Abstract
Motivation: Spial (Specificity in alignments) is a tool for the comparative analysis of two alignments of evolutionarily related sequences that differ in their function, such as two receptor subtypes. It highlights functionally important residues that are either specific to one of the two alignments or conserved across both alignments. It permits visualization of this information in three complementary ways: by colour-coding alignment positions, by sequence logos and optionally by colour-coding the residues of a protein structure provided by the user. This can aid in the detection of residues that are involved in the subtype-specific interaction with a ligand, other proteins or nucleic acids. Spial may also be used to detect residues that may be post-translationally modified in one of the two sets of sequences.
Availability: http://www.mrc-lmb.cam.ac.uk/genomes/spial/; supplementary information is available at http://www.mrc-lmb.cam.ac.uk/genomes/spial/help.html
Contact: ajv@mrc-lmb.cam.ac.uk
1 INTRODUCTION
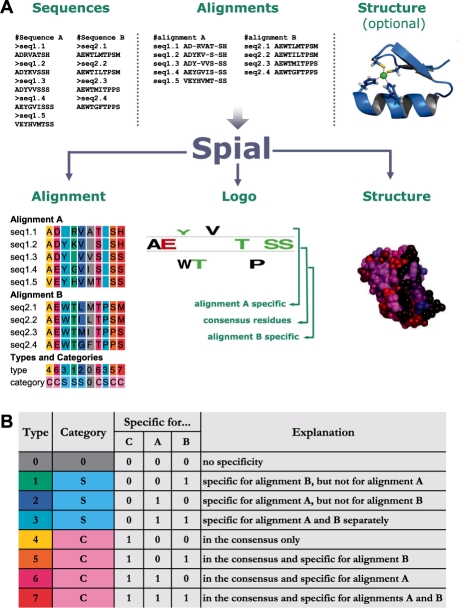
Identifying residues in proteins that are associated with specific functions is a recurring task in molecular biology. To assist this, we have developed Spial (Specificity in alignments), a web-based tool that allows the comparative analysis of two related protein subtypes (Fig. 1A). Spial differs from other related tools by allowing the simultaneous identification of residues that are (i) either specific to one of the two subtypes but not the other and/or (ii) conserved across the two subtypes. It also permits visualization using sequence logo and coloured alignments and mapping this information on to a representative structure, if available.
Fig. 1.
(A) Spial input and output. (B) Types and categories for a position in the alignment.
For example, when comparing the alignments of two related receptor subtypes that bind two different ligands, Spial allows the identification of residues that are specific to the binding of each of the ligands. For this, Spial takes two related sets of sequences or alignments as input and assigns each residue to one of eight possible types, depending on whether it is specific to the first alignment, the second alignment, the consensus or any combination of those (Fig. 1B).
Spial accepts sequences or multiple sequence alignments as input. In the case of submitting sequences, they are first combined together, aligned using MUSCLE (Edgar, 2004) and then split into two separate sets of alignments. In the case of submitting alignments, they are accepted in FASTA and SELEX formats. The sequences in the two input alignments should originate from related protein subfamilies. The alignments have to be of the same length and the positions in both alignments have to correspond. In rest of this article, we refer to these two alignments as alignments A and B, respectively. Additionally, Spial accepts a protein structure in the Protein Data Bank (PDB) format as input. In this case, the sequence of the structure should be present and indicated in the input.
For each position in the two input alignments A and B, Spial decides whether the residue is in consensus or not. In order for an amino acid to be in consensus, it has to be present above a user-specified threshold proportion in both alignments. In Figure 1A, a consensus threshold value of 0.35 (default value) was used. Next, Spial decides whether there are amino acids that are specific for one of the two alignments, but not for the consensus. For this, a non-consensus amino acid has to be present above a user-specified threshold proportion in one of the alignments. In Figure 1A, a specificity threshold value of 0.35 (default value) was used. As long as the sum of the consensus threshold and the specificity threshold is lower than one, a position can be in the consensus and specific to one of the alignments at the same time (Supplementary Material at http://www.mrc-lmb.cam.ac.uk/genomes/spial/help.html). Therefore, there are eight possible combinations of specificity for alignment A, alignment B or the consensus (Fig. 1B). We here refer to these eight combinations as types. Each position in an alignment can also be one of three possible categories, which indicate whether the position is in the consensus (C), specific to one or both of the input alignments but not in the consensus (S), or not specific at all (0). The one-letter codes that specify the types and categories of each residue are located in two rows below the Spial output alignment (Fig. 1A).
Spial's output consists of coloured alignments as described above, of sequence logos (Crooks et al., 2004; Schneider and Stephens, 1990), and of coloured protein structures (Fig. 1). The logos produced by Spial appear similar to those produced by the program Two Sample Logo (Vacic et al., 2006), which treats one alignment as the background and then computes whether there are residues that are enriched or depleted in the other alignment. Spial logos differ from this by visualizing how frequent a residue is in either alignment, or, if it is a consensus residue, how frequent it is in the consensus. In the output protein structures, the default colouring scheme differs from that used in the alignments. The colour of each protein residue reflects whether it is specific to alignment A (red), specific to alignment B (blue), specific to both (pink) or specific to neither (black). The structure, coloured using this scheme, can be viewed either directly in the browser using Jmol (http://www.jmol.org/), or by loading the structure into the PyMol (http://www.pymol.org/) structure viewer and then running a script that is provided by Spial. Another option offered by Spial is the colouring of residues according to residue type as defined above.
Spial is a versatile tool with a number of potential applications. Scenarios in which Spial may be useful include: (i) Of a number of homologous proteins, some bind a certain ligand or drug while others do not. Spial can assist in identifying surface patches that are specific to the proteins that bind the ligand. (ii) A protein has homologues in two different evolutionary lineages. Spial can assist in identifying residues that are specific to either lineage, and those that are conserved in both. (iii) Of a number of paralogues in a genome, some have a specific function while others do not. Spial can assist in identifying the residues that are specific to the proteins that have the function of interest. (iv) Spial can assist in identifying residues that undergo post-translational modifications by running an alignment of sequences that are commonly modified against an alignment of related sequences that are not.
Here, we use Spial to elucidate the differences in coordination of retinal between vertebrate and cephalopod rhodopsin. Opsins are a family of seven-helix membrane receptors that activate G proteins in a light-dependent manner via the photo-isomerization of retinal in the protein. Vertebrate and invertebrate rhodopsins are two subgroups of opsins. Though related, they differ in their molecular properties and function. While vertebrate rhodopsin activates the cGMP signalling pathway through the Gt-type G-protein (Li et al., 2004), invertebrate rhodopsin activates the I3P pathway via a Gq-type G protein (Murakami and Kouyama, 2008). The molecular causes of these functional differences still remain unclear. We used the alignments of rhodopsin sequences as available from GPCR database (http://www.gpcr.org/7tm/) as input for Spial and mapped the results to the structure of squid rhodopsin (Murakami and Kouyama, 2008). The result indicates that although some residues that coordinate retinal are conserved between vertebrate and cephalopod rhodopsin, this does not apply to all of them. For example, Lys 305, which covalently binds retinal, is conserved between both vertebrate and cephalopod rhodopsin. Other hydrophobic residues in the retinal binding pocket, including Phe 120 and Phe 188 are specific to cephalopod rhodopsin and are not conserved in vertebrate rhodopsin. Phe 205, although it is part of the binding pocket in the squid rhodopsin structure used here, is generally not conserved in cephalopods but conserved in vertebrates. The Spial output for this example is available at: http://www.mrc-lmb.cam.ac.uk/genomes/spial/examples/example_rhodopsin.html.
In conclusion, Spial can be used as a tool for the detection and visualization of information about the specificity of protein residues. This can aid in understanding protein function, protein–small molecule, protein–nucleic acid and protein–protein interactions.
ACKNOWLEDGEMENTS
We thank Cyrus Chothia, Derek Wilson, Daniel Hebenstreit, Rouslan Moukhametzianov and Madhav Sukumaran at the MRC Laboratory of Molecular Biology for testing Spial, Gabriella Frigerio and Guilhem Chalancon for comments on the manuscript. M.M.B. is an EMBO YI.
Funding: Medical Research Council; Karn Fund Postdoctoral Fellowship (to A.W.); LMB-Cambridge scholarship and St. John's College Benefactor Scholarship (to A.J.V.); Darwin College and Schlumberger (to M.M.B.).
Conflict of Interest: none declared.
REFERENCES
- Crooks GE, et al. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, et al. Structure of bovine rhodopsin in a trigonal crystal form. J. Mol. Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, et al. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics. 2006;22:1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]