Abstract
Malate dehydrogenase (MDH) catalyzes a reversible NAD+-dependent-dehydrogenase reaction involved in central metabolism and redox homeostasis between organelle compartments. To explore the role of mitochondrial MDH (mMDH) in Arabidopsis (Arabidopsis thaliana), knockout single and double mutants for the highly expressed mMDH1 and lower expressed mMDH2 isoforms were constructed and analyzed. A mmdh1mmdh2 mutant has no detectable mMDH activity but is viable, albeit small and slow growing. Quantitative proteome analysis of mitochondria shows changes in other mitochondrial NAD-linked dehydrogenases, indicating a reorganization of such enzymes in the mitochondrial matrix. The slow-growing mmdh1mmdh2 mutant has elevated leaf respiration rate in the dark and light, without loss of photosynthetic capacity, suggesting that mMDH normally uses NADH to reduce oxaloacetate to malate, which is then exported to the cytosol, rather than to drive mitochondrial respiration. Increased respiratory rate in leaves can account in part for the low net CO2 assimilation and slow growth rate of mmdh1mmdh2. Loss of mMDH also affects photorespiration, as evidenced by a lower postillumination burst, alterations in CO2 assimilation/intercellular CO2 curves at low CO2, and the light-dependent elevated concentration of photorespiratory metabolites. Complementation of mmdh1mmdh2 with an mMDH cDNA recovered mMDH activity, suppressed respiratory rate, ameliorated changes to photorespiration, and increased plant growth. A previously established inverse correlation between mMDH and ascorbate content in tomato (Solanum lycopersicum) has been consolidated in Arabidopsis and may potentially be linked to decreased galactonolactone dehydrogenase content in mitochondria in the mutant. Overall, a central yet complex role for mMDH emerges in the partitioning of carbon and energy in leaves, providing new directions for bioengineering of plant growth rate and a new insight into the molecular mechanisms linking respiration and photosynthesis in plants.
Plant tissues contain multiple isoforms of malate dehydrogenase (l-malate-NAD-oxidoreductase [MDH]; EC 1.1.1.37) that catalyze the interconversion of malate and oxaloacetate (OAA) coupled to reduction or oxidation of the NAD pool. These isoforms are encoded by separate genes in plants and have been shown to possess distinct kinetic properties as well as subcellular targeting and physiological functions (Gietl, 1992). While the MDH reaction is reversible, it strongly favors the reduction of OAA. The direction of the reaction in vivo depends on substrate/product ratios and the NAD redox state, and it can vary even in the same tissue due to prevailing physiological conditions. Isoforms operate in mitochondria, chloroplasts, peroxisomes, and the cytosol, but due to the ready transport and utilization of malate and OAA and the availability of NAD, this reaction can cooperate across compartments and is the basis for malate/OAA shuttling of reducing equivalents in many different metabolic schemes of plant cellular function (Krömer, 1995). It is clear, however, that the exchange through the membranes is strictly controlled, since large redox differences in NAD(H) pools exist between compartments (Igamberdiev and Gardeström, 2003).
The mitochondrial MDH (mMDH) is thought to operate in at least three different pathways in plants. First, it is a classical tricarboxylic acid (TCA) cycle enzyme that oxidizes the malate product from the fumarase reaction to OAA for the citrate synthase-dependent condensation with acetyl-CoA to form citrate. Second, it is considered to operate in the reverse direction during the conversion of Gly to Ser by reducing OAA to malate and providing a supply of NAD+ for Gly decarboxylase (Journet et al., 1981). Third, in a more specialized pathway in C4 plants, it provides a supply of CO2 for fixation in bundle sheath chloroplasts by reducing OAA (generated from Asp transported from mesophyll cells) into malate that is then decarboxylated by NAD-malic enzyme (NAD-ME) to CO2 and pyruvate (Hatch and Osmond, 1976). Plant mitochondria can support TCA cycle activity with malate as the sole substrate due to MDH and NAD-ME, both ubiquitous in plants (Palmer, 1984). OAA is readily transported both into and out of isolated plant mitochondria (Douce and Bonner, 1972), in contrast to mammalian mitochondria, which are essentially impermeable to this organic acid.
While these three mMDH schemes and metabolic schemes for other MDH isoforms are plausible, widely accepted, and consistent with a range of biochemical studies, the depletion, removal, and overexpression of specific MDH isoforms in plants have led to surprising insights into MDH roles in vivo. For example, the peroxisomal MDH (PMDH) was until recently generally considered to be involved in the synthesis of NADH for hydroxypyruvate reduction in the photorespiratory cycle and for the oxidation of NADH generated during β-oxidation of fatty acids, but its potential role in the oxidation of malate in the glyoxylate cycle was unclear. However, studies of the double knockout of PMDH in Arabidopsis (Arabidopsis thaliana) showed that while PMDH is essential for β-oxidation, its removal does not impair glyoxylate cycle activity (Pracharoenwattana et al., 2007) and has only a limited impact on hydroxypyruvate reduction (Cousins et al., 2008).
Changes in mMDH have been reported both through the study of spontaneous mutants and the expression of antisense constructs. Spontaneous null mutants of mMDH1 in soybean (Glycine max) are linked to a yellow foliage phenotype and are associated with the removal of two of the three mMDH isoforms (Imsande et al., 2001). Expression of an antisense fragment of mMDH in tomato (Solanum lycopersicum), driven by the 35S promoter, lowered mMDH protein in mitochondria, decreased total cellular MDH by approximately 60%, but had a positive impact on photosynthetic activity, CO2 assimilation rate, and total plant dry matter in long-day-grown plants (Nunes-Nesi et al., 2005). A range of carbohydrates also accumulated in the tomato antisense plants, as did redox-related compounds such as ascorbate. The increase in ascorbate content may be linked to the enhancement of photosynthesis, as ascorbate feeding to leaves can also increase photosynthetic performance (Nunes-Nesi et al., 2005). This link is not absolute, however, given that short-day-grown antisense tomato plants had stunted growth, which was potentially due to impaired photosynthesis, but still had elevated levels of ascorbate due to a higher ratio of reduction of the ascorbate pool compared with the wild type (Nunes-Nesi et al., 2008). Analysis of roots from these antisense tomato plants revealed a negative impact of mMDH loss, leading to a lower root dry weight and lower root respiratory rate (van der Merwe et al., 2009). This implies a distinct impact of mMDH loss on roots and shoots. Overexpression of cytosolic MDH led to a 4-fold elevation of root organic acids in alfalfa (Medicago sativa) plants and high rates of organic acid exudation that increased aluminum tolerance through metal chelation in the soil (Tesfaye et al., 2001). These studies imply that there is a complex form of functional redundancy between MDH isoforms in different compartments, allowing MDH in separate locations to maintain specific pathways via malate/OAA shuttling, or that a range of redox requirements that have been linked to MDH in accepted metabolic schemes are incorrect and other reactions couple NAD/NADH pool homeostasis. In addition, these studies clearly show that changes in the amount of MDH isoforms can alter metabolic flux into a range of organic acids and have far-reaching effects on plant growth and development.
To better understand the importance of the mMDH and to determine if plants are viable without any mMDH isoforms due either to the role of NAD-ME and/or malate/OAA shuttling to other compartments, we have constructed and analyzed mMDH mutants in Arabidopsis. A major and a minor MDH isoform exist in Arabidopsis mitochondria, evidenced by differing levels of gene expression and differing protein abundance (Lee et al., 2008). We hypothesized that if mMDH works in concert with other MDH isoforms and is responsible for the reduction of OAA to malate for export from the mitochondrion, then if we remove mMDH, not only would the loss of extramitochondrial malate and the slowing of Gly decarboxylation limit photorespiratory carbon flux, but oxidation of NADH remaining in the mitochondrion could lead to elevated leaf respiration and alteration in plant growth. We found that not only did mutants have low photorespiratory flux, but they also increased respiration and had slow growth due to lowered net CO2 assimilation. The previously established correlation between mMDH abundance, photosynthetic performance, and foliar ascorbate levels was also investigated. Elevated levels of the metabolite were found in Arabidopsis, consolidating the work done in tomato (Nunes-Nesi et al., 2005). Proteomic analyses, followed by immunodetection studies, unearthed altered abundance of the terminal enzyme of the ascorbate biosynthetic pathway, galactono-1,4-lactone dehydrogenase (GLDH), as a mechanistic element in the phenomenon linked directly to mitochondrial function.
RESULTS
T-DNA Knockout of Mitochondrial mMDHs
The genome of Arabidopsis carries eight genes encoding isoforms of NAD-dependent MDH (NAD-MDH). Two of these are targeted to peroxisomes, PMDH1 and PMDH2 (Pracharoenwattana et al., 2007; Eubel et al., 2008), and one to chloroplasts (Berkemeyer et al., 1998; Peltier et al., 2006); three have no apparent targeting sequences and are assumed to be cytosolic, and two have predicted mitochondrial targeting sequences and are the only MDH isoforms identified in purified mitochondrial samples (Heazlewood et al., 2004). The two putative mitochondrial enzymes will be referred to as mMDH1 (At1g53240) and mMDH2 (At3g15020). The analysis of microarray data available on the Genevestigator Web site (Hruz et al., 2008) reveals nine times more transcript for mMDH1 than mMDH2, which is also consistent with mMDH1 being the dominant protein isoform observed in mitochondrial extracts (Millar et al., 2001; Lee et al., 2008).
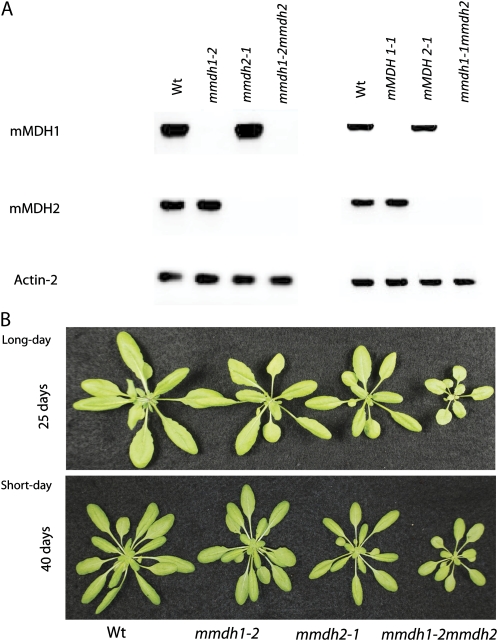
Homozygous T-DNA insertion lines were isolated: two independent lines for mmdh1 (mmdh1-1 [GABI_097C10] and mmdh1-2 [GABI_540F04]) and one for mmdh2 (SALK_126994; Supplemental Fig. S1). A second independent line for T-DNA insertion in mMDH2 is unavailable among public insertion seed banks. Crossing of the three lines and selection of double mutants provided two double mutants, mmdh1-1mmdh2 and mmdh1-2mmdh2, independent for the mmdh1 insertion (Fig. 1; Supplemental Fig. S1). Genotyping and biochemical analyses of isolated mitochondria from the two double knockout lines indicated that mMDH activity was effectively absent in both double mutant combinations (Supplemental Fig. S1). The mmdh1-2mmdh2 mutant and its constituent genotypes were chosen for the subsequent experiments, and it is referred to as mmdh1mmdh2 in all other experiments in this paper. The absence of the mMDH mRNAs from mature leaves in the mutants was established by reverse transcription (RT)-PCR using gene-specific primers (Fig. 1A). Plant growth was unaffected in all single insertion lines but was reduced markedly in the double mutants (Fig. 1B; Supplemental Fig. S1). The whole-plant phenotypes were observed in both short- and long-day growth conditions, as outlined in “Materials and Methods” (Fig. 1B). This shows that the growth defect displayed in the double knockouts is not significantly affected by lighting regimes, as opposed to observations of growth enhancement under long-day cycles (Nunes-Nesi et al., 2005) and growth inhibition under short-day cycles for tomato antisense MDH plants (Nunes-Nesi et al., 2008).
Figure 1.
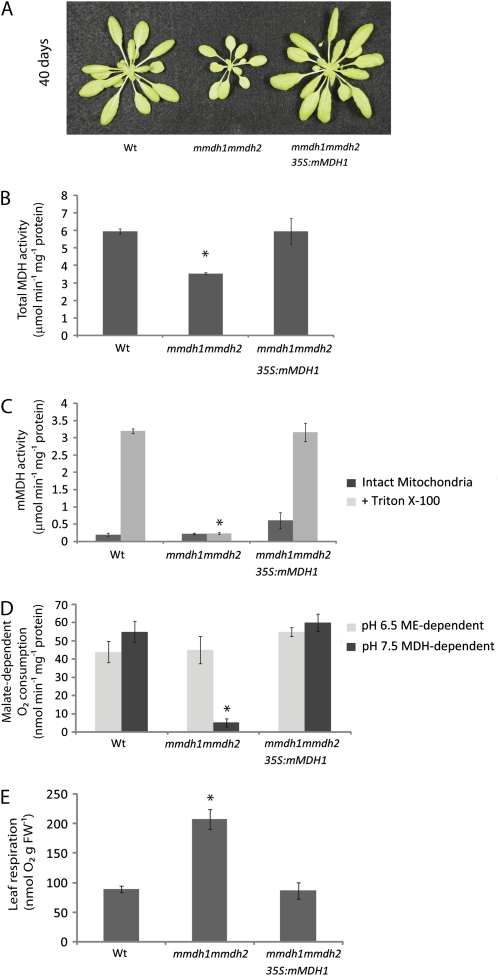
mmdh1mmdh2 knockout plants in Arabidopsis show reduced plant growth. A, RT-PCR from total RNA with primers for mMDH1 and mMDH2 compared with Actin2 control for mmdh mutants. B, Plant growth of single mmdh1, mmdh2, and mmdh1-2mmdh2 plants compared with the wild type (Wt) in long-day and short-day growth conditions at 25 and 40 d of growth, respectively.
Total leaf extracted MDH activity from long-day-grown plants was decreased 30% in the mmdh1-2 mutant and more than 40% in mmdh1mmdh2 but was not different from the wild type in the mmdh2 single mutant (Fig. 2A). These reductions show the significant contribution of mMDH activity to the total NAD-MDH activity in mature leaves of Arabidopsis.
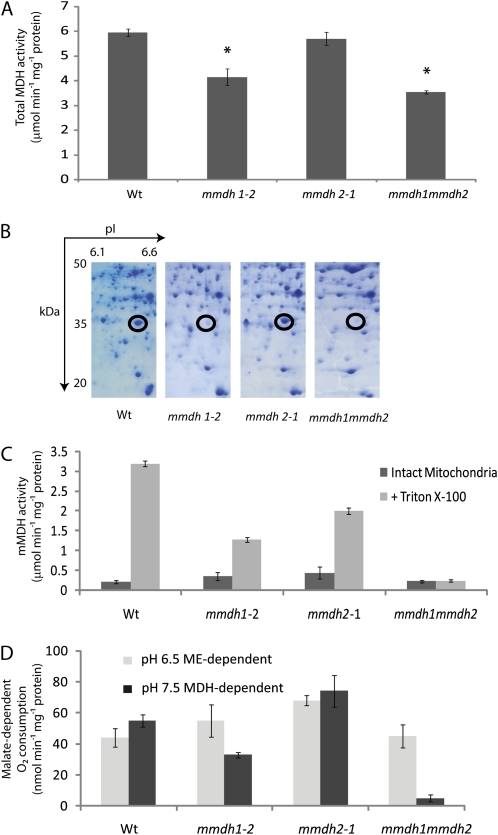
Figure 2.
mMDH protein abundance and activity are altered in mmdh knockout plants. A, Changes in total cellular MDH activity. Bars are means ± se (n = 4), and asterisks indicate significant differences (P < 0.05) from the wild type (Wt). B, Loss of mMDH1 on two-dimensional gels of mitochondrial proteins from mmdh1 and mmdh1mmdh2. C, Total MDH activity in isolated mitochondria, showing activity in intact mitochondria and activity after organelle rupture with Triton X-100. Bars are means ± se (n = 3). D, Malate-dependent respiratory rate of isolated intact mitochondria at pH 6.5 to maximize NAD-ME activity and at pH 7.5 to maximize MDH activity. Bars are means ± se (n = 3).
Mitochondria were purified from mature leaves of the wild type and single and double mutants grown hydroponically under short-day conditions, and total mitochondrial protein extracts were separated by two-dimensional gels to reveal the known major mMDH1 protein spot (Fig. 2B). The mMDH1 protein spot was absent in mmdh1 and mmdh1mmdh2. The mMDH2 protein spot was very faint in comparison and was best observed in later experiments (see below). Total mMDH activity was recorded in freshly isolated intact mitochondria and after mitochondrial solubilization with Triton X-100, showing the high latency of MDH in mitochondria and the effectively complete loss of mMDH latent activity in mmdh1mmdh2 (Fig. 2C; Supplemental Fig. S1).
Altered Malate-Dependent Respiratory Rate and Metabolic Compensation in mmdh1mmdh2
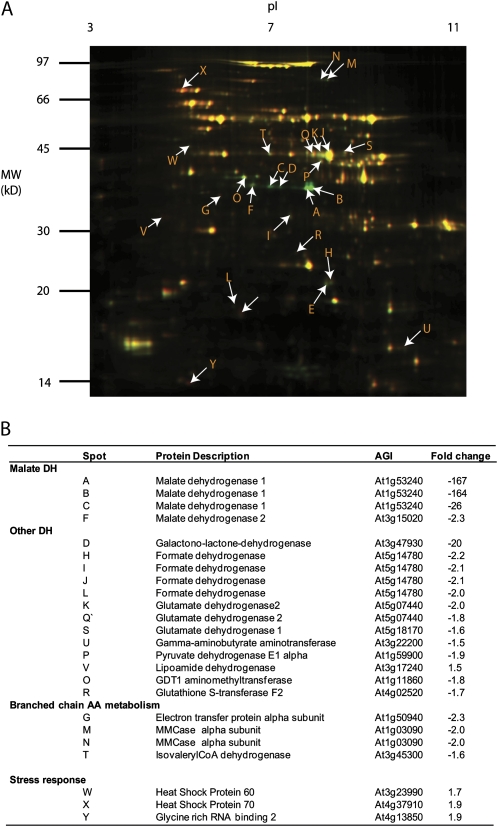
Measurements of malate-dependent oxygen consumption by isolated mitochondria showed that oxidation at pH 7.5 (at which MDH activity is maximized and NAD-ME activity is minimized) was half in the mmdh1 single mutant and 11 times lower in the double mutant compared with the wild type. Malate oxidation at pH 6.5 (at which NAD-ME is maximized and MDH is minimized) was not significantly modified in the mutants (Fig. 2D; Supplemental Fig. S1). Compensation in the mitochondrial proteome due to the loss of mMDH was investigated using differential in-gel electrophoresis (DIGE) analysis (Fig. 3A). While the majority of proteins were unaltered in abundance in mmdh1mmdh2, a set of decreased and increased protein spots were quantified and identified by peptide mass spectrometry (Fig. 3B; Supplemental Table S1). This analysis confirmed the loss of both mMDH1 and mMDH2 in mmdh1mmdh2. It also highlighted the lowered abundance of protein spots for formate dehydrogenase, GLDH, Glu dehydrogenase 1 and 2, two components of the pyruvate dehydrogenase (PDC E1a and E3), and a set of proteins in the branched chain amino acid degradation pathway (IVD, MMCase, and EFT). Notably, these proteins are all involved or closely linked to NAD-dependent metabolism. No changes in respiratory chain components were noted. Two heat shock proteins (HSP60 and HSP70) were subtly increased in abundance, consistent with this genetic modification being a stressed state for the mitochondrial proteome (Taylor et al., 2009).
Figure 3.
Differential in-gel analysis of mitochondria shows compensatory changes in the mitochondrial proteome in mmdh1mmdh2. A, Overlay of fluorescence images of typical two-dimensional gels showing common proteins in yellow, protein decreased in mmdh1mmdh2 in green, and protein enhanced in mmdh1mmdh2 in red. B, Proteins identified from protein spots marked in A with fold changes (n = 3; P < 0.05). AGI, Arabidopsis Genome Initiative number.
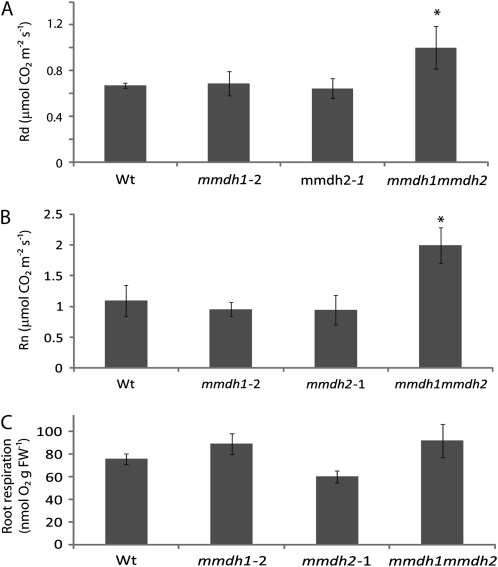
mmdh1mmdh2 Has Elevated Whole-Leaf Respiratory Rates
Given the impact of the loss of mMDH on malate oxidation rate of isolated mitochondria, we then assayed the impact of mMDH loss on total leaf respiration from short-day-grown plants. When mitochondrial respiration was calculated in gas phase as the rate of CO2 release in the light, as determined by the Laisk method (Laisk, 1977), it was 50% higher in the double mutant that in the wild type or single mutants (Fig. 4A). When respiration was measured directly in gas phase as the rate of CO2 release in the dark (Rn), mmdh1mmdh2 respiratory rate was nearly double the rate in the wild type or the single mutants (Fig. 4B). Analysis of root respiration rates as oxygen consumption showed no difference between the wild type and mmdh1mmdh2 (Fig. 4C), implying either a lack of a role for mMDH or a compensatory mechanism.
Figure 4.
Impact of MDH loss on respiratory rate of leaves and roots. A, Respiratory CO2 production of lighted leaves in gas phase measured by the method of Laisk. Bars are means ± sd (n > 3), and asterisks indicate significant differences (P < 0.05) from the wild type (Wt). B, CO2 production in darkened leaves in gas phase. Bars are means ± sd (n > 3), and asterisks indicate significant differences (P < 0.05) from the wild type. C, Root respiration as oxygen consumption rate in liquid phase. Bars are means ± se (n = 3). FW, Fresh weight. Plants were grown under short-day conditions.
Slow Growth Rates in mmdh1mmdh2 Are Linked to Altered Net CO2 Assimilation Rates But Not to Changes in Photosynthetic Capacity or Efficiency
In controlled conditions, the growth of the double mutant was reduced compared with the wild type, with a reduction of 50% in rosette diameter and 42% in leaf number after 6 weeks (Supplemental Fig. S2). The double mutant also showed delayed seed germination rate and frequency (Supplemental Fig. S2). While all lines produced sufficient seeds under controlled conditions for study and did not appear to differ in seed set, a quantification of seed production in plants grown outside showed reduced seed set in the single mutants, while all mmdh1mmdh2 plants died (Supplemental Fig. S2).
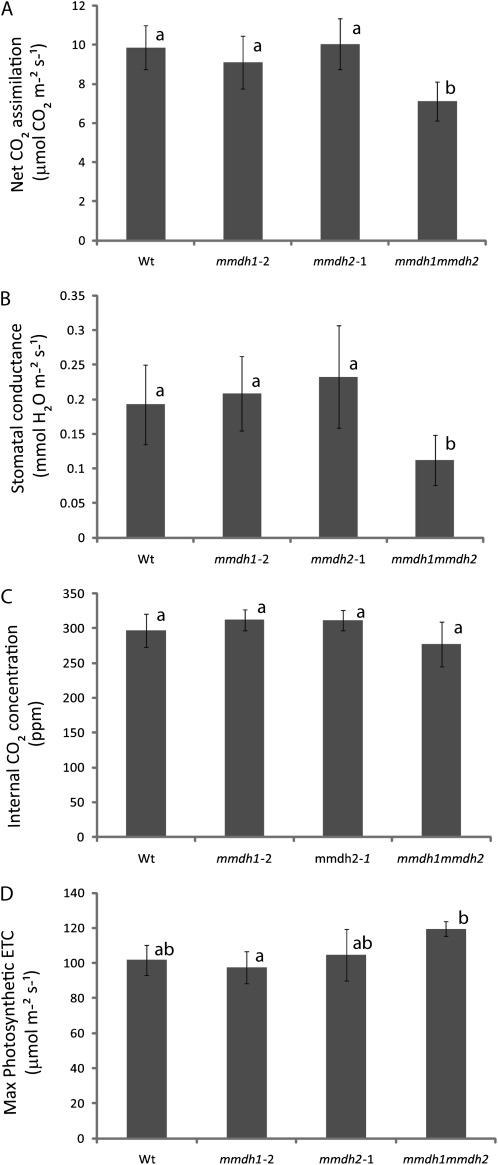
To further investigate the reasons for the growth reduction observed in the double mutant, other physiological parameters were measured in mature rosette leaves. The rate of CO2 assimilation and the stomatal conductance, as measured under growth conditions with limiting photosynthetic photon flux density (PPFD), were 25% and 46% lower, respectively, in the double mutant compared with the wild type (Fig. 5, A and B), while internal CO2 concentration was unaltered in the mutants (Fig. 5C). Under saturating PPFD, the CO2 assimilation at 400 μL L−1 CO2 was reduced by 32% in the double mutant (Supplemental Fig. S4). In addition, the double mutant showed a small reduction of average chlorophyll content, but this was not statistically significant (Supplemental Fig. S4). However, despite this, photosynthetic capacity of the double mutant did not seem to be impaired, as both the maximum CO2 assimilation rate (measured at saturating PPFD and CO2) and the maximum photosynthetic electron transport rate (inferred from CO2 assimilation/intercellular CO2 [A/Ci] curves) tended to be higher in the double mutant than in the wild type (Fig. 5D). The decrease in CO2 assimilation under growth conditions in the double mutant represents a shortfall of 2.5 μmol CO2 m−2 s−1, while the stimulation of mitochondrial respiration in the light (Fig. 4A) would only account for a loss of 0.4 μmol CO2 m−2 s−1. So while the decrease in CO2 assimilation rate might not be caused by the increased mitochondrial respiration alone, the increased respiratory rate during both light and dark respiration (Fig. 4, A and B) would elevate the role that respiratory CO2 loss plays in the lower net growth rate of mmdh1mmdh2.
Figure 5.
Changes in net CO2 assimilation rate and photosynthetic performance measurements in mmdh mutants. A, Net CO2 assimilation rate under growth conditions. B, Stomatal conductance to water vapor under growth conditions. C, Internal CO2 concentration under growth conditions. D, Maximum photosynthetic electron transport rate (ETC) inferred from A/Ci curves. Bars are means ± sd (n > 3), and letters designate groups that significantly differ (P < 0.05). Wt, Wild type. Plants were grown under short-day conditions.
mMDH Has a Role in Maximizing Photorespiratory Rate
A potential role for mMDH in the photorespiratory pathway has long been proposed in metabolic schemes associated with redox transfer between mitochondrial, peroxisomal, and cytosolic reactions (Krömer, 1995). As photorespiration is another pathway leading to CO2 losses in the light, changes in photorespiratory rate could be involved in the decrease in net CO2 assimilation observed in mmdh1mmdh2.
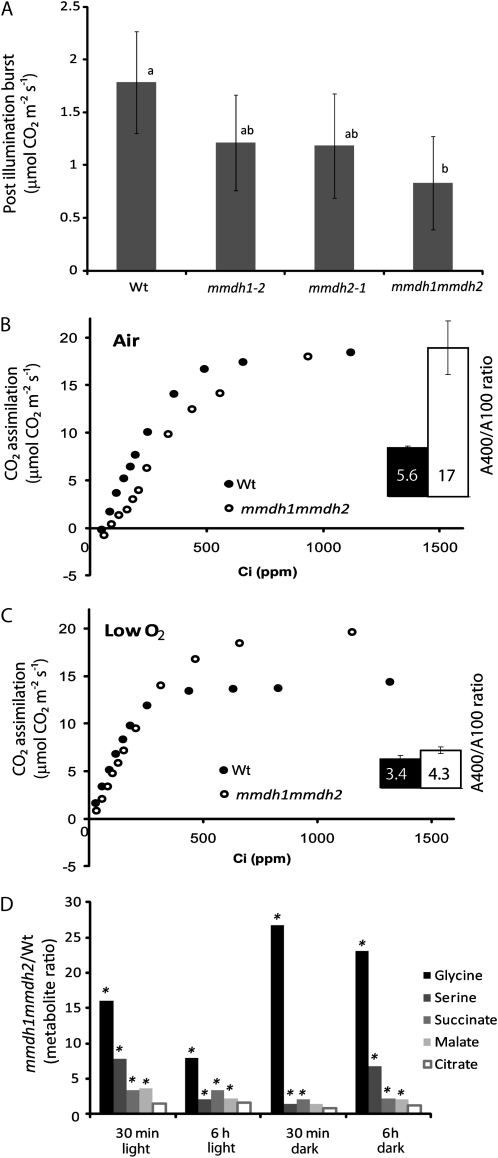
A series of different measurements indicate an impact on photorespiration in mmdh1mmdh2. First, the kinetics of the immediate CO2 postillumination burst (PIB), often reported as respiration of residual photorespiratory substrates (Kebeish et al., 2007), was significantly different in the double mutant and in the wild type. This change can be observed in the lowered height of the PIB peak compared with the wild type (Fig. 6A) but also in the extended duration of the PIB event in mmdh1mmdh2 (Supplemental Fig. S3). Second, photosynthetic rates measured under saturating PPFD at ambient CO2 (400 μL L−1) were 5.6 times higher than at low CO2 (100 μL L−1) in the wild type, while they were 17 times higher in mmdh1mmdh2 (Fig. 6B). For the single mutants, mmdh2 was similar to the wild type, whereas mmdh1 was intermediate between the wild type and mmdh1mmdh2 (data not shown). Furthermore, the double mutant exhibited a distinct break in the initial part of the A/Ci curves, the slope of the linear part of the curve being lower at low CO2 (50–150 μL L−1) than at higher CO2 (150–350 μL L−1) concentrations. The break in the initial part of the A/Ci curve was suppressed when the measurements were performed in very low oxygen, under conditions that restricted photorespiration, and the difference in the A400/A100 ratio was greatly reduced (Fig. 6C). The initial slope of the curve was lower in the double mutant than in the wild type in normal air but was restored in nonphotorespiratory conditions to the same slope as in the wild type. In low oxygen, the photosynthetic rates under saturating light and ambient CO2 hardly increased in the wild type (+7%) but were markedly enhanced in the double mutant (+50%) compared with the values recorded in normal air. Analysis of the single mutants showed no differences between the wild type and mmdh2, but the mmdh1 data were intermediate between mmdh1mmdh2 and the wild type (Supplemental Fig. S4). Metabolite analysis 30 min and 6 h into the day and into the night showed that Gly and Ser levels were higher in mmdh1mmdh2 than in the wild type (Fig. 6D). Smaller elevations of succinate and malate content were recorded, while citrate levels were not significantly different between the genotypes during the day and night. Altogether, these results indicate a deficiency in the functioning of the photorespiratory cycle in mmdh1mmdh2.
Figure 6.
Changes in photorespiration in mmdh mutants. A, PIB. Bars are means ± sd (n = 4). B and C, A/Ci curves in air and low oxygen in the wild type (Wt) and mmdh1mmdh2. The inset graphs show A400/A100 ratios. Bars are mean ratios ± sd (n = 3). D, Metabolite ratios between the wild type and mmdh1mmdh2 at different times during the day and night. Fold changes with significant differences (P < 0.05; n = 5) are indicated with asterisks. Plants were grown under short-day conditions.
The observed alterations to photorespiratory parameters prompted the examination of growth in mmdh1mmdh2 and its constituent genotypes under high atmospheric CO2 conditions (5,000 μL L−1). In conditions normally beneficial to “classical” photorespiratory mutants, growth was not enhanced in mmdh1mmdh2 (Supplemental Fig. S1). This suggests the overriding role of the high-respiration phenotype in maintaining the stunted growth of mmdh1mmdh2 even under conditions that should limit photorespiration and enhance plant growth. This shows that mmdh1mmdh2 is likely not a true photorespiratory mutant but rather has affected photorespiration due to alterations in metabolism.
Complementation of mmdh1mmdh2 with mMDH cDNA
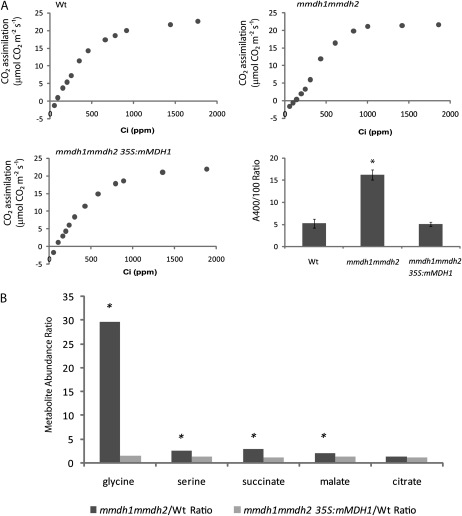
To confirm if the loss of MDH activity in the double mutant and subsequent changes in respiration and plant growth were definitively linked, we used a cDNA for mMDH1 to complement mmdh1mmdh2. The Arabidopsis mMDH1 was chosen as a generic MDH enzyme to determine if characteristics of double mutants could be recovered by the expression of a functional cDNA for MDH. As the mmdh2 mutant has no phenotype, implying functional redundancy with mMDH1, and given that only in the double mutant did lack of mMDH2 cause a phenotype, we determined that recovery of MDH activity via the expression of mMDH1 in the mutant should be sufficient to complement the mutant. Six independent complemented transformants were isolated, and PCR was used to determine that the complemented line used in the experiments contained both T-DNA insertions and the MDH1 cDNA (Supplemental Fig. S5). The complemented double mutant recovered a wild-type-like phenotype in both short-day (Fig. 7A) and long-day conditions at all stages of growth. This indicated that mMDH activity alone was required to recover plant growth. Measurement of total MDH activity in leaves of the complemented line showed recovery to wild-type levels (Fig. 7B), and work on isolated mitochondria highlighted the recovery of latent mMDH activity (Fig. 7C) in addition to oxygen consumption attributed to the oxidation of malate (Fig. 7D). Measurement of whole-leaf respiration as oxygen consumption in the dark showed a reversal of the enhanced respiratory phenotype of the double mutant back to wild-type levels (Fig. 7E). A/Ci curves of the complemented line versus mmdh1mmdh2 showed an amelioration of the break of the curve at low intercellular CO2 concentrations (Fig. 8A), highlighting a reversal of the mmdh1mmdh2 photorespiratory defect in the complemented line. In addition to this, comparison of the ratio of photosynthetic rates measured at saturating PFFD and ambient CO2 (A400) to low CO2 (A100) show the complemented line to have a restored A400/A100 ratio when compared with the high ratio of mmdh1mmdh2 (17 times higher than the wild type). Further consolidating rescue of the photorespiratory defect in mmdh1mmdh2, metabolite analyses of 4-week-old mmdh1mmdh2 and mmdh1mmdh2 35S:mMDH1 plants (sampled 3 h into the day) showed that, in comparison with the wild type, the high levels of Gly, Ser, succinate, and malate seen in mmdh1mmdh2 were restored to wild-type levels in the complemented line (Fig. 8B).
Figure 7.
Growth and biochemical characterization of mmdh1mmdh2 complemented with mMDH. A, Plant growth in short-day growth conditions at 40 d of age in the wild type (Wt), mmdh1mmdh2, and a complemented mmdh1mmdh2 35S:mMDH line. B, Changes in total cellular MDH activity. Bars are means ± se (n = 4). C, Total MDH activity in isolated mitochondria, showing activity in intact mitochondria and activity after organelle rupture with Triton X-100. Bars are means ± se (n = 3). D, Malate-dependent respiratory rate of isolated intact mitochondria at pH 6.5 to maximize NAD-ME activity and at pH 7.5 to maximize MDH activity. Bars are means ± se (n = 3). E, Changes in leaf respiration rate measured as oxygen consumption rate in the dark. Bars are means ± se (n = 5). FW, Fresh weight.
Figure 8.
Physiological and metabolite characterization of mmdh1mmdh2 complemented with mMDH. A, A/Ci curves of the wild type (Wt), mmdh1mmd2, and mmdh1mmdh2 35S:mMDH1 and A400/A100 ratios, where bars are mean ratios ± sd (n = 3). B, Metabolite ratios between the wild type and mmdh1mmdh2 and between the wild type and mmdh1mmdh2 35S:mMDH1 at 3 h into the day. Fold changes with significant differences (P < 0.05; n = 5) are indicated with asterisks. Plants were grown under short-day conditions.
mMDH Affects GLDH Abundance and Foliar Ascorbate Concentration
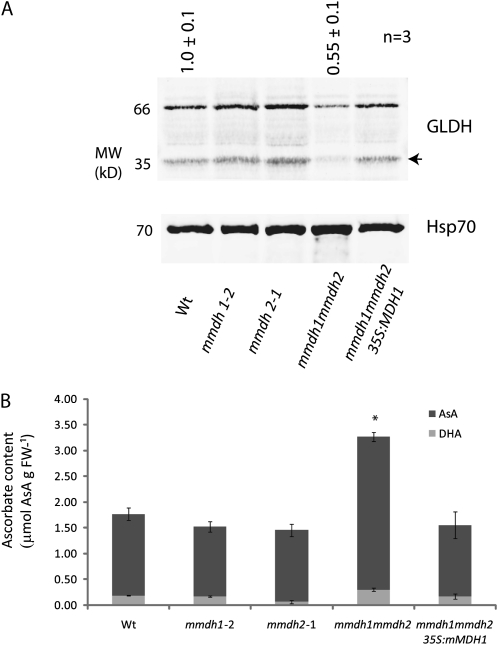
Antisense inhibition of MDH and aconitase in tomato plants has been reported to elevate the cellular ascorbate pool, and this has been linked to enhanced photosynthetic performance in long days (Nunes-Nesi et al., 2005; van der Merwe et al., 2009) but to stunted growth in short days (Nunes-Nesi et al., 2008). In our initial DIGE analysis, we noted that a truncated product of the enzyme that generates ascorbate, GLDH, was very low in abundance in mmdh1mmdh2 (Fig. 3). This prompted a further investigation of this enzyme and ascorbate levels in our knockout lines to determine if the links to plant growth and respiratory and photosynthetic performance reported in tomato could be confirmed in Arabidopsis. Antibodies raised to GLDH identified two bands on western blots of isolated mitochondria, one the expected size of GLDH (approximately 66 kD) and the other the size of the truncated product identified in the DIGE analysis (approximately 35 kD; Fig. 9A). These immunoblots, loaded on a total mitochondrial protein basis, revealed that mmdh1mmdh2 had only approximately 60% of the wild-type abundance of GLDH and very low levels of the 35-kD truncated product, but wild-type levels of GLDH were regained in the complemented mmdh1mmdh2. For comparison, antibodies against HSP70 were used to show that the abundance of this protein was slightly increased in mmdh1mmdh2, consistent with the DIGE analysis. Analysis of the total cellular ascorbate pool from long-day-grown leaves also revealed a doubling of ascorbate content in mmdh1mmdh2 that returned to wild-type levels in the complemented mutant (Fig. 9B). The oxidation state of ascorbate was not significantly different across the genotypes. Interestingly, this increased ascorbate level did not manifest with enhanced photosynthetic performance, with the mmdh1mmdh2 double mutant showing unchanged photosynthetic capacity and a clearly visible growth defect across different lighting regimes (Fig. 1B). These data indicate either an effect of MDH abundance or activity on the steady-state abundance of GLDH, inversely impacting the ascorbate pool size, or high ascorbate levels having a negative feedback on GLDH abundance.
Figure 9.
Change in GLDH protein amount and total cellular ascorbate levels in mmdh1mmdh2. A, Immunoreactivity of GLDH in different mutants on a mitochondrial protein basis. A representative gel is shown, and mean abundance of the immuonreactive band ± se is shown above the lanes (n = 3). The arrow indicates a truncated product of GLDH found in DIGE (Fig. 3) and also immunodetected. B, Reduced and oxidized ascorbate (AsA) content of leaf tissue. Bars are means ± se (n = 4), and asterisks indicate significant differences (P < 0.05) from the wild type. FW, Fresh weight; Wt, wild type. Plants were grown under short-day conditions.
DISCUSSION
Removal of mMDH Reveals Metabolic Plasticity
Our data reveal that mMDH can be effectively eliminated from plant mitochondria without a loss of plant viability under controlled conditions, albeit with a significant growth deficit. This implies that the TCA cycle function of mMDH can be compensated by other cellular reactions to a certain extent. Multiple levels of explanation for this can be found in our data sets. At the level of malate oxidation in isolated mitochondria, it is clear that mmdh1mmdh2 can sufficiently drive maximal malate-dependent respiratory flux through the electron transport chain using NAD-ME alone, in the absence of mMDH, and thus a classical TCA cycle. TCA cycle function in vivo could be maintained by OAA/malate transport to the cytosol, allowing other cytosolic MDH reactions to support a cycle and moving a portion of the NADH production to the cytosol. However, the altered abundance of other mitochondrial matrix dehydrogenases when mMDH is lacking in mmdh1mmdh2 may indicate that the balance of NAD-linked reactions has been altered. These decreases (rather than compensatory increases) in NAD-linked enzyme abundance also hint that maintaining the redox balance between mitochondrial matrix carbon metabolism and electron transport activities in mmdh1mmdh2 requires decreases in other NAD+-reducing activities. This would be consistent with mMDH significantly operating in the direction of NADH oxidation, reducing OAA to malate. Further analysis of the mutants confirmed some expected roles of MDH but also highlighted some surprising impacts on cellular function.
Under outdoor conditions in the open air, a decreased mMDH capacity had much more severe effects when compared with climate chamber conditions. The double knockout mmdh1mmdh2 plants did not survive in outdoor conditions, and the mmdh1 plants with more than 50% decrease in MDH capacity showed a significant 60% reduction in seed production (Supplemental Fig. S2B). This implies that under the outdoor conditions, the compensatory mechanisms that operate are not sufficient to support normal metabolism. A reason for this is likely to be the rapid fluctuations in light intensity and temperature that occur in open-air conditions. If the compensatory mechanism involves cytosolic MDH (as suggested above), a shuttling of OAA/malate between compartments must occur. Changes in the fluxes of such a mechanism are likely to be comparably slow, since several metabolic pools are involved. Consequently, negative effects on metabolism can occur if the changes in redox regulation cannot keep pace with the demands set by the external fluctuations.
Carbon and Energy Are Directed Away from Respiration by mMDH
A decrease in the rate of mitochondrial decarboxylations linked to TCA cycle activity is often observed in the light (Hurry et al., 2005). This light inhibition of respiration has been directly linked to mitochondrial photorespiratory reactions, resulting in increased redox state and adenylate phosphorylation level in the mitochondrial matrix (Bykova et al., 2005). The effect is understood to include restrictions at the pyruvate dehydrogenase complex and at other reactions within the TCA cycle. The mutant plants studied here also show light inhibition of TCA cycle-linked respiration. In the wild type and single mutants, the inhibition was about 40% (Fig. 4, A and B), and mmdh1mmdh2 showed a similar level of inhibition (about 50%), although from a higher level in the dark.
What we note here that is different is the clear elevation of respiratory rate in terms of direct measurements of oxygen consumption and CO2 evolution in darkened leaves and calculations of leaf CO2 respiration in light in the mmdh1mmdh2 plants (Figs. 4, A and B, and 7E). This implies that in wild-type Arabidopsis plants, mMDH has a degree of metabolic control in limiting respiratory rate in leaves. This connection is confirmed by the analysis of the complemented mmdh1mmdh2, where respiratory rates are returned to wild-type levels by the expression of mMDH1 (Fig. 7). Notably, this is a leaf-specific effect, as analysis of root respiration suggests no change in intact tissue respiratory rates (Fig. 4C). One explanation of this effect is that in leaves, mMDH uses OAA from the cytosol to generate malate, competing with the electron transport chain for NADH and slowing TCA cycle function by competing with citrate synthase for OAA. When considering the negative impact that mMDH knockout has on net CO2 assimilation (Fig. 5A) and consequently in germination, growth, and seed set (Fig. 1B; Supplemental Fig. S2), it appears that this respiratory change has broad downstream consequences for plant biomass production.
However, this result of an elevated respiration by mMDH removal is not, at face value, consistent with reports on antisense inhibition of mMDH in tomato. In the primary report, lowering mMDH caused a decline in respiratory rates measured as 14CO2 release from [14C]Glc in whole leaves (Nunes-Nesi et al., 2005). Work on isolated mitochondria from fruits from the same plants showed a decrease in TCA cycle flux when analyzed by NMR but no change in respiratory capacity when measured as oxygen consumption. The same workers analyzed root respiration rates and showed dramatic reductions in respiratory rate (van der Merwe et al., 2009), which is distinct from our data in Arabidopsis mutants (Fig. 4C). However, direct gas-exchange measurements of respiration in tomato plants are not reported, and although the antisense clearly reduces mMDH, it is unclear how much residual mMDH was present in the tomato antisense plants to compare with the single or double mutant Arabidopsis data set. So, while it is not clear how to directly reconcile the differences noted at this time, the hint that the phenotype in tomato is very conditional on daylength might show that substantial dynamics operate in the translation of mMDH changes to resultant changes in respiration rate and plant growth (Nunes-Nesi et al., 2005, 2008). Our data in Arabidopsis, irrespective of daylength, are most similar to those reported in tomato for short-day-grown rather than long-day-grown plants (Nunes-Nesi et al., 2005, 2008).
A Role of mMDH in Photorespiration
Metabolic schemes of photorespiration note that to balance the pathway as a whole, the reducing equivalents from Gly oxidation in mitochondria need to be returned to the peroxisome to allow hydroxpyruvate reduction to generate glycerate for reentry to the chloroplast. Most often, it is assumed that a malate/OAA shuttle including both mMDH and PMDH is responsible; however, the real mechanism of this linkage has been a matter of conjecture. Recently, we noted that PMDH double mutants are not classical photorespiratory mutants, as they can grow in normal air, albeit with a slight decrease in photosynthetic rates compared with the wild type (Cousins et al., 2008). However, we did report alterations of the rate of photorespiration and the CO2 PIB, suggesting that the ratio of CO2 release to ribulose 1,5-bisphosphate oxygenation was altered. Here, we note that the double mutants for mMDH have a modified CO2 PIB, a distinct break in the initial part of the A/Ci curves, and elevated Gly and to a lesser extent Ser levels (Figs. 6 and 8; Supplemental Figs. S3 and S4). This clearly implies some role of mMDH in photorespiration, perhaps mainly through a limitation on Gly oxidation rate.
Respiration, mMDH, Ascorbate Synthesis, and Ascorbate Pool Size
A role of plant mitochondria in ascorbate synthesis and metabolism was noted with the discovery that the terminal step of the main biosynthetic pathway, GLDH, is a mitochondrial inner membrane protein (Siendones et al., 1999), a plant-specific ancillary subunit of complex I (Millar et al., 2003; Pineau et al., 2008), and directly donates electrons to cytochrome c (Bartoli et al., 2000; Millar et al., 2003). A direct role for ascorbate in mitochondrial function was enhanced by the discovery that enzymes of the ascorbate/glutathione cycle are dual targeted to mitochondria and chloroplasts (Jimenez et al., 1997; Chew et al., 2003). The report that antisense of mMDH can elevate ascorbate pool size in tomato leaves and increase GL-dependent ascorbate biosynthesis rate has been interpreted as linked to these respiratory chain connections and the redox poise of the mitochondrial matrix (Nunes-Nesi et al., 2005). It has been reported that the abundance and activity of GLDH is not well correlated with foliar ascorbate levels (Bartoli et al., 2005), although RNA interference and antisense suppression of the enzyme have previously been reported to greatly reduce cellular ascorbate pools (Tabata et al., 2001; Alhagdow et al., 2007). Interestingly, a recent report has shown the capacity for GLDH to be redox regulated, particularly by glutathionylation of a particular amino acid residue (Leferink et al., 2009). We observed here that mMDH loss is associated with a decrease in the abundance of GLDH in mitochondria and elevated abundance of total cellular ascorbate. Currently, this does not directly explain the tantalizing connection between the TCA cycle and ascorbate content per se, but it does provide a clear molecular linkage between the machinery of the two pathways. It also shows that GLDH is just one of a number of NAD+- and peripheral electron transport chain-linked components that decrease in the mmdh1mmdh2 background. Other notable examples are NAD-dependent Glu and formate dehydrogenase and the enzymes for the NAD+- and ubiquinone-linked oxidation of branched chain amino acids. Hence, the ascorbate link to mMDH may be part of a shift in the steady-state abundance of enzymes to maintain the redox poise of the mitochondrial matrix in the absence of mMDH.
CONCLUSION
Overall, a complex and respiration-controlling role for mMDH is revealed by this mutant analysis in Arabidopsis. Changes in the abundance of isoforms of mMDH in mitochondria between tissues in different plants (Bardel et al., 2002; Lee et al., 2008), in different nuclear backgrounds in cytoplasmic mail sterility (Witt et al., 1997), and across inbred lines in crop genetic studies (Yang et al., 1977; Imsande et al., 2001) that have been noted over time may indicate that mMDH is not always expressed at the same level but could be used as a significant regulator of respiratory rate in plants. This may provide new directions for bioengineering of plant growth rate and a new insight into the molecular mechanisms underpinning to multifaceted link between respiration and photosynthesis in plants.
MATERIALS AND METHODS
Plant Lines and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana) ecotype Columbia wild type and T-DNA insertion lines (mmdh1 and mmdh2 single mutants as well as mmdh1mmdh2 double mutants) were typically sown on a 1:3 perlite:soil mix (Hasselfors Garden, P-Jord; nitrogen:phosphorus:potassium 14:7:18, pH 6, magnesium 250 g m−3). Seedlings were first covered with transparent acrylic hoods and regularly sprayed. After 1 week, the hoods were removed and the seedlings were transplanted into individual pots filled with the same 1:3 perlite:soil mix. Plants were grown under controlled conditions as follows: a short-day photoperiod (8 h of light/16 h of dark), an irradiance of 250 μmol m−2 s−1 PPFD, a relative humidity of 75%, and a temperature cycle of 22°C day/17°C night; a long-day photoperiod (16 h of light/8 h of dark), an irradiance of 200 μmol m−2 s−1 PPFD, a relative humidity of 60%, and a temperature cycle of 22°C day/17°C night; and a medium-day photoperiod (12 h of light/12 h of dark), an irradiance of 150 μmol m−2 s−1 PPFD, a relative humidity of 60%, and a temperature cycle of 22°C day/17°C night or in a Biochambers controlled growth cabinet with identical medium-day conditions and a heightened atmospheric CO2 concentration of 5,000 μL L−1. The outdoor experiments were performed in the experimental garden at Umeå University (63°50′N, 20°20′E). Growth conditions were similar to those described by Külheim et al. (2002). Seeds were germinated indoors and transferred to pots with soil at the seedling stage and then moved to the outdoor garden. For the wild type, mmdh1, and mmdh2, 40 plants were used, and for mmdh1mmdh2, 30 plants were used.
Isolation of mmdh T-DNA Insertion Mutants
The insertion lines GABI_097C10 (referred to as mmdh1-1) and GABI_540F04 (referred to as mmdh1-2) were obtained from the GABI-Kat population (Rosso et al., 2003). The insertion line SALK_126994 (referred to as mmdh2-1) was obtained from the Nottingham Arabidopsis Stock Center (Alonso et al., 2003). PCR-based screening was used to identify individuals homozygous for T-DNA insertions in mMDH1 (At1g53240) and mMDH2 (At3g15020) genes. The gene-specific primers mMDH1-1_F (5′-TAGTAACCCCAGCATGACCAC-3′), mMDH1-1_R (5′-CTATGCTCGTCCGATCTTCTG-3′), mMDH1-2_F (5′-CGGGATTCAAATTGTGATCAC-3′), mMDH1-2_R (5′-GATTGCTTCAGATTCGTCAGC-3′), mMDH2_F (5′-ATGACCACCAACAACTGGAAC-3′), and mMDH2_R (5′-AATCCCATAACTTTCCCAACG-3′) were used in combination with GABI-Kat (o8409, 5′-ATATTGACCATCATACTCATTGC-3′) and SALK (Lba1, 5′-TGGTTCACGTAGTGGGCCATCG-3′) T-DNA left border primers.
Complementation of mmdh1mmdh2
The mmdh1-2mmdh2 double mutant was transformed via the floral dip method with the reconstructed pGreen binary vector p0179 harboring a double cauliflower mosaic virus 35S promoter, full-length mMDH1 cDNA, and a cauliflower mosaic virus terminator cassette (Pracharoenwattana et al., 2005) transfected by Agrobacterium tumefaciens. Seeds collected from 35S:mMDH1 and 35S:empty vector-transformed plants were screened for hygromycin resistance.
RT-PCR
RNA extraction, DNase treatment, and first-strand cDNA synthesis were performed according to the manufacturer’s instructions using the Qiagen RNeasy kit and SuperScript III reverse transcriptase (Invitrogen). PCR was performed for 30 cycles with gene-specific primers as follows: for Actin-2 (At3g18780), ACT2-F (5′-GTATCGCTGACCGTATGAG-3′) and ACT2-R (5′-CTGCTGGAATGTGCTGAG-3′); for mMDH1 (At1g53240), mMDH1-F (5′-CAGCCTCTTGCTCTCCTCAT-3′) and mMDH1-R (5′-TCAAGAACCTCCTCCACACC-3′); for mMDH2 (At3g15020), mMDH2-F (5′-GACGTCGGTCACATCAACAC-3′) and mMDH2-R (5′-TCTGCCTTGAGGGCTTCTAA-3′). PCR products were visualized on an agarose gel stained with ethidium bromide. Fragment sizes for mMDH1 (1,512 bp) and mMDH2 (1,404 bp) were of the expected size.
Gas Exchange
Gas-exchange parameters were measured on 6-week-old plants using a LI-6400 XT infrared gas analyzer (Li-Cor). All measurements were done after at least 2 h of illumination. One fully developed leaf per plant was enclosed in a 6-cm2 leaf chamber. Data were recorded after the leaf acclimated in the leaf chamber to the desired conditions of light, temperature, relative humidity, and CO2 concentration. Data were adjusted to the enclosed leaf area determined with a LI-3000 leaf area meter (Li-Cor).
Gas Exchange in Growth Conditions
Gas-exchange parameters, including CO2 assimilation rate and stomatal conductance, were recorded after the leaf acclimated in the gas-exchange system’s leaf chamber under conditions similar to growth conditions (PPFD of 250 μmol m−2 s−1, relative humidity of 60%–70%, temperature of 22°C, and CO2 concentration of 400 μL L−1).
A/Ci Curves
After acclimation to the following conditions (saturating PPFD of 1,000 μmol m−2 s−1, relative humidity of 60% to 70%, and temperature of 25°C), the rate of CO2 assimilation was measured at intervals of CO2 concentration decreasing from 2,000 to 50 μL L−1. At each step, the rate of CO2 evolution was recorded after steady-state conditions were reached. The maximum rate of photosynthetic electron transport and the maximum carboxylation rate of Rubisco were determined from A/Ci response curves as described previously (Sharkey et al., 2007).
Determination of Mitochondrial CO2 Production
The rate of mitochondrial respiration in the light (day respiration [Rd]) was determined using the Laisk method as described by Laisk (1977) or more recently by Villar et al. (1994). This method determines the internal CO2 concentration (Ci) at which the rate of photosynthesis equals that of photorespiration. At this Ci (γ*), the rate of CO2 evolution represents Rd. For each leaf, the photosynthetic rate was recorded at decreasing Ci values (in the range 120–45 μL L−1) and at three different photon flux densities (25, 45, and 70 μmol m−2 s-l PPFD) at 22°C and 60% to 70% relative humidity. For each PPFD, the linear regression of photosynthetic rate versus Ci was calculated. For each leaf, the three linear regressions intersected at one point, the coordinates of which represent Rd and γ*.
The rate of mitochondrial respiration in the dark (Rn) was determined by measuring the rate of CO2 evolution in the dark. Leaves were dark acclimated in conditions similar to growth conditions (relative humidity of 60%–70%, temperature of 22°C, and CO2 concentration of 400 μL L−1). Dark respiration was also measured on detached leaf discs using a Clark-type oxygen electrode (Hansatech Instruments). Oxygen consumption was monitored at 25°C in aqueous phase.
PIB
The CO2 PIB was measured under photorespiratory conditions (saturating PPFD of 1,000 μmol m–2 s–1, CO2 concentration of 100 μL L−1, temperature of 25°C, relative humidity of 60%–70%) as described by Kebeish et al. (2007).
Gas Exchange in Low Oxygen
Gas-exchange parameters and CO2 responses curves were determined as described above, feeding the gas-exchange system with N2 containing only traces of oxygen (less than 0.05%; AGA Gas).
Purification of Mitochondria from Hydroponic Shoot Tissue
All relevant Arabidopsis genotypes were grown and maintained in exact accordance with a previously published protocol (Lee et al., 2008). Shoot mitochondria were isolated from 3-week-old hydroponically grown Arabidopsis using a method adapted from Lee et al. (2008) with slight revisions. Approximately 100 g of shoot material was homogenized with a Polytron blender (Kinematica) in 300 mL of cold grinding medium (0.3 m Suc, 25 mm tetrasodium pyrophosphate, 1% [w/v] polyvinylpyrrolidone-40, 2 mm EDTA, 10 mm KH2PO4, 1% [w/v] bovine serum albumin [BSA], and 20 mm sodium-ascorbic acid, pH 7.5) for 10 s twice with 10-s intervals between bursts. The homogenate was filtered through four layers of Miracloth and centrifuged at 1,500g for 5 min, and the resulting supernatant was then centrifuged at 24,000g for 15 min. The organelle pellet was washed by repeating the 1,500g and 24,000g centrifugation steps twice in Suc wash medium (0.3 m Suc, 0.1% [w/v] BSA, and 10 mm TES, pH 7.5). The resulting pellet of crude organelles was carefully resuspended in Suc wash medium and gently layered over a 35-mL continuous 28% Percoll density gradient consisting of 0% to 4.4% polyvinylpyrrolidone-40. The gradient was then centrifuged at 40,000g for 45 min. The mitochondrial band was seen as a yellow-brown band near the bottom of the centrifuge tube. The upper layers of the density gradient were removed, and the mitochondrial band was collected. The mitochondrial fraction was diluted approximately 5-fold with Suc wash medium devoid of BSA and centrifuged at 24,000g for 10 min. The pellet was resuspended in residual Suc wash medium and loaded onto a 45% Percoll gradient, which was consequently centrifuged at 40,000g for 45 min. Mitochondria formed a white-yellow band near the top of the gradient and were carefully removed by pipette, with maximal effort exercised to avoid coremoval of the adjacent green band containing chloroplasts and plastids. The mitochondria obtained were then diluted 5-fold in wash buffer (no BSA) and centrifuged at 24,000g for 10 min. This step was repeated three times, with the final pellet retained in a small volume of wash buffer for usage in later experiments.
Leaf, Root, and Isolated Mitochondria Oxygen Consumption
All oxygen consumption measurements were performed using a Clarke-type oxygen electrode, and the data feed was collected by Oxygraph Plus version 1.01 software (Hansatech Instruments). The electrode was calibrated via the addition of excess sodium dithionite for depletion of all oxygen in a 1-mL volume of deionized water housed in the electrode chamber. Equal-sized leaf discs or root tissues were removed from plants, immersed in incubation medium (10 mm HEPES, 10 mm MES, and 2 mm CaCl2, pH 7.2), and incubated in the dark for a period of 30 min. Total respiration was measured at 22°C in the dark following the addition of approximately 20 to 50 mg of leaf tissue or 50 to 80 mg of root tissue to 2 mL of incubation medium.
Respiration rates on isolated mitochondria were performed at 25°C on 100 μg of mitochondrial proteins suspended in 1 mL of respiration buffer (0.3 m Suc, 5 mm KH2PO4, 10 mm TES, 10 mm NaCl, 2 mm MgSO4, and 0.1% [w/v] BSA, pH 7.2). Oxygen consumption attributed to malate-to-OAA conversion by ME (respiration buffer pH 6.5) or MDH (respiration buffer pH 7.5) was measured in the presence of NAD+ (2 mm), CoA (12 μm), TPP (0.2 mm), Glu (10 mm), ADP (0.3 mm), and malate (10 mm) in order to initiate NADH production through the TCA cycle and also to obtain state III respiration rates. KCN (0.1 mm) was added to halt the reaction. Note that Glu was used as a respiratory substrate to prevent the accumulation of OAA to drive the reaction in favor of malate oxidation. Mitochondrial integrity was measured following oxygen uptake after the addition of ascorbate (10 mm), cytochrome c (25 μm), and Triton X-100 (0.05%, w/v). In all experiments, membrane integrity exceeded 90%.
Two-Dimensional Gel Electrophoresis
Mitochondrial proteins (250 μg) from control, mmdh1-2, mmdh2-1, mmdh1mmdh2, and complemented plants were precipitated overnight at −20°C in 100% (v/v) acetone. After centrifugation at 20,000g and 4°C for 20 min, pellets were resuspended in 450 μL of rehydration buffer (8 m urea, 4% [w/v] CHAPS, 0.5% [v/v] pH 3–10 nonlinear immobilized pH gradient IPG buffer, 18 mm dithiothreitol [DTT], and 0.001% [w/v] bromphenol blue). The buffer was loaded onto a 24-cm-long strip with an immobilized nonlinear pH gradient of 3 to 10 (Immobiline DryStrip; GE Healthcare). Rehydration of the strips and the first isoelectric focusing (IEF) dimension electrophoresis were performed on an IPGphor Unit (GE Healthcare) using the following settings: 12 h at 30 V (rehydration step), 1 h at 500 V, 1-h gradient from 500 to 1,000 V, 1-h gradient from 1,000 to 3,000 V, 2-h gradient from 3,000 to 8,000 V, and 5 h at 8,000 V. After IEF, strips were incubated for 15 min in an equilibration buffer (6 m urea, 2% [w/v] SDS, 26% [v/v] glycerol, 65 mm DTT, 0.001% [w/v] bromphenol blue, and 50 mm Tris-HCl, pH 8.8) and then for 15 min in an equilibration buffer containing iodoacetamide (6 m urea, 2% SDS [w/v], 26% [v/v] glycerol, 135 mm iodoacetamide, 0.001% [w/v] bromphenol blue, and 50 mm Tris-HCl, pH 8.8). The equilibrated strips were then loaded on top of a 12% (w/v) acrylamide gel. Following separation, gels were immersed for 1 h in fixing solution (40% methanol and 10% acetic acid) and stained with blue silver.
Two-Dimensional DIGE
Mitochondrial proteins from control and mmdh1mmdh2 plants were precipitated overnight at −20°C in 100% (v/v) acetone. After centrifugation at 20,000g and 4°C for 20 min, pellets were resuspended in 10 μL of DIGE lysis buffer (8 m urea, 4% [w/v] CHAPS, and 40 mm Tris, pH 8.5) and centrifuged again at 12,000g and 4°C for 10 min in order to remove insoluble material. Fifty micrograms of proteins from each sample were labeled with a different Cy dye (Cy-3 or Cy-5) by the addition of 400 pmol of dye (freshly prepared in dimethylformamide according to the manufacturer’s instructions) on ice in the dark for 30 min. The reaction was stopped by the addition of Lys (10 mm) for 10 min on ice in the dark. An equal volume (12 μL) of DIGE lysis buffer with 22 mm DTT was added to each sample. Each of the labeled protein samples was mixed, and rehydration buffer (8 m urea, 4% [w/v] CHAPS, 0.5% [v/v] 3 to 10 NL IPG buffer, 18 mm DTT, and 0.001% [w/v] bromphenol blue) was added to give a final volume of 450 μL. The mix was loaded onto a 24-cm-long strip with an immobilized nonlinear pH gradient of 3 to 11 (Immobiline DryStrip; GE Healthcare). Rehydration of the strips and the first IEF dimension electrophoresis were performed on an IPGphor Unit (GE Healthcare) using the following settings: 12 h at 30 V (rehydration step), 1 h at 500 V, 1-h gradient from 500 to 1,000 V, 1-h gradient from 1,000 to 3,000 V, 2-h gradient from 3,000 to 8,000 V, and 5 h at 8,000 V. After IEF, strips were incubated for 15 min in an equilibration buffer (6 m urea, 2% [w/v] SDS, 26% [v/v] glycerol, 65 mm DTT, 0.001% [w/v] bromphenol blue, and 50 mm Tris-HCl, pH 8.8) and then for 15 min in an equilibration buffer containing iodoacetamide (6 m urea, 2% [w/v] SDS, 26% [v/v] glycerol, 135 mm iodoacetamide, 0.001% [w/v] bromphenol blue, and 50 mm Tris-HCl, pH 8.8). The equilibrated strips were then loaded on top of a 12% (w/v) acrylamide gel. Following separation, gels were scanned using the Typhoon Trio Variable Mode Imager at a resolution of 100 (pixel size), and the photomultiplier tube was set to 500 V. Proteins were processed (quantification) using the DeCyder 2-D Differential Analysis software version 6.5 (GE Healthcare). In order to get statistical significance from these experiments, three sets of proteins from three independent experiments were labeled and subjected to electrophoresis. Standard gels were also used, and precipitation, IEF, and SDS-PAGE were run in parallel with labeled samples. These standard gels were loaded with a mix of 250 μg of each protein sample. After electrophoresis, proteins were visualized by colloidal Coomassie Brilliant Blue G250 staining. The aim of the standard gel is to allow the identification of proteins by excision of gel spots followed by mass spectrometry.
Identification of Proteins by Liquid Chromatography-Tandem Mass Spectrometry
Gel spots to be analyzed were cut from colloidal Coomassie Brilliant Blue-stained gels. Proteins were digested with trypsin (trypsin sequencing grade; Roche Diagnostic) according to Lee et al. (2008). Prior to mass spectrometry, peptides were resuspended in 5% acetonitrile and 0.1% formic acid. The samples were analyzed with an Agilent 1100 series capillary liquid chromatography system and an Agilent Technologies XCT Ultra IonTrap with an electrospray ionization source equipped with a low-flow nebulizer in positive mode controlled by Chemstation (Rev B.01.03 [204]; Agilent Technologies) and MSD Trap Control version 6.0 (Build 38.15) software (Bruker Daltonik). Samples were loaded with an Agilent 1100 series capillary liquid chromatography system onto a 0.5- × 50-mm C18 (5 μm, 100 Å) reverse-phase column (Higgins Analytical) with a C18 OPTI-GUARD guard column (Optimize Technologies) at 10 μL min−1 equilibrated with 5% acetonitrile and 0.1% formic acid under a regulated temperature of 50°C. Peptides were eluted from the C18 reverse-phase column at 10 μL min−1 into the XCT Ultra IonTrap by a 9-min acetonitrile gradient (5%–60%) in 0.1% formic acid at 50°C. Initial ion detection utilized a mass range of 200 to 1,400 mass-to-charge ratio (m/z) with a scan mode set to 8,100 m/z per s. Ions reaching an intensity of 80,000 cps were sent to tandem mass spectrometry (MS/MS) analysis, and two precursors were selected for the initial MS scan. MS/MS conditions employed SmartFrag for ion fragmentation (scan range of 70–2,200 m/z). The resulting MS/MS spectra were exported from the Data Analysis for LC/MSD Trap version 3.3 (Build 149) software package (Bruker Daltonik) using default parameters for AutoMS(n) and compound export. For protein identification, data generated from MS/MS spectra were matched against The Arabidopsis Information Resource (release 8) Arabidopsis database supplemented with chloroplast- and mitochondria-encoded protein sequences (the combined database contained a total of 30,700 protein sequences with 12,656,682 residues) using the Mascot search engine version 2.1.04 (Matrix Sciences). Searches were conducted at parameters set at error tolerances of ±1.2 for MS and ±0.6 for MS/MS, with “Max. missed cleavages” set to 1. Results were filtered using “Standard scoring,” “Max. number of hits” set to 20, and “Significance threshold” at P < 0.05. Details of matches are shown in Supplemental Table S1.
Extraction of Total Leaf Proteins
In accordance with a previously established protocol (Ritte et al., 2003), leaves of relevant Arabidopsis genotypes were ground in extraction buffer (50 mm HEPES-KOH, pH 7.5, 1 mm EDTA, 5 mm DTT, 10% [v/v] glycerol, 0.5 mm phenylmethylsulfonyl fluoride, 2 mm benzamidine, and 2 mm ε-aminocaproic acid). The homogenates were centrifuged at 14,000g and 4°C for 10 min. The supernatant (designated crude extract) was quantified and used for the determination of total MDH activity.
Assay of Total and Mitochondrial Specific MDH Activity
The activity of MDH was measured by following NADH oxidation to NAD+ at 340 nm. The assay was carried out using the following reaction mixture: 90 mm KH2PO4-KOH (pH 7.4), 0.05% (v/v) Triton X-100, 5 mm MgCl2, and 2 μm NADH. The reaction was initiated by the addition of 750 μm OAA. Freshly isolated mitochondria (25 μg) were used for the determination of latent mMDH activity in much the same way, albeit in a reaction mixture comprising mitochondrial reaction buffer (0.3 m Suc, 5 mm KH2PO4, 10 mm TES, 10 mm NaCl, 2 mm MgSO4, and 0.1% [w/v] BSA, pH 7.4), 50 μm n-propylgallate, and 1 mm KCN. NADH oxidation rates before and after the addition of 0.05% Triton X-100 were obtained to determine the latency of mMDH activity in the mitochondria of each genotype.
Gas Chromatography-MS Analysis of Organic and Amino Acids
Metabolites were extracted from frozen Arabidopsis leaves that had been harvested at the times indicated according to a method adapted from that described by Roessner-Tunali et al. (2003), and derivatized metabolite samples were analyzed on an Agilent Technologies GC/MSD system (http://www.home.agilent.com). Detailed methods are given by Sappl et al. (2009). Raw gas chromatography (GC)-MS data preprocessing and statistical analysis were performed using Metabolome-Express software (Carroll et al., 2010; version 1.0; http://www.metabolome-express.org). Full data are available via the aforementioned Web site, and processed data are provided in Supplemental Tables S2 and S3.
Ascorbic Acid Measurements
Ascorbic acid content was measured by the ascorbate oxidase assay (Rao and Ormrod, 1995). In accordance with an established protocol (Zhang et al., 2009), plant extracts were made using liquid N2-cooled tissue ground in 6% (w/v) meta-phosphoric acid and centrifuged at 15,000g for 15 min. Reduced ascorbic acid was determined by measuring the decrease in A265 (ε = 14.3 mm−1 cm−1) after the addition of 1 unit of Cucurbita-derived ascorbate oxidase (Sigma) to 1 mL of reaction medium containing the plant extract and 100 mm KH2PO4 (pH 6.9). Oxidized ascorbic acid was measured by the increase in A265 following the addition of 1 μL of 0.2 mm DTT and incubating at room temperature for 15 min. Total ascorbate represents the addition of reduced and oxidized ascorbate concentrations.
Immunodetection of Mitochondrial Proteins
For western blotting with one-dimensional SDS-PAGE, Bio-Rad Criterion precast gels (10%–20% [w/v] acrylamide, Tris-HCl, 1 mm thick, 18-well comb) were used with equal amounts of total protein loaded in each lane. Gel electrophoresis was performed at 25 mA per gel for 3 h. Polyacrylamide gels were then incubated in a transfer solution for 1 h. Protein (quantified mitochondrial protein extracts) transfer onto a Hybond-C Extra nitrocellulose membrane (GE Healthcare) was performed using a Hoefer SemiPhor (GE Healthcare) instrument according to the manufacturer’s instructions. Transferred proteins were probed with primary antibodies against mtHSP70 (from T. Elthon) and GLDH (commercial antibody from Agrisera). Chemiluminescence detection linked to horseradish peroxidase was used to visualize a secondary antibody, and quantitative light emission was recorded using ImageQuant-RT ECL (Amersham Biosciences).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. mMDH genes in Arabidopsis and T-DNA mutant characterizations.
Supplemental Figure S2. Additional phenotypic characterization of mmdh mutants.
Supplemental Figure S3. Kinetics of CO2 evolution during the postillumination period in mmdh mutants.
Supplemental Figure S4. A/Ci curves for mmdh mutants, light-saturated CO2 assimilation rates, and chlorophyll content index measurements.
Supplemental Figure S5. PCR amplification of intact genes, t-DNA inserts, and inserted mMDH1 cDNA in single and double mutants and the complemented mutant, used in Figure 7 and 8 experiments.
Supplemental Table S1. Identification of proteins from protein spots changing in abundance in mitochondrial extracts from the wild type (WT) and mmdh1mmdh2 mutants using nano-liquid chromatography-MS/MS.
Supplemental Table S2. GC-MS metabolite analysis of mmdh1mmdh2 versus the wild type (WT) for Figure 6.
Supplemental Table S3. GC-MS metabolite analysis for mmdh1mmdh2 versus the wild type (WT) and mmdh1mmdh2 35S:mMDH versus the wild type for Figure 8.
Supplementary Material
References
- Alhagdow M, Mounet F, Gilbert L, Nunes-Nesi A, Garcia V, Just D, Petit J, Beauvoit B, Fernie AR, Rothan C, et al. (2007) Silencing of the mitochondrial ascorbate synthesizing enzyme l-galactono-1,4-lactone dehydrogenase affects plant and fruit development in tomato. Plant Physiol 145: 1408–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bardel J, Louwagie M, Jaquinod M, Jourdain A, Luche S, Rabilloud T, Macherel D, Garin J, Bourguignon J. (2002) A survey of the plant mitochondrial proteome in relation to development. Proteomics 2: 880–898 [DOI] [PubMed] [Google Scholar]
- Bartoli CG, Guiamet JJ, Kiddle G, Pastori GM, Di Cagno R, Theodoulou FL, Foyer CH. (2005) Ascorbate content of wheat leaves is not determined by maximal L-galactono-1,4-lactone dehydrogenase (GalLDH) activity under drought stress. Plant Cell Environ 28: 1073–1081 [Google Scholar]
- Bartoli CG, Pastori GM, Foyer CH. (2000) Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol 123: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkemeyer M, Scheibe R, Ocheretina O. (1998) A novel, non-redox-regulated NAD-dependent malate dehydrogenase from chloroplasts of Arabidopsis thaliana L. J Biol Chem 273: 27927–27933 [DOI] [PubMed] [Google Scholar]
- Bykova NV, Keerberg O, Pärnik T, Bauwe H, Gardeström P. (2005) Interaction between photorespiration and respiration in transgenic potato plants with antisense reduction in glycine decarboxylase. Planta 222: 130–140 [DOI] [PubMed] [Google Scholar]
- Carroll AJ, Badger MR, Harvey Millar A. (2010) The MetabolomeExpress Project: enabling Web-based processing, analysis and transparent dissemination of GC/MS metabolomics datasets. BMC Bioinformatics 11: 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew O, Whelan J, Millar AH. (2003) Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J Biol Chem 278: 46869–46877 [DOI] [PubMed] [Google Scholar]
- Cousins AB, Pracharoenwattana I, Zhou W, Smith SM, Badger MR. (2008) Peroxisomal malate dehydrogenase is not essential for photorespiration in Arabidopsis but its absence causes an increase in the stoichiometry of photorespiratory CO2 release. Plant Physiol 148: 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R, Bonner WD., Jr (1972) Oxalacetate control of Krebs cycle oxidations in purified plant mitochondria. Biochem Biophys Res Commun 47: 619–624 [DOI] [PubMed] [Google Scholar]
- Eubel H, Meyer EH, Taylor NL, Bussell JD, O’Toole N, Heazlewood JL, Castleden I, Small ID, Smith SM, Millar AH. (2008) Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol 148: 1809–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietl C. (1992) Malate dehydrogenase isoenzymes: cellular locations and role in the flow of metabolites between the cytoplasm and cell organelles. Biochim Biophys Acta 1100: 217–234 [DOI] [PubMed] [Google Scholar]
- Hatch M, Osmond C. (1976) Transport in Plants III. Springer, Berlin [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurry V, Igamberdiev AU, Keerberg O, Pärnik T, Atkin OK, Zaragoza-Castells J, Gardeström P. (2005) Respiration in photosynthetic cells. Lambers H, Ribas-Carbo M, , Plant Respiration: From Cell to Ecosystem. Springer, Dordrecht, The Netherlands, pp 43–61 [Google Scholar]
- Igamberdiev AU, Gardeström P. (2003) Regulation of NAD- and NADP-dependent isocitrate dehydrogenases by reduction levels of pyridine nucleotides in mitochondria and cytosol of pea leaves. Biochim Biophys Acta 1606: 117–125 [DOI] [PubMed] [Google Scholar]
- Imsande J, Pittig J, Palmer RG, Wimmer C, Gietl C. (2001) Independent spontaneous mitochondrial malate dehydrogenase null mutants in soybean are the result of deletions. J Hered 92: 333–338 [DOI] [PubMed] [Google Scholar]
- Jimenez A, Hernandez JA, Del Rio LA, Sevilla F. (1997) Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiol 114: 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, Neuburger M, Douce R. (1981) Role of glutamate-oxaloacetate transaminase and malate dehydrogenase in the regeneration of NAD for glycine oxidation by spinach leaf mitochondria. Plant Physiol 67: 467–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebeish R, Niessen M, Thiruveedhi K, Bari R, Hirsch HJ, Rosenkranz R, Stäbler N, Schönfeld B, Kreuzaler F, Peterhänsel C. (2007) Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat Biotechnol 25: 593–599 [DOI] [PubMed] [Google Scholar]
- Krömer S. (1995) Respiration during photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 46: 47–70 [Google Scholar]
- Külheim C, Agren J, Jansson S. (2002) Rapid regulation of light harvesting and plant fitness in the field. Science 297: 91–93 [DOI] [PubMed] [Google Scholar]
- Laisk A. (1977) Kinetics of Photosynthesis and Photorespiration in C3-Plants. Nauka, Moscow [Google Scholar]
- Lee CP, Eubel H, O’Toole N, Millar AH. (2008) Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol Cell Proteomics 7: 1297–1316 [DOI] [PubMed] [Google Scholar]
- Leferink NGH, van Duijn E, Barendregt A, Heck AJR, van Berkel WJH. (2009) Galactonolactone dehydrogenase requires a redox-sensitive thiol for optimal production of vitamin C. Plant Physiol 150: 596–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Mittova V, Kiddle G, Heazlewood JL, Bartoli CG, Theodoulou FL, Foyer CH. (2003) Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiol 133: 443–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giegé P, Leaver CJ. (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AMO, Loureiro ME, Ratcliffe RG, Sweetlove LJ, Fernie AR. (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Sulpice R, Gibon Y, Fernie AR. (2008) The enigmatic contribution of mitochondrial function in photosynthesis. J Exp Bot 59: 1675–1684 [DOI] [PubMed] [Google Scholar]
- Palmer J. (1984) The mechanism and regulation of malate oxidation in isolated plant mitochondria. Physiol Veg 22: 665–673 [Google Scholar]
- Peltier JB, Cai Y, Sun Q, Zabrouskov V, Giacomelli L, Rudella A, Ytterberg AJ, Rutschow H, van Wijk KJ. (2006) The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol Cell Proteomics 5: 114–133 [DOI] [PubMed] [Google Scholar]
- Pineau B, Layoune O, Danon A, De Paepe R. (2008) L-Galactono-1,4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. J Biol Chem 283: 32500–32505 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM. (2005) Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17: 2037–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM. (2007) Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J 50: 381–390 [DOI] [PubMed] [Google Scholar]
- Rao MV, Ormrod DP. (1995) Ozone exposure decreases UVB sensitivity in a UVB-sensitive flavonoid mutant of Arabidopsis. Photochem Photobiol 61: 71–78 [DOI] [PubMed] [Google Scholar]
- Ritte G, Steup M, Kossmann J, Lloyd JR. (2003) Determination of the starch-phosphorylating enzyme activity in plant extracts. Planta 216: 798–801 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR. (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sappl PG, Carroll AJ, Clifton R, Lister R, Whelan J, Millar AH, Singh KB. (2009) The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J 58: 53–68 [DOI] [PubMed] [Google Scholar]
- Sharkey TD, Bernacchi CJ, Farquhar GD, Singsaas EL. (2007) Fitting photosynthetic carbon dioxide response curves for C(3) leaves. Plant Cell Environ 30: 1035–1040 [DOI] [PubMed] [Google Scholar]
- Siendones E, Gonzalez-Reyes JA, Santos-Ocana C, Navas P, Cordoba F. (1999) Biosynthesis of ascorbic acid in kidney bean: l-galactono-γ-lactone dehydrogenase is an intrinsic protein located at the mitochondrial inner membrane. Plant Physiol 120: 907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata K, Oba K, Suzuki K, Esaka M. (2001) Generation and properties of ascorbic acid-deficient transgenic tobacco cells expressing antisense RNA for L-galactono-1,4-lactone dehydrogenase. Plant J 27: 139–148 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Tan YF, Jacoby RP, Millar AH. (2009) Abiotic environmental stress induced changes in the Arabidopsis thaliana chloroplast, mitochondria and peroxisome proteomes. J Proteomics 72: 367–378 [DOI] [PubMed] [Google Scholar]
- Tesfaye M, Temple SJ, Allan DL, Vance CP, Samac DA. (2001) Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiol 127: 1836–1844 [PMC free article] [PubMed] [Google Scholar]
- van der Merwe MJ, Osorio S, Moritz T, Nunes-Nesi A, Fernie AR. (2009) Decreased mitochondrial activities of malate dehydrogenase and fumarase in tomato lead to altered root growth and architecture via diverse mechanisms. Plant Physiol 149: 653–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar R, Held AA, Merino J. (1994) Comparison of methods to estimate dark respiration in the light in leaves of two woody species. Plant Physiol 105: 167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt U, Lührs R, Buck F, Lembke K, Grüneberg-Seiler M, Abel W. (1997) Mitochondrial malate dehydrogenases in Brassica napus: altered protein patterns in different nuclear mitochondrial combinations. Plant Mol Biol 35: 1015–1021 [DOI] [PubMed] [Google Scholar]
- Yang NS, Sorenson JC, Scandalios JG. (1977) Genetic control of mitochondrial malate dehydrogenases: evidence for duplicated chromosome segments. Proc Natl Acad Sci USA 74: 310–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Lorence A, Gruszewski HA, Chevone BI, Nessler CL. (2009) AMR1, an Arabidopsis gene that coordinately and negatively regulates the mannose/l-galactose ascorbic acid biosynthetic pathway. Plant Physiol 150: 942–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.