Abstract
Aluminum (Al) is the most abundant metal in the Earth's crust, but its trivalent ionic form is highly toxic to all organisms at low concentrations. How Al enters cells has not been elucidated in any organisms. Herein, we report a transporter, Nrat1 (Nramp aluminum transporter 1), specific for trivalent Al ion in rice. Nrat1 belongs to the Nramp (natural resistance-associated macrophage protein) family, but shares a low similarity with other Nramp members. When expressed in yeast, Nrat1 transports trivalent Al ion, but not other divalent ions, such as manganese, iron, and cadmium, or the Al–citrate complex. Nrat1 is localized at the plasma membranes of all cells of root tips except epidermal cells. Knockout of Nrat1 resulted in decreased Al uptake, increased Al binding to cell wall, and enhanced Al sensitivity, but did not affect the tolerance to other metals. Expression of Nrat1 is up-regulated by Al in the roots and regulated by a C2H2 zinc finger transcription factor (ART1). We therefore concluded that Nrat1 is a plasma membrane-localized transporter for trivalent Al, which is required for a prior step of final Al detoxification through sequestration of Al into vacuoles.
Aluminum (Al) is the third most abundant terrestrial element and ubiquitously distributed throughout the environment (1). The solubility of Al increases markedly under acidic conditions, resulting in the mobilization of trivalent Al3+ ion, which is toxic to all living cells at low concentrations (2–4). In plants, ionic Al rapidly inhibits root elongation by targeting multiple cellular sites and subsequently the uptake of water and nutrients (5, 6), resulting in poor growth. Al toxicity has therefore been recognized as a major factor limiting crop production on acid soils, which account for 30% to 40% of the world's arable soils (7).
However, some plants have evolved mechanisms to detoxify Al, both externally and internally (5, 6). The most-documented and general mechanism of Al tolerance in both monocots and dicots is release of organic acid anions, including malate, citrate, and oxalate, from the roots in response to Al (5, 6). These anions are able to chelate Al to form nonphytotoxic Al form. Genes responsible for the secretion of Al-induced malate (ALMT1) in wheat and citrate (HvAACT1 and SbMATE) in barley and sorghum have been identified (8–10). On the other hand, by using Al-sensitive mutants, several Al-tolerance genes have been identified in Arabidopsis and rice. The ALS1 and ALS3 genes from Arabidopsis (11, 12), STAR1 and STAR2 from rice (13), encode ATP-binding cassette (ABC) proteins. Although functions of Arabidopsis ALS1 and ALS3 proteins were not elucidated, it is speculated that ALS3 involved in redistributing Al from root apexes to other less sensitive tissues (11), and ALS1 is responsible for the sequestration of Al into the vacuoles (12), respectively. A bacterial-type ABC transporter complex, STAR1–STAR2, transports UDP-glucose, which may be used for modification of the cell wall (13).
Recently, a transcription factor (ART1) for Al tolerance was identified in rice (14). ART1 is a unique C2H2 zinc finger-type transcription factor and regulates a total of 31 genes (14). One of the genes (Os02g0131800) is annotated to encode a protein belonging to the Nramp (natural resistance-associated macrophage protein) family. Functional analysis of this gene in the present study was unique in finding that this gene encodes a plasma membrane-localized transporter specific for trivalent Al and is required for Al tolerance in rice.
Results and Discussion
Sequence Analysis of an Nramp Gene.
The rice gene (Os02g0131800) consists of 13 exons and 12 introns, encoding a protein with 545 amino acids, and belongs to the Nramp. In the rice genome, there are seven Nramp members (Fig. S1), but the protein encoded by Os02g0131800 shows a low similarity with other members, sharing 36 to 59% identity at the amino acid level. None of the rice Nramp genes has been functionally characterized, but some of Arabidopsis Nramp genes have been reported to encode divalent metal ion transporters (15–18).
Nrat1 Functions as a Transporter Specific for Trivalent Al.
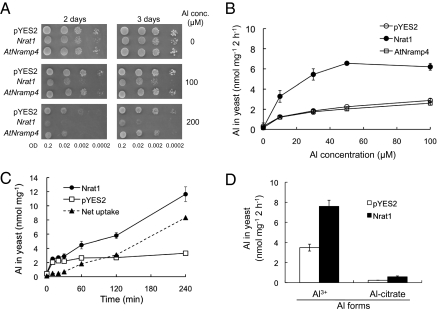
We tested the transport substrates for the protein encoded by Os02g0131800 in yeast by using AtNramp4 as a positive control, which has been shown to transport Fe2+, Mn2+, and Cd2+ (17). Unlike AtNramp4, when Os02g0131800 was expressed in the ferrous iron transport-deficient yeast strain (fet3fet4), it could not complement the iron uptake by the yeast (Fig. S2). Os02g0131800 also could not restore the growth of a yeast mutant (smf1) defective in Mn uptake (Fig. S2). Furthermore, expression of AtNramp4 resulted in enhanced uptake of Cd in yeast, but the expression of Os02g0131800 had no such effect (Fig. S3). However, the expression of Os02g0131800 increased the sensitivity of yeast to Al toxicity (Fig. 1A) and its Al uptake at pH 4.2, at which most Al is present in the form of trivalent ion (Fig. 1B). In contrast, the expression of AtNramp4 had no effect on Al tolerance or uptake in the yeast (Fig. 1 A and B). A time-course experiment showed that Al uptake increased linearly with time in the yeast expressing Os02g0131800 (Fig. 1C). Furthermore, the Al uptake was not affected by the presence of equimolar concentration of divalent ions including Cd and Mn (Fig. S4). Taken together, these results indicate that, unlike other Nramp members, Os02g0131800 encodes a transporter for trivalent Al3+ ion, but not for divalent metals such as Cd2+, Mn2+, and Fe2+. We therefore name this gene Nrat1 (Nramp aluminum transporter 1). Among the seven rice Nramp members, only Nrat1 shows a transport activity for aluminum in yeast (Fig. S5).
Fig. 1.
Transport activity of Nrat1 for aluminum in yeast. (A) Effect of Nrat1 expression on Al tolerance. Yeast cells (BY4741) carrying empty vector pYES2, Nrat1, or AtNramp4 were spotted on LPM-ura medium (pH 4.2) buffered with 5 mM succinic acid with or without AlCl3 at different dilutions. The plates were incubated at 30 °C for 2 to 3 d. (B) Transport activity of Nrat1 for trivalent Al ion. Yeast cells carrying empty vector pYES2, Nrat1, or AtNramp4 were exposed to 10, 30, 50, or 100 μM AlCl3 at pH 4.2 for 2 h. (C) Time-dependent transport of Nrat1 for Al. Yeast cells carrying empty vector pYES2 and Nrat1 were exposed to 50 μM AlCl3 at pH 4.2 for different times. Net uptake was the difference between Al uptake from yeast carrying Nrat1 and empty vector. (D) Transport activity of Nrat1 for different Al forms. Yeast cells transformed with Nrat1 were exposed for 2 h to a solution (pH 4.2) containing 50 μM AlCl3, or an Al–citrate complex prepared by mixing 50 μM AlCl3 with 500 μM citrate. After uptake, the yeast cells were washed and digested with HCl. The aluminum concentration in the digest solution was determined by atomic absorption spectrophotometer. Data in B, C, and D are means ± SD of three biological replicates.
Rice roots secrete citrate in response to aluminum exposure, although the amount secreted is much lower compared with other cereal crops, such as rye and wheat (19). Therefore, there is a possibility that the Al–citrate complex is the substrate taken up by Nrat1. To examine this possibility, we compared Al uptake by Nrat1 from ionic Al3+ and Al–citrate complex. Nrat1 shows a transport activity only for Al3+ ion, and not for the Al–citrate complex (Fig. 1D).
Expression Analysis of Nrat1.
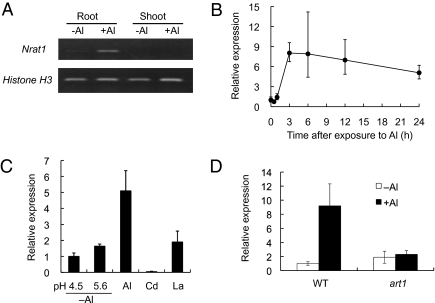
We investigated the expression pattern of Nrat1, which is expressed only in the roots, not in the shoots (Fig. 2A). Furthermore, the expression of Nrat1 is up-regulated rapidly by Al, with the level of expression reaching the maximum at 3 h after the exposure to Al (Fig. 2B). The expression of Nrat1 is specifically induced by Al, but not by other metals, including Cd and La, and also not by low pH (Fig. 2C). In a rice mutant defective in the Al response C2H2-type zinc finger transcription factor (art1), the expression of Nrat1 was not induced by Al (Fig. 2D), confirming that Nrat1 expression is regulated by ART1 (14).
Fig. 2.
Expression pattern of Nrat1. (A) Expression of Nrat1 in different tissues. Rice seedling (cv. Nipponbare) was exposed to a solution containing 0 or 30 μM AlCl3 for 6 h. Histone H3 was used as an internal standard. (B) Time-dependent expression of Nrat1 in rice roots. Rice seedlings were exposed to a solution containing 20 μM Al for different time. (C) Expression of Nrat1 in response to other metals. Rice seedlings were exposed to a solution containing 0, 30 μM Cd, 10 μM La, or 50 μM Al at pH 4.5 or containing 0 Al at pH 5.6 for 6 h. (D) Expression of Nrat1 in the art1 mutant. Both wild-type rice and the art1 mutant were exposed to 20 μM Al for 4 h. The expression of Nrat1 in the roots were determined by quantitative real-time PCR and relative expression to Histone H3 (internal standard) is shown. Data are means ± SD of three biological replicates.
Cellular and Subcellular Localization of Nrat1.
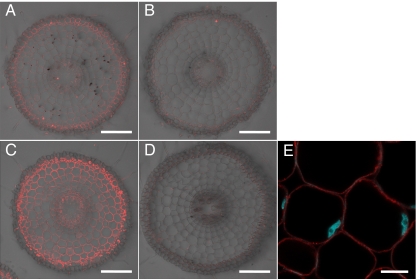
We then investigated the cellular and subcellular localization of Nrat1. Immunostaining showed that Nrat1 was expressed in all root cells except epidermal cells in the wild-type rice and the expression was enhanced by Al (Fig. 3 A and C). The absence of signal in the knockout line indicates the specificity of the antibody (Fig. 3 B and D). Costaining with DAPI showed that Nrat1 was localized at the plasma membrane (Fig. 3E).
Fig. 3.
Localization of Nrat1. (A and B) Immunostaining of Nrat1 in the roots (2 mm from the root tip) of wild-type rice (A) and knockout line (B) without Al treatment. (C and D) Immunostaining of Nrat1 in the roots (2 mm from the root tip) of wild-type rice (C) and knockout line (D) exposed to 30 μM Al for 12 h. (E) Subcellular localization of Nrat1 (red color) costained with DAPI (cyan color). (Scale bars, 100 μm in A–D; 10 μm in E.)
Furthermore, when Nrat1 fused with GFP was transiently introduced into the epidermal cells of onion, we also found that Nrat1 is localized to the plasma membrane in contrast to GFP alone, which is localized at the cytoplasm and nucleus (Fig. S6 A–C). Immunostaining with an antibody against GFP in a transgenic rice plant carrying GFP under the control of Nrat1 promoter also showed that Nrat1 is localized in all cells except the epidermal cells in both the root tip region and the mature zone of the roots (Fig. S6 D–H).
Role of Nrat1 in Al Tolerance of Rice.
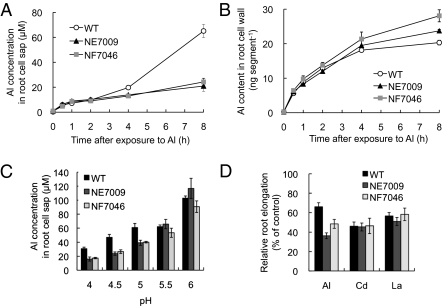
To elucidate the function of Nrat1 in rice, we obtained two independent retrotransposon (Tos-17) insertion lines of Nrat1 (Fig. S7). No expression of Nrat1 was found in either line. There was no difference in other cation uptake between wild-type rice and the knockout line (Table S1). A time-course experiment showed that the difference in Al concentration of root-cell sap between the knockout lines and the wild-type rice was observed at 4 h after the exposure to Al (Fig. 4A). In contrast, the Al content in the cell wall was higher in the knockout lines than in the wild-type rice (Fig. 4B). Introducing Nrat1 into one of the knockout lines increased Al concentration in the root-cell sap to the level similar to the wild-type rice (Fig. S8), indicating that Nrat1 is responsible for the phenotype in the knockout line.
Fig. 4.
Transport of aluminum in rice roots. (A and B) Time-dependent concentration of Al in the root-cell sap (A) and time-dependent accumulation of Al in the root-cell wall (B) of wild-type rice and two Nrat1 knockout lines (NE7009 and NF7046). The roots were exposed to a solution containing 30 μM Al for different times up to 8 h. (C) pH-dependent concentration of Al in the root-cell sap of wild-type rice and two Nrat1 knockout lines (NE7009 and NF7046). The roots were exposed to 30 μM Al solution buffered with homopipes at different pHs for 8 h. Al was determined by atomic absorption spectrophotometer. Data in A to C are means ± SD of three biological replicates. (D) Sensitivity of Nrat1 knockout line to metals. Seedlings of wild-type rice (WT) and two Nrat1 knockout lines (NE7009 and NF7046) were exposed to a solution containing 30 μM Al, 10 μM Cd, or 5 μM La for 24 h. The root length was measured before and after the treatment and elongation relative to the root growth without Al was shown. Data are means ± SD (n = 10).
Because Al speciation depends on pH (20), we investigated pH-dependent uptake of Al between the wild-type and knockout lines. At pH below 5.0, most Al is present in the form of trivalent ion (Al3+), but at pH above 5.0, Al is present in the form of Al(OH)2+, Al(OH)2+, and Al(OH)4− (20). The Al concentration was significantly higher in the wild-type rice than the knockout lines at pHs below 5.0 (Fig. 4C), but was similar at pHs above 5.5. These results are consistent with those of yeast (Fig. 1), further demonstrating that Nrat1 is a transporter for trivalent Al ion.
We also compared the effect of temperatures on Al uptake between wild-type rice and two knockout lines. At 4 °C, there was no difference in the Al concentration of root-cell sap and cell wall between wild-type rice and the knockout lines (Fig. S9). However, at 25 °C, the Al concentration in the root-cell sap was higher in the wild-type rice than in the knockout line (Fig. S9A), whereas Al in the cell wall was lower in the wild-type rice (Fig. S9B). This result indicates that Nrat1-mediated Al uptake is an active process.
Knockout of Nrat1 resulted in increased sensitivity to Al, but did not affect the sensitivity to other metals, including Cd and La (Fig. 4D), consistent with the metal specificity of Nrat1 (Fig. 1 and Figs. S2 and S3). The increased Al sensitivity in the Nrat1 knockout rice is opposite to the effect observed in the yeast assay (Fig. 1A). This is because plant cells possess cellular Al detoxification mechanisms of chelation with organic acid anions and sequestration into the vacuoles (6). The ABC transporter ALS1 in Arabidopsis has been suggested to be involved in the sequestration of Al into vacuoles (12). There is a homolog of ALS1 in rice, which also has been suggested to be involved in Al tolerance and regulated by ART1 (14), although the function of these genes has not been characterized in both Arabidopsis and rice. The strategy of Al3+ uptake mediated by Nrat1 followed by cellular complexation and sequestration is more effective in detoxifying Al than allowing Al to accumulate in the root apoplast (mainly cell wall) because Al accumulation in the root apoplast is inhibitory to root growth by limiting cell wall extensibility (21). Indeed, more Al was found in the cell wall of the knockout lines of Nrat1 (Fig. 4B and Fig. S9B).
Our results indicate that Nrat1, an Nramp member, is a plasma membrane-localized transporter specific for trivalent aluminum ion in rice. Nramp proteins are evolutionarily conserved, with homologs in bacteria, algae, plants, and animals (22). Previous studies have shown that Nramp proteins have a broad range of substrates including Fe2+, Zn2+, Mn2+, Co2+, Cd2+, Cu2+, Ni2+, and Pb2+ (23). They play important roles in metal ion homeostasis, especially iron uptake and recycling in mammals and manganese uptake in yeast and bacteria. For example, Arabidopsis AtNramp3 and AtNramp4 are localized at tonoplast and function in the mobilization of iron from the vacuolar store to feed the developing plant (24). AtNramp6 is found to be targeted to a vesicular-shaped endomembrane compartment and function as an intracellular metal transporter (17). Recently, AtNramp1 was reported to function as a high-affinity transporter for Mn (18). However, the transport substrates of Nramp proteins identified so far are limited to divalent metal ions. Our study is unique in reporting that an Nramp protein transports trivalent aluminum ion. This difference in transport substrates is not surprising given the low similarity between Nrat1 and other Nramp proteins (Fig. S1), although further work on the relationship between the protein structure and metal transport selectivity is required in the future.
It may be questioned why plants develop a transport system specific for toxic metal, Al. However, considering that Al is the most abundant metal in the earth's crust and plants always have the possibility to be exposed to Al stress, it is not surprising that plants have a transporter specific for Al for the detoxification. This result may be different from other toxic elements in soil, such as Cd and As, which have low abundance and plants are rarely exposed to toxic level of these elements; therefore, there are no specific transport systems for these toxic metals. For example, it is known that the uptake of Cd and As into the root cells is meditated through transporters for essential elements (25, 26).
Soil and water acidification is increasing globally (27), heightening the concern on Al toxicity in the environment. Identification of this unique Al transporter in the present study may help further effort to identify other transporters involved in Al transport in different cells of plants, humans, and other organisms.
Materials and Methods
Plant Materials and Growth Conditions.
Two Tos-17 insertion lines of rice (Oryza sativa L.); NE7009 and NF7046 for Nrat1, were obtained from the Rice Genome Resource Center in Japan. The homozygous lines were screened by PCR using OsNrat1-specific primers (5′-TGATGACATTCCTGAAGGTTGA-3′ and 5′-CGGAAACAAGATAGGGTCAAAC-3′ for NE7009, 5′-ATCAAGGGTGAGCACTACGG-3′ and 5′-GGCTGCTTGCAGATACTTCC-3′ for NF7046) and a left-border Tos-17 primer (5′-ATTGTTAGGTTGCAAGTTAGTTAAGA-3′). Seeds of both wild-type rice and two Tos-17 homozygous lines mutant were soaked in deionized water overnight at 30 °C in the dark, and then transferred to a net floating on a 0.5 mM CaCl2 solution in a 1.5-L plastic container. Seedlings were grown for 4 to 7 d at 25 °C before being used for various experiments. For root elongation measurement, the roots were exposed to a 0.5 mM CaCl2 solution (pH 4.5) containing 30 μM Al, 10 μM Cd, or 5 μM La for 24 h. The root length of each seedling was measured with a ruler before and after the treatments. Relative root elongation was calculated as follows: (root elongation with Al treatment)/(root elongation without metal) × 100. The concentration of cations in the roots and shoots was also determined by atomic absorption spectrophotometer after the plants of wild-type rice and the knockout lines were grown in a nutrient solution for 1 mo.
Transport Activity in Yeast Cells.
The cDNA fragment containing an entire ORF for Nrat1 and AtNramp4 were amplified by RT-PCR using the primers 5′-GGTACCAAAATGGAAGGGACTGGTGAGATGA -3′ and 5′-CTACATGGAAGCATCGGCAA-3′ for Nrat1, and 5′-GGATCCGAAATATGTCGGAGACTGATAGAG-3′ and 5′-TCACTCATCATCCCTCTGTGGT-3′ for AtNramp4. These primers contained a KpnI or BamHI site to facilitate cloning of the amplified cDNA. The fragment was first cloned into the pGEM-T vector (Promega). After sequence confirmation, the Nrat1 cDNA with KpnI and NotI and AtNramp4 cDNA with BamHI and NotI were excised for cloning into pYES2 (Invitrogen). The resulting plasmid was introduced into yeast strain.
The yeast strain used in this study was BY4741 (MATa his2Δ0 met15Δ0 ura3Δ0). Nrat1, AtNramp4 vector construct, or the empty vector pYES2 were introduced into BY4741 strain according to the manufacturer's protocols (SC easy comp transformation kit; Invitrogen). Transfomants were selected on uracil-deficient medium and grown in synthetic complete (SC-uracil) yeast medium containing 2% glucose, 0.67% yeast nitrogen base without amino acids (Difco), 0.2% appropriate amino acids, and 2% agar at pH 6.0. One colony was selected in each transformation strain and grown in the liquid SC-uracil medium. For measurement of Al concentration, cells at midexponential phase were harvested and transferred to a LPM medium containing 2% galactose for induction of the GAL promoter and 5 mM succinic acid adjusted to pH 4.2. Cells were cultured for 2 h. The precultured yeast was adjusted to an OD600 value of 3.0 by reducing the amount of liquid. AlCl3 was then added to the medium at a concentration of 10, 30, 50, or 100 μM. After 2 h of incubation with gentle shaking, cells were harvested by centrifugation and washed three times with deionized water (MilliQ; Millipore) and then digested with 2 N HCl. In a time-course experiment, cells were collected at different times after the exposure to 50 μM Al. Competition experiments were performed by exposing the yeast cells to a solution with or without equal concentration of Cd, Mn, in the presence of 50 μM Al for 2 h. The concentration of Al in the digest solution was determined by atomic absorption spectrophotometer. For measurement of Cd2+, LPM medium was replaced by liquid SC-uracil medium containing 2% galactose at pH 4.6. Three replicates for each treatment were made.
Functional Complementation in Yeast.
The strains used in this study were smf1 (MATa his2Δ0 met15Δ0 ura3Δ0 YOL122c::KanMX4) for Mn complementation and DDY4 (fet3fet4) (MATa ade6 can1 his3 leu2 trp1 ura3 fet3-2::HIS3 fet4-LEU) (28) for Fe complementation. DDY4 was grown in media supplemented with 0.2 mM FeCl3.
Complementation of the smf1 phenotype was tested on a synthetic medium containing 2% galactose, 0.67% yeast nitrogen base without metals (BIO 101 Systems), 0.2% appropriate amino acids, and 2% agar buffered at pH 6 with 50 mM Mes and supplemented with or without 2 mM EGTA.
Complementation of the DDY4 phenotype was tested on synthetic medium containing 2% galactose, 0.67% yeast nitrogen base without metals (BIO 101 Systems), 0.2% appropriate amino acids, and 2% agar buffered at pH 5.5 with 50 mM Mes in the presence or absence of 8 and 10 μM 4,7-biphenyl-1,10-phenanthroline-disulphonic acid (BPDS).
After spotting at three yeast-cell dilutions (optical densities at 600 nm of 0.2, 0.02, 0.002, and 0.0002), plates were incubated for 3 to 5 d at 30 °C.
Generation of Transgenic Rice.
For complementation test of Nrat1, we amplified a 6.915-kb DNA fragment containing the Nrat1 promoter region (2.1 kb before ATG), the entire ORF and the 3′-untranslated region (1 kb after TGA) from Nipponbare genomic DNA by PCR. The DNA fragment was inserted into pPZP2H-lac vector and then transformed into Agrobacterium tumefaciens (Strain EHA101). Calluses derived from Tos-17 insertion line NE7009 were transformed by Agrobacterium-mediated transformation.
To investigate the cellular localization of Nrat1, we introduced a construct consisting of the promoter (2.1 kb) of Nrat1 fused with GFP to calluses (cv. Nipponbare) using an Agrobacterium-mediated transformation system (29). The 2.1-kb region upstream of the initiation codon of Nrat1 was amplified by PCR from Nipponbare genomic DNA using primer 5′-GGTACCAACACGTCTGACGCTTGTT-3′ and 5′-CTCGAGATTCTATGTTGCTAATGCACCTTGT -3′. Using KpnI and SalI, the amplified fragment was cloned into pPZP2H-lac carrying GFP and the terminator of the nopaline synthase gene, producing the Nrat1 promoter-GFP construct. We selected transformed calluses by hygromycin resistance, and from them regenerated plants.
RNA Isolation and RT-PCR.
To examine the expression pattern of Nrat1, we exposed seedlings of the wild-type rice to different Al concentrations (0–50 μM) for different times, to different pHs, and other metals including Cd and La. Both the roots and shoots were sampled with three replicates and subjected to RNA extraction. Total RNA was extracted using the RNeasy Mini Kit (Qiagen). One microgram of total RNA was used for first strand cDNA synthesis using a SuperScript II kit (Invitrogen), following the manufacturer's instructions with an oligo(dT)12–18 primer. The expression was determined with SYBR Premix Ex Taq (Takara) by Mastercycler ep realplex (Eppendorf). The primer sequences for RT-PCR of Nrat1 were 5′-TCGCATTGGCTCGCACCCT-3′ and 5′-TCGTCTTCTTCAGCCGCACGAT-3′. HistoneH3 (Forward primer, 5′-AGTTTGGTCGCTCTCGATTTCG-3′; Reverse primer, 5′-TCAACAAGTTGACCACGTCAC G-3′) was used as an internal control.
GFP Fusion and Subcellular Localization.
To construct the Nrat1-GFP fusion protein, Nrat1 cDNA containing a SalI restriction site, but not the stop codon, was amplified by RT-PCR using the primers 5′-GTCGACAAGGTGCATTAGCAACATAG-3′ and 5′ GTCGACAGCATCGGCAAGGTCCTTCCTG-3′. The amplified cDNA fragment was then cloned in a frame in front of the GFP coding region in pBluescript vector, producing the Nrat1-GFP construct under the control of the 35S promoter.
To construct the GFP-Nrat1 fusion protein, Nrat1 cDNA was amplified by RT-PCR using the primers 5′-TGTACAAGATGGAAGGGACTGGTGAGATGA-3′ and 5′-GCGGCCGCCTACATGGAAGCATCGGCAAGGT-3′. The amplified cDNA fragment was then cloned in a frame after the GFP coding region in pBluescript vector, producing GFP-Nrat1 construct under the control of the 35S promoter.
Onion epidermal cells were bombarded with 1-μm gold particles coated with plasmid DNA Nrat1-GFP, GFP-Nrat1, or GFP and incubated in the dark at 25 °C for 20 h. We observed fluorescence by confocal laser scanning microscopy (LSM700; Carl Zeiss).
Immunohistological Staining.
Antibodies against Nrat1 were obtained by immunizing rabbits with the synthetic peptide MEGTGEMREVGRETLHGG-C (positions 1–18 of Nrat1). We performed immunostaining with the roots of wild-type rice, the mutant line, and overexpressed lines exposed to 30 μM Al or not for 12 h. The procedures for immunostaining were followed according to ref. 30. Fluorescence was observed by a laser-scanning confocal microscope (LSM700; Carl Zeiss).
To further observe the localization of Nrat1, we also performed an immunostaining using an antibody against GFP (A11122; Molecular Probes) in the transgenic plant carrying Nrat1 promoter-GFP prepared as described above. The seedlings were exposed to a solution with or without Al (50 μM) for 6 h.
Root-Cell Sap Preparation and Al Determination.
Five-day-old seedlings of both wild-type rice and two Nrat1 knockout lines were exposed to 30 μM Al (pH 4.2) for 0.5, 1, 2, 4, and 8 h. To investigate the effect of pH on Al uptake, the seedlings of all lines were exposed to 30 μM Al in a 0.5 mM CaCl2 solution buffered with 10 mM Homopipes at different pHs ranging from 4.0 to 6.0 The effect of temperature on the Al uptake was investigated by exposing the seedlings to 30 μM Al in a 0.5 mM CaCl2 solution (pH 4.2) at 4 °C and 25 °C. After the treatment, the root segments (0–1 cm, 20 roots each) were excised after washing three times with 0.5 mM CaCl2 and then put in a Ultra free-MC Centrifugal filter units (Millipore) and centrifuged at 3,000 × g for 10 min at 4 °C to remove apoplastic solution. The roots were then frozen at −80 °C overnight. The root-cell sap solution was obtained by thawing the samples at room temperature, and then centrifuging at 20,600 × g for 10 min. The residual cell wall were washed with 70% ethanol three times and then immersed in 0.5 mL of 2 N HCl for at least 24 h with occasional vortex. The Al in the symplastic solution and cell wall extracts was determined by atomic absorption spectrophotometer.
Supplementary Material
Acknowledgments
We thank the Rice Genome Resource Center for providing Tos17 seeds. This research was supported by Grant Genomics for Agricultural Innovation IPG-0006 from the Ministry of Agriculture, Forestry and Fisheries of Japan (to J.F.M.) and Grant-in-Aid for Scientific Research 21248009 and 22119002 on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to J.F.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004949107/-/DCSupplemental.
References
- 1.Yokel RA. In: Elements and their Compounds in the Environment. 2nd Ed. Anke MM, Inhat M, Stoeppler M, editors. Weinheim: Wiley-VCH; 2004. pp. 635–658. [Google Scholar]
- 2.Yokel RA. The toxicology of aluminum in the brain: A review. Neurotoxicology. 2000;21:813–828. [PubMed] [Google Scholar]
- 3.Crapper DR, Krishnan SS, Dalton AJ. Brain aluminum distribution in Alzheimer's disease and experimental neurofibrillary degeneration. Science. 1973;180:511–513. doi: 10.1126/science.180.4085.511. [DOI] [PubMed] [Google Scholar]
- 4.Driscoll CT. Aluminum in acidic surface waters: Chemistry, transport, and effects. Environ Health Perspect. 1985;63:93–104. doi: 10.1289/ehp.856393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kochian LV, Pineros MA, Hoekenga OA. The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil. 2005;274:175–195. [Google Scholar]
- 6.Ma JF. Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int Rev Cytol. 2007;264:225–252. doi: 10.1016/S0074-7696(07)64005-4. [DOI] [PubMed] [Google Scholar]
- 7.von Uexkull HR, Mutert E. Global extent, development and economic impact of acid soils. Plant Soil. 1995;171:1–15. [Google Scholar]
- 8.Sasaki T, et al. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004;37:645–653. doi: 10.1111/j.1365-313x.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- 9.Furukawa J, et al. An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 2007;48:1081–1091. doi: 10.1093/pcp/pcm091. [DOI] [PubMed] [Google Scholar]
- 10.Magalhaes JV, et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet. 2007;39:1156–1161. doi: 10.1038/ng2074. [DOI] [PubMed] [Google Scholar]
- 11.Larsen PB, Geisler MJB, Jones CA, Williams KM, Cancel JD. ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 2005;41:353–363. doi: 10.1111/j.1365-313X.2004.02306.x. [DOI] [PubMed] [Google Scholar]
- 12.Larsen PB, Cancel J, Rounds M, Ochoa V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta. 2007;225:1447–1458. doi: 10.1007/s00425-006-0452-4. [DOI] [PubMed] [Google Scholar]
- 13.Huang CF, et al. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell. 2009;21:655–667. doi: 10.1105/tpc.108.064543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamaji N, et al. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell. 2009;21:3339–3349. doi: 10.1105/tpc.109.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J. 2000;347:749–755. [PMC free article] [PubMed] [Google Scholar]
- 16.Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cailliatte R, Lapeyre B, Briat JF, Mari S, Curie C. The NRAMP6 metal transporter contributes to cadmium toxicity. Biochem J. 2009;422:217–228. doi: 10.1042/BJ20090655. [DOI] [PubMed] [Google Scholar]
- 18.Cailliatte R, Schikora A, Briat JF, Mari S, Curie C. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell. 2010;22:904–917. doi: 10.1105/tpc.109.073023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma JF, et al. Response of rice to Al stress and identification of quantitative trait Loci for Al tolerance. Plant Cell Physiol. 2002;43:652–659. doi: 10.1093/pcp/pcf081. [DOI] [PubMed] [Google Scholar]
- 20.Martin RB. The chemistry of aluminum as related to biology and medicine. Clin Chem. 1986;32:1797–1806. [PubMed] [Google Scholar]
- 21.Ma JF, Shen R, Nagao S, Tanimoto E. Aluminum targets elongating cells by reducing cell wall extensibility in wheat roots. Plant Cell Physiol. 2004;45:583–589. doi: 10.1093/pcp/pch060. [DOI] [PubMed] [Google Scholar]
- 22.Courville P, Chaloupka R, Cellier MFM. Recent progress in structure-function analyses of Nramp proton-dependent metal-ion transporters. Biochem Cell Biol. 2006;84:960–978. doi: 10.1139/o06-193. [DOI] [PubMed] [Google Scholar]
- 23.Gunshin H, et al. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 24.Lanquar V, et al. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J. 2005;24:4041–4051. doi: 10.1038/sj.emboj.7600864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clemens S. Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie. 2006;88:1707–1719. doi: 10.1016/j.biochi.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Ma JF, et al. Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc Natl Acad Sci USA. 2008;105:9931–9935. doi: 10.1073/pnas.0802361105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pannatier GE, Luster J, Zimmermann S, Blaser P. Monitoring of water chemistry in forest soils: an indicator for acidification. Chimia (Aarau) 2005;59 [Google Scholar]
- 28.Dix DR, Bridgham JT, Broderius MA, Byersdorfer CA, Eide DJ. The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J Biol Chem. 1994;269:26092–26099. [PubMed] [Google Scholar]
- 29.Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 30.Yamaji N, Ma JF. Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol. 2007;143:1306–1313. doi: 10.1104/pp.106.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.