Abstract
SgrAI is a type II restriction endonuclease that cuts an unusually long recognition sequence and exhibits allosteric self-modulation of DNA activity and sequence specificity. Precleaved primary site DNA has been shown to be an allosteric effector [Hingorani-Varma & Bitinaite, (2003) J. Biol. Chem. 278, 40392-40399], stimulating cleavage of both primary (CR|CCGGYG, | indicates cut site, R=A,G, Y=C,T) and secondary (CR|CCGGY(A/C/T) and CR|CCGGGG) site DNA sequences. The fact that DNA is the allosteric effector of this endonuclease suggests at least two DNA binding sites on the functional SgrAI molecule, yet crystal structures of SgrAI [Dunten, et al., (2008) Nucleic Acids Res. 36, 5405–5416] show only one DNA duplex bound to one dimer of SgrAI. We show that SgrAI forms species larger than dimers or tetramers (High Molecular Weight Species, HMWS) in the presence of sufficient concentrations of SgrAI and its primary site DNA sequence, that are dependent on the concentration of the DNA bound SgrAI dimer. Analytical ultracentrifugation indicates that the HMWS is heterogeneous, has sedimentation coefficients of 15–20 s, and is composed of possibly 4–12 DNA bound SgrAI dimers. SgrAI bound to secondary site DNA will not form HMWS itself, but can bind to HMWS formed with primary site DNA and SgrAI. Uncleaved, as well as precleaved, primary site DNA is capable of stimulating HMWS formation. Stimulation of DNA cleavage by SgrAI, at primary as well as secondary sites, is also dependent on the concentration of primary site DNA (cleaved or uncleaved) bound SgrAI dimers. SgrAI bound to secondary site DNA does not have significant stimulatory activity. We propose that the oligomers of DNA bound SgrAI (i.e. HMWS) are the activated, or activatable, form of the enzyme.
Type II restriction endonucleases are bacterial enzymes thought to protect their host from phage infection, by cleaving phage DNA injected into the cell prior to replication1. Sequence specific endonucleases capable of cleaving longer unique recognition sequences are highly sought for genomic work, since longer sequences occur less frequently and allow the manipulation of larger DNA fragments. Most of the type II restriction endonucleases characterized to date cleave 4–6 bp recognition sites in DNA, however, SgrAI, a type IIF restriction endonuclease from Streptomyces griseus, cleaves an 8 base pair recognition sequence, CR|CCGGYG2 (R=A,G, Y=C,T, | indicates site of cleavage), known as the SgrAI cognate or primary site sequence. Interestingly, SgrAI cleaves plasmids bearing two copies of its recognition sequence faster than those bearing only a single site3, 4. In addition, plasmid assays revealed that SgrAI will also cleave the sequences CR|CCGGY(A,C,T) and CR|CCGGGG, known as secondary sites, but only substantially in plasmids containing primary site sequences5. Secondary sites are distinct from star sites, in that secondary sites are cleaved appreciably under solution conditions optimal for primary sequence cleavage. In contrast, star site sequences are sequences that are cleaved appreciably only under special reaction conditions, such as high enzyme concentrations or the presence of organic solvents or Mn2+, and are discriminated against under optimal enzyme conditions by 2–4 orders of magnitude6. The self-activation with self-modulation of sequence specificity exhibited by SgrAI is quite unusual and has not been detected before in type II restriction endonucleases.
Type II restriction endonucleases typically bind and recognize palindromic sequences as dimers1, 6, but the unusual biochemical properties exhibited by SgrAI suggest the formation of a higher order oligomer containing altered enzymatic properties. For example, at low enzyme concentrations, SgrAI cleaves plasmids bearing one or two sites at equal rates, but higher concentrations of enzyme result in the faster cleavage of the two site plasmid4, 7. Similarly, the DNA cleavage turnover number, kcat, of SgrAI with its primary sequence shows a sigmoidal dependenon on SgrAI concentration, consistent with the formation of an activated oligomer at the higher enzyme concentrations4, 7. The stimulation of DNA cleavage activity must occur through three-dimensional space, as the accelerated and concerted cleavage also occurs with plasmids each bearing a single site but connected by catenation7. Cleavage at primary and secondary site sequences in plasmids can also be stimulated by the addition of oligonucleotides containing the primary site sequence, intact or mimicking the cleavage products of SgrAI5, 7, 8, indicating that DNA is the allosteric effector, and that the functional unit of stimulated SgrAI has at least two DNA binding sites. (An alternative mechanism would involve the memory of the binding of cleaved primary site after its dissociation. Such mechanisms typically require a covalent modification if oligomerization is absent.) Analytical ultracentrifugation shows that SgrAI exists as a dimer in the absence of DNA, but forms both DNA bound dimers and high molecular mass aggregates in the presence of a 20 base pair DNA containing a primary site4. The stoichiometry of this mixture of species has been determined by titration of DNA with SgrAI in analytical untracentrifuge sedimentation velocity experiments showing 1 dimer of SgrAI per DNA duplex4. A reasonable model for the oligomerization of DNA bound dimers (DBD), which will bring two DNA binding sites together into a single functional unit of the enzyme, is the formation of a tetramer, since the evolutionarily related enzymes NgoMIV, Cfr10I, and Bse634I form tetramers that bind to two duplexes of DNA. However, we show here that rather than tetramers, SgrAI forms larger oligomers of DBD (HMWS for High Molecular Weight Species) containing possibly as many as 4–12 DNA bound SgrAI dimers. The HMWS is formed with primary site DNA, which need not be cleaved. SgrAI dimers bound to secondary site DNA will not form HMWS unless sufficient concentrations of SgrAI dimers bound to primary site are also present. Since the stimulation of the single turnover rate of DNA cleavage (on primary or secondary site DNA) also depends on the presence of sufficient concentrations of SgrAI enzyme bound to primary site DNA, conditions where HMWS form, we conclude that the HMWS is composed of many DBD, and is the activated, or activatable, form of SgrAI.
Experimental Procedures
Protein Purification
Wild type SgrAI was prepared as described9. Briefly, SgrAI was expressed in E. coli strain ER2566 in the presence of the MspI methyltransferase (New England Biolabs). The enzyme was purified using FPLC (GE Healthcare Biosciences) chromatography and the following chromatographic resins: Heparin FF Sepharose (Pharmacia), SP FF Sepharose (GE Healthcare Biosciences), Q FF Sepharose (GE Healthcare Biosciences), and then a second Heparin FF Sepharose (GE Healthcare Biosciences) chromatographic step. Finally, SgrAI enzyme was dialyzed into storage buffer (20 mM Tris-OAc, (pH 8.0), 50 mM KOAc, 0.1 mM EDTA, 1 mM DTT, 50% glycerol), aliquoted into single use aliquots, flash frozen in liquid nitrogen, and stored at −80°C.
DNA Preparation
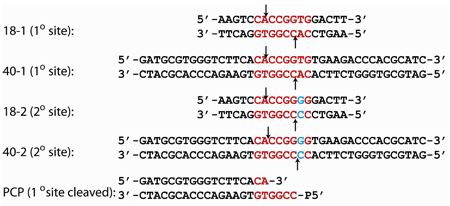
The oligonucleotides were made synthetically and purified using C18 reverse phase HPLC10. The concentration was measured spectrophotometrically, with an extinction coefficient calculated from standard values for the nucleotides11, and fluorophore where appropriate. Fluorophore labeled DNA utilized a 6-(3',6'-dipivaloylfluoresceinyl-6-carboxamido)-hexyl (FLO) group attached to the 5’ phosphate of the top strand of precleaved 40 bp primary site containing DNA (FLO-PCP), or 6- (4,7,2’,4’,5’,7’-hexachloro-(3’,6’-dipivaloylfluoresceinyl)-6-carboxamido)-hexyl (HEX) group attached to the 5’ phosphate of both strands (HEX-18-1) or only the top strands of the 18 bp secondary site DNA (HEX-18-2) and 40 bp primary site containing DNA (HEX-40-1), and were obtained from a commercial synthetic source (Sigma Genosys, Inc.) and contain a 6 carbon spacer between the fluorophore and the 5’ phosphate. The self-complementary DNA, or equimolar quantities of complementary DNA, were annealed by heating to 90°C for 10 minutes at a concentration of 1 mM, followed by slow-cooling to 4°C over 4–5 hours in a thermocycler. Sequences of the DNA used are:
Because freeze-thawing altered the concentration of double stranded DNA used in the assays, DNA used for stimulation of HMWS formation or in single turnover assays was treated very carefully to minimize this problem. Such DNA samples were either reannealed immediately prior to the assay, or carefully annealed, assessed for concentration, aliquoted into small amounts, flash frozen in liquid nitrogen, stored at −20°C (in water), and used only once after removing from the freezer. DNA was 5’ end labeled with 32P using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]-ATP (Perkin-Elmer, Inc.), and excess ATP removed using G-30 spin columns (Biorad Laboratories, Inc.).
Binding Assays
Fluorescence polarization anisotropy (FPA)
The equilibrium dissociation constant, KD, of SgrAI-DNA complexes was measured using a fluorescence polarization anisotropy technique (FPA)12. DNA oligonucleotides (1 nM in 2 ml binding buffer: 20 mM Tris-OAc (pH 8.0), 50 mM KOAc, 10 mM Ca(OAc)2, 1 mM DTT, 10% glycerol at 4°C) containing a fluorophore (HEX or FLO) ligated to the 5' end, were titrated with increasing amounts of SgrAI enzyme (1 nM – 1 µM), and the polarization of the emitted fluorescence monitored. Excitation occurred at 537 nm (HEX) or 492 nm (FLO) in a PC1 (ISS) fluorimeter with T format, automatic polarizers and temperature control. The emitted intensities were measured using a 50.8 mm diameter 570 nm cut-on filter with 580-2750 nm transmittance range (ThermoOriel Inc., no. 59510) and 1 mm slit widths. The polarization of the emitted light as a function of added enzyme was fit to 1:1 binding using Kaleidagraph software and the following12:
where A is the polarization at a given protein concentration, Amax is the predicted polarization of fully bound DNA, Amin is the polarization with no protein binding, PT is the total concentration of protein, OT is the total concentration of the DNA, and KD is the dissociation constant to be determined.
Gel shift
The gel shift assay13 was also used to measure binding affinities of SgrAI to DNA. DNA oligonucleotides were 5’ end labeled14 with 32P and held constant at a concentration of 10 pM in 20 µl binding buffer (20 mM Tris-OAc (pH 8.0), 50 mM KOAc, 10 mM Ca(OAc)2, 1 mM DTT, 10% glycerol at 4°C). Separate incubations were performed with the DNA and varied concentrations of SgrAI enzyme (10 pM – 1 µM). These concentrations were chosen carefully to give a well defined binding curve. Native PAGE (8–10% 19:1 or 29:1 acrylamide:bisacrylamide, 89 mM Tris, 89 mM boric acid, and 10 mM Ca2+) was used to separate the bound and unbound DNA, as the DNA bound to SgrAI will have reduced electrophoretic mobility. Care was taken to prevent heating of the gel, by running at 4°C at low voltage (190 V). The electrophoresis buffer (89 mM Tris, 89 mM boric acid, and 10 mM Ca2+) was recirculated during electrophoresis. Gels were loaded while undergoing electrophoresis at 300 V, and the voltage returned to 190 V five minutes after the loading of the last sample. Gels were then subjected to electrophoresis for an additional 2 hours at 4°C. Autoradiography of gels was performed without drying with a phosphor image plate exposed at 4°C for 12–17 hours. Densitometry of phosphor image plates was performed with a Typhoon Scanner (GE Healthcare Life Sciences), and integration using ImageQuant (GE Healthcare Life Sciences) or ImageJ15. The equilibrium dissociation constant, KD, was determined as above for the FPA measurements, using normalized values of the amount of shifted DNA for each concentration of SgrAI.
Single turnover DNA cleavage assays
Single turnover measurements of DNA cleavage were performed using chemical rapid quench techniques and 5' end 32P labeled oligonucleotide substrates (typically 1 nM), under conditions of excess enzyme (1 µM), with and without the addition of unlabeled DNA. All reactions were performed at 37°C in 20 mM Tris-OAc (pH 8.0), 50 mM KOAc, 10 mM Mg(OAc)2, and 1 mM DTT. For sampling by hand, 5 µl aliquots were withdrawn at specific time intervals after mixing the enzyme and labeled DNA (100 µl total reaction volume), quenched by addition to 5 µl of quench (80% formamide, 50 mM EDTA), and electrophoresed on 20% denaturing polyacrylamide (19:1 acrylamide:bisacrylamide, 4 M urea, 89 mM Tris, 89 mM boric acid, 2 mM EDTA) gels. Autoradiography of gels was performed without drying with a phosphor image plate exposed at 4°C for 12–17 hours. Densitometry of phosphor image plates was performed with a Typhoon Scanner (GE Healthcare Life Sciences), and integration using ImageQuant (GE Healthcare Life Sciences) or ImageJ15. The percent of product formed as a function of time was determined by integrating both cleaved and uncleaved DNA bands. The single turnover DNA cleavage rate constant was determined from the data using a single exponential function:
where C1 is a constant fitting the baseline, C2 is the total percent of DNA predicted to be cleaved by SgrAI, k is the rate constant, and t is the length of incubation in minutes. To achieve time points faster than 10 seconds, a RQF-3 rapid quench flow instrument (KinTek Inc.), was used according to a method modified from that of the instrument manual. Because dilution of the samples was a concern using samples in the sample loops, the enzyme and DNA solutions were instead placed in the drive syringes A and B, respectively. Quench solution was used in the C syringe. For the reactions, loop 3 (40 µl) was used in a two step push. Each step was made at 500 rpm (7,475 µl/sec) to ensure good mixing (Re = 11,900). Each step pushed 64 µl from each syringe (128 µl total). The reaction time was set by the time between the two steps and was controlled by the instruments' computer. The step size was chosen such that the sample loop would be completely filled with one step and ejected in the second step.
Native gel analysis of HMWS formation
Formation of HMWS was monitored using native PAGE and the method described above for gel shift measurements with the following modifications. The acrylamide composition was 8% 29:1 acrylamide:bisacrylamide, and samples were prepared with 1 µM SgrAI, 1 nM 32P labeled DNA, and varied concentrations of unlabeled DNA in binding buffer (20 mM Tris-OAc (pH 8.0), 50 mM KOAc, 10 mM Ca(OAc)2, 1 nM DTT, 10% glycerol) in 20 µl. The effect of increased ionic strength was tested using buffers with varied concentrations (0–300 mM) of KOAc or NaCl in place of the 50 mM KOAc. Prepared samples were incubated for 30 minutes at 4°C prior to electrophoresis. Integrated band intensities were normalized using the sum of the DNA bound species (DBD and HMWS) to determine the percent HMWS.
Sedimentation velocity
Sedimentation velocity experiments were performed in a Beckman Coulter XL-I equipped with monochrometer and interference scanning optics (632 nm) using a Ti-50 rotor. Two sector sedimentation velocity cells were loaded with approximately 400 µl sample containing 3 µM fluorophore labeled DNA with or without 6 µM SgrAI in one sector, 425 µl buffer in the other. The buffers used in the measurements contained 20 mM Tris-OAc (pH 8.0), 1 mM DTT, and either 50 mM KOAc or 150 mM NaCl and either 10 mM Ca(OAc)2 or 10 mM Mg(OAc)2. After sealing the cell, loading, balancing and installing the rotor, the system was allowed to equilibrate for at least 1 hour after reaching the target temperature (4°C or 37°C). The sample was spun at 40,000 rpm (115,000 × g) and absorbance scans were taken at 495 nm (FLO-PCP) or 537nm (HEX-18-2) continuously, with a 0.003 cm step stize, until the last of the boundaries had moved to the bottom of the solution column (at least 12–17 hours). Data from the scans were fit to a sedimentation coefficient distribution, c(s), using the software SEDFIT16. The SgrAI dimer partial specific volume was calculated using SEDNTERP17. Only a single partial specific volume can be used in the data analysis with SEDFIT, and that of the 1:1 SgrAI dimer:DNA duplex was used (calculated using a weighted, by their respective molecular weights, average of the partial specific volumes of the protein (0.74 ml/g) and DNA (0.55 ml/g)). This same partial specific volume was also used in the analysis of data from samples containing only DNA, in order to allow for the assignment of the peaks corresponding to the unbound DNA in the SgrAI/DNA mixtures. Viscosities and densities were calculated using SEDNTERP17. .Due to limited component selection, SEDNTERP calculations utilized the parameters of potassium sulfate, magnesium sulfate, and calcium chloride for potassium acetate, magnesium acetate, and calcium acetate of the buffers. Calculated values for similar buffers as used in these studies have been found to be within 1% error of the values measured experimentally using a Mettler-Toledo five place densitomer and Canon-Fenske viscometer.
Sedimentation equilibrium
Sedimentation equilibrium experiments were performed as above for sedimentation velocity with the following modifications. Samples (110 µl) containing varied concentrations of SgrAI (3–12 µM) and fluorophore labeled DNA (1.5–6 µM) were loaded into three chambers of a six chamber centerpiece, with buffer occupying the remaining three. The system was allowed to equilibrate for at least 1 hour after reaching the target temperature (4 or 37°C). Samples were then subjected to centrifugal speeds from 4,000 to 18,000 rpm for a period of 72–96 hours. High resolution scans were made at 492 nm (FLO-PCP) or 537 nm (HEX-18-2) every 4 hours, with a step size of 0.001 cm. The establishment of equilibrium was determined when a reasonable (<0.07) and constant RMSD was found between sequential scans. The data was analyzed using global fitting in SEDFIT16 with a single species. Density and partial specific volumes were determined as above.
Results
DNA binding assays
The equilibrium dissociation binding constants (KD) of SgrAI and various DNA oligonucleotide constructs containing intact primary site DNA (18-1 and 40-1), secondary site DNA (18-2 and 40-2), or precleaved 40 bp primary site containing DNA (PCP or PC) were measured using a fluorescence polarization assay (FPA, Fig. 1), or a gel shift assay (Table 1). The measurements were made at 4°C in 20 mM Tris-OAc (pH 8.0), 50 mM KOAc, 10 mM Ca(OAc)2, 1 mM DTT, 10% glycerol (unless otherwise noted), and are presented as the average of at least three independent measurements ± the standard deviation. Ca2+ was used as a substitute for Mg2+, as numerous studies show that this substitution retains binding specificity without conferring DNA cleavage activity12, 18–21. All of the binding isotherms fit very well to a model for 1:1 binding (see Methods). Differences in KD measured by the two methods can be explained by the difficulty in measuring KD smaller than 1 nM using FPA, since DNA concentrations below 1 nM are too weak to detect using this method. In such cases, the gel shift measurements (using 10 pM 32P-labeled DNA) are likely to be more reliable. The results show that SgrAI binds to primary site containing DNA with KD= 0.6±0.2 nM (by gel shift) when embedded in an 18 bp DNA (18-1), and tighter, KD= 0.057±0.009 nM (by gel shift), when embedded in a 40 bp DNA (40-1). The measurements show that a primary site with more flanking DNA binds tighter to SgrAI. The values are comparable to previously published measurements, for example, Daniels, et al.4 measured a KD for SgrAI and a 20 bp duplex containing a primary site of 1.8±0.4 nM, comparable to our measurement with 18 bp primary site containing DNA (18-1) (0.6±0.2 nM). Hingorani-Varma & Bitinaite8 measured 0.016±0.002 nM for the primary site in an 80 bp duplex, and 0.019±0.002 nM when in a 30 bp duplex, only about two-fold tighter than our measurement for the 40 bp primary site containing DNA (40-1) (0.057±0.009 nM). Differences in the measured KD values could be due to slightly different buffer composition and the temperature of the measurements (room temperature vs. 4°C).
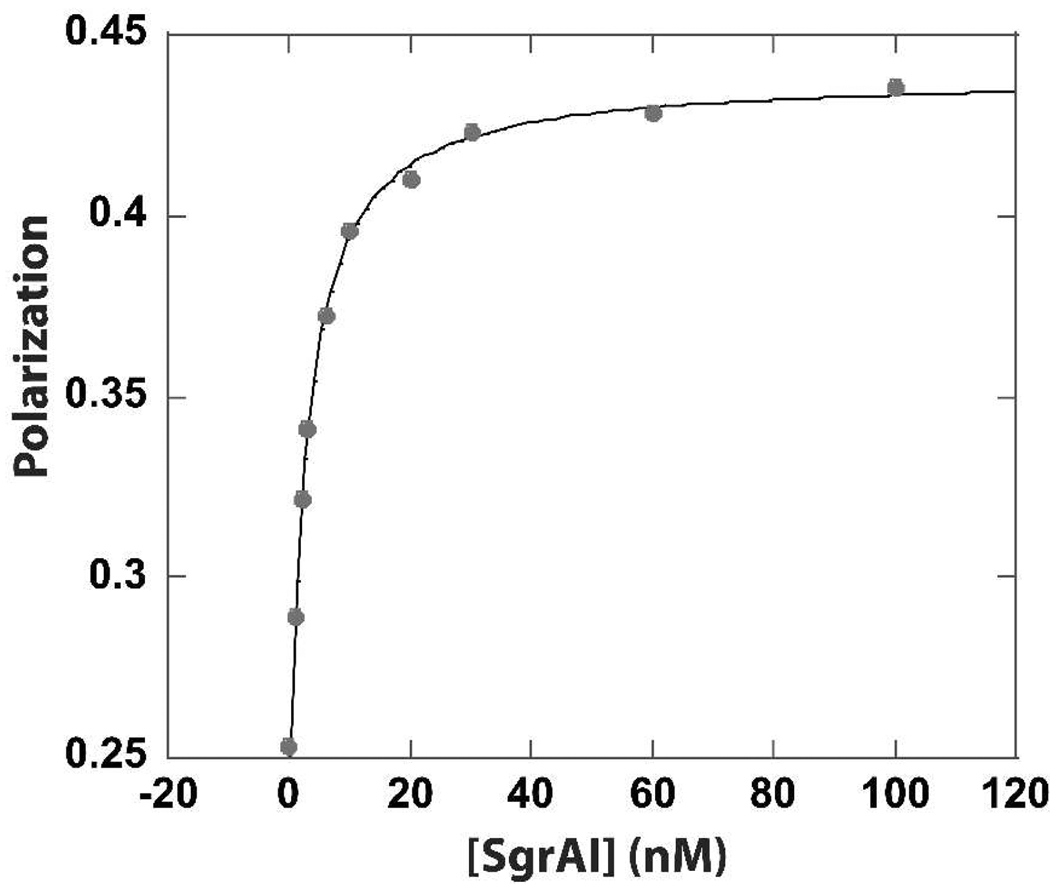
Figure 1.
Fluorescence polarization anisotropy (FPA) measurement (circles) of SgrAI binding to FLO labeled PCP in Buffer 3 (Table 1 legend) at 4°C. The line shows a fit (see Methods) to a 1:1 binding model giving a KD of 10.8±0.5 nM with R=0.99965.
Table 1.
Equilibrium dissociation constants for 1µM wild type SgrAI dimer and DNA sequences at 4°C.
| DNAa | Bufferb | KDc (nM) FPA |
KDc (nM) Gel shift |
|---|---|---|---|
| 18-1 | 1 | 2.5±0.9 | 0.6±0.2 |
| 18-2 | 1 | 1.5±0.2 | 2.6±1.2 |
| 40-1 | 1 | 0.9±0.2 | 0.057±0.009 |
| PCP | 1 | 6±2 | ND |
| PC | 1 | 5±1 | ND |
| PCP | 2 | 3±1 | ND |
| PCP | 3 | 14±4 | ND |
FPA utilized HEX labeled 18-1, 40-1, 18-2 and FLO labeled PCP, and PC.
Buffer 1: 20 mM Tris-Acetate (pH 8.0), 50 mM KOAc, 10 mM Ca(OAc)2, 1 mM DTT, 10% glycerol.
Buffer 2: 20 mM Tris-Acetate (pH 8.0), 50 mM KOAc, 10 mM Ca(OAc) 2, 1 mM DTT.
Buffer 3: 20 mM Tris-Acetate (pH 8.0), 50 mM KOAc, 10 mM Mg(OAc) 2, 1 mM DTT.
Equilibrium dissociation constants, KD given as the average of at least three different measurements ± the standard deviation, and assuming 1:1 binding.
The affinity to secondary site DNA (in an 18 bp DNA) is surprisingly strong, with KD=1.5±0.2 (by FPA) or 2.6±1.2 nM (by gel shift). Therefore the lower activity of SgrAI on secondary site DNA does not appear to derive from weakened binding. Our measured affinity for SgrAI to secondary site DNA is comparable to that measured previously for a different secondary site, CACCGGCT, embedded in an 80 bp duplex, of 0.7±0.1 nM8, with the tighter binding possibly due to the longer flanking DNA sequences.
The affinity to a mimic of cleaved 40 bp primary site DNA (PCP) is also surprisingly strong (KD=6±2 nM by FPA). This DNA has been synthesized to contain the 5’phosphate that would be present after cleavage of a primary site by SgrAI. The construct PC is identical with PCP but missing the 5’ phosphate. The binding affinity of PC to SgrAI has been measured with a KD= 5±1 nM (by FPA). Therefore the presence of the 5’phosphate at the cleavage site affects binding affinity very little consistent with previously published observations that the absence of the 5’ phosphate does not affect the stimulatory activity of the precleaved primary site5, 7, 8. Using a competition method and the gel shift assay, the affinity to a 38 bp duplex with two “SgrAI cleaved ends” (although no 5’phosphate on these ends) was determined to be 0.36±0.04 nM8, ~17 fold tighter than our measurement by FPA, however the DNA construct contains twice as many “cleaved ends” as PCP. The “cleaved end” of PCP contains a single stranded overhang which would be left after cleavage of a primary site by SgrAI, 5’-CCGG-3’, and also contains the phosphate at the 5’ end of this overhang to better mimic the naturally cleaved ends.
The affinity of SgrAI to PCP in buffer without glycerol (but with 10 mM Ca2+) was measured with a KD=3±1 nM (by FPA), indicating that the 10% glycerol of the binding buffer also has little affect on the binding affinity. Since PCP cannot be cleaved further by SgrAI, the binding affinity to this DNA could also be measured in the presence of Mg2+, rather than Ca2+, and was found to have KD=14±4 nM (by FPA). Therefore, use of Ca2+ in place of Mg2+ appears to have the effect of tightening the DNA binding affinity by ~3 fold.
Stoichiometric measurements were performed with SgrAI and the 18 bp DNA containing the prinary site sequence (18-1), as well as the 18 bp DNA containing the secondary site sequence (18- 2), using FPA or the gel shift method. In this method, 75-100 nM DNA is titrated with SgrAI until a break is seen (Fig. 2) giving the concentration of SgrAI dimer which fully saturates all binding sites on the DNA. The expected 1:1 binding is seen for secondary site DNA using hexachlorofluoescein (HEX) labeled 18-2 and FPA (Fig. 2A), and 18-1 using 32P labeled 18-1 and the gel shift assay (Fig. 2B). However, two breaks are seen in the plot of polarization vs. SgrAI concentration with HEX labeled 18-1, one at 1:1 binding, and one at 2:1 SgrAI dimers per 18-1 duplex (Fig. 2C). Hence a second dimer, binding more weakly than the first, appears to add to the SgrAI dimer/HEX-18-1 complex. The binding of additional SgrAI dimers to 1:1 saturated primary site has been seen before, also using FPA, although the second binding was not titrated to completion4.
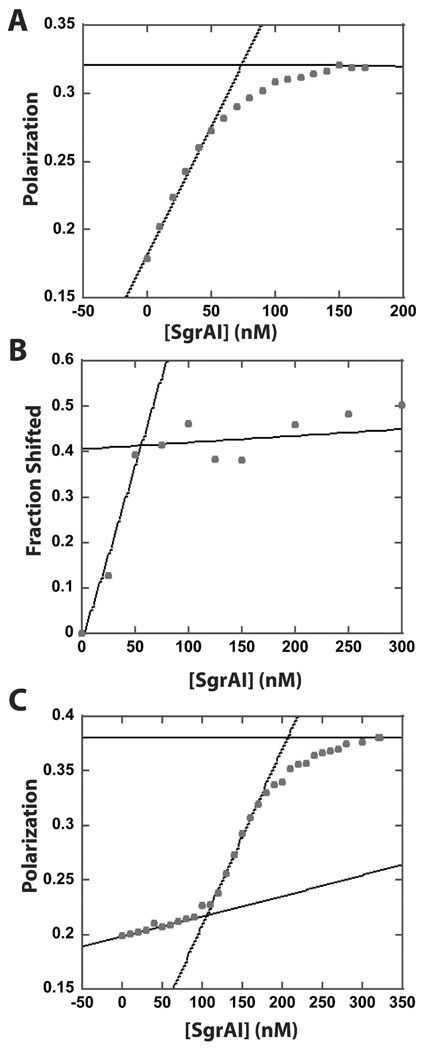
Figure 2.
Determination of the stoichiometry of binding of SgrAI to 18-2 and 18-1. A. Titration of 75 nM HEX labeled 18-2 in Buffer 2 (Table 1) at 4°C. The intercept of the two lines is 69 nM SgrAI dimer, indicating a 1:1 stoichiometry of SgrAI dimer to 18-2 duplex with 92% activity of SgrAI enzyme. B. Native PAGE gel shift of 100 nM 32P labeled 18-1 (of which 40% is double stranded). The intercept occurs at 55 nM SgrAI, suggesting 1:1 binding of 18-1 to SgrAI dimer with 73% enzyme activity. C. Titration of 100 nM HEX labeled 18-1 in Buffer 2 (Table 1) at 4°C. The first intercept at 109 nM SgrAI dimer indicates a 1:1 stoichiometry of SgrAI dimer to 18-1 duplex with 92% activity of SgrAI enzyme, the second intercept at 207 nM SgrAI dimer indicates a second SgrAI dimer binds the original 1:1 complex.
All of the binding isotherms used to measure the KD (with 1 nM DNA), fit very well to a model for 1:1 binding without cooperativity (Fig. 1), most likely indicating that the additional SgrAI dimers do not bind at the low nanomolar concentrations of the SgrAI:DNA complex. The fit to the 1:1 binding model also suggests that only a single duplex of DNA binds to one SgrAI binding site, which was surprising in the case of PCP, since two molecules of PCP are equivalent to one complete primary site DNA. Since unbound SgrAI is dimeric under the conditions of the binding measurements (as determined by analytical ultracentrifugation4), the fit to 1:1 binding without any evidence of cooperativity could mean that one molecule of PCP binds to one dimer of SgrAI. Alternatively, it could mean that two molecules of PCP bind uncooperatively to the SgrAI dimer having two independent binding sites, but this appears intuitively unlikely since the single stranded 5’-CCGG-3’ overhangs would be expected to anneal in the enzyme binding site and therefore interact cooperatively. Still a third possibility is that SgrAI binds to one molecule of DNA, composed of two PCP molecules. Since the binding measurements were performed at 4°C, and the Tm of the single stranded overhangs of PCP, 5’-CCGG-3’, is calculated to be 16°C, the binding data determined at 4°C likely represents the binding of SgrAI to one molecule of annealed PCP, comprised of two PCP molecules.
Characterization of HMWS using native gel electrophoresis
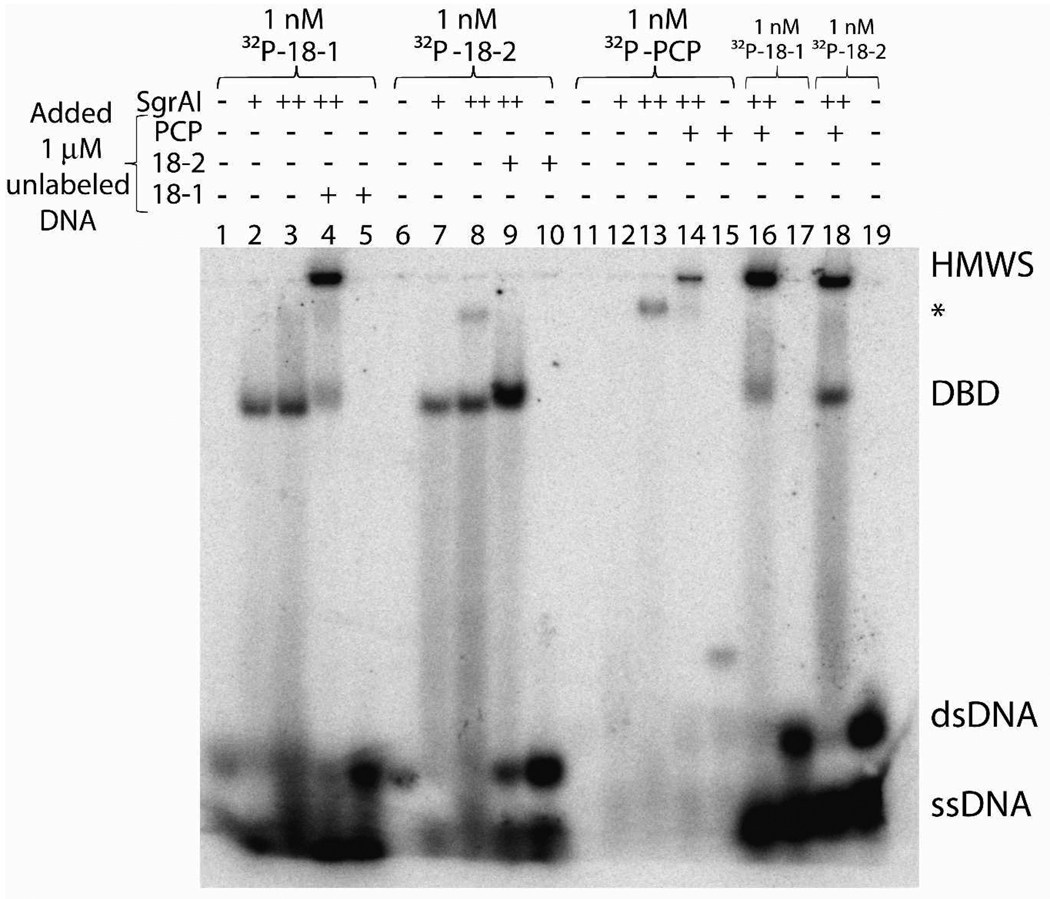
We have determined conditions to separate DNA bound dimers of SgrAI (DBD) from a high molecular weight species (HMWS, Fig. 3) composed of SgrAI and DNA using native polyacrylamide gel electrophoresis at 4°C. The binding reactions were prepared in binding buffer (20 mM Tris-OAc (pH 8.0), 50 mM KOAc, 1 mM DTT, 10 mM Ca(OAc)2, and 10% glycerol) with 1 nM 32P labeled DNA and varied concentrations (typically 1 µM) SgrAI. Additional unlabeled DNA was added in these experiments, in the range of 1 – 3000 nM. Using the native PAGE, we observed two shifted bands, the faster moving band was identified as dimeric SgrAI bound to DNA (i.e., DBD), since analytical ultracentrifugation (see below and Daniels, et al.4) have shown that only DBD is formed with secondary site DNA, and lane 9 in Fig. 3, mimicking the conditions used in the analytical ultracentrifugation, shows only this species. The HMWS is so named because it runs slower than DBD, however the exact composition of HMWS cannot be determined from the gel (see below for analytical ultracentrifugation of HMWS). Using this native gel assay we have discovered that the formation of HMWS depends upon the concentration of DBD, as well as the sequence of bound DNA. Only DBD form with 1 nM 32P labeled uncleaved DNA and 1 µM SgrAI dimer, whether the DNA is the primary or secondary site DNA (lanes 2–3 and 7–8, Fig. 3), however, with primary site DNA at concentrations of 1 µM DNA (with 1 nM 32P labeled DNA, and 1 µM SgrAI dimer), both DBD and HMWS are found (lane 4, Fig. 3). HMWS is never formed with only secondary site DNA, regardless of the concentration of DNA or SgrAI (lanes 6–10, Fig. 3), or length (18 or 40 bp, Fig. 4C). However, HMWS will form with 1 nM 32P labeled secondary site DNA in the presence of 1 µM SgrAI and 1 µM unlabeled pre-cleaved 40 bp primary site DNA (PCP) (lane 18, Fig. 3). PCP binds with nanomolar affinity to SgrAI (Table 1), however at 50 nM SgrAI and 1 nM 32P labeled PCP, the complex appears to be too labile to form a shifted band (lane 12, Fig. 3). This could be due to differences in the temperature at which the two measurements were performed; both were performed at 4°C, however local heating can occur in the gel during electrophoresis. PCP contains a 4 nucleotide self-complementary single stranded overhang (5’-CCGG-3’) that anneals to itself with a calculated Tm of 16°C to form a 40 bp duplex, which may bind tighter to SgrAI than the unannealed form. In addition, a band running more slowly than the DNA bound dimer appears at 1 µM SgrAI dimer and 1 nM 32P labeled PCP (lane 13, Fig. 3), suggesting a propensity for additional SgrAI dimers to bind to the DNA bound dimer of SgrAI (DBD) at these concentrations, as is seen with 1 µM SgrAI dimer and 1 nM 18 bp DNA containing the secondary site sequence (18-2) (lane 8, Fig. 3), and 1 nM 18 bp DNA containing the primary site sequence (18-1) when analyzed by FPA (see above). At 1 µM SgrAI dimer and 1 µM PCP (of which 1 nM is 32P labeled), only HMWS is observed (lane 14, Fig. 3).
Figure 3.
Autoradiogram of native PAGE with 1 nM 32P labeled DNA as indicated and varied additions including 1 µM unlabeled 18-1, 18-2, or PCP DNA (+), 50 nM SgrAI dimer (+), and/or 1 µM SgrAI dimer (++). HMWS: high molecular weight species, DBD: DNA bound dimer, *: DBD bound to one or more additional SgrAI dimers.
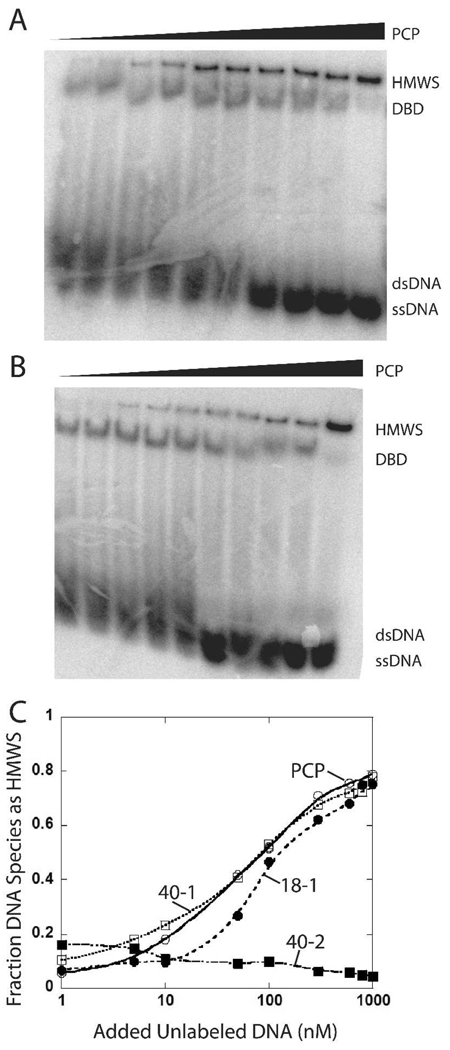
Figure 4.
Native gel electrophoresis of 1 nM 32P labeled 18 bp DNA and 1 µM SgrAI dimer and varied concentrations of unlabeled DNA. A. 1 nM 32P labeled primary site (18-1) with added unlabeled PCP (10, 30, 60, 100, 200, 300, 400, 500, 600, 1000 nM). B. 1 nM 32P labeled secondary site (18-2) with added unlabeled PCP (10, 30, 60, 100, 200, 300, 400, 500, 600, 1000 nM). C. Plot of percent of DNA (1 nM 32P-18-1) bound species with 1 µM SgrAI found in HMWS vs. concentration of added unlabeled PCP (open circles), 40-1 (open boxes), 18-1 (filled circles), or 40-2 (filled boxes).
The precleaved 40 bp primary site containing DNA (PCP) induces HMWS with 1 nM 32P labeled uncleaved primary site containing DNA (18-1, lane 16, Fig. 3) and with an 18 bp secondary site containing DNA (18-2, lane 18, Fig. 3) in a concentration dependent manner (Fig. 4A–B). Side by side comparisons of HMWS formation on 32P labeled 18-1 (with 1 µM SgrAI) as a function of DNA concentration shows that PCP, 40-1, and 18-1 induce HMWS similarly (Fig. 4C). This is somewhat unexpected as one might expect that twice as much precleaved 40 bp primary site containing DNA (PCP) as uncleaved (40-1) would be required to induce the same amount of HMWS if two PCP bind to one dimer. This result may indicate 1:1 binding of PCP to SgrAI dimer, or that SgrAI bound to PCP is better able to stimulate the HMWS.
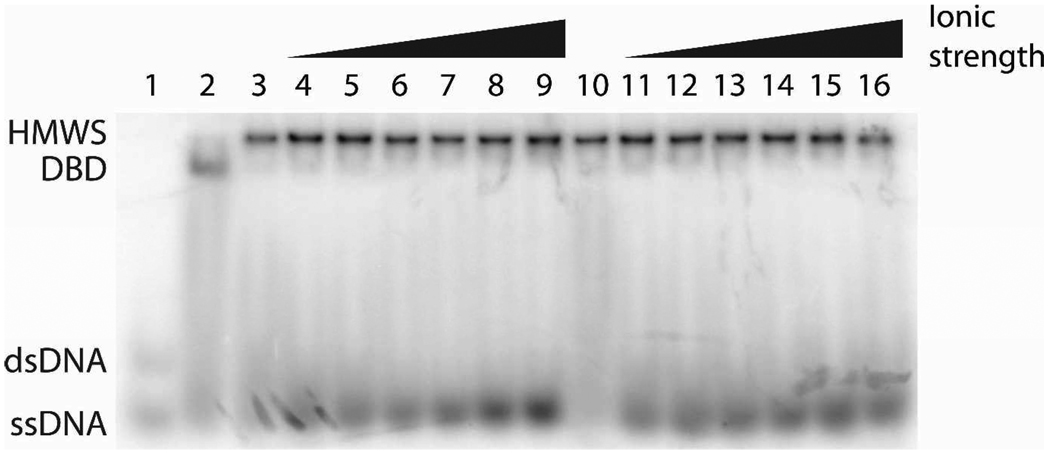
The effect of increased ionic strength on the high molecular weight species (HMWS) stability was tested using increasing concentrations of either KOAc or NaCl in buffer containing 20 mM Tris-OAc (pH8.0), 10 mM Ca(OAc)2, 10% glycerol, and 1 mM DTT (Fig. 5). The samples were electrophoresed at 4°C as described above after 30 minutes incubation at 4°C. Lanes 1 and 2 contain samples that mark the positions of unbound DNA (lane 1, containing 1 nM 32P labeled 18 bp primary site containing DNA (18-1) only) and DBD (DNA bound SgrAI dimer, lane 2, containing 1 nM 32P- 18-1 and 1 µM SgrAI). Samples loaded into lanes 3–16 each contained 1 nM 32P labeled 18-1, 1 µM SgrAI and 1 µM precleaved 40 bp primary site containing DNA (PC) in the different buffer conditions, and all lanes show a clear HMWS band. The samples loaded into lanes 3 and 10 contained no added KOAc or NaCl, while lanes 4–9 contain samples incubated in buffer with increasing concentration of KOAc from 50 mM to 300 mM, in 50 mM increments. Similarly, lanes 11–16 show the results from samples containing increasing concentrations of NaCl, from 50–300 mM. Clearly, the higher salt concentrations of the incubation buffer do not eliminate the appearance of the HMWS.
Figure 5.
Native gel electrophoresis of samples in binding buffer containing KOAc or NaCl concentrations from 0 – 300 mM. All samples contain 1 nM 32P-18-1 DNA, 1 µM SgrAI, and 1 µM PC DNA with the exception of that loaded in lane 1, which contains only the 32P-18-1, and lane 2, which contains 32P-18-1 and SgrAI. All samples were incubated in buffer containing 20 mM Tris-OAc (pH 8.0), 10 mM Ca(OAc)2, 10% glycerol and 1 mM DTT and varied concentrations of KOAc (lanes 4-9, with 50, 100, 150, 200, 250, 300 mM respectively) or NaCl (lanes 11–16, with 50, 100, 150, 200, 250, 300 mM respectively). Samples loaded in lanes 1–3 and 10 contain no KOAc or NaCl.
Analytical Ultracentrifugation
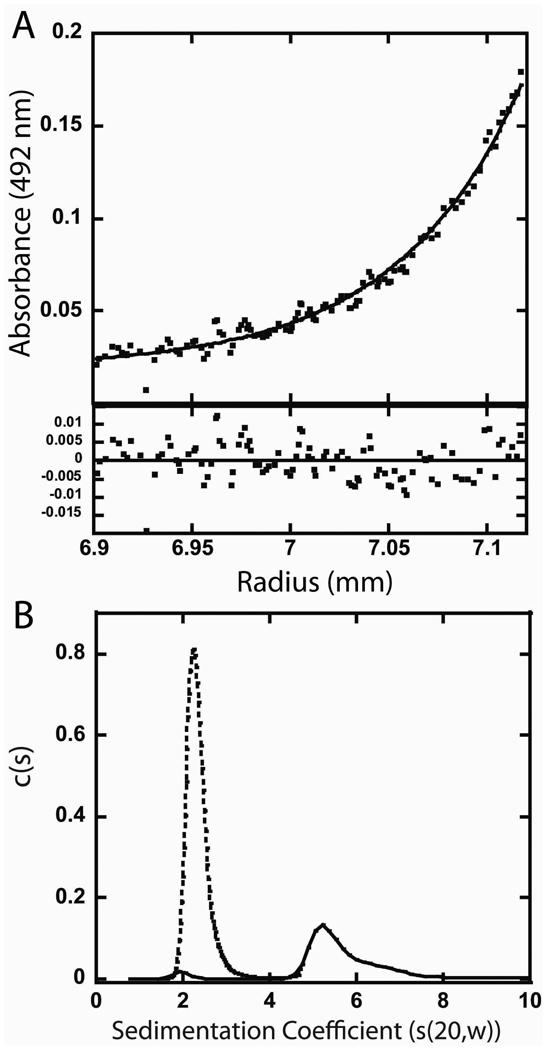
Using fluorophore labeled DNA and both the sedimentation equilibrium and velocity methods, we have been able to characterize SgrAI/DNA complexes formed with secondary site DNA and precleaved 40 bp primary site containing DNA (Fig. 6–7). Figure 6A shows the data from sedimentation equilibrium of 7 µM SgrAI and 3.6 µM fluorophore labeled 18 bp secondary site containing DNA (HEX-18-2) in binding buffer without glycerol (20 mM Tris-OAc, 50 mM KOAc, 10 mM Ca(OAc)2, 1 mM DTT) at 4°C. The binding buffer contains Ca2+ which inhibits DNA cleavage by SgrAI. A global fit to a single species using data from two speeds (13,500 rpm and 18,000 rpm) and two concentrations of SgrAI/DNA (7.0 µM/3.6 µM and 5.4 µM/2.7 µM) resulted in a molecular weight of 90.7 kDa, very close to the calculated molecular weight of the DNA bound SgrAI dimer (DBD, 87.5 kDa). Hence, SgrAI forms only DBD with this secondary site DNA. The excess SgrAI was added in these experiments to ensure complete binding of the fluorophore labeled DNA, yet the fitted molecular weight from the data indicates that additional SgrAI dimers did not bind to the DNA bound SgrAI dimer (DBD). Figure 6B shows the c(s) distribution from the sedimentation velocity experiment performed in the same buffer and same temperature as the sedimentation equilibrium experiment shown in Figure 6A, with either HEX-18-2 alone (3 µM, dotted line, Fig. 6B) or with SgrAI (6 µM, solid line, Fig. 6B). The DNA alone has an s value of 2.2 s, while that in the presence of SgrAI occurs at 5.2 s. Since the sedimentation equilibrium data indicate that the SgrAI/HEX-18-2 complex is predominantly in the form of the DNA bound SgrAI dimer (DBD), the 5.2 s peak must correspond to the DBD.
Figure 6.
Analytical ultracentrifugation of SgrAI with secondary site containing DNA (HEX-18-2). A. Sedimentation equilibrium experiment with 7 µM SgrAI and 3.6 µM HEX-18-2 in 20 mM Tris-OAc (pH 8.0), 50 mM KOAc, 1 mM DTT, 10 mM Ca(OAc)2 at 4°C at 13,500 rpm. Upper panel: Data shown as filled circles, the line corresponds to a fit of a single species with molecular weight of 90.7 kDa. Lower panel: residuals between fit and data. B. Sedimentation velocity c(s) distribution of 6 µM SgrAI and 3 µM HEX-18-2 in 20 mM Tris-OAc (pH 8.0), 50 mM KOAc, 1 mM DTT, 10 mM Ca(OAc)2 at 4°C.
Figure 7.
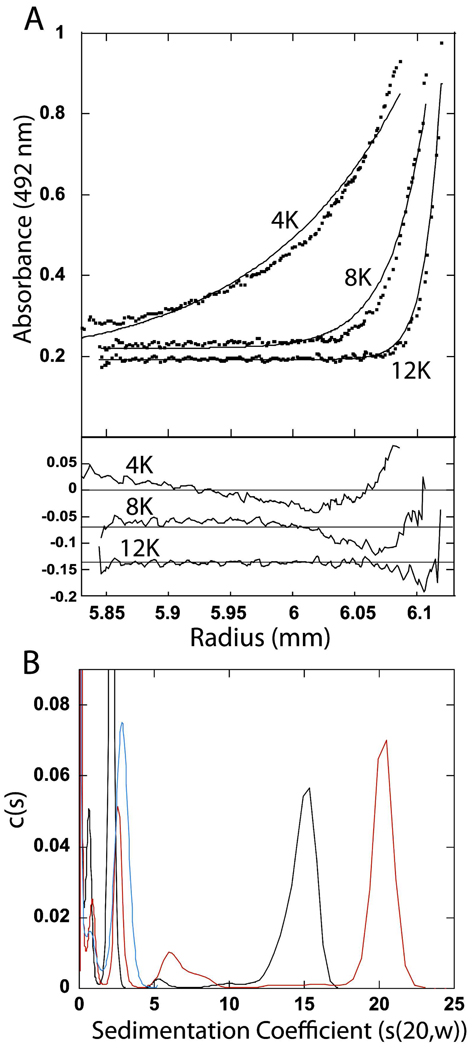
Analytical ultracentrifugation of SgrAI with precleaved 40 bp primary site containing DNA (FLO-PCP). A. Sedimentation equilibrium experiment with 3 µM SgrAI and 1.5 µM FLO-PCP in 20 mM Tris-OAc (pH 8.0), 50 mM KOAc, 1 mM DTT, 10 mM Mg(OAc)2 at 37°C at 4,000, 8,000, and 12,000 rpm. Upper panel: Data shown as filled circles, the line corresponds to a global fit of a single species with molecular weight of 732 kDa. Lower panel: residuals between fit and data. B. Sedimentation velocity experiments with 3 µM FLO-PCP only (blue line, c(s) values scaled by a factor of 0.5) or with 6 µM SgrAI (red and black lines) at 37°C in 20 mM Tris-OAc (pH8.0), 1 mM DTT, 10 mM Mg(OAc)2 and either 50 mM KOAc (red and blue lines) or 150 mM NaCl (black line).
Figure 7A shows the result of sedimentation equilibrium experiments performed at three different speeds with 3 µM SgrAI and 1.5 µM fluorophore labeled precleaved 40 bp primary site containing DNA (FLO-PCP) in the buffer used for the DNA cleavage assays (20 mM Tris-OAc, 50 mM KOAc, 10 mM Mg(OAc)2, 1 mM DTT) and at 37°C. Data was also collected at two other concentrations of SgrAI (6 and 12 µM) and DNA (3 and 6 µM), and the same three speeds. Again, excess SgrAI enzyme was included to ensure complete binding of the fluorophore labeled DNA. The line in Figure 7A (upper panel) corresponds to a fit for a single species of approximately 732 kDa, derived from global fitting of all of the data. The residuals between the fits and the data are shown in the lower panel of Figure 7A. Clear trends in the residuals can be seen indicating a poor fit of the single species model to the data, hence the SgrAI/FLO-PCP complexes must be heterogeneous. In addition, the raised baseline of the data suggests the presence of significantly smaller species that are not sedimented significantly at these speeds (likely the unbound DNA).
Figure 7B shows the c(s) distribution from the sedimentation velocity measurement of either 3 µM FLO-PCP DNA only (blue line, Fig. 7B), or with 6 µM SgrAI (red line, Fig. 7B), in the same buffer conditions and temperature as the sedimentation equilibrium experiment of Figure 7A. In addition, the sedimentation of a SgrAI/FLO-PCP mixture in a buffer with higher ionic strength (150 mM NaCl in place of the 50 mM KOAc) (black line, Fig. 7B) was also analyzed. The data from the FLO-PCP DNA only sample (blue line, Fig. 7B) marks the sedimentation of the free DNA, the peaks with sedimentation coefficients less than 4 s. The 5–6 s peaks, more prevalent in the lower ionic strength buffer (red line, Fig. 7B), is similar in sedimentation coefficient to the DNA bound SgrAI dimer (DBD) seen with secondary site DNA in Fig. 6B (5.2 s). A shoulder is found with higher s values adjacent to this peak, which may indicate additional species, however the predominant SgrAI bound DNA species runs with s values of either 15 or 20 s, depending on the ionic strength of the buffer. The s value of this species is consistent with the large molecular weights derived from the sedimentation equilibrium measurements, and therefore corresponding to the HMWS.
Single turnover DNA cleavage assays
Single turnover DNA cleavage rate constants were measured by mixing solutions of 32P labeled DNA (1 nM) with a solution of enzyme (typically 1 µM) at 37°C, in buffer supporting DNA cleavage containing 10 mM Mg2+ (20 mM mM Tris-OAc, 50 mM KOAc, 10 mM Mg(OAc)2, 1 mM DTT). Some reactions were also performed at 4°C. The amount of DNA cleaved with time was measured by quenching aliquots of the reaction mixtures at different times after mixing the SgrAI enzyme with the DNA, separating cleaved from uncleaved DNA via denaturing PAGE, and quantitating by autoradiography. The amount of cleaved DNA was normalized (using either the total intensity of the lane, or the sum of the amount of cleaved and uncleaved DNA in the same lane), then plotted vs. time. Unlabeled DNA was added in some reactions to test for its stimulatory capacity, and was added to the solution with the labeled DNA prior to mixing with the enzyme solution. The resulting single turnover rate constants measured for DNA cleavage of various sequences by SgrAI are presented in Table 2. Most data fit well (R>0.99) to a single exponential equation (see Methods), while the data with 0.9 µM 40 bp uncleaved primary site DNA (40-1) (Table 2) required two exponential functions in order to fit the data. The total amount of the labeled DNA that was cleaved in each reaction varied, in some cases due to the melting of the duplex into single strands (which are not cleavable by SgrAI) as a consequence of repeated freeze-thawing of the labeled DNA stock solution. However, the amount of secondary site DNA cleaved did show a systematic trend where a greater fraction was cleaved with increasing amounts of added unlabeled precleaved 40 bp primary site containing DNA (PCP). The percent of the DNA cleaved was then very carefully measured in triplicate, in single turnover reactions performed simultaneously.
Table 2.
Single turnover DNA cleavage rate constants using 1 µM SgrAI and 37°C (unless otherwise noted)
|
32P labeled DNA (1 nM) |
Conc. Added unlabeled DNA | WT SgrAI Rate constant (min−1) |
|---|---|---|
| 1° site (18-1, 18 bp) |
0 | 0.094±0.015 |
| 10 nM PCP | 0.18±0.06 | |
| 100 nM PCP | 0.30±0.03 | |
| 1 µM PCP | >20 | |
| 1 µM PCP (QF) | 22±7 | |
| 2 µM PCP | >20 | |
| 0.9 µM 18-1 | 0.063±0.006 | |
| 0.9 µM 40-1 | 10.0±1.4 (56±2%) 0.032±0.012 (44±2%) |
|
| 0.9 µM 40-2 | 0.16±0.06 | |
| 0, 4°C | <4×10−5b | |
| 0.9 µM 18-1, 4°C | 0.118±0.014 | |
| 0.9 µM PC, 4°C | 2.2±0.04 | |
| 1° site (40-1, 40 bp) | 0 | 0.14±0.05 |
| 0.9 µM 40-1 | >20 (22±9%) 0.024±0.008 (78±9%) |
|
| 2° site (18-2, 18 bp) | 0 | 0.020±0.006 (2.8%±0.6%) |
| 10 nM PCP | 0.012±0.001a (3.4%±0.1%) a |
|
| 100 nM PCP | 0.016±0.005 (19%±5%) |
|
| 1 µM PCP | 0.05±0.01 (24%±1%) |
Only 2 repetitions.
QF=measurement made in the Kintek RQF-3 Rapid Quench Flow Instrument.
No cutting detected after 21 hours of incubation. Rate constant estimated assuming 5% cleavage or less after 21 hours.
The rate constants for cleavage of primary site DNA embedded in constructs with different amounts of flanking DNA, 18-1 and 40-1, are very similar: 0.094±0.015 min−1 and 0.14±0.05 min−1, respectively. The rate constant for cleavage of secondary site DNA (18-2), in the absence of added precleaved 40 bp primary site containing DNA (PCP), was only ~5 fold smaller than that for primary site DNA (18-1): 0.020±0.006 min−1. Added PCP increased the rate constant for cleavage of 1 nM 32P labeled primary site (18-1) dramatically, from 0.094 min−1 to >20 min−1 with 1 µM PCP, a rate too fast to measure by hand. Using a quench flow instrument, this rate constant was measured as 22±7 min−1, representing a >200 fold increase. Similarly, added 40-1 (0.9 µM) also stimulated cleavage of 1 nM 32P labeled primary site (18-1 or 40-1), to a rate too fast to measure by hand, >20 min−1, although only a portion of the total amount of cleaved DNA was cleaved with an accelerated rate. In contrast, added unlabeled 18-1 (0.9 µM) did not stimulate primary site DNA cleavage by SgrAI (at 37°C), and neither did unlabeled 0.9 µM 40-2 (0.16±0.06 min−1), the secondary site embedded in the same oligonucleotide as 40-1.
The cleavage of secondary site DNA (18-2) was also stimulated by added precleaved 40 bp primary site containing DNA (PCP), from 0.020±0.006 min−1 in the absence of PCP, to 0.05±0.01 min−1 in the presence of 1 µM PCP, an increase by a factor of ~2.5. More significantly, the percentage of the secondary site DNA cleaved by SgrAI was very low in the absence of PCP, and is increased, from about 2% to ~24% with 1 µM PCP. The total amount of DNA cleaved reaches a maximum at ~25% cleavage rather than 100%, most likely due to the dissociation of duplex DNA into single strands during freezing for storage of the DNA.
Cleavage reactions were also performed at 4°C with 1 nM 32P labeled 18 bp primary site DNA (18-1) and 1 µM SgrAI, in the presence or absence of the addition of 0.9 µM unlabeled DNA, either 18-1 or precleaved 40 bp primary site DNA (PC, as in PCP without the 5’phosphate at the cleavage site). No cleavage of the labeled DNA was detected in the absence of the additional DNA, even after 21 hours, suggesting a rate constant less than 4×10−5 min−1 (assuming a 5% cleavage limit of detection). However, the presence of 0.9 µM 18-1 stimulates cleavage, with a rate constant of 0.118±0.014 min−1. Therefore, although 18-1 fails to stimulate DNA cleavage at 37°C, stimulation of DNA cleavage by 18-1 does occur at 4°C. The cleavage of 1 nM 18-1 at 4°C is also stimulated by PC to 2.2±0.04 min−1, indicating that the stimulatory capacity of 18-1 is less than that of PC DNA.
Discussion
DNA binding and stoichiometric measurements indicated very tight binding (nM or better) to the primary and secondary site DNA constructs used in this study, with 1:1 binding of the SgrAI dimer to the 18 bp duplex DNA (Table 1, Results).
In addition, a method developed for separating different DNA bound forms of SgrAI using native gel electrophoresis showed the presence of two DNA bound SgrAI forms(Fig. 3–4). The faster moving species is identified as the DNA bound SgrAI dimer (DBD), since sedimentation measurements (see below) show that only DBD are formed with the secondary site DNA 18-2, and only this band is seen with 18-2 (lanes 7-9, Fig. 3). The slower moving species, which barely enters the gel, has been termed HMWS for high molecular weight species, as it has been determined to be quite large by analytical ultracentrifugation (Fig. 7). The HMWS forms only under certain conditions, namely that sufficient concentrations of both SgrAI and primary site DNA (cleaved or uncleaved) are present. DNA containing only secondary site, whether in an 18 bp or 40 bp duplex (ie. 18-2 or 40-2), will not form HMWS without the presence of any primary site (Fig. 3). Secondary site DNA bound to SgrAI can, however, become part of the HMWS in the presence of primary site DNA (Fig. 3–4). Precleaved 40 bp primary site containing DNA (PCP) also stimulates HMWS formation. Comparison of the ability of uncleaved primary site containing DNA, (with different lengths of flanking DNA, 18-1, 40-1), and PCP to induce HMWS formation with a 32P labeled 18-1 show similar activities (Fig. 4C). Analysis of the concentration dependence of HMWS formation shows a midpoint to saturation at ~100 nM DNA, which corresponds to ~100 nM DBD, since at 1 µM SgrAI nearly all DNA will be bound. These results also explain why no cooperativity was seen in the FPA and gel shift assays used to measure DNA binding affinity, since at 1 nM DNA concentration, the concentration of DBD would be too low to form HMWS.
Analytical ultracentrifugation was used to analyze the size of the high molecular weight species (HMWS). Fluorophore labeled DNA was used to limit detection to only DNA containing species. However, unlike the native gel electrophoresis described above, the higher concentrations of DNA necessary for detection, with 2.7–3.6 µM DNA (and 5.4–7.0 µM SgrAI) used in this study. Figure 7A shows the data from a sedimentation equilibrium experiment with 1.5 µM fluorophore labeled precleaved 40 bp primary site containing DNA (FLO-PCP) and 3 µM SgrAI, in the same buffer used for the DNA cleavage assays and at 37°C. Data from Fig. 7A, as well as at two higher concentrations of SgrAI and DNA, were globally fit to an equation for a single species, resulting in a fitted molecular weight of 723 kDa. Clearly the data do not fit well to this single species model, indicating heterogeneity(Fig. 7A). Fits to individual sedimentation equilibrium curves lead to molecular weight estimates ranging from 400–1200 kDa. Hence the species formed by SgrAI and FLO-PCP are large and heterogeneous, with DBD associating in numbers in the range of 4–12 (the calculated molecular weight of the SgrAI dimer bound to two molecules of FLO-PCP is approximately 101.5 kDa). The large size is consistent with the native PAGE that indicated it to be a species larger than the DNA bound SgrAI dimer (DBD). A large, heterogeneous species was also detected in a previous report4, using SgrAI and a 20 bp primary site containing DNA, with a rough estimate of ~7 DBD reported4. The previous study utilized a buffer containing Ca2+, rather than Mg2+, and the experiments were performed at 4°C rather than 37°C4. Therefore, formation and size of the HMWS appears to be relatively independent of the temperature, the type of divalent cation (Ca2+ or Mg2+), the length of the DNA flanking the primary site sequence, as well as whether or not the site is cleaved.
In contrast to the results with FLO-PCP, the sedimentation equilibrium experiment with secondary site containing DNA (HEX-18-2) indicates that only DBD are present (the fit provides a molecular weight of 90.7 kD, while the predicted DBD molecular weight is 87.5 kDa)(Fig. 6A). Sedimentation distributions with only HEX-18-2 DNA (3 µM, dotted line, Fig. 6B) or with HEX-18- 2 and SgrAI (3 and 6 µM, respectively)(solid line, Fig. 6B), in identical buffer conditions and temperature, show free DNA having a sedimentation coefficient of 2.2 s, and 5.2 s for the SgrAI/DNA complex. Since the sedimentation equilibrium experiment indicates that the SgrAI bound DNA is predominantly in the form of DBD (Fig. 6A), the single SgrAI bound DNA peak in the c(s) distribution at 5.2 s (solid line, Fig. 6B) must correspond to the DBD.
The c(s) distribution from the sedimentation velocity experiment with SgrAI (6 µM) and precleaved 40 bp primary site containing DNA (FLO-PCP) (3 µM) shows several species (Fig. 7B). In this case the buffer conditions and temperature are those used in the DNA cleavage measurements (red line, Fig. 7B) or in a buffer with higher ionic strength (150 mM NaCl in place of the 50 mM KOAc of the DNA cleavage buffer) (black line, Fig. 7B). The two peaks with the smallest s values correspond to free DNA (blue line, Fig. 7B). A peak appears at 5-6 s, similar in size to the DBD formed by SgrAI and HEX-18-2 (5.2 s). The majority of the SgrAI bound DNA is found sedimenting with coefficients of 15 and 20 s. The sedimentation coefficients differ depending on the ionic strength of the buffer, perhaps due to dissociation22, effects on size, shape, or solvation or from simply from variation sample to sample in fitting multipeaked data.
The excess SgrAI (over DNA) concentration used in the sedimentation experiments with FLO-PCP could result in additional SgrAI dimers binding to the SgrAI/FLO-PCP complex. However, the species with high sedimentation coefficients found in this study (20 s in the buffer with 150 mM NaCl) is similar in sedimentation coefficient (20 s) to that of the previous study4 that used a 1:1 molar ratio of SgrAI:DNA in a buffer with 150 mM KOAc, and a DNA with very short flanking sequences (a 20 bp uncleaved primary site containing DNA). The additional excess SgrAI enzyme was also used in the sedimentation experiments with HEX-18-2, yet did not lead to additional SgrAI dimers binding to the SgrAI/DNA complex. Native gels do indicate that additional SgrAI dimers bind to the SgrAI/PCP complex (lane 13,Fig. 3), however, these complexes are distinct from the HMWS (high molecular weight species) in that they migrated separately and faster, indicating that they were smaller than the HMWS. This faster migrating species only occurred with lower concentrations of the DNA bound SgrAI dimer (DBD), and once the concentration of DBD increased above ~100 nM, only HMWS were observed. The native PAGE results argue that the HMWS is composed of oligomers of DBD, since the formation of HMWS is dependent on the concentration of DBD and not merely SgrAI dimer concentration, however, they do not rule out the binding of additional SgrAI dimers to the oligomerized SgrAI/PCP complex. Thus the estimated size of the HMWS formed by SgrAI and PCP, in buffer conditions supporting DNA cleavage (and at 37°C, where the DNA cleavage measurements have been made), of 4–12 DBD may include the presence of additional SgrAI dimers to the oligomerized DBD. In any event, the DNA cleavage measurements also typically have an excess of SgrAI dimer over concentration of the DNA, and therefore this measurement of the size of the HMWS may actually be more relevant than a measurement with an alternative ratio of SgrAI to DNA.
The single turnover rate constants for DNA cleavage of the primary site in the two different length duplexes (18-1 and 40-1) and with a secondary site DNA (18-2) were measured (Table 2). The reactions were performed at 37°C (unless otherwise specified) with 10 mM Mg2+ (rather than Ca2+) and in the absence of glycerol, but the buffer components were otherwise the same as those for the FPA and native PAGE, The concentration of SgrAI in the measurements was 1 µM, to ensure complete saturation of the 32P labeled DNA, which was 1 nM at the initiation of the reactions. The rate constants for cleavage of the primary site DNAs were found to be similar, 0.094±0.015 min−1 and 0.14±0.05 min−1 for 18-1 and 40-1, respectively, indicating that the added flanking DNA of 40-1 does not greatly influence the DNA cleavage rate constant. These numbers compare well to a previously published single turnover cleavage rate constant on plasmids with single primary sites4, 0.04 min−1. The rate constant for cleavage of the secondary site, 18-2, was measured to be 0.020±0.006 min−1 (Table 2), only five fold slower than that of the primary site in the same length DNA. This rate constant is similar to that found using steady state kinetics for a secondary site in an 80 bp duplex8, 0.012 min−1. Cleavage of both primary and secondary site DNA was found to be stimulated under the conditions of the assay, by the addition of primary site DNA (40-1, PCP) with sufficient flanking sequences. Both 40-1 and PCP (precleaved 40 bp primary site containing DNA) stimulated cleavage of 18-1 by 1 µM SgrAI to a rate constant >20 min−1, too fast to measure by hand. The rate constant was measured by quench flow as 22±7 min−1 (with 1 µM PCP), a stimulatory factor of >200 fold. Stimulation of cleavage of 20 and 30 bp oligonucleotides with primary and secondary sites has been observed previously, using a 30 bp oligonucleotide with a primary site sequence, although rate constants were not reported7. Surprisingly, the 18-1 DNA could not stimulate its own cleavage (at 37°C), even at 0.9 µM (and 1 µM SgrAI), similar to that reported using a 20 bp primary site duplex4. The requirement for sufficient flanking DNA to provide stimulation was seen in a previous study7, as well as the ability of both uncleaved and cleaved primary site to stimulate DNA cleavage by SgrAI5, 7, 8. Flanking DNA itself however is not sufficient, as a construct with the same flanking DNA but a secondary, rather than a primary site, (40-2), did not stimulate DNA cleavage (at a concentration of 0.9 µM with 1 µM SgrAI) (Table 3). Previous measurements of DNA cleavage under stimulated conditions at 37°C include a single turnover DNA cleavage rate constant measured with a plasmid containing two recognition sites4 of 0.5–0.7 min−1, and with an 80 bp duplex using steady state kinetics8 2.4 min−1, and the same duplex but added precleaved primary site containing DNA 16.2 min−1 (using steady state kinetics). Our measurement of the maximally stimulated primary site DNA cleavage by SgrAI compares best to the latter example.
The stimulation of primary site DNA cleavage increased with increasing concentration of added precleaved 40 bp primary site containing DNA (PCP) by at least 200 fold, however, stimulation of secondary site DNA increased by only ~2.5 fold, to 0.05±0.01 min−1. This value is similar to that found by Wood, et al.7, in the single turnover cleavage of secondary sites on plasmid DNA with primary sites, 0.08 min−1. However more significantly, the overall percent of secondary site DNA that was cleaved by the end of the reaction was much greater with higher concentrations of PCP. The total amount of DNA cleaved appears to reach a maximum at only 24%, probably due to a majority of it being single stranded after repeated freeze-thaw of the labeled DNA. In the absence of PCP, only 2%, or 10% of the final fully cleavable amount, is cleaved by SgrAI. The stimulated rate constants for cleavage of the secondary site we measure, 0.05±0.01 min−1, is 5 times slower than that measured using steady state kinetics with an 80 bp duplex (0.35 min−1)8, however, a different secondary site sequence was used in our study (CACCGGGG compared to CACCGGCT).
The effects of buffer conditions such as ionic strength, temperature, and divalent cation cofactor were tested on the high molecular weight species (HMWS) formation using either the native gel electrophoresis analysis or the sedimentation velocity method. First, the formation of HMWS at 4°C analyzed using the native gel electrophoresis assay with samples incubated in buffers containing 10 mM Ca2+ and either 0–300 mM KOAc or 0-300 mM NaCl (Fig. 5) showed that HMWS formed with radiolabeled 18 bp DNA containing the primary site sequence (18-1) and precleaved 40 bp primary site containing DNA (PC) was stable at all ionic strengths tested. Hence the HMWS is not an artifact of low ionic strength. Sedimentation velocity experiments carried out at 37°C with 10 mM Mg2+, 6 µM SgrAI and 3 µM fluorophore labeled precleaved 40 bp primary site containing DNA (FLO-PCP) in buffer containing either 50 mM KOAc or 150 mM NaCl (Fig. 7B) show changes in the centering of the peak designated as HMWS, indicating altered sedimentation coefficients, again, the higher ionic strength does not eliminate the presence of the HMWS. In addition, the fact that HMWS was detected in the sedimentation experiments performed with Mg2+, and at 37°C (Fig. 7) show that the HMWS is not an artifact of Ca2+ or temperature (4°C) used in the native gel electrophoresis assays.
In a previous study, steady state kinetic measurements of the cleavage of 80 bp DNA constructs containing primary or secondary site DNA showed Hill coefficients of 2–48. These Hill coefficients are reduced to ~1 when an added pre-cleaved primary site DNA is added, supporting the cooperativity on DNA concentration in enhancing DNA cleavage. The cooperativity seen with secondary site DNA, in the absence of any primary site DNA, may appear contradictory to the results presented here that show no DNA cleavage stimulation in single turnover cleavage assays by the addition of secondary site DNA. However, upon cleavage of secondary site DNA, two cleaved ends are produced, one of which is exactly the same as a cleaved primary site. With multiple enzyme turnovers, as in the steady state kinetic assay, these species can accumulate to sufficient concentrations to influence the reaction kinetics. Therefore, the cooperativity seen in the steady state kinetics with secondary site DNA and the lack of stimulation by secondary site DNA seen in the current work are consistent if the species mediating the cooperativity in the steady state kinetics is the product of secondary site DNA cleavage.
The self-activation of DNA cleavage by SgrAI implies allostery, where at a minimum a low and high activity conformation exist in an equilibrium that can be shifted by the interaction with an allosteric effector. The fact that the addition of pre-cleaved 40 bp primary site containing DNA (PCP) can enhance the DNA cleavage rate by SgrAI suggests the existence of more than one DNA binding site on the functional SgrAI molecule. Type II restriction endonucleases typically bind to duplex DNA as dimers, in a 1:1 complex, however several type II restriction endonucleases form tetramers in solution, and bind to two DNA duplexes. SgrAI shares amino acid sequence similarity and DNA cleavage sequence similarity with three such enzymes: NgoMIV, Cfr10I, and Bse634I, therefore it was reasonable to consider that SgrAI might also form a tetramer with two DNA binding sites under activating conditions. Analytical ultracentrifugation of the SgrAI enzyme showed it to exist as a dimer in solution in the absence of DNA4; with DNA it formed a 1:1 complex of SgrAI dimer to DNA duplex that appeared to aggregate into a very large species, large enough to accommodate at least seven SgrAI dimers bound to duplex DNA4. Still, the model for SgrAI activity included slow cleavage of DNA by SgrAI dimers, and association of dimers bound to cleaved DNA with other dimers bound to uncleaved DNA to form an activated tetramer4, 7, 8. The activated tetramer conformation is predicted by the model to exhibit very fast cleavage of DNA. In addition, the activated DNA bound dimers could dissociate from the tetramer and associate with other DNA bound dimers thus activating them, and hence a chain reaction would result in the overall observed accelerated DNA cleavage rate. The addition of precleaved primary site DNA presumably accelerated the rate of DNA cleavage by inducing the activated conformation and stimulating dimers to form tetramers with the activated conformation. Structural studies reported to date show a dimer of SgrAI bound to a single duplex of primary site containing DNA (the 18-1 construct used in this study), which is considered to be in a low activity conformation9. A structure of the activated conformation has not yet been reported, and no direct data exists to support a tetrameric form of SgrAI as the activated species.
Native PAGE and analytical ultracentrifugation were successful in unambiguously demonstrating the presence of DNA bound species of SgrAI larger than dimers (Fig. 3–5, Fig. 7). Identified as high molecular weight species (HMWS) on native gels, the sedimentation measurements indicate that HMWS composed of SgrAI and PCP is heterogeneous containing possibly as many as 4–12 DNA bound SgrAI dimers (DBD). We found that formation of the HMWS species by SgrAI is dependent on DNA sequence (requiring the primary site sequence), dependent on the concentration of DNA bound SgrAI dimer (DBD) (forming significantly with DBD of 100 nM or higher in our assays), and can form with either cleaved or uncleaved primary site containing DNA. We also found that HMWS formation did not depend on the divalent cation or temperature used in the assays, as HMWS is observed in assays with either Ca2+ or Mg2+, and at both 4°C and 37°C. Initially we used the pre-cleaved primary site (PCP) DNA to induce HMWS and to stimulate DNA cleavage, although later determined that uncleaved 40 bp primary site containing DNA (40-1) can also induce HMWS. We found that added PCP stimulated both HMWS formation (in 10 mM Ca2+ or Mg2+, 4°C) and DNA cleavage (10 mM Mg2+, 37°C) with 1 nM uncleaved (primary or secondary site) DNA at similar concentrations of PCP (100–1000 nM). Since both DNA cleavage activity and the oligomerization of DBD into HMWS are dependent on the concentrations of added PCP, we propose that the HMWS is the activated (or activatable) form of the enzyme. Activation by oligomerization is a very unusual mechanism, although has been proposed in one other system, that of Acetyl-CoA carboxylase23.
The simplest model of DNA cleavage by SgrAI includes two conformations of SgrAI, a low activity form, and a high activity form. In such a model, intermediate rate constants for DNA cleavage should not occur, since there are no conformations with intermediate levels of activation. Instead, in cases where only a subset of the total population of enzymes is stimulated, the amount of cleaved DNA with time would be the sum of that from two independent first order processes from the two enzyme states, and therefore the data of cleaved DNA with time should fit to two exponential functions. This was true in the assays with 0.9 µM 40 bp primary site containing DNA (40-1), where a portion of the labeled DNA was cleaved at a much stimulated rate, and a portion cleaved at the lower, unstimulated rate. However, the experimental data using precleaved 40 bp primary site containing DNA (PCP) as the stimulating DNA is best fit not with multiple exponential functions, but rather only a single. The measured rate constants increase with the concentration of added PCP, such that with intermediate concentrations of PCP, intermediate values of the rate constant are exhibited (Table 2). If the rate constants measure the actual rate of the chemical step in DNA cleavage, they then indicate that intermediate levels of enzyme activation exist. However, because these first order rate constants vary with the concentration of added PCP, they could also be pseudo-first order rate constants for a second order process that is dependent on PCP concentration, which occurs prior to DNA cleavage. For example, the binding of DNA to SgrAI, or the association of the SgrAI dimer bound to 32P labeled 18 bp primary site containing DNA (18-1) with SgrAI dimers bound to PCP. The association of DNA binding proteins with DNA is typically very fast24, therefore the association of the DNA bound dimers could be the second order process that governs the observed kinetics. In the case of the cleavage of the secondary site DNA, the observed kinetics appear more like the contributions from two enzyme states, one that is completely inactive, and another that has a maximal rate of cleavage that is similar to the slow cleavage rate of primary site DNA. Roughly the same cleavage rate constant is measured in the absence and presence of stimulation, but a greater percentage of the secondary site DNA is cleaved under stimulatory conditions. Therefore the apparent alteration of sequence specificity in SgrAI appears to involve the switching on of DNA cleavage at a sequence already bound tightly by the enzyme, through shifting the enzyme conformation to a more active form. However, the cleavage rate on secondary site DNA is never stimulated to nearly the same extent as that on primary site DNA, suggesting that the secondary site sequence may impede the attainment of the fully activated conformation by SgrAI or the SgrAI/DNA complex.
We found that uncleaved primary site DNA, if present in a 40 bp duplex (40-1), is capable of stimulating both high molecular weight species (HMWS) formation (using native PAGE) and DNA cleavage (in single turnover assays). However, 40-1 when cleaved creates two molecules of PCP, which also have stimulatory properties, and we wonder if it is the PCP that induces the activated conformation, and HMWS is a form required for the activated conformation but is actually capable of both low and high activity conformations. However, the unstimulated cleavage of 40-1 is 0.14±0.05 min−1, while the activated cleavage rate is >20 min−1, too fast for significant amounts of PCP to be created by DNA cleavage. Therefore it may be that the HMWS is the activated conformation itself, which is capable of forming with Ca2+, and forms with sufficient concentrations of DNA bound SgrAI dimer regardless of the presence or absence of cleavage, explaining the ability of 40-1 to both induce HMWS and stimulate prior to its own cleavage. Alternatively, within the HMWS, a small amount of cleavage of 40-1 into PCP could be rapidly communicated, and cleavage of all DNA in the HMWS rapidly performed. If true, the HMWS would be capable of both the low and high activity conformations, cleaved DNA would be the signal to shift conformation, and the communication of DNA cleavage in one dimer would induce the conformational change in all associated SgrAI enzymes in the HMWS. Further studies with uncleavable substrates could address this question.
In contrast to the results with the 40 bp primary site containing DNA (40-1) and the precleaved 40 bp primary site containing DNA (PCP or PC), the primary site in an 18 bp duplex (18- 1) does not stimulate DNA cleavage under single turnover cleavage assay conditions (ie. 37°C and 10 mM Mg2+), but does stimulate high molecular weight species (HMWS) formation under binding assay conditions (ie. 4°C and 10 mM Ca2+). The lack of correlation of HMWS formation and stimulatory capacity with 18-1 could be due to the different conditions of the two assays, for example, the lower temperature and/or Ca2+ may stabilize HMWS that is not stable at 37°C with Mg2+. Our binding affinity measurements of SgrAI with DNA suggested a 5 fold lower KD in the presence of Ca2+ relative to Mg2+. If the lack of stimulatory activity by 18-1 is due to diminished HMWS stability, then the role of the flanking DNA in stabilizing HMWS suggests perhaps an additional, albeit non-sequence-specific DNA binding site on SgrAI. To investigate this issue further, single turnover DNA cleavage assays were performed at 4°C. No cleavage of 1 nM 32P labeled 18-1 by 1 µM SgrAI was found, even after 21 hours of incubation. However, with 0.9 µM unlabeled 18-1 added to the reaction, the labeled DNA was found to be cleaved with a rate constant of 0.118±0.014 min−1. PC DNA was found to stimulate the cleavage more effectively, with a measured rate constant of 2.2±0.04 min−1. The failure of 18-1 to stimulate cleavage at 37°C, but successfully at 4°C, is consistent with the proposal that the lower temperature stabilizes HMWS. The stabilization may occur through altered thermodynamic properties of the complex, or alternatively through preventing dissociation of cleaved 18-1 into single strands, if cleaved DNA is the signal for activation. The cleavage rate constants obtained with 40-1 (see above) argue against cleaved DNA as the activation signal unless the signal is propagated within the HMWS. We did find that SgrAI behaved differently with pre-cleaved primary site containing DNA compared to 40-1, its uncleaved version, in HMWS formation and in the cleavage assays; while PCP stimulated accelerated cleavage of all of the labeled DNA in the assays, 40-1 stimulated the cleavage of only a portion. However recent structural studies of SgrAI bound to cleaved primary site DNA argue against cleaved primary site itself as the stimulatory signal25, this structure contains the 18 bp primary site DNA and the SgrAI-DNA complex is in the DNA bound dimer form. Structural characterization of the HMWS will be key to understanding the origin of the activation signal.
The unusual DNA cleavage activity of SgrAI may be a consequence of the large genome of S. griseus, from which it is derived. Restriction endonucleases are coexpressed with a methyltransferase enzyme having the same sequence specificity, which functions to protect the host genome from the cleavage activity of the endonuclease. Hence the SgrAI methyltransferase must methylate all SgrAI recognition sequences within the genome before cleavage by the endonuclease can occur, and this requirement may be difficult due to the large size of the genome (over 8 million bp). The relatively long sequence recognized by SgrAI, 8 bp versus the usual 4–6, may have evolved due to this pressure, since the longer sequence greatly reduces the number of sites to be methylated in the host DNA. In addition, the inherently low cleavage activity of SgrAI in the absence of cleaved primary site DNA also reduces the pressure on host DNA, as well as the methyltransferase enzyme. However, such a long recognition sequence will also occur far less frequently in the phage DNA, and hence place selective pressure on the enzyme for increased activity in order for adequate protection of the host from phage infection. It appears that one way in which the SgrAI enzyme activity is increased is through the stimulation of its cleavage activity. Another way is through its secondary site cleavage activity, which will induce more cleavages in the phage DNA than at the primary sites alone, and hence could better protect the host. These secondary sites of the host are normally protected from SgrAI cleavage activity, since cleavable primary sites are required to induce secondary site cleavage, and primary sites are protected by methylation in the host. However, activation of SgrAI by invading phage could result in cleavage of the host DNA, in addition to phage DNA, at the secondary sites. Therefore, the formation of aggregates, or oligomers, of DNA bound dimers may have an important role in sequestering activated SgrAI on the phage DNA and away from the host genomic DNA.
Acknowledgements
This work was supported by the National Institutes of Health [GM066805 to NCH] and the Howard Hughes Medical Institute [number 52005889 to the University of Arizona supporting MIG) We also acknowledge the Analytical Biophysics Core Facility at the University of Arizona for the analytical ultracentrifugation data and its interpretation.
Abbreviations
- DBD
DNA bound SgrAI dimer
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- FLO
fluorescein moiety
- FPA
fluorescence polarization anisotropy
- HEX
hexachlorofluorescein moiety
- HMWS
high molecular weight species
- OAc
acetate
- PAGE
polyacrylamide gel electrophoresis
- PC
pre-cleaved primary site DNA without 5’end at the cleavage site phosphorylated
- PCP
pre-cleaved primary site DNA with 5’end at the cleavage site phosphorylated
- RMSD
root mean square deviation
References
- 1.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and function. Cell. Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 2.Tautz N, Kaluza K, Frey B, Jarsch M, Schmitz GG, Kessler C. SgrAI, a novel class- II restriction endonuclease from Streptomyces griseus recognizing the octanucleotide sequence 5' - CR/CCGGYG-3' [corrected] Nucleic Acids Res. 1990;18(10):3087. doi: 10.1093/nar/18.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilcock DT, Daniels LE, Bath AJ, Halford SE. Reactions of type II restriction endonucleases with 8-base pair recognition sites. J Biol Chem. 1999;274(51):36379–36386. doi: 10.1074/jbc.274.51.36379. [DOI] [PubMed] [Google Scholar]
- 4.Daniels LE, Wood KM, Scott DJ, Halford SE. Subunit assembly for DNA cleavage by restriction endonuclease SgrAI. J Mol Biol. 2003;327(3):579–591. doi: 10.1016/s0022-2836(03)00143-8. [DOI] [PubMed] [Google Scholar]
- 5.Bitinaite J, Schildkraut I. Self-generated DNA termini relax the specificity of SgrAI restriction endonuclease. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(3):1164–1169. doi: 10.1073/pnas.022346799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29(18):3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wood KM, Daniels LE, Halford SE. Long-range communications between DNA sites by the dimeric restriction endonuclease SgrAI. J Mol Biol. 2005;350(2):240–253. doi: 10.1016/j.jmb.2005.04.053. [DOI] [PubMed] [Google Scholar]
- 8.Hingorani-Varma K, Bitinaite J. Kinetic analysis of the coordinated interaction of SgrAI restriction endonuclease with different DNA targets. J Biol Chem. 2003;278(41):40392–40399. doi: 10.1074/jbc.M304603200. [DOI] [PubMed] [Google Scholar]
- 9.Dunten PW, Little EJ, Gregory MT, Manohar VM, Dalton M, Hough D, Bitinaite J, Horton NC. The structure of SgrAI bound to DNA; recognition of an 8 base pair target. Nucleic Acids Res. 2008;36:5405–5416. doi: 10.1093/nar/gkn510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aggarwal AK. Crystallization of DNA binding proteins with oligodeoxynucleotides. Methods: A Companion to Methods in Enzymology. 1990;1:83–90. doi: 10.1016/S1046-2023(05)80150-1. [DOI] [PubMed] [Google Scholar]
- 11.Fasman GD. CRC Handbook of Biochemistry and Molecular Biology. 3rd ed. Cleveland, OH: CRC; 1975. [Google Scholar]
- 12.Reid SL, Parry D, Liu HH, Connolly BA. Binding and recognition of GATATC target sequences by the EcoRV restriction endonuclease: a study using fluorescent oligonucleotides and fluorescence polarization. Biochemistry. 2001;40(8):2484–2494. doi: 10.1021/bi001956p. [DOI] [PubMed] [Google Scholar]
- 13.Carey J. Gel retardation. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning : a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1989. p. 3. v. [Google Scholar]
- 15.Abramoff MD. Image Processing with ImageJ. Biophotonics International. 2009;11(7):36–42. [Google Scholar]
- 16.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78(3):1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge [UK]: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 18.Vipond IB, Halford SE. Specific DNA recognition by EcoRV restriction endonuclease induced by calciumions. Biochemistry. 1995;34(4):1113–1119. doi: 10.1021/bi00004a002. [DOI] [PubMed] [Google Scholar]
- 19.Martin AM, Horton NC, Lusetti S, Reich NO, Perona JJ. Divalent metal dependence of site-specific DNA binding by EcoRV endonuclease. Biochemistry. 1999;38(26):8430–8439. doi: 10.1021/bi9905359. [DOI] [PubMed] [Google Scholar]
- 20.Etzkorn C, Horton NC. Ca2+ binding in the active site of HincII: implications for the catalytic mechanism. Biochemistry. 2004;43(42):13256–13270. doi: 10.1021/bi0490082. [DOI] [PubMed] [Google Scholar]
- 21.Joshi HK, Etzkorn C, Chatwell L, Bitinaite J, Horton NC. Alteration of sequence specificity of the type II restriction endonuclease HincII through an indirect readout mechanism. J Biol Chem. 2006;281(33):23852–23869. doi: 10.1074/jbc.M512339200. [DOI] [PubMed] [Google Scholar]
- 22.Schuck P. Diffusion of the reaction boundary of rapidly interacting macromolecules in sedimentation velocity. Biophys J. 98(11):2741–2751. doi: 10.1016/j.bpj.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brownsey RW, Boone AN, Elliott JE, Kulpa JE, Lee WM. Regulation of acetyl- CoA carboxylase. Biochem Soc Trans. 2006;34(Pt 2):223–227. doi: 10.1042/BST20060223. [DOI] [PubMed] [Google Scholar]
- 24.Halford SE. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem Soc Trans. 2009;37(Pt 2):343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 25.Little EJ, Dunten PW, Bitinaite J, Horton NC. Structures of SgrAI Bound to Cleaved Primary Site DNA and Uncleaved Secondary Site DNA. Submitted for publication. 2010 doi: 10.1107/S0907444910047785. [DOI] [PMC free article] [PubMed] [Google Scholar]