Abstract
Plant calcium-dependent protein kinases (CDPKs) may function as calcium sensors and play important roles in the regulation of plant growth and development and in plant responses to biotic and abiotic stresses. The Arabidopsis (Arabidopsis thaliana) genome encodes 34 CDPKs, and most of them have not been functionally characterized. Here, we report the functional characterization of CPK10 in Arabidopsis response to drought stress. The cpk10 mutant, a T-DNA insertion mutant for the Arabidopsis CPK10 gene, showed a much more sensitive phenotype to drought stress compared with wild-type plants, while the CPK10 overexpression lines displayed enhanced tolerance to drought stress. Induction of stomatal closure and inhibition of stomatal opening by abscisic acid (ABA) and Ca2+ were impaired in the cpk10 mutants. Using yeast two-hybrid methods, a heat shock protein, HSP1, was identified as a CPK10-interacting protein. The interaction between CPK10 and HSP1 was further confirmed by pull-down and bimolecular fluorescence complementation assays. The HSP1 knockout mutant (hsp1) plants showed a similar sensitive phenotype under drought stress as the cpk10 mutant plants and were similarly less sensitive to ABA and Ca2+ in regulation of stomatal movements. Electrophysiological experiments showed that ABA and Ca2+ inhibition of the inward K+ currents in stomatal guard cells were impaired in the cpk10 and hsp1 mutants. All presented data demonstrate that CPK10, possibly by interacting with HSP1, plays important roles in ABA- and Ca2+-mediated regulation of stomatal movements.
Plants are subjected to various environmental stresses during their growth and development and have developed various mechanisms to adapt to these stresses. As an important cytoplasmic second messenger, Ca2+ plays critical roles in plant responses to environmental stresses (Rudd and Franklin-Tong, 2001; Sanders et al., 2002; Kudla et al., 2010). Specific calcium signatures may be recognized by different sensor proteins. Three major families of Ca2+ sensors have been identified in higher plants: calmodulins (CaMs) and CaM-like proteins (McCormack et al., 2005); calcineurin B-like (CBL) proteins (Kolukisaoglu et al., 2004; Luan, 2009; Weinl and Kudla, 2009); and calcium-dependent protein kinases (CDPKs; Harmon et al., 2000; Cheng et al., 2002; Harper et al., 2004; Harper and Harmon, 2005). CaMs and CBLs are small proteins and transmit the Ca2+ signal through interacting target proteins and regulating their activities. The CBLs not only regulate the activities of CBL-interacting protein kinases, but at least some of them are also involved in recruiting the kinases to different membranes (Luan, 2009). CDPKs are activated upon binding Ca2+ to their CaM-like domain and then relay the signaling to their downstream targets (Harmon et al., 2000; Cheng et al., 2002; Harper et al., 2004; Harper and Harmon, 2005).
CDPKs are found in a wide range of vascular and nonvascular plants as well as in green algae and certain protozoa (Harmon et al., 2001), suggesting their potential importance in Ca2+ signaling in plant cells. The CDPKs are encoded by multigene families and have been identified in various plant species, such as Arabidopsis (Arabidopsis thaliana; Harmon et al., 2001; Cheng et al., 2002), rice (Oryza sativa; Asano et al., 2005; Wan et al., 2007), cotton (Gossypium hirsutum; Huang et al., 2008), and wheat (Triticum aestivum; Li et al., 2008). Some CDPKs are expressed ubiquitously, whereas others are present in specific tissues or their expression is regulated by different stimuli (Hrabak et al., 2003). It is also known that different CDPKs have different subcellular locations, including cytosol, nucleus, the plasma membrane, endoplasmic reticulum, peroxisomes, mitochondrial outer membrane, and oil bodies (Harper et al., 2004), indicating their possible diverse functions. A number of studies have demonstrated that CDPKs play important roles in plant responses to various abiotic stresses, including cold, salt, drought, wounding, etc. (Cheng et al., 2002; Ludwig et al., 2004; Klimecka and Muszyńska, 2007; DeFalco et al., 2010). Transcription of AtCPK10 and AtCPK11 can be rapidly induced by drought and high-salt stresses (Urao et al., 1994). Overexpression of OsCDPK7 in rice plants enhanced plant tolerance to cold, salt, and drought stresses (Saijo et al., 2000, 2001). In ice plant (Mesembryanthemum crystallinum), transcription of McCPK1 was increased after exposure to high-salt and dehydration stresses (Chehab et al., 2004). Arabidopsis AtCPK32 is involved in abscisic acid (ABA)/stress responses through phosphorylating ABA-induced transcription factor ABF4 (Choi et al., 2005). Two Arabidopsis guard cell-expressed CDPK genes, CPK3 and CPK6, have been identified as important components in the regulation of guard cell ion channels and in ABA-regulated stomatal signaling (Mori et al., 2006). AtCPK23 was reported to play roles in Arabidopsis responses to drought and salt stresses (Ma and Wu, 2007). Arabidopsis CPK4 and CPK11 may be two positive regulators in CDPK/calcium-mediated ABA signaling (Zhu et al., 2007). Recently, Mehlmer et al. (2010) reported that CPK3 is involved in Arabidopsis acclimation to salt stress, and Xu et al. (2010) demonstrated that CPK6 functions as a positive regulator in Arabidopsis responses to salt/drought stress.
Although many previous studies have shown the importance of CDPKs for plant signaling in response to various environmental stresses, biological functions of most CDPKs have not been characterized so far. Obviously, to identify potential targets of CDPKs is becoming an important task for further understanding of the CDPK-involved plant signaling mechanisms. Various approaches have been employed to screen CDPK-interacting proteins, such as a phage display library screening (Shao and Harmon, 2003), yeast two-hybrid screening (Patharkar and Cushman, 2000; Lee et al., 2003; Choi et al., 2005; Rodriguez Milla et al., 2006; Uno et al., 2009), and a chemical-genetic approach (Böhmer and Romeis, 2007).
Here, we report that CPK10 (At1g18890) is involved in plant responses to drought stress via modulation of ABA- and Ca2+-regulated stomatal movements. Following the observation that plants of the CPK10 T-DNA insertion mutant (cpk10) were much more sensitive to drought stress, we have identified a heat shock protein, HSP1 (At4g14830), as a CPK10-interacting protein. Functions of CPK10 and HSP1 in plant responses to drought stress via modulation of ABA and Ca2+ signaling are discussed.
RESULTS
Disruption of CPK10 Transcription in the cpk10 Mutants Increased Arabidopsis Sensitivity to Drought Stress
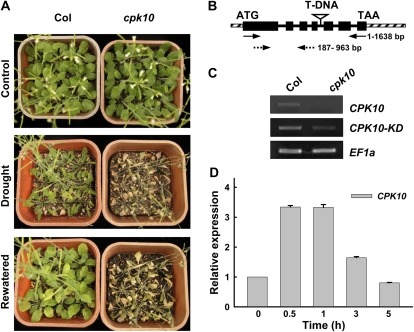
In total, 23 T-DNA insertion lines corresponding to 19 Arabidopsis CDPK genes (expressed in guard cells from GUS staining results; data not shown) were obtained from the Arabidopsis Biological Resource Center (ABRC; http://www.arabidopsis.org/abrc/) and were tested for their sensitivity to the drought stress treatment. Among several CDPK knockout mutants that showed sensitivity to drought stress (data not shown), the cpk10 mutants (SALK_082441) showed a much more sensitive phenotype compared with wild-type plants (Fig. 1A, middle panel). There was no obvious morphological or developmental difference between the cpk10 plants and wild-type plants under normal growth conditions (Fig. 1A, top panel). After 1 week of growth in a growth chamber, both the wild-type plants and mutants had irrigation withheld for drought stress treatment. After a 20-d period of drought treatment, the cpk10 mutants showed a much more sensitive phenotype compared with the wild-type plants. After drought stress treatment, the cpk10 plants wilted and the rosette leaves became chlorotic, whereas the wild-type plants were turgid and their leaves remained green (Fig. 1A, middle panel). After rewatering for 3 d, the wild-type plants quickly recovered, while the cpk10 mutants could not survive (Fig. 1A, bottom panel).
Figure 1.
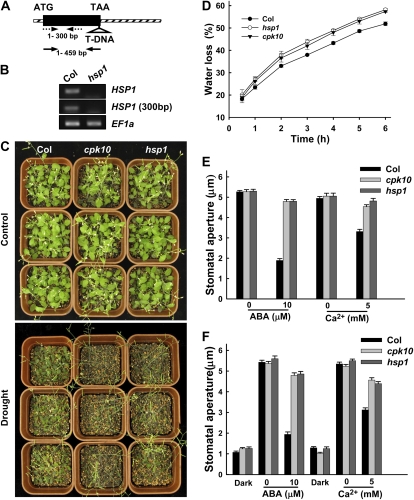
Experimental analysis of the cpk10 mutants. A, Phenotype analysis of Arabidopsis cpk10 mutants (SALK_082441) compared with wild-type plants under normal and drought stress conditions. Wild-type (Col) and cpk10 mutant plants were grown in pots containing soil mixture (rich soil:vermiculite, 2:1, v/v) and kept in a growth chamber at 22°C with illumination at 120 μmol m−2 s−1 for a 16-h daily light. For the drought stress treatment (middle panel), the photograph was taken after withholding water for 20 d. For the rewatering treatment (bottom panel), the photograph was taken 3 d after rewatering the same plants as shown in the middle panel for drought stress. The experiments were repeated seven times with similar results. B, T-DNA insertion site in the cpk10 mutant. The T-DNA was inserted in the fourth intron of the CPK10 genomic DNA. Black boxes, solid lines, and diagonal striped boxes denote exons, introns, and untranslated regions, respectively. Solid and dashed arrows indicate the primer locations for CPK10 full transcript and CPK10 kinase domain transcript, respectively. C, RT-PCR analysis of CPK10 full transcript and CPK10 kinase domain transcript in wild-type plants and the cpk10 mutant. Four-week-old leaves were used for RNA extraction. EF1a was used as a loading control. The primer sequences are described in “Materials and Methods.” D, qRT-PCR analysis of CPK10 transcription under drought stress. Total RNA was extracted from the detached leaves placed on the laboratory bench for the indicated times. The 18S rRNA was used as an internal control. The primer sequences are described in “Materials and Methods.” Each data point represents the mean ± se (n = 3).
To confirm the disruption of CPK10 transcription in the cpk10 mutants, reverse transcription (RT)-PCR experiments were conducted. The T-DNA insertion in the cpk10 mutant was located in the fourth intron of CPK10 genomic DNA (Fig. 1B). Although the partial CPK10 transcripts (777 bp, encoding the kinase domain) in the cpk10 mutant can be detected when using primers to amplify the cDNA fragment upstream of the insertion site (Fig. 1C, middle panel), no transcript was observed in the homozygous cpk10 plants when using specific primers to amplify the full-length cDNA (1,638 bp) of CPK10 (Fig. 1C, top panel).
Urao et al. (1994) showed that CPK10 transcription was rapidly induced by drought and salt stress. In contrast with this previous report, our quantitative real-time (qRT)-PCR results presented in Figure 1D showed that the transcription of CPK10 was quickly induced by a dehydration treatment within 30 min and gradually decreased to the initial level after 3 h.
Overexpression of CPK10 Enhanced Arabidopsis Tolerance to Drought Stress
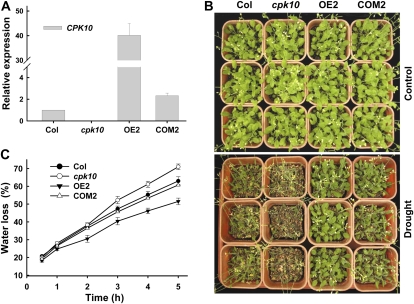
To further confirm that the increased sensitivity of the cpk10 mutants to drought stress resulted from the disruption of CPK10 transcription, the CPK10 overexpression lines (ecotype Columbia [Col] + CPK10; OE) and the complementation lines (cpk10 + CPK10; COM) were generated and analyzed. The expression levels of CPK10 in different plant materials were analyzed by qRT-PCR (Fig. 2A). The CPK10 transcription was much higher in line OE2 than in the wild-type plants, and the CPK10 transcription in the complementation line (COM2) was comparable to that in wild-type plants (Fig. 2A). When subjected to the drought stress, the COM2 plants showed a similar phenotype as the wild-type plants (Fig. 2B), indicating a full complementation of the cpk10 mutant. The CPK10-overexpressing plants did not show any difference in their phenotype compared with the wild-type plants under the control conditions, but they did show much stronger tolerance to drought stress than wild-type plants under the drought stress (Fig. 2B).
Figure 2.
Analysis of CPK10 transcriptional expression and phenotype tests for the cpk10 mutant and the CPK10 overexpression and complementation lines. A, qRT-PCR analysis of CPK10 expression in wild-type plants (Col), the cpk10 mutant, overexpression line 2 (OE2), and complementation line 2 (COM2). The 18S rRNA was used as an internal control. The primer sequences are described in “Materials and Methods.” Each data point represents the mean ± se (n = 3). B, Phenotype tests of various Arabidopsis plants under drought stress. One-week-old seedlings were grown for a 20-d treatment with (control; top panel) or without (drought; bottom panel) irrigation. The photographs were taken at the end of the 20-d water stress treatment. The experiments were repeated three times with similar results. C, Time course of water loss from the detached leaves of various plant materials. Water loss is expressed as a percentage of the initial fresh weight. The experiments were repeated three times with similar results. Each data point represents the mean ± se (n = 3).
The assays of water loss from detached leaves showed that the cpk10 plants lost water much more quickly than the wild-type plants and that the OE2 plants lost water much more slowly than both wild-type and cpk10 plants (Fig. 2C). These results suggest that CPK10 may function as a positive regulator in plant responses to drought stress.
Expression Patterns of CPK10
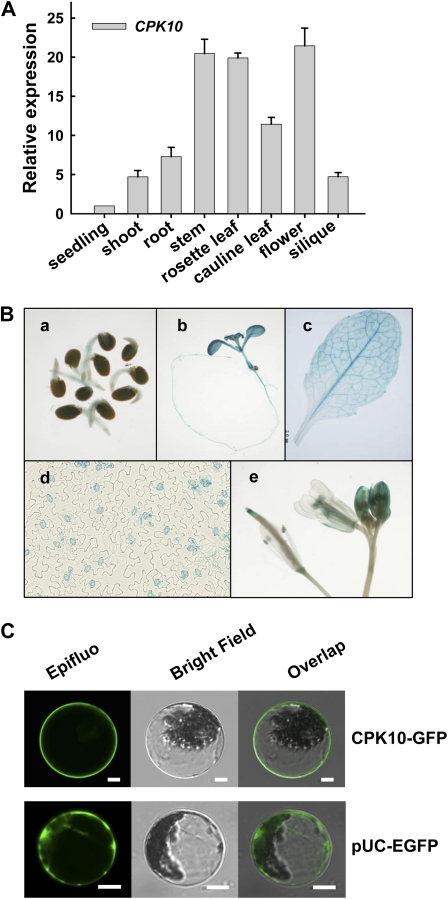
The qRT-PCR results showed that CPK10 mRNA was detectable in all tested tissues or organs, including seedling, root, leaf, stem, flower, and silique (Fig. 3A). To get a further insight into the expression patterns of CPK10, transgenic plants harboring a GUS reporter gene fusion with the CPK10 promoter were generated. GUS staining of the CPK10::GUS transgenic lines confirmed the universal expression patterns of the CPK10 gene (Fig. 3B). High GUS activities were detected in the stomata of leaf epidermis (Fig. 3B), suggesting the potential importance of CPK10 in the regulation of stomatal movements.
Figure 3.
Expression patterns and subcellular localization of CPK10. A, qRT-PCR analysis of CPK10 expression in different tissues as indicated. The 18S rRNA was used as an internal control. The primer sequences are described in “Materials and Methods.” Each data point represents the mean ± se (n = 3). B, Expression patterns of CPK10 as determined by CPK10::GUS transgenic plants. Transgenic plants were stained with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid solution for 12 h. GUS staining is shown in a 2-d-old seedling (a), a 10-d-old seedling (b), a mature rosette leaf (c), stomata (d), and flowers (e). C, Subcellular localization of CPK10. Confocal images of Arabidopsis mesophyll protoplasts transiently expressed with CPK10-GFP construct are shown. The protoplast expressed with pUC-EGFP was used as the control. Bars = 10 μm.
The transient expression of CPK10-GFP in the mesophyll protoplasts of Arabidopsis was tested to determine the subcellular localization of CPK10 proteins. As shown in Figure 3C, CPK10 proteins were localized in the plasma membrane, consistent with the prediction that CPK10 has an N-terminal myristoylation site that can promote membrane association (Cheng et al., 2002).
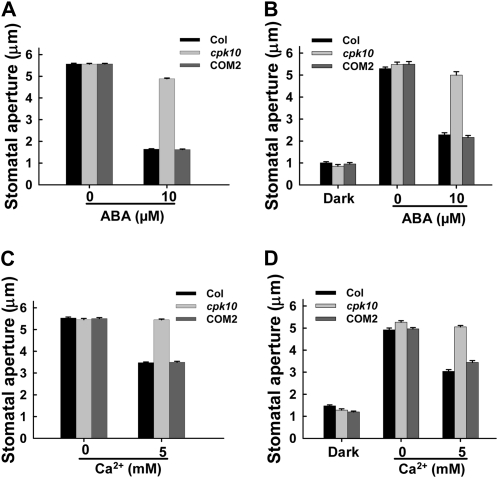
ABA- and Ca2+-Induced Stomatal Closure and Inhibition of Stomatal Opening Were Impaired in the cpk10 Mutant
Stomatal movements are finely regulated to control water loss through transpiration in response to environmental changes. It is well known that ABA and Ca2+ play essential roles in the regulation of stomatal movements (Blatt, 2000; Evans et al., 2001; Schroeder et al., 2001; Fedoroff, 2002; Desikan et al., 2004). Addition of 10 μm ABA in the incubation medium induced the closure of the opened stomata for wild-type plants, which is consistent with numerous previous reports, whereas ABA-induced stomatal closure was remarkably impaired in the cpk10 mutant (Fig. 4A). ABA is a drought-inducible plant hormone and can induce cytoplasmic Ca2+ elevation in guard cells, and Ca2+, as an essential second messenger in stomatal regulation, can induce stomatal closure (McAinsh et al., 2000). Addition of 5 mm Ca2+ in the incubation medium induced the closure of the opened stomata for wild-type plants, whereas Ca2+-induced stomatal closure was impaired in the cpk10 mutant (Fig. 4C). Furthermore, ABA or Ca2+ inhibition of stomatal opening was also remarkably impaired in the cpk10 mutants (Fig. 4, B and D). These results demonstrated that CPK10 plays an important role in transducing ABA and Ca2+ signals in the regulation of stomatal movements.
Figure 4.
ABA- and Ca2+-induced stomatal closing and inhibition of stomatal opening are impaired in the cpk10 mutant. Stomatal apertures were measured on epidermal peels of the wild-type plants, the cpk10 mutant, and the complementation line COM2. The experiments were repeated three times. Each data point represents the mean ± se (n = 50). A, ABA-induced stomatal closing measurements. Stomata were preopened in the light for 2.5 h and then incubated for 2 h with or without the addition of 10 μm ABA in the incubation medium. B, ABA inhibition of light-induced stomatal opening. The epidermis was pretreated in darkness and then incubated for 2 h in the light with or without the addition of 10 μm ABA in the incubation medium. C, Extracellular Ca2+-induced stomatal closing measurements. Stomata were preopened in the light for 2.5 h and then incubated for 2 h with or without the addition of 5 mm Ca2+ in the incubation medium. D, Extracellular Ca2+ inhibition of light-induced stomatal opening. The epidermis was pretreated in darkness and then incubated for 2 h in the light with or without the addition of 5 mm Ca2+ in the incubation medium. The detailed procedures are described in “Materials and Methods.”
Identification of CPK10-Interacting Proteins
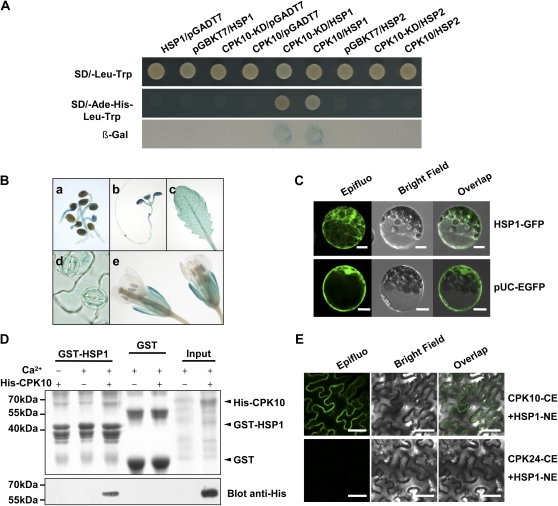
Yeast two-hybrid methods were further applied to screen a cDNA library for the identification of candidate proteins that interact with CPK10. The full length and kinase domain (KD) of CPK10 were constructed as baits and then transformed into Y187 yeast strain. The cDNA library prepared from Arabidopsis leaves of drought-stressed plants was constructed and used for yeast mating with CPK10 baits according to the manufacturer’s protocol (Clontech). Among several positive clones identified from yeast two-hybrid experiments, one cDNA clone, encoding heat shock protein 20-like protein 1 (At4g14830 [HSP1]; Uno et al., 2009), showed strong interaction with CPK10 (Fig. 5A). However, HSP2 (At3g22530; Uno et al., 2009), as a homolog of HSP1, did not interact with CPK10 (Fig. 5A).
Figure 5.
Interaction assays between CPK10 and HSP1. A, Yeast two-hybrid assay. The kinase domain (KD) and full length of CPK10 were used as bait constructs. The blank pGADT7 and pGBKT7 vectors were used as negative controls. Transformants were then patched on selection medium and grown for 4 d before β-galactosidase (β-Gal) assay. B, Expression patterns of HSP1 determined by HSP1::GUS transgenic plants. Transgenic plants were stained with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid solutions for 12 h. GUS staining is shown in a 2-d-old seedling (a), a 10-d-old seedling (b), a mature rosette leaf (c), stomatal guard cells (d), and flowers (e). C, Subcellular localization of HSP1. Confocal images of Arabidopsis mesophyll protoplasts transiently expressed with HSP1-GFP construct are shown. The protoplast expressed with pUC-EGFP was used as the control. Bars = 10 μm. D, The pull-down assay showing interaction between CPK10 and HSP1. The GST-HSP1 fusion protein was used as a bait to pull down the prey His-tagged CPK10 in the presence (+) or absence (−) of 0.2 mm calcium. For a negative control, GST was used as bait. The top panel shows a Coomassie Brilliant Blue-stained SDS-PAGE gel indicating the amount of bait proteins used in each pull-down assay. E, BiFC assays of CPK10 interaction with HSP1 in vivo. The C-terminal half of the GFP was fused to CPK10 (CE), and the N-terminal half of the GFP was fused to the HSP1 (NE). Photographs were taken with a confocal laser-scanning microscope (Nikon EZ-C1). Coexpression of CPK24 and HSP1 was used as a negative control. Bars = 50 μm.
The GUS-staining assays showed that the expression pattern of HSP1 (Fig. 5B) was similar to that of CPK10 (Fig. 3B). Subcellular localization analysis showed that HSP1 proteins are located in cytosol and nucleus (Fig. 5C; Supplemental Fig. S1). The interaction between CPK10 and HSP1 was further tested using in vitro pull-down assay with His-CPK10 and glutathione S-transferase (GST)-HSP1. As shown in Figure 5D, His-CPK10 interacted specifically with HSP1 in the presence of Ca2+, whereas His-CPK10 signal was undetectable with the anti-His antibody on the resulting western blot in the absence of Ca2+ (Fig. 5D). This finding indicates that CPK10 interaction with HSP1 is Ca2+ dependent. The bimolecular fluorescence complementation (BiFC) assays (Fig. 5E) showed that the interaction between CPK10 and HSP1 occurred in the plasma membrane, suggesting that CPK10 may recruit HSP1 to the plasma membrane and form a functional complex.
The hsp1 Mutant Plants Showed Similar Sensitivity to Drought Stress
The HSP1 T-DNA insertion mutant (hsp1; SALK_017461) was obtained from the ABRC and tested for its sensitivity to drought stress. There is only one exon in HSP1 genomic DNA, and the T-DNA insertion site is indicated in Figure 6A. As shown in Figure 6B, either the full length of HSP1 transcript or a 300-bp fragment upstream of the T-DNA insertion site of HSP1 was not detectable in the hsp1 homozygous mutants by RT-PCR analysis, indicating that the expression of HSP1 was completely disrupted in the mutant.
Figure 6.
Analysis of the hsp1 and cpk10 mutants compared with wild-type Arabidopsis plants. A, T-DNA insertion site in the hsp1 (SALK_017461) mutant. The T-DNA was inserted in the exon of the hsp1 genomic DNA. The exon is shown by a black rectangle, and diagonal striped boxes denote untranslated regions. Solid and dashed arrows indicate the primer locations for HSP1 full transcript and HSP1 fragment transcript upstream of the T-DNA insertion site, respectively. B, RT-PCR verification of HSP1 expression in the hsp1 mutant. The expression of full length (top panel) and fragment upstream of the insertion site (middle panel) of HSP1 was detected. EF1a was used as a loading control (bottom panel). C, Phenotype tests of the hsp1 mutants compared with Arabidopsis cpk10 mutants and wild-type plants under drought stress. One-week-old seedlings were grown for a 20-d treatment with (control; top panel) or without (drought; bottom panel) irrigation. The experiments were repeated five times with similar results. The photographs were taken at the end of the 20-d treatment. D, Time courses of the water loss from the detached leaves of Col, cpk10, and hsp1. The experiments were repeated three times with similar results. Each data point represents the mean ± se (n = 3). E, ABA- and Ca2+-induced stomatal closing. Stomatal apertures were measured on epidermal peels of the wild type and the cpk10 and hsp1 mutants. The tested epidermis was pretreated in the light for 2.5 h and then incubated for 2 h with or without the addition of 10 μm ABA or 5 mm Ca2+ in the incubation medium. Each data point represents the mean ± se (n = 50). F, ABA- and Ca2+-inhibited stomatal opening. Concentrations of 10 μm ABA or 5 mm Ca2+ were added into the incubation medium in the dark, and then the tested epidermis was incubated in the above buffer for 2 h in the light. Each data point represents the mean ± se (n = 50).
After a 20-d period of drought treatment, the hsp1 mutant plants exhibited a similar sensitive phenotype under the drought stress as the cpk10 plants (Fig. 6C), and the leaves of the hsp1 mutants wilted and their overall growth was inhibited. The water-loss assays showed that the detached leaves of the hsp1 plants lost water faster than leaves of wild-type plants did (Fig. 6D). Similar to the CPK10 mutants shown in Figure 4, the hsp1 mutants also showed impaired ABA- and Ca2+-induced stomatal closure (Fig. 6E) and inhibition of stomatal opening (Fig. 6F). These results further support the notion that HSP1 may function together with CPK10 in response to drought stress by the regulation of stomatal movements.
ABA and Ca2+ Inhibition of the Inward K+ Currents Was Impaired in the cpk10 and hsp1 Mutants
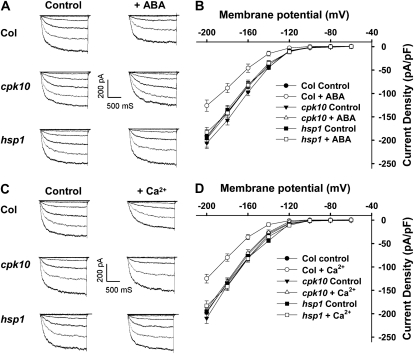
During stomatal closure, the ion channel-mediated inward K+ currents were inhibited by elevations of ABA and cytosolic Ca2+ in stomatal guard cells (Pandey et al., 2007). Increases in cytosolic Ca2+ concentration in guard cells inhibit inward K+ currents and activate anion channels, facilitating solute efflux and stomatal closure (Blatt, 2000). In contrast to the response of wild-type plants, the cpk10 and hsp1 mutants showed decreased sensitivity to ABA- and Ca2+-induced stomatal closure (Figs. 4 and 6, E and F). It was further hypothesized that CPK10 and HSP1 may function in the regulation of stomatal movements by modulation of the inward K+ currents in stomatal guard cells. To test this hypothesis, the patch-clamp method was applied to analyze inward K+ currents in guard cell protoplasts. As shown in Figure 7, the basal (control) inward K+ currents displayed no difference between wild-type plants and the cpk10 and hsp1 mutants. In contrast to wild-type plants, ABA and Ca2+ inhibition of the inward K+ currents was not observed in guard cell protoplasts of the cpk10 and hsp1 mutants. These results demonstrated that ABA and Ca2+ inhibition of the inward K+ channels requires the presence of functional CPK10 as well as HSP1, and CPK10 and HSP1 function as important regulatory components involved in ABA- and Ca2+-mediated regulation of the inward K+ channels in stomatal guard cells.
Figure 7.
Impairment of ABA and Ca2+ inhibition on the inward K+ currents in the cpk10 and hsp1 mutants. A, Patch-clamp whole-cell recordings of the inward K+ currents in guard cell protoplasts isolated from different plant materials (Col, cpk10, and hsp1) with or without the addition of 50 μm ABA in the bath solution. B, Current density-voltage curves of the steady-state whole-cell inward K+ currents in guard cell protoplasts isolated from different plant materials with or without the addition of 50 μm ABA in the bath solution. The data are derived from the recordings as shown in A and are presented as means ± se (Col, n = 11; Col + ABA, n = 11; cpk10, n = 10; cpk10 + ABA, n = 10; hsp1, n = 8; hsp1 + ABA, n = 8). C, Patch-clamp whole-cell recordings of the inward K+ currents in guard cell protoplasts isolated from different plant materials with or without the addition of 2 μm free Ca2+ in the pipette solutions. D, Curves of the steady-state whole-cell inward K+ currents in guard cell protoplasts isolated from different plant materials with or without the addition of 2 μm free Ca2+ in the pipette solution. The data are derived from the recordings as shown in C and are presented as means ± se (Col, n = 11; Col + Ca2+, n = 12; cpk10, n = 13; cpk10 + Ca2+, n = 15; hsp1, n = 10; hsp1 + Ca2+, n = 13).
DISCUSSION
In this study, CPK10 was identified and characterized as an important regulatory component involved in plant response to drought stress through the modulation of ABA- and Ca2+-mediated stomatal movements (Figs. 1, 2, and 4). HSP1 was identified as a CPK10-interacting protein (Fig. 5), and disruption of HSP1 expression in the hsp1 mutants resulted in similar effects on plant responses to drought stress as disruption of the CPK10 gene in the cpk10 mutants (Fig. 6). Furthermore, ABA and Ca2+ inhibition of the inward K+ currents was similarly impaired in both the cpk10 and hsp1 mutants (Fig. 7). The presented results demonstrate that CPK10 and HSP1 function in the regulation of stomatal movements via ABA and Ca2+ signaling pathways. The results also indicate that CPK10 and HSP1 may play important roles in plant responses to drought stress.
Regulation of Ion Channels by CDPKs
A previous study reported that Arabidopsis CPK1 (AK1) activated a tonoplast Cl− channel in isolated vacuoles from Vicia faba guard cells (Pei et al., 1996). The CDPK from Vicia guard cells phosphorylated the K+ channel KAT1 (At5g46240) protein in a Ca2+-dependent manner (Li et al., 1998). Guard cells of Arabidopsis cpk3/cpk6 double mutants showed impaired Ca2+-induced activation of S-type anion currents and reduced sensitivity to ABA regulation of these channels (Mori et al., 2006). The cpk3/cpk6 double mutants also displayed reduced sensitivity of stomatal closure to ABA and Ca2+ (Mori et al., 2006). The guard cell anion channel SLAC1 is regulated by CPK21 and CPK23 with distinct Ca2+ affinities (Geiger et al., 2010). The results in this study demonstrated that CPK10 as well as its interacting protein HSP1 are involved in ABA and Ca2+ inhibition of the inward K+ channels (Fig. 7). All these results indicate that the CDPKs, at least some of them predominantly expressed in stomatal guard cells, may function as Ca2+ sensors and play important roles in the regulation of stomatal movements by the regulation of ion channels.
CDPKs, as Ca2+ Sensors, Play Important Roles in Ca2+-Mediated Stomatal Regulation
As discussed previously, several CDPKs have been shown to be involved in Ca2+-mediated stomatal regulation, such as AtCPK1 (Pei et al., 1996), AtCPK3 and AtCPK6 (Mori et al., 2006), and AtCPK4 and AtCPK11 (Zhu et al., 2007). This study demonstrates that CPK10 functions in the regulation of stomatal movements via ABA- and Ca2+-mediated signaling. Although many previous studies have shown Ca2+ regulation of ion channel activity in guard cells (Pei et al., 2000; Mori et al., 2006; Geiger et al., 2010), our study provides direct molecular-genetic (Figs. 2 and 4) and electrophysiological (Fig. 7) evidence for CPK10 involvement in ABA- and Ca2+-mediated stomatal signaling.
The results of our pull-down assay (Fig. 5D) showed that CPK10 can recruit HSP1 in a Ca2+-dependent manner. The plasma membrane localization of CPK10 (Fig. 3C) and interaction between CPK10 and HSP1 (Fig. 5, A and E) suggest that CPK10 may recruit HSP1 to the plasma membrane. Furthermore, our patch-clamp data (Fig. 7) revealed the important roles of CPK10 and HSP1 in the ABA and Ca2+ regulation of inward K+ channels in stomatal guard cells.
CPK10 Functions Together with HSP1 in Drought Stress Signaling
Several Arabidopsis CDPKs have been functionally characterized as important regulators involved in plant responses to drought and salt stresses (Ma and Wu, 2007; Zhu et al., 2007; Mehlmer et al., 2010; Xu et al., 2010), in ABA signaling (Choi et al., 2005; Mori et al., 2006; Zhu et al., 2007), and in the regulation of pollen tube growth (Myers et al., 2009). Obviously, to identify targets or substrates of these CDPKs becomes an important task for further understanding the molecular mechanisms of CDPK functions. Increasing evidence for the identification of CDPK substrates supports the notion that CDPKs are multifunctional kinases involved in the regulation of diverse cellular functions (Harper and Harmon, 2005). Efforts have been made to identify CDPK-interacting proteins, such as ABF4 for AtCPK32 (Choi et al., 2005), AtDi19-1 for CPK11 (Rodriguez Milla et al., 2006), StRBOHB for StCDPK5 (Kobayashi et al., 2007), ABF1 and ABF4 for AtCPK4 and AtCPK11 (Zhu et al., 2007), and RSG for NtCDPK1 (Ishida et al., 2008). In this study, HSP1 was identified and also functionally characterized as CPK10-interacting protein. HSP1 was previously demonstrated to interact with CPK4 and CPK11 (Uno et al., 2009). The interactions between HSP1 and other CDPKs as CPK10 homologs (Supplemental Fig. S2) indicate that HSP1 may interact with different CDPKs. To further validate these interactions in vivo and investigate the functions of these protein-protein interactions in a particular biological process may reveal diverse functions of HSP1. HSP1 belongs to class I or II of small heat shock proteins (sHSPs) for its low molecular mass (16.9 kD) and subcellular localization (cytosol and nucleus; Sun et al., 2002). The HSP1 mRNA was highly expressed in stomatal guard cells (Leonhardt et al., 2004). Similar to the transgenic CPK10::GUS plants, high GUS activity was also detected in stomatal guard cells of the transgenic HSP1::GUS plants (Fig. 5B). The results from GST pull-down and BiFC assays (Fig. 5, D and E) indicated that CPK10 may recruit HSP1 to the plasma membrane and form a complex to regulate stomatal movements under drought stress.
Considering the fact that the sHSPs play important roles in plant tolerance to environmental stresses (for review, see Sun et al., 2002; Wang et al., 2004), it is not surprising that the HSP1 functions together with CPK10 in plant responses to drought stress. Several studies have demonstrated that sHSPs may be involved in plant responses to water stress, such as the induction of sHSPs by water stress in sunflower (Helianthus annuus; Almoguera et al., 1993) and the increased Arabidopsis osmotolerance by overexpression of a class II sHSP (Sun et al., 2001). Desiccation tolerance of the resurrection plant (Craterostigma plantagineum) seems related to high and continuous expression of sHSPs in all vegetative tissues (Alamillo et al., 1995).
Although the molecular mechanisms by which sHSPs are involved in plant responses to abiotic stresses remain unclear, a number of findings support the notion that these small proteins may function as molecular chaperones to prevent their partners (such as CPK10 proteins in this study) from deactivation or degradation under various stresses (for review, see Sun et al., 2002; Wang et al., 2004). Alternatively, phosphorylation of sHSPs has been demonstrated before, such as phosphorylation of a class I heat shock protein by a SNF1-related protein kinase (Slocombe et al., 2004) and in vivo-detected phosphorylation of a maize (Zea mays) mitochondrial HSP22 at its N terminus (Lund et al., 2001), suggesting possible HSP1 phosphorylation by CPK10.
Considering all the previously reported results together with the evidence presented in this study, we conclude that the interaction between CPK10 and HSP1 is physiologically important in plant responses to drought stress, particularly in stomatal regulation under water stress.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) T-DNA insertion mutants of CPK10 (cpk10; SALK_082441) and HSP1 (hsp1; SALK_017461) were obtained from the ABRC. The seeds were surface sterilized with mixed solutions of NaClO (0.5%, w/v) and Triton X-100 (0.01%, v/v) for 10 min followed by washing with sterile water four times. Seeds were placed in petri dishes containing Murashige and Skoog agar (0.8%, w/v) medium and incubated for 2 d at 4°C before being transferred to 22°C for germination. After 7 d of growth on plates under constant illumination at 60 μmol m−2 s−1 at 22°C, the seedlings were transplanted to pots containing soil mixture (rich soil:vermiculite, 2:1, v/v) and kept in growth chambers at 22°C with illumination at 120 μmol m−2 s−1 for a 16-h daily light period. The relative humidity was approximate 70% ± 5%.
To obtain the homozygous mutant lines, the individual plants were identified by PCR using T-DNA left border primer LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) and CPK10 gene-specific primers (forward primer 5′-ATCCTGATCCGACTAAGCG-3′ and reverse primer 5′-CCAACAATCCGACTCAGAA-3′) or HSP1 gene-specific primers (forward primer 5′-CTTAACAAAGCGAAGGTCGC-3′ and reverse primer 5′-AATTTTGACCCATTCGCTTC-3′).
Plasmid Construction and Arabidopsis Transformation
CPK10::GUS and HSP1::GUS were generated by fusing the promoter fragments of CPK10 (1.57 kb) or HSP1 (927 bp) in front of the GUS coding sequence in pCAMBIA1381 vector, respectively. The sequences for CPK10 promoter-specific primers are 5′-AAGCACTTAAGCTCTGTGTGG-3′ and 5′-TTTAGTTCATATGGACGCCG-3′, and the sequences for HSP1 promoter-specific primers are 5′-CGGCGTATACAATTGAATTT-3′ and 5′-GTGTGAGAGTGATAAGAAGCG-3′. The GUS staining assays were carried out as described previously (Xu et al., 2006).
For the EGFP constructs, the CPK10 coding sequence was amplified by the forward primer 5′- TTTCTAGAATGGGTAACTGTAACGCCTGT-3′ (XbaI site underlined) and the reverse primer 5′-TTGGTACCAACAGGAACAGTTTGTCCAGT-3′ (KpnI site underlined), and the HSP1 coding sequence was amplified by the forward primer 5′-TATAGTCGACATGAAAATCCACCCATTACC-3′ (SalI site underlined) and the reverse primer 5′-TATACTCGAGCTGTACAAGCACTATCAAATCTC-3′ (XhoI site underlined). The PCR products of CPK10 and HSP1 were verified by DNA sequencing by cloning them into pUC-EGFP vector (Xu et al., 2006). To generate the CPK10 overexpression plants, the coding sequence of CPK10 was cloned and introduced into Super1300 vector (Chen et al., 2009). For the generation of complementation lines of the cpk10 mutant, a 4,682-bp genomic DNA sequence of CPK10 (including 1,690 bp upstream of the ATG codon and 723 bp downstream of the stop codon) was amplified by PCR with CPK10-specific primers (forward primer 5′-TTGAGCTCTACTTAATCCACCACATGTCCCT-3′ and reverse primer 5′-TTGTCGACTGATGGTGTCTGCAATTGTAGAAC-3′; SacI and SalI restriction sites underlined, respectively). The PCR product was verified by DNA sequencing and cloned into pCAMBIA1300 vector.
The constructs were introduced into Agrobacterium tumefaciens (strain GV3101) and transferred into Arabidopsis plants (wild type or mutant) by the floral dip method (Clough and Bent, 1998). All transgenic lines used in this study were T3 homozygous plants with single copy insertion.
Drought Stress and Water Loss Experiments
For the phenotype tests under drought stress, seedlings grown on Murashige and Skoog medium for 7 d were transferred to mixed soil (rich soil:vermiculite, 2:1, v/v) and grown for 1 week with sufficient watering. Each pot had nine seedlings planted. Then the plants were subjected to drought stress treatment by withholding irrigation, and the well-watered plants were taken as the control. To ensure reproducibility for the phenotype tests, three or six pots of plants were grown for each line (genotype) and each treatment in one experiment (e.g. for wild-type plants, at least three pots for control and three pots for water stress treatment). All pots were placed under the same conditions in a growth chamber in a random order, and the position of each pot was changed every day in a random way to exclude “position effect.” The experiments were repeated seven times (Fig. 1A), three times (Fig. 2B), and five times (Fig. 6C), and similar results were observed for each group of experiments, although slight difference appeared from one experiment to another. The photographs were taken after withholding water or rewatering for the indicated times.
For water loss measurement, rosette leaves were detached from 4-week-old plants, weighed immediately on a piece of weighing paper, and then placed on the laboratory bench (the aerial relative humidity was between 40% and 50% and the temperature was between 22°C and 23°C). The weight losses of the samples were measured at designated time points (as indicated in Figs. 2C and 6D). The proportion of water loss was calculated on the basis of the initial fresh weight of the samples.
Stomatal Aperture Measurements
Stomatal apertures were measured as described previously (Pei et al., 1997) with slight modification. Briefly, plants were grown in a growth chamber with illumination at 120 μmol m−2 s−1 for a 10-h daily light period and day/night temperatures were 22°C/20°C, respectively. The relative humidity was between 80% and 90%. For stomatal opening assays (Figs. 4, B and D, and 6F), rosette leaves from 4- to 5-week-old plants were harvested in darkness at the end of the night and then floated in solutions containing 50 mm KCl, 10 mm MES-KOH (pH 6.15), and 100 μm CaCl2 at 22°C followed by the indicated treatments. For stomatal closing assays (Figs. 4, A and C, and 6E), rosette leaves from 4- to 5-week-old plants were harvested in darkness at the end of the night and then floated in the same solutions as for stomatal opening assays and kept at 22°C under light (150 μmol m−2 s−1) for 2.5 h before the indicated treatments. The abaxial epidermis was peeled from the treated leaves, and stomata on the epidermal strips were photographed using a Leica microscope (Leica DFC320). Stomatal apertures were measured using ImageJ software (National Institutes of Health).
Subcellular Localization of CPK10 and HSP1
For generation of the GFP fusion protein, the coding sequences of CPK10 and HSP1 were cloned into the XbaI-KpnI and SalI-XhoI sites of pUC-EGFP (Xu et al., 2006), respectively. Mesophyll protoplasts were isolated from 5-week-old wild-type plants (Col) and transformed with the constructs as described (http://genetics.mgh.harvard.edu/sheen web/protocols_reg.html). The protoplasts expressed with the blank GFP vector (pUC-EGFP) were used as the control. Fluorescence of GFP in the transformed protoplast was imaged using a confocal laser-scanning microscope (LSM 510; Carl Zeiss) after the protoplasts were incubated at 23°C for 12 to 18 h.
RT-PCR and qRT-PCR Analysis
For RT-PCR analysis, total RNA was extracted using Trizol reagents according to the manufacturer’s instructions (Invitrogen) and then treated with DNase I (Takara) to eliminate genomic DNA contamination. The cDNA was synthesized from treated RNA by SuperScript II reverse transcriptase (Invitrogen) using oligo(dT)15 primer (Promega). The full transcript of CPK10 was amplified using the forward primer 5′-ATGGGTAACTGTAACGCCTG-3′ and the reverse primer 5′-TTAAACAGGAACAGTTTGTCCA-3′. The transcript of CPK10-KD was amplified using the forward primer 5′-TACATCTTAGGTCGTGAATTAGGTC-3′ and the reverse primer 5′-TATCCATGGGTGAGCTAACACT-3′. The full transcript of HSP1 was amplified using the forward primer 5′-ATGAAAATCCACCCATTACC-3′ and the reverse primer 5′-TTACTGTACAAGCACTATCAAATCTC-3′, and the transcript of HSP1 fragment was amplified using the forward primer 5′-ATGAAAATCCACCCATTACC-3′ and reverse primer 5′-AGGCCTAGTTGATTCCGG-3′. The EF1a (At5g60390) transcript served as a control and was amplified by EF1a-specific primers 5′-ATGCCCCAGGACATCGTGATTTCAT-3′ and 5′-TTGGCGGCACCCTTAGCTGGATCA-3′.
For qRT-PCR analysis, total RNA extraction and treatment were performed as described above. The cDNA was synthesized from total RNA by SuperScript II reverse transcriptase (Invitrogen) using Random Hexamer Primer (Promega). qRT-PCR was performed as described previously (Chen et al., 2009). Relative quantitative results were calculated by normalization to 18S rRNA. qRT-PCR was conducted with CPK10-specific primers (forward primer 5′-GCTTCAGAAGGTCGGTTCAC-3′ and reverse primer 5′-AGCTTCCCGTAGCTCATCAA-3′).
Yeast Two-Hybrid and GST Pull-Down Assays
Bait cloning and yeast two-hybrid screening were performed following the manufacturer’s instructions (Clontech). The coding sequences of CPK10 and CPK10-KD were cloned into pGBKT7 (Clontech) as baits and transformed into Y187 yeast strain. The cDNA library was constructed using total RNA isolated from leaves of 4-week-old plants after a dehydration treatment and transformed into AH109 yeast strain for yeast mating. The putative positive clones from yeast two-hybrid screening were further confirmed by yeast two-hybrid tests and assayed for β-galactosidase activity according to the manufacturer’s instructions (Clontech).
For GST pull-down assays, the coding region of CPK10 cDNA was amplified by using the forward primer 5′-TTGGTACCATGGGTAACTGTAACGCCTGT-3′ and the reverse primer 5′-TTGAGCTCTTAAACAGGAACAGTTTGTCCAG-3′ and cloned into KpnI and SacI sites of pET-30a(+) vector. HSP1 was amplified by using the forward primer 5′-TCTAGTCGACTCATGAAAATCCACCCATTACC-3′ and the reverse primer 5′-TATAGCGGCCGCTTACTGTACAAGCACTATCAAATCTC-3′ and cloned into SalI and NotI sites of pGEX-4T-1 vector. Fusion protein was expressed in the BL21 (DE3) strain of Escherichia coli by induction with 0.5 mm isopropyl β-d-thiogalactoside. GST-HSP1 fusion protein was immobilized on Glutathione-Sepharose 4B beads and incubated with His-tag CPK10 extracts (soluble fraction; Shi et al., 1999). After 2 h of incubation at 4°C on a rotary incubator, the beads were washed five times with ice-cold phosphate-buffered saline, resuspended in SDS gel-loading buffer, and then loaded on SDS-PAGE (10%, w/v) gels. For two SDS-PAGE gels in each experiment, one of them was stained with Commassie Brilliant Blue R 250 and the other was analyzed by protein gel blotting against anti-His antibody.
BiFC Assays
The cDNAs of CPK10 and HSP1 were cloned into vector kanII-SPYCE(MR) and vector hygII-SPYNE(R)173, respectively, as described by Waadt and Kudla (2008). The sequences for the CPK10-specific primers are 5′-TATAACTAGTATGGGTAACTGTAACGCCTG-3′ (SpeI site underlined) and 5′-TATACTCGAGAACAGGAACAGTTTGTCCAGT-3′ (XhoI site underlined), and the sequences for the HSP1-specific primers are 5′-TATAACTAGTATGAAAATCCACCCATTACC-3′ (SpeI site underlined) and 5′-TATACTCGAGCTGTACAAGCACTATCAAATCTCTAC-3′ (XhoI site underlined). For transient expression, A. tumefaciens strain GV3101 carrying the BiFC constructs was used to infiltrate 5-week-old Nicotiana benthamiana leaves. The photographs were taken after 24 to 72 h of transformation using a confocal laser-scanning microscope (Nikon EZ-C1).
Patch-Clamp Experiments
Arabidopsis guard cell protoplasts were isolated as described previously (Pei et al., 1997; Wang et al., 2001). Standard whole-cell recording techniques were applied in this study (Hamill et al., 1981). All experiments were conducted at room temperature (approximately 22°C) under dim light. The bath solutions contained 30 mm KCl, 2 mm MgCl2, 10 mm MES-Tris (pH 5.6), and 1 mm CaCl2, with osmolality of 485 mmol kg−1. The pipette solutions contained 3.35 mm CaCl2, 6.7 mm EGTA, 2 mm MgCl2, 10 mm HEPES-Tris (pH 7.1), 70 mm K-Glu, and 30 mm KCl, with osmolality of 500 mmol kg−1. The final osmolalities in both bath solution and pipette solution were adjusted with d-sorbitol. The final free Ca2+ concentration in the pipette solution was 200 nm, calculated using Chelator software Max Chelator version 5.60 (developed by Dr. Chris Patton at Stanford University). The fresh ATP (Mg-ATP; 5 mm) was added into the pipette solution before use. For the internal high-Ca2+ treatment, the final free Ca2+ concentration in the pipette solution was adjusted to 2 μm by addition of CaCl2 (calculated using Chelator software Max Chelator version 5.60), and the whole-cell currents were recorded 5 min after the whole-cell configuration was achieved using an Axopatch-200B amplifier (Axon Instruments) connected to a computer via an interface (TL-1 DMA Interface; Axon Instruments). For the ABA treatment, ABA was added into the bath solution and resulted in a final ABA concentration of 50 μm after the whole-cell configuration was achieved, and the whole-cell currents were recorded 10 min after the whole-cell configuration was achieved. The pCLAMP software (version 6.0.4; Axon Instruments) was used to acquire and analyze the whole-cell currents. SigmaPlot software was used to draw current density-voltage plots and for data analysis.
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under accession numbers At1g18890 (CPK10), At4g14830 (HSP1), At3g22530 (HSP2), and At5g60390 (EF1a).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Subcellular localization of CPK10 and HSP1.
Supplemental Figure S2. Yeast two-hybrid assays for the identification of HSP1-interacting CDPKs.
Supplementary Material
References
- Alamillo J, Almoguera C, Bartels D, Jordano J. (1995) Constitutive expression of small heat shock proteins in vegetative tissues of the resurrection plant Craterostigma plantagineum. Plant Mol Biol 29: 1093–1099 [DOI] [PubMed] [Google Scholar]
- Almoguera C, Coca MA, Jordano J. (1993) Tissue-specific expression of sunflower heat shock proteins in response to water stress. Plant J 4: 947–958 [Google Scholar]
- Asano T, Tanaka N, Yang G, Hayashi N, Komatsu S. (2005) Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol 46: 356–366 [DOI] [PubMed] [Google Scholar]
- Blatt MR. (2000) Ca2+ signalling and control of guard-cell volume in stomatal movements. Curr Opin Plant Biol 3: 196–204 [PubMed] [Google Scholar]
- Böhmer M, Romeis T. (2007) A chemical-genetic approach to elucidate protein kinase function in planta. Plant Mol Biol 65: 817–827 [DOI] [PubMed] [Google Scholar]
- Chehab EW, Patharkar OR, Hegeman AD, Taybi T, Cushman JC. (2004) Autophosphorylation and subcellular localization dynamics of a salt- and water deficit-induced calcium-dependent protein kinase from ice plant. Plant Physiol 135: 1430–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH. (2009) The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3554–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Willmann MR, Chen HC, Sheen J. (2002) Calcium signaling through protein kinases: the Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129: 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HI, Park HJ, Park JH, Kim S, Im MY, Seo HH, Kim YW, Hwang I, Kim SY. (2005) Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol 139: 1750–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- DeFalco TA, Bender KW, Snedden WA. (2010) Breaking the code: Ca2+ sensors in plant signalling. Biochem J 425: 27–40 [DOI] [PubMed] [Google Scholar]
- Desikan R, Cheung MK, Bright J, Henson D, Hancock JT, Neill SJ. (2004) ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J Exp Bot 55: 205–212 [DOI] [PubMed] [Google Scholar]
- Evans NH, McAinsh MR, Hetherington AM. (2001) Calcium oscillations in higher plants. Curr Opin Plant Biol 4: 415–420 [DOI] [PubMed] [Google Scholar]
- Fedoroff NV. (2002) Cross-talk in abscisic acid signaling. Sci STKE 2002: re10. [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KA, Grill E, et al. (2010) Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA 107: 8023–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. (1981) Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch 391: 85–100 [DOI] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Gubrium E, Harper JF. (2001) The CDPK superfamily of protein kinases. New Phytol 151: 175–183 [DOI] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Harper JF. (2000) CDPKs: a kinase for every Ca2+ signal? Trends Plant Sci 5: 154–159 [DOI] [PubMed] [Google Scholar]
- Harper JF, Breton G, Harmon A. (2004) Decoding Ca2+ signals through plant protein kinases. Annu Rev Plant Biol 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Harper JF, Harmon A. (2005) Plants, symbiosis and parasites: a calcium signalling connection. Nat Rev Mol Cell Biol 6: 555–566 [DOI] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al. (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang QS, Wang HY, Gao P, Wang GY, Xia GX. (2008) Cloning and characterization of a calcium dependent protein kinase gene associated with cotton fiber development. Plant Cell Rep 27: 1869–1875 [DOI] [PubMed] [Google Scholar]
- Ishida S, Yuasa T, Nakata M, Takahashi Y. (2008) A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 20: 3273–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimecka M, Muszyńska G. (2007) Structure and functions of plant calcium-dependent protein kinases. Acta Biochim Pol 54: 219–233 [PubMed] [Google Scholar]
- Kobayashi M, Ohura I, Kawakita K, Yokota N, Fujiwara M, Shimamoto K, Doke N, Yoshioka H. (2007) Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19: 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolukisaoglu Ü, Weinl S, Blazevic D, Batistic O, Kudla J. (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134: 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Batistič O, Hashimoto K. (2010) Calcium signals: the lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Cho HS, Yoon GM, Ahn JW, Kim HH, Pai HS. (2003) Interaction of NtCDPK1 calcium-dependent protein kinase with NtRpn3 regulatory subunit of the 26S proteasome in Nicotiana tabacum. Plant J 33: 825–840 [DOI] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AL, Zhu YF, Tan XM, Wang X, Wei B, Guo HZ, Zhang ZL, Chen XB, Zhao GY, Kong XY, et al. (2008) Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol Biol 66: 429–443 [DOI] [PubMed] [Google Scholar]
- Li J, Lee YR, Assmann SM. (1998) Guard cells possess a calcium-dependent protein kinase that phosphorylates the KAT1 potassium channel. Plant Physiol 116: 785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S. (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 14: 37–42 [DOI] [PubMed] [Google Scholar]
- Ludwig AA, Romeis T, Jones JD. (2004) CDPK-mediated signalling pathways: specificity and cross-talk. J Exp Bot 55: 181–188 [DOI] [PubMed] [Google Scholar]
- Lund AA, Rhoads DM, Lund AL, Cerny RL, Elthon TE. (2001) In vivo modifications of the maize mitochondrial small heat stress protein, HSP22. J Biol Chem 276: 29924–29929 [DOI] [PubMed] [Google Scholar]
- Ma SY, Wu WH. (2007) AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol Biol 65: 511–518 [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Gray JE, Hetherington AM, Leckie CP, Ng C. (2000) Ca2+ signalling in stomatal guard cells. Biochem Soc Trans 28: 476–481 [PubMed] [Google Scholar]
- McCormack E, Tsai YC, Braam J. (2005) Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci 10: 383–389 [DOI] [PubMed] [Google Scholar]
- Mehlmer N, Wurzinger B, Stael S, Hofmann-Rodrigues D, Csaszar E, Pfister B, Bayer R, Teige M. (2010) The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J 63: 484–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, et al. (2006) CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+ -permeable channels and stomatal closure. PLoS Biol 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers C, Romanowsky SM, Barron YD, Garg S, Azuse CL, Curran A, Davis RM, Hatton J, Harmon AC, Harper JF. (2009) Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J 59: 528–539 [DOI] [PubMed] [Google Scholar]
- Pandey S, Zhang W, Assmann SM. (2007) Roles of ion channels and transporters in guard cell signal transduction. FEBS Lett 581: 2325–2336 [DOI] [PubMed] [Google Scholar]
- Patharkar OR, Cushman JC. (2000) A stress-induced calcium-dependent protein kinase from Mesembryanthemum crystallinum phosphorylates a two-component pseudo-response regulator. Plant J 24: 679–691 [DOI] [PubMed] [Google Scholar]
- Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI. (1997) Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9: 409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Murata Y, Benning G, Thomine S, Klüsener B, Allen GJ, Grill E, Schroeder JI. (2000) Calcium channels activated by hydrogen peroxide mediated abscisic acid signalling in guard cells. Nature 406: 731–734 [DOI] [PubMed] [Google Scholar]
- Pei ZM, Ward JM, Harper JF, Schroeder JI. (1996) A novel chloride channel in Vicia faba guard cell vacuoles activated by the serine/threonine kinase, CDPK. EMBO J 15: 6564–6574 [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Milla MA, Uno Y, Chang IF, Townsend J, Maher EA, Quilici D, Cushman JC. (2006) A novel yeast two-hybrid approach to identify CDPK substrates: characterization of the interaction between AtCPK11 and AtDi19, a nuclear zinc finger protein. FEBS Lett 580: 904–911 [DOI] [PubMed] [Google Scholar]
- Rudd JJ, Franklin-Tong VE. (2001) Unravelling response-specificity in Ca2+ signalling pathways in plant cells. New Phytol 151: 7–33 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23: 319–327 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Kinoshita N, Ishiyama K, Hata S, Kyozuka J, Hayakawa T, Nakamura T, Shimamoto K, Yamaya T, Izui K. (2001) A Ca2+-dependent protein kinase that endows rice plants with cold- and salt-stress tolerance functions in vascular bundles. Plant Cell Physiol 42: 1228–1233 [DOI] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF. (2002) Calcium at the crossroads of signaling. Plant Cell (Suppl) 14: S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. (2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52: 627–658 [DOI] [PubMed] [Google Scholar]
- Shao J, Harmon AC. (2003) In vivo phosphorylation of a recombinant peptide substrate of CDPK suggests involvement of CDPK in plant stress responses. Plant Mol Biol 53: 691–700 [DOI] [PubMed] [Google Scholar]
- Shi J, Kim KN, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J. (1999) Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11: 2393–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocombe SP, Beaudoin F, Donaghy PG, Hardie DG, Dickinson JR, Halford NG. (2004) SNF1-related protein kinase (snRK1) phosphorylates class I heat shock protein. Plant Physiol Biochem 42: 111–116 [DOI] [PubMed] [Google Scholar]
- Sun W, Bernard C, van de Cotte B, Van Montagu M, Verbruggen N. (2001) At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J 27: 407–415 [DOI] [PubMed] [Google Scholar]
- Sun W, Van Montagu M, Verbruggen N. (2002) Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta 1577: 1–9 [DOI] [PubMed] [Google Scholar]
- Uno Y, Rodriguez Milla MA, Maher E, Cushman JC. (2009) Identification of proteins that interact with catalytically active calcium-dependent protein kinases from Arabidopsis. Mol Genet Genomics 281: 375–390 [DOI] [PubMed] [Google Scholar]
- Urao T, Katagiri T, Mizoguchi T, Yamaguchi-Shinozaki K, Hayashida N, Shinozaki K. (1994) Two genes that encode Ca2+-dependent protein kinases are induced by drought and high-salt stresses in Arabidopsis thaliana. Mol Gen Genet 244: 331–340 [DOI] [PubMed] [Google Scholar]
- Waadt R, Kudla J. (2008) In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). Cold Spring Harb Protoc 2008: doi/10.1101/pdb.prot4995 [DOI] [PubMed] [Google Scholar]
- Wan B, Lin Y, Mou T. (2007) Expression of rice Ca2+-dependent protein kinases (CDPKs) genes under different environmental stresses. FEBS Lett 581: 1179–1189 [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A. (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9: 244–252 [DOI] [PubMed] [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM. (2001) G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Weinl S, Kudla J. (2009) The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol 184: 517–528 [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH. (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360 [DOI] [PubMed] [Google Scholar]
- Xu J, Tian YS, Peng RH, Xiong AS, Zhu B, Jin XF, Gao F, Fu XY, Hou XL, Yao QH. (2010) AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta 231: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ, et al. (2007) Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.