Abstract
Some forms of learning and memory, and their electrophysiologic correlate, long-term potentiation (LTP), require dendritic translation. We demonstrate that Pin1, a peptidyl-prolyl isomerase, is present in dendritic spines and shafts and inhibits protein synthesis induced by glutamatergic signaling. Pin1 suppression increased dendritic translation, possibly through eIF4E binding proteins 1 and 2 (4E-BP1/2) and eukaryotic translation initiation factor 4E (eIF4E). Consistent with increased protein synthesis, hippocampal slices from Pin−/− mice had normal early LTP (E-LTP) but significantly enhanced late LTP (L-LTP) compared to wild-type controls. Protein kinase C ζ (PKCζ) and protein kinase M ζ (PKMζ) were increased in Pin1−/− mouse brain and their activity was required to maintain dendritic translation. PKMζ interacted with and inhibited Pin1 by phosphorylating Ser16. Therefore, glutamate-induced, dendritic protein synthesis is sequentially regulated by Pin1 and PKMζ signaling.
INTRODUCTION
Long-term memory and long-lasting forms of synaptic plasticity, such as late-phase long-term potentiation (L-LTP), require the synthesis of new protein on dendritic polyribosomes [1–3] at or near activated synapses [1]. This is distinct from short-term forms of synaptic plasticity such as early phase LTP (E-LTP) which are protein synthesis independent. Multiple dendritic kinases, including calcium-calmodulin-dependent protein kinase II (CaMKII), mitogen-activated protein kinase (MAPK), mitogen-activated or extracellular signal-regulated protein kinase kinase (MEK1), protein kinase C ζ (PKCζ), and protein kinase M ζ (PKMζ) have been implicated in the induction or maintenance of hippocampal L-LTP [4–7]. Blockade of PKC, MEK1, or MAPK prevents the induction of L-LTP and the initial phases of learning and memory consolidation [8,9]. In contrast, PKMζ is essential for the maintenance of L-LTP and the persistence of spatial memory storage in the hippocampus [7,8,10]. PKMζ is an atypical PKC isoform that is unique in lacking an autoinhibitory, pseudosubstrate regulatory domain [11] and is activated by phosphoinositide-dependent protein kinase-1 (PDK1) [12,13]. How PKMζ maintains L-LTP and spatial memory is largely unknown but may involve AMPA-type glutamate receptor (AMPAR) phosphorylation and trafficking, with subsequent changes in excitatory postsynaptic potential (EPSP) amplitude, and likely requires on-going dendritic translation [14,15].
The regulation of dendritic translation is predominantly at the level of initiation through eukaryotic translation initiation factor 4E (eIF4E), the component of the multi-subunit eIF4F complex that binds to the 5' methylguanosine cap of mRNAs [16]. eIF4F assembly is inhibited through direct interaction of eIF4E with partially phosphorylated eIF4E binding proteins (4E-BPs), preventing cap-dependent translation [16,17]. Additional phosphorylation of 4E-BPs on Thr37 or Thr46 by mammalian target of rapamycin [mTOR, also known as FKBP-12-rapamycin-associated protein (FRAP)], S6 kinase p70 (S6K), MAPKs, or PKC causes release of 4E-BP1 from eIF4E, leading to the subsequent assembly of the translation initiation complex [7–19]. Both Thr37 and Thr46 are immediately N-terminal to a proline and are thus potential recognition sites for Pin1 (protein interacting with NIMA 1) [20], a peptidyl-prolyl isomerase (PPIase). Pin1, which is highly abundant in the cytoplasm and nucleus of neurons [20–22], is composed of an N-terminal type IV WW-domain connected by a flexible linker to a C-terminal PPIase domain. The WW-domain mediates specific interactions with target substrates containing dipeptide Ser-Pro or Thr-Pro motifs, distinguishing its actions from those of the related but less selective PPIases cyclophilin A and FK506-binding protein (FKBP) [20]. Pin1 preferentially isomerizes the Ser/Thr-Pro peptide bond after phosphorylation of Ser or Thr, leading to alterations in target protein conformation and biological activity [20,22]. Pin1 has been linked to cellular transformation and tumorigenesis, apoptosis, neurodegeneration, and the transcriptional regulation of cytokines [22–24].
FKBPs and cyclophilins have been implicated in protein synthesis in dendrites and in L-LTP [25–28]. FKBP12 binds to mTOR, the inhibition of which with rapamycin inhibits dendritic protein synthesis and LTP [19,25,28]. Deletion of FKBP in rodents leads to enhanced L-LTP that is resistant to rapamycin but sensitive to inhibitors of protein synthesis [25]. Knockout of cyclophilinD improves learning, memory, and synaptic function in a mouse model of Alzheimer's disease [26,27]. Thus, PPIases likely play a role in plasticity and memory by regulating the initiation of dendritic protein synthesis.
We now demonstrate that Pin1 is present and constitutively active in dendritic shafts and spines, where it interacts with 4E-BP and eIF4E and, under basal conditions, suppresses protein synthesis. Pin1 activity was rapidly decreased by glutamatergic signaling. Genetic ablation or pharmacologic inhibition of Pin1 increased dendritic translation. Both PKMζ and the related PKCζ increased in abundance in dendrites after Pin1 inhibition or knock-out and functioned in feed-forward loops to maintain translation and feed-back loops to inhibit Pin1 activity. Hippocampal slices from Pin1−/− mice displayed normal E-LTP but enhanced protein synthesis-dependent L-LTP. These results implicate Pin1 and PKMζ in the regulation of glutamate-induced dendritic protein synthesis and clarify how PKMζ functions to modulate synaptic plasticity.
RESULTS
Pin1 is present and catalytically active in dendrites
Pin1 has been identified in the nucleus and cytoplasm of neurons [21,22,29] but not in dendritic shafts or spines. Immunofluorescence analysis of hippocampal sections revealed strong Pin1 staining in CA1, CA3, and the dentate gyrus (Fig. 1A). Synaptoneurosomes (SN, resealed vesicles containing pre- and post-synaptic structures) prepared from total mouse cortex were positive for Pin1 by Western blot (Fig. 1B). SN include ribosomes, as well as functional glutamate receptors, making them a convenient and powerful system to study dendritic protein synthesis [30,31]. Confocal analysis of cultured cortical neurons prepared from the brains of wild-type (WT) embryonic day 17 to 18 (E17 to E18) mouse embryos showed punctate Pin1 staining along dendritic shafts, which colocalized with postsynaptic density protein 95 (PSD95) (Fig. 1C). Immunoblot (Fig. 1B) and immunofluorescence (fig. S1A) analyses of Pin1−/− brain tissue were negative, indicating that the antisera were specific for Pin1. These results suggest Pin1 is present in dendritic spines and shafts of rodent neurons and that SN are markedly enriched for pre- and post-synaptic structures.
Fig. 1. Pin1 is present and active in post-synaptic terminals.
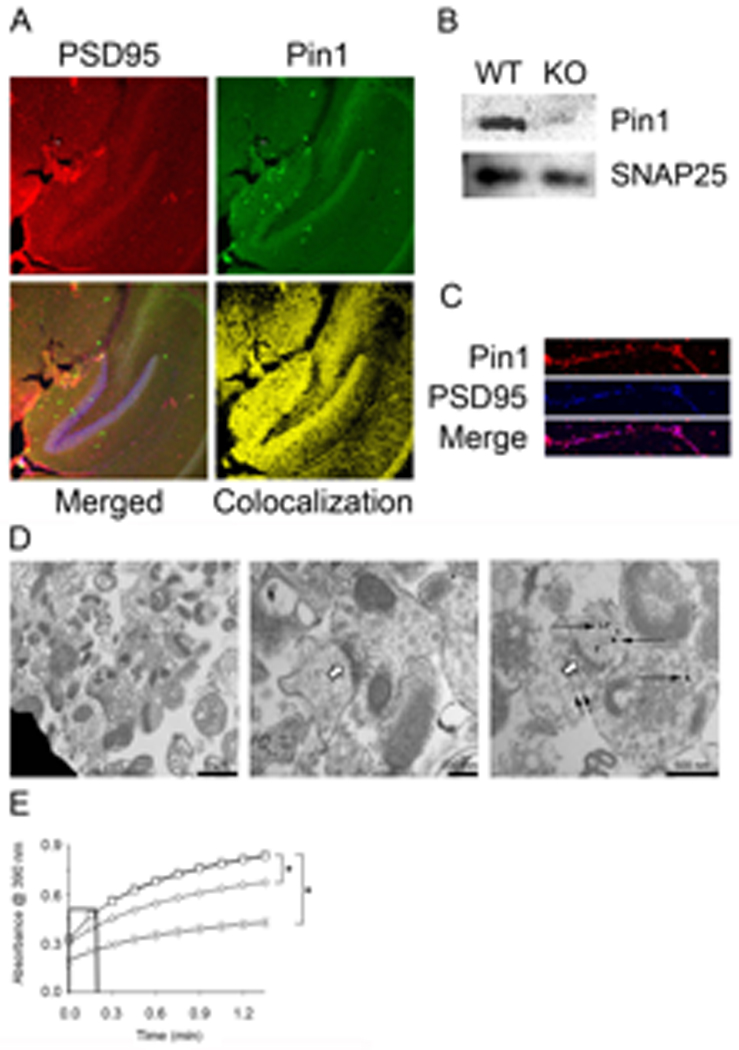
(A) Representative 100× confocal images of 30 µm hippocampal slices from P21 to P25 mice immunostained with anti-PSD95-Rhodamine, anti-Pin1-FITC, and To-Pro3. Merge panel shows uncolocalized (Pin1-red, PSD95-green, and To-Pro3-blue) and colocalized (yellow and magenta) points, whereas colocalization panel shows only points of Pin1 and PSD95 colocalization (yellow), (B) Pin1 and SNAP25 were detected by Western blot of 10 µg SN protein from WT and Pin1−/− mice. (C) Representative confocal image (600×) of E17 cortical neuron dendrites, 18 DIV. Cells were labeled with anti-Pin1-Rhodamine and anti-PSD95-Cy5 to identify colocalization (magenta). (D) EM of SN stained with anti-Pin1 coupled to gold beads [61]. Left and Center: vesicles (bars:1 µm left, 200 nm right) containing pre- and post-synaptic terminals. Right: representative region (bar 500 nm) stained with anti-Pin1-gold. Open arrows denote post-synaptic densities. Long, narrow arrows denote Pin1 staining. Small, narrow arrows denote synaptic vesicles. (E) SN were untreated (□), or pre-treated with 1 µM juglone (◊), or 1 µM CsA (○) for 10 min prior to lysis and isomerase activity assay. No SN control (X) contained complete reaction except SN, n=3, ± SEM, * denotes p<0.001. Initial slopes were calculated to determine K (see Table 1A).
To precisely identify the subcellular location of Pin1, we performed immuno-electron microscopy. SN from Pin1+/+ mice (C57BL6 background, age P12–P22) were isolated by percoll-gradient centrifugation, fixed, and stained with antibody directed against Pin1 (anti-Pin1) coupled to gold beads. As typical for SN preparations, various organelles, including mitochondria, were present along with the resealed vesicles, which contained pre- and post-synaptic terminals (Fig. 1D). Immunogold reactivity in vesicles was most pronounced post-synaptically, with much weaker pre-synaptic staining (Fig. 1D, right-hand panel). In contrast, mitochondria failed to stain with anti-Pin1. We thus conclude that Pin1 is present in dendritic shafts and spines in rodent cortex and hippocampus, and is preferentially located in the post-synaptic region.
Pin1 activity can be measured in SN or cell lysates by means of a protease-coupled, cis to trans isomerization assay [24,32,33]. Conversion of the peptide substrate (Suc-Ala-Glu-Pro-Phe-pNA) from cis to trans permits cleavage of the C-terminal nitroaniline by chymotrypsin and detection at 390nm. Untreated SN contained substantial basal PPIase activity (Fig. 1E, Table 1). Although Suc-Ala-Glu-Pro-Phe-pNA is isomerized by Pin1 [21,33,34,35], other PPIases, including cyclophilinA, may also act on it. Therefore, we performed isomerase assays on SN in the presence of the selective PPIase inhibitors cyclosporineA (CsA), FK506 and juglone. We also tested the effects of a membrane permeable, catalytically inactive form of Pin1 created by fusion of a trans-activator of transcription (TAT) penetratin tag to the N-terminus and substitution of lysine 63 with alanine (TAT-Pin1-K63A). CsA inhibits cyclophilinA, FK506 blocks FKBP, and juglone and TAT-Pin1-K63A specifically inhibit endogenous Pin1 by covalent modification and preventing substrate access, respectively [32,34,35,36]. Neither CsA (Fig. 1E, Table 1A) nor FK506 (Table 1A) reduced the rate of substrate isomerization. However, TAT-Pin1-K63A or the Pin1 inhibitor juglone [32] dose-dependently inhibited PPIase activity (Fig. 1E, Table 1B). Recombinant TAT-Pin1-K63A also dose-dependently prevented recombinant WT-Pin1 from isomerizing substrate, supporting the conclusion that the former can function as a dominant-negative form of Pin1 (fig. S2E). Therefore, we conclude that constitutively active Pin1 is present in dendrites and that the observed isomerization of target substrate is independent of cyclophilinA or FKBP. Thus, unlike the immune system where Pin1 is inactive in unstimulated cells [24], dendritic Pin1 shows elevated activity under basal conditions.
Table 1. PPIase activity in SN is mediated by Pin1 and regulated by glutamate and PKCζ/PKMζ.
SN were untreated (untreated control), or pre-treated with the indicated drugs prior to lysis and assay of isomerase activity. The initial slopes were calculated to determine K (K = kobs−k0/k0 [36]). No SN control (k0) contained the complete reaction mixture including protease but excluding SN, n=3, ± SEM. Individual groups are compared to no SN control (p valuec) in all cases; p compared to CsA, βgal or myr-peptide controls shown as appropriate. (A) SN were untreated (control), or pre-treated with 1 µM juglone or 1 µM CsA for 10 min. (B) SN were untreated (control), or incubated with 1 µM CsA for 5 min alone or prior to the addition of TAT-Pin1-K63A or TAT-βGal for 5 additional min.. (C) SN were untreated or treated with Glu for 30 min. (D) SN were untreated or treated with 10 µM myr-PKC/PKMζ or myr-control peptide for 5 min.
| Set | K | SEM | p valuec |
p valueCsA | |
|---|---|---|---|---|---|
| A | Untreated Control | 1.290 | 0.038 | ||
| CsA (1 µM) | 1.303 | 0.039 | 0.523 | ||
| FK-506 (1 µM) | 1.467 | 0.019 | 0.015 | 0.011 | |
| Juglone (1 µM) | 0.586 | 0.039 | 0.0001 | 0.00002 | |
| K | SEM | p valuec | p valueβGal | ||
| B | Untreated Control | 0.984 | 0.085 | ||
| CsA only | 0.986 | 0.095 | 0.893 | ||
| CsA + TAT-βGal (300 nM) | 0.995 | 0.097 | 0.840 | ||
| CsA + TAT-βGal (600 nM) | 0.994 | 0.128 | 0.874 | ||
| CsA + TAT-Pin1 K63A (300 nM) | 0.593 | 0.031 | 0.014 | 0.017 | |
| CsA + TAT-Pin1 K63A (600 nM) | 0.378 | 0.090 | 0.023 | 0.026 | |
| K | SEM | p valuec | |||
| C | Control | 1.854 | 0.075 | ||
| Glu | 1.064 | 0.248 | 0.027 | ||
| K | SEM | p valuec | p valueMyr-PepC | ||
| D | Control | 1.593 | 0.288 | ||
| myr-Peptide Control | 1.523 | 0.233 | 0.935 | ||
| myr-PKCζ/PKMζ | 4.813 | 0.381 | 0.003 | 0.005 |
Pin1 inhibits protein synthesis and is itself inhibited by glutamate stimulation
Dendritic translation is required for the induction of L-LTP and for memory consolidation, which presumably reflects a requirement for the creation of new proteins that underlie synaptic remodeling [37]. We modeled in vivo events by evaluating SN translation by measuring 35S-methionine incorporation into protein after addition of glutamate. SN were translationally active, producing a broad spectrum of proteins, the abundance of which increased significantly in response to treatment with 50 µM glutamate/10 µM glycine (Glu) (Fig. 2A,B,C) and decreased in response to cycloheximide (fig. S2A), and which were not apparent after treatment with anisomycin (Aniso) (Fig. 2A,B). Translation was progressively reduced after treatment with increasing concentrations of Triton-X100, consistent with the notion that protein synthesis occurred within a membrane-encased organelle (fig. S2D). The rate of protein synthesis in unstimulated SN transduced with dominant-negative TAT-Pin1-K63A was nearly identical with that in untransduced SN treated with Glu (Fig. 2A,B). Similar effects were seen after treatment of SN with the Pin1 inhibitor juglone (fig. S2B). We also evaluated protein synthesis in SN from Pin1−/− mouse brains at postnatal day 16 (P16) to day 22 (P22). SN isolated from Pin1−/− mice showed twice the basal rate of 35S-Met incorporation than did those from Pin1+/+ littermate controls (Fig. 2C,D). Moreover, glutamate had no effect on translation in Pin1−/− SN. Thus, these results suggest that Pin1 normally suppresses dendritic translation, an inhibition that can be overcome through glutamate-mediated signaling or Pin1 ablation.
Fig. 2. Pin1 suppresses dendritic translation.
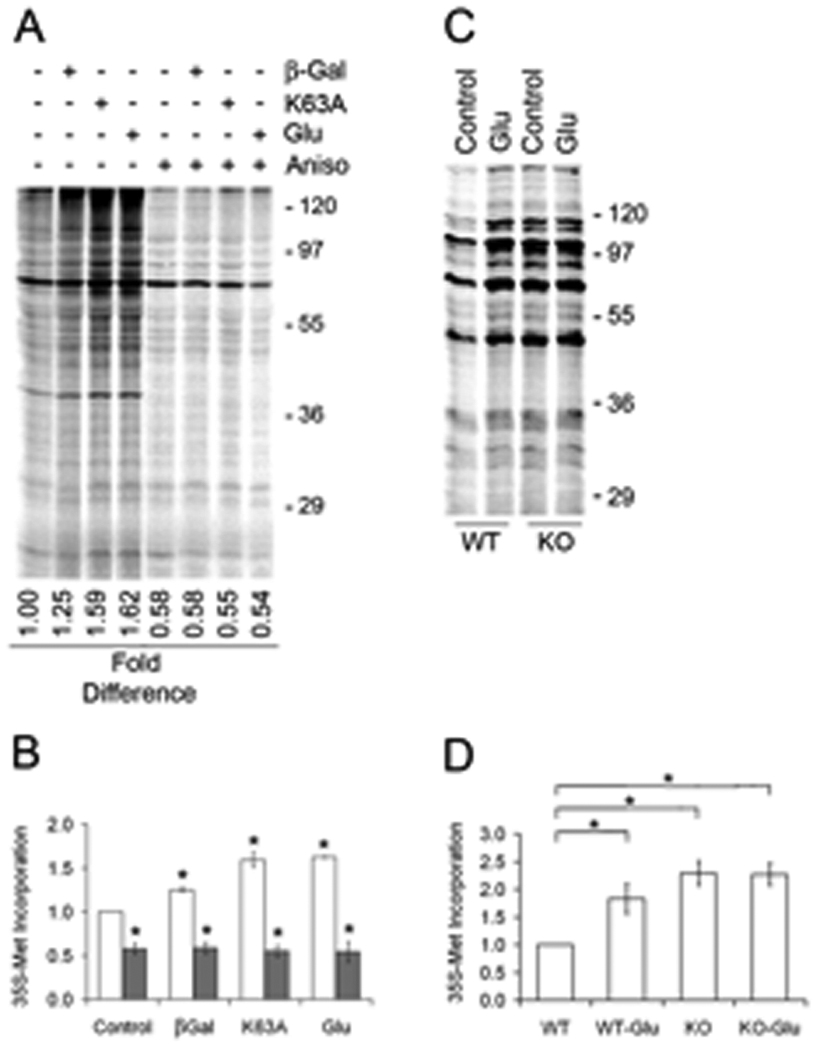
(A) SN were preincubated for 15 min. without (lanes 1–4) or with 40 µM anisomycin (Aniso, lanes 5–8) prior to addition of 35S-Met and no additional treatment, transduction with 50 nM TAT-βGal (βGal) or 50 nM TAT-Pin1-K63A (K63A), or treatment with Glu for 30 min prior to lysis and SDS-PAGE. (B) Total 35S-Met incorporation into protein was quantitated by phosphorimaging. Relative translation compared to control SN (□) was 1.25 ± 0.03 for βGal, 1.59 ± 0.09 for K63A and 1.62 ± 0.01 for Glu, 0.58 ± 0.06 for Aniso (■), 0.58 ± 0.07 for Aniso plus βGal, 0.55 ± 0.07 for Aniso plus K63A, 0.54 ± 0.12 for Aniso plus Glu, n=3, ± SEM, * denotes p<0.019 between K63A and βGal, p<0.003 between K63A and control. (C) WT or Pin1 KO SN were untreated (Control) or treated with Glu for 30 min. Total 35S-Met incorporation was quantitated (D) n=3, ± SEM, * denotes p<0.04.
Glutamatergic signaling could activate dendritic translation by inducing Pin1 catabolism or by suppressing Pin1 PPIase activity. Western analysis revealed no significant changes in the abundance of Pin1 after glutamate stimulation of SN (fig. S2C), although Pin1 abundance significantly increased in dendritic spines of cultured cortical neurons treated with glutamate (Fig. 3B, fig. S3A), demonstrating Pin1 can be locally synthesized in response to glutamatergic signaling. However, PPIase activity was significantly reduced in Glu-treated SN compared to that in untreated controls (Table 1C). The degree of Glu-induced suppression of Pin1 isomerase activity and stimulation of Pin1 translation was comparable to that seen after TAT-Pin1-K63A transduction (Table 1B, Fig. 2A). Therefore, Pin1 appears to repress translation under basal conditions through mechanisms that require its isomerase activity. Glutamatergic signaling rapidly suppresses Pin1 activity as a prerequisite for translational upregulation.
Fig. 3. Pin1 associates with signaling proteins, and Pin1−/− hippocampal slices show increased L-LTP.
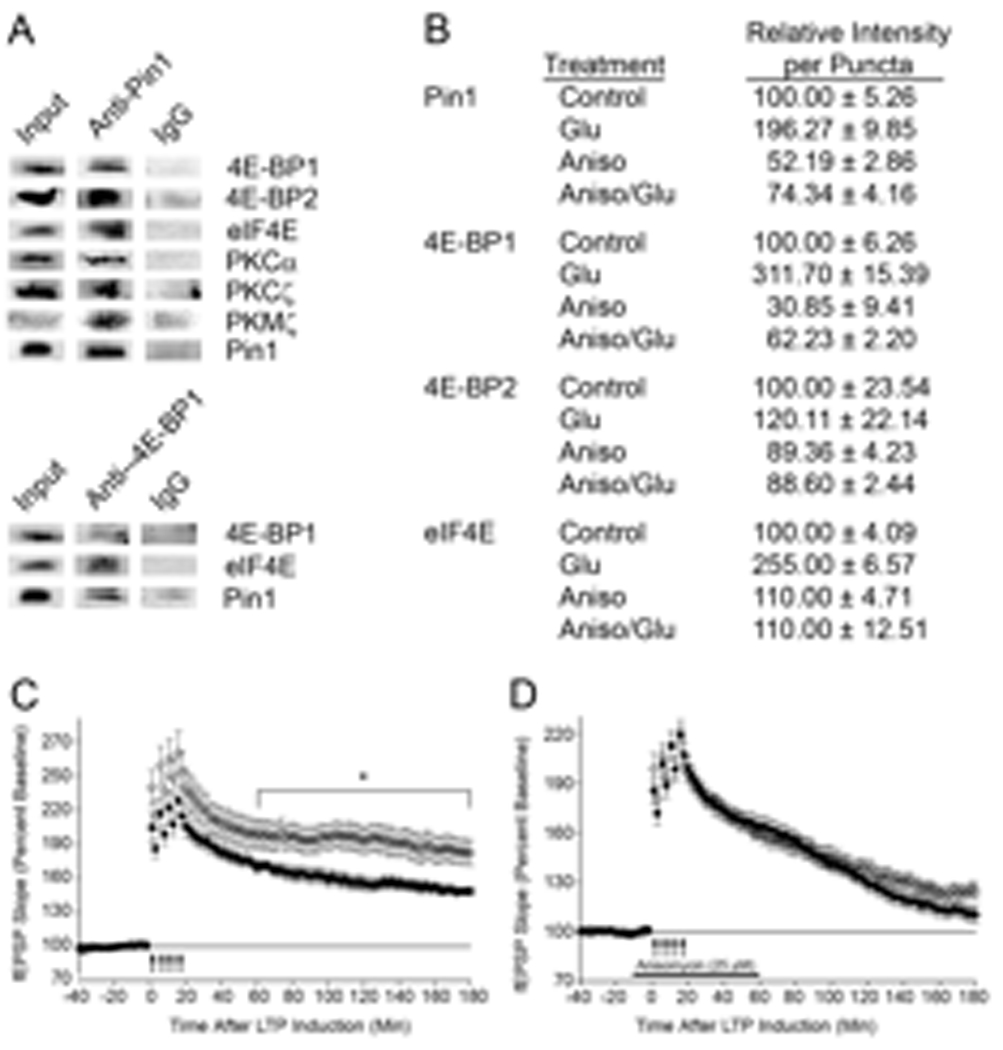
(A) WT brain homogenates (600 µg) were lysed and immunoprecipitated with pre-immune IgG, anti-Pin1, or anti-4E-BP1 followed by immunoblot, n=3. Homogenate (50–200 µg, input) was used as a positive control. (B) E17 cortical neurons, DIV18, were untreated (Control), stimulated with Glu, or pretreated with 40 µM Aniso before Glu, and then fixed, stained, and the dendrites visualized. Fluorescence of ~300 random puncta were quantitated and SEM determined. (C) L-LTP was induced in Pin1−/− (O; n=11 slices, n= 3 mice) or Pin1+/+ (●; n=9 slices, n=3 mice, p =0.0295) hippocampal slices with four trains of high frequency stimulation. (D) L-LTP was induced in anisomycin-treated hippocampal slices from Pin1+/+ [●; n = 12 slices, n = 3 mice, p = 0.0053 (WT with and without Aniso)] and Pin1−/− (O; n = 11 slices, n = 3 mice, p = 0.0003 (KO with and without Aniso)).
Pin1 interacts with 4EBP1/2 and eIF4E, but not with mTOR or p70-S6K
4E-BP1 and 2 (4E-BP1/2), mTOR, ribosomal p70-S6K, and eIF4E all contain Ser/Thr-Pro sites that can potentially be recognized and isomerized by Pin1. Immunoprecipitation of cortical lysates with antibodies directed against Pin1 or 4E-BP1 followed by immunoblot revealed that Pin1 interacts with 4E-BP1/2 and eIF4E (Fig. 3A) but not p70-S6K, mTOR or MEK1 (fig. S3B). The interaction of Pin1 with 4E-BP1/2 was unaffected by brief (10 min) treatment of SN with Glu, rapamycin, or a combination of the two (fig. S3C). To visualize the location of the interactions between Pin1 and translational regulators, we analyzed cultured cortical neurons prepared from E17 embryos by confocal microscopy. Pin1 was present in a punctate pattern along dendrites, where its abundance was modestly increased by Glu, and decreased by Aniso (Fig. 3B, fig. S3A). eIF4E, 4E-BP1/2, and p70-S6K displayed similar staining patterns along the dendritic shafts and spines as Pin1 (fig. S3A). Overlay of the images with ImageJ [38] demonstrated considerable colocalization of Pin1 with eIF4E, 4E-BP1/2, and p70-S6K, which was decreased after Aniso treatment (fig. S3A). These results demonstrate that Pin1 is present in dendritic shafts and spines, where it is associated with 4E-BP1/2 and eIF4E, known regulators of global protein synthesis, suggesting that Pin1 modulates these proteins to influence translation initiation.
Pin1−/− mice show enhanced L-LTP
Protein synthesis, especially in dendrites, is essential for the formation of long-term memory and the maintenance of long-term forms of synaptic plasticity, such as L-LTP [2,9]. Thus far, our data indicate that Pin1 is a mediator of glutamate-induced dendritic protein synthesis. Therefore, we investigated the possibility that Pin1 was involved in LTP. Although basal synaptic transmission, as defined by the field excitatory postsynaptic potential (fEPSP) slope versus voltage were equivalent in hippocampal slices from WT and Pin1−/− mice (fig. S4A), paired-pulse facilitation, a form of short- term synaptic plasticity, was enhanced at 10 and 20 ms interstimulus intervals in Pin1−/− slices (10 msec interstimulus interval, ***p<0.001; 20 ms interstimulus interval, **p<0.01) (fig. S4B), indicating that Pin1 may affect neurotransmitter release. Next we evaluated the role of Pin1 during a protocol designed to induce L-LTP (four high-frequency trains of stimuli). Slices isolated from Pin1−/− animals showed a similar degree of LTP induction compared to those from WT mice, and showed a similar fEPSP slope for the first 60 minutes (Fig. 3C). After one hour, Pin1−/− slices showed a significant increase in fEPSP slope compared to that in WT slices (P=0.0214; repeated measures ANOVA, 60 minutes to 180 minutes post-stimulation), which was prevented by protein synthesis inhibitors (Fig. 3D). In contrast, when L-LTP was induced with a Theta Burst Stimulation (TBS) protocol, we found no significant differences between WT and KO slices in LTP induction or the magnitude of the fEPSP slope (fig. S4C). The differences found in Pin1 KO slices between the two stimulation protocols may reflect the different biochemical pathways induced by high-frequency stimulation and TBS [41–44]. In addition, our results are distinct from those observed in mice lacking 4E-BP2, the eIF2a kinase GCN2, or MEK1 KO mice. In these mice, a single stimulus which normally leads to E-LTP induces plasticity that mimics L-LTP [9,39,40].
PKMζ and PKCζ are increased in abundance in Pin1−/− mice
Multiple kinases (PKC, MEK1, MAPK, PKCζ and PKMζ) and translational regulators (4E-BP2 and GCN2) have been implicated in the induction or maintenance (or both) of L-LTP [8,9,10,12,39,40] or persistent memory storage [45]. Given the increased maintenance of L-LTP we observed in the Pin1−/− hippocampal slices, we evaluated PKCζ/PKMζ abundance in brain lysates. Western blot revealed that the abundance of both PKC isoforms was increased by up to 200% in the cortex and hippocampus (Fig. 4A) of Pin1−/− mice compared to that in the cortex and hippocampus of WT mice. In contrast, mGluR1, GABAβR1 and actin abundance were unchanged (Fig. 4A). PKMζ rapidly increases by ~50% after the induction of L-LTP in wild type hippocampal slices [15,45].
Fig. 4. A–F. PKCζ/PKMζ show increased abundance in Pin−/− brain and control Pin1 activity through Ser16 phosphorylation.
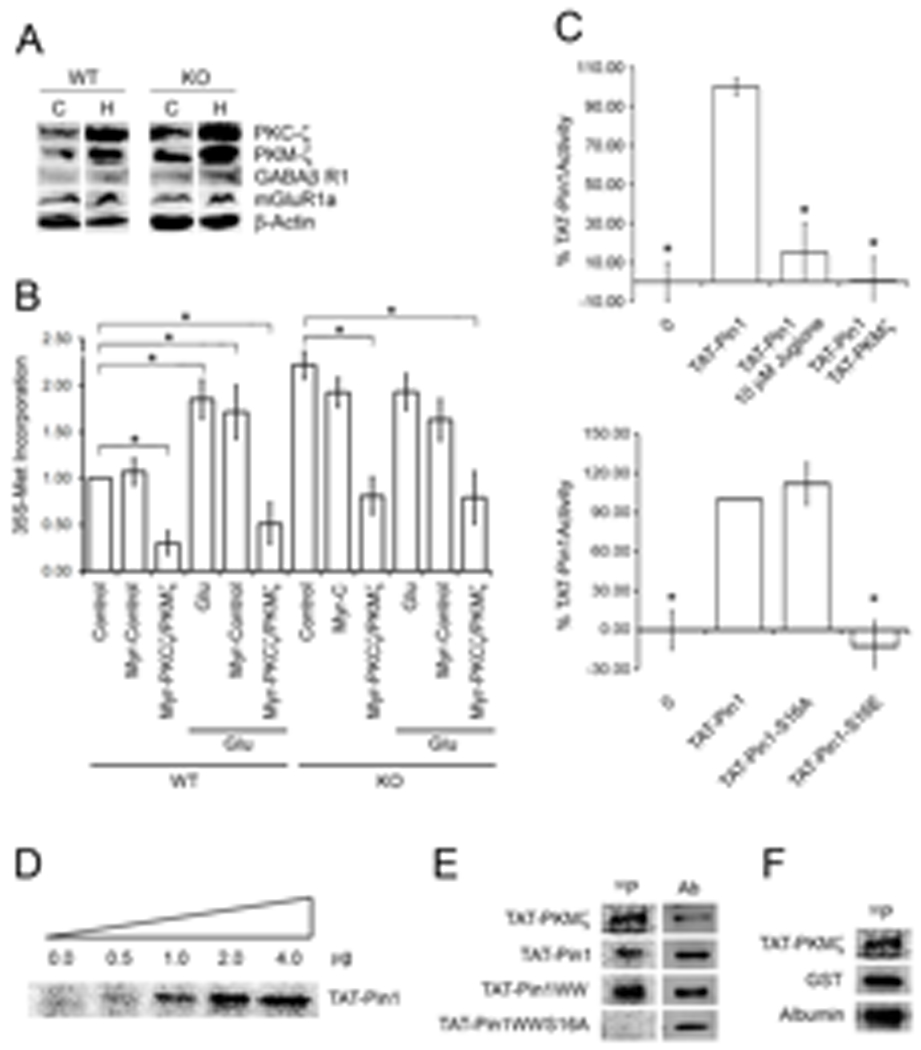
(A) Cortex (C) and hippocampus (H) were dissected from 4 week-old WT and Pin1−/− mouse brains and lysates analyzed by Western blot. (B) WT and Pin1 KO SN were pretreated with 10 µM myr-control or myr-PKCζ/PKMζ inhibitor peptides and left untreated or treated with Glu for 30 min. Total 35S-Met incorporation was quantitated. * denotes p<0.018. (C) Upper Panel: Isomerase activity of recombinant TAT-Pin1 (1 µg), plus 1 µM juglone or plus 400 ng TAT-PKMζ and ATP, after a 2 min pre-incubation, n=3, ± SEM, * denotes p<0.01. Lower Panel: Isomerase activity of recombinant TAT-Pin1, TAT-Pin1-S16A, TAT-Pin1-S16E (2 µg each), n=3, ±SEM, * denotes p<0.01. (D) Kinase activity assays were performed with 2 µg TAT-PKMζ, 32γ-ATP and increasing amounts of TAT-Pin1 (0–4 µg), n=3. (E) Full-length TAT-Pin1, TAT-Pin1-WW or TAT-Pin1-WW-S16A was incubated with TAT-PKMζ, 32γ-ATP for 15 min prior to SDS-PAGE, Western blotting with anti-His tag (Ab), and autoradiography, n=3. (F) Recombinant GST, albumin, or no additions were incubated with TAT-PKMζ, 32γ-ATP for 15 min prior to SDS-PAGE,
PKCζ/PKMζ Inhibit Pin1 and Promote Protein Synthesis
The induction of L-LTP can be prevented by inhibitors of protein synthesis, whereas L-LTP maintenance is sensitive to inhibition of PKCζ/PKMζ [9,10]. Whether PKCζ/PKMζ also participate in the regulation of dendritic translation is unknown. We treated SN from Pin1−/− and Pin1+/+ mouse brains with a membrane-permeable myristoylated-PKCζ peptide inhibitor (myr-PKCζ/PKMζ) or an irrelevant myristoylated control peptide (myr-control) and assessed translation. Myr-PKCζ/PKMζ mimics the pseudosubstrate domain that binds the PKCζ catalytic domain and suppresses its catalytic activity [10,45]. Because PKCζ/PKMζ share a common catalytic domain [11], the same myr-peptide inhibits both isoforms. Basal or glutamate- induced translation by Pin1+/+ SN was unaffected by myr-control but reduced by >75% by myr-PKCζ/PKMζ (Fig. 4B). Translation by Pin1−/− SN was also sensitive to myr-PKCζ/PKMζ (Fig. 4B), suggesting that PKCζ/PKMζ are downstream of Pin1 and, once produced, are required to maintain translation. The requirement for PKCζ/PKMζ activity in the maintenance of L-LTP may reflect their involvement in the regulation of dendritic protein synthesis.
Because Pin1 activity is inhibited by reversible phosphorylation [24,47], we asked if Pin1 was a PKCζ/PKMζ target. (Fig 3A), Both PKCζ and PKMζ immunoprecipitated with Pin1 (Fig. 3A); moreover, myr-PKCζ/PKMζ markedly increased Pin1 activity (Table 1D). Therefore, we evaluated the ability of recombinant PKMζ to inactivate recombinant Pin1. In the presence of ATP, recombinant PKMζ was capable of phosphorylating itself, glutathione-S-transferase and albumin (Fig. 4F). When incubated with ATP and PKMζ, recombinant Pin1 was phosphorylated and its activity was significantly decreased to a similar degree as with juglone treatment (Fig. 4C–E). Phosphorylation of the WW domain of Pin1 at Ser16 prevents its interaction with Ser/Thr-Pro substrates, leading to a functional loss of Pin1 PPIase activity [47]. Therefore, we performed PKMζ kinase activity assays with different TAT-linked, Pin1 recombinant proteins including full length wild type (TAT-Pin1), the N-terminal WW domain (TAT-Pin1WW), or a mutant WW domain with Ala substituted for Ser16 (TAT-Pin1WWS16A). Both TAT-Pin1 and TAT-Pin1WW were phosphorylated, whereas TAT-Pin1WWS16A was not (Fig. 4E). To determine whether Ser16 phosphorylation affected Pin1 activity, we compared the in vitro PPIase activity of recombinant TAT-Pin1 constructs with Ala16 (TAT-Pin1-S16A) or Glu16 (TAT-Pin1-S16E) to TAT-Pin1. The Glu substitution mimics phosphorylated Ser whereas Ala can not be phosphorylated. TAT-Pin1-S16A and TAT-Pin1 showed equivalent PPIase activity, whereas TAT-Pin1-S16E showed none (Fig. 4C). Therefore, we conclude that PKMζ phosphorylates Pin1 on Ser16. This modification is sufficient to prevent the WW-domain of Pin1 from binding substrate and leads to a functional loss of PPIase activity.
DISCUSSION
Pin1 is a molecular switch that binds and isomerizes phosphorylated Ser/Thr-Pro containing proteins, thereby regulating their biological activity [20]. The function of Pin1, in neurons in general and at synapses in particular, is largely unknown. Here, we show that Pin1, by regulating translation and inhibiting PKMζ production, is involved in LTP and synaptic plasticity.
Pin1 abundance is increased during cell division and in continuously dividing tumor cell lines and is an independent risk factor for increased tumor grade and metastatic potential [48]. Pin1 abundance has been implicated in the pathogenesis Alzheimers disease, possibly by mislocalization to hyperphosphorylated tau [22,29]. Compared to wild type animals, accelerated apoptotic neurodegeneration occurs in Pin−/− mice, likely through misregulation of Bim-EL, a BH3 containing, proapoptotic member of the Bcl-2 family [20]. These effects of Pin1 depend on its cytoplasmic or nuclear location; here, we show that Pin1 is highly abundant and constitutively active in post-synaptic terminals of rodent brain where, under basal conditions, it suppresses protein synthesis. These results suggest that Pin1 may also antagonize neurodegeneration by decreasing the synthesis of potentially toxic proteins such as the transcription factor E-26-like protein 1 (Elk-1), whose dendritic translation induced neuronal apoptosis [49].
Under basal conditions, 4E-BP is partially phosphorylated [52,53] which is associated with maximal affinity for eIF4E and suppression of translation [17–19,52,53]. Glutamate activates p70-S6K, PKC and mTOR [50,51] all of which phosphorylate 4E-BP at multiple sites [17–19]. Akt does not directly act on 4E-BP although it is required for 4E-BP inhibition after growth factor mediated signaling [18]. Therefore, we propose that under basal conditions in dendrites, active Pin1 binds to and isomerizes minimally (hypo) phosphorylated 4E-BP1, preventing additional phosphorylation and facilitating its inhibitory interactions with eIF4E. Pin1 isomerization inhibits hyperphosphorylation of tau by restricting its access to protein phosphatase 2A (PP2A), and neurons from Pin1−/− mice accumulate hyperphosphorylated proteins [35]. Glutamatergic signaling rapidly increases dendritic translation by inducing phosphorylation of Pin1 at Ser16 and 4E-BP at multiple sites including Thr37, Thr46, Ser65, Ser101 and Ser112 [17,18,19,52] which inactivate Pin1 and 4E-BP, respectively. Despite glutamatergic signaling, we did not observe a change in the degree of interaction between Pin1 and 4E-BP. Similar events were observed between Pin1 and heterogenous nuclear ribonucleoprotein (hnRNP D) in activated immune cells [24]. Because PKMζ-mediated Ser16 phosphorylation of Pin1 clearly prevents Pin1 interactions with its targets, these results suggest Pin1 PPIase activity may be inactivated through additional mechanisms or that distinct pools of differentially modified Pin1 exist in dendritic compartments.
Classical PKCs indirectly cause the dephosphorylation of 4E-BP1/2 [54,55] and consequently suppress translation. After eIF4E dissociates from 4E-BP, it is activated by PKC-mediated phosphorylation [56]. It is also possible that Pin1 isomerizes hypophosphorylated eIF4E, enhancing its binding to hypophosphorylated 4E-BP1/2 and suppressing translation. Either model is consistent with the increase in basal translation and insensitivity to glutamate activation of translation found in SN derived from Pin1−/− mice. Because Pin1 has not been implicated in translational control in other cell types, these functional attributes may be unique to neurons.
PKMζ, an atypical PKC, is transcribed independently under the control of an internal promoter within the PKCζ gene [11], and its mRNA is subsequently transported to dendrites and dendritic spines [57]. We show that PKCζ/PKMζ, known regulators of synaptic plasticity, influence Pin1 function. Consistent with a direct regulatory role, Pin1 activity was increased in SN treated with myristoylated-PKMζ inhibitor peptides and Pin1 activity was directly inhibited by PKMζ in vitro. Ser16, which has previously been shown to inactivate Pin1 substrate binding activity, is one likely site of Pin1 modification by PKMζ [47].
In addition to regulating Pin1, PKCζ/PKMζ promote protein synthesis in SN. Specific blockade of PKCζ/PKMζ reduced both basal and Glu-induced translation in WT and Pin1 KO preparations. These results suggest that PKCζ/PKMζ function independently and downstream from Pin1 to maintain dendritic translation after glutamatergic signaling. These results may also explain how PKCζ/PKMζ blockade can markedly and quickly reverse the maintenance phase of L-LTP, as well as memory persistence [7,12,45]. The classical view that protein synthesis is not required for the maintenance of L-LTP [58,59] is at odds with this interpretation. However, memory can last a lifetime, whereas the turnover of dendritic proteins is usually on the order of several hours, indicating that on-going protein synthesis must be involved [60]. Thus, the physiologic significance of this previously unrecognized function of PKMζ is yet to be fully resolved.
PKCζ/PKMζ has been implicated in several cellular signaling cascades, but little is known about their specific substrates. Our data identify Pin1 as a PKCζ/PKMζ substrate and possibly 4E-BP or eIF4E. We propose that Pin1 regulates LTP through bidirectional interactions with PKMζ, whereby GluR-mediated signaling decreases Pin1 activity, leading to an increase in PKMζ abundance. PKMζ maintains translation and suppresses Pin1 through phosphorylation on Ser16. Thus, PKMζ likely contributes to the maintenance of L-LTP through the induction of dendritic translation. Pin1, as an upstream modulator of PKMζ abundance, plays an integral role in the maintenance of L-LTP.
MATERIALS AND METHODS
Materials
Reagents were obtained from the following companies: Antibodies, see Table S1 for manufacturers and antibody dilutions used for immunoblots and immunofluorescence; anisomycin, anti-βActin, cycloheximide, glutamate, glycine, mouse and rabbit IgG, okadaic acid and protease inhibitor cocktail were from Sigma Chemical Company; juglone from Calbiochem; anti-rabbit and anti-mouse HRP conjugated secondary anti bodies, ECL+ detection reagents, percoll, Redivue Pro-Mix-L [35S] from Amersham Pharmacia Corp.; goat anti-mouse rhodamine-conjugated antibody and Prolong Gold Antifade from Invitrogen; MagnaBind Protein A and G beads, PAGEprep Advance Kit, micro BCA protein assay reagent kit, and Seize™ Primary Mammalian Immunoprecipitation kit from Pierce Biotechnology, Inc.; myristoylated-PKCζ peptide (P-219, N-myr-SIYRRGARRWRKL), myristoylated-PKC peptide (P-205, N-myr-FARKGALRQ) from Biomol.; myristoylated Control peptide (#1776, N-myr-QPPASNPRVR) from Tocris.
Recombinant TAT proteins
The cDNA encoding Pin1 (provided by K.P. Lu, Harvard University), WW-Pin1 and PKMζ were cloned in-frame into pHisTAT (provided by S. Dowdy, UCSD). The cDNA was mutated using QuikChange® XL Site-Directed Mutagenesis Kit (Stratagene), as described in the manufacturer's manual, to produce TAT-Pin1-K63A, TAT-Pin1-S16A, TAT-Pin1-S16E, TAT-WW-S16A and TAT-WW-S16E. Proteins were expressed in Escheria coli and purified on a Ni2+ chelate column (Qiagen) as described by the manufacturer with and without urea. TAT-linked proteins were more than 95% pure based on Ponceau S, Coomassie and Western blots.
Mouse Husbandry
C57BL/6 mice were used for all experiments described herein except where it was specified Pin1−/− and Pin1+/+ were used. WT and Pin1−/− mice in the C57BL/6J background were a gift from Dr. Anthony Means of Duke University Medical Center. All husbandry and euthanasia procedures were performed in accordance with NIH and an approved University of Wisconsin Madison animal care protocol through the Research Animal Resources Center, as described previously [31]. Pin1 genotypes were determined by PCR analysis of DNA extracted from tail biopsies.
SN Preparation and Treatment
SN were prepared from WT and Pin1−/− mouse cortical tissue as described [31,65,66] (pups age 16–21 days). Briefly, P16 to P21 mouse pups were killed by carbon dioxide asphyxiation followed by removal of the brain cortices. The cortices were washed in ice-cold gradient medium (GM buffer: 0.25 M sucrose, 5 mM Tris [pH 7.5], and 0.1 mM EDTA), transferred to a glass dounce homogenizer containing ice-cold GM buffer, and gently homogenized with seven strokes of the loose pestle followed by five strokes of the tight pestle. The homogenate was spun at 1000g for 10 min at 4 °C in round-bottom tubes to pellet cellular debris and nuclei. The supernatant (2 ml aliquots) was applied to percoll gradients (layers = 2 ml each of 23%, 15%, 10%, and 3% isomotic percoll) and spun at speed (32,500g) for 5 min at 4°C. The third band from the top of the gradient (the 23%/15% interface) containing intact SNs was removed and pooled for the experiments. The two higher-molecular-weight bands at the 15%/10% and 10%/3% interfaces contain broken membranes. The salt concentration of the SNs was adjusted by adding one-tenth volume of 10X stimulation buffer (100 mM Tris [pH 7.5], 5 mM Na2HPO4, 4 mM KH2PO4, 40 mM NaHCO3, 800 mM NaCl). In addition, CaCl2 was added to a final concentration of 12 nM. To suppress nonspecific excitation, 1 µM tetrodotoxin was added. The protein concentration of the SNs was determined by BCA assay and ranged from 200–500 ng/µl.
SN were pretreated with 40 µM Aniso, 100 nM Rap, 10 µ M myr-proteins, TAT-proteins or appropriate solvent control and equilibrated at 37°C for 10–15 min on a nutator prior to treatment with 50 µM glutamate and 10 µM glycine (Glu) or no additive (and S35-Met for translation experiments). Samples were mixed at 37°C in 1.5 mL Eppendorf tubes for 5, 10, 15 or 30 min and then snap-frozen. To characterize the SN preparation, we performed immunoblots of cortical homogenate, post-nuclear supernatants and post-percoll-gradient SN for nuclear, cytoplasmic, mitochondrial, astrocytic, and synaptic markers. The protein concentration was determined by the BCA assay and ranged from 200–500 ng/µL. Synaptic and neuronal markers were maintained in purified SN fractions (fig. S1A) but the abundance of all other markers was significantly reduced.
Immunogold-Electron Microscopy
SN prepared from C57BL/6 mice (age P15–P22) were fixed with paraformaldehyde, inactivated with sodium borohydride to neutralize the aldehyde groups, permeablilized with Triton X-100, blocked with normal goat serum, incubated with anti-Pin1 antibody, and stained with secondary antibody conjugate, which was ultrasmall gold-conjugated F(ab')2 fragments as previously described [61].
35S Methionine Incorporation
For radiolabeling, 500 µL of SNs were pretreated for 15 min with drug prior to addition of 35S Methionine, 15 µL Redivue Pro-Mix-L [35S], plus or minus Glu. To analyze new protein synthesis, SN lysates were cleared of free isotope, percoll, and sucrose by purification with the PAGEprep Advance kit per the manufacturers' instructions. PAGEprep samples were denatured and run on 15% SDS gels. The gels were dried, exposed to a phosphorimager screen and scanned on a Storm 860 phosphorimager (Molecular Dynamics). The bands from 3 independent experiments were quantitated with ImageQuant software.
Pin1 Activity Assays
Pin1 activity was measured as previously described [24], with a few modifications. Briefly, for analysis of Pin1 inhibitors, SN were pretreated for 15 min at 37°C with drugs before lysates were prepared by addition of Triton X-100 to a final concentration of 1%, followed by three freeze-thaw cycles and 20 min incubation on ice. Lysed SN were diluted five to ten-fold with 50 mM HEPES and 100 mM NaCl, pH 7.0 and allowed to further incubate on ice. Inhibitors were added back to retain the original concentration prior to assay of protein lysates (0.05 µg). For analysis of PKCζ/PKMζ inhibitors, 10 µM okadaic acid was added prior to Triton X-100 addition, freeze-thaw cycles were excluded, and incubations on ice were reduced to 5 min each. The substrate, Suc-AEPF-pNA was prepared in a LiCl/triflouroethane (TFE) solution, as described previously [24] to preserve the cis conformation. The final reaction concentration was 12.8 µM. Readings were taken for the first 1–2 min of the reaction and plotted as OD390 versus time (min). Spontaneous cis-trans isomerization of the substrate (k0) was measured using the complete reaction mixture minus SN protein. The initial reaction slopes were used to determine K’s as described by Küllertz, et al using the equation (1) K= (kobs − k0)/k0 [36].
Kinase Activity Assays
PKMζ activity was measured as previously described [62], with a few modifications. Briefly, for analysis of phosphorylation activity, TAT-PKMζ was added to 0 to 4 µg of various TAT-Pin1 substrates. The final reaction buffer was 50 mM Tris, pH 7.5, 5 mM MgCl2, 1 mM NaVaO4, 1 mM NaF, 500 µM β-glycerol phosphate, 1× protease inhibitor cocktail, and 0.1 mg/mL BSA. 10 µ Ci of [γ-32P]–ATP was added to initiate reactions, which were incubated at 37°C for 60 min. Phosphorylation was stopped by addition of reducing Laemmli buffer and proteins were separated on 15 or 20% SDS-PAGE, transferred to nitrocellulose, and analyzed by phophorimaging and Western blotting.
Immunoprecipitation
After drug treatment, SN were snap-frozen at −80 °C and protein concentration was determined by BCA assay (Pierce). For immunoprecipitation, 10 µg antibody was added to each SN lysate (600 µg), followed by rocking for 2 hr at 4°C. Protein A or G beads were added and rotation continued overnight. Beads were washed three times with lysis buffer. SDS loading buffer was added directly to the beads and the sample analyzed by Western blot. For immunoprecipitation of phosphoproteins, okadaic acid (10 µM) was added to SN.
Neuronal Cell Culture, Confocal Microscopy and Image Analysis
Primary neuronal embryonic cultures were prepared as described [31]. After 18–19 days in vitro (DIV), the primary neurons were pretreated with or without 40 µM Aniso at 37°C for 15 min,, and then treated with 300 µM Glu, 50 nM TAT-βGal, or 50 nM TAT-Pin1-K63A. Cells were stained and imaged as described [31] with rhodamine- labeled anti-Pin1 (anti-Pin1-Rhodamine), FITC-labeled anti-eIF4E (anti-eIF4E-FITC), FITC-labeled anti-4E-BP1 (anti-4E-BP1-FITC), FITC-labeled anti-4E-BP2 (anti-4E-BP2-FITC), Cy5-labeled (anti-PSD95-Cy5) and/or To-Pro 3-iodide (To-Pro3). See Table S1 for antibody concentrations. Colocalization images were generated by ImageJ RG2B Colocalization plug-in.
Hippocampal Slices and Immunostaining
Hippocampal slices from mice ranging from 21 to 25 days old were prepared and stained as described previously [63]. In brief, mice ranging from P21 to P25 were sacrificed by CO2 asphyxiation and the brains removed and immersion fixed in 10% formalin overnight. The fixed brains were then cyroprotected in 30% sucrose/0.1 M phosphate-buffered saline overnight for at least 2 days. Frozen 30-µm thick coronal sections of the hippocampus were cut in a microtome cryostat and placed in PBS for storage. Slices were permeabilized in blocking solution for 2 h with 2 to 10% FBS, 0.4% Triton X-100, and 0.02% sodium azide in 0.1 M phosphate-buffered saline [63]. The subsequent staining and imaging were similar to that described above for primary neurons except the wash buffer used was PBS containing 2% FBS and 0.4% Triton X-100.
Hippocampal Slice Electrophysiology
Hippocampal slices were isolated from WT and Pin1−/− mice, and electrophysiology performed as described [64]. Slices were subjected to high frequency stimulation consisting of a 100 Hz, 1 second-long titanic stimulus, repeated 4 times, with an interstimulus interval of 2 minutes to generate L-LTP in the presence or absence of 25 µM Aniso. Theta burst stimulation was induced by 3 trains of theta burst stimulation (10 bursts at 5 Hz per train, 4 pulses (1 sec) at 100 Hz per burst) separated by 20 sec.
Statistical Analysis
P-values were calculated using the Student's t-Test with a two-tailed distribution on samples, except for LTP studies, which used two-way ANOVA with repeated measures (mixed model) and Bonferroni post-tests.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the members of the laboratory for their comments and suggestions, Benjamin K. August from the UW-Madison Electron Microscope Facility for the immuno-EM, Dr. Anthony Means of Duke University Medical Center for generously providing the C57BL/6J Pin1+/− mice, Emily Whitesel for constructing Pin1 mutant cDNAs, and Keith A. Hanson for helping construct TAT-PKMζ cDNA.
FUNDING: This work was supported by the National Institutes of Health grants R01-DA026067 and P30-HD03352 (to J.S.M.).
Footnotes
AUTHOR CONTRIBUTIONS: PRW, JL, CB and JSM generated the hypotheses; PRW, CJW, SW, JL, KJO and CB all performed experiments; PRW, KJO, CB and JSM analyzed data and PRW, JL, CB and JSM wrote the manuscript.
COMPETING INTEREST STATEMENT: The authors declare that they have no competing financial interests.
REFERENCES and NOTES
- 1.Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3'-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc Natl Acad Sci U S A. 1996;93(23):13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourne JN, Sorra KE, Hurlburt J, Harris KM. Polyribosomes are increased in spines of CA1 dendrites 2 h after the induction of LTP in mature rat hippocampal slices. Hippocampus. 2007;17(1):1–4. doi: 10.1002/hipo.20238. [DOI] [PubMed] [Google Scholar]
- 3.Ostroff LE, Fiala JC, Allwardt B, Harris KM. Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron. 2002;35(3):535–545. doi: 10.1016/s0896-6273(02)00785-7. [DOI] [PubMed] [Google Scholar]
- 4.Klann E, Chen SJ, Sweatt JD. Persistent protein kinase activation in the maintenance phase of long-term potentiation. J Biol Chem. 1991;266(36):24253–24256. [PubMed] [Google Scholar]
- 5.Klann E, Chen SJ, Sweatt JD. Mechanism of protein kinase C activation during the induction and maintenance of long-term potentiation probed using a selective peptide substrate. Proc Natl Acad Sci U S A. 1993;90(18):8337–8341. doi: 10.1073/pnas.90.18.8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hrabetova S, Sacktor TC. Bidirectional regulation of protein kinase M zeta in the maintenance of long-term potentiation and long-term depression. J Neurosci. 1996;16(17):5324–5333. doi: 10.1523/JNEUROSCI.16-17-05324.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sacktor TC, Osten P, Valsamis H, Jiang X, Naik MU, Sublette E. Persistent activation of the zeta isoform of protein kinase C in the maintenance of long-term potentiation. Proc Natl Acad Sci U S A. 1993;90(18):8342–8346. doi: 10.1073/pnas.90.18.8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehm J, Kang MG, Johnson RC, Esteban J, Huganir RL, Malinow R. Synaptic incorporation of AMPA receptors during LTP is controlled by a PKC phosphorylation site on GluR1. Neuron. 2006;51(2):213–225. doi: 10.1016/j.neuron.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Kelleher RJ, III, Govindarajan A, Jung H-, Kang H, Tonegawa S. Translational control by MAPK signalling in long-term synaptic plasticity and memory. Cell. 2004;116:467–479. doi: 10.1016/s0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 10.Ling DS, Benardo LS, Serrano PA, Blace N, Kelly MT, Crary JF, Sacktor TC. Protein kinase mzeta is necessary and sufficient for LTP maintenance. Nat Neurosci. 2002;5(4):295–296. doi: 10.1038/nn829. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez AI, Blace N, Crary JF, Serrano PA, Leitges M, Libien JM, Weinstein G, Tcherapanov A, Sacktor TC. Protein kinase M zeta synthesis from a brain mRNA encoding an independent protein kinase C zeta catalytic domain. implications for the molecular mechanism of memory. J Biol Chem. 2003;278(41):40305–40316. doi: 10.1074/jbc.M307065200. [DOI] [PubMed] [Google Scholar]
- 12.Kelly MT, Crary JF, Sacktor TC. Regulation of protein kinase M: Synthesis by multiple kinases in long-term potentiation. J Neurosci. 2007;27(13):3439–3444. doi: 10.1523/JNEUROSCI.5612-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastalkova E, Serrano P, Pinkhasova D, Wallace E, Fenton AA, Sacktor TC. Storage of spatial information by the maintenance mechanism of LTP. Science. 2006;313(5790):1141–1144. doi: 10.1126/science.1128657. [DOI] [PubMed] [Google Scholar]
- 14.Osten P, Valsamis L, Harris A, Sacktor TC. Protein synthesis-dependent formation of protein kinase m zeta in long-term potentiation. J Neurosci. 1996;16(8):2444–2451. doi: 10.1523/JNEUROSCI.16-08-02444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao Y, Kelly MT, Sajikumar S, Serrano P, Tian D, Bergold PJ, Frey Ju, Sacktor TC. PKM zeta maintains late long-term potentiation by N-ethylmaleimide-sensitive factor/GluR2-dependent trafficking of postsynaptic AMPA receptors. J Neurosci. 2008;28(31):7820–7827. doi: 10.1523/JNEUROSCI.0223-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Provenzani A, Fronza R, Loreni F, Pascale A, Amadio M, Quattrone A. Global alterations in mRNA polysomal recruitment in a cell model of colorectal cancer progression to metastasis. Carcinogenesis. 2006;27(7):1323–1333. doi: 10.1093/carcin/bgi377. [DOI] [PubMed] [Google Scholar]
- 17.Loreni F, Thomas G, Amaldi F. Transcription inhibitors stimulate translation of 5' TOP mRNAs through activation of S6 kinase and the mTOR/FRAP signalling pathway. Eur J Biochem. 2000;267(22):6594–6601. doi: 10.1046/j.1432-1327.2000.01753.x. [DOI] [PubMed] [Google Scholar]
- 18.Gingras AC, Kennedy SG, O'Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the akt(PKB) signaling pathway. Genes Dev. 1998;12(4):502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272(42):26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 20.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, Cantley LC, Lu KP. Sequence-specific and phosphorylation-dependent proline isomerization: A potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 21.Becker EB, Bonni A. Pin1 mediates neural-specific activation of the mitochondrial apoptotic machinery. Neuron. 2006;49(5):655–662. doi: 10.1016/j.neuron.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 22.Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP. The prolyl isomerase Pin1 restores the function of alzheimer-associated phosphorylated tau protein. Nature. 1999;399:784–788. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- 23.Shen J, Dahmann C. Extrusion of cells with inappropriate dpp signaling from drosophila wing disc epithelia. Science. 2005;307(5716):1789–1790. doi: 10.1126/science.1104784. [DOI] [PubMed] [Google Scholar]
- 24.Shen ZJ, Esnault S, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat Immunol. 2005;6(12):1280–1287. doi: 10.1038/ni1266. [DOI] [PubMed] [Google Scholar]
- 25.Hoeffer CA, Tang W, Wong H, Santillan A, Patterson RJ, Martinez LA, Tejada-Simon MV, Paylor R, Hamilton SL, Klann E. Removal of FKBP12 Enhances mTOR-Raptor Interactions, LTP, Memory, and Perseverative/Repetitive Behavior. Neuron. 2008;60:832–845. doi: 10.1016/j.neuron.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mouri A, Noda Y, Shimizu S, Tsujimoto Y, Nabeshima T. The role of Cyclophilin D in learning and memory. Hippocampus. 2009 May 12; doi: 10.1002/hipo.20625. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 27.Du H, Guo L, Zhang W, Rydzewska M, Yan S. Cyclophilin D deficiency improves mitochondrial function and learning/memory in aging Alzheimer disease mouse model. Neurobiol Aging. 2009 Apr 10; doi: 10.1016/j.neurobiolaging.2009.03.003. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci U S A. 2002;99(1):467–472. doi: 10.1073/pnas.012605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamdane M, Dourlen P, Bretteville A, Sambo AV, Ferreira S, Ando K, Kerdraon O, Bégard S, Geay L, Lippens G, Sergeant N, Delacourte A, Maurage CA, Galas MC, Buée L. Pin1 allows for differential tau dephosphorylation in neuronal cells. Mol Cell Neurosci. 2006;32(1–2):155–160. doi: 10.1016/j.mcn.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Eyman M, Cefaliello C, Ferrara E, De Stefano R, Crispino M, Giuditta A. Synaptosomal protein synthesis is selectively modulated by learning. Brain Res. 2007;1132(1):148–157. doi: 10.1016/j.brainres.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 31.Westmark CJ, Malter JS. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLOS Biol. 2007;5(3):e52. doi: 10.1371/journal.pbio.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hennig L, Christner C, Kipping M, Schelbert B, Rucknagel KP, Grabley S, Küllertz G, Fischer G. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry. 1998;37(17):5953–5960. doi: 10.1021/bi973162p. [DOI] [PubMed] [Google Scholar]
- 33.Janowski B, Wollner S, Schutkowski M, Fischer G. A protease-free assay for peptidyl prolyl cis/trans isomerases using standard peptide substrates. Anal Biochem. 1997;252(2):299–307. doi: 10.1006/abio.1997.2330. [DOI] [PubMed] [Google Scholar]
- 34.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283(5406):1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 35.Liou YC, Sun A, Ryo A, Zhou XZ, Yu ZX, Huang HK, Uchida T, Bronson R, Bing G, Li X, Hunter T, Lu KP. Role of the prolyl isomerase Pin1 in protecting against age-dependent neurodegeneration. Nature. 2003;424(6948):556–561. doi: 10.1038/nature01832. [DOI] [PubMed] [Google Scholar]
- 36.Kullertz G, Luthe S, Fischer G. Semiautomated microtitre plate assay for monitoring peptidyl cis/trans isomerase activity in normal and pathological human sera. Clinical Chem. 1998;44(3):502–508. [PubMed] [Google Scholar]
- 37.Levenson J, Endo S, Kategaya LS, Fernandez RI, Brabham DG, Chin J, Byrne JH, Eskin A. Long-term regulation of neuronal high-affinity glutamate and glutamine uptake in aplysia. Proc Natl Acad Sci U S A. 2000;97(23):12858–12863. doi: 10.1073/pnas.220256497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 39.Banko JL, Poulin F, Hou L, DeMaria CT, Sonenberg N, Klann E. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J Neurosci. 2005;25(42):9581–9590. doi: 10.1523/JNEUROSCI.2423-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costa-Mattioli M, Gobert D, Harding H, Herdy B, Azzi M, Bruno M, Bidinosti M, Ben Mamou C, Marcinkiewicz E, Yoshida M, Imataka H, Cuello AC, Seidah N, Sossin W, Lacaille JC, Ron D, Nader K, Sonenberg N. Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature. 2005;436(7054):1166–1173. doi: 10.1038/nature03897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patterson SL, Pittenger C, Morozov A, Martin KC, Scanlin H, Drake C, Kandel ER. Some forms of cAMP-mediated long-lasting potentiation are associated with release of BDNF and nuclear translocation of phospho-MAP kinase. Neuron. 2001;32(1):123–140. doi: 10.1016/s0896-6273(01)00443-3. [DOI] [PubMed] [Google Scholar]
- 42.Stäbli U, Scafidi J, Chun D. GABA-B receptor antagonism: facilitatory effects on memory parallel those on LTP induced by TBS but not HFS. J Neurosci. 1999;19(11):4609–4615. doi: 10.1523/JNEUROSCI.19-11-04609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volianskis A, Jensen MS. Transient and sustained types of long-term potentiation in the CA1 area of the rat hippocampus. J Physiol. 2003;550(Pt 2):459–492. doi: 10.1113/jphysiol.2003.044214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zakharenko SS, Patterson SL, Dragatsis I, Zeitlin SO, Siegelbaum SA, Kandel ER, Morozov A. Presynaptic BDNF required for a presynaptic but not postsynaptic component of LTP at hippocampal CA1–CA3 synapses. Neuron. 2003;39(6):975–990. doi: 10.1016/s0896-6273(03)00543-9. [DOI] [PubMed] [Google Scholar]
- 45.Shema R, Sacktor TC, Dudai Y. Rapid erasure of long-term memory associations in the cortex by an inhibitor of PKM zeta. Science. 2007;317(5840):951–953. doi: 10.1126/science.1144334. [DOI] [PubMed] [Google Scholar]
- 46.Naik MU, Benedikz E, Hernandez I, Libien J, Hrabe J, Valsamis M, Dow-Edwards D, Osman M, Sacktor TC. Distribution of protein kinase mzeta and the complete protein kinase C isoform family in rat brain. J Comp Neurol. 2000;426(2):243–258. doi: 10.1002/1096-9861(20001016)426:2<243::aid-cne6>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 47.Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J Biol Chem. 2002;277(4):2381–2384. doi: 10.1074/jbc.C100228200. [DOI] [PubMed] [Google Scholar]
- 48.Yeh ES, Means AR. PIN1, the cell cycle and cancer. Nat Rev Cancer. 2007;7(5):381–388. doi: 10.1038/nrc2107. [DOI] [PubMed] [Google Scholar]
- 49.Barrett LE, Sul JY, Takano H, Van Bockstaele EJ, Haydon PG, Eberwine JH. Region-directed phototransfection reveals the functional significance of a dendritically synthesized transcription factor. Nat Methods. 2006;3(6):455–460. doi: 10.1038/nmeth885. [DOI] [PubMed] [Google Scholar]
- 50.Lenz G, Avruch J. Glutamatergic regulation of the p70S6 kinase in primary mouse neurons. J Biol Chem. 2005;280(46):38121–38124. doi: 10.1074/jbc.C500363200. [DOI] [PubMed] [Google Scholar]
- 51.Banko JL, Hou L, Poulin F, Sonenberg N, Klann E. Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2006;26(8):2167–2173. doi: 10.1523/JNEUROSCI.5196-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Li W, Parra JL, Beugnet A, Proud CG. The C terminus of initiation factor 4E-binding protein 1 contains multiple regulatory features that influence its function and phosphorylation. Mol Cell Biol. 2003;23(5):1546–1557. doi: 10.1128/MCB.23.5.1546-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fletcher CM, McGuire AM, Gingras AC, Li H, Matsuo H, Sonenberg N, Wagner G. 4E binding proteins inhibit the translation factor eIF4E without folded structure. Biochemistry. 1998;37(1):9–15. doi: 10.1021/bi972494r. [DOI] [PubMed] [Google Scholar]
- 54.Guan L, Song K, Pysz MA, Curry KJ, Hizli AA, Danielpour D, Black AR, Black JD. Protein kinase C-mediated down-regulation of cyclin D1 involves activation of the translational repressor 4E-BP1 via a phosphoinositide 3-kinase/Akt-independent, protein phosphatase 2A-dependent mechanism in intestinal epithelial cells. J Biol Chem. 2007;282(19):14213–14225. doi: 10.1074/jbc.M610513200. [DOI] [PubMed] [Google Scholar]
- 55.Hizli AA, Black AR, Pysz MA, Black JD. Protein kinase C alpha signaling inhibits cyclin D1 translation in intestinal epithelial cells. J Biol Chem. 2006;281(21):14596–14603. doi: 10.1074/jbc.M601959200. [DOI] [PubMed] [Google Scholar]
- 56.Whalen SG, Gingras AC, Amankwa L, Mader S, Branton PE, Aebersold R, Sonenberg N. Phosphorylation of eIF-4E on serine 209 by protein kinase C is inhibited by the translational repressors, 4E-binding proteins. J Biol Chem. 1996;271(20):11831–11837. doi: 10.1074/jbc.271.20.11831. [DOI] [PubMed] [Google Scholar]
- 57.Muslimov IA, Nimmrich V, Hernandez AI, Tcherepanov A, Sacktor TC, Tiedge H. Dendritic transport and localization of protein kinase mzeta mRNA: Implications for molecular memory consolidation. J Biol Chem. 2004;279(50):52613–52622. doi: 10.1074/jbc.M409240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frey U, Krug M, Reymann KG, Matthies H. Anisomycin, an inhibitor of protein synthesis, blocks late phases of LTP phenomena in the hippocampal CA1 region in vitro. Brain Res. 1988;452(1–2):57–65. doi: 10.1016/0006-8993(88)90008-x. [DOI] [PubMed] [Google Scholar]
- 59.Fonseca R, Nägerl UV, Bonhoeffer T. Neuronal activity determines the protein synthesis dependence of long-term potentiation. Nat Neurosci. 2006;9(4):478–480. doi: 10.1038/nn1667. [DOI] [PubMed] [Google Scholar]
- 60.Aslam N, Kubota Y, Wells D, Shouval HZ. Translational switch for long-term maintenance of synaptic plasticity. Mol Syst Biol. 2009;5:284. doi: 10.1038/msb.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yi H, Leunissen J, Shi G, Gutekunst C, Hersch S. A novel procedure for pre-embedding double immunogold-silver labeling at the ultrastructural level. J Histochem Cytochem. 2001;49(3):279–284. doi: 10.1177/002215540104900301. [DOI] [PubMed] [Google Scholar]
- 62.Umar S, Sellin JH, Morris AP. Increased nuclear translocation of catalytically active PKC-zeta during mouse colonocyte hyperproliferation. Am J Physiol Gastrointest Liver Physiol. 2000;279(1):G223–G237. doi: 10.1152/ajpgi.2000.279.1.G223. [DOI] [PubMed] [Google Scholar]
- 63.McCormack K, Connor JX, Zhou L, Ho LL, Ganetzky B, Chiu SY, Messing A. Genetic analysis of the mammalian K+ channel beta subunit kvbeta 2 (Kcnab2) J Biol Chem. 2002;277(15):13219–13228. doi: 10.1074/jbc.M111465200. [DOI] [PubMed] [Google Scholar]
- 64.O'Riordan KJ, Huang IC, Pizzi M, Spano P, Boroni F, Egli R, Desai P, Fitch O, Malone L, Ahn HJ, Liou HC, Sweatt JD, Levenson JM. Regulation of nuclear factor kappaB in the hippocampus by group I metabotropic glutamate receptors. J Neurosci. 2006;26(18):4870–4879. doi: 10.1523/JNEUROSCI.4527-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dunkley PR, Heath JW, Harrison SM, Jarvie PE, Glenfield PJ, Rostas JA. A rapid Percoll gradient procedure for isolation of synaptosomes directly from an S1 fraction: Homogeneity and morphology of subcellular fractions. Brain Res. 1988;441:59–71. doi: 10.1016/0006-8993(88)91383-2. [DOI] [PubMed] [Google Scholar]
- 66.L, Dotti CG, Amaldi F. Chemical stimulation of synaptosomes modulates alpha-Ca2+/calmodulin-dependent protein kinase II mRNA association to polysomes. J Neurosci. 2000;20:RC76. doi: 10.1523/JNEUROSCI.20-10-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


