Abstract
Chromosomal ends are protected by a high-order structure called telomere. Maintenance of correct telomere length and structure is critically important for the viability of both dividing and non-dividing cells. Notably, targeted deletion of a component of the multi-protein telomere-capping complex, TRF1 (telomeric repeat binding factor 1), causes lethality at embryonic day 5-6 without apparent telomere deficiency1, raising the possibility that TRF1 may also moonlight outside the telomere. Further reinforcing the extra-telomeric tie of TRF1, two studies from our group have reported the findings that TRF1 can be bound and modulated by two nucleolar GTP-binding proteins, nucleostemin (NS) and guanine nucleotide binding protein-like 3 (GNL3L), which exhibit apparently opposite effects on the protein degradation of TRF1. In particular, GNL3L is able to stabilize TRF1 protein during mitosis and promote the metaphase-to-anaphase transition. This manuscript extends the discussion on how this GNL3L-mediated TRF1 regulation creates a novel dynamic control on telomere and cell cycle, and extrapolates its evolutionary significance by contrasting the activities of NS and GNL3L.
Keywords: Nucleostemin, GNL3L, TRF1, Telomere, Cell Cycle, Nucleolus, Mitosis, Dynamic
In 2006, we reported the ability of NS, a stem cell-enriched nucleolar factor, to interact with and promote the degradation of TRF1 protein.2 In 2009, we extended this NS-based regulation of TRF1 to GNL3L, which is the closest homologue of NS in mammal.3 TRF1 has been shown to be modified by a number of post-translational mechanisms, including ubiquitylation, SUMOylation, ADP-ribosylation, and phosphorylation.4-7 In this recent study published in the Journal of Cell Biology in 2009, we demonstrated that GNL3L is capable of promoting the homodimerization and telomeric association of TRF1, preventing the PML-body recruitment of telomere-bound TRF1, and stabilizing TRF1 protein by inhibiting its ubiquitylation and binding to FBX4, an E3 ubiquitin ligase for TRF1.3 The biological significance of the TRF1-stabilizing activity of GNL3L resides in its role in mediating the mitotic increase of TRF1 protein and the metaphase-to-anaphase cell cycle transition. The latter observation resonates with the idea that TRF1 may have an obligatory role outside the telomere. This Extra View will discuss the importance of equipping cells with a nucleolus or GTP-based regulatory mechanism for TRF1 and the potential meaning of the TRF1’s involvement in the mitotic transit.
Of NS, GNL3L and TRF1
The NS family contains three vertebrate members, i.e. NS, GNL3L, and Ngp-1, and is defined by the unique combination of a nucleolus-predominant distribution and a MMR_HSR1 domain.8, 9 The MMR_HSR1 domain is featured by five GTP-binding motifs arranged in a circularly permuted order, where a conserved G5* variant motif (DAR×P) and a G4 (NK×DL) motif are positioned N-terminally to the G1 (G×PNVGKSS), G2 (G×T), and G3 (D×PG) motifs.10 Among the three NS family proteins, NS and GNL3L are most closely related to one another in protein sequence when compared with Ngp-1. NS was first identified as a neural stem cell (NSC)-enriched factor11 and later found to be highly expressed by embryonic stem (ES) cells, several types of somatic stem/progenitor cells, and human cancer cells.11-14 In adult tissues, a high level of NS expression can only be found in the testis. In contrast, GNL3L is abundantly expressed by several adult tissues, including the brain and the eye.15
Our initial interest in the connection between NS and TRF1 stems from the thinking that both NS and telomere play pivotal roles in the maintenance of stem and cancer cell self-renewal. In the first study on this subject, we provided evidence showing that NS and TRF1 do indeed interact with each other in vivo, and, as a result of this binding, the degradation of TRF1 is enhanced.2 To expand the biological relevance of the NS-TRF1 interaction, we investigated the possibility that TRF1 may bind other NS family proteins. It turns out that the interaction of NS family proteins with TRF1 appears to follow their protein homology, with only NS and GNL3L showing the ability to bind TRF1. Somewhat surprisingly, we found that GNL3L has the ability to stabilize TRF1 protein, opposite to that of NS.3 This observation, in addition to revealing a novel extra-telomeric mechanism in TRF1 regulation, also represents the first evidence for the opposite biological activities of NS and GNL3L on regulating the same target. Besides protein stability, GNL3L also exerts a multitude of effects on TRF1, which include promoting its homodimerization and telomeric residence and reducing the ALT-associated PML body (APB) formation. The TRF1 homodimerization effect of GNL3L is not a prerequisite for its TRF1 stabilization activity, but may be related to the increased telomeric residence of TRF1 by GNL3L. On the other hand, the TRF1 stabilization activity of GNL3L is responsible for the mitotic increase and mitotic function of TRF1. So far, it remains to be determined whether the GNL3L’s ability to reduce APB formation correlates with its TRF1 homodimerization, stabilization, or other activities, and whether NS plays a similar or opposite role in the other aspects of TRF1 regulation.
From Nucleoli to Telomeres
Although the nucleolus and the telomere form separate subnuclear domains, they are both non-membrane-bound and, therefore, are open to a vast sea of proteins diffusing through the nucleoplasm. The molecular connection between the nucleolus and telomere regulation was first suggested by the nucleolar transit of the telomerase reverse transcriptase, TERT.16, 17 Although the functional significance of this phenomenon is still a much debated issue18, 19, the nucleolar presence of both TERT and the telomerase RNA component (TERC)20-22 may imply the nucleolus as one of the potential sites for telomerase ribonucleoprotein assembly. Some telomere-capping proteins have also been found accumulated in the nucleolus as apparently unexpected visitors, e.g. TRF2 and, to a less extent, TRF1 (Fig. 1).23, 24 While the physiological role of a nucleolar transit of these telomere-associated components remains unclear, the movement of TRF2 to and from the nucleolus seems to be cell cycle-dependent.23 If biologically relevant, these observations may signify the nucleolus as a passive storage or an active processing site for these proteins. The fact that NS and GNL3L function as TRF1 modulators reinforces the idea that the nucleolus does indeed participate in the regulation of telomeric proteins. One key cell biological finding regarding the binding between GNL3L and TRF1 is that their interaction takes place neither in the nucleolus nor at the telomere but within the nucleoplasm, and that there is no evidence to support the GNL3L’s presence at the telomere, either transient or stable. Therefore, we raise the model that GNL3L may act as a chaperon protein that regulates the availability and readiness of TRF1 before it hops on the telomere to conduct its telomere-capping activity.
Figure 1.
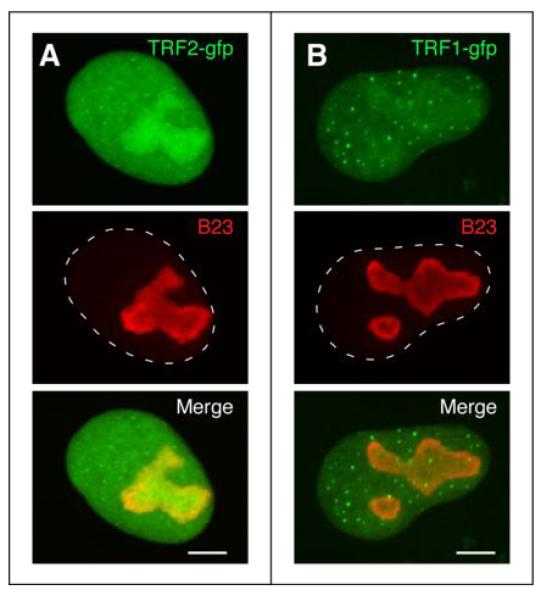
Nucleolar distributions of TRF2 and TRF1.
Some telomere-capping proteins, e.g. TRF2 (A) and TRF1 (B), can be found accumulated in the nucleolus. The subcellular distributions of TRF2 and TRF1 are revealed here by a fused GFP tag. The nucleolus and nucleo-cytoplasmic boundary are marked by B23 immunofluorescence (red) and dashed lines, respectively. Scale bar: 5 um.
Since the majority of the endogenous TRF1 protein is found outside the nucleolus, it is logical to assume that the nucleolar “sequestration” of NS and GNL3L would render them inactive in modulating TRF1 protein. As a result, the mobilization of NS and GNL3L to and from the nucleolus may constitute a dynamic mechanism for TRF1 regulation. It should be noted that in addition to the constant protein exchange between the nucleolar and the nucleoplasmic pools of NS and GNL3L, these two proteins can also be vacated en masse from the nucleolus during mitosis and nucleolar stress. Therefore, one utility of the nucleolar storage of NS and/or GNL3L may be to allow cells to respond to stress in a regulated manner via TRF1 modulation.
The Omnipresent GTP Switch
Nucleolar compartmentalization is not the only mechanism for NS and GNL3L to exercise their activities dynamically. These two proteins are also endowed with the ability to bind GTP. Proteins with GTP-binding capacity have been known to utilize GTP as a molecular switch to turn on or off their biological functions, and, in the case of NS and GNL3L, it has been demonstrated that their GTP binding essentially acts as a key to unlock the nucleoplasm-docking activity of a nucleoplasmic localization signal (NpLS) that prevents the nucleolar entry of NS and GNL3L.25, 8 Furthermore, the GTP-binding domain itself also works in a GTP-independent fashion as a novel nucleolar retention signal that cooperates with the basic residue-enriched nucleolar localization signal (NoLS) to facilitate the nucleolar accumulation of NS.25 In this regard, GTP binding and dissociation represents a critical signal that drives the rapid nucleolar-nucleoplasmic cycling of NS and GNL3L, and this constant shuttling between the nucleolus and the nucleoplasm allows individual NS and GNL3L protein the opportunity to interact with others residing in separate subcellular compartments without changing their overall protein distributions.25
One should not overlook the possibility that GTP binding may in fact control the activities of NS and GNL3L in a distribution-independent manner. This point is best illustrated by contrasting the activities of a non-GTP-binding mutant of GNL3L, N166I, which loses its GTP-binding capability by a point mutation that replaces Asn 166 with Ile in the conserved G4 domain, and a nucleoplasmic mutant of GNL3L, G3dB, which is deleted of the NoLS but remains capable of binding GTP.3 We demonstrated that this non-GTP-binding N166I mutant is still able to bind TRF1 and retains the wild-type activity in promoting TRF1 homodimerization. Yet, it loses its ability to prevent TRF1 ubiquitylation and degradation. In contrast, the GTP-binding G3dB mutant maintains the wild-type TRF1-stabilization activity despite its nucleoplasmic distribution, thereby supporting the idea that, regardless of the subcellular distribution, the GTP-binding state of GNL3L per se can control its ability to regulate TRF1 ubiquitylation and degradation.
GNL3L-mediated TRF1 Regulation and Cell Cycle Control
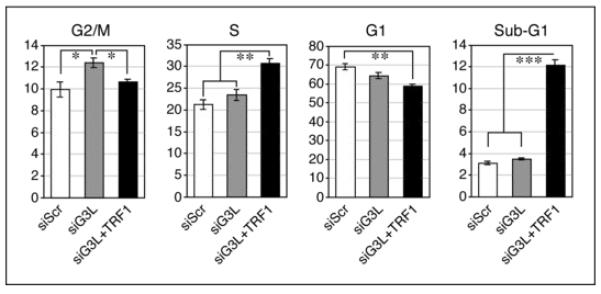
An obligatory role of TRF1 in cell cycle control has been proposed26 based on the observations that TRF1 dysregulation perturbs the mitotic transit of dividing cells 27, and that overexpression of TRF1 can increase the G2/M cell percentage27 and the apoptotic cell percentage.28. This idea is in consistence with the finding that TRF1-null embryos die at E5-6 without discernible telomere length defect.1 A notable finding in our study is that the protein level of GNL3L increases during the mitotic window, which not only coincides with but also is required for the mitotic increase of TRF1 protein. Depletion of GNL3L itself affects G2/M progression primarily at the metaphase-to-anaphase transition2, which is partially rescued by TRF1 putback (Fig. 2). How TRF1 promotes the transition between the metaphase and anaphase is unclear, but ought not to occur at the mitotic telomeres for two reasons: first, when TRF1 protein is increased during mitosis by tankyrase 1 knockout, the transition from metaphase to anaphase is blocked by tethered sister chromatids at the telomere29, 30, and second, high-resolution confocal analyses reveal that the TRF1 signals in the metaphase, anaphase, and early telophase cells are less concentrated at the telomere compared to that in the interphase cells, despite its protein level increase.3 Therefore, we propose that TRF1 may display dual activities at different phases of the cell cycle. In interphase cells, the majority of TRF1 proteins are associated with telomeric DNAs, serving its role in capping the chromosomal end. During this cell cycle window, the GNL3L protein is largely stored in the nucleolus and segregated from the telomeric TRF1. The remaining part of GNL3L diffuses through the nucleoplasm, serving the role of promoting TRF1 homodimerization and its subsequent formation of telomere-capping complex. When dividing cells enter the mitotic stage, the telomere-bound TRF1 protein becomes diffuse in the nucleoplasm to allow proper resolution of sister chromatids. Concomitantly, the nucleolar GNL3L is released as a result of the mitotic disassembly of the nucleolar structure. As such, the TRF1 protein is stabilized, which may be necessary to build up an effective concentration of TRF1 protein for its mitotic function. Finally, it should be pointed out that GNL3L knockdown also causes a G2/M blockage, but to a less extent compared to its metaphase-to-anaphase transit effect, which may not be related to its TRF1 regulatory activity.
Figure 2.
GNL3L promotes the G2/M transition via TRF1 stabilization.
Propidium iodide-labeled cell cycle analyses showed that GNL3L knockdown in HeLa cells (siG3L, grey bars) induces a significant increase in the G2/M cell percentage compared to the control knockdown cells (siScr, white bars), and this G2/M arrest phenotype can be reversed by restoring TRF1 expression (siG3L+TRF1, black bars). The siG3L+TRF1 cells show additional S-phase blockage and increased sub-G1 apoptotic cells compared to the siG3L sample. , p < 0.01; , p < 0.001.
Evolution of Telomere and Cell Cycle Regulation
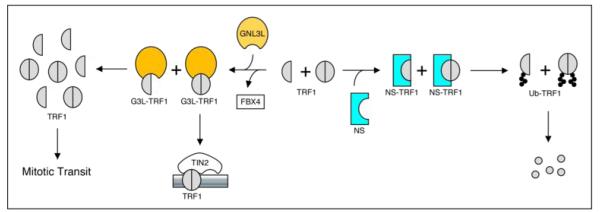
During evolution, NS and GNL3L start to appear as separate genes only in the vertebrate lineages. In all invertebrate genomes, only one common orthologue (hereafter named as GNL3) is found for both NS and GNL3L (Table 1). This observation suggests that NS and GNL3L may have been created from a common ancestor gene at or around the inception of vertebrate evolution. In the event of gene duplication, multiple paralogues in one species either share redundant functions or evolve distinct activities. Multiple lines of evidence now indicate that NS and GNL3L may have been functionally diverged from each other. First, these two genes are differentially expressed in the developing and adult mammalian tissues.11, 15 Second, NS and GNL3L exhibit distinct static and dynamic nucleolar dynamics and slightly different nucleolar distributions.25, 31, 8 Third, they interact with overlapping but non-identical protein targets.9 Fourth, they show distinct activities in complementing the loss-of-function phenotype of their common invertebrate orthologues, e.g. GNL3/Grn1p in S. pombe32 and GNL3/nst-1 (K01C8.9) in C. elegans.33 Finally, these two proteins exert opposite effects on controlling TRF1 protein stability (Fig. 3). In view of the last result, we propose that the creation of NS during vertebrate evolution may be installed to provide self-renewing cells (i.e. ES, somatic stem cells, and cancer cells) with a way to regulate telomere and/or cell cycle in a cell context-specific manner. As TRF1 functions negatively in regulating the telomere length by blocking the access of telomerase 34, GNL3L is expected to prevent telomere elongation in the telomerase+ cells, which has been shown in another study from the angle of telomerase regulation by GNL3L.35 In telomerase− cells, GNL3L reduces the APB formation. As the formation of APB positively correlates with the homologous recombination-based mechanism for telomere elongation, GNL3L is also predicted to decrease the telomere length in telomerase− cells. Although NS appears not to affect telomere length in telomerase+ cells35, we predict that it may increase the telomere length in a telomerase-independent manner, which has been shown to be the dominant mechanism during early embryogenesis.36
Table 1.
Overall protein sequence identity (%) between members of the NS family in Homo sapiens and other species.
| Organism | hNS | hGNL3L | hNgp-1 | |
|---|---|---|---|---|
| NS | B.taurus | 78.22 | 30.22 | 17.98 |
| NS | R.norvegicus | 74.13 | 28.86 | 16.15 |
| NS | M.musculus | 71.22 | 28.86 | 17.49 |
| NS | X.laevis | 45.45 | 27.67 | 19.15 |
| NS | D.rerio | 44.21 | 26.86 | 15.87 |
| NS | G.gallus | 41.16 | 24.93 | 18.80 |
| GNL3L | B.taurus | 28.52 | 91.58 | 18.01 |
| GNL3L | R.norvegicus | 27.92 | 87.84 | 18.12 |
| GNL3L | M.musculus | 27.75 | 86.82 | 18.52 |
| GNL3L | X.laevis | 36.10 | 50.43 | 19.21 |
| GNL3L | D.rerio | 35.28 | 45.63 | 18.54 |
| GNL3 | D.melanogaster | 31.15 | 31.83 | 20.03 |
| GNL3 | A.gambiae | 30.62 | 30.05 | 16.58 |
| GNL3 | C.elegans | 30.43 | 32.45 | 17.53 |
| GNL3 | A.thaliana | 31.69 | 32.03 | 19.47 |
| Nug1p | S.cerevisiae | 22.50 | 19.59 | 14.00 |
| Grn1p | S.pombe | 21.17 | 23.03 | 13.20 |
| Ngp-1 | B.taurus | 18.01 | 19.97 | 87.06 |
| Ngp-1 | R.norvegicus | 12.04 | 18.87 | 83.45 |
| Ngp-1 | M.musculus | 16.64 | 18.09 | 85.36 |
| Ngp-1 | X.laevis | 17.53 | 19.15 | 69.03 |
| Ngp-1 | G.gallus | 15.66 | 17.61 | 68.29 |
| Ngp-1 | D.rerio | 18.67 | 20.37 | 65.55 |
| Ngp-1 | D.melanogaster | 19.51 | 20.06 | 44.31 |
| Ngp-1 | A.gambiae | 18.69 | 19.94 | 47.56 |
| Ngp-1 | C.elegans | 18.55 | 21.47 | 40.05 |
| Ngp-1 | A.thaliana | 19.43 | 18.98 | 36.19 |
| Ngp-1 | S.cerevisiae | 11.10 | 18.00 | 34.10 |
Figure 3.
A model for opposite regulation of TRF1 by NS and GNL3L.
Our work describes a novel TRF1 regulatory mechanism by a pair of vertebrate paralogues, NS and GNL3L, that exert opposite effects on modulating TRF1 protein.
Whether NS modifies the cell cycle activity of TRF1 in a GNL3L-opposing way is not yet determined. At the protein level, unlike GNL3L, the NS protein remains constant throughout different phases of the cell cycle, and NS does not appear to be required for the mitotic increase of TRF1 protein. At the functional level, NS deletion does not affect the metaphase-to-anaphase transition, but instead leads to a G2/M arrest by blocking cell cycle entry into the mitotic prophase, which partially overlaps with that of GNL3L deletion. Therefore, it is mostly likely that the opposite effects of NS and GNL3L on TRF1 affect primarily its telomere rather than mitotic function.
Perspective
As the dynamic and versatile properties of various subnuclear organelles become an intense research focus, the identification of nucleolar proteins such as NS and GNL3L as novel TRF1 modulators provides a molecular platform to investigate the interplay between the two physically discrete organelles, the nucleolus and the telomere. While the regulatory network of telomere and cell cycle control, via TRF1 regulation or not, may seem already far more complex than what has been known, one should still be mindful of the cell type-dependent variation of these complex networks. From the evolutionary standpoint, an intriguing question remains unanswered: why the newly born NS, with distinctive expression and contrasting activity from its invertebrate ancestor (GNL3) and vertebrate twin (GNL3L), gets created in the first place and selected throughout evolution? Finally, as TRF1 has been shown to be more versatile than simply maintaining the length and integrity of telomere, the possibility that it may conduct moonlighting jobs while in the nucleolus remains open and awaits future investigation.
Acknowledgements
This work is supported by NIH grant R01CA113750 to R.Y.T.
References
- 1.Karlseder J, Kachatrian L, Takai H, Mercer K, Hingorani S, Jacks T, et al. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol Cell Biol. 2003;23:6533–41. doi: 10.1128/MCB.23.18.6533-6541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Q, Yasumoto H, Tsai RY. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol Cell Biol. 2006;26:9279–90. doi: 10.1128/MCB.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Q, Meng L, Hsu JK, Lin T, Teishima J, Tsai RY. GNL3L stabilizes the TRF1 complex and promotes mitotic transition. J Cell Biol. 2009;185:827–39. doi: 10.1083/jcb.200812121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–7. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 5.Kishi S, Zhou XZ, Ziv Y, Khoo C, Hill DE, Shiloh Y, et al. Telomeric protein Pin2/TRF1 as an important ATM target in response to double strand DNA breaks. J Biol Chem. 2001;276:29282–91. doi: 10.1074/jbc.M011534200. [DOI] [PubMed] [Google Scholar]
- 6.Chang W, Dynek JN, Smith S. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 2003;17:1328–33. doi: 10.1101/gad.1077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007;14:581–90. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- 8.Meng L, Zhu Q, Tsai RY. Nucleolar trafficking of nucleostemin family proteins: common versus protein-specific mechanisms. Mol Cell Biol. 2007;27:8670–82. doi: 10.1128/MCB.00635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai RY, Meng L. Nucleostemin: A latecomer with new tricks. Int J Biochem Cell Biol. 2009 doi: 10.1016/j.biocel.2009.05.020. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vernet C, Ribouchon MT, Chimini G, Pontarotti P. Structure and evolution of a member of a new subfamily of GTP-binding proteins mapping to the human MHC class I region. Mamm Genome. 1994;5:100–5. doi: 10.1007/BF00292335. [DOI] [PubMed] [Google Scholar]
- 11.Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baddoo M, Hill K, Wilkinson R, Gaupp D, Hughes C, Kopen GC, et al. Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J Cell Biochem. 2003;89:1235–49. doi: 10.1002/jcb.10594. [DOI] [PubMed] [Google Scholar]
- 13.Liu SJ, Cai ZW, Liu YJ, Dong MY, Sun LQ, Hu GF, et al. Role of nucleostemin in growth regulation of gastric cancer, liver cancer and other malignancies. World J Gastroenterol. 2004;10:1246–9. doi: 10.3748/wjg.v10.i9.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kafienah W, Mistry S, Williams C, Hollander AP. Nucleostemin is a marker of proliferating stromal stem cells in adult human bone marrow. Stem Cells. 2006;24:1113–20. doi: 10.1634/stemcells.2005-0416. [DOI] [PubMed] [Google Scholar]
- 15.Yasumoto H, Meng L, Lin T, Zhu Q, Tsai RY. GNL3L inhibits activity of estrogen-related receptor {gamma} by competing for coactivator binding. J Cell Sci. 2007;120:2532–43. doi: 10.1242/jcs.009878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Etheridge KT, Banik SS, Armbruster BN, Zhu Y, Terns RM, Terns MP, et al. The nucleolar localization domain of the catalytic subunit of human telomerase. J Biol Chem. 2002;277:24764–70. doi: 10.1074/jbc.M201227200. [DOI] [PubMed] [Google Scholar]
- 17.Wong JM, Kusdra L, Collins K. Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat Cell Biol. 2002;4:731–6. doi: 10.1038/ncb846. [DOI] [PubMed] [Google Scholar]
- 18.Yang Y, Chen Y, Zhang C, Huang H, Weissman SM. Nucleolar localization of hTERT protein is associated with telomerase function. Exp Cell Res. 2002;277:201–9. doi: 10.1006/excr.2002.5541. [DOI] [PubMed] [Google Scholar]
- 19.Lin J, Jin R, Zhang B, Chen H, Bai YX, Yang PX, et al. Nucleolar localization of TERT is unrelated to telomerase function in human cells. J Cell Sci. 2008;121:2169–76. doi: 10.1242/jcs.024091. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999;19:567–76. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narayanan A, Lukowiak A, Jady BE, Dragon F, Kiss T, Terns RM, et al. Nucleolar localization signals of box H/ACA small nucleolar RNAs. Embo J. 1999;18:5120–30. doi: 10.1093/emboj/18.18.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukowiak AA, Narayanan A, Li ZH, Terns RM, Terns MP. The snoRNA domain of vertebrate telomerase RNA functions to localize the RNA within the nucleus. Rna. 2001;7:1833–44. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S, Hemmerich P, Grosse F. Nucleolar localization of the human telomeric repeat binding factor 2 (TRF2) J Cell Sci. 2004;117:3935–45. doi: 10.1242/jcs.01249. [DOI] [PubMed] [Google Scholar]
- 24.Yoo JE, Oh BK, Park YN. Human PinX1 mediates TRF1 accumulation in nucleolus and enhances TRF1 binding to telomeres. J Mol Biol. 2009;388:928–40. doi: 10.1016/j.jmb.2009.02.051. [DOI] [PubMed] [Google Scholar]
- 25.Tsai RY, McKay RD. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol. 2005;168:179–84. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou XZ, Perrem K, Lu KP. Role of Pin2/TRF1 in telomere maintenance and cell cycle control. J Cell Biochem. 2003;89:19–37. doi: 10.1002/jcb.10496. [DOI] [PubMed] [Google Scholar]
- 27.Shen M, Haggblom C, Vogt M, Hunter T, Lu KP. Characterization and cell cycle regulation of the related human telomeric proteins Pin2 and TRF1 suggest a role in mitosis. Proc Natl Acad Sci U S A. 1997;94:13618–23. doi: 10.1073/pnas.94.25.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kishi S, Wulf G, Nakamura M, Lu KP. Telomeric protein Pin2/TRF1 induces mitotic entry and apoptosis in cells with short telomeres and is down-regulated in human breast tumors. Oncogene. 2001;20:1497–508. doi: 10.1038/sj.onc.1204229. [DOI] [PubMed] [Google Scholar]
- 29.Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- 30.Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S. Protein requirements for sister telomere association in human cells. Embo J. 2007;26:4867–78. doi: 10.1038/sj.emboj.7601903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng L, Yasumoto H, Tsai RY. Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm. J Cell Sci. 2006;119:5124–36. doi: 10.1242/jcs.03292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du X, Rao MR, Chen XQ, Wu W, Mahalingam S, Balasundaram D. The homologous putative GTPases Grn1p from fission yeast and the human GNL3L are required for growth and play a role in processing of nucleolar pre-rRNA. Mol Biol Cell. 2006;17:460–74. doi: 10.1091/mbc.E05-09-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudron MM, Reinke V. C. elegans nucleostemin is required for larval growth and germline stem cell division. PLoS Genet. 2008;4:e1000181. doi: 10.1371/journal.pgen.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–3. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 35.Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell. 2007;28:773–85. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L, Bailey SM, Okuka M, Munoz P, Li C, Zhou L, et al. Telomere lengthening early in development. Nat Cell Biol. 2007;9:1436–41. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]