902256b. [DOI] [PubMed] [Google Scholar]
- Szczypka MS, Wemmie JA, Moye-Rowley WS, Thiele DJ. A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J Biol Chem. 1994;269:22853–22857. [PubMed] [Google Scholar]
- Tommasini R, Evers R, Vogt E, et al. The human multidrug resistance-associated protein functionally complements the yeast cadmium resistance factor 1. Proc Natl Acad Sci U S A. 1996;93:6743–6748. doi: 10.1073/pnas.93.13.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmie JA, Moye-Rowley WS. Mutational analysis of the Saccharomyces cerevisiae ATP-binding cassette transporter protein Ycf1p. Mol Microbiol. 1997;25:683–694. doi: 10.1046/j.1365-2958.1997.5061868.x. [DOI] [PubMed] [Google Scholar]
- Wemmie JA, Szczypka MS, Thiele DJ, Moye-Rowley WS. Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J Biol Chem. 1994;269:32592–32597. [PubMed] [Google Scholar]
- Westlake CJ, Cole SP, Deeley RG. Role of the NH2-terminal membrane spanning domain of multidrug resistance protein 1/ABCC1 in protein processing and trafficking. Mol Biol Cell. 2005;16:2483–2492. doi: 10.1091/mbc.E04-12-1113. Epub 2005 Mar 2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake CJ, Qian YM, Gao M, Vasa M, Cole SP, Deeley RG. Identification of the structural and functional boundaries of the multidrug resistance protein 1 cytoplasmic loop 3. Biochemistry. 2003;42:14099–14113. doi: 10.1021/bi035333y. [DOI] [PubMed] [Google Scholar]
- Yang Y, Liu Y, Dong Z, Xu J, Peng H, Liu Z, Zhang JT. Regulation of function by dimerization through the amino-terminal membrane-spanning domain of human ABCC1/MRP1. J Biol Chem. 2007;282:8821–8830. doi: 10.1074/jbc.M700152200. Epub 2007 Jan 8830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.